The three main actors of iron acquisition in Arabidopsis root, namely IRT1, FRO2, and AHA2, assemble into a dedicated protein complex.
Abstract
In plants, iron uptake from the soil is tightly regulated to ensure optimal growth and development. Iron absorption in Arabidopsis root epidermal cells requires the IRT1 transporter that also allows the entry of certain non-iron metals, such as Zn, Mn, and Co. Recent work demonstrated that IRT1 endocytosis and degradation are controlled by IRT1 non-iron metal substrates in a ubiquitin-dependent manner. To better understand how metal uptake is regulated, we identified IRT1-interacting proteins in Arabidopsis roots by mass spectrometry and established an interactome of IRT1. Interestingly, the AHA2 proton pump and the FRO2 reductase, both of which work in concert with IRT1 in the acidification-reduction–transport strategy of iron uptake, were part of this interactome. We confirmed that IRT1, FRO2, and AHA2 associate through co-immunopurification and split-ubiquitin analyses, and uncovered that they form tripartite direct interactions. We characterized the dynamics of the iron uptake complex and showed that FRO2 and AHA2 ubiquitination is independent of the non-iron metal substrates transported by IRT1. In addition, FRO2 and AHA2 are not largely endocytosed in response to non-iron metal excess, unlike IRT1. Indeed, we provide evidence that the phosphorylation of IRT1 in response to high levels of non-iron metals likely triggers dissociation of the complex. Overall, we propose that a dedicated iron-acquisition protein complex exists at the cell surface of Arabidopsis root epidermal cells to optimize iron uptake.
Iron is essential for plant growth and development as it plays fundamental roles in many cellular processes, including photosynthetic and respiratory electron transfer reactions. However, iron is also toxic when present in excess because it induces oxidative stress. Iron bioavailability to plants is often limited, such as in calcareous soils in which iron is present in the form of insoluble complexes (Briat et al., 2015). Iron is a limiting factor for plant biomass production and hence is considered as an important component of agriculture productivity. To maintain iron homeostasis, plants must tightly regulate iron absorption from the soil. In nongraminaceous plants, including the model plant Arabidopsis (Arabidopsis thaliana), iron absorption by root epidermal cells is achieved through the so-called strategy I, which requires three successive steps. Firstly, soil ferric chelates are solubilized by local rhizosphere acidification via the release of protons by the proton pump H+-ATPase2 (AHA2). Solubilized Fe3+ ions are then reduced to Fe2+ by the Ferric Reduction Oxydase2 (FRO2) reductase and finally transported into the cell by the iron transporter Iron Regulated Transporter1 (IRT1; Palmer and Guerinot, 2009; Thomine and Vert, 2013; Jeong et al., 2017). During this acidification-reduction–transport mechanism, reduction of Fe3+ ions by FRO2 has been proposed to be the rate-limiting step in iron acquisition (Robinson et al., 1999; Connolly et al., 2003). In Arabidopsis, the expression of IRT1, FRO2, and AHA2 genes is activated under iron-limited conditions through the direct action of the basic helix–loop–helix transcription factor FER-like iron deficiency-induced transcription factor that can form heterodimers with other basic helix–loop–helix proteins (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2008). Apart from the predominant role of IRT1, FRO2, and AHA2, another membrane protein called ATP-Binding Cassette G37/Pleiotropic Drug Resistance 9 (ABCG37/PDR9), was demonstrated to be involved in Arabidopsis iron acquisition by exporting coumarins in the rhizosphere under iron deficiency (Fourcroy et al., 2014). These excreted phenolic compounds chelate Fe3+ and facilitate iron availability for reduction by FRO2 (Fourcroy et al., 2016). Similar to the other components of the Arabidopsis iron-acquisition machinery, the ABCG37/PDR9 gene is transcriptionally induced in response to iron deficiency in a FER-like iron deficiency-induced transcription factor-dependent manner (Rodríguez-Celma et al., 2013).
IRT1 is a major player in the regulation of plant iron homeostasis, as attested by the severe chlorosis and the lethality of the irt1-1 knock-out mutant (Vert et al., 2002). Interestingly, IRT1 is a broad-spectrum transporter, which also allows the absorption of metals such as zinc (Zn), manganese (Mn), cobalt (Co), and cadmium (Cd), in addition to iron (Fe; Rogers et al., 2000; Vert et al., 2001, 2002). The dynamics of IRT1 protein and its role in the maintenance of metal homeostasis in Arabidopsis have been widely investigated. IRT1 localizes to early endosomes in root epidermal cells and rapidly cycles between this compartment and the cell surface to perform iron absorption (Barberon et al., 2011). Importantly, IRT1 dynamics requires clathrin-mediated endocytosis and is controlled by ubiquitination on two cytosol-exposed Lys residues (Barberon et al., 2011), a process mediated by the E3 ubiquitin (Ub) ligase IRT1 DEGRADATION FACTOR1 (IDF1; Shin et al., 2013; Dubeaux et al., 2018). Surprisingly, IRT1 ubiquitination and endocytosis are not regulated by iron, the primary substrate of the IRT1 transporter, but rather by its secondary metal substrates (Zn, Mn, and Co, herein called “non-iron metal substrates”). These non-iron metal substrates, which are highly reactive and toxic when present in excess in plant cells, were recently demonstrated to regulate IRT1 endocytosis (Barberon et al., 2014). In the presence of physiologically relevant concentrations of non-iron metal substrates, a functional IRT1-mCitrine fusion protein is localized in early endosomes and to some extent at the plasma membrane (PM) of root epidermal cells. Interestingly, in the presence of an excess of non-iron metal substrates, IRT1-mCitrine is targeted to late endosomes and then reaches the vacuole for degradation, whereas it is exclusively localized to the PM in the absence of such metals. The sensing of the excess of non-iron metal substrates is directly mediated by IRT1 through the binding of metals on a His-rich stretch located in IRT1 large cytosolic loop (Dubeaux et al., 2018). Hence, IRT1 was proposed to act as a transceptor, combining transporter and receptor properties (Cointry and Vert, 2019). Non-iron metal binding to IRT1 allows the recruitment of Calcineurin B-like-interacting Ser/Thr-protein kinase23 (CIPK23) and subsequent phosphorylation of IRT1. This in turn allows the Lys-63 polyubiquitination of IRT1 by IDF1, triggering IRT1 endocytosis and targeting to the vacuole (Dubeaux et al., 2018). The control of IRT1 degradation by its secondary substrates certainly constitutes a protective mechanism allowing limitation of the absorption of readily available Zn2+, Mn2+, and Co2+ ions that, contrary to Fe3+, do not need prior reduction by FRO2 to be transported (Zelazny et al., 2011). Interestingly, the PM pool of IRT1 is present at the outer polar domain of root epidermal cells, which is necessary for proper radial transport of metals in the root (Barberon et al., 2014). IRT1 physically interacts with FYVE1, a protein recruited to the Endosomal Sorting Complexes Required for Transport in late endosomes (Barberon et al., 2014; Gao et al., 2014). FYVE1 drives the maintenance of IRT1 lateral polarity, through a mechanism that is not fully understood. Interference with FYVE1 expression induces apolar localization of IRT1 at the PM and disturbs metal uptake in Arabidopsis (Barberon et al., 2014). IRT1 recycling from endosomes to the PM was shown to require Sorting Nexin1 (Ivanov et al., 2014). Recently, the peripheral membrane protein ENHANCED BENDING1 was demonstrated to interact with IRT1 in a calcium-dependent manner and was proposed to act as an inhibitor of IRT1-mediated iron transport (Khan et al., 2019).
Because IRT1 is an essential determinant of root metal uptake in low-iron conditions, we sought to identify additional proteins involved in IRT1 dynamics/activity or working in concert with IRT1 to better characterize how plant metal homeostasis is controlled. Using co-immunopurification (co-IP) of IRT1 and identification of IRT1-interacting proteins by mass spectrometry, we shed light on the existence of a dedicated protein complex composed of IRT1, FRO2, and AHA2 that likely optimizes iron uptake in root epidermal cells. We also uncovered that IRT1 is selectively removed from the complex in response to non-iron metal excess, in a process involving its phosphorylation.
RESULTS
IRT1 Interactome in Arabidopsis Root Epidermal Cells
To better understand how the non-iron metal-mediated internalization of IRT1 is controlled in root epidermal cells, we searched for proteins interacting with IRT1 upon non-iron metal excess. To this purpose, a functional IRT1-mCitrine fusion protein expressed under the control of the IRT1 promoter in Arabidopsis irt1-1 null mutant (Dubeaux et al., 2018) was immunopurified, and co-purified proteins were identified by mass spectrometry. irt1-1/IRT1::IRT1-mCitrine transgenic line and Ws wild-type plants, used as a negative control, were initially grown for 9 d on one-half strength Murashige and Skoog (MS/2) medium containing iron, transferred for 5 d onto a medium lacking iron and containing physiological concentrations of non-iron metals to induce IRT1-mCitrine expression, and finally transferred to a medium without iron and containing an excess of non-iron metal substrate for 2 d. To purify the transmembrane IRT1-mCitrine protein, root protein extracts from IRT1-mCitrine–expressing plants were solubilized with a soft nonionic detergent, n-dodecyl β-d-maltoside (DDM), to preserve the interactions between IRT1 and its partners. Then, IRT1-mCitrine and the associated proteins were immunopurified with anti-GFP antibodies coupled to magnetic microbeads, and the same procedure was performed on DDM-solubilized proteins from wild-type plant roots. Immunopurified proteins from IRT1-mCitrine and wild-type plants were separated by SDS-PAGE (Supplemental Fig. S1) and analyzed by mass spectrometry. Proteins were considered as interacting with IRT1 when specifically identified in the IRT1-mCitrine immunopurified fraction with at least two different peptides. This approach allowed the identification of 142 putative IRT1 interactants (Supplemental Dataset S1). Among these, 31 were found in the two independent experiments whereas 111 were detected in only one of the two replicates. Because the control of IRT1 trafficking is of the utmost importance for the regulation of this transporter, we specifically looked for IRT1-interacting proteins that are linked to the secretory or endocytic pathways (Table 1). Clathrin was found as putatively interacting with IRT1, which is in accordance with our previous results showing that IRT1 undergoes clathrin-mediated endocytosis (Barberon et al., 2011, 2014). Interestingly, SEC13a and SEC31b proteins were identified as IRT1 putative interactants, suggesting a role of coat protein complex II (COPII) machinery in the export of IRT1 from the endoplasmic reticulum (ER) to the Golgi apparatus (Chung et al., 2016). Conversely, IRT1 probably also undergoes retrograde transport from the Golgi to the ER because it interacts with components of COPI vesicles (Table 1; Yorimitsu et al., 2014). Another interesting group of IRT1 interactants are proteins linked to Arabidopsis metal homeostasis such as the Pleiotropic drug resistance8/Penetration3 protein that was proposed to act as a cadmium extrusion pump (Table 1; Kim et al., 2007). IRT1 also associates with two iron transporters from the vacuolar iron transporter (VIT) family; however, the relevance of this interaction remains unclear because these proteins were described as localizing to the ER body membrane (Yamada et al., 2013). Recently, rhizosphere-excreted coumarins were shown to be important for iron acquisition in an IRT1-dependent manner (Fourcroy et al., 2016). Interestingly, the Feruloyl-Coenzyme A 6′-Hydroxylase1 (F6′H1; Schmid et al., 2014) and the Cytochrome P450/CYP82C4 (Rajniak et al., 2018), which are both involved in coumarin biosynthesis, were identified as putative IRT1-interacting proteins (Table 1). In addition, FRO2 and AHA2, which are known to both act in concert with IRT1 in the acidification-reduction–transport strategy for iron uptake in Arabidopsis, were also identified (Table 1). Mass-spectrometry analyses performed on IRT1 co-immunopurified fractions indeed identified peptides specific to the AHA2 isoform, but also identified peptides common to AHA2 and other AHA proteins, mostly AHA1. However, because no peptide specific to other AHA proteins than AHA2 were found, and because rhizosphere acidification is chiefly mediated by AHA2 in lack of iron (Santi and Schmidt, 2009), we decided to focus our attention on AHA2. These observations indicate that IRT1, FRO2, and AHA2 likely associate to drive iron uptake. To gain further insight into the regulation of the iron uptake machinery, we characterized in more detail the interaction and the spatial organization of the three major actors of iron acquisition, namely FRO2, AHA2, and IRT1.
