The development of marginal ray flowers in Asteraceae flower heads is regulated by a network of CINCINNATA- and SEPALLATA-like transcription factors upstream of a CYCLOIDEA-like gene.
Abstract
The large sunflower family, Asteraceae, is characterized by compressed, flower-like inflorescences that may bear phenotypically distinct flower types. The CYCLOIDEA (CYC)/TEOSINTE BRANCHED1–like transcription factors (TFs) belonging to the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) protein family are known to regulate bilateral symmetry in single flowers. In Asteraceae, they function at the inflorescence level, and were recruited to define differential flower type identities. Here, we identified upstream regulators of GhCYC3, a gene that specifies ray flower identity at the flower head margin in the model plant Gerbera hybrida. We discovered a previously unidentified expression domain and functional role for the paralogous CINCINNATA-like TCP proteins. They function upstream of GhCYC3 and affect the developmental delay of marginal ray primordia during their early ontogeny. At the level of single flowers, the Asteraceae CYC genes show a unique function in regulating the elongation of showy ventral ligules that play a major role in pollinator attraction. We discovered that during ligule development, the E class MADS-box TF GRCD5 activates GhCYC3 expression. We propose that the C class MADS-box TF GAGA1 contributes to stamen development upstream of GhCYC3. Our data demonstrate how interactions among and between the conserved floral regulators, TCP and MADS-box TFs, contribute to the evolution of the elaborate inflorescence architecture of Asteraceae.
The Asteraceae family is the largest family of flowering plants. Phylogenetically, Asteraceae is a late-branching family on the angiosperm tree, and its reproductive structures are among the most complex within flowering plants (Broholm et al., 2014; Mandel et al., 2019). With its species-richness and morphological diversity, Asteraceae provides an excellent target for evolutionary developmental (evo-devo) studies to understand how developmental regulatory networks have diversified in the evolution of novelty. The species in this family develop head-like inflorescences that may assemble distinct flower types into a single structure that resembles a flower itself. This unique architecture is considered as a key innovation for the evolutionary success of this widely spread family (Broholm et al., 2014). In simple radiate heads, the marginal ray flowers are bilaterally symmetrical, female or neuter, and develop large and showy ventral ligules (lips of fused petals). The hermaphroditic disc flowers are less conspicuous and radially symmetrical, and are located in the center of the head. Across eudicots, bilateral flower symmetry has evolved independently a number of times, and is known to be regulated by the CYCLOIDEA/TEOSINTE BRANCHED1 (CYC/TB1)-like transcription factors (TF) belonging to the TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) protein family (Hileman 2014; Spencer and Kim, 2018). However, in Asteraceae these TFs have been recruited to regulate ray flower identity (Broholm et al., 2008; Chapman et al., 2008; Kim et al., 2008; Tähtiharju et al., 2012; Juntheikki-Palovaara et al., 2014; Garcês et al., 2016; Huang et al., 2016; Chen et al., 2018). This function is specifically attributed to the CYC2 subclade of the TCP gene family, which has been expanded in Asteraceae (Chapman et al., 2008; Tähtiharju et al., 2012; Chen et al., 2018). The expression of CYC2 clade genes localizes to the periphery of flower heads, to emerging ray (and trans) flower primordia, but is absent in the central disc primordia (Broholm et al., 2008; Chapman et al., 2008; Tähtiharju et al., 2012; Chen et al., 2018).
Ray flower identity is controlled by different CYC2 clade paralogs in distinct Asteraceae lineages (Chapman et al., 2008; Tähtiharju et al., 2012; Garcês et al., 2016), providing molecular support that ray identity itself evolved multiple times independently in the family (Panero and Funk, 2008). In Gerbera hybrida (gerbera), overexpression of CYC2 clade genes GhCYC2, GhCYC3, or GhCYC4 in disc flowers converted them into ray-like with elongated petals and disrupted stamen development (Broholm et al., 2008; Juntheikki-Palovaara et al., 2014). In contrast, disc flower development was not affected by overexpression of ray-specific CYC2 genes in Chrysanthemum spp. (Huang et al., 2016) or Senecio spp. (Kim et al., 2008). Furthermore, in ray flowers, the length of the ventral ligule was differentially affected by genetic transformation in distinct species (Broholm et al., 2008; Kim et al., 2008; Huang et al., 2016, Garcês et al., 2016). The regulatory networks upstream of CYC2 clade genes, or in fact, TCP genes in general, are poorly understood. There are indications that CYC-like TFs show both auto- and cross-regulation as a mechanism to maintain their highly specific expression domains (Yang et al., 2012; Yuan et al., 2020). TCP genes, as regulators of organ size and shape, have also been associated with MADS-box genes, which regulate organ identities but also organ growth and differentiation at late developmental stages (Dornelas et al., 2011). In Arabidopsis (Arabidopsis thaliana), for example, TCP genes were identified as targets of the SEPALLATA3 (SEP3) and APETALA1 (AP1) MADS-box proteins (Kaufmann et al., 2009; Wellmer et al., 2006).
Among the six CYC2 clade genes in gerbera, GhCYC3 is the strongest candidate gene to be responsible for the regulation of ray flower identity as well as the growth of the ventral ligule. GhCYC3 is exclusively expressed in marginal ray flower primordia (Tähtiharju et al., 2012). It also shows the greatest expression in elongating ligules in contrast with the other five CYC2 clade genes that show constant and low expression levels throughout ligule development (Juntheikki-Palovaara et al., 2014). Here, by identifying upstream regulators of GhCYC3 in gerbera, we show how conserved floral regulators (i.e. TCP and MADS-box TFs), form dynamic regulatory networks in specifying flower type identity, and their subsequent morphological differentiation in Asteraceae.
RESULTS
Identification of Putative Upstream Transcriptional Regulators of GhCYC3
We performed a yeast one-hybrid (Y1H) screen to identify putative upstream TFs interacting with the gerbera GhCYC3 cis-regulatory region. We first performed a sequence analysis on 21 promoter regions of selected Asteraceae CYC2 clade genes (Supplemental Table S1). MEME analysis (http://meme-suite.org) identified a 27-bp consensus element shared by all promoters except by two Helianthus annuus promoters (HaCYC2e and HaCYC2e-like; Supplemental Fig. S1). The corresponding 27-bp region of the GhCYC3 promoter (Fig. 1A) was cloned in three tandem repeats into a Y1H bait plasmid (pHTT873) that was used to screen the heterologous Arabidopsis TF prey library (Mitsuda et al., 2010). Of 28 candidate Arabidopsis TFs, five were selected for further studies as the corresponding gerbera homologs showed differential expression between the distinct flower types (Supplemental Table S2; Supplemental Fig. S2). These five Arabidopsis TFs belonged to the Apetala2 (AP2)/Ethylene response factor, NAC domain, DNA binding with one finger domain, Homeodomain-Leu zipper, and TCP TF gene families, and ten gerbera homologs corresponding to these TFs were identified in BLAST searches (Supplemental Table S2).
Figure 1.
The schematic structure of the GhCYC3 gene and transient agroinfiltration assay in Nicotiana benthamiana leaves. A to C, The position weight matrix (PWM) of the conserved 27-bp motif identified from the Asteraceae CYC2 clade promoter sequences. The position of the 27-bp motif within the promoter is marked with a yellow box (A). The identified TCP TFBSs, TCP1 and TCP2 (B), and CArG boxes, CArG1 and CArG2 (C), within the 1900 bp regulatory region of GhCYC3 are shown. The locations of TCP TFBSs are marked with green boxes, and CArG TFBSs with blue boxes. The introduced mutations are indicated in (B) and (C). The nucleotide positions are counted from the TSS set as 1. d to F, Activation of PGhCYC3:LUCIFERASE (LUC) reporter construct using selected effector constructs. The binding activities of CIN-like TCP TFs GhCIN1, GhCIN2, and GhCIN3 (D); of SEP-like MADS-box TFs GRCD4, GRCD5, and GRCD8 (E); and of B- (GGLO1, GDEF1/2) and C-class (GAGA1/2) as well as FUL-like (GSQUA2) MADS box TFs (F) are shown. Error bars represent se from three biological replicates. Asterisks indicate statistically significant differences (***P < 0.001).
