Triple-localized WHIRLY2 plays a vital role in carbon reallocation from maternal tissue to filial tissue through controlling sucrose transporter gene expression.
Abstract
Coordination of gene expression in mitochondria, plastids, and nucleus is critical for plant development and survival. Although WHIRLY2 (WHY2) is involved in mitochondrial genome repair and affects the DNA copy number of the mitochondrial genome, the detailed mechanism of action of the WHY2 protein is still elusive. In this study, we found that WHY2 was triple-localized among the mitochondria, plastids, and the nucleus during Arabidopsis (Arabidopsis thaliana) aging. Overexpressing WHY2 increased starch granule numbers in chloroplasts of pericarp cells, showing a partially dry, yellowing silique and early senescence leaves. Accordingly, WHY2 protein could directly activate the expression of jasmonic acid carboxyl methyltransferase and senescence associated gene 29 (SWEET15) gene expression and repress SWEET11 gene expression in the nucleus, leading to alteration of starch accumulation and transport in pericarp cells. In contrast, loss of WHY2 decreased starch and sugar content in pericarp cells but promoted starch accumulation in leaves and seeds. These phenotypes of WHY2-overexpressing plants were enhanced in response to methyl jasmonate. Our results suggest that WHY2 in plastids, mitochondria, and the nucleus plays a vital role in alteration of carbon reallocation from maternal tissue to filial tissue.
There are three organelles in plant cells, which possess and maintain genetic information: nucleus, plastids, and mitochondria. Coordination of gene expression in these organelles is critical for plant development and survival (Nott et al., 2006; Liu and Butow, 2006). In addition to the crosstalk between chloroplasts/mitochondria and nucleus, functional interactions between chloroplasts and mitochondria have been established during plant evolution to coordinate the activities of these two organelles, which exhibit a high degree of metabolic interdependence (Leister 2005; Bailleul et al., 2015). Therefore, communication among multiple organelles in the cell is important for cell fate and survival.
The WHIRLY protein family is a small family of single-stranded DNA/RNA binding proteins located in organelles and the nucleus. The first identified WHIRLY family member was the transcription activator p24/PBF2 (PR10a binding factor 2), isolated by Desveaux and coauthors from potato (Solanum tuberosum). PBF2 binds to the elicitor inductive response element and activates the expression of PR10a, a pathogen-related gene of potato, and was later named StWHIRLY1 (Desveaux et al., 2000). Subsequently, WHIRLY family members were found in many plant species (Desveaux et al., 2005). WHIRLY1 (WHY1) and WHIRLY2 (WHY2) exist in all plants; WHIRLY3 (WHY3) is found only in dicotyledonous plants. Two members of the WHIRLY family in Arabidopsis (Arabidopsis thaliana), WHY1 and WHY3, were found in chloroplasts and the nucleus (Krause et al., 2005; Xiong et al., 2009), whereas WHY2 was localized in mitochondria (Krause et al., 2005; Maréchal et al., 2008; Cai et al., 2015). The functions of WHY1 have been well documented in various plant species and can be summarized in four categories: (1) a transcription factor in the nucleus regulating the expression of downstream target genes involved in the pathogen response pathway and plant senescence, such as PR10a in potato, WRKY53 and kinesin in Arabidopsis, HvS40 in barley (Desveaux et al., 2004; Xiong et al., 2009; Miao et al., 2013; Krupinska et al., 2019); (2) an auxiliary factor for homologous recombination and double-strand DNA break repair in organelles, playing a role in maintaining organelle genome stability (Maréchal et al., 2008); (3) a DNA/RNA-binding protein interacting with the telomere repeat sequence in plastids, such as psbA and the TAC complex, playing the role of a “chaperone” protein in RNA processing (Pfalz et al., 2006; Prikryl et al., 2008; Melonek et al., 2010; Świda-Barteczka et al., 2018; Zhuang et al., 2018); (4) its enrichment at the thylakoid membrane appears to have an effect on photosynthesis, redox stress, and the hormone signaling system (Isemer et al., 2012; Foyer et al., 2014; Comadira et al., 2015; Huang et al., 2017; Guan et al., 2018; Lin et al., 2019). WHY1 could also interact with WHY3 synergistically in maintaining organelle genome stability and protein metabolism (Maréchal et al., 2008; Guan et al., 2018). WHY2 functions have not been studied as extensively. Overexpressing WHY2 affected the mitochondrial complex I/V of the respiratory transport chain, and it was proposed that WHY2 plays a role in Arabidopsis mitochondrial genome repair and protects against mitochondrial genome dysfunction (Maréchal et al., 2008; Cappadocia et al., 2010). Later, the why2 null mutant showed low copy number DNA of the mitochondrial genome, whereas WHY2 overexpression in pollen tube cells caused abnormalities in respiration (Cai et al., 2015). However, the detailed mechanisms of action of the WHY2 protein in different organelles are still elusive.
In this study, we found that WHY2 was localized in all three DNA-containing organelles: mitochondria, plastids, and the nucleus, during plant aging. It could bind both to a DNA/RNA fragment of the mitochondrial genome and to a DNA fragment of the nuclear genome, and alter gene expression of the Suc transporter SWEET11/15 and starch allocation in the pericarp cell during leaf senescence and silique development. This phenomenon was enhanced in response to methyl jasmonate (MeJA). These results suggest that WHIRLY2 plays an important role in carbon reallocation between organelles and the nucleus, working as a potential retrograde signal connecting mitochondria, plastids, and the nucleus.
RESULTS
Overexpression of WHY2 Accelerates Leaf Senescence and Causes Abnormal Silique Development
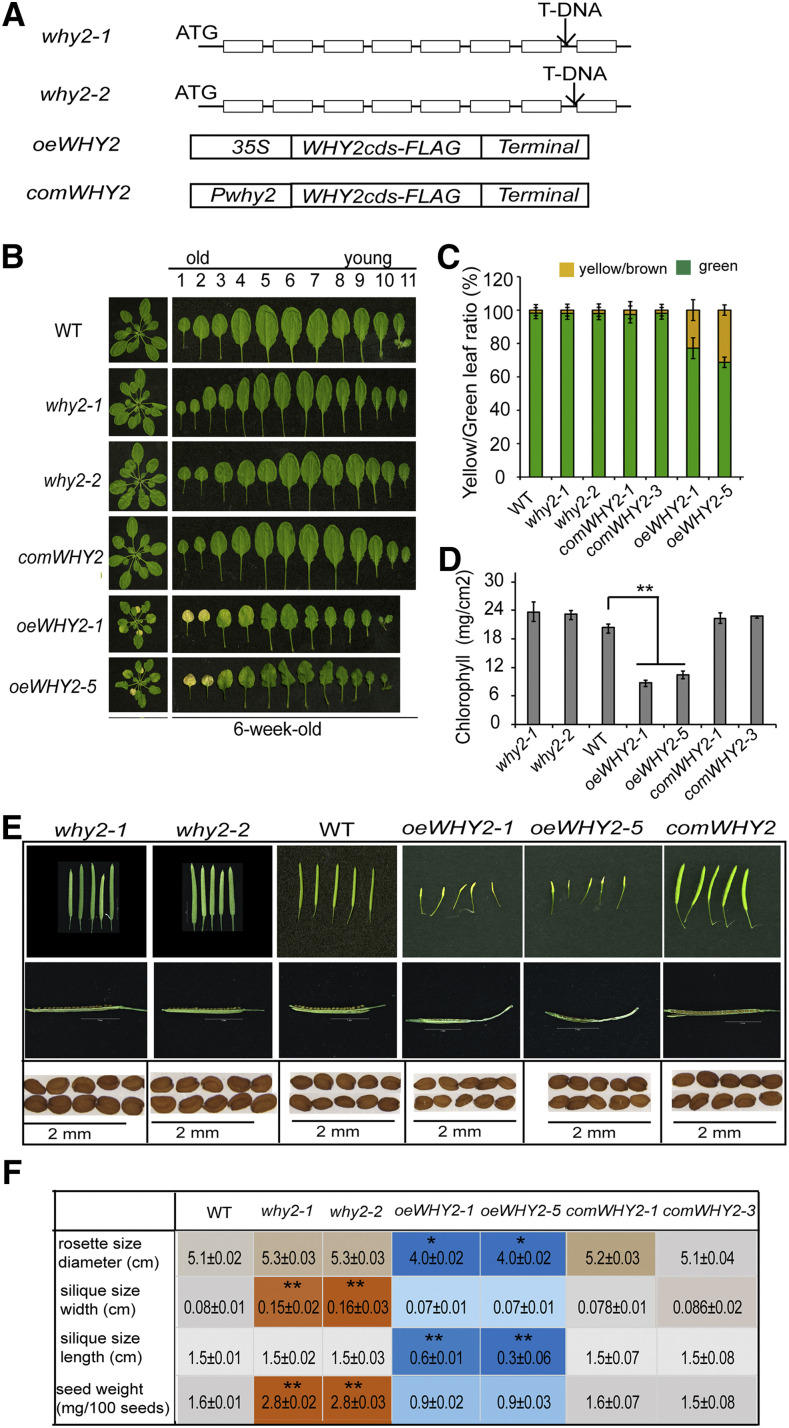
To systemically investigate the function of WHIRLY2 (WHY2) protein during the development of Arabidopsis plants, we produced gain-of WHY2, loss-of WHY2, and complemented WHY2 transgenic plants. We identified them by northern blot, semi reverse transcription quantitative PCR, reverse transcription-quantitative PCR (RT-qPCR), and Western blotting (Supplemental Fig. S1). After we screened eight overexpressing lines, three T-DNA insertion knockout lines, and four complement lines by RT-qPCR, we selected two homozygous WHY2 overexpressing lines (oeWHY2-FLAG L1 and oeWHY2-FLAG L5), two complemented lines that harbors the WHY2 coding sequence (CDS) fused to FLAG driven by its own promoter (Pwhy2:WHY2-FLAG) in the why2 background (comWHY2-1 and comWHY2-3), and two why2 knockout mutants (why2-1 and why2-2) with different T-DNA insertion sites (Fig. 1A). The results of the northern blot and semiquantitative PCR showed that the mRNA level of WHY2 in these two knockout lines (why2-1, why2-2) is barely detectable, but a 600-bp mRNA signal can be detected in wild-type plants and the comWHY2-FLAG line and they had the same WHY2 mRNA level (Supplemental Fig. S1A). Furthermore, the expression level of WHY2 in oeWHY2-L1 and oeWHY2-L5 was significantly higher than that in wild-type plants (Supplemental Fig. S1B). Simultaneously, WHY2 protein levels in oeWHY2, why2, comWHY2, and wild type were immunodetected with an antibody against a WHY2 peptide (Supplemental Fig. S2), which showed a strong signal in the two oeWHY2 lines (Lines 1 and 5), a weak signal in comWHY2 and wild-type plants, and no signal in the two why2 lines (Supplemental Fig. S1C). Next, we assessed the phenotypes of the above-mentioned transgenic plants. Interestingly, we found a small rosette, earlier leaf senescence, 4 to 5 d earlier bolting, and an abnormal silique development phenotype occurring in the overexpressing WHY2 lines, and larger siliques and seeds in the why2 lines. Twelve 5-week-old plants of the oeWHY2, why2, and comWHY2 lines and wild-type plants were systematically and statistically analyzed. The rosette size of oeWHY2 plants was significantly smaller compared to the wild type (Fig. 1, B and F). Additionally, the rosette of 6-week-old oeWHY2 plants showed a much severer yellowing and senescence phenotype up to leaf number 3, whereas all leaves of the why2 plants stayed green (Fig. 1B). A statistical analysis of 12 plants was performed by categorizing the leaves into four groups according to their leaf color (green; green/yellow; fully yellow; brown/dry; Fig. 1C); the yellowing leaves ratio was much higher, and the chlorophyll content of rosettes in the oeWHY2 plants was coordinately lower by 30% (Fig. 1D). Intriguingly, the width and length of siliques in the oeWHY2 plants were significantly shorter compared with wild-type plants; in contrast, the width of siliques in the why2 line was larger relative to wild type, and the weight per 100 seeds in why2 was significantly increased by nearly twofold (Fig. 1F). The complemented lines rescued the why2 phenotype, similar to wild type (Fig. 1, D–F). The microarray data taken from public datasets of the The Arabidopsis Information Resource Web site (Winter et al., 2007) clearly showed that WHY2 was highly expressed in imbibed seeds, flowers, young siliques, and the shoot apex (Supplemental Fig. S3), but expressed at low levels in rosette leaves, roots, mature siliques, and pollen (Supplemental Fig. S3).