Table 1. IRT1 interactants involved in intracellular trafficking or metal homeostasis.
Among the 142 putative IRT1 interacting-proteins identified by co-IP combined with mass spectrometry, we present in this table proteins that are associated to intracellular trafficking or metal homeostasis according to gene ontogeny annotations from The Arabidopsis Information Resource. The maximum number of unique peptides identified for each protein from the two independent experiments (see Supplemental Dataset S1) is indicated.
| Category | Accession | Protein Name | Maximum Number of Unique Peptides |
|---|---|---|---|
| Intracellular Trafficking | AT3G11130 | Clathrin H chain | 24 |
| AT1G04820 | Tubulin alpha-4 chain | 6 | |
| AT1G29310 | SecY protein transport family protein | 2 | |
| AT5G09810 | Actin 7 | 4 | |
| AT4G33650 | Dynamin-related protein 3A | 2 | |
| AT3G63460 | SEC31b, COPII component | 6 | |
| AT4G18800 | RAB GTPase homolog A1D | 3 | |
| AT1G62020 | Coatomer alpha subunit, COPI component | 2 | |
| AT4G31480 | Coatomer beta subunit, COPI component | 2 | |
| AT4G17530 | RAB GTPase homolog 1C | 2 | |
| AT1G49240 | Actin 8 | 2 | |
| AT5G05010 | Clathrin adaptor complexes medium subunit family protein | 2 | |
| AT2G21390 | Coatomer, alpha subunit, COPI component | 2 | |
| AT5G44340 | Tubulin beta chain 4 | 2 | |
| AT2G30050 | SEC13a, COPII component | 2 | |
| Metal Homeostasis | AT4G30190 | H+-ATPase 2 | 13 |
| AT4G31940 | Cytochrome P450/CYP82C4 | 3 | |
| AT3G13610 | F6′H1 | 2 | |
| AT4G27860 | VIT family protein | 11 | |
| AT5G24290 | VIT family protein | 8 | |
| AT2G01530 | MLP-like protein 329 | 5 | |
| AT1G59870 | Pleiotropic drug resistance 8/Penetration 3 | 4 | |
| AT1G01580 | Ferric reduction oxidase 2 | 3 | |
| AT1G07890 | Ascorbate peroxidase 1 | 2 |
IRT1 Directly Interacts with FRO2 and AHA2
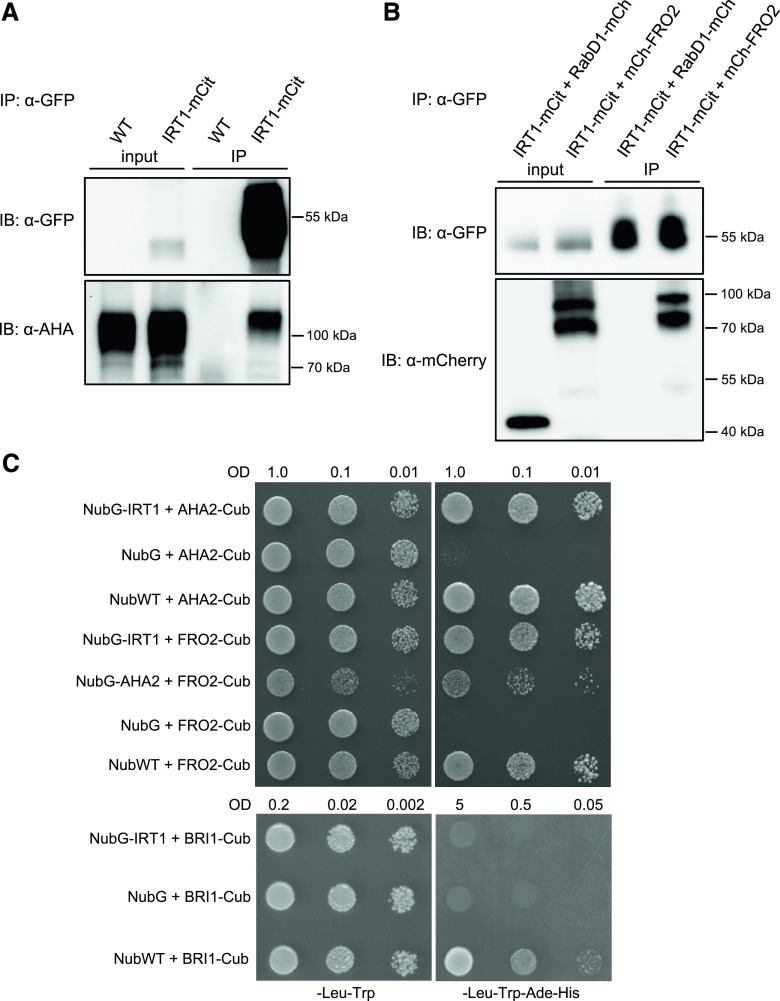
To validate the observations obtained from mass-spectrometry analyses, the interactions between IRT1 and AHA2/FRO2 were first investigated in Arabidopsis roots by performing co-IP combined with immunodetections. We analyzed the interaction between IRT1-mCitrine, expressed under the control of IRT1 promoter, and endogenous AHA2 protein by using a previously described antibody raised against PM H+-ATPase2 (PMA2) from Nicotiana plumbaginifolia that recognizes AHA2 and also other Arabidopsis AHA proteins. When the IRT1-mCitrine IP fraction was probed with anti-PMA2 antibodies, a strong signal corresponding to the expected size of AHA2 (104 kD) was detected, whereas no signal was observed in the IP fraction from wild-type plant roots used as a negative control (Fig. 1A). Although this signal may correspond in part to other AHA isoforms than AHA2 due to a lack of specificity of the anti-PMA2 antibodies, this result suggests that endogenous AHA2 likely associates with IRT1 in root epidermal cells. The existence of an IRT1–AHA2 complex is substantiated by other protein–protein interaction assays described hereafter. Because no antibody raised against FRO2 was available, we generated a translational fusion of FRO2 expressed under control of the FRO2 promoter in the previously described fro2 mutant named frd1-1 (Robinson et al., 1999). Expression of FRO2 fused at its N-terminal end to the mCherry fluorescent protein (mCherry-FRO2) complemented the hypersensitivity of frd1-1 to low iron, even for transgenic lines expressing low levels of mCherry-FRO2 (Supplemental Fig. S2, A and B). This clearly shows that mCherry-FRO2 fusion protein is fully functional. Consistently, mCherry-FRO2 protein was only detected in root epidermal cells (Supplemental Fig. S2C), highlighting the specificity of FRO2 promoter, as reported in Connolly et al. (2003). To analyze the interaction between FRO2 and IRT1 in Arabidopsis roots by co-IP, we generated transgenic lines co-expressing mCherry-FRO2 and IRT1-mCitrine under control of the FRO2 and IRT1 promoters, respectively. Transgenic lines coexpressing IRT1-mCitrine and RabD1-mCherry, the latter co-localizing with IRT1 in endosomes (Supplemental Fig. S3), were used as a negative control. mCherry-FRO2 was co-immunopurified with IRT1-mCitrine whereas RAbD1-mCherry was not, proving that FRO2 and IRT1 form a protein complex in root epidermal cells (Fig. 1B; Supplemental Fig. S4).
Figure 1.
FRO2 and AHA2 are part of an IRT1–protein complex. A, Endogenous AHA2 is co-immunopurified with IRT1-mCitrine in Arabidopsis root cells. IP were performed using anti-GFP antibodies on solubilized root protein extracts from irt1-1/IRT1::IRT1-mCitrine and wild-type (WT) plants (negative control). Inputs and IP fractions were subjected to immunoblotting (IB) with anti-GFP (top) and anti-AHA/PMA2 antibodies (bottom). B, mCherry-FRO2 is co-IP with IRT1-mCitrine in Arabidopsis root cells. IP were performed using anti-GFP antibodies on solubilized root protein extracts from irt1-1/IRT1::IRT1-mCitrine plants co-expressing FRO2::mCherry-FRO2 or UBQ10::RabD1-mCherry, this protein co-localizing with IRT1 in endosomes (negative control). Inputs and IP fractions were subjected to IB with anti-GFP (top) and anti-mCherry antibodies (bottom). Note that mCherry-FRO2 migrates at the expected size (top band) but also at a lower molecular weight (bottom band). For the co-IP experiments in A and B, roots were harvested from plants grown for 11 d on MS/2 medium containing 50 μm of Fe-EDTA, and then were transferred for 4 d on MS/2 medium lacking iron and supplemented with 300 μm of the iron chelator 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate, in the presence of physiologically relevant concentrations of non-iron metal substrates. Representative immunoblots are shown. C, IRT1 directly interacts with FRO2 and AHA2 in a split-Ub assay. Yeasts co-expressing Cub-PLV fusion proteins with NubG fusion proteins or NubG (negative control of interaction) or NubWT (positive control of interaction) were dropped in serial dilutions on a synthetic medium without Leu and Trp (control medium) or without Leu, Trp, His, and Ade (selective medium). For the interaction tests between IRT1, FRO2, and AHA2, yeast growth on control and selective media was recorded after 24 h and 48 h, respectively. Besides internal negative interaction tests performed by co-expressing Cub fusion proteins with NubG, an additional negative control was introduced in this assay by co-expressing NubG-IRT1 and BRI1-Cub. For this IRT1-BRI1 interaction test, yeast growth on control and selective media was recorded after 48 and 72 h, respectively. A representative assay is shown.