We also applied in silico analysis to identify putative transcription factor binding sites (TFBSs) within the 1000 bp GhCYC3 promoter region. Considering potential auto- and cross-regulatory feedback loops of CYC2 clade genes (Yang et al., 2012; Yuan et al., 2020), we identified two putative TCP TFBSs at positions −278 to −282 bp and −920 to −924 bp from the translation start site (TSS; Fig.1B). These elements correspond to the core motif (GGNCC) that is over-represented in many TCP-regulated genes (Martín-Trillo and Cubas, 2010). Moreover, the GTGCCC motif within the 27-bp consensus sequence strongly resembles the GC-rich core motif (Fig. 1A). We also focused on MADS-box TF binding sites, called as CArG boxes, CC(A/T)6GG, as MADS-box proteins have been suggested to operate in the same regulatory cascades as TCP factors (Kaufmann et al., 2009). We identified two candidate CArG boxes within the GhCYC3 promoter, CCTAAAAGAG at −155 to −164 bp, and CCAATTCTGA at −192 to −201 bp (Fig. 1C).
GhCIN1/2, GRCD5, and GAGA1 TFs Activate the PGhCYC3:LUC Reporter
We tested the ability of the ten candidate TFs of gerbera (Supplemental Table S2) to transcriptionally activate the PGhCYC3:LUC reporter by transient agroinfiltration in N. benthamiana. We did not observe reporter activation with any of the candidate proteins of the AP2/Ethylene response factor, NAC, DNA binding with one finger, or Homeodomain- Leu zipper families (Supplemental Fig. S3A). However, two out of the three CIN-like TCP domain TFs, GhCIN1 and GhCIN2, activated the reporter construct (Fig. 1D). Both GhCIN1 and GhCIN2 showed activation only when fused to the VP16 domain, suggesting that they may bind the target DNA, but require (an)other cofactor(s) for transcriptional activation. In contrast, GhCIN3 did not show any transcriptional activity even when fused with the VP16 domain. We also tested ten previously identified gerbera CYC/TB1-like TFs (GhCYC1-10; Supplemental Table S3; Tähtiharju et al., 2012); however, none of them could individually activate the PGhCYC3:LUC reporter (Supplemental Fig. S3B).
Additionally, we tested candidate MADS-box TFs based on their known functional roles during flower primordia and/or petal and stamen development in gerbera (Supplemental Table S3). These included B (GGLO1 and GDEF1/2), C (GAGA1/2), and E (GRCD4/5/8) class as well as FUL-like (GSQUA2) genes. Of these, the SEP3-like MADS-box TFs GRCD5 and its close paralog GRCD8, as well as the C class TF GAGA1, showed reporter activation in the agroinfiltration assay (Fig. 1, E and F). None of the other tested MADS-box proteins activated the reporter, including the SEP1/2/4-like GRCD4, GAGA2 (a close paralog of GAGA1), GSQUA2, or the B protein heterodimer combinations (GGLO1/DEF1 and GGLO1/DEF2).
Next, we explored different regions of the GhCYC3 promoter (Fig. 2A, constructs 1–5) in combination with the candidate upstream TFs GhCIN1, GhCIN2, GAGA1, GRCD5, and GRCD8. All candidate proteins activated the reporter constructs including either the 1900, 878, or 367 bp 3′ fragments of the promoter (constructs 2, 4, and 5, respectively), while the lack of the 3′ region (construct 3) abolished activation (Fig. 2, B and C). This result corresponds with the presence of the TCP TFBS and the two CArG boxes within the 367-bp sequence upstream of the TSS (Fig. 1).
Figure 2.
Activation of PGhCYC3:LUC reporter constructs by CIN and MADS box proteins in transient luciferase assay in N. benthamiana. A, Tested deletion constructs of GhCYC3 promoter (constructs 2–5), and constructs with mutated TCP and MADS transcription factor binding sites marked by red asterisks (constructs 6–8). Detailed mutated sequences are shown in Figure 1. B and C, The effector constructs GhCIN1:VP16 and GhCIN2:VP16 (B), and GAGA1:VP16, GRCD5, and GRCD8 (C) show strong activation with the full-length PGhCYC3 promoter (construct 2) and with constructs containing the intact 3′ end of the promoter (constructs 4 and 5). Deletion of the 3′ end (construct 3) abolishes activation. The reporter activation by CIN and MADS-box TFs was abolished when the TCP2 (construct 6 shown in B) and CArG1 (construct 7 shown in C) binding sites were mutated. Red connecting lines refer to intact and mutated reporter constructs, respectively. Empty reporter construct without the GhCYC3 promoter fragment was used as a control (construct 1). Error bars represent se from three biological replicates. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001).
Mutations in the TCP and MADS-Box Binding Motifs Abolish GhCYC3 Activation
To verify the DNA binding activity of the CIN-like factors with the core TCP binding motif GGtCC at position −272-bp upstream of the 27 bp consensus, we mutated the site into GGtTG within the 367-bp promoter sequence (mTCP2; Figs. 1 and 2). Using the mutated reporter (Fig. 2A, construct 6), GhCIN1:VP16 and GhCIN2:VP16 fusion proteins failed to induce luciferase activity above the background level (Fig. 2B). Our results show that the conserved GGNCC motif mediates GhCIN1/2 activation.
Similarly, to understand the specificity of the two CArG boxes to MADS TF binding within the 367-bp GhCYC3 promoter sequence, we mutated them individually: CCAATTCTGA to AAAATTCTGA (mCArG1) and CCTAAAAGAG to TTTAAAAGAG (mCArG2; Figs. 1 and 2). Transient LUC assays indicated that the CArG1 box (construct 7; Fig. 2A), but not the CArG2 box (construct 8; Fig. 2A), is necessary for transcriptional activation by GAGA1 and GRCD5/8 (Fig. 2C). Mutated CArG boxes did not affect the reporter activation by GhCIN1/2 TFs, and neither did the TCP mutations affect activation by MADS TFs (Supplemental Fig. S4).
GhCIN1/2 and GRCD5 Expression Colocalizes with GhCYC3 mRNA During Ray Primordia and Ligule Development
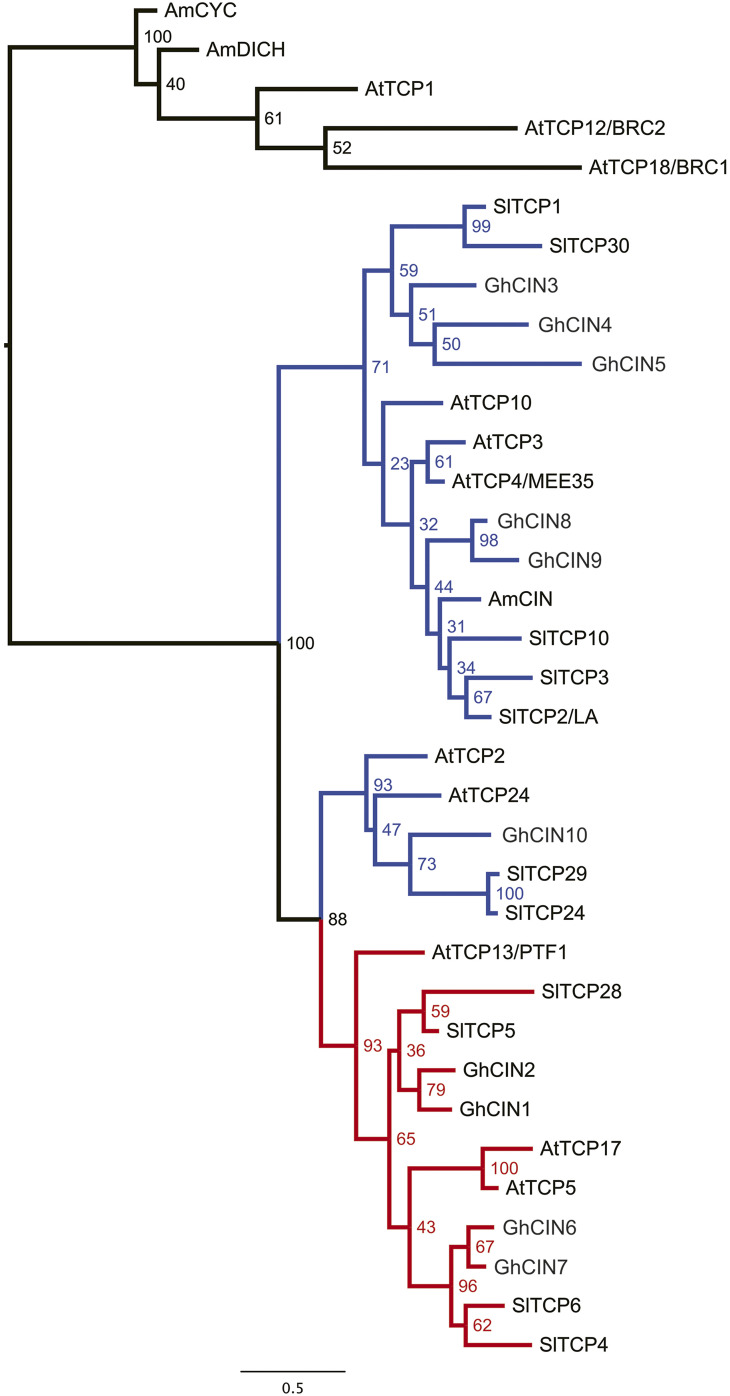
The identified TCP genes are homologs of class II, CIN-like TF genes. For phylogenetic analysis, we identified ten CIN-like genes from the gerbera RNA Sequence (RNASeq) database (Supplemental Table S4). The reconstructed phylogeny indicates that these genes are grouped into the previously identified subclades (Fig. 3; Martín-Trillo and Cubas, 2010; Parapunova et al., 2014). GhCIN1 and GhCIN2 appear as duplicated paralogs representing the TCP5-like clade, and GhCIN3 groups with the JAGGED AND WAVY (JAW)–like genes. Based on RNAseq read counts, both GhCIN1 and GhCIN2 are up-regulated in ray flower primordia, similar to GhCYC3 (Tähtiharju et al., 2012), while GhCIN3 expression is up-regulated in disc flowers (Supplemental Fig. S2, A–C).