Figure 1.
Phenotyping of overexpressing WHY2 (oeWHY2), WHY2 knockout (why2), and complemented WHY2 (comWHY2) lines. A, Scheme of oeWHY2, why2, and comWHY2 constructs. B, Observation of rosette leaf senescence of the oeWHY2, why2, comWHY2, and wild type (WT) lines. C, Senescence leaf ratio in the oeWHY2, why2, comWHY2, and wild-type lines. D, Chlorophyll content in the oeWHY2, why2, comWHY2, and wild-type lines. E, Observation of silique development and seed size in the oeWHY2, why2, comWHY2, and wild-type lines; seed size represents the length of five seeds, with why2 lines >2 mm, and wild type and oeWHY2 line ≤2 mm. F, Calculation of rosette size (diameter), silique size (width and length), and seed weight (weight per 100 seeds) in the oeWHY2, why2, comWHY2, and wild-type lines. Error bars represent the sd of triplicates. Asterisks denote statistically significant differences from the wild type, calculated using Student’s t test: *P < 0.05; **P < 0.01. Orange colum indicates increasing; blue column indicates decreasing.
Overexpressing WHY2 Increases the Number of Starch Granules in Chloroplasts of Pericarp Cells
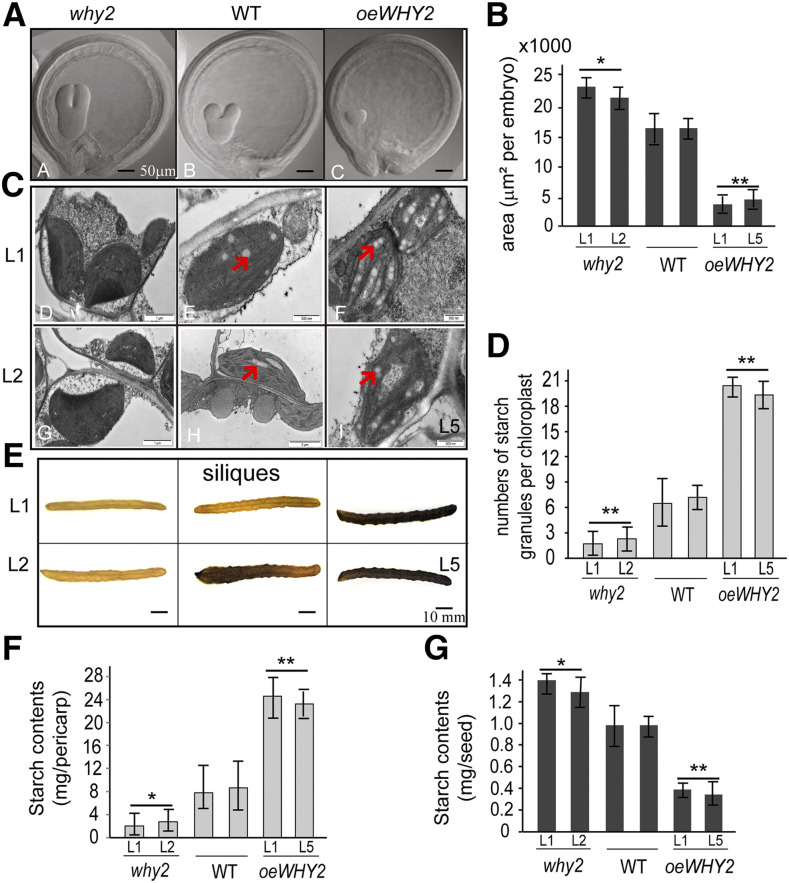
We further observed the development of siliques in three different WHY2 expression level backgrounds of 7-week-old WHY2 transgenic plants by cytological observation. We did not find any significant defects in germinated pollen. However, we observed delayed embryo development in oeWHY2 and slightly earlier development in the why2 line, compared with the wild type (Fig. 2A). We calculated and measured the area of 20 embryos per line and showed 90% embryos were fivefold smaller in the two oeWHY2 lines and slightly larger in the two why2 lines compared with the wild type (Fig. 2B). The results of ultrastructural observation did not reveal any structural changes in the chloroplast grana, stromal thylakoids, and mitochondria of pericarp cells of the silique from the oeWHY2 lines that displayed abnormal embryo development. However, a significant difference in starch granule number was observed. The accumulation of starch granules in chloroplasts of pericarp cells was significantly higher in oeWHY2. In contrast, the number of starch granules was significantly lower in the why2 line, compared with the wild type (Fig. 2, C and D). Further, starch content was analyzed in siliques, including seeds and pericarp cells of oeWHY2, why2, and the wild type using Lugol’s iodine, which stained starch blue/black (Fig. 2E). Starch was barely detectable in why2 pericarp cells, whereas a dramatically stronger blue signal was observed in the oeWHY2 pericarp cells, relative to the wild type, indicative of reduced delivery of Suc to the seeds of oeWHY2 siliques (Fig. 2E). Consistent with the optical analysis of the starch staining, enzymatic quantitation of starch showed that the pericarp cells of oeWHY2 siliques accumulated up to three times more starch, compared to the wild type. However, the seeds of oeWHY2 accumulated up to five times less starch relative to the wild type (Fig. 2, F and G). The accumulation of starch in the pericarp cells implicated a block in sugar release from the pericarp cells as a key step in embryo development and seed filling. Therefore, the abnormal development of silique caused by overexpressing WHY2 may be related to the disturbance of starch output from chloroplasts of maternal tissue pericarp cells or leaf cells.
Figure 2.
Cytological and histochemical analysis of siliques of oeWHY2, why2, and wild-type (WT) plants. A, Representative images of developing embryo in the seeds (7-week-old plant) are shown above the panels. Scale bars = 50 μm (as shown in the top left). B, Calculation of the embryo size in the silique of oeWHY2, why2, and wild-type plants. Twenty embryos per line were used. C, Observation of chloroplast ultrastructure. The red arrows indicate the starch granules. Scale bars = 250 nm. D, Calculation of the starch granules in the pericarp cells of oeWHY2, why2, and wild-type plants. Ten cells/silique and 5 chloroplasts/cell were used. Error bars represent the sd of triplicates. E, Starch staining of siliques including pericarp and seed in 9-week-old oeWHY2, why2, and wild-type plants. The red arrows indicate the seeds. Scale bars =10 mm. The siliques were separated to pericarp and seed to digitally extract for comparison. F and G, Starch content of siliques including pericarp (F) and seed (G) in 9-week-old oeWHY2, why2, and wild-type plants. Three plants, three siliques/plant, and 30 seeds/silique were used. Error bars represent the sd of triplicate reactions. Asterisks denote statistically significant differences from the wild type, calculated using Student’s t test: *P < 0.05; **P < 0.01; and ***P < 0.001.
WHY2 Alters the Expression Level of Starch Metabolism and Senescence-Related Genes
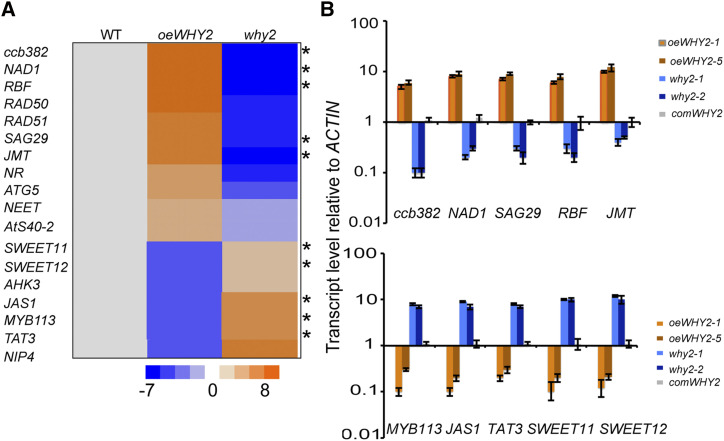
Based on the above visible phenotype, we screened the gene expression level of a total of 81 genes, including genes related to starch metabolism, sugar transport, energy metabolism, silique development, and senescence (Supplemental Table S1). The gene expression levels of most of the genes, such as meiotic recombination 11 (MRE11), phytosphingosine (PHS), alpha amylase 3 (AMY3), and genes related to starch granule initiation or starch degradation and synthesis, and stress, were not altered in response to varying levels of WHY2 (Supplemental Fig. S4). The gene expression profiles of senescence associated gene 29 (SAG29), jasmonic acid carboxyl methyltransferase (JMT), RecA homologs (RAD50, RAD51), and RNA binding factor genes, as well as NADH dehydrogenase subunit 1 (NAD1) and cytochrome c biogenesis 382 (ccb382), were significantly up-regulated in the oeWHY2 line and down-regulated in the why2 line, compared to the wild type, but the ATPase subunit 9 (ATP9) gene was up-regulated, and the MYB34 gene was down-regulated in both overexpression and knockout lines (Supplemental Fig. S4). In contrast, the gene expression of pollen-specific aquaporin NIP4, Tyr aminotransferase 3 (TAT3), MYB113, jasmonate-associated (JAS), His kinase 3 (AHK3), and Suc transporters SWEET11 and SWEET12 was down-regulated in the oeWHY2 and up-regulated in the why2 lines (Fig. 3). Interestingly, NAD1, ccb382, and ATP9 are encoded by the mitochondrial genome, whereas the others, such as JMT, RAD50, RAD51, SAG29, and SWEET11 proteins, are encoded by the nuclear genome. It has previously been reported that NAD1, ccb382, and ATP9 function in the respiratory electron transport chain (complex I) and are also involved in silique development (Leu et al., 2016). The nuclear genes listed above are mostly related to Suc transport and leaf senescence (Supplemental Table S2; Zabaleta et al., 1996; Chen et al., 2012, 2015). These results suggest that WHY2 has dual functions in the mitochondria and the nucleus.