The interaction between IRT1 and AHA2/FRO2 was also confirmed by a split-Ub assay, which allows the detection of direct interactions between membrane proteins in yeast (Saccharomyces cerevisiae). Such an approach has been successfully implemented with IRT1 and IDF1 (Shin et al., 2013; Dubeaux et al., 2018). IRT1 fused to the mutated N-terminal half of ubiquitin (NubG), which is unable to spontaneously reassemble with the C-terminal part of ubiquitin (Cub), was co-expressed in yeast with AHA2 or FRO2 fused to Cub linked to the chimeric transcription factor ProteinA-LexA-VP16 (PLV; Fig. 1C). Physical interactions between IRT1 and AHA2/FRO2 were tested through the ability to rescue yeast growth on a selective medium. Yeast co-expressing NubG-IRT1 with AHA2-Cub or FRO2-Cub grew on a selective medium, similarly to the respective positive controls expressing NubWT with AHA2-Cub/FRO2-Cub. However, no growth was observed for the respective negative controls expressing NubG with AHA2-Cub/FRO2-Cub. As an additional negative control for split-Ub, NubG-IRT1 was co-expressed with an unrelated transmembrane protein, specifically the brassinosteroid receptor BRI1, fused to Cub. Whereas yeast co-expressing NubWT with BRI1-Cub (positive control of interaction) grew on a selective medium, no growth was observed when BRI1 and IRT1 were co-expressed, indicating that these two proteins do not interact. Interestingly, our split-Ub assay also revealed that AHA2 and FRO2 could physically associate with each other (Fig. 1C). This result was confirmed in plants by co-IP approaches showing that endogenous AHA2 is co-immunopurified with mCherry-FRO2 from Arabidopsis root protein extracts (Supplemental Fig. S5). Altogether, these observations validate the existence of a protein complex in root epidermal cells that gathers the different actors of the high-affinity iron uptake machinery in close proximity.
Differential Regulation of the Iron Uptake System by Ubiquitination
We recently demonstrated that IRT1 endocytosis is controlled by the non-iron metal substrates of IRT1 (Dubeaux et al., 2018). Upon an excess of these metals, IRT1 ubiquitination strongly increases, leading to the endocytosis and the degradation of IRT1 in the vacuole. Interestingly, proteomic analyses allowed the identification of AHA2 and FRO2 as part of the Arabidopsis ubiquitinome (Kim et al., 2013; Johnson and Vert, 2016; Walton et al., 2016). Because AHA2 and FRO2 belong to an IRT1-containing protein complex, we wondered whether ubiquitination of these proteins could be co-regulated by non-iron metal availability. First, we analyzed the ubiquitination profile of AHA2 and FRO2 by performing IP of AHA2-GFP and mCherry-FRO2 expressed in Arabidopsis roots, followed by the immunodetection of ubiquitination with the P4D1 general anti-Ub antibodies. In the presence of physiological concentrations of non-iron metal substrates, AHA2-GFP and mCherry-FRO2 IP fractions probed with P4D1 antibodies exhibited high-molecular–weight smears that are typical of ubiquitinated proteins as observed for IRT1-mCitrine used as a control (Fig. 2). To quantify the effect of non-iron metal regime on the ubiquitination of the investigated proteins, the signal intensity observed for the anti-Ub immunoblots performed on IRT1-mCitrine, AHA2-GFP, or mCherry-FRO2 immunopurified proteins was measured and normalized to the corresponding immunopurified proteins (Fig. 2C). As previously observed (Dubeaux et al., 2018), a short treatment with an excess of non-iron metals led to a strong increase in IRT1-mCitrine ubiquitination (Fig. 2, A and C). By contrast, the pool of ubiquitinated AHA2-GFP and mCherry-FRO2 remained unchanged in both metal regimes (Fig. 2; Supplemental Fig. S6). Hence, although AHA2, FRO2, and IRT1 belong to the same complex involved in a common mechanism i.e. iron acquisition, ubiquitination of these proteins is differentially regulated by metal availability.
Figure 2.
FRO2 and AHA2 are ubiquitinated in Arabidopsis root cells in a metal-independent manner. IP were performed using anti-GFP antibodies on solubilized root protein extracts from wild-type (WT), irt1-1/IRT1::IRT1-mCitrine, and Col0/35S::AHA2-GFP plants (A) or using anti-mCherry antibodies on solubilized root protein extracts from wild-type and frd1/FRO2::mCherry-FRO2 plants (B). Inputs and IP fractions were subjected to immunoblotting (IB) with anti-Ub antibody (P4D1; A and B, top), anti-GFP (A, bottom), or anti-mCherry antibodies (B, bottom). Non-ubiquitinated and ubiquitinated forms of the studied proteins are indicated by an arrow and a bar (−Ubn), respectively. Roots were harvested from plants grown for 11 d on MS/2 medium lacking iron (−), in the presence of physiologically relevant concentrations of non-iron metal substrates (+), and then transferred for 2 h to the same medium (control) or to a medium lacking iron and containing an excess of non-iron metals (+++). Wild-type plants were used as negative controls for IP. In A, due to very low expression level, AHA2-GFP is not detected in inputs but solely in IP fractions after enrichment. C, Quantification of IRT1-mCitrine, AHA2-GFP, and mCherry-FRO2 ubiquitination in the presence of physiologically relevant concentrations of non-iron metal substrates (+) or an excess of these metals (+++). The intensity of the Ub signal from IRT1-mCitrine, AHA2-GFP, and mCherry-FRO2 IP shown in A and B was measured using the program Image Lab and normalized to the quantity of immunopurified proteins. Results are shown as a ratio relative to the (+) condition for each protein to reveal the influence of metal treatment.
Selective Endocytosis of IRT1 in Response to Non-Iron Metal Excess
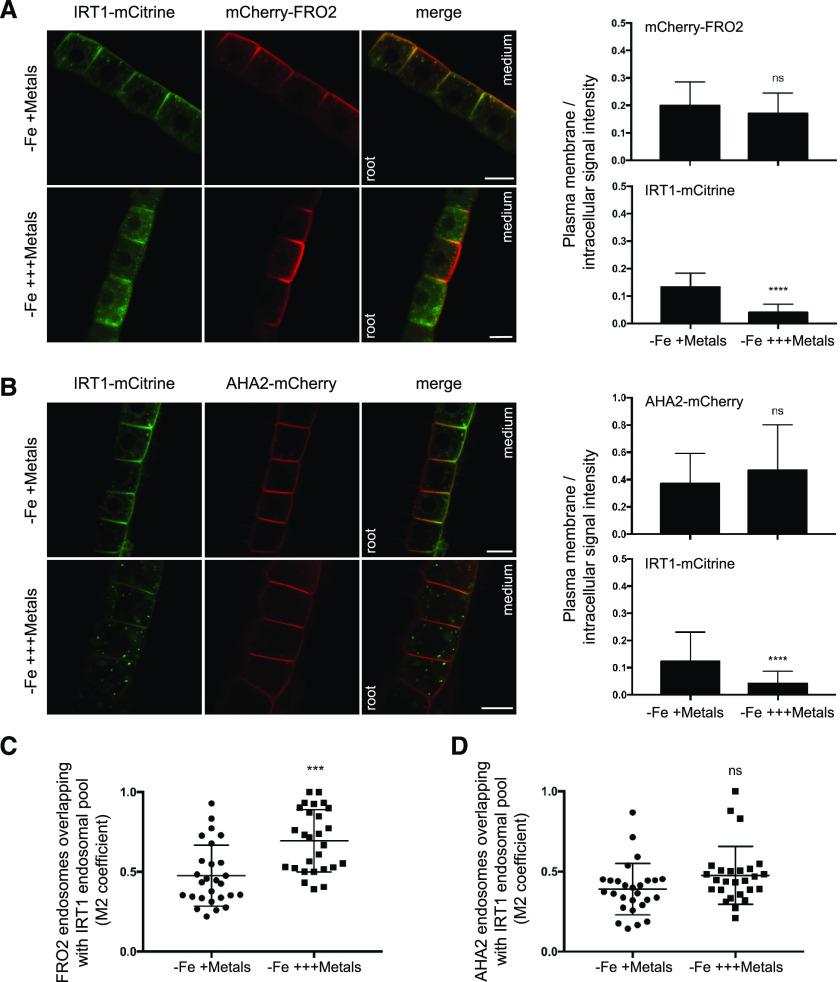
Although the intracellular dynamics of IRT1 and AHA2 have been independently investigated (Barberon et al., 2011; Dubeaux et al., 2018; Haruta et al., 2018), the subcellular distribution of FRO2 has not been determined. Thus, the respective localizations of IRT1-mCitrine and FRO2/AHA2 expressed as mCherry fusion proteins were investigated. We used root-tip epidermal cells, as these cells are well suited to analyze the precise localization of endocytosed PM proteins, and the metal-triggered endocytosis of IRT1 was already characterized in such cells (Dubeaux et al., 2018). Interestingly, in the absence of iron and in the presence of physiologically relevant amounts of non-iron metal substrates, mCherry-FRO2 was present at the PM in the outer polar domain facing the rhizosphere, as observed for IRT1-mCitrine (Fig. 3A, left; Barberon et al., 2014; Dubeaux et al., 2018). FRO2 also co-localized with IRT1 in intracellular vesicles (Mander’s coefficient, M2 = 0.48; Fig. 3, A, left, and C), which correspond to early endosomes because IRT1 constitutes a marker of these compartments in such metal conditions (Barberon et al., 2011; Dubeaux et al., 2018). In contrast to IRT1 and FRO2, AHA2-mCherry displayed an apolar PM localization in epidermal cells in the same metal conditions and although AHA2-mCherry was observed in a limited number of endosomes, these were mostly co-labeled with IRT1-mCitrine (M2 = 0.39; Fig. 3, B, left, and D).
Figure 3.
The endocytosis of IRT1 and FRO2/AHA2 is differentially regulated by non-iron metal substrates in root tip epidermal cells. Confocal microscopy analyses of root epidermal cells from irt1-1/IRT1::IRT1-mCitrine plants co-expressing IRT1::mCherry-FRO2 (A, left) or IRT1::AHA2-mCherry (B, left). Plants were grown for 11 d on MS/2 medium lacking iron, in the presence of physiologically relevant concentrations of non-iron metal substrates, and then transferred for 2 h to the same medium (−Fe +Metals control) or to a medium lacking iron and containing an excess of non-iron metals (−Fe +++Metals). To standardize microscopy analysis at the root tip, the same cells located just above the lateral root cap were systematically analyzed. Scale bars, 10 μm. Representative images are shown. Right representations in (A) and (B) show the ratio of PM to intracellular signal intensities for IRT1-mCitrine, mCherry-FRO2, and AHA2-mCherry from experiments performed as in A and B, left. M2 values of mCherry-FRO2 (C) and AHA2-mCherry (D) endosomal structures showing overlap with IRT1-mCitrine–labeled endosomes were calculated from experiments performed as in A and B, left. All the quantifications shown in this figure were carried out in triplicates on stacks encompassing epidermal cells. In total, 27 cells were analyzed for each condition and genotype. Error bars represent sd (n = 27) for each genotype. The asterisks indicate significant differences to –Fe +Metals by unpaired t test (*P < 0.0001). ns, Not significant.
The intracellular dynamics of FRO2 and AHA2 were then investigated after a short-term treatment with an excess of non-iron metal substrates that triggers IRT1 endocytosis and its subsequent degradation (Dubeaux et al., 2018). Upon non-iron metal excess, IRT1-mCitrine was depleted from the cell surface and accumulated in late endosomes (Fig. 3, A and B, left). mCherry-FRO2 and AHA2-mCherry were, however, mostly detected at the PM, even though they were also found to co-localize with IRT1-mCitrine in late endosomes (Fig. 3, A and B, left). To quantify the response to metals, the ratio of PM to intracellular fluorescence signal intensities was measured for the three fusion proteins under physiological non-iron metal provision or in the presence of an excess of these metals (Fig. 3, A and B, right). As previously reported, the PM/intracellular ratio highly decreased for IRT1-mCitrine in response to non-iron metal excess (Dubeaux et al., 2018). For mCherry-FRO2 and AHA2-mCherry, however, no significant difference was observed, indicating that FRO2 and AHA2 are not largely endocytosed in response to non-iron metal excess. However, the level of co-localization of FRO2 and IRT1 in endosomes significantly increased with non-iron metal excess (M2 = 0.69) compared to control conditions (M2 = 0.48; Fig. 3C), suggesting a minor effect of non-iron metal status on FRO2 endocytosis. On the other hand, the co-localization level between AHA2 and IRT1 in endosomes was not significantly modified by non-iron metal substrates (Fig. 3D).
The absence of a massive internalization of FRO2 and AHA2 in response to non-iron metal excess suggests that the IRT1/FRO2/AHA2 complex dissociates before IRT1 endocytosis. Because the phosphorylation of residues in the large cytosolic loop of IRT1 was shown to be the trigger of IRT1 Ub-dependent endocytosis upon non-iron metal substrate excess (Dubeaux et al., 2018), we investigated whether phosphomimic (IRT1S/TxD) or nonphosphorylatable (IRT1S/TxA) variants of IRT1 would display modified association with AHA2 and FRO2. Using split-Ub approaches, we observed that yeast co-expressing NubG-IRT1S/TxD and AHA2- or FRO2-Cub grew slower on the selective medium than yeast co-expressing NubG-IRT1 or NubG-IRT1S/TxA together with AHA2- or FRO2-Cub (Fig. 4, A and B). To better quantify the effect of the respective IRT1 variants, we measured yeast growth in liquid cultures. We consistently observed reduced growth for yeast co-expressing FRO2 or AHA2 with IRT1S/TxD compared to their wild-type or IRT1S/TxA counterparts (Fig. 4, C and D), although NubG-IRT1S/TxD protein accumulated to the same extent as wild type or IRT1S/TxA when expressed in yeast cells (Supplemental Fig. S7). These results show that the phosphomimic variant of IRT1 displays a reduced interaction with AHA2 and FRO2. Altogether, these observations clearly indicate a role for IRT1 phosphorylation in dissociation of the high-affinity iron uptake complex upon non-iron metal excess.
Figure 4.
Phosphorylation of IRT1 decreases its interaction with AHA2 and FRO2. Tentative model for the functioning of the iron-acquisition complex. A to D, Effect of IRT1 phospho-status on its interaction with AHA2 and FRO2. Split-Ub interaction tests were performed between wild-type (WT) IRT1, IRT1 variants for phosphorylation sites (phosphomimic S/TxD and nonphosphorylatable S/TxA), and AHA2 (A) or FRO2 (B). Yeasts were dropped in serial dilutions on a synthetic medium without Leu and Trp (control medium) or without Leu, Trp, His, and Ade (selective medium). NubG and NubWT were used as negative and positive controls of interaction, respectively. Yeast growth on control and selective medium was monitored after 48 h and 72 h, respectively. Representative assays are shown. To quantify interactions, the same tests were performed in liquid cultures as shown in C and D. The O.D. measured for transformed yeast grown in the selective medium for 24 h was normalized to the O.D. measured for the same yeast grown in the control medium for 16 h (relative growth). Error bars represent sem, n = 8 in C and n = 12 in D. E, Putative functioning of the iron-acquisition complex. IRT1, FRO2, and AHA2 proteins interact at the outer polar PM domain of root epidermal cells to form a specialized complex that likely optimizes iron acquisition by creating a local environment with low pH and high Fe2+ concentration. Outside this optimal zone for iron acquisition, we propose that Fe2+ is constantly reoxidized to Fe3+, which in turn forms insoluble iron complexes. In addition to be PM-localized, the iron-acquisition complex is also probably present in early endosomes, reflecting endocytic events of the complex. Note that contrary to IRT1 and FRO2, AHA2 is distributed at both PM polar domains. FRO2 might be not exclusively associated with IRT1. This model depicts the localization of IRT1, FRO2, and AHA2 in the absence of iron and in the presence of physiologically relevant concentrations of non-iron metal substrates (–Fe +Metals). Although IRT1, FRO2, and AHA2 interact all together, this feature is not represented to simplify the scheme.
DISCUSSION
IRT1 Interactome: New Insight into IRT1 Dynamics and Metal Uptake
IRT1 mediates the absorption of iron but also non-iron metal substrates such as Zn, Mn, and Co in root epidermal cells and is a major actor of metal nutrition in Arabidopsis. Previous studies revealed that the tight control of IRT1 intracellular dynamics by IDF1, CIPK23, FYVE1, or Sorting Nexin1 is essential to maintain metal homeostasis and to ensure optimal plant growth and development (Shin et al., 2013; Barberon et al., 2014; Ivanov et al., 2014; Dubeaux et al., 2018). To better understand how IRT1 is controlled in the cell, we searched for further IRT1-interacting proteins by performing IP of the functional IRT1-mCitrine fusion protein expressed under the control of IRT1 promoter coupled to mass-spectrometry analyses. Co-IP has the great advantage to reveal physiologically relevant protein–protein interactions in plant cells; and in regard to false positives, co-IP does not generate more than the yeast two-hybrid or the split-Ub techniques, and actually results in less than by using bimolecular fluorescence complementation (Xing et al., 2016). Additionally, co-IP was successfully used in the past to identify interactants of channels and receptors (Karlova et al., 2006; Bellati et al., 2016). One must consider that an interactome provides an overview of proteins putatively interacting with a bait, but that each candidate protein has to be validated by an independent protein–protein interaction technic before further investigation to avoid putative artifacts. Here, we decided to use the IRT1 endogenous promoter to maximize the detection of interactions between IRT1 and its interactants in relevant cell types, i.e. root epidermal cells, and validate each candidate of interest using the split-Ub technic. Two independent co-IP/mass-spectrometry experiments were performed, which provides qualitative data on proteins interacting with IRT1. Using another replicate would have certainly allowed us to obtain more quantitative data. Because several very interesting candidates such as FRO2 were identified in one of the two experiments, we decided to include the proteins that were identified in only one replicate in the IRT1 interactome. Among 142 proteins putatively interacting with IRT1, a group of proteins related to intracellular trafficking emerged, which included clathrin, tubulin, and actin (Table 1). An association between clathrin and IRT1 is in agreement with the previously reported clathrin-mediated internalization of IRT1 from the PM (Barberon et al., 2014). The co-IP approach does not allow the determination of whether two proteins directly interact or not; however, according to what is known for other cargo proteins, it is likely that the interaction between IRT1 and clathrin is mediated by an unknown adaptor protein. So far, the role of actin and tubulin in IRT1 dynamics is poorly understood; however, because these proteins are known to be involved in plant endocytosis (Fan et al., 2015), their presence in the IRT1-interactome also opens interesting perspectives. Two small G proteins from the Rab class, namely RAB GTPase homolog A1D and RAB GTPase homolog 1C, were also identified as putative interactants of IRT1 (Table 1). Although the role of these two proteins in intracellular trafficking is still elusive, similar Rab proteins were described to act in plant endocytic pathways (Qi and Zheng, 2013). Intriguingly, the IDF1 E3 Ub ligase and the CIPK23 kinase that are important for IRT1 endocytosis and were demonstrated to interact with IRT1 in split-Ub and yeast-two hybrid candidate approaches, respectively (Shin et al., 2013; Dubeaux et al., 2018), were not co-purified with IRT1 in this study. Given the functions of IDF1 and CIPK23, their interactions with IRT1 are probably very transient and may be lost during the co-IP procedure—contrary to other systems such as split-Ub and yeast-two hybrid, where these associations would be stabilized (Xing et al., 2016). Until now, studies of IRT1 trafficking in plant cells mainly described endocytic mechanisms including IRT1 internalization from the PM in an Ub-dependent process or IRT1 recycling from endosomes to the PM. How IRT1 traffics along the secretory pathway to reach the PM remains largely unknown, even though Malus xiaojinesis IRT1 was proposed to exit the ER in COPII vesicles (Tan et al., 2018). The Arabidopsis IRT1 interactome provides interesting clues on these aspects because IRT1 was found to putatively interact with several components of the COPII machinery that are sequentially recruited to the surface of the ER membrane to induce the formation of transport vesicles and ensure the delivery of cargo proteins to the Golgi apparatus (Table 1; Chung et al., 2016). Membrane proteins exit the ER via the recognition of specific cytoplasmic export motifs by the COPII machinery; these signals include diacidic motifs corresponding to (D/E)x(D/E), with x representing any amino acid residue (Zelazny et al., 2009). A diacidic motif (EDD) located in the large cytosolic loop of IRT1 at the position 180 may be involved in the packaging of IRT1 in COPII vesicles and its export from the ER, although this remains to be experimentally determined.
Besides the IRT1 interactants linked to intracellular trafficking, proteins involved in metal homeostasis represent a very interesting group of candidates (Table 1). Recently, coumarins, which are excreted in the rhizosphere by PDR9, were demonstrated to be important for Arabidopsis iron acquisition by chelating Fe3+ and as a result facilitating iron availability for FRO2 (Fourcroy et al., 2014; Fourcroy et al., 2016). Intriguingly, IRT1 potentially interacts with F6′H1 and Cytochrome P450/CYP82C4 that are both involved in coumarin biosynthesis (Schmid et al., 2014; Rajniak et al., 2018), but the meaning of these interactions remains unclear, notably because PDR9 was not identified as interacting with IRT1. However, solubilization of this large transmembrane protein may not be optimal in the conditions we used for the co-IP and other approaches will be necessary to determine whether PDR9 and IRT1 could work in concert among a common protein complex. Interestingly, FRO2 and AHA2, which act with IRT1 in the acidification-reduction–transport strategy for iron acquisition, were found to interact with IRT1.
IRT1, FRO2, and AHA2 Form an Iron-Acquisition Complex to Optimize Iron Uptake in Arabidopsis Roots
By combining co-IP analyses and split-Ub assays, we showed that IRT1 associates with FRO2 and AHA2 proteins, and also that FRO2 and AHA2 interact together (Fig. 1). Importantly, the tripartite physical interactions among IRT1, FRO2, and AHA2 are direct, suggesting the existence of a dedicated protein complex at the cell surface for iron uptake. Interaction between IRT1 and FRO2/AHA2 is probably not required for the intrinsic activity of IRT1, because heterologous expression of IRT1 alone allows phenotypic complementation of the fet3 fet4 yeast mutant defective in iron uptake (Eide et al., 1996). Similar to IRT1, FRO2 was also observed at the outer polar domain of the PM in epidermal cells from the root tip (Fig. 3) and from the differentiated zone of the root (Supplemental Fig. S8A). The co-polarity between FRO2 and IRT1 proteins in this domain of the PM highlights the specificity of their functions achieved at the interface between the root surface and the rhizosphere. On the other hand, AHA2 distribution in the PM of root epidermal cells was homogenous (Fig. 3; Supplemental Fig. S8B), suggesting that AHA2 does not obligatory associate with FRO2 and IRT1. This result is in accordance with AHA2 function not being restricted to iron acquisition (Yuan et al., 2017; Hoffmann et al., 2019; Pacifici et al., 2018). Even though the performed interaction tests do not provide any information on the localization of the IRT1/FRO2/AHA2 complex, co-localization analyses suggest that IRT1 may interact with FRO2 and AHA2 in the PM and in endosomes (Fig. 3; Supplemental Fig. S8). In the future, Förster resonance energy transfer-fluorescence lifetime imaging microscopy, which allows the detection of protein–protein interactions in living plant cells with a high spatial and temporal resolution (Zelazny et al., 2007), may reveal where IRT1 interact with FRO2 and AHA2 in the cell.
IRT1 intracellular dynamics is regulated by ubiquitination notably in response to an excess of non-iron metal substrates, after which IRT1 ubiquitination is enhanced, thus triggering its endocytosis and degradation in the vacuole (Dubeaux et al., 2018). Our biochemical analyses revealed that FRO2 and AHA2 are also ubiquitinated; however, their ubiquitination is not modulated by the non-iron metal provision (Fig. 2; Supplemental Fig. S6). This is consistent with the endocytosis of both proteins being rather constitutive and not regulated by non-iron metals (Fig. 3). In this context, the ubiquitination of FRO2 and AHA2 appears to be involved in a nondegradative process. First, FRO2 and AHA2 may undergo mono-ubiquitination that is known to promote internalization from the cell surface but to be insufficient for vacuolar targeting (Lauwers et al., 2010). Second, the internalized pool of AHA2 and FRO2 may be rapidly de-ubiquitinated in endosomes, allowing their recycling to the PM. Third, because ubiquitination is not only involved in endocytosis and degradation but is also implicated in other processes such as the allosteric regulation of proteins (Komander, 2009), we may speculate that FRO2 and AHA2 ubiquitination may convey this type of regulation. Although the identity of the E3 Ub ligase at stake is unknown, IDF1 represents a possible candidate for constitutive ubiquitination of AHA2 and FRO2 that will have to be tested in the future. The fact that AHA2 and FRO2 likely carry out other functions independent of iron nutrition may provide an explanation for their not being degraded upon non-iron metal excess. Indeed, AHA2 was previously reported to contribute to acidic growth or phosphorus uptake (Yuan et al., 2017; Hoffmann et al., 2019; Pacifici et al., 2018). FRO2 may play a role in copper reduction because frd1-1 mutants lack low iron-inducible copper chelate reductase activity and 35S::FRO2 plants display elevated copper reduction (Yi and Guerinot, 1996; Robinson et al., 1999; Connolly et al., 2003). The absence of internalization of FRO2 and AHA2 from the PM in root epidermal cells in response to non-iron metal excess indicates that the IRT1/FRO2/AHA2 complex must disassemble before IRT1 endocytosis to release AHA2 and FRO2 pools that may engage in other processes. In response to non-iron metal excess, IRT1 is phosphorylated by CIPK23 in its large cytosolic loop, boosting the interaction with the E3 Ub ligase IDF1 and yielding polyubiquitinated IRT1 (Dubeaux et al., 2018). We demonstrated in this study that phosphorylation of IRT1 at the same residues also controls the disassembly of the root high-affinity iron uptake complex (Fig. 4). Phosphorylation of IRT1 therefore has two opposite effects: dissociation of the IRT1/AHA2/FRO2 complex and recruitment of IDF1. Although phosphorylation is often considered as a post-translational modification allowing the recruitment of downstream factors, there is mounting evidence that it also controls the disassembly of protein complexes (Zhang et al., 2010; Couto et al., 2016). The fact that AHA2 and FRO2 were identified in our co-IP/mass-spectrometry analysis as interacting with IRT1 in non-iron metal excess is rather surprising, because IRT1 phosphorylation in response to non-iron metal excess induces the dissociation of the IRT1/FRO2/AHA2 complex. However, this may not induce total dissociation of the complex, as evidenced by the residual interaction observed between the phosphomimic IRT1 and AHA2/FRO2. This is also supported by the partial co-localization of FRO2 and AHA2 with IRT1 in late endosomes under non-iron metal excess. In addition, we showed that the level of co-localization between FRO2 and IRT1 in late endosomes slightly increased with non-iron metal excess, suggesting that a small proportion of FRO2 is still able to associate with IRT1 in these conditions. Therefore, our co-IP/mass-spectrometry analysis carried out when plants experience non-iron metal excess, likely allowed us to identify the small pool of FRO2 and AHA2 still interacting with IRT1 in endosomes. This may explain why a limited number of FRO2 peptides were identified by proteomics.
The biological significance of iron uptake using a specific platform at the cell surface is unclear, but likely relies in the chemistry of iron. Although iron is abundant in most soils, its bioavailability to plants is often limited. This is especially true for calcareous soils, which represent one-third of cultivated lands, where iron is present under the form of insoluble complexes (Briat et al., 2015). During the iron-acquisition process, rhizosphere acidification by the root is essential to increase iron availability; indeed, the solubility of iron increases 1,000-fold for every one-unit drop in pH (Olsen et al., 1981). However, this acidification process, mainly mediated by AHA2 under iron deficiency (Santi and Schmidt, 2009), is very local, which likely impacts on the efficiency of iron uptake. Moreover, the presence of oxygen in most soils likely provokes the rapid reoxidation of Fe2+ produced by FRO2 into Fe3+ that is not transported by IRT1. Thus, we propose that the tripartite protein complex gathering IRT1, FRO2, and AHA2 together creates a local environment of pH and Fe2+ concentration in the rhizosphere that favors an optimal acquisition of iron (Fig. 4E). To experimentally validate this model, mutated versions of these proteins that do not interact with each other but that conserve their activities need to be generated to evaluate the functional outcome on iron uptake. However, this requires deep knowledge about the structure or interaction domains between these highly hydrophobic membrane proteins, which is currently missing. Alternatively to the local environment theory, we can also speculate that reduced iron is directly transferred from FRO2 to IRT1 by a channeling mechanism, similar to what has been described for metabolic pathways. Channeling consists in the transfer of the product of a proximal activity as substrate to a distal activity without equilibration with bulk solvent, which increases the efficiency of the kinetic process (Kwok et al., 2006). Such a mechanism requires the close proximity of the donor and acceptor sites. Interestingly, channeling of iron was described in yeast between the multicopper oxidase Fet3p, which oxidizes Fe2+ to Fe3+ and the iron permease Ftr1p, which transports Fe3+ into the cells, with both proteins forming a hetero-oligomeric complex (Kwok et al., 2006; Singh et al., 2006). Further work will elucidate whether FRO2 and IRT1 use a similar mechanism. Whether the formation of an iron-acquisition complex comprising IRT1, FRO2, and AHA2 is conserved in plants other than Arabidopsis remains to be determined. This complex is probably not present in rice (Oryza sativa) that combines two strategies to take up iron from the soil: a phytosiderophore-based system allowing the acquisition of Fe3+ (Inoue et al., 2009; Lee et al., 2009), and the use of Fe2+ transporters such as OsIRT1 (Ishimaru et al., 2006). Indeed, Fe3+ chelate reductase activity has been shown to not be required for Fe2+ uptake under iron deficiency in rice, suggesting that OsIRT1 works independently of OsFRO2-like proteins (Ishimaru et al., 2006). In paddy fields, where rice plants are grown, Fe2+ is abundant due to the low redox potential and therefore rice plants do not need to reduce Fe3+ to Fe2+ (Ishimaru et al., 2007). So far, the description of protein complexes aimed at optimizing nutrient uptake in plant remains scarce. To our knowledge, only the interaction between Gln synthase, the principal ammonia assimilatory enzyme and the aquaglyceroporin Nodulin 26, a transporter of NH3, was proposed to promote efficient assimilation of nitrogen in soybean (Masalkar et al., 2010). Although experimental evidence is still needed, the co-localization between FRO2/AHA2 and IRT1 in early endosomes in the presence of physiologically relevant levels of non-iron metals suggests that the iron-acquisition complex may exist in this compartment in addition to the PM (Fig. 4E). This complex may help plant metal uptake by translocating iron from endocytic vesicles to the cytosol. Alternatively, the AHA2/FRO2/IRT1 complex may simply cycle between early endosomes and the PM in a constitutive manner or in response to some undetermined stimulus. Finally, because early endosomes/trans-Golgi network constitute a crossroad between endocytic and secretory pathways in plants (Dettmer et al., 2006), the presence of the IRT1/FRO2/AHA2 complex in early endosomes may also reflect a step in the delivery of a preformed complex to the PM. Future work will be needed to discriminate among these different scenarios.