Figure 3.
Maximum likelihood–based phylogenetic tree of class II TCP genes based on the nucleotide alignment of the TCP domain. The sequences used for the analysis are listed in Supplemental Table S4. The TCP5-like CIN genes are indicated in red, and the JAW-like in blue. CYC/TB1-like genes were used as an outgroup (black). One thousand bootstrap replicates were generated to assess support for the inferred relationships. The scale for the branch lengths refers to the expected number of nucleotide substitutions per site.
We performed reverse transcription quantitative PCR (RT-qPCR) for expression analysis of GhCIN1, GhCIN2, GRCD5, and GRCD4. In parallel, the expression pattern of GhCYC3 was verified (Fig. 4). In general, the expression of both GhCIN1 and GhCIN2 overlaps with GhCYC3, although GhCIN1 is expressed at a much higher level than GhCIN2 (Fig. 4, A–D and F). During early development, both GhCYC3 and GhCIN1 are up-regulated in ray flower primordia compared with disc flower primordia (Fig. 4, A and C). This confirms our previous data showing that GhCYC3 is exclusively expressed in ray primordia (Tähtiharju et al., 2012). Although GhCIN2 also shows differential expression in ray versus disc flower primordia, it is predominantly expressed in involucral bract primordia that surround and protect the growing head (Fig. 4B). The expression of GhCYC3 and GhCIN1 also overlaps during ray flower ligule development (stages 2–10; Fig. 4, D–F). As previously detected for GhCYC3 (Juntheikki-Palovaara et al., 2014), both genes show the greatest expression at stage 2, after which their expression gradually decreases. Similar to the early primordia stage, GhCIN2 shows strongest expression in mature involucral bracts (Fig. 4, B and D), while its expression is low and constant during ligule development (Fig. 4D). Our data suggests that GhCIN1 is likely to be involved in ray primordia and ligule development while GhCIN2 may affect bract development.
Figure 4.
Expression patterns of GhCIN1, GhCIN2, GhCYC3, GRCD4, and GRCD5. A to C, During early flower primordia development, GhCIN1 (A), GhCIN2 (B), and GhCYC3 (C) show overlapping expression domains being up-regulated in ray primordia (RP) compared with disc primordia (DP). GhCIN2 (B) expression is highest in early bract primordia (BrP). d to F, During ray flower ligule development, GhCIN1 (D), GRCD5 (E), and GhCYC3 (F) expression is strongest at the early stages in elongating ligules, and gradually decreases along the development. GhCIN2 expression is strongest in mature involucral bracts (Br; D). GRCD4 (E) is up-regulated during late ligule development. The ray and disc flower primordia samples represent stages 2, 4, and 6 (S2, S4, S6; Laitinen et al., 2006). The undifferentiated inflorescence meristem (IM) samples correspond to three developmental stages (Zhang et al., 2017). Ray flower ligule samples correspond to developmental stages 2, 4, 6, 8, and 10 (S2, S4, S6, S8, S10; Laitinen et al., 2007). Other samples include leaf (Lf) and root (Rt). The relative expression levels of given genes are normalized to the GhACTIN gene, and are comparable with each other by the ΔCt method. Error bars represent se from three biological replicates.
For the MADS-box genes, we focused on GRCD4 and GRCD5 as we have previously defined their functions in ray flower ligule development (Zhang et al., 2017). At this stage, we omitted GRCD8 from further analyses. When compared with its paralog GRCD5 that is predominantly expressed in ligules, GRCD8 shows more ubiquitous expression in all floral organs, and we still lack transgenic lines to verify its function (Zhang et al., 2017). Regarding GAGA1, our previous data indicates that it represents a classical C class gene being expressed only in stamens and carpels (Yu et al., 1999). Silencing of GAGA1 in transgenic gerbera led to homeotic conversion of stamens into petals and carpels into sepal-like structures (Yu et al., 1999; Kotilainen et al., 2000).
Here, we analyzed the expression of GRCD5 and GRCD4 during ray flower ligule development (Fig. 4E). GRCD5 follows a pattern similar to GhCYC3, peaking at stage 2 and gradually decreasing along the developmental sequence, while GRCD4 is up-regulated during late ligule development (stage10). Our data suggest that the specific function of GRCD5 affecting ray flower ligule elongation (Zhang et al., 2017) may occur through GhCYC3. Our previous functional data indicated that GRCD4 would instead control the specification of epidermal cells in ligules (Zhang et al., 2017).
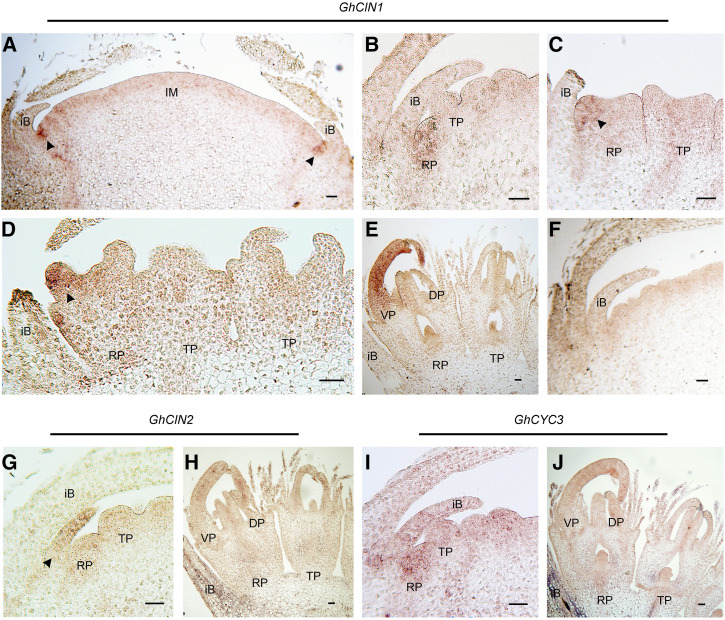
We further conducted in situ hybridization to compare the tissue-specific expression domains of GhCIN1, GhCIN2, and GhCYC3 during early flower primordia development (Fig. 5). GhCIN1 expression is absent from the undifferentiated inflorescence meristem but is restricted to the axils of involucral bracts, localizing to the positions of emerging ray primordia (Zhao et al., 2016; Fig. 5A). This pattern continues when the ray primordia initiate (Fig. 5B). From stage 2, GhCIN1 is exclusively expressed at the ventral side of ray primordia, in association with elongation of the ventral ligule (Fig. 5, C–E). We did not detect any expression at the dorsal side of ray primordia or in trans- or disc-flower primordia. In contrast, GhCIN2 expression localizes to the involucral bract primordia and weakly in initiating ray primordia, corresponding to our RT-qPCR results (Figs. 4B and 5, G and H). Similar to GhCIN1, GhCYC3 shows strongest expression within the initiating ray primordia (Fig. 5I), as well as in the elongating ventral ligules of ray flowers (Fig. 5J).
Figure 5.
Localization of GhCIN1, GhCIN2, and GhCYC3 expression by in situ hybridization. A, GhCIN1 localizes at the axils of involucral bracts (iB; arrows) that surround the inflorescence meristem (IM). B, GhCIN1 marks the initiation of ray primordia (RP) but is absent from adjacent trans primordia (TP). C to E, GhCIN1 is expressed at the ventral side of the RP (arrow; C and D), and later it localizes to the ventral ligular petal (VP) but is absent from the dorsal petal (DP; E). F, Negative (sense) control of GhCIN1. G and H, GhCIN2 shows high expression in young bract primordia (iB, arrow). I and J, GhCYC3 expression is localized to the incipient RP (I) and to the ventral and dorsal petals (VP, DP) of ray primordia (J). Scale bars = 50 µm.
Silencing of GhCIN1/2 Affects Ray Primordia Development in Association with Reduced GhCYC3 Expression
For functional studies, we generated transgenic lines using the GhCIN1 and GhCIN2 RNA interference (RNAi) constructs. Four independent lines for GhCIN1 and three for GhCIN2 were analyzed. All transgenic lines showed down-regulation of both genes, however, to a different extent (Supplemental Fig. S5, A and B). As expected, loss of GhCIN1 and GhCIN2 expression caused reduced GhCYC3 expression in ray primordia samples (Supplemental Fig. S5, A and B). Although GhCIN1 and GhCIN2 share perfect 20-mer sequence stretches only with each other, and not with the other gerbera CIN-like genes (Supplemental Table S5), we verified the expression GhCIN3, GhCIN8, GhCIN9, and GhCIN10 for possible cross-silencing in the transgenic lines (Supplemental Fig. S5C). All these genes are expressed in flower primordia of wild-type gerbera. Two GhCIN1 RNAi lines showed down-regulation of GhCIN3 expression, whereas two other lines did not, indicating that GhCIN3 very unlikely contributes to the early ray primordia phenotype that was consistent in all of these lines. The other tested genes did not show cross-silencing in the transgenic lines (Supplemental Fig. S5C).
In transgenic lines, the phenotypes of mature inflorescences especially regarding ligule growth were minor and variable; however, phenotypic changes during early primordia initiation were obvious and were observed in all four GhCIN1 RNAi lines and in one GhCIN2 RNAi line (TR15; Fig. 6). We have previously shown that development of ray primordia in wild-type gerbera is temporally delayed compared with their neighboring trans-flower primordia (Zhao et al., 2016). At an early stage, the ray primordia are undifferentiated and bump-shaped, whereas the adjacent trans-primordia already start to initiate ring-shaped petal primordia (Fig. 6, A and B). Later, when ray primordia become ring-shaped, the neighboring trans-primordia already initiate petal and stamen primordia (Fig. 6, C and D). In GhCIN1 and GhCIN2 RNAi lines, such delay in organogenesis was not observed (Fig. 6, E–L). In fact, ray primordia development was accelerated compared with that of the trans-primordia (Fig. 6, F, H, J, and L). This suggests that GhCIN1 and GhCIN2 contribute to the early ontogeny of ray primordia, and regulate their delayed early development through upregulation of GhCYC3 expression.
Figure 6.
Phenotypes of the transgenic GhCIN1 and GhCIN2 RNAi lines compared with nontransgenic, wild-type gerbera plants (WT). A to d, Two consecutive developmental stages of early head development in wild-type gerbera. Ray primordia (shaded in yellow) show delayed organogenesis compared with neighboring trans-primordia (shaded in red). E to l, Corresponding developmental stages in GhCIN1 RNAi lines (E–H) and GhCIN2 RNAi lines (I–L). In contrast with wild type, the ray primordia in GhCIN1 RNAi and GhCIN2 RNAi plants show faster organogenesis than neighboring trans-primordia. Scale bars = 500 µm.
GRCD5 RNAi Transgenic Lines Show Reduced GhCYC3 Expression
We have previously produced transgenic RNAi lines showing that GRCD4 and GRCD5 encode partially redundant proteins that affect ray flower ligule development (Zhang et al., 2017). Loss of GRCD4 function resulted in trichome formation on the petal epidermis, whereas loss of GRCD5 reduced the ligule length in ray flowers. The luciferase reporter and expression analyses presented above suggest that GRCD5, rather than GRCD4, is a putative upstream regulator of GhCYC3. Therefore, we determined the GhCYC3 expression levels in early ray flower ligule samples (stage 2 and stage 4) of gene-specific GRCD4 RNAi and GRCD5 RNAi as well as GRCD4 and GRCD5 double RNAi lines (Supplemental Fig. S5D). The selected lines are specific and do not show cross downregulation of other SEP-like gene family members (Zhang et al., 2017). We observed that in GRCD4 RNAi lines, GhCYC3 transcript levels were not affected, whereas GhCYC3 expression was significantly down-regulated in association with reduced GRCD5 expression in both the GRCD5 RNAi lines and in the GRCD4 and GRCD5 double RNAi lines. These data indicate that GhCYC3 acts downstream of GRCD5 to affect ligule elongation.
DISCUSSION
CYC2 clade proteins are conserved regulators of bilateral flower symmetry across angiosperms. In Asteraceae this gene family has, through gene duplications and diversification, evolved new functions in defining ray flower identity, thereby contributing to the diversity of inflorescence form as well as floral architecture among flowering plants. The Asteraceae CYC2 clade genes are expressed in the inflorescence margins, in emerging ray flower primordia but also later during floral organ differentiation. However, we still lack knowledge of how their highly localized early expression domain is defined, and the regulatory networks that they are involved in. Here, we discovered previously unidentified regulatory links among TCP and between TCP and MADS-box TFs and showed functional evidence that they contribute to flower type differentiation in gerbera. We showed that CIN-like genes, whose functions have previously been studied only in Antirrhinum majus and Arabidopsis, have been recruited to regulate the early ontogeny of ray flowers, and likely also the development of involucral bracts. Moreover, during late development of ray flowers, MADS-box TF complexes target the GhCYC3 gene during both petal and stamen differentiation (Fig. 7).
Figure 7.
The involvement of CIN- and SEP-like TFs in defining ray flower development in gerbera. A, During early stage of inflorescence development, GhCIN1 and GhCIN2 expression marks the positions of emerging ray flowers and involucral bracts, respectively. B to D, During flower differentiation, GhCIN1 activates GhCYC3 expression in the ventral domain of marginal ray flowers. The SEP-like GRCD5 expression extends to disc flowers but is colocalized with GhCYC3 in emerging ray primordia. E to G, During late ray primordia development, GhCIN1, GRCD5, and GhCYC3 expression colocalizes to the elongating ventral ligule (Vp). Ca, Carpel; Dp, dorsal petal; IM, inflorescence meristem; Ov, ovary; Pa, pappus; St, stamen.
GhCIN1 Affects Early Ray Flower Development Through GhCYC3
Our data demonstrate regulatory interactions among the class II TCP genes, between the CIN-like TFs and the CYC2 clade gene GhCYC3. We showed using agroinfiltration assays that both TCP5-like CIN TFs, GhCIN1:VP16 and GhCIN2:VP16, activated the PGhCYC3:LUC reporter construct while the JAW-like GhCIN3:VP16 did not (Fig. 1D). The core sequence motif (GGNCC) of the TCP binding site was shown to be necessary for binding (Fig. 2).
So far, the known CIN functions have been related to leaf and petal lobe development. In A. majus, the strong cin mutant develops larger leaves with concave and curled edges as well as reduced petal lobes, indicating that AmCIN can both promote and arrest growth by affecting cell division (Crawford et al., 2004). In Arabidopsis, eight highly redundant CIN-like genes are divided into two subclades: the microRNA-regulated (miR319) JAW-like (TCP2/3/4/10/24) and the TCP5-like (TCP5/13/17) genes. Loss of JAW-like function in Arabidopsis promotes cell divisions at leaf margins resulting in highly crinkled leaves (Efroni et al., 2008; Nicolas and Cubas, 2015). Additionally, reproductive tissues such as petals, sepals, and siliques show wavy surfaces and serrated margins (Koyama et al., 2007; Nag et al., 2009). The triple mutant of TCP5-like genes (tcp5tcp13tcp17) has larger leaves and wider petals, whereas overexpression of TCP5 results in smaller petals (Efroni et al., 2008; Huang and Irish, 2015; van Es et al., 2018). In contrast with Asteraceae with several CYC2 clade genes, Arabidopsis harbors only a single CYC2 clade gene, AtTCP1, which controls elongation of petioles, leaf blades, and inflorescence stems (Koyama et al., 2010). No putative TCP binding site has been detected in the AtTCP1 promoter, suggesting that the link between CIN-like proteins and CYC2 clade genes may not exist in Arabidopsis (Yang et al., 2012).
CYC2 clade genes have independently been recruited to control bilateral flower symmetry across angiosperms. Delayed initiation of floral organs was associated with CYC2 clade gene expression in Antirrhinum majus (Luo et al., 1996), Torenia fournieri (Su et al., 2017), and Saintpaulia ionantha (Hsu et al., 2018). In these species, CYC2-like gene expression is restricted to the dorsal domains of the flowers suppressing the growth of the petal and stamen primordia. When CYC2-like gene expression is lost in Antirrhinum spp. or Saintpaulia spp., all petal and stamen primordia, respectively, appear at approximately the same time (Luo et al., 1996; Hsu et al., 2018). In Asteraceae, including gerbera, CYC2 gene expression localizes in marginal ray flower primordia, which show developmental delay compared with their neighboring trans- or disc-flowers (Harris, 1995; Tähtiharju et al., 2012; Zhao et al., 2016). Our data indicates that in gerbera, the CIN-like TFs regulate this delay, upstream of the ray-specific GhCYC3.
GhCIN1 is expressed in the incipient ray primordia at the axils of the involucral bracts (Figs. 5 and 7A). Our previous data showed that the flower meristem identity gene GhLFY is also expressed at the same location (Zhao et al., 2016). Zhao et al. (2016) showed that suppression of GhLFY expression converted ray initials into branched structures, suggesting that marginal ray flowers evolved as axillary structures with suppressed branching properties. It is tempting to speculate that GhCIN1 and/or GhCYC3 function in parallel or downstream of GhLFY to promote ray flower differentiation. We also showed that GhCIN2, a paralog of GhCIN1, activates the PGhCYC3:LUC reporter. However, GhCIN2 is expressed in involucral bract primordia that lack GhCYC3 expression (Figs. 4, B and C, 5, G and I, and 7A), and thus GhCIN2 may act as a repressor, most likely through interaction with yet unknown factors, to exclude GhCYC3 from bracts. Because the transgenic gerbera RNAi lines did not show any bract phenotypes, the specific role of GhCIN2 still remains open.
At the level of single flowers, asymmetrical ventralized expression of CYC2 clade genes is characteristic for Asteraceae (Broholm et al., 2008; Juntheikki-Palovaara et al., 2014; Garcês et al., 2016), but has also been detected in Zingiberales and Commelinales lineages of monocots (Bartlett and Specht, 2011; Preston and Hileman, 2012). The expression of GhCIN1 in gerbera colocalizes with GhCYC3 to the ventral side of initiating ray flower primordia, as well as to the expanding ventral ligule (Figs. 5, A–E, and 7, B and C), suggesting a direct regulatory link. Similar transient ventral expression at the onset of ray primordia has earlier been shown for the gerbera B genes GGLO1 and GDEF2 (Yu et al., 1999) and for the E gene GRCD1 (Kotilainen et al., 2000). We propose that this pattern is a response to a yet unknown signal across capitulum development contributing to establishment of bilateral symmetry of ray flowers (Yu et al., 1999). Ectopic activation of GhCYC3 in transgenic gerbera promoted ligule elongation in disc flowers because of increased cell numbers (Juntheikki-Palovaara et al., 2014). Yet, during late developmental stages, the GhCIN1 RNAi lines did not show consistent phenotypes in ligules. We anticipate that the other CYC2 genes redundantly contribute to ligule growth necessitating additional studies to understand their specific roles.
MADS Box Proteins Target GhCYC3 During Ligule Elongation and Staminode Development in Ray Flowers
Our data suggest the involvement of GRCD5, and possibly also its paralog GRCD8, in ligule development and GAGA1 in staminode development as potential upstream regulators of GhCYC3. First, we showed that the SEP3-like proteins GRCD5 and GRCD8 could effectively activate the PGhCYC3:LUC reporter, whereas GRCD4 did not (Fig. 1E). The expression of GRCD5 closely overlaps with GhCYC3 specifically during ray flower ligule development (Figs. 4, E and F, and 7, D and E). We also showed that GhCYC3 expression was significantly reduced in GRCD5 RNAi and in GRCD4 and GRCD5 double RNAi lines (Supplemental Fig. S5D) in association with reduced ligule length (Zhang et al., 2017). In the future, the functional role of GRCD8 should be clarified. Our previous studies have shown that GRCD4 and GRCD5 interact both pairwise and with many other MADS box proteins in yeast 2-hybrid assays (Ruokolainen et al., 2010a). Among the fourteen MADS box proteins tested, they were the only ones forming homodimers that may explain the functional specificity observed here. Yet, protein complex formation should be verified in planta and include DNA binding assays.
In Arabidopsis, TCP genes (including PCF-like and JAW-like, as well as the CYC-like gene TCP18) were significantly overrepresented among SEP3 targets (Kaufmann et al., 2009). The regulatory link between SEP3 and target TCP gene(s) may thus be conserved, although it is likely to be connected with distinct biological functions in diverse species. Kaufmann et al. (2009) also showed that the DNA-binding motifs of both MADS-box and TCP TFs are enriched in regions bound by SEP3. Similarly, we identified two CArG boxes and two conserved TCP TFBSs in the regulatory region of GhCYC3 (Fig. 1). Whether the CIN-like TCPs and GRCD5 interact or function in the same protein complexes needs to be verified.
Previous studies in Antirrhinum spp. suggested that the maintenance of CYC expression during late petal development depends on the B class MADS-box gene DEFICIENS (Clark and Coen, 2002). Our luciferase assay with heterodimeric combinations of gerbera B genes did not activate the GhCYC3 reporter construct (Fig. 1F). The gerbera B class proteins, however, form higher order complexes with AP1/FUL, SEP, and C class proteins (Broholm et al., 2010; Ruokolainen et al., 2010a). As shown in yeast three-hybrid assays, the gerbera B function GGLO1-GDEF2 dimer forms protein complexes with both GRCD5 and GAGA1 (Ruokolainen et al., 2010a). Therefore, we cannot rule out the possibility that the B proteins, as members of higher order protein complexes, may also contribute to GhCYC3 regulation during petal and stamen development.
Both CYC2 clade and MADS box genes have been shown to affect stamen development in gerbera. Although stamen primordia in gerbera initiate similarly in both flower types, their development stops in ray flowers leading to development of sterile staminodes. The C class MADS box proteins GAGA1 and GAGA2 form higher order complexes with the E class protein GRCD1 in yeast 3-hybrid assays, and all of them have previously been shown to regulate staminode development (Yu et al., 1999; Kotilainen et al., 2000; Ruokolainen et al., 2010a). Suppression of these genes led to similar phenotypes, respectively, and converted the staminodes into petals. Here we showed that GAGA1 is able to activate the GhCYC3 reporter construct, whereas GAGA2 is not (Fig. 1F). On the other hand, overexpression of CYC2 clade genes in gerbera, including GhCYC3, disrupted stamen development in modified disc flowers (Broholm et al., 2008; Juntheikki-Palovaara et al., 2014). As GhCYC3 is not expressed in staminodes (Fig. 5; Juntheikki-Palovaara et al., 2014), it is possible that this phenotype is caused by other CYC2 clade genes activated by GhCYC3. Based on the data shown here, we propose that the GAGA1-GRCD1 pair, possibly together with B proteins, may be involved in a protein complex that suppresses GhCYC3 expression in ray flower staminodes.
In summary, we show here that TCP and MADS-box TFs cooperate to control flower type identity at the inflorescence level, and their morphological differentiation at flower organ level. Our results emphasize the importance of future studies to explore whether the observed interactions are specific to Asteraceae, and to identify the in planta protein complexes, as well as the detailed downstream processes.
MATERIALS AND METHODS
Plant Materials
Gerbera hybrida (gerbera; Asteraceae) ‘Terra Regina’ and transgenic gerbera lines derived from it were grown under standard greenhouse conditions (Ruokolainen et al., 2010b). Nicotiana benthamiana plants were germinated and grown as previously described (Bashandy et al., 2015).
Genome Walking for GhCYC Promoter Regions
We applied genome walking for identification of promoter sequences for the gerbera CYC clade genes (Tähtiharju et al., 2012). Genomic DNA was extracted by the cetyl-trimethyl-ammonium bromide method (Chang et al., 1993), and depending on the given gene sequence, digested with a restriction enzyme (either EcoRV, DraI, PuvII, StuI, HindII, SspI, NaeI, or Eco47III). The Genome Walker Adaptor was ligated according to the instructions of the GenomeWalker Universal kit (Clontech Laboratories). The first PCR was performed with the corresponding adaptor primer1 (AP1, GER1) and gene-specific primer1 (GSP1), and the second PCR with the AP2 (GER2) and gene-specific primer2 (Supplemental Table S6), using the recommended conditions and the Advantage 2 Polymerase Mix (Clontech Laboratories). The PCR products for each GhCYC promoter were cloned into pGEM-T Easy Vector (Promega).
In Silico Analysis of the Asteraceae CYC2 Clade Promoter Sequences
We performed in silico analyses using MEME (http://meme-suite.org; Bailey and Elkan, 1994) and DiAlign (http://www.genomatix.de/cgi-bin/dialign/dialign.pl; Morgenstern et al., 1996) to search for conserved cis-elements among the Asteraceae CYC2 clade promoter sequences. In addition to gerbera promoters, we included promoter sequences of CYC2 clade genes from Helianthus annuus (nine sequences provided by Mark Chapman, John Burke, and Nicolas B. Langlade, and confirmed in https://sunflowergenome.org/), Berkheya purpurea (three sequences provided by Mark Chapman and John Burke), and Lactuca sativa (three sequences obtained from http://lgr.genomecenter.ucdavis.edu/; Supplemental Table S1). For the analyses, we used 1000 bp sequences upstream of the TSS, except for those where only shorter sequence stretches were available (891 bp for HaCYC2d, 579 bp for LsCYC2a, 273 bp for LsCYC2c2, 364 bp for BpCYC2a, 558 bp for BpCYC2b, and 423 bp for BpCYC2c). We also identified additional putative TFBSs within the GhCYC3 regulatory region by using PlantPAN (http://plantpan2.itps.ncku.edu.tw/; Chow et al., 2016), CIS-BP (http://cisbp.ccbr.utoronto.ca/; Weirauch et al., 2014), and JASPAR (http://jaspar.genereg.net/; Khan et al., 2017).
Y1H Assays
Based on the MEME results, a conserved 27-bp element was identified from the Asteraceae CYC2 clade promoters (Supplemental Fig. S1). We cloned the corresponding 27-bp element from the gerbera GhCYC3 promoter (from -151 to −178 bp) in three tandem repeats into a yeast reporter vector pAbAi (PT4091-5; Clontech) conferring resistance to Aureobasidin A (AbA, Clontech), following the protocol of Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech). This construct, named as pHTT873, was used as a bait in Y1H screening. Cloning primers are listed in Supplemental Table S6.
To generate the yeast bait strain, the bait plasmid was linearized with BstBI, transformed into the Y1H Gold strain (Clontech), and plated on SD-Ura growth medium. Integration into the yeast genome was confirmed by colony PCR combining a vector-specific forward primer and an insert-specific reverse primer (Supplemental Table S6). The bait strain was tested for the minimal inhibitory concentration of AbA. We also integrated the pAbAi vector without any insert into Y1HGold, and used it as a negative control bait strain. All yeast transformations were done following either small- or library-scale LiAc-transformation protocols described in Yeastmaker Yeast transformation system 2 manual (PT1172-1, Clontech).
A Y1H screening was performed using the Arabidopsis (Arabidopsis thaliana) AtTF prey library expressing c. 1500 AtTFs (Mitsuda et al., 2010), and using the bait pHTT873. Approximately two million colonies were screened and selected on SD-Leu-Ura/900 ng mL−1 AbA selection plates. The candidate Arabidopsis AtTF gene sequences obtained from the library screen were used in BLAST searches against the gerbera RNASeq databases (T. Teeri and P. Elomaa, unpublished data) to identify the gerbera homologs (Supplemental Table S2). In addition, we defined the expression patterns for the gerbera homologs based on the read counts in our RNASeq data and identified candidate genes that are coexpressed with GhCYC3 (Supplemental Fig. S2). Their ability to activate the GhCYC3 reporter construct was examined in planta using agroinfiltration into N. benthamiana.
Isolation of Gerbera Homologs, Genetic Transformation of Gerbera, and Phenotypic Analysis of Transgenic Lines
Based on the Y1H result, the sequences of corresponding gerbera homologs were identified from the gerbera RNAseq database using BLAST searches (Supplemental Table S2). The full-length cDNAs (with and without stop codon) were cloned into Gateway entry vector pDONR221 (Invitrogen) and were verified by sequencing. Two CIN-like genes (GhCIN1 and GhCIN2) were selected for further functional studies in transgenic gerbera. For genetic transformation, we used the Gateway binary vectors pK7GWIWG2D(II; Karimi et al., 2002) to generate RNAi constructs with full-length GhCIN1 and GhCIN2 cDNAs, respectively. The gene constructs were electroporated into the Agrobacterium tumefaciens strain C58C1 harboring pGV3101. Cloning primers are listed in Supplemental Table S5. Transformation of gerbera ‘Terra Regina’ with GhCIN1 RNAi (PAT73) and GhCIN2 RNAi (PAT75) constructs was done as previously described (Elomaa and Teeri, 2001). Four independent RNAi lines for GhCIN1 and three lines for GhCIN2 were included for phenotypic analyses, and 2 to 6 heads from each line were analyzed. Scanning electron microscopy (SEM) was conducted as in Uimari et al. (2004) and Zhang et al. (2017).
Transient Agroinfiltration Analyses
For the effector constructs (Supplemental Table S2), we used the Gateway destination vector pDEST35SVP16HSP (Oshima et al., 2013) to fuse the TF genes with the VP16 activation domain. The fusion fragments (TF:VP16) were then cloned into pK7WG2D (Karimi et al., 2002) using a gene-specific adaptor forward primer and the VP16 reverse primer GER956 (35S:TF:VP16). The TFs known to show autoactivation, including GRCD4 and GRCD5 (Ruokolainen et al., 2010a), GRCD8 as a close paralog of GRCD5 (Zhang et al., 2017), and all GhCYC proteins except GhCYC5 and GhCYC7 (Tähtiharju et al., 2012) were tested without the VP16 domain in pK7WG2D (Karimi et al., 2002; 35S:TF; Supplemental Table S3). For the reporter constructs used for agroinfiltration, different fragments of GhCYC3 promoter sequences (−1900/−878/−367 bp to 1 bp, and −1900 to −800 bp) were first cloned into the Gateway entry vector pDONR221 (Invitrogen) using gene-specific primers. The promoter fragments were further cloned into the Gateway destination vector pKGWL7.0 (VIB, Ghent University), and thus fused with the luciferase (LUC) reporter (PGhCYC3:LUC). To obtain the mutated reporter constructs, two parallel PCR amplifications from the original plasmid templates were performed using interval site-directed mutagenesis forward and reverse primers with 3′ and 5′ gene-specific adaptor primers GER115/GER1014, respectively. To fuse the two PCR fragments, full-length fragments were amplified from a mixture of the two PCR products using adaptors GER29+GER30, cloned into pDONR221, and then into pKGWL7.0. The negative control (pKGWL7.0-SalI) is the backbone of the luciferase reporter plasmid pKGWL7.0 without the attR1-attR2 fragment that is removed by SalI digestion and ligated by T4 ligation (Thermo Fisher Scientific). All cloning primers are listed in Supplemental Table S6.
All constructs were electroporated into the A. tumefaciens strain C58C1 (pGV2260). The agroinfiltration in N. benthamiana was conducted as in Bashandy et al. (2015) except that we used a final bacterial density of OD600 = 1 in the infiltration medium. Six-week-old N. benthamiana plants were used for agroinfiltration and sampled after 3 d for luciferase activity.
Determination of Luciferase Activity
Infiltrated leaves were sampled and punched using the cap of 2 mL Eppendorf tubes. Each leaf sample was added to tubes containing 100 µL of cold sampling buffer (50 mm Na-phosphate pH 7.0, 4% [w/v] soluble polyvinylpyrrolidone MW 360,000, 2 mm EDTA, 20 mm dithiothreitol). Soluble proteins were homogenized by grinding using a mixer mill (Retsch MM400) and collected by centrifuging 13,300 rpm 15 min at 4°C. To test luciferase activities, 20 µL of the supernatant was added into 50 µL of enzyme substrate (Luciferase 1000 Assay System, no. E4550, Promega) in cuvettes (PP, SARSTEDT), fast vortexed, and by counting the photons for 1 s in the luminometer (Luminoskan TL plus, Generation II, Thermo Labsystems). Two to five replicates were analyzed for luciferase activity, and the experiment was repeated at least two times. Statistical differences in luciferase activities between the control and test samples were analyzed using the general linear model in SPSS.
RT-qPCR for Expression Analyses
RT-qPCR was applied for expression analysis in wild-type gerbera tissues and transgenic samples. The wild-type tissues consist of flower primordia of ray and disc flowers at developmental stages 2, 4, and 6 (Laitinen et al., 2006), ray flower ligule samples from stages 2, 4, 6, 8, and 10 (Laitinen et al., 2007), undifferentiated inflorescence meristem samples (Zhang et al., 2017), and vegetative samples including early involucral bract primordia, mature bracts, young leaf, and root. For the GhCIN1 and GhCIN2 RNAi transgenic lines, ray primordia samples were collected at stage 3. For the GRCD4 and/or GRCD5 RNAi lines the samples were collected from ray flower petals at staged 2 and 4. Two to three biological replicates for each sample were used. The RT-qPCR primers used for expression analyses are listed in Supplemental Table S6. GhACTIN was used as an internal control. Statistical differences in expression levels between the control and the transgenic samples were analyzed using the independent samples t test.
In Situ Hybridization
The preparation of the plant samples, sectioning, and hybridization steps were performed as previously described (Elomaa et al., 2003). Gene-specific probes for GhCIN1 (181 bp), GhCIN2 (196 bp), and GhCYC3 (253 bp) were synthesized using a PCR-amplified fragment of the target gene with primers containing a few extra nucleotides (uppercase) and a T7 overhang (lowercase; CAtaatacgactcactataGGG) at the 5′ end (Supplemental Table S6), and labeled following the instructions of the DIG RNA Labeling Kit (Roche; Juntheikki-Palovaara et al., 2014). Sections were examined and photographed using the Leitz Laborlux S Microscope equipped with the Leica DFC420 C Digital Camera (Wetzlar, Germany).
Phylogenetic Analysis
Sequences for the CIN-like TCP genes, as well as CYC/TB1 genes used as outgroup, were identified from the gerbera RNASeq database using BLAST searches and the National Center for Biotechnology Information GenBank (http://www.ncbi.nlm.nih.gov/; Supplemental Table S4). An alignment of the TCP domain was generated using Clustal Omega and was converted into a corresponding nucleotide alignment. The resulting codon alignment was then subjected to phylogenetic analysis. The best-fit substitution model GTR+I+G was determined by the Bayesian Information Criterion using the program jModeltest v2.1.6 (Darriba et al., 2012). Maximum-likelihood phylogenetic reconstruction was then conducted using RAXML-HPC v.8.2.10 (Stamatakis, 2006) in CIPRES (Miller et al., 2010) with 1000 bootstrap replicates.
Accession Numbers
Sequence information for the ten GhCIN genes are available in GenBank under accession numbers MT294113 to MT294122.
Supplemental Data
The following supplemental data are available.
Supplemental Figure S1. Analysis of the selected Asteraceae CYC2 clade regulatory sequences.
Supplemental Figure S2. Expression patterns of the ten gerbera homologs identified in yeast one-hybrid screening.
Supplemental Figure S3. Transient luciferase assay by N. benthamiana agroinfiltration.
Supplemental Figure S4. Transient luciferase assay using the mutated reporter constructs.
Supplemental Figure S5. Expression analysis of the transgenic GhCIN1 and GhCIN2 RNAi lines.
Supplemental Table S1. Selected CYC2 clade regulatory regions in G. hybrida, H. annuus, L. sativa, and B. purpurea.
Supplemental Table S2. Candidate upstream TF identified in the Y1H screen of the Arabidopsis TF prey library.
Supplemental Table S3. Selected gerbera MADS-box and TCP TFs for agroinfiltration assays.
Supplemental Table S4. The sequences used in the phylogenetic analysis.
Supplemental Table S5. Number of shared perfect 20-mers between GhCIN transcripts.
Supplemental Table S6. The list of primers used in this study.
Acknowledgments
Mark Chapman and John Burke (University of Georgia) and Nicolas Langlade (INRA-CNRS) are thanked for their kind help in providing promoter sequences used in this study. We also thank Nobutaka Mitsuda (National Institute of Advanced Industrial Science and Technology) for providing the Arabidopsis TF library. Anu Rokkanen (University of Helsinki) is thanked for excellent technical assistance in cloning and expression analyses, and the glasshouse team led by Sanna Peltola (University of Helsinki) for taking care of the plants.
Footnotes
This work was supported by the Academy of Finland (grant nos. 139092 and 310318 to P.E.), the Jenny and Antti Wihuri Foundation, and the Finnish Cultural Foundation (to Y.Z.).
Articles can be viewed without a subscription.
References
- Bailey TL, Elkan C(1994) Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Bartlett ME, Specht CD(2011) Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am J Bot 98: 227–243 [DOI] [PubMed] [Google Scholar]
- Bashandy H, Jalkanen S, Teeri TH(2015) Within leaf variation is the largest source of variation in agroinfiltration of Nicotiana benthamiana. Plant Methods 11: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Pöllänen E, Ruokolainen S, Tähtiharju S, Kotilainen M, Albert VA, Elomaa P, Teeri TH(2010) Functional characterization of B class MADS-box transcription factors in Gerbera hybrida. J Exp Bot 61: 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Tähtiharju S, Laitinen RAE, Albert VA, Teeri TH, Elomaa P(2008) A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc Natl Acad Sci USA 105: 9117–9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broholm SK, Teeri TH, Elomaa P(2014) Molecular control of inflorescence development in Asteraceae. Adv Bot Res 72: 297–333 [Google Scholar]
- Chang S, Puryear J, Cairney J(1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11: 113–116 [Google Scholar]
- Chapman MA, Leebens-Mack JH, Burke JM(2008) Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol Biol Evol 25: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Chen J, Shen CZ, Guo YP, Rao GY(2018) Patterning the Asteraceae capitulum: Duplications and differential expression of the flower symmetry CYC2-like genes. Front Plant Sci 9: 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CN, Zheng HQ, Wu NY, Chien CH, Huang HD, Lee TY, Chiang-Hsieh YF, Hou PF, Yang TY, Chang WC(2016) PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res 44(D1): D1154–D1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JI, Coen ES(2002) The cycloidea gene can respond to a common dorsoventral prepattern in Antirrhinum. Plant J 30: 639–648 [DOI] [PubMed] [Google Scholar]
- Crawford BCW, Nath U, Carpenter R, Coen ES(2004) CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 135: 244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D(2012) jModelTest 2: More models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas MC, Patreze CM, Angenent GC, Immink RGH(2011) MADS: The missing link between identity and growth? Trends Plant Sci 16: 89–97 [DOI] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y(2008) A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20: 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomaa P, Teeri TH(2001) Transgenic gerbera In Bajaj YPS, ed, Biotechnology in Agriculture and Forestry 48, Transgenic Crops III. Springer Verlag, Berlin, pp 139–154 [Google Scholar]
- Elomaa P, Uimari A, Mehto M, Albert VA, Laitinen RAE, Teeri TH(2003) Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol 133: 1831–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcês HMP, Spencer VMR, Kim M(2016) Control of floret symmetry by RAY3, SvDIV1B, and SvRAD in the capitulum of Senecio vulgaris. Plant Physiol 171: 2055–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.(1995) Inflorescence and floral ontogeny in Asteraceae: A synthesis of historical and current concepts. Bot Rev 61: 93 [Google Scholar]
- Hileman LC.(2014) Bilateral flower symmetry--how, when and why? Curr Opin Plant Biol 17: 146–152 [DOI] [PubMed] [Google Scholar]
- Hsu H-J, He C-W, Kuo W-H, Hsin K-T, Lu J-Y, Pan Z-J, Wang C-N(2018) Genetic analysis of floral symmetry transition in African violet suggests the involvement of trans-acting factor for CYCLOIDEA expression shifts. Front Plant Sci 9: 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Li X, Sun M, Zhang T, Pan H, Cheng T, Wang J, Zhang Q(2016) Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front Plant Sci 7: 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Irish VF(2015) Temporal control of plant organ growth by TCP transcription factors. Curr Biol 25: 1765–1770 [DOI] [PubMed] [Google Scholar]
- Juntheikki-Palovaara I, Tähtiharju S, Lan T, Broholm SK, Rijpkema AS, Ruonala R, Kale L, Albert VA, Teeri TH, Elomaa P(2014) Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J 79: 783–796 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A(2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC(2009) Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, et al. (2017) JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Cui M-L, Cubas P, Gillies A, Lee K, Chapman MA, Abbott RJ, Coen E(2008) Regulatory genes control a key morphological and ecological trait transferred between species. Science 322: 1116–1119 [DOI] [PubMed] [Google Scholar]
- Kotilainen M, Elomaa P, Uimari A, Albert VA, Yu D, Teeri TH(2000) GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 12: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M(2007) TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M(2010) A role of TCP1 in the longitudinal elongation of leaves in Arabidopsis. Biosci Biotechnol Biochem 74: 2145–2147 [DOI] [PubMed] [Google Scholar]
- Laitinen RAE, Broholm S, Albert VA, Teeri TH, Elomaa P(2006) Patterns of MADS-box gene expression mark flower-type development in Gerbera hybrida (Asteraceae). BMC Plant Biol 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen RAE, Pöllänen E, Teeri TH, Elomaa P, Kotilainen M(2007) Transcriptional analysis of petal organogenesis in Gerbera hybrida. Planta 226: 347–360 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E(1996) Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Mandel JR, Dikow RB, Siniscalchi CM, Thapa R, Watson LE, Funk VA(2019) A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proc Natl Acad Sci USA 116: 14083–14088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Trillo M, Cubas P(2010) TCP genes: A family snapshot ten years later. Trends Plant Sci 15: 31–39 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, pp 1–8
- Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, Fujita M, Shinozaki K, Matsui M, Ohme-Takagi M(2010) Efficient yeast one-/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana. Plant Cell Physiol 51: 2145–2151 [DOI] [PubMed] [Google Scholar]
- Morgenstern B, Dress A, Werner T(1996) Multiple DNA and protein sequence alignment based on segment-to-segment comparison. Proc Natl Acad Sci USA 93: 12098–12103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, King S, Jack T(2009) miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci USA 106: 22534–22539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M, Cubas P(2015) The role of TCP transcription factors in shaping flower structure, leaf morphology, and plant architecture In Gonzalez DH, ed, Plant Transcription Factors: Evolutionary, Structural and Functional Aspects. Academic Press, Elsevier, London, pp 247–267 [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M(2013) MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panero JL, Funk VA(2008) The value of sampling anomalous taxa in phylogenetic studies: Major clades of the Asteraceae revealed. Mol Phylogenet Evol 47: 757–782 [DOI] [PubMed] [Google Scholar]
- Parapunova V, Busscher M, Busscher-Lange J, Lammers M, Karlova R, Bovy AG, Angenent GC, de Maagd RA(2014) Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol 14: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston JC, Hileman LC(2012) Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. Evodevo 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH(2010a) Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol 10: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruokolainen S, Ng YP, Broholm SK, Albert VA, Elomaa P, Teeri TH(2010b) Characterization of SQUAMOSA-like genes in Gerbera hybrida, including one involved in reproductive transition. BMC Plant Biol 10: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer V, Kim M(2018) Re“CYC”ling molecular regulators in the evolution and development of flower symmetry. Semin Cell Dev Biol 79: 16–26 [DOI] [PubMed] [Google Scholar]
- Stamatakis A.(2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690 [DOI] [PubMed] [Google Scholar]
- Su S, Xiao W, Guo W, Yao X, Xiao J, Ye Z, Wang N, Jiao K, Lei M, Peng Q, et al. (2017) The CYCLOIDEA-RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol 215: 1582–1593 [DOI] [PubMed] [Google Scholar]
- Tähtiharju S, Rijpkema AS, Vetterli A, Albert VA, Teeri TH, Elomaa P(2012) Evolution and diversification of the CYC/TB1 gene family in Asteraceae--a comparative study in Gerbera (Mutisieae) and sunflower (Heliantheae). Mol Biol Evol 29: 1155–1166 [DOI] [PubMed] [Google Scholar]
- Uimari A, Kotilainen M, Elomaa P, Yu D, Albert VA, Teeri TH(2004) Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc Natl Acad Sci USA 101: 15817–15822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es SW, Silveira SR, Rocha DI, Bimbo A, Martinelli AP, Dornelas MC, Angenent GC, Immink RGH(2018) Novel functions of the Arabidopsis transcription factor TCP5 in petal development and ethylene biosynthesis. Plant J 94: 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellmer F, Alves-Ferreira M, Dubois A, Riechmann JL, Meyerowitz EM(2006) Genome-wide analysis of gene expression during early Arabidopsis flower development. PLoS Genet 2: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Pang H-B, Liu B-L, Qiu Z-J, Gao Q, Wei L, Dong Y, Wang Y-Z(2012) Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24: 1834–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Kotilainen M, Pöllänen E, Mehto M, Elomaa P, Helariutta Y, Albert VA, Teeri TH(1999) Organ identity genes and modified patterns of flower development in Gerbera hybrida (Asteraceae). Plant J 17: 51–62 [DOI] [PubMed] [Google Scholar]
- Yuan C, Huang D, Yang Y, Sun M, Cheng T, Wang J, Pan H, Zhang Q(2020) CmCYC2-like transcription factors may interact with each other or bind to the promoter to regulate floral symmetry development in Chrysanthemum morifolium. Plant Mol Biol 103: 159–171 [DOI] [PubMed] [Google Scholar]
- Zhang T, Zhao Y, Juntheikki I, Mouhu K, Broholm SK, Rijpkema AS, Kins L, Lan T, Albert VA, Teeri TH, Elomaa P(2017) Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol 216: 939–954 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang T, Broholm SK, Tähtiharju S, Mouhu K, Albert VA, Teeri TH, Elomaa P(2016) Evolutionary co-option of floral meristem identity genes for patterning of the flower-like Asteraceae inflorescence. Plant Physiol 172: 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]