Figure 3.
The expression profile of select genes in the rosette leaf of oeWHY2, why2, and wild-type (WT) plants. A, heatmap showing the changes in expression of select genes (see Supplemental Fig. S4) in oeWHY2, why2, and wild type by RT-qPCR analysis. B, Confirmation of the expression of select genes in the two oeWHY2, two why2, and two comWHY2 lines, compared with wild type by RT-qPCR analysis. Error bars represent the sd of three biological replicates.
WHY2 Is Triply Localized Among Mitochondria, Plastids, and the Nucleus during Plant Development
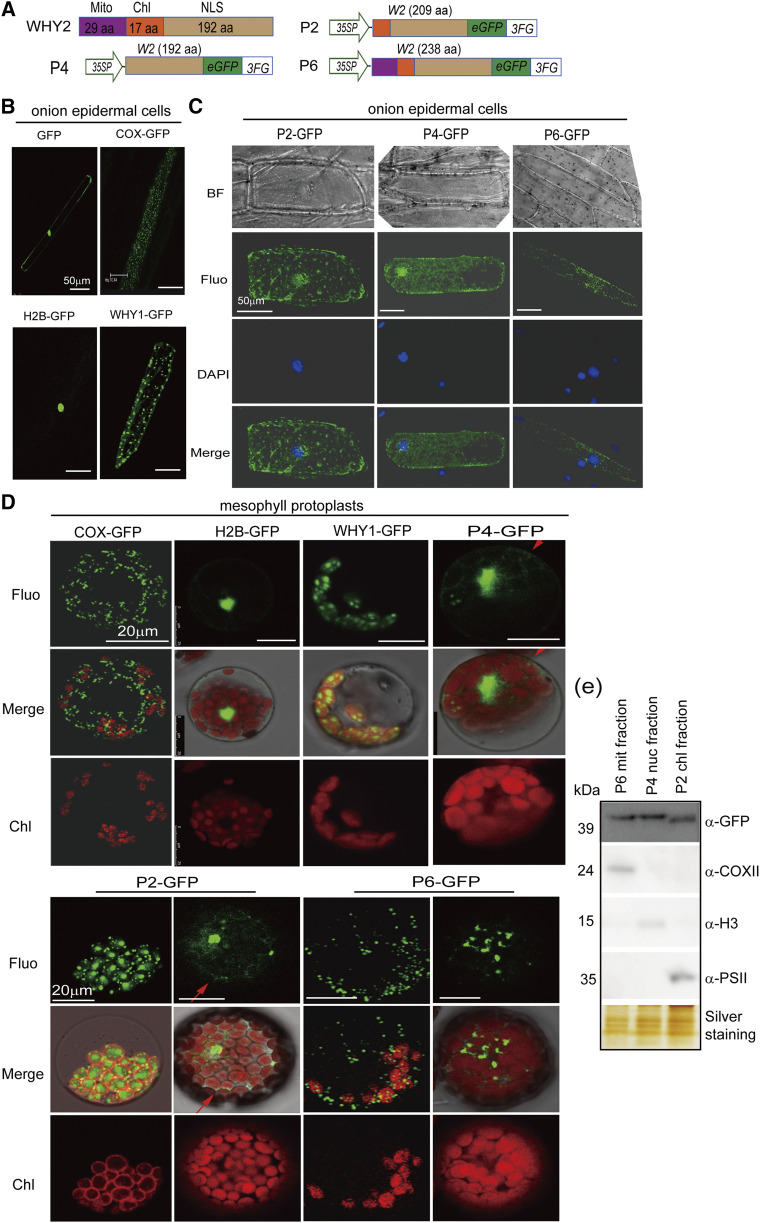
Until now, there was no direct evidence that WHY2 is localized in the nucleus or plastids. Because overexpressing WHY2 altered the expression of many mitochondrial and nuclear genes, we investigated the subcellular localization of WHY2. The WHY2 protein sequence was analyzed by a signal peptide analysis program (http://www.signalpeptide.de/). The primary structure of WHY2 is shown in Figure 4A. There is a putative plastid transit peptide and a putative mitochondrial transit peptide in the N-terminal portion of the WHY2 protein, indicating it may be localized in mitochondria and/or plastids (Fig. 4A). To clarify its localization, we produced a series of WHY2 constructs fused to GFP and transformed them into protoplasts by a transient assay or into onion epidermal cells by biolistic transformation. Mitochondrial cyclooxygenase (COX) protein, nuclear histone (H2B) protein, and chloroplast-localized WHY1 GFP fusions were used as subcellular localization positive controls (Fig. 4B). The results of the protoplast transit assay showed that the full-length WHY2 (P6) was localized in the mitochondria, which surprisingly revealed many aggregated particles that were not observed in chloroplasts. The deletion mutant (P4), lacking both the putative mitochondrial transit peptide and the putative plastid transit peptide, was localized in the nucleus; and the deletion mutant (P2), missing only the putative mitochondrial transit peptide, was localized both in the nucleus and in plastids (Fig. 4, C and D). We further collected the transformed protoplasts, then isolated and immunodetected mitochondrial protein, nuclear protein, and plastid protein in P6, P4, and P2 transformants using an antibody against GFP. The GFP signals appeared in the mitochondria fraction of P6 transformants, in the plastid fraction of P4 transformants, and in the nuclear fraction of P2 transformants. This result confirmed the imaging results (Fig. 4E).
Figure 4.
Detection of WHY2 triple localization. A, Schematic of a series of WHY2 deletion constructs. P2: WHY2 with deleted mitochondrion transit peptide fused to GFP. P4: WHY2 with deleted plastid and mitochondrial transit peptide fused to GFP. P6: full-length WHY2 fused to GFP. Scale bars = 50 μm. B, Positive control plasmids (COX-GFP, H2B-GFP, WHY1-GFP) expressed in onion epidermal cells by gene gun biolistic assay. C, Observation of P2, P4, and P6 subcellular localization in onion epidermal cells by gene gun biolistic assay. BF, bright field; Fluo, Fluorescence; DAPI, 4′,6-diamino-phenylindole staining the nucleus; scale bars = 50 μm. D, Observation of P2, P4, and P6 subcellular localization in Arabidopsis mesophyll cells by protoplast transit assay. The plasmids of COX-GFP, H2B-GFP, and WHY1-GFP were used as positive controls. Scale bars = 20 μm. E, Western blot detection of WHY2 expression in the mitochondria (Mito) fractions isolated from P6, nuclear fraction isolated from P4, and chloroplast (Chl) fraction isolated from P2 using an antibody against GFP. Anti-PSII was used as a chloroplast protein control, anti-H3 as a nuclear protein control, and anti-COXII as a mitochondrial protein control. Silver staining of the protein gel was used to indicate loading.
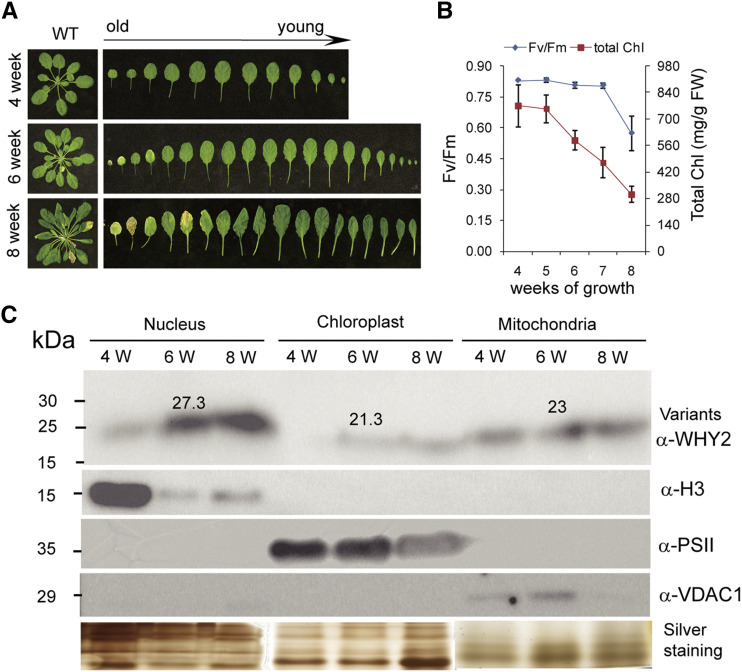
We subsequently used a cytochemistry assay to detect the cellular compartmental distribution of WHY2 protein. The proteins isolated from mitochondria, plastids, and nucleus of wild-type plants during development were subjected to immunoblot analysis using an antibody against a WHY2 peptide. Antibodies against histone 3 (H3), voltage-dependent anion channel 1 (VDAC1), and a PSII peptide were used as controls to assess the purity of nuclear, mitochondrial, and plastid protein preparations, respectively. Silver staining was used for loading control. The results showed that WHY2 protein increasingly accumulated in the nucleus, compared to the loading control and positive control protein, over the course of development from young (4 weeks) to old (8 weeks) plants (Fig. 5A), as indicated by the decrease in chlorophyll content and fluorescence activity (Fv/Fm) of PSII (Fig. 5B). In contrast, WHY2 protein was relatively stable in mitochondria during plant aging (Fig. 5C), whereas WHY2 protein in plastids was lower at week 4, increased at week 6, and slightly declined again at week 8, when normalized to the loading control. Therefore, WHY2 is localized in all three cellular compartments, and its location and distribution among cellular compartments is developmentally dependent.
Figure 5.
Detection of WHY2 triple localization during plant aging. A, Indication of three stages of wild-type (WT) plant development [4, 6, and 8 weeks (W)] by visible rosette leaf senescence. B, Indication of three stages of wild-type plant development (4, 6, and 8 weeks) and photosystem II fluorescence efficiency (Fv/Fm) and chlorophyll content. Error bars represent the sd of six biological replicates. C, WHY2 protein distribution among mitochondrion, chloroplast (Chl), and nucleus by immunodetection. Anti-PSII was used as a chloroplast protein control, anti-H3 as a nuclear protein control, and anti-VDAC1 (voltage-dependent anion channel 1) as a mitochondrial protein control. Silver staining of the protein gel was used to indicate loading.
WHY2 Directly Activates NAD1 and ccb382 Gene Expression in the Mitochondrial Genome
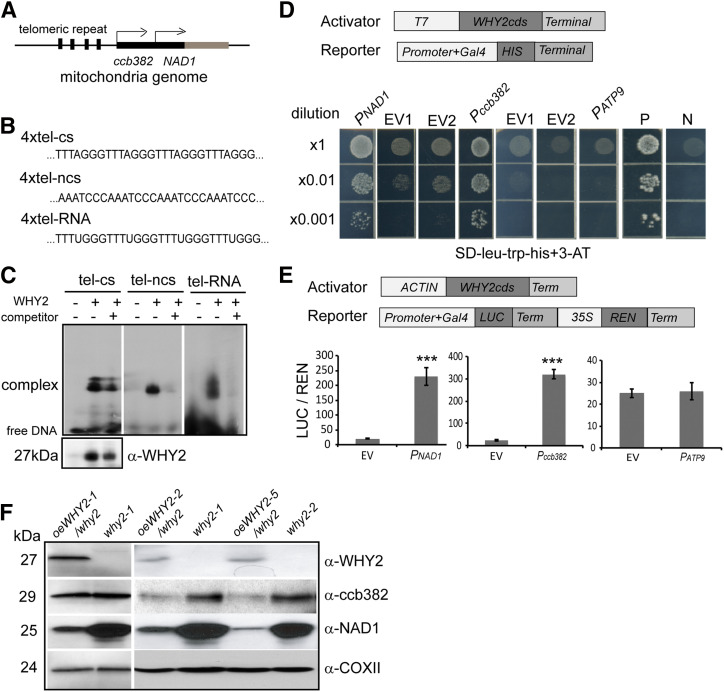
It is well documented that WHIRLY1 binds to the elicitor response element (ERE) and AT-rich region (…AAAT…AAAT) repeat motifs and the telomere repeat of downstream gene promoters (Desveaux et al., 2002; Yoo et al., 2007; Miao et al., 2013). Because WHIRLY proteins share the same ssDNA binding domain, it is possible that WHY2 binds directly to similar fragments. To address this possibility, we first screened for these sequence motifs in the whole mitochondrial genome. Interestingly, there are several telomere repeats existing in the upstream regions of NAD1 and ccb382 (Fig. 6A). We chemically synthesized the DNA fragments of upstream regions including four repeat telomere sequences as probes for coding chain (4xTel-cs), noncoding chain (4xTel-ncs), and RNA chain (4×Tel-RNA; Fig. 6B). The recombinant proteins were expressed in E. coli and detected by Western blot with an antibody against a WHY2 peptide. Electrophoretic mobility shift assay (EMSA) was used to detect the binding affinity of WHY2 to upstream regions of NAD1 and ccb382 genes. The probes were labeled with 32P and incubated with WHY2. The unlabeled fragment was used as a competitive probe. As more competitive probe was added, the binding signal became weaker (Fig. 6C). These results indicated that WHY2 protein bound to 4xTel-ncs and 4xTel-RNA fragments of the upstream regions of the NAD1 and ccb382 genes of the mitochondrial genome in vitro, but the 4xTel-cs did not show good competition and had weaker binding. A similar result was obtained for WHY1 binding to telomere-ncs (Yoo et al., 2007), and to noncoding stranded DNA chain of the WRKY53 promoter in the nucleus (Miao et al., 2013).
Figure 6.
WHY2 binds to the upstream region of NAD1 and ccb382 of the mitochondrial genome and promotes their expression. A, Diagram of the upstream regions of NAD1 and ccb382 genes. B, The sequences of the upstream regions of NAD1 and ccb382 genes. tel-cs, coding strand; tel-ncs, noncoding strand; tel-RNA, RNA sequence. C, EMSA. The tel-cs, tel-ncs, and tel-RNA fragments were used as the probe (labeled with biotin). Twentyfold excess competitor probe (without biotin) was added as a specificity control. The recombinant WHY2, expressed and isolated from E. coli, used in the reaction was detected by Western blot with anti-WHY2. D, Yeast one-hybrid assay results. The pNAD1, pccb382, and pATP9 fragments were inserted into the pHIS2 expression vector. The various dilutions of colonies on the selective medium showed the activation of expression by WHY2 of the HIS reporter gene driven by the indicated fragments in yeast. EV1, the plasmid GAD-WHY2 with the empty pHIS2; EV2, the empty plasmid GAD with promoter fragment fused HIS2 plasmid; they were cotransformed into the yeast strain AH109 as negative controls: P, positive, N, negative. E, Luciferase (LUC)/Renilase (REN) dual activation assay. Agrobacterium cells containing the vectors expressing WHY2-FLAG (ACTIN:WHY2-FLAG) and the Agrobacterium cells containing the vectors expressing fragments: LUC-REN were coinjected into Nicotiana benthamiana leaves. ATP9 promoter was used as a negative control. Shown are mean and SE of six biological replicates. Asterisks denote statistically significant differences from the empty vector, calculated using Student’s t test: *P < 0.05; **P < 0.01; and ***P < 0.001. F, Western blot detection of NAD1 and ccB382 protein levels in the oeWHY2 and why2 lines. COXII was used as a loading control.
Furthermore, we confirmed a direct interaction of WHY2 with the upstream regions of the NAD1 and ccb382 genes using yeast one-hybrid system. The upstream fragments of NAD1, ccb382, and ATP9 were cloned into the pHIS2 reporter vector, and WHY2 CDS was subcloned into the pGADT7 vector under the control of a T7 promoter. The two plasmids were then cotransformed into the yeast strain AH109, incubated on SD-trp-leu-his medium plus 50 mm 3-amino-1,2,4-triazole. In the presence of the plasmid encoding WHY2-BD, both PNAD1:HIS2 and Pccb382:HIS2 were activated, as indicated by colony growth on selective medium (Fig. 6D). In contrast, no growth was observed with PATP9:HIS2 (Fig. 6D).
The binding of WHY2 to the upstream regions of NAD1, ccb382, and ATP9 was examined using the transient dual-luciferase assay system in Nicotiana benthamiana leaves (Hellens et al., 2005). The promoter sequences of NAD1, ccb382, and ATP9 were cloned into dual-luciferase vectors. The WHY2 coding sequence, fused to FLAG-tag and under the control of the Arabidopsis ACTIN1 promoter (ACTIN:WHY2-HA), was coinfiltrated with reporter vector containing the above putative promoter sequence of the tested genes fused to the luciferase (LUC) and rennilase (REN) reporter (Hellens et al., 2005). We then measured the LUC and REN luminescence ratio (LUC/REN ratio) in infiltrated leaves. To assess any basal activation or repression of putative promoters, a mini-GAL4 promoter vector was used in each coinfiltration experiment as a control; ATP9 promoter was used as a negative control. The results showed high LUC/REN ratio with the promoters of NAD1 and ccb382 in the presence of WHY2 (Fig. 6E). Therefore, WHY2 directly activates NAD1 and ccb382 expression. Surprisingly, NAD1 protein substantially accumulated in the why2 plants, whereas ccb382 protein maintained the same level or slightly increased in the why2 plants (Fig. 6F). Additionally, NAD1 dramatically declined upon overexpression of WHY2 in the why2 background (oeWHY2/why2), as assessed by immunodetection (Fig. 6F). We hypothesize that NAD1 and ccb382 protein levels were affected by WHY2 at the posttranscriptional level.
Taken together, our results show that WHY2 functions as a DNA/RNA-binding protein to activate NAD1 and ccb382 gene transcription. However, WHY2 suppresses NAD1 and ccb382 protein accumulation in the mitochondrion.
Nuclear WHY2 Directly Binds to the Promoter Region of JMT, SAG29, and SWEET11 Genes and Alters Their Expression Level and Carbon Allocation
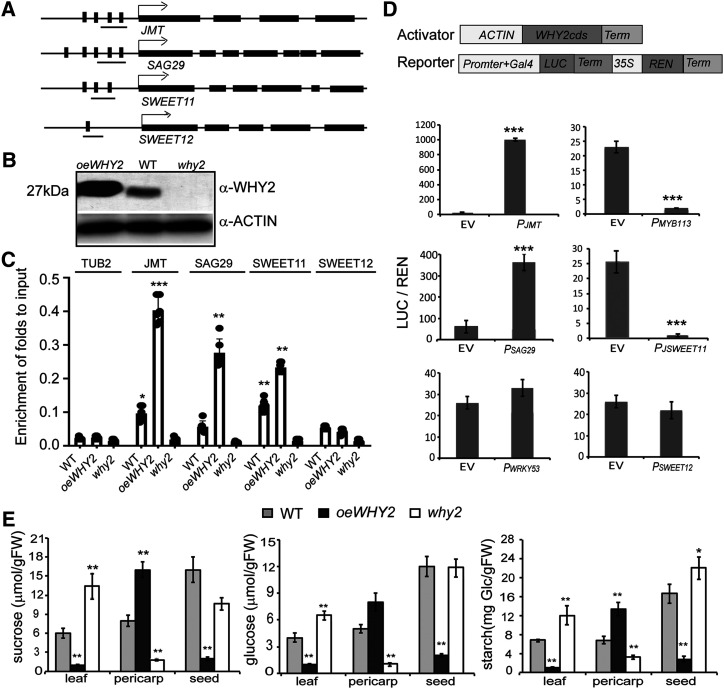
As shown above, WHY2 is also localized in the nucleus. As WHY2 is an ssDNA-binding protein in the nucleus, it is possible that WHY2 directly binds to the promoter region of the downstream genes. To investigate this possibility, promoter analysis was done for several selected genes: JMT, MYB113, SWEET11, SWEET12, and SAG29 (SWEET15), by using the PlantCARE program (Lescot et al., 2002). Only the promoters of JMT, MYB113, SAG29, SWEET12, and SWEET11 contain ERE and AT-rich motifs. A chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) assay was performed to test whether WHY2 directly bind to a fragment of its downstream genes JMT, MYB113, SWEET11, SWEET12, and SAG29 promoters (Fig. 7A). The ChIP-qPCR results showed that the binding signal of the fragment was considerably enriched in the anti-WHY2 peptide ChIP samples of the oeWHY2 plants, compared with the samples of the why2 line or wild type (Fig. 7B). A strong binding signal of JMT, SAG29, and SWEET11 was detected in the samples from the oeWHY2 line, and a modest signal was detected in the wild type samples (Fig. 7C). These results suggested that WHY2 could directly bind to these regions of JMT, SAG29, and SWEET11, but not MYB113 and SWEET12. The promoter region of tubulin beta chain 2 (TUB2; −246 to −362 bp), which was used as negative control, was not enriched in the ChIP samples (Fig. 7D).
Figure 7.
WHY2 binds to upstream regions of JMT, SAG29, and SWEET11 and alters their expression. A, Diagram of the promoters of JMT, SAG29, SWEET11, and SWEET12 genes. B, Western blot detection of WHY2 protein in the rosette of 7-week-old oeWHY2, why2, and wild-type (WT) plants. C, Chromatin immunoprecipitation (ChIP) assay in rosette leaves of 7-week-old Arabidopsis oeWHY2, why2, and wild-type plants. Antibody against WHY2 peptide was used. TUB2 was used as a negative control. The fold enrichment of ChIP is relative to input. Data represent mean ± sd of five biological replicates. Asterisks denote statistically significant differences from the enrichment of TUB2, calculated using Student’s t test: (*P < 0.05; **P < 0.01, ***P < 0.001). D, LUC/REN dual activation assay, as above. Agrobacterium cells containing the vectors expressing WHY2-FLAG (ACTIN:WHY2-FLAG) or vectors expressing candidate promoter fragments plus GAL4: LUC-REN were coinjected into Nicotiana benthamiana leaves. The WRKY53 promoter was used as a negative control. E, Suc, Glc, and starch content in the leaf, pericarp, and seeds of 7-week-old oeWHY2, why2, and wild-type plants. Shown are mean ± se of six biological replicates. Asterisks denote statistically significant differences from the empty vector or wild type calculated using Student’s t test: *P < 0.05; **P < 0.01; and ***P < 0.001.
Similarly, the binding activity of WHY2 to the upstream region of JMT, SAG29, SWEET12, and SWEET11 was examined by a transient dual-luciferase assay system in Nicotiana benthamiana leaves. The promoter sequences of JMT, SAG29, SWEET12, and SWEET11 were cloned into a dual-luciferase vector; the promoter of WRKY53 was used as a control. The plasmid encoding ACTIN:WHY2-FLAG was coinfiltrated with the reporter vector containing the putative promoter sequences of the above tested genes fused to the LUC reporter. A LUC/REN ratio of 350 was detected in infiltrated leaves with the SAG29 promoter in the presence of WHY2; however, a 20-fold drop in LUC/REN ratio was detected with the SWEET11 promoter in the presence of WHY2 (Fig. 7D). Surprisingly, a 1000-fold increase was observed with the JMT promoter, whereas no change was observed with SWEET12 and WRKY53. These results suggest that WHY2 directly binds to the promoter regions of JMT, SAG29, and SWEET11, and activates JMT and SAG29 gene expression, whereas repressing SWEET11 gene expression, consistent with the gene expression profile detected by RT-qPCR (Fig. 3).
SAG29 was first identified as a jasmonic acid (JA)–mediated senescence marker gene, highly expressed in senescent leaves (Seo et al., 2001; Qi et al., 2015). It is a membrane protein that is able to transport Suc, which led to it later being renamed SWEET15. SWEETs were prime candidates to play roles in sugar secretion from maternal tissues (leaf, pericarp, seed coat) during seed development (Chen et al., 2015). Because WHY2 activated SAG29 (SWEET15) gene expression and repressed SWEET11 gene expression, WHY2 should affect Suc transport from chloroplast of leaf and pericarp to other compartments or tissues. To test this, we measured the starch accumulation and sugar (Suc and Glc) content of leaf, pericarp, and seed of 7-week-old oeWHY2, why2, and wild-type plants. Interestingly, the starch content and sugar content were significantly increased in the pericarp cells of siliques but decreased in the seeds of the oeWHY2 line. In contrast, the starch content and sugar (Suc and Glc) content declined fivefold in the pericarp cells of the why2 line. Although the starch content significantly increased in the seeds of the why2 plants, the sugar content was not changed in the seeds of the why2 line, compared with the wild type. The starch accumulation and sugar content in stems did not change significantly in response to varying levels of WHY2. However, the content of starch and sugar in leaves decreased significantly in the oeWHY2 plants, and increased in the why2 plants (Fig. 7E). Therefore, it seems that WHY2 affects Suc transport from leaf to pericarp cells via SWEET15 and from pericarp cells to seeds via SWEET11.
WHY2 Levels Affect JMT and SAG29 Levels and Starch Content in the Seedling in Response to JA
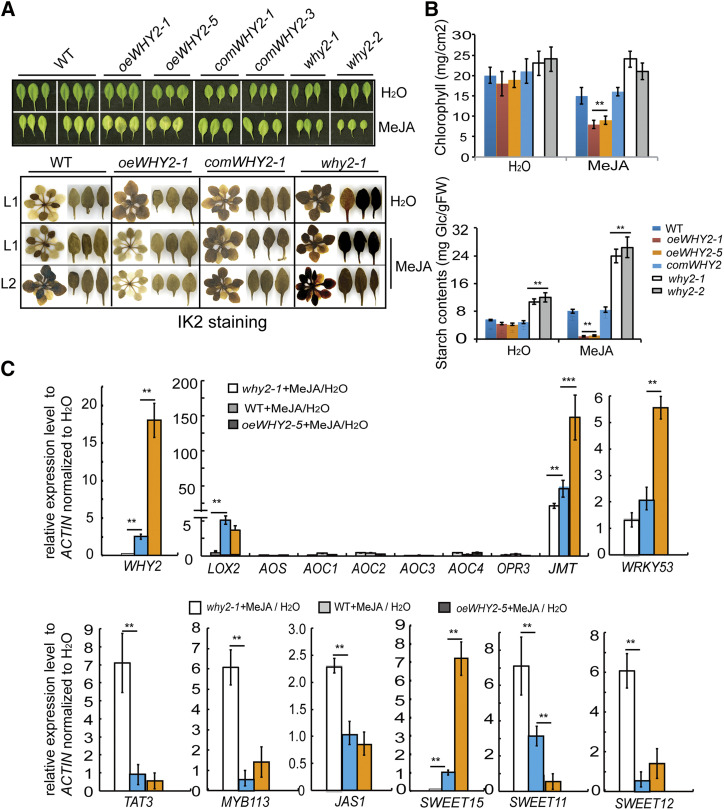
In addition to SAG29 and SWEET11, WHY2 could activate JMT expression and repress JAS and TAT3 gene expression (Fig. 3). Therefore, WHY2 may be involved in the JA responsive signal pathway. To test this possibility, we treated 4-week-old oeWHY2, why2, and wild-type seedlings with 100 mm MeJA for 1, 2, 3, and 4 h (Supplemental Fig. S5), and observed the senescence phenotype of seedlings and the starch content of leaves (in the fifth, sixth, and seventh leaves). Iodine staining of seedlings showed that a blue signal was enhanced in why2 seedlings and declined in oeWHY2 seedlings, compared with wild type, after MeJA treatment for 4 h, demonstrating an accelerated early senescence phenotype (Fig. 8A). Starch content and chlorophyll content declined significantly in the leaves of the oeWHY2 line in response to MeJA (Fig. 8B). Surprisingly, the gene expression levels of WHY2, JMT, and SWEET15 were largely increased in the oeWHY2 and wild-type plants, but that of SWEET11 was significantly decreased after MeJA treatment (Fig. 8C). This result suggests that WHY2 accelerates Suc transport out of leaves, but inhibits Suc transport from pericarp cells to seeds in response to MeJA. Thus, sugar starvation might cause early leaf senescence and at the same time delay (embryo) seed development.
Figure 8.
WHY2 alters carbon reallocation and leaf senescence in response to methyl jasmonate. A, Photograph of 4-week-old seedlings and iodine-stained rosettes of the oeWHY2, why2, comWHY2, and wild-type (WT) seedlings after methyl jasmonate (MeJA) treatment for 4 h. The degree of blue staining of the leaves (leaf 5, 6, 7) reflects the accumulation of starch. B, Determination of chlorophyll and starch content. The results were repeated in three independent experiments. Shown are the mean ± sd of three biological replicates. Asterisks denote statistically significant differences from the wild type calculated using Student’s t test: **P < 0.01. C, The expression levels of jasmonic acid-responsive genes and SWEET 11, 12, 15 genes in response to MeJA by RT-qPCR analysis. Shown are the mean ± sd of three biological replicates. Asterisks denote statistically significant differences from the wild type, calculated using Student’s t test: **P < 0.01 and ***P < 0.001.
DISCUSSION
Effective degradation and remobilization of macromolecules is important for senescence and successful reproduction (Gepstein et al., 2003). In this study, we found that WHY2, a protein triple-localized among mitochondrion, plastids, and the nucleus, has altered localization during plant aging. It works as a DNA/RNA binding protein both in mitochondria and in the nucleus, controlling NAD1/ccb382 and SWEET11/SWEET15 levels, respectively. Overexpressing WHY2 enhanced starch accumulation in chloroplasts of pericarp cells, leading to withered, yellowed, and premature senescence phenotypes of leaves and siliques.
One of the WHIRLY family proteins, WHIRLY1, has dual locations and functions (Krause et al., 2005; Grabowski et al., 2008; Ren et al., 2017b). It has been reported, using an in vitro import assay, that the WHY2 protein could be imported into mitochondria and plastids (Krause et al., 2005). This study further addresses the subcellular localization of WHY2, and shows that it is localized in three compartments (mitochondria, plastids, and the nucleus) during plant aging (Fig. 5), with nuclear localization mainly in senescent leaf cells. In mitochondria, WHY2 could directly bind to the 4× telomere repeat DNA/RNA fragments of the upstream regions of NAD1 and ccb382 genes, and activate NAD1 and ccb382 gene transcription, but suppress NAD1 protein accumulation (Fig. 6). NAD1 is known to be a critical component of mitochondrial respiration complex I, with mutants demonstrating retarded plant growth and defects in postembryonic development and/or seed germination during plant development (Leu et al., 2016; Sun et al., 2018). In particular, R300W and P313S mutations in the mitochondrial NAD1 protein (Ren et al., 2017a) caused reduced levels of the mitochondrial protein complex I (Solotoff et al., 2015). A failure in nad1 intron splicing exhibited a severe reduction in complex I assembly and activity, mitochondrial structural disturbances, and an increase in alternative oxidase AOX2 and AOX3 levels in maize (Keren et al., 2012; Ren et al., 2017a; Sloan et al., 2018). Cytochrome c biogenesis 382 (ccb382) is also involved in the mitochondrial energy redox pathway (Welchen et al., 2012; Liang et al., 2015). ATP9 is related to splicing and editing of intron RNA (Forner et al., 2007), and overexpressing an unedited form of ATP9 (u-atp9) can induce male sterility in Arabidopsis (Gómez-Casati et al., 2002). ATP9 is a core component of the F0 moiety of the mitochondrial ATP synthase, with a major role in mitochondrial function (Naumenko et al., 2017). ATP9 mRNA interference using a tRNA mimic strategy altered the carbon metabolism pathway and chlorophyll metabolism in the chloroplast (Busi et al., 2011a; Niazi et al., 2019). This suggests that NAD1, ccb382, and ATP9 are involved in the mitochondrial respiratory electron transmission chain. Interestingly, mitochondrial protein complexes I and V were reduced in the oeWHY2 line (Maréchal et al., 2008). In transgenic plants, with WHY2 under the control of the pollen vegetative cell-specific promoter Lat52 (LYWHY2), it was found that the WHY2 level impacted the ATP/ADP ratio and the energy allocation of pollen cells during pollen development (Cai et al., 2015). On the one hand, binding of WHY2 in the mitochondrial genome activates NAD1 and ccb382 gene expression at the transcriptional level, whereas on the other hand this binding might indirectly alter NAD1-82 and ATP9 RNA editing or splicing. Although WHY2 is not a direct RNA editing factor (Maréchal et al., 2008), WHY2 might interact with another mitochondrial-localized RNA binding protein (Law et al., 2015). It is hypothesized that mtWHY2 takes part in maintaining the respiratory electron transport chain by controlling NAD1 and ccb382 levels (Kühn et al., 2005). High mtWHY2 levels may inhibit electron transfer and cause redox imbalance, possibly as a retrograde signal linking carbon metabolism in the nucleus and chloroplast. This hypothesis is supported by the results of the transcriptome analysis in Arabidopsis plants with mitochondrial dysfunction, which revealed important modifications in the expression of some genes from the carbon metabolic pathways with inhibition of glycolysis and the induction of the Malate Dehydrogenase (MDH) alternative pathways (Busi et al., 2011b).
Although the WHY2 protein sequence lacks a nuclear localization peptide signal, we could detect WHY2 in the nucleus by GFP fusion transformation and protein immunodetection of organelles. In the nucleus, nWHY2 could directly bind to the upstream ERE plus AAAT-rich regions of SAG29 and SWEET11 by ChIP-qPCR (Fig. 6), activate the expression of SAG29, and repress SWEET11 gene expression (Figs. 6 and 7). SAG29 was identified as SWEET15, which was used as a JA-inducible senescence marker because its transcript level was found to increase 22-fold in leaves during senescence (Seo et al., 2001; Qi et al., 2015). SWEET15 may function in mobilization of carbohydrates, including Gal, Fru, and Glc, during senescence (Seo et al., 2001; Chen et al., 2010). It was previously suggested that SWEET15 is unlikely to play a major role in phloem loading, and is likely responsible for mediating Suc efflux from the outer integument into the apoplasm. SWEET11 is a key player in seed coat Suc efflux. SWEET11 and SWEET15 are both expressed in leaves (Chen et al., 2012; Le Hir et al., 2015) and seeds (Chen et al., 2015). A sweet11/12/15 triple mutant exhibited retarded embryo development, reduced seed weight and lipid content, and “wrinkled” seeds (Chen et al., 2015). The seed coat of sweet11/12/15 mutants accumulated more starch, whereas the embryos had reduced starch content compared with the wild type (Chen et al., 2010; Dean et al., 2011; Belmonte et al., 2013; Chen et al., 2015). However, from our study, WHY2 seemed to affect Suc transport from leaf to pericarp cells via SWEET15 and from pericarp to seeds via SWEET11 (Fig. 8). Thus, it appears that high levels of WHY2 may accelerate Suc transport out of the leaf, such that sugar starvation causes early senescence in leaves, whereas the observed seed phenotype is likely due to a combination of reduced Suc supply from leaves and reduced import into filial tissues (Chen et al., 2015; Durand et al., 2016; Gebauer et al., 2017).
WHY2 also altered JMT, JAS, and TAT3 gene expression (Fig. 3). MeJA treatment enhanced the effects of overexpressing WHY2: inducing a leaf early senescence phenotype and increasing SAG29 expression, but repressing SWEET11 gene expression. JMT is a key enzyme for jasmonate–regulated plant responses (Seo et al., 2001). In Arabidopsis, the expression of jasmonic acid O-methyltransferase (JMT), which is responsible for JA methylation and MeJA formation, was developmentally regulated, displaying constitutively up-regulated expression of defense and JA biosynthesis-related genes (Seo et al., 2001). At the same time, overexpressing AtJMT in tomato, thus creating a metabolic sink, had tissue-specific consequences for the JA metabolic network, which was associated with the quick biochemical activation of specific lipases and oxidizing enzymes (Stitz et al., 2011). Stitz and coauthors also reported Gene Ontology (GO) enrichment analysis of JA-deficient plants highlighted strong transcriptional modulations in floral energy metabolism, and indicated that JA substantially regulated carbohydrate metabolism during floral maturation. In JA biosynthesis-deficient flowers, they found major alterations in limb carbohydrate and energy metabolism, whereby soluble sugars like Fru or combined Glc/hexoses and the intermediates of the TCA cycle were particularly affected (Stitz et al., 2014; Qi et al., 2015). In this study, we found that WHY2 is an upstream regulator of both SAG29/SWEET15 and SWEET11 and is feedback-enhanced in response to MeJA. This provides evidence for a connection between JA signaling and Suc transport. This suggests that the WHY2 function in the nucleus affects JA-mediated carbohydrate metabolism (Fig. 9).
Figure 9.
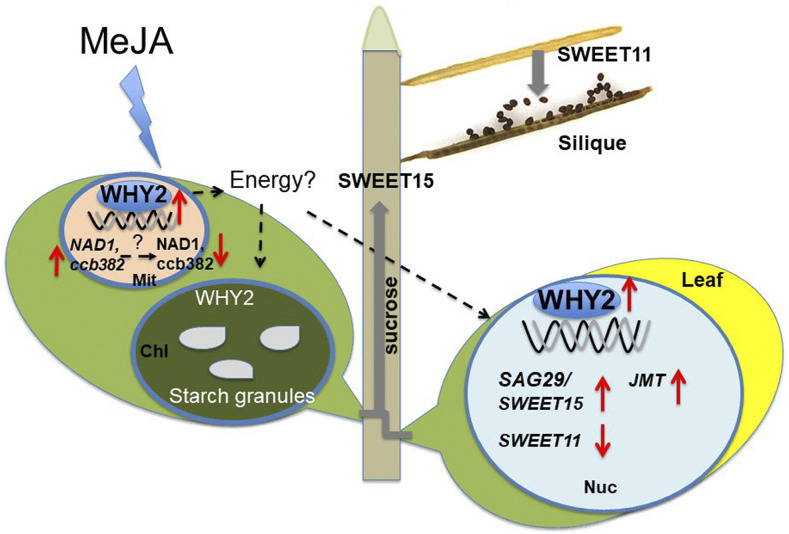
Triple-localized WHY2 protein affects leaf senescence and silique development via carbon allocation. Model: (1) Overexpressing WHY2 up-regulation of NAD1 and ccb382, and down-regulation of NAD1 and ccb382 protein accumulation might lead to the interruption of mitochondrial (Mit) functional activity. (2) The nWHY2 protein in the nucleus (Nuc) directly up-regulates the expression of SWEET15 and down-regulates the expression of SWEET11, thus promoting Suc transport out of the leaf to the pericarp cells of the silique, but preventing transport of Suc derived from photosynthesis out of the chloroplast (Chl; Busi et al., 2011b), leading to an increase in starch granule numbers in chloroplasts of pericarp cells but a decline in starch content in the seed. (3) Overexpressing WHY2 in leaf cells enhances JMT and SWEET15 expression. This enhancement is up-regulated by MeJA and is consistent with the carbon reallocation in nectary cell and pollen mother cells, leading to early cell death due to carbon reallocation (Busi et al., 2011b; Cai et al., 2015). Our results suggest that WHY2 may mediates mitochondrial dysfunction, affect carbon reallocation, accelerate leaf and silique senescence, and operate on the three different plant genomes.
In addition to its function in mitochondria and the nucleus, in this study we show that WHY2 is also localized in plastids, as was previously shown using an in vitro import assay (Krause et al., 2005). Based on the characterization of WHY2 primary sequence, there is a plastid transit peptide following the mitochondria transit peptide, and it is cleaved at a Cys site according to the TargetP1.1 prediction program (Supplemental Fig. S6). From the size of the detected protein, we can see that WHY2 in chloroplasts is the smallest (∼21 kD; Fig. 5C), which was approximately the same size as shown in the in vitro import assay (Krause et al., 2005). We hypothesize that WHY2 might be directly imported from the cytoplasm to the chloroplast or from mitochondria to chloroplasts under yet unknown conditions. Although this study further showed that WHY2 affected starch granule numbers in chloroplasts of pericarp cells (Fig. 2), specific functions of WHY2 in the chloroplast still remain elusive.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The seeds of Arabidopsis (Arabidopsis thaliana) Columbia and transgenic plants were germinated and seedlings were grown on half-strength Murashige–Skoog (MS) medium supplemented with 0.7% (w/v) agar. Ten-day-old seedlings were transplanted to the vermiculite, watering with half-strength MS medium in climate rooms under controlled conditions (22.5°C, 13/11 h of light/dark photoperiod with a light intensity of 80 μmol photons m−2 s−1, 60% relative humidity). The T-DNA insertion mutants Salk_118900 and Salk_118907 of WHY2 were purchased from the Nottingham Arabidopsis Stock Center. The homozygous WHY2 T-DNA insertion knockout mutants were obtained by genome insertion screening with the primers described in Supplemental Table S1.
Chlorophyll Measurements
The chlorophyll content of leaf 7 was measured according to the method described previously (Porra et al., 1989). At least 12 individual plants were used for analysis, and the mean chlorophyll content was calculated.
Plasmid Construction and Plant Transformation
A series of WHY2 CDS constructs including the full-length (714 bp) mtWHY2, the deleted mitochondrial transit peptide (627 bp) ptWHY2, and the deleted mitochondrial and plastid transit peptide (576 bp) nWHY2 were amplified with primers adding CACC sequence at the 5′ end and a FLAG tag at the 3′ end (Supplemental Table S1), and cloned in a pENTR-TOPO vector using the Gateway system (Invitrogen). The sequence of WHY2 was verified by sequencing. Then WHY2 was transferred into the destination vector pB2WG7 by recombination using the Gateway system (Invitrogen). A positive clone of Agrobacterium tumefaciens GV3101 was obtained and transformed into the why2-1 mutant and wild-type plants. More than 10 positive lines were screened by 0.1% (v/v) Basta in the T1 generation. The homozygous transgenic WHY2 overexpressing (oeWHY2) plants were obtained in the T3 generation by genetic ratio test. In the complemented WHY2 transgenic plant, WHY2 was under the control of the WHY2 promoter, instead of the 35S promoter, and transformed into the why2-1 background plants. Several homozygous transgenic lines were confirmed by RT-qPCR (Supplemental Fig. S1).
Northern Blotting
The 10 μg of total RNA was run on a denaturing agarose gel in the presence of formaldehyde, and then transferred to a membrane. The membrane was prehybridized and hybridized with probe of the WHY2 CDS labeled with 32P; then the results were obtained by x-ray film exposure using a Phosphor Imager, as described previously (Miao and Zentgraf, 2007).
Western Blotting
The procedure of protein isolation and immunodetection was described previously by Ren et al. (2017b). An antibody against the WHY2 peptide was prepared by Faan company, the specificity of which is shown in Supplemental Fig. S2. Antibodies against COXII, NAD1, ccb382, VDAC1, and PSII were purchased from Agrisera, and antibodies against H3 and ACTIN were purchased from Cell Signaling Technology.
Phenotype Observation and Documentation
For phenotype observation, thirty siliques of three plants were used. Silique size (width and length) was measured using a measuring stick. Seed weight was calculated as the weight per 100 seeds. Siliques were opened on a microscope slide right after collection. Thirty seeds were mounted in water, and gentle pressure was applied to the coverslip on the seeds to release the embryos. Afterward, nondamaged embryos were documented under a Leica stereomicroscope. Twenty embryos per line were observed, and the area of embryos were calculated using a width times length measurement.
Tissue Section and Ultrastructural Observation
The silique of a 7-week-old plant was fixed in 100 mL formaldehyde-acetic acid [containing 38% (v/v) formaldehyde, glacial acetic acid, and 70% (v/v) alcohol (0.5: 0.5: 9)] for 12 h, followed by alcohol gradient dehydration, clearing, wax immersion, paraffin embedding, and cutting with a slice thickness 5–8 µm, and organizing the structure. Photographs were taken using a Leica microscope.
For transmission electron microscopy, the fresh fruit were placed in 2.5% (v/v) glutaraldehyde for 2 h, then rinsed with 1% (v/v) osmium tetroxide. After 1 h of fixation, the specimen was rinsed with phosphate buffer and dehydrated with acetone gradient. After embedding with epoxy resin, the specimen was prepared by hand and then ultrathin sections were prepared. The ultrathin sections were incubated with 1% (w/v) uranyl acetate and lead citrate and examined with an electron microscope (TECNAI G2 20; FEI) at an accelerating voltage of 120 kV.
RT-qPCR Analysis
The total RNA extracted from the siliques and rosette leaves of 7-week-old oeWHY2, why2, and wild-type plants were treated with DNase I. Then the first strand of complementary DNA was synthesized by RevertAid First Strand complementary DNA Synthesis Kit (Thermo Fisher Scientific). RT-qPCR amplification and detection of related genes were carried out by the Connect Real-Time PCR Detection System (Bio-RAD). The Arabidopsis Actin2 or GAPC gene was used as an internal reference. The average and sd were calculated from the results of three biological replicates. The primers used in RT-qPCR are listed in Supplemental Table S1.
EMSA
Total protein was extracted from wild-type, oeWHY2, and why2 plants, as described previously by Miao et al. (2013). The concentration of total protein was measured by the Bradford assay.
DNA/RNA fragments of the upstream region of mitochondrial NAD1: coding region (Tel-CS), noncoding region (Tel-NCS), and transcribed RNA (Tel-RNA) probes were synthesized in vitro and radioactively labeled according to the method of Miao et al. (2013). The probe was labeled using 32P-ATP and T4 ligase. The WHY2 protein and labeled probe were incubated in a 10 µL binding reaction at room temperature for 30 min and then run on a 4% nondenaturing polyacrylamide gel. A 20-fold excess of unlabeled probe was used as a competitor.
ChIP-qPCR
For ChIP-qPCR determination of WHY2-FLAG occupancies on promoter regions in the downstream target genes, JMT and SWEET11/15, we used 4-week-old rosettes from oeWHY2-1, wild-type, and why2-1 plants. The cross-linked DNA fragments ranging from 200 to 1000 bp in length were immunoprecipitated by an antibody against the WHY2 peptide (Faan), whose specificity is shown in Supplemental Fig. S2. The enrichment of the selected promoter regions of both genes was determined by comparing the amounts in the precipitated and nonprecipitated (input) DNA samples, by qPCR using designed region-specific primers (Supplemental Table S1). The same quantification in the why2 mutant line served as a control for the respective overexpressing line and wild type, and was used for normalization to give the fold enrichment factors over the mock-treated samples. The experiments were performed in triplicate.
Dual-Luciferase Activity Assay
Nicotiana benthamiana plants were grown in climate rooms (22°C, 16/8 h of light/dark, 85μmol photons m-2 s-1 light condition). Plants were grown until they had four leaves and then infiltrated with Agrobacterium tumefaciens GV3101. Plants were maintained in the climate rooms and, after 4 to 5 d, 1-cm discs were collected from the fourth and fifth leaves of each plant. Six biological replicates with their respective negative controls were used per assay. The experiment was performed as previously described by Hellens et al. (2005) with minor changes. In the dual luciferase assays, WHY2 was in the 1α1 vectors, which are based on a pGREENII backbone. For generating the WHY2 overexpression construct, a 712-bp coding sequence was amplified, subcloned into pGEM-T Easy (Promega), cut with BamHI and SalI restriction enzymes, and cloned under the Cauliflower mosaic virus-35S promoter into pFLAP, before restriction enzyme digest with PacI and AscI and ligation into the pBIN+ binary vector.
The Agrobacterium strain was grown overnight in LB and brought to a final O.D.600 of 0.2 in infiltration buffer. Coinfiltrated Agrobacterium carried separate plasmids:900 μL of an empty cassette or one that contains WHY2 CDS driven by the Arabidopsis ACTIN 2kb promoter, and 100 μL of the reporter cassette carrying one of the promoters. Leaf discs were homogenized in 300 μL of a passive lysis buffer. Then 25 μL of a 1/100 dilution of the crude extract were assayed in 125 μL of Luciferase assay buffer, and LUC and REN chemiluminescence of each sample was measured in separate wells on the same plate. REN and LUC Unit was measured in a Turner 20/20 luminometer, with a 5-s delay and 15-s measurement. Raw data were collected and the LUC/REN ratio was calculated for each sample. Biological samples were pooled together, and a Student's t test was performed against a background control for each experiment, as described in “Results.”. The entire experiment was repeated a second time under similar conditions to confirm the regulatory effect of transcription factors.
Yeast One-Hybrid Assay
Yeast one-hybrid screening was performed using MATCHMAKER One-Hybrid Library Construction Kit (Clontech). The full length CDS of WHY2 was cloned into the pGADT7 vector. The promoter region-containing upstream region fragments (−160 bp) of NAD1, ccb382, and ATP9 were cloned into the pHIS2 reporter vector. Positive clones were identified by sequencing with AD sequencing primers. Empty pHIS2 reporter vector plus an appropriate concentration (50 mM) of 3-amino-1,2,4-triazole was used to inhibit HIS leakage.
Starch Staining
Whole rosettes of 4-week-old plants were either harvested or covered with black trays in the late afternoon. In the early afternoon of the next day, rosettes of covered plants were harvested. Starch staining was performed right after rosette harvesting. Samples were cleared in 80% (v/v) ethanol plus 5% (v/v) formic acid at 22°C, stained in KI2 Lugol's iodine solution (43.4 mm KI/5.7 mM) and washed twice in water. Starch staining was performed with plants grown in the presence of MeJA for 1, 2, 3, and 4 h (Supplemental Fig. S5).
Metabolite Content Measurement
Starch content was measured according to the procedures described previously by Smith and Zeeman (2006), with some changes. Three siliques or thirty seeds, or three leaves were collected from one 7-week-old plant for one analysis. Thirty seeds were rapidly dissected from each freshly harvested silique. Three siliques or leaves were collected from each individual plant. In total, three individual mutant or Col-0 plants were used. The samples were pooled, homogenized, and transferred to 300 mL of ice-cold 0.7 m perchloric acid, then centrifuged at 11,000 g for 45 min at 4°C. Pellets were washed once in 1 mL distilled water and twice in 1 mL 80% (v/v) ethanol, then vacuum dried. Dried pellets were resuspended in 40 mL distilled water by boiling for 6 min twice. A starch measurement kit (R-Biopharm) was used to quantify starch content. Briefly, starch was hydrolyzed to d-Glc by amyloglucosidase. d-Glc was enzymatically assayed in the presence of hexokinase and Glc 6-phosphate dehydrogenase by determining the amount of NADPH (A340).
Soluble sugars in the leaves and developing siliques were measured on samples of three leaves and five siliques pooled from five 7-week-old plants, according to Critchley et al. (2001) and Durand et al. (2018). Previously lyophilized tissue sample were washed in methanol:chloroform:water (12:5:3, by volume). Supernatants containing soluble sugars were pooled and mixed with 0.6 volume of water and centrifuged. Soluble sugars were quantified using a Suc/d-Glc Assay Kit (Megazyme).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of WHY2 transgenic plants.
Supplemental Figure S2. Specific detection of antibody against WHY2 peptide.
Supplemental Figure S3. WHY2 expression profile in different tissue/organs according to public datasets from the TAIR Web site (Winter et al., 2007).
Supplemental Figure S4. Gene expression changes in oeWHY2, why2, and WT by RT-qPCR.
Supplemental Figure S5. Starch staining in oeWHY2, why2, and WT following treatment of methyl jasmonate or water for 0, 2, 3, and 4 h.
Supplemental Figure S6. Illustration of WHY2 primary sequence analyzed by the Target1.1 program.
Supplemental Table S1. List of candidate genes for screening in oeWHY2, why2, and WT plants.
Supplemental Table S2. List of primer sequences for RT-qPCR, ChIP-qPCR, and subcloning.
Acknowledgments
We thank Roger P. Hellens (HortResearch, Mt Albert Research Centre) for providing the vectors for dual luciferase assay. We thank Nottingham Arabidopsis Stock Center for T-DNA insertion lines.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31770318 to Y.M. and 31400260 to Y.R.).
Articles can be viewed without a subscription.
References
- Bailleul B, Berne N, Murik O, Petroutsos D, Prihoda J, Tanaka A, Villanova V, Bligny R, Flori S, Falconet D, et al. (2015) Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369 [DOI] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013)) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci U S A 110: E435-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi MV, Gomez-Lobato ME, Araya A, Gomez-Casati DF(2011a) Mitochondrial dysfunction affects chloroplast functions. Plant Signal Behav 6: 1904–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi MV, Gomez-Lobato ME, Rius SP, Turowski VR, Casati P, Zabaleta EJ, Gomez-Casati DF, Araya A(2011b) Effect of mitochondrial dysfunction on carbon metabolism and gene expression in flower tissues of Arabidopsis thaliana. Mol Plant 4: 127–143 [DOI] [PubMed] [Google Scholar]
- Cai Q, Guo L, Shen ZR, Wang DY, Zhang Q, Sodmergen(2015) Elevation of pollen mitochondrial DNA copy number by WHIRLY2: Altered respiration and pollen tube growth in Arabidopsis. Plant Physiol 169: 660–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L, Maréchal A, Parent JS, Lepage E, Sygusch J, Brisson N(2010) Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 22: 1849–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468: 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Lin IW, Qu XQ, Sosso D, McFarlane HE, Londoño A, Samuels AL, Frommer WB(2015) A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB(2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Comadira G, Rasool B, Kaprinska B, García BM, Morris J, Verrall SR, Bayer M, Hedley PE, Hancock RD, Foyer CH(2015) WHIRLY1 functions in the control of responses to nitrogen deficiency but not aphid infestation in barley. Plant Physiol 168: 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley JH, Zeeman SC, Takaha T, Smith AM, Smith SM(2001) A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G(2011) Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 4: 1074–1091 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Allard J, Brisson N, Sygusch J(2002) A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat Struct Biol 9: 512–517 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Després C, Joyeux A, Subramaniam R, Brisson N(2000) PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 12: 1477–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desveaux D, Maréchal A, Brisson N(2005) Whirly transcription factors: Defense gene regulation and beyond. Trends Plant Sci 10: 95–102 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Subramaniam R, Després C, Mess JN, Lévesque C, Fobert PR, Dangl JL, Brisson N(2004) A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev Cell 6: 229–240 [DOI] [PubMed] [Google Scholar]
- Durand M, Mainson D, Porcheron B, Maurousset L, Lemoine R, Pourtau N(2018) Carbon source-sink relationship in Arabidopsis thaliana: The role of sucrose transporters. Planta 247: 587–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M, Porcheron B, Hennion N, Maurousset L, Lemoine R, Pourtau N(2016) Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiol 170: 1460–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J, Weber B, Thuss S, Wildum S, Binder S(2007) Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-Elements contribute to 5′ and 3′ end formation. Nucleic Acids Res 35: 3676–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Karpinska B, Krupinska K(2014) The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: A hypothesis. Philos Trans R Soc Lond B Biol Sci 369: 20130226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer P, Korn M, Engelsdorf T, Sonnewald U, Koch C, Voll LM(2017) Sugar accumulation in leaves of Arabidopsis sweet11/sweet12 double mutants enhances priming of the salicylic acid-mediated defense response. Front Plant Sci 8: 1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, Dor C, Bassani M(2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Gómez-Casati DF, Busi MV, Gonzalez-Schain N, Mouras A, Zabaleta EJ, Araya A(2002) A mitochondrial dysfunction induces the expression of nuclear-encoded complex I genes in engineered male sterile Arabidopsis thaliana. FEBS Lett 532: 70–74 [DOI] [PubMed] [Google Scholar]
- Grabowski E, Miao Y, Mulisch M, Krupinska K(2008) Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol 147: 1800–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Wang W, Yu X, Lin W, Miao Y(2018) Comparative proteomic analysis of coregulation of CIPK14 and WHIRLY1/3 mediated pale yellowing of leaves in Arabidopsis. Int J Mol Sci 19: 2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA(2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Lin W, Deng B, Ren Y, Miao Y(2017) Dual-Located WHIRLY1 interacting with LHCA1 alters photochemical activities of Photosystem I and is involved in light adaptation in Arabidopsis. Int J Mol Sci 18: 2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araújo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O(2012) nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J 71: 413–426 [DOI] [PubMed] [Google Scholar]
- Krause K, Kilbienski I, Mulisch M, Rödiger A, Schäfer A, Krupinska K(2005) DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett 579: 3707–3712 [DOI] [PubMed] [Google Scholar]
- Krupinska K, Braun S, Nia MS, Schäfer A, Hensel G, Bilger W(2019) The nucleoid-associated protein WHIRLY1 is required for the coordinate assembly of plastid and nucleus-encoded proteins during chloroplast development. Planta 249: 1337–1347 [DOI] [PubMed] [Google Scholar]
- Kühn K, Weihe A, Börner T(2005) Multiple promoters are a common feature of mitochondrial genes in Arabidopsis. Nucleic Acids Res 33: 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isemer R, Krause K, Grabe N, Kitahata N, Asami T, Krupinska K(2012) Plastid Located WHIRLY1 enhances the responsiveness of Arabidopsis seedlings toward abscisic acid. Front Plant Sci 3: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law YS, Zhang R, Guan X, Cheng S, Sun F, Duncan O, Murcha MW, Whelan J, Lim BL(2015) Phosphorylation and dephosphorylation of the presequence of pMORF3 during import into mitochondria from Arabidopsis thaliana. Plant Physiol 169: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir R, Spinner L, Klemens PA, Chakraborti D, de Marco F, Vilaine F, Wolff N, Lemoine R, Porcheron B, Géry C, et al. (2015) Disruption of the sugar transporters AtSWEET11 and AtSWEET12 affects vascular development and freezing tolerance in Arabidopsis. Mol Plant 8: 1687–1690 [DOI] [PubMed] [Google Scholar]
- Leister D.(2005) Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354: 110–116 [DOI] [PubMed] [Google Scholar]
- Lescot M, Deohais P, Thijs G, Marchal K, Moreau Y, Van de Peer P, Rouz P, Rombauts S(2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu KC, Hsieh MH, Wang HJ, Hsieh HL, Jauh GY(2016) Distinct role of Arabidopsis mitochondrial P-type pentatricopeptide repeat protein-modulating editing protein, PPME, in nad1 RNA editing. RNA Biol 13: 593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Zhang Y, Cheng S, Osorio S, Sun Y, Fernie AR, Cheung CYM, Lim BL(2015) Impacts of high ATP supply from chloroplasts and mitochondria on the leaf metabolism of Arabidopsis thaliana. Front Plant Sci 6: 922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Huang D, Shi X, Deng B, Ren Y, Lin W, Miao Y(2019) H2O2 as a feedback signal on dual-located WHIRLY1 associates with leaf senescence in Arabidopsis. Cells 8: 1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Butow RA(2006) Mitochondrial retrograde signaling. Annu Rev Genet 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Maréchal A, Parent JS, Sabar M, Véronneau-Lafortune F, Abou-Rached C, Brisson N(2008) Overexpression of mtDNA-associated AtWhy2 compromises mitochondrial function. BMC Plant Biol 8: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melonek J, Mulisch M, Schmitz-Linneweber C, Grabowski E, Hensel G, Krupinska K(2010) Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232: 471–481 [DOI] [PubMed] [Google Scholar]
- Miao Y, Jiang J, Ren Y, Zhao Z(2013) The single-stranded DNA-binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis. Plant Physiol 163: 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Zentgraf U(2007) The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the Jasmonic and salicylic acid equilibrium. The Plant Cell 19: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumenko N, Morgenstern M, Rucktäschel R, Warscheid B, Rehling P(2017) INA complex liaises the F1Fo-ATP synthase membrane motor modules. Nat Commun 8: 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niazi AK, Delannoy E, Iqbal RK, Mileshina D, Val R, Gabryelska M, Wyszko E, Soubigou-Taconnat L, Szymanski M, Barciszewski J, et al. (2019) Mitochondrial transcriptome control and intercompartment cross-talk during plant development. Cells 8: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J(2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759 [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R(2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18: 176–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE(1989) Deteermination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectrometry. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A(2008) A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res 36: 5152–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Wang J, Huang H, Liu B, Gao H, Liu Y, Song S, Xie D(2015) Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 27: 1634–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Pan Z, Zhao H, Zhao J, Cai M, Li J, Zhang Z, Qiu F(2017a) EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J Exp Bot 68: 4571–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Li Y, Jiang Y, Wu B, Miao Y(2017b) Phosphorylation of WHIRLY1 by CIPK14 shifts its localization and dual functions in Arabidopsis. Mol Plant 10: 749–763 [DOI] [PubMed] [Google Scholar]
- Soo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD(2001) Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate- regulated plant responses. Processing National Academic Science 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD(2001) Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Wu Z, Sharbrough J(2018) Correction of persistent errors in Arabidopsis reference mitochondrial genomes. Plant Cell 30: 525–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Zeeman SC(2006) Quantification of starch in plant tissues. Nat Protoc 1: 1342–1345 [DOI] [PubMed] [Google Scholar]
- Solotoff V, Moseler R, Schulte U(2015) Two pentatricopeptide repeat domain proteins are required for the synthesis of respiratory complex I. Curr Genet 61: 19–29 [DOI] [PubMed] [Google Scholar]
- Stitz M, Gase K, Baldwin IT, Gaquerel E(2011) Ectopic expression of AtJMT in Nicotiana attenuata: Creating a metabolic sink has tissue-specific consequences for the jasmonate metabolic network and silences downstream gene expression. Plant Physiol 157: 341–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitz M, Hartl M, Baldwin IT, Gaquerel E(2014) Jasmonoyl-L-isoleucine coordinates metabolic networks required for anthesis and floral attractant emission in wild tobacco (Nicotiana attenuata). Plant Cell 26: 3964–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Huang J, Zhong S, Gu H, He S, Qu LJ(2018) Novel DYW-type pentatricopeptide repeat (PPR) protein BLX controls mitochondrial RNA editing and splicing essential for early seed development of Arabidopsis. J Genet Genomics 45: 155–168 [DOI] [PubMed] [Google Scholar]
- Świda-Barteczka A, Krieger-Liszkay A, Bilger W, Voigt U, Hensel G, Szweykowska-Kulinska Z, Krupinska K(2018) The plastid-nucleus located DNA/RNA binding protein WHIRLY1 regulates microRNA-levels during stress in barley (Hordeum vulgare L.). RNA Biol 15: 886–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Hildebrandt TM, Lewejohann D, Gonzalez DH, Braun HP(2012) Lack of cytochrome c in Arabidopsis decreases stability of Complex IV and modifies redox metabolism without affecting Complexes I and III. Biochim Biophys Acta 1817: 990–1001 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ(2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JY, Lai CX, Qu Z, Yang XY, Qin XH, Liu GQ(2009) Recruitment of AtWHY1 and AtWHY3 by a distal element upstream of the kinesin gene AtKP1 to mediate transcriptional repression. Plant Mol Biol 71: 437–449 [DOI] [PubMed] [Google Scholar]
- Yoo HH, Kwon C, Lee MM, Chung IK(2007) Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J 49: 442–451 [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Mouras A, Hernould M, Suharsono, Araya A(1996) Transgenic male-sterile plant induced by an unedited atp9 gene is restored to fertility by inhibiting its expression with antisense RNA. Proc Natl Acad Sci USA 93: 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang K, Kong F, Zhang S, Meng C, Yang M, Liu Z, Wang Y, Ma N, Meng QW(2018) Whirly1 enhances tolerance to chilling stress in tomato via protection of photosystem II and regulation of starch degradation. New Phytol 221: 1998–2012 [DOI] [PubMed] [Google Scholar]