MATERIAL AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) wild-type plants (Col-0, Col-gl1, and Ws), the fro2 loss-of-function mutant named frd1-1 (Robinson et al., 1999), the previously described irt1-1/IRT1::IRT1-mCitrine line (Dubeaux et al., 2018), and the various transgenic plants generated in this study were vertically grown in sterile conditions at 21°C with 16-h light/8-h dark cycles with a light intensity of 90 μmol m−2 s−1 using 7W F17T8/TL741 bulbs (Philips). The plant growth medium used was MS/2 medium containing 1% (w/v) Suc, 1% (w/v) agar, and various concentrations of metals. Hence, depending on the experiment (see below), plants were grown in the absence of iron and in the presence of physiological concentrations of IRT1 secondary substrates Zn (15 μm), Mn (50 μm), and Co (0.05 μm; −Fe +Metals) or in the presence of 10-fold more Zn, Mn, and Co (−Fe +++Metals) corresponding to an excess of non-iron metal substrates, as described in Dubeaux et al. (2018). Plants have also been grown in iron-replete conditions using MS/2 medium containing 50 μm or 100 μm of Fe-EDTA (+Fe).
For IPs followed by mass-spectrometry analyses, irt1-1/IRT1::IRT1-mCitrine transgenic lines and Ws wild-type plants were initially grown for 9 d on MS/2 medium containing 50 μm of Fe-EDTA, transferred for 5 d onto a −Fe +Metals medium to induce IRT1-mCitrine expression, and then finally subjected to a −Fe +++Metals treatment for 48 h. To confirm the interactions between IRT1, FRO2, and AHA2 by co-IPs, the various genotypes were grown for 11 d on MS/2 medium containing 50 μm of Fe-EDTA, and then transferred for 4 d on a −Fe +Metals medium supplemented with 300 μm of the iron chelator 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate (Ferrozine) to ensure a rapid and strong expression of genes under the control of IRT1 and FRO2 promoters.
To analyze mCherry-FRO2, AHA2-GFP, and IRT1-mCitrine ubiquitination profiles, the appropriate transgenic lines as well as wild-type plants used as negative controls were grown for 11 d on −Fe +Metals MS/2 solid medium. Then, plants were transferred for 2 h in −Fe +Metals (control) or −Fe +++Metals MS/2 liquid medium as described in Dubeaux et al. (2018).
For microscopy analyses, transgenic lines expressing IRT1/AHA2/FRO2 fusion proteins under the control of IRT1 promoter, were first grown for 11 d on a −Fe +Metals MS/2 medium to ensure protein expression. Then, before observation, plants were transferred for 2 h in −Fe +Metals (control) or −Fe +++Metals MS/2 liquid medium. The localization of mCherry-FRO2 protein in frd1-1/FRO2::mCherry-FRO2 transgenic lines was performed on plants grown for 11 d in −Fe +Metals condition.
For an mCherry-FRO2 functionality test, the frd1-1/FRO2::mCherry-FRO2 transgenic lines, the frd1-1 mutant, and the Col-gl1 wild-type plants were grown for 11 d on MS/2 lacking iron (−Fe +Metals) or on MS/2 supplemented with 100 μm of Fe-EDTA (control conditions). Roots from iron-starved transgenic lines and frd1-1 (negative control) were collected to analyze mCherry-FRO2 protein accumulation by western-blot analysis, as detailed below.
Constructions and Generation of Arabidopsis Transgenic Lines
All the constructions described in this section were obtained using the MultiSite Gateway Three-Fragment Vector Construction system. The FRO2 promoter corresponding to a sequence of 1,845 bp upstream of the FRO2 start-codon was amplified from Arabidopsis genomic DNA using the attB4.promoFRO2 forward and attB1r.promoFRO2 reverse primers (Supplemental Table S1) and was subsequently cloned into the pDONR.P4P1R entry vector. The FRO2 open reading frame (ORF) was amplified from Arabidopsis cDNAs with the attB2r.FRO2 forward and attB3.FRO2 reverse primers (Supplemental Table S1) and was cloned into the pDONR.P2RP3 entry vector. The AHA2 ORF without the stop-codon was amplified from Arabidopsis cDNA with the AHA2.F forward and AHA2.R reverse primers and was cloned into the pDONR.221 entry vector. The mCherry sequence without stop-codon was amplified with attB1.mCherry forward and attB2.mCherry reverse primers and also cloned into the pDONR.221 entry vector (Supplemental Table S1). Entry vectors carrying the IRT1 and 35S promoters (pDONR.P4P1R-IRT1 and pDONR.P4P1R-35S) or the GFP and the mCherry coding sequence allowing C-terminal fusions (pDONR.P2RP3-GFP and pDONR.P2RP3-mCherry) were described in Marquès-Bueno et al. (2016) and Dubeaux et al. (2018). Final destination vectors for expression in plants were obtained by multisite Gateway recombination using the entry vectors described above and the pH7m34GW and pK7 m34GW destinations vectors used for mCherry and GFP fusions, respectively. The following constructs were generated: FRO2::mCherry-FRO2, IRT1::mCherry-FRO2, IRT1::AHA2-mCherry, and 35S::AHA2-GFP.
The irt1-1/IRT1::IRT1-mCitrine line described in Dubeaux et al., 2018) was transformed with FRO2::mCherry-FRO2, IRT1::mCherry-FRO2, and IRT1::AHA2-mCherry constructions by the floral-dipping technique using Agrobacterium tumefaciens. The frd1-1 mutant and Col-0 plants were transformed with the FRO2::mCherry-FRO2 and 35S::AHA2-GFP constructs, respectively. The irt1-1/IRT1::IRT1-mCitrine transgenic line was crossed with the Wave marker line no. 25 expressing RabD1-mCherry fusion protein under the control of UBQ10 promoter (Geldner et al., 2009).
IPs
IPs were performed on ∼500 mg of Arabidopsis roots, mostly as described in Dubeaux et al. (2018). Briefly, for IRT1-mCitrine IP followed by mass spectrometry, for co-IP analyses between IRT1-mCitrine and mCherry-FRO2, as well as for co-IP analyses between mCherry-FRO2 and endogenous AHA2, roots were ground in liquid nitrogen and resuspended in IRT1 solubilization buffer (50 mm of Tris-HCl at pH 7.4, 150 mm of NaCl, 5 mm of EDTA, 1% [w/v] DDM, and plant-specific protease inhibitors [Sigma-Aldrich]). For co-IP analyses between IRT1-mCitrine and endogenous AHA2, roots were ground and resuspended in RIPA buffer (50 mm Tris-HCl at pH 7.5, 150 mm of NaCl, 0.5% [w/v] sodium deoxycholate, 1% [v/v] IGEPALCA-630, 0.1% [w/v] SDS, and plant-specific protease inhibitors [Sigma-Aldrich]). After two successive centrifugations at 3,800g for 10 min at 4°C, the resultant supernatants were collected and solubilization of membrane proteins was continued for 1 h 30 min at 4°C on a rotating wheel. Samples were then centrifuged at 100,000g for 1 h at 4°C to remove unsolubilized material and supernatants containing solubilized proteins were recovered for IPs. This ultracentrifugation step avoids the immuno-capture of proteins present in patches of residual nonsolubilized membranes, allowing the IP to be carried out on solubilized protein complexes only. IPs of GFP and mCitrine fusion proteins were performed using an μMACS GFP isolation kit (Miltenyi Biotec), whereas IP of mCherry fusion proteins was performed using RFP-Trap_MA magnetic beads (Chromotek), following the instructions of the manufacturers. Before elution, extensive washes were performed with IRT1 solubilization buffer or RIPA buffer, depending on the IP type. Co-IP analyses followed by mass spectrometry were performed twice. Co-IP combined with immunodetections were performed thrice with similar results.
To analyze the ubiquitination profile of AHA2-GFP and IRT1-mCitrine, the solubilization of fusion proteins as well as the IP procedure were performed exactly as described in Dubeaux et al. (2018). The same protocol was used for mCherry-FRO2, except that the protein was immunopurified with RFP-Trap_MA magnetic beads (Chromotek). Three independent analyses of ubiquitination profiles were performed.
Mass-Spectrometry Analysis
For sample preparation, proteins from each eluate were separated by SDS-PAGE to fractionate the protein samples into two fractions, including proteins 10 to 63 kD and >63 kD, respectively, to exclude abundant contaminating IRT1-mCitrine protein at 63 kD. After Coomassie-Blue staining, each gel fraction was cut into bands and subjected to in-gel trypsin digestion with the Progest robot (Genomic Solutions) using standard conditions including reduction and alkylation as described in Blanchet et al. (2014). Tryptic peptides extracted from the different bands of each gel fraction were pooled, vacuum-dried, and resuspended in 0.1% (v/v) formic acid before nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. The same cutting pattern of the SDS-PAGE lane was performed for each eluate.
Tryptic peptides from the two or three SDS-PAGE fractions from each eluate were analyzed separately by nanoLC-MS/MS with the Triple-TOF 4600 mass spectrometer (ABSciex) coupled to the nano rapid sceparation LC ultra performance LC system (Thermo Fisher Scientific) equipped with a trap column (Acclaim PepMap100C18, 75 μm internal diameter × 2 cm, 3 μm; Thermo Fisher Scientific) and an analytical column (Acclaim PepMapRSLCC18, 75 μm internal diameter × 50 cm, 2 μm, 100 Å; Thermo Fisher Scientific). Peptides were loaded at 5 μL min−1 with 0.05% (v/v) trifluoroacteic acid in 5% (v/v) acetonitrile, and peptide separation was performed at a flow rate of 300 nL/min with a 5% to 35% (v/v) solvent B gradient in 40 min. Solvent A was 0.1% (v/v) formic acid in water, and solvent B was 0.1% (v/v) formic acid in 100% (v/v) acetonitrile. NanoLC-MS/MS experiments were conducted in a data-dependent acquisition method by selecting the 20 most intense precursors for collision-induced dissociation fragmentation with Q1 quadrupole set at low resolution for better sensitivity.
Protein identification was performed by processing raw data with the software MS Data Converter (ABSciex) for generating .mgf data files and protein identification were performed using the search engine MASCOT (Matrix Science) against the SwissProt (www.uniprot.org) and The Arabidopsis Information Resource (TAIR; https://www.arabidopsis.org) databases with carbamidomethylation of cysteines set as fixed modification and oxidation of methionines as variable modifications. Peptide and fragment tolerance were set at 20 μL L−1 and 0.05 Da, respectively. Results were analyzed with the software Scaffold v.3.6.5 (Proteome Software). Proteins were validated when identified with at least two unique peptides and 95% probability levels for both peptides and proteins.
Extraction of Total Proteins and Immunoblots
Total proteins were extracted from ∼100 mg of Arabidopsis roots ground in liquid nitrogen and directly resuspended in 2× SDS sample buffer. Samples were heated at 65°C for 10 min, centrifuged 10 min at 20,000g, and finally supernatants were then collected and directly used for SDS-PAGE. Protein extraction from yeast (Saccharomyces cerevisiae) was carried out as described in von der Haar (2007).
Immunoblot analyses were performed as described in Barberon et al. (2011). Immunodetection of GFP and mCitrine fusion proteins was performed using an anti-GFP antibody conjugated to horseradish peroxidase (HRP; catalog no. 130-091-833, 1/5,000; Miltenyi Biotec). mCherry fusion proteins were monitored with a rabbit anti-DsRed antibody (catalog no. 632496, 1/5,000; Clontech). Endogenous AHA2 protein was immunodetected using a rabbit antibody initially raised against PMA2 from Nicotiana plumbaginifolia diluted 1/15,000 (Morsomme et al., 1998). Ub modifications were detected with the P4D1 mouse anti-Ub antibody (catalog no. 05–944, 1/4,000; Millipore). The detection of NubG-IRT1 protein from yeast used anti-HA antibodies (catalog no. 130–091–972, 1/7,000; Miltenyi Biotec). Anti-tubulin antibodies were used as loading control (catalog no. AS10 681, 1/5,000; Agrisera). The anti-rabbit IgG or anti-mouse IgG secondary antibodies coupled to HRP were both diluted 1/20,000. Detection of HRP chemiluminescence was performed using SuperSignal West Dura Extended Duration Substrate (Thermo Fisher Scientific) in a Chemidoc Touch Imaging system (Bio-Rad). Stain-Free Protein Staining (Bio-Rad) was used as a loading control as described in Dubeaux et al. (2018). To quantify IRT1-mCitrine, AHA2-GFP, and mCherry-FRO2 ubiquitination levels under different metal regimes, signal intensity observed with anti-Ub immunoblots performed on IRT1-mCitrine, AHA2-GFP, or mCherry-FRO2 immunopurified proteins was measured using the software Image Lab (v.6.0.1; www.bio-rad.com/en-us/product/image-lab-software) and normalized to the quantity of immunopurified proteins detected in IP with anti-GFP or anti-DsRed antibodies. To facilitate comparisons, the ubiquitination level measured in the presence of physiological concentrations of non-iron metal substrates was arbitrarily fixed to 1.
Constructions and Split-Ub Assay
Split-Ub vectors were generated using the Gateway technology. First, the FRO2 ORF without the stop-codon and the AHA2 ORF with the stop-codon were amplified with FRO2.F/FRO2.R and AHA2.F/AHA2stop.R primers, respectively (Supplemental Table S1), and were both cloned into the pDONR.221 entry vector. pDONR.221-BRI1 without the stop-codon was previously generated (Martins et al., 2015) and pDONR.221-AHA2 without the stop-codon was created in this study, as mentioned above. Then, FRO2, AHA2, and BRI1 ORFs without the stop-codon were inserted into pMetYC-DEST destination vector (Hachez et al., 2014) to produce Met-repressible constructs FRO2-Cub-PLV, AHA2-Cub-PLV, and BRI1-Cub-PLV, respectively, where Cub corresponds to the C-terminal part of Ub and PLV to a chimeric transcription factor. The AHA2 ORF with the stop-codon was cloned into the pNX35-DEST destination vector to generate the NubG-AHA2 fusion wherein NubG corresponds to the mutated N-terminal part of Ub. The NubG-IRT1, NubG-IRT1S/TxD, and NubG-IRT1S/TxA constructs were described in Dubeaux et al. (2018). It is important to note that NubG and Cub were fused to a cytosolic part of IRT1, FRO2, AHA2, and BRI1, according to the known or predicted topology of these proteins. The wild-type Ub N-terminal fragment (NubWT) expressed by the pNubWT-Xgate vector and the NubG fragment expressed by the non-recombined pNX35-DEST vector were used as positive and negative controls, respectively (Hachez et al., 2014).
Split-Ub assay was performed as described in Dubeaux et al. (2018). Briefly, the THY.AP4 yeast strain was co-transformed with the Nub and Cub constructs of interest, and co-transformed cells were selected on synthetic defined (SD) medium lacking Leu and Trp. Then, yeast co-expressing Cub-PLV fusion proteins with NubG fusion proteins or NubG (negative control of interaction) or NubWT (positive control of interaction) were dropped in serial dilutions (optical density [O.D.], 1, 0.1, 0.01) onto SD medium without Leu and Trp (control medium) or onto SD medium lacking Leu, Trp, His, and Ade (selective medium) supplemented with 500 μM of Met (250 μM of Met for IRT1/BRI1 interaction test) to limit the expression of the Cub-PLV fusion proteins. Yeast growth on control and selective medium was recorded after 24 h and 48 h at 30°C, respectively. Besides internal negative interaction tests performed by co-expressing Cub-PLV fusion proteins with NubG, co-expression of NubG-IRT1 and BRI1-Cub-PLV was used as an additional negative control. Quantification of interactions were carried out using liquid yeast cultures and by measuring O.D. over time. Three independent split-Ub interaction tests were performed with similar results.
Confocal Microscopy
Microscopy was performed with an SP8 upright confocal laser scanning microscope (Leica). For mCitrine and mCherry imaging, the 514- and 561-nm lasers were used, respectively. Before observation, plants were mounted in MS/2 liquid medium containing the proper metal composition (−Fe +Metals or −Fe +++Metals). Representative images are shown. For quantifications, z-stacks encompassing the whole cell volume were imaged and then subjected to maximum projection. The M2 of mCherry-FRO2 and AHA2-mCherry endosomal structures showing overlap with IRT1-mCitrine–labeled endosomes were determined using the ImageJ plug-in Coloc2 (https://imagej.nih.gov/ij/). Twenty-seven cells (three independent cells from three different plants among three independent experiments) were analyzed for each condition and genotype. The M2 coefficient is the ratio of the summed intensities of pixels from the red image, for which the intensity in the green channel is above zero, to the total intensity in the red channel. M2 coefficients vary from 0 to 1, the former value corresponding to nonoverlapping endosomes and the latter reflecting 100% co-localization between both channels. A ratio of 0.5 indicates a 50% overlap between the two channels. Here, we used M2 to reflect the co-localization between mCherry-FRO2 and IRT1-mCitrine, or AHA2-mCherry and IRT1-mCitrine. An unpaired t test was used to determine whether the overlapping was different in response to metal excess (−Fe +++Metals). The ratios of PM over intracellular signal content were obtained by selecting whole-cell and intracellular content mean fluorescence with the software ImageJ.
Statistical Analyses
For confocal microscopy experiments, a representative image is shown. Statistical analyses were performed using the software GraphPad Prism 7. The sample size and statistical tests used are mentioned in the figure legends.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: IRT1 (AT4G19690), FRO2 (AT1G01580), AHA2 (AT4G30190), and BRI1 (AT4G39400).
Supplemental Data
The following materials are available as supplemental data.
Supplemental Figure S1. Immunoprecipitation of IRT1-mCitrine and the associated proteins.
Supplemental Figure S2. Expression of mCherry-FRO2 complements the frd1-1 mutant phenotype in iron-deficient conditions.
Supplemental Figure S3. IRT1-mCitrine and RabD1-mCherry co-localize in early endosomes in Arabidopsis root epidermal cells.
Supplemental Figure S4. mCherry-FRO2 protein is not cross immunopurified with anti-GFP antibody.
Supplemental Figure S5. FRO2 and AHA2 interact in Arabidopsis root cells.
Supplemental Figure S6. FRO2 and AHA2 are ubiquitinated in Arabidopsis root cells in a metal-independent manner, replicate.
Supplemental Figure S7. Phosphomimic and nonphosphorylatable mutations in IRT1 do not influence IRT1 protein accumulation in yeast.
Supplemental Figure S8. Co-localization of IRT1 and FRO2/AHA2 in differentiated root cells.
Supplemental Table S1. List of primers used in this study.
Supplemental Dataset S1. IRT1 interactome determined by co-immunopurification of IRT1-mCitrine combined with mass-spectrometry analyses.
Acknowledgments
We thank Marc Boutry for the anti-PMA2 antibody, Mary Lou Guerinot for providing the frd1-1 mutant, François Chaumont for the split-Ub vectors, and Alexandre Martinière and Alexander Johnson for their help in image analysis. We also thank Sébastien Thomine for interesting scientific discussions. We would also like to thank the facilities and expertise of the Institute for Biology of the Cell (I2BC) proteomic platform named Service d’Identification et de Caractérisation des Protéines par Spectrométrie de masse (SICaPS; supported by Infrastructure Biologie Santé Agronomie [IBiSA]), Ile de France Region, Plan Cancer, Centre National de la Recherche Scientifique (CNRS), Paris-Sud University, the expertise of David Cornu, and Imagerie‐Gif core facility (supported by Agence Nationale de la Recherche).
Footnotes
This work was supported by the French National Research Agency (grant nos. ANR–10–LABX–0040–SPS, ANR–11–IDEX–0003–02, and ANR–18–CE20–0008, project "NUTRIR", to E.Z. and ANR–13–JSV2–0004–01 to G.V.), the Marie Skłodowska-Curie Actions (grant no. PCIG–GA–2012–334021 to G.V.), the French Laboratory of Excellence (project “TULIP” grant nos. ANR–10–LABX–41 and ANR–11–IDEX–0002–02 to G.V.), and the European Molecular Biology Organization long-term post-doctoral fellowship (grant no. ALTF334–2018 to G.D.).
References
- Barberon M, Dubeaux G, Kolb C, Isono E, Zelazny E, Vert G(2014) Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc Natl Acad Sci USA 111: 8293–8298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G(2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellati J, Champeyroux C, Hem S, Rofidal V, Krouk G, Maurel C, Santoni V(2016) Novel aquaporin regulatory mechanisms revealed by interactomics. Mol Cell Proteomics 15: 3473–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet S, Cornu D, Argentini M, Namy O(2014) New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res 42: 10061–10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Dubos C, Gaymard F(2015) Iron nutrition, biomass production, and plant product quality. Trends Plant Sci 20: 33–40 [DOI] [PubMed] [Google Scholar]
- Chung KP, Zeng Y, Jiang L(2016) COPII paralogs in plants: Functional redundancy or diversity? Trends Plant Sci 21: 758–769 [DOI] [PubMed] [Google Scholar]
- Cointry V, Vert G(2019) The bifunctional transporter-receptor IRT1 at the heart of metal sensing and signalling. New Phytol 223: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML(2004) The essential basic helix–loop–helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML(2003) Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol 133: 1102–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Niebergall R, Liang X, Bücherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D, et al. (2016) The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog 12: e1005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K(2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubeaux G, Neveu J, Zelazny E, Vert G(2018) Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol Cell 69: 953–964 e955 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML(1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Li R, Pan J, Ding Z, Lin J(2015) Endocytosis and its regulation in plants. Trends Plant Sci 20: 388–397 [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Alvarez-Fernández A, Briat JF(2014) Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol 201: 155–167 [DOI] [PubMed] [Google Scholar]
- Fourcroy P, Tissot N, Gaymard F, Briat JF, Dubos C(2016) Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe2+ transport system. Mol Plant 9: 485–488 [DOI] [PubMed] [Google Scholar]
- Gao C, Luo M, Zhao Q, Yang R, Cui Y, Zeng Y, Xia J, Jiang L(2014) A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr Biol 24: 2556–2563 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J(2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C, Laloux T, Reinhardt H, Cavez D, Degand H, Grefen C, De Rycke R, Inzé D, Blatt MR, Russinova E, Chaumont F(2014) Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Tan LX, Bushey DB, Swanson SJ, Sussman MR(2018) Environmental and genetic factors regulating localization of the plant plasma membrane H+-ATPase. Plant Physiol 176: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann RD, Olsen LI, Ezike CV, Pedersen JT, Manstretta R, Lopez-Marques RL, Palmgren M(2019) Roles of plasma membrane proton ATPases AHA2 and AHA7 in normal growth of roots and root hairs in Arabidopsis thaliana. Physiol Plant 166: 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK(2009) Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem 284: 3470–3479 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Kim S, Tsukamoto T, Oki H, Kobayashi T, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, et al. (2007) Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proc Natl Acad Sci USA 104: 7373–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, et al. (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45: 335–346 [DOI] [PubMed] [Google Scholar]
- Ivanov R, Brumbarova T, Blum A, Jantke AM, Fink-Straube C, Bauer P(2014) SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 26: 1294–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P(2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Jeong J, Merkovich A, Clyne M, Connolly EL(2017) Directing iron transport in dicots: Regulation of iron acquisition and translocation. Curr Opin Plant Biol 39: 106–113 [DOI] [PubMed] [Google Scholar]
- Johnson A, Vert G(2016) Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiol 171: 1808–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S(2006) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 18: 626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, Gratz R, Denezhkin P, Schott-Verdugo SN, Angrand K, Genders L, Basgaran RM, Fink-Straube C, Brumbarova T, Gohlke H, et al. (2019) Calcium-promoted interaction between the C2-domain protein EHB1 and metal transporter IRT1 inhibits Arabidopsis iron acquisition. Plant Physiol 180: 1564–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY, Bovet L, Maeshima M, Martinoia E, Lee Y(2007) The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J 50: 207–218 [DOI] [PubMed] [Google Scholar]
- Kim DY, Scalf M, Smith LM, Vierstra RD(2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25: 1523–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D.(2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37: 937–953 [DOI] [PubMed] [Google Scholar]
- Kwok EY, Severance S, Kosman DJ(2006) Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry 45: 6317–6327 [DOI] [PubMed] [Google Scholar]
- Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B(2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol 20: 196–204 [DOI] [PubMed] [Google Scholar]
- Lee S, Chiecko JC, Kim SA, Walker EL, Lee Y, Guerinot ML, An G(2009) Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol 150: 786–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquès-Bueno MDM, Morao AK, Cayrel A, Platre MP, Barberon M, Caillieux E, Colot V, Jaillais Y, Roudier F, Vert G(2016) A versatile multisite Gateway-compatible promoter and transgenic line collection for cell type-specific functional genomics in Arabidopsis. Plant J 85: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S, Dohmann EM, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaître B, Jaillais Y, Chory J, Geldner N, et al. (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6: 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masalkar P, Wallace IS, Hwang JH, Roberts DM(2010) Interaction of cytosolic glutamine synthetase of soybean root nodules with the C-terminal domain of the symbiosome membrane nodulin 26 aquaglyceroporin. J Biol Chem 285: 23880–23888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P, Dambly S, Maudoux O, Boutry M(1998) Single point mutations distributed in 10 soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J Biol Chem 273: 34837–34842 [DOI] [PubMed] [Google Scholar]
- Olsen RA, Clark RB, Bennett JH(1981) The enhancement of soil fertility by plant roots. Am Sci 69: 378–384 [Google Scholar]
- Pacifici E, Di Mambro R, Dello Ioio R, Costantino P, Sabatini S(2018) Acidic cell elongation drives cell differentiation in the Arabidopsis root. EMBO J 37: e99134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CM, Guerinot ML(2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Zheng H(2013) Rab-A1c GTPase defines a population of the trans-Golgi network that is sensitive to endosidin1 during cytokinesis in Arabidopsis. Mol Plant 6: 847–859 [DOI] [PubMed] [Google Scholar]
- Rajniak J, Giehl RFH, Chang E, Murgia I, von Wirén N, Sattely ES(2018) Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat Chem Biol 14: 442–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML(1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, López-Millán AF, Schmidt W(2013) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol 162: 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Eide DJ, Guerinot ML(2000) Altered selectivity in an Arabidopsis metal transporter. Proc Natl Acad Sci USA 97: 12356–12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W(2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schmid NB, Giehl RF, Döll S, Mock HP, Strehmel N, Scheel D, Kong X, Hider RC, von Wirén N(2014) Feruloyl-CoA 6′-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol 164: 160–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin LJ, Lo JC, Chen GH, Callis J, Fu H, Yeh KC(2013) IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 25: 3039–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Severance S, Kaur N, Wiltsie W, Kosman DJ(2006) Assembly, activation, and trafficking of the Fet3p.Ftr1p high affinity iron permease complex in Saccharomyces cerevisiae. J Biol Chem 281: 13355–13364 [DOI] [PubMed] [Google Scholar]
- Tan S, Zhang P, Xiao W, Feng B, Chen LY, Li S, Li P, Zhao WZ, Qi XT, Yin LP(2018) TMD1 domain and CRAC motif determine the association and disassociation of MxIRT1 with detergent-resistant membranes. Traffic 19: 122–137 [DOI] [PubMed] [Google Scholar]
- Thomine S, Vert G(2013) Iron transport in plants: Better be safe than sorry. Curr Opin Plant Biol 16: 322–327 [DOI] [PubMed] [Google Scholar]
- Vert G, Briat JF, Curie C(2001) Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J 26: 181–189 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C(2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T.(2007) Optimized protein extraction for quantitative proteomics of yeasts. PLoS One 2: e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton A, Stes E, Cybulski N, Van Bel M, Iñigo S, Durand AN, Timmerman E, Heyman J, Pauwels L, De Veylder L, et al. (2016) It’s time for some “site”-seeing: novel tools to monitor the ubiquitin landscape in Arabidopsis thaliana. Plant Cell 28: 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Wallmeroth N, Berendzen KW, Grefen C(2016) Techniques for the analysis of protein–protein interactions in vivo. Plant Physiol 171: 727–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nagano AJ, Nishina M, Hara-Nishimura I, Nishimura M(2013) Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol 161: 108–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML(1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Sato K, Takeuchi M(2014) Molecular mechanisms of Sar/Arf GTPases in vesicular trafficking in yeast and plants. Front Plant Sci 5: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Zhang D, Song T, Xu F, Lin S, Xu W, Li Q, Zhu Y, Liang J, Zhang J(2017) Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J Exp Bot 68: 1731–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ(2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zelazny E, Barberon M, Curie C, Vert G(2011) Ubiquitination of transporters at the forefront of plant nutrition. Plant Signal Behav 6: 1597–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E, Borst JW, Muylaert M, Batoko H, Hemminga MA, Chaumont F(2007) FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc Natl Acad Sci USA 104: 12359–12364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E, Miecielica U, Borst JW, Hemminga MA, Chaumont F(2009) An N-terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2;4 and ZmPIP2;5 to the plasma membrane. Plant J 57: 346–355 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010) Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301 [DOI] [PubMed] [Google Scholar]