A lipase plays essential roles in anther cuticle and pollen exine development and contributes to male fertility in maize.
Abstract
Anther cuticle and pollen exine are two physical barriers protecting plant reproductive cells against environmental stresses; defects in either often cause male sterility. Here, we report the characterization of a male-sterile mutant irregular pollen exine2 (ipe2) of maize (Zea mays), which displays shrunken anthers and no starch accumulation in mature pollen grains. We cloned the causal gene IPE2 and confirmed its role in male fertility in maize with a set of complementary experiments. IPE2 is specifically expressed in maize developing anthers during stages 8 to 9 and encodes an endoplasmic-reticulum–localized GDSL lipase. Dysfunction of IPE2 resulted in delayed degeneration of tapetum and middle layer, leading to defective formation of anther cuticle and pollen exine, and complete male sterility. Aliphatic metabolism was greatly altered, with the contents of lipid constituents, especially C16/C18 fatty acids and their derivatives, significantly reduced in ipe2 developing anthers. Our study elucidates GDSL function in anther and pollen development and provides a promising genetic resource for breeding hybrid maize.
Maize (Zea mays) is one of the crops with substantial heterosis, and hybrids have been widely utilized to maximize yield. Detasseling of the female parent is the prerequisite for hybrid seed production. Manual and mechanical detasseling are two commonly used methods in maize, which are often labor-/time-consuming, weather-dependent, and detrimental to plant growth. Male sterility is an efficient alternative to female parent detasseling. Two types of male sterility occur in maize, cytoplasmic male sterility (CMS) and genic/nuclear-encoded male sterility (GMS). In practice, the CMS system has not been extensively used due to its serious potential risks of certain disease susceptibilities (Siedow et al., 1995). Lack of restorer genes of male parental lines also limits the use of CMS. GMS can theoretically overcome the drawbacks of CMS. However, the bottleneck of GMS is the difficulty in maintaining pure male sterile lines (Xie et al., 2018). In recent years, pure GMS lines have been developed through transgenic approaches, which opens the door for GMS application in maize hybrid seed production (Zhang et al., 2018).
Male reproductive development is a complex biological process. In maize, the male inflorescence exists as the apically positioned tassel, in which the stamen primordial cell specifies and differentiates to generate mature anthers, which coordinately release pollen grains. The processes from anther formation to mature pollen grain release undergo several critical events, including specification of various anther cell layers, meiotic cytokinesis, microspore development, maturation, and release of pollen grains. Male sterility may occur if any of these processes is disrupted. In recent years, dozens of male-sterile genes have been studied in maize. These genes encode various proteins including redox proteins, secreted proteins, lipid transfer proteins, transcription factors, and enzymes (Supplemental Table S1). Among them, MALE STERILE CONVERTED ANTHER1 encodes a glutaredoxin that acts as a redox regulator of target proteins and contributes to the initiation of archesporial cells (Albertsen et al., 2011). MULTIPLE ARCHESPORIAL CELLS1 encodes a small secreted protein that is the ortholog of rice (Oryza sativa) TAPETUM DETERMINANT1 protein homolog 1A and is involved in the proliferation of archesporial cells (Wang et al., 2012). OUTER CELL LAYER4, MALE STERILE23 (MS23), and MS32 all encode transcription factors regulating the differentiation and development of anther wall layers (Vernoud et al., 2009; Moon et al., 2013; Nan et al., 2017). OUTER CELL LAYER4 encodes a HD-ZIP IV transcription factor required for trichome patterning and subepidermal development (Vernoud et al., 2009). MS23 and MS32 encode basic helix-loop-helix transcription factors responsible for tapetal specification and development (Moon et al., 2013; Nan et al., 2017). MS88 encodes a β-1,3-galactosyltransferase mediating the epidermal and tapetal differentiation during early anther development in maize (Wang et al., 2013). MS9 encodes a R2/R3 MYB transcription factor that may be the control point for the entry into meiosis (Albertsen et al., 2016). INDETERMINATE GAMETOPHYTE1 encodes a LATERAL ORGAN BOUNDARIES domain protein essential for embryo sac development and leaf development (Evans, 2007). All the above genes function at the early stage of anther development and can be grouped as premeiotic GMS genes.
In contrast to the premeiotic GMS genes, the following nine GMS genes function at the later stages of anther development and can be categorized as postmeiotic GMS genes in maize. MS45 encodes a strictosidine synthase contributing to pollen wall initiation after tetrad stage (Skibbe and Schnable, 2005). MS26 encodes a cytochrome P450 monooxygenase regulating pollen exine formation (Djukanovic et al., 2013). MS44 is the only dominant GMS gene and encodes a lipid transfer protein involved in postmeiotic tapetum secretion of proteins from tapetal cells into locule, which is critical for pollen development (Fox et al., 2017). MS7 encodes a PHD-finger transcription factor, playing important roles in microspore wall formation and programmed cell death (PCD) of tapetal cells (Zhang et al., 2018). Collectively, IRREGULAR POLLEN EXINE1 (IPE1)/MS20, ABNORMAL POLLEN VACUOLATION1 (APV1), MS6021, MS33, and MS30 all encode enzymes participated in lipid or fatty acid metabolism, and mutation of any of them results in defective anther cuticle and pollen exine formation (Chen et al., 2017; Somaratne et al., 2017; Tian et al., 2017; Xie et al., 2018; An et al., 2019; Wang et al., 2019). Among them, MS30 was recently identified as a GDSL lipase regulating male fertility in maize (An et al., 2019).
The plant anther wall, from the outside to the inside, is composed of four successive cell layers: epidermis, endothecium, middle layer, and tapetum. The outer epidermis is the protective layer for all inner anther layers and microspores. The outermost layer of anther epidermis is the anther cuticle, which mainly consists of polymerized cutin and wax (Yeats and Rose, 2013). The pollen wall comprises exine and intine, and sporopollenin is the major component of exine, which is structurally divided into sexine and nexine. In general, sexine consists of tectum and bacula, while nexine consists of foot layer and endexine. All these structures, especially anther cuticle and pollen exine, are biological barriers for protecting male gametophytes against external environmental stresses. The essential role of endothecium is secreting materials for pollen grain development in the late stage of anther development, when tapetum and middle layer are not visible in anther lumen (Somaratne et al., 2017). The middle layer is involved in the transport of calcium ions or other secreted materials to the inner tapetum. It has also been reported to regulate the fate of microspore in kiwifruit (Actinidia deliciosa; Falasca et al., 2013). Tapetum, as the innermost somatic cell layer of anther lobes, provides enzymes, proteins, lipids, and nutrients for the reproductive cell development (Ariizumi and Toriyama, 2011). These components are secreted and further transferred to the surface of anther and pollen grain with the help of transporters and the specialized structures called “Ubisch bodies” (Chen et al., 2017). During pollen maturation, tapetum degeneration occurs via a coordinated PCD to provide nutrients for pollen development. Disruption of anther and pollen wall development may lead to male sterility.
In this study, we characterized a postmeiotic male-sterile mutant irregular pollen exine2 (ipe2) in maize. IPE2 was predicted to encode a GDSL lipase. Although IPE2 and MS30 are paralogues that are essential for anther cuticle and pollen exine formation, they share only 24.69% of amino acid identity and are functionally distinct in lipid metabolism. The identification of IPE2 would provide insight to uncover the molecular mechanism of male sterility in maize and facilitate its future application in maize hybrid production.
RESULTS
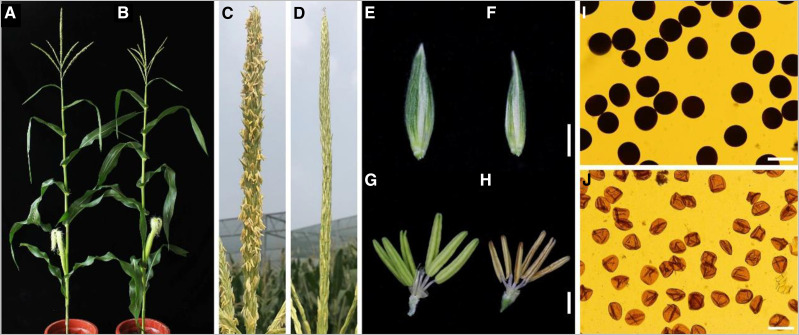
ipe2 Exhibits Irregular Anther Cuticle and Pollen Nexine
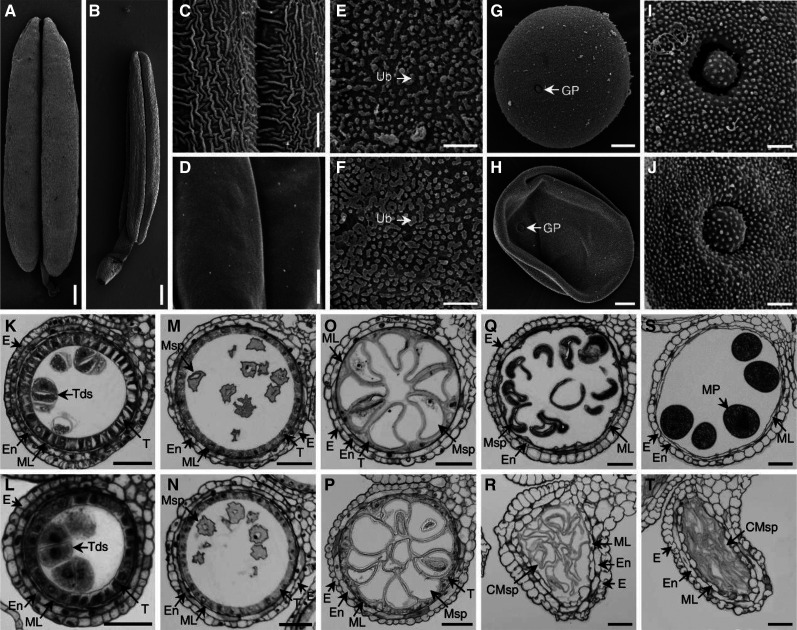
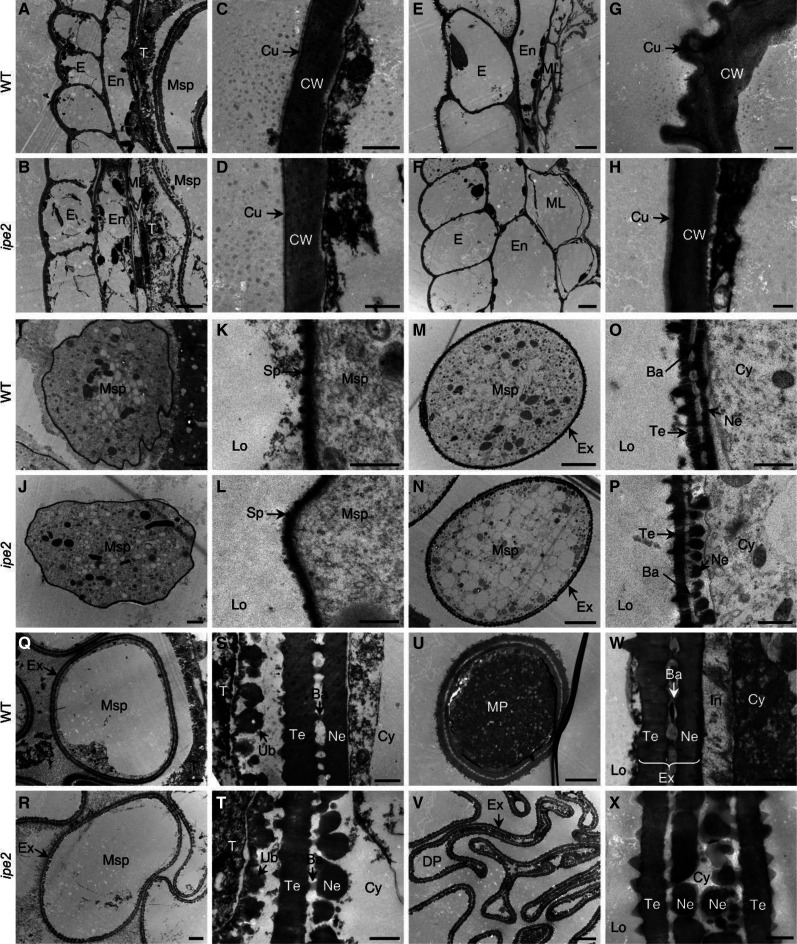
The maize ipe2 and its putative allelic male-sterile mutants ipe2-a, ipe2-b, and ipe2-c were provided by the Maize GDB Stock Center (516Cms5, 503Hms5-6048, 503Ims5-6061 and 503Jms5-6062, respectively; https://www.maizegdb.org/data_center/stock). We confirmed their allelic relationship by genetic allelic testing (Supplemental Table S2). At flowering stage, compared with the wild-type siblings, ipe2 anthers failed to shed pollen and exhibited complete male sterility with no anther extruding out of glumes (Fig. 1, A–D). Both the glumes and anthers of ipe2 were smaller than those of the wild-type siblings (Fig. 1, E–H). The mature pollen grains of ipe2 were shrunken and much lighter in color under I2-KI staining compared to the round-shaped and dark-blue stained ones of wild type (Fig. 1, I and J). Under scanning electron microscopy (SEM), ipe2 anthers showed smooth epidermal surface compared with the latticed-waxy crystal epidermal surface of wild type (Fig. 2, A–D). No substantial difference was detected in the inner surface of anthers between ipe2 and wild type (Fig. 2, E and F). At maturity, the wild-type anthers produced round pollen grains while ipe2 generated shrunken ones (Fig. 2, G–J). Transverse section was then conducted to examine the details inside anthers. The four cell layers of ipe2 anther were normally developed and no difference between ipe2 and wild type was observed in meiosis and microspores released from tetrads (Fig. 2, K–N). Subsequently, tapetum degeneration occurred and microspores were vacuolated and round-shaped in the locules of wild type (Fig. 2O). By contrast, ipe2 tapetum remained almost intact, indicative of delayed degeneration (Fig. 2P). After the vacuolated stage, both anthers and microspores were severely destroyed in ipe2, exhibiting shrunken anther wall, collapsed microspores, and an undegenerated or even swelling middle layer at the binucleate stage (Fig. 2R). With the proper degeneration of middle layer and tapetum, mature pollen grains were well-developed and accumulated in the anther locule of wild type (Fig. 2, Q and S). In contrast, the pollen grain defects extended beyond abnormal starch accumulation in ipe2 (Fig. 2T). Transmission electron microscopy (TEM) was implemented to gain insights into the elaborate structures of ipe2 anthers. We observed that the wild-type anther cuticle exhibited a solid thick band while the mutant displayed a narrow thin band at uninucleate stage (Fig. 3, A–D). At mature pollen stage, the wild-type anther was covered with wax crystals while the mutant cuticle was smooth without the rugged structures (Fig. 3, E–H). We further detected an irregular deposition of sporopollenin on the ipe2 microspore outer surface after the release of microspores from the tetrads (Fig. 3, I–L). Nexine, the inner layer of exine, was discontinuous, exhibiting a dashed structure in ipe2 (Fig. 3, M–P). Compared with wild type (Fig. 3, Q, S, U, and W), the distinct dashed nexine of ipe2 occurred at the vacuolated stage (Fig. 3, R, T, V, and X), even though we could not detect obvious defects in the microspore wall through transverse sections (Fig. 2P). All three allelic mutants showed complete male sterility and the same defects in anther cuticle and pollen nexine as ipe2 (Supplemental Fig. S1). Nevertheless, we could not detect the regular accumulation of Ubisch bodies in the allelic mutants (Supplemental Fig. S1, J, K, and P).
Figure 1.
Phenotypic comparison between wild type and ipe2. A and B, Mature plants of wild type (A) and ipe2 (B). C and D, Tassels of the wild type (C) and ipe2 (D). E and F, Comparison of spikelets of wild type (E) and ipe2 (F). G and H, Comparison of anthers of wild type (G) and ipe2 (H) at flowering stage. I and J, Mature pollen grains of wild type (I) and ipe2 (J) stained with 1% I2-KI solutions. Scale bars = 2 mm (E–H) and 100 μm (I–J).
Figure 2.
SEM analysis and transverse section of wild-type and ipe2 anthers. A and B, Mature anthers of wild type (A) and ipe2 (B). C and D, Mature anther outer surfaces of wild type (C) and ipe2 (D). E and F, Mature anther inner surfaces of wild type (E) and ipe2 (F). G and H, Mature pollen grains of wild type (G) and ipe2 (H). I and J, Mature pollen grains outer surfaces with germination apertures of wild type (I) and ipe2 (J). K and L, Anthers of wild type (K) and ipe2 (L) at tetrad stage. M and N, Anthers of wild type (M) and ipe2 (N) at microspore release stage. O and P, Anthers of wild type (O) and ipe2 (P) at large vacuole stage. Q and R, Anthers of wild type (Q) and ipe2 (R) at binucleate stage. S and T, Anthers of wild type (S) and ipe2 (T) at mature pollen stage. CMSp, cCollapsed microspore; E, epidermis; En, endothecium; GP, germination pore; ML, middle layer; MP, mature pollen; MSp, microspore; T, tapetum; Tds, tetrads; Ub, Ubisch body. Scale bars= 400 μm in (A and B), 6 μm (C and D), 4 μm (E and F), 10 μm (G and H), 2 μm (I and J), and 50 μm (K–T).
Figure 3.
TEM analysis of wild-type (WT) and ipe2 anthers. A and B, Anther wall of wild type (A) and ipe2 (B) at large vacuole stage. E and F, Anther wall of wild type (E) and ipe2 (F) at mature pollen stage. I and J, Microspores of wild type (I) and ipe2 (J) at early uninucleate stage. M and N, Microspores of wild type (M) and ipe2 (N) at microspore release stage. Q and R, Microspores of wild type (Q) and ipe2 (R) at large vacuole stage. U and V, Mature pollen grains of wild type (U) and ipe2 (V). In C, D, G, H, K, L, O, P, S, T, W, and X, are the close-up views of identical-stage tissues in A, B, E, F, I, J, M, N, Q, R, U, and V, respectively. Ba, Bacula; Cu, cuticle; CW, cell wall; Cy, cytoplasm; DP, defective pollen; E, epidermis; En, endothecium; Ex, exine; In, intine; Lo, locule; ML, middle layer; MP, mature pollen; MSp, microspore; Ne, nexine; Sp, sporopollenin; T, tapetum; Te, tectum; Ub, ubisch body. Scale bars = 5 μm (A, B, E, F, M, N, Q, R, U, and V), 2 μm (I and J), and 500 nm (C, D, G, H, K, L, O, P, S, T, W, and X).
Aliphatic Metabolism Alteration of ipe2 Anthers
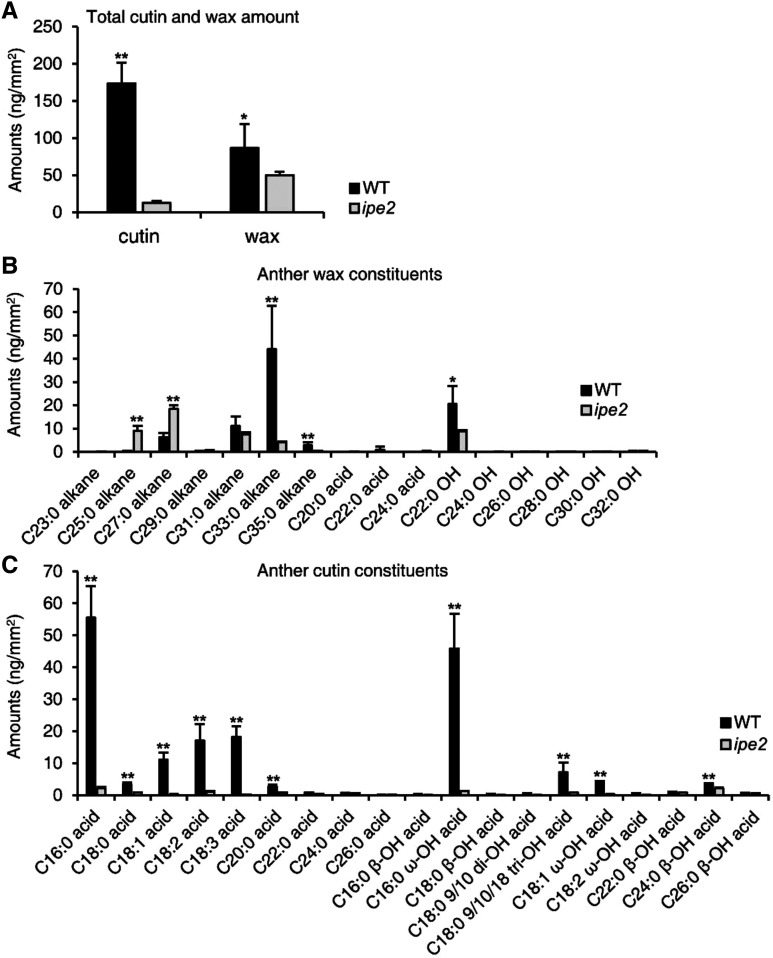
The phenotypic defects of anther cuticle and pollen wall in ipe2 reveal that abnormalities may occur in the metabolic process of aliphatic compounds. To decipher the possible mechanisms, we first extracted cuticle wax, cutin, and soluble fatty acid from ipe2 and wild-type anthers, and then analyzed their compositions by chromatography-mass spectrometry. Surface area against fresh weight of anthers is shown in Supplemental Figure S2. Compared with wild type, the total cutin content of ipe2 mature anthers significantly decreased by 92.6% (P < 0.01) with 12.88 ng/mm2 in ipe2 and 173.43 ng/mm2 in wild type. The total wax content decreased by 42.2% (0.01 < P < 0.05) with 50.16 ng/mm2 in ipe2 anthers and 86.85 ng/mm2 in wild type (Fig. 4A). For wax constituents, the contents of C22 hydroxyl alcohol, alkanes of C33 and C35, and some fatty acids were significantly decreased, while the contents of C25 and C27 alkanes increased in ipe2 (Fig. 4B), indicating that the elongation of fatty acids is disturbed in ipe2. Meanwhile, all aliphatic monomer compositions were significantly reduced in ipe2, especially the cutin monomers like C16:0, C18:0, C18:1, C18:2, C18:3, C20:0, C16:0 ω-OH, C18:0 tri-OH, and C18:1 ω-OH acids (Fig. 4C), which suggests that IPE2 is involved in the synthesis of cutin monomers. Moreover, the content of total soluble fatty acids was 3.53 μg/mg in wild-type anthers and the value was reduced to 1.14 μg mg−1 in ipe2 (Supplemental Fig. S3A). Of them, the contents of C16:0, C18:0, C18:2, C18:3, and C20:0 acids were significantly reduced, while C22:0, C24:0, and C28:0 acids were significantly increased in ipe2 (Supplemental Fig. S3B). Taken together, we predict that IPE2 is involved in lipid metabolism that is essential for anther development in maize.
Figure 4.
The contents of wax and cutin constituents in wild type (WT) and ipe2 mature anthers. A, Anther total cutin and wax amounts per unit surface area in wild type (black bars) and ipe2 (gray bars). B, The amounts of anther wax constituents per unit surface area in wild type and ipe2. C, The amounts of anther cutin constituents per unit surface area in wild type and ipe2. Error bars represent sd (n = 5). Asterisks represent significance between wild type and ipe2 by Student’s t test (*0.01 < P < 0.05 and ** P < 0.01). C16:0 acid, Hexadecanoic acid; C18:0 acid, octadecanoic acid; C18:1 acid, oleic acid; C18:2, linoleic acid; C18:3 acid, linolenic acid; C20:0 acid, eicosanoic acid; C22:0 acid, docosanoic acid; C24:0 acid, tetracosanoic acid; C26:0 acid, hexacosanoic acid; C16:0 β-OH acid, 2-hydroxy-hexadecanoic acid; C16:0 ω-OH acid, 16-hydroxy-hexadecanoic acid; C18:0 β-OH acid, 2-hydroxy-octadecanoic acid; C18:0 9/10 di-OH acid, 9,10-dihydroxy-octadecanoic acid; C18:0 9/10/18 tri-OH acid, 9,10,18-trihydroxy-octadecanoic acid; C18:1 ω-OH acid, 18-hydroxy-oleic acid; C18:2 ω-OH acid, 18-hydroxy-linoleic acid; C22:0 β-OH acid, 2-hydroxy-docosanoic acid; C24:0 β-OH acid, 2-hydroxy-tetrasanoic acid; C26:0 β-OH acid, 2-hydroxy-hexasanoic acid.
Map-Based Cloning of IPE2
To study the genetic nature of ipe2, the mutant was crossed with inbred line B73. All F1 progenies were fertile. The 3,230 F2 individuals segregated at a ratio of 3:1 (fertile to sterile; χ2 = 0.9,911, P > 0.05) and the 3,939 BC1F1 (ipe2 as the recurrent parent) individuals segregated at a ratio of 1:1 (fertile to sterile; χ2 = 1.0726, P > 0.05), suggesting that ipe2 male sterility is controlled by a single recessive gene.
Map-based cloning was conducted to isolate ipe2 using both F2 and BC1F1 segregating populations. The gene was initially mapped between markers B1 and B11 on the long arm of chromosome 5 (Supplemental Fig. S4A). Additional markers including insertion or deletion, simple sequence repeats, and single-nucleotide polymorphisms (SNPs) were developed for fine mapping. A total of 22,792 F2 and BC1F1 individuals were screened for recombinants, and the gene was eventually narrowed down to a 2.66-Mb region between markers C3 and B7. There are 27 putative protein encoding genes within the mapped region based on the B73 reference genome (B73_RefGen_v4, http://plants.ensembl.org/Zea_mays/Info/Index; Supplemental Fig. S4A). All lines of evidence including their expression profiles, genomic sequences, and the nature of predicted proteins indicate that Zm00001d015960 is most likely the candidate gene. Putative gene structure and amino acid sequence of Zm00001d015960 are shown in Supplemental Figure S4, B and C. Compared with the male fertile siblings, Zm00001d015960 in ipe2 has a 12-bp deletion (four amino acids deletion) in exon1. The three allelic mutants all have a “G” deletion at the 259-bp site in the first exon, leading to a frame shift and premature stop codon (Supplemental Figs. S4B, S5, and S6). Additional mutations in ipe2 include a 30-bp deletion at the 168-bp upstream of the 5′ untranslated region, a 314-bp insertion in the 3′ untranslated region, and three nonsynonymous SNP mutations (C to T in exon1, A to G in exon4, and C to G in exon5; Supplemental Figs. S4B, S5, and S6). Genetic complementation experiment was performed to validate that Zm00001d015960 is the causal gene of ipe2. A 7,524-bp genomic fragment including 2,869-bp upstream, Zm00001d015960 genomic DNA, and 1,773-bp downstream was ligated into the expression vector and then transformed into the maize inbred line Zong31. Eight transgenic events were obtained and four of them were chosen for further testing. When introduced into ipe2 homozygous background by crossing/selfing and marker-assisted selection, all four transgenic events were able to fully restore ipe2 male fertility, confirming that Zm00001d015960 is IPE2 (Fig. 5 ).
Figure 5.
Genetic complementation of IPE2. A, Flow chart of genetic complementation of IPE2. A/a, Alleles of IPE2/ipe2 in the innate locus; B/b, alleles of IPE2/ipe2 in the transgenic insertion site. AaBb individuals from T1 generation were selected for self-crossing. B, PCR detection of T0 transgenic lines. Lane 1 is a positive control, lane 2 is a negative control, and lanes 3 to 10 are positive lines. C, Pollen grains from positive lines with “aaB_” genotype are stained with 1% I2-KI solution. D, Pollen grains from negative lines with “aabb” genotype are stained with 1% I2-KI solution. Scale bars = 100 μm. E, G, I, and K, Positive transgenic plants with “aaB_” genotype from T2 individuals of the four events. F, H, J, and L, Negative transgenic plants with “aabb” genotype from T2 individuals of the four events.
IPE2 Is Expressed in Developing Anthers and IPE2 Is Localized to Endoplasmic Reticulum
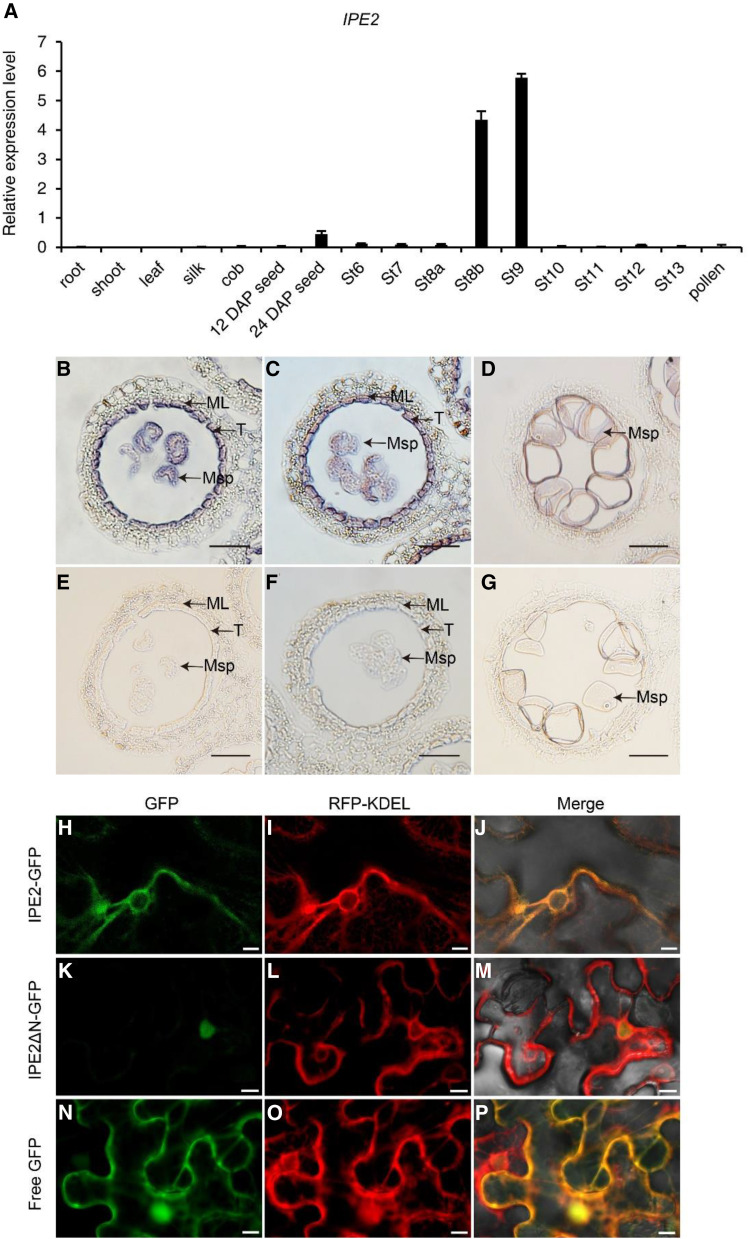
To study the detailed expression pattern of IPE2, we first established an anther-length/anther-development stage that corresponds. We then collected anther samples of 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, and 5 mm (±0.03 mm) in length corresponding to pollen/microspore mother cell stage (St6), meiotic cell/meiocytes stage (St7), tetrad stage (St8b), microspore release stage (St9), and large vacuole stage (St10), bicellular stage (St11), trinucleate stage (St12), mature pollen stage (St13), and pollen/pollen grain release stage (St14), respectively. Reverse transcription quantitative PCR (RT-qPCR) results showed that IPE2 was preferentially expressed at tetrad stage (stage 8b) and microspore release stage (stage 9) of developing anthers (Fig. 6A). This is consistent with the phenotypic differences observed at postmeiotic stages between wild type and ipe2. In situ hybridization was performed to investigate the tissue specificity of IPE2. Strong signals were detected in microspores, tapetum, and middle layer during microspore release stage (Fig. 6, B–G).
Figure 6.
Spatiotemporal expression pattern and subcellular localization of IPE2. A, Expression pattern of IPE2 by RT-qPCR. DAP, Days after pollination. Error bars indicate sd (n = 3). B to G, RNA in situ hybridization of the wild-type anthers using an IPE2-specific antisense (B–D) and a negative control sense probe (E–G). Hybridization signal at stage 9, early uninucleate microspore stage, with winkled microspores (B) and rounded microspores (C). Hybridization signal at stage 10 (D). ML, Middle layer; Msp, microspore; T, tapetum. Scale bars = 50 μm. H to P, Subcellular localization of IPE2 with tobacco transformation showing IPE2-GFP and RFP-KDEL colocalization (H–J), nuclear localization of IPE2ΔN-GFP (K–M), and the control empty construct with GFP (N–P). Scale bars = 10 μm.
We predicted that a fragment including 22 amino acids at the N terminus of IPE2 is the signal peptide by using the SignalP 4.0 program (Supplemental Fig. S7). To reveal the subcellular localization of IPE2, two constructs driven by 35S were generated: Pro-35S::IPE2-GFP containing the full-length IPE2 coding sequence fused to the 5′ terminus of GFP and Pro-35S::IPE2ΔN-GFP without the putative signal peptide (Supplemental Fig. S8). These two constructs and Pro-35S::GFP were cotransformed with red fluorescent protein (RFP)-KDEL into the tobacco (Nicotiana benthamiana) mesophyll cells, respectively. Free GFP signal had distributed itself throughout the entire cell (Fig. 6, N–P), while IPE2-GFP signal was merged with the signal of the endoplasmic reticulum (ER)-specific marker (Fig. 6, H–J), implying that IPE2 is localized to the ER. On the contrary, IPE2ΔN-GFP fluorescence was only localized to the nucleus (Fig. 6, K–M), confirming that the signal peptide is required for IPE2 ER localization. These results provide a clue for the putative function of IPE2 in fatty acids metabolism involved in cutin monomer, wax, and sporopollenin biosynthesis occurring in anther tapetum ER.
Phylogenetic Studies of IPE2 and GDSL Lipase Gene Family
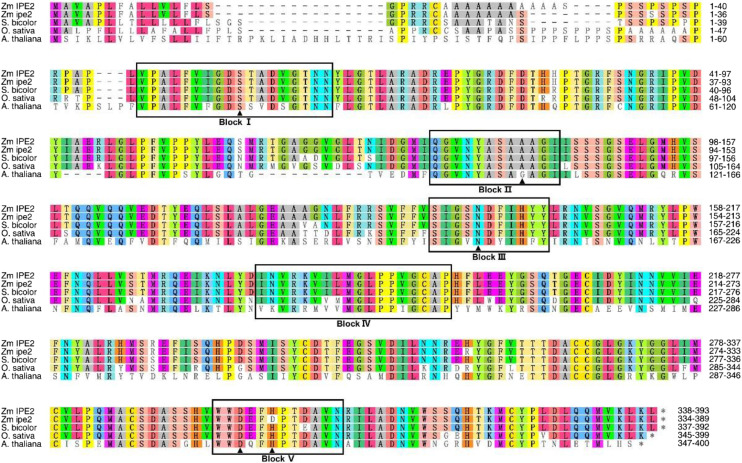
We predicted that IPE2 encodes a SGNH hydrolase-type esterase by maize genome annotation in EnsemblPlants (http://plants.ensembl.org/Zea_mays/Info/Index). Further Pfam testing against the Hidden Markov model (HMM) profile confirmed that IPE2 possesses accession PF00657, a GDSL-like lipase/acylhydrolase. Amino acid sequences of IPE2 homologs in maize, sorghum (Sorghum bicolor), rice, and Arabidopsis (Arabidopsis thaliana) were aligned to explore the conserved domains and active sites in IPE2 (Fig. 7). IPE2 amino acid sequences were divided into five blocks (I to V). The Ser in block I, Asp in block III, and His in block V were conserved active sites in the four species, implying the conserved function of IPE2 in both monocots and dicots (Upton and Buckley, 1995). In ipe2, we detected an His to Asp transformation in block V (Fig. 7), which was reported to be critical for enzyme activity (Volokita et al., 2011). To further understand the evolutionary relationship of maize IPE2 with other species, we constructed an unrooted neighbor-joining tree using amino sequences of 17 IPE2 homologs representing monocots and eudicots. Maize IPE2 was grouped into the monocot clade and was the closest to sorghum homolog XP_002451978 (Supplemental Fig. S9).
Figure 7.
Amino acid alignment of IPE2 homologs in maize, sorghum, rice, and Arabidopsis. Black boxes indicate the five conserved blocks. Amino acids marked by the solid black triangle indicate the putative active sites. Among them, S in Block I and D and H in Block V indicate the catalytic triad; and S in Block I, A/G in Block II, and N in Block III indicate the oxyanion hole. Accession numbers for homologs in sorghum, rice, and Arabidopsis are XP 002451978.1, XP_015623170.1, and NP_567372.1, respectively.
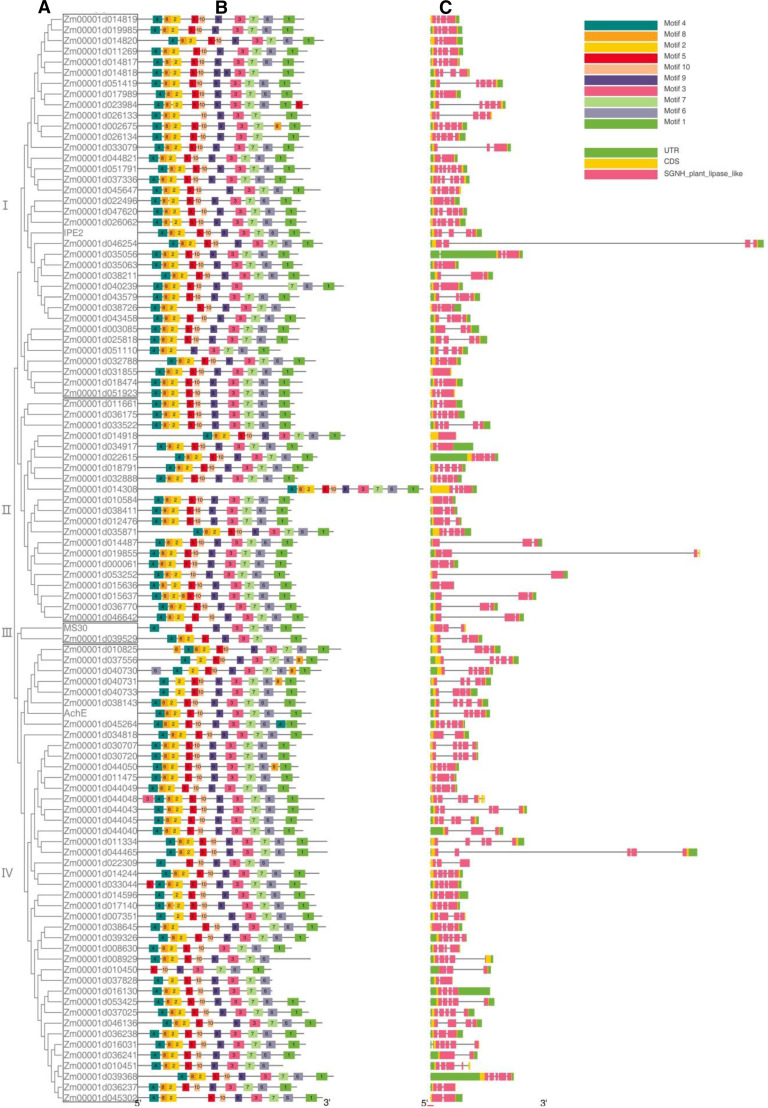
A total of 102 putative members of GDSL-type lipase gene family were proposed from maize reference genome (B73_RefGen_v4). Phylogenetic analysis showed that maize GDSL lipase gene family could be classified into four clades (Fig. 8). The phylogenetic analysis (Fig. 8A), combined with protein motifs (Fig. 8B), gene structures (Fig. 8C), and expression profiling (Supplemental Fig. S10) were used to predict the function of GDSL genes. The results demonstrated that the motifs were much conserved in most of the family members, especially in the same clade. However, the gene structures were substantially diverged among members, as further confirmed by the huge sequence diversity of the gene family. Aside from IPE2, only two members of GDSL lipase gene family have been functionally studied in maize; one is AchE (Zm00001d021961; Yamamoto et al., 2011), and the other is MS30 (Zm00001d052403; An et al., 2019). By amino acid sequence alignment, we found that the three proteins shared very low identity with each other, i.e. 19.66% between MS30 and AchE, 21.55% between IPE2 and AchE, and 24.69% between IPE2 and MS30 (Supplemental Fig. S11). This is consistent with the phylogenetic analysis in which IPE2, MS30, and AchE were respectively classified into clades I, III, and IV in the maximum likelihood (ML) tree (Fig. 8).
Figure 8.
The phylogenetic tree, protein motifs, and gene structures of GDSL gene family in maize. A, Phylogenetic tree. B, Protein motifs. C, Gene structures.
Genes Involved in Lipid Metabolism Are Disrupted in ipe2 Mutants
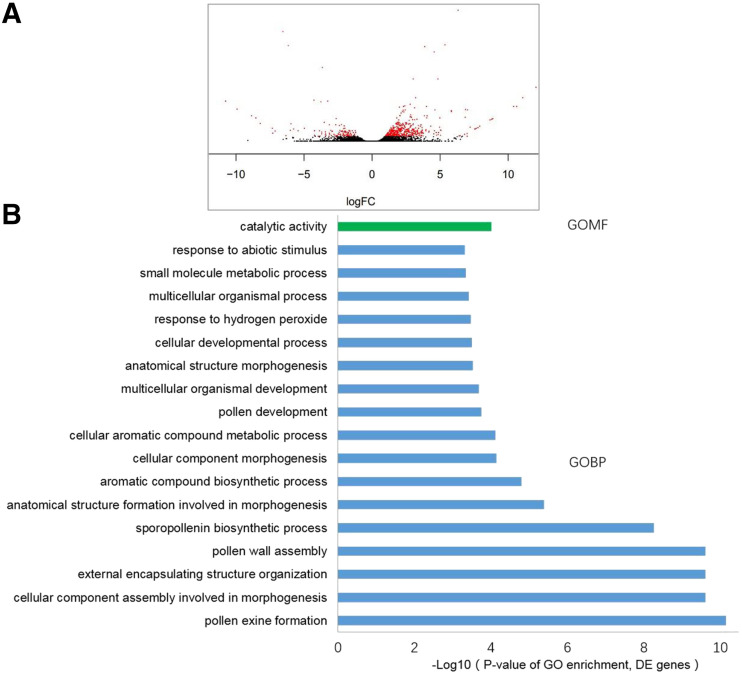
To better understand the molecular mechanism involved in ipe2 male sterility, transcriptome profiles were generated from uninucleate stage anthers of wild type and ipe2 mutant. High-throughput sequencing was performed based on the Hiseq2500 platform (Illumina). For each sample, three biological replicates were set, and clean reads were mapped to the maize genome reference (B73_RefGen_v4). A total of 25,864 transcripts were detected in wild type and ipe2. We identified 588 differentially expressed genes (DEGs) in ipe2 anthers with 119 genes upregulated while 469 downregulated (Fig. 9A). The results were validated by the high correlation of the log2-fold change values between RNA sequencing (RNA-seq) and RT-qPCR results of six randomly selected genes (Zm00001d020737, Zm00001d020867, Zm00001d021168, Zm00001d024403, Zm00001d034506, and Zm00001d043868; Supplemental Fig. S12).
Figure 9.
Transcriptome analysis of wild-type and ipe2 anthers at the early uninucleate stage. A, The volcano plot of the DEGs. Red dots indicate DEGs, and black dots indicate false. B, Significantly enriched GO terms of the DEGs.
Gene ontology (GO) analysis indicated that the DEGs are involved in multiple biological processes including development of the pollen (GO:0009555), the aromatic compound biosynthetic process (GO:0019438), assembly of the pollen wall (GO:0010208), formation of the pollen exine (GO:0010584), and the sporopollenin biosynthetic process (GO:0080110; Fig. 9B). Of the most significant DEGs (Supplemental Table S3), Zm00001d019478 and Zm00001d013991 were proposed to encode fewer adhesive pollen proteins required for pollen exine formation in Arabidopsis (Dobritsa et al., 2010). Zm00001d020970 encodes a dihydroflavonol 4-reductase-like1 protein essential for male fertility in Arabidopsis (Tang et al., 2009). Zm00001d003702 encodes Acyl-CoA synthetase5, which plays important roles in sporopollenin precursor synthesis (de Azevedo Souza et al., 2009). Moreover, Zm00001d046537 encodes ABCG26, which acts as a sporopollenin precursor transporter in Arabidopsis (Quilichini et al., 2014). Because IPE2 was categorized as postmeiotic, we next checked the expression patterns of the nine post-meiotic male-sterile genes (Supplemental Table S1) in ipe2. Transcriptome data showed that the expressions of MS26, MS7, IPE1, APV1, and MS6021 were significantly downregulated in ipe2, while the expression levels of MS45, ms44, and MS33 showed no differences between ipe2 and wild type (Supplemental Table S3). MS30 was upregulated in ipe2 (fragments per kilobase million 3,535.32 in wild type versus FPKM 5,694.32 in ipe2; Supplemental Table S3) but did not reach the standard of the 2-fold variation in FPKM value. The significant downregulation of MS26, MS7, IPE1, APV1, and MS6021 in ipe2 may indicate that they are coexpressed in up/downstream pathways during late stages of anther development. The upregulation of MS30 in ipe2 mutant may indicate a compensation effect between IPE2 and MS30 in lipid metabolic processes. Taken together, the expression data of the postmeiotic male sterile genes may provide insights into the gene regulatory network involved in postmeiotic male sterility in maize.
The main components of sporopollenin and pollen wall are long-chain fatty acids, polyketides, and aromatic compounds such as phenolics (Ariizumi and Toriyama, 2011; Wang et al., 2018). The phenolics are mainly comprised of stilbenes, coumarins, flavonoids, and lignin that are produced from Phe metabolism (Dobritsa et al., 2010). Kyoto Encyclopedia of Genes and Genomes analysis demonstrates that a series of biological processes including cutin and wax biosynthesis, unsaturated fatty acid biosynthesis, fatty acid elongation, α-linolenic acid metabolism, flavonoid biosynthesis, glycerolipid/glycerophospholipid, phenylpropanoid biosynthesis, and GPI-anchor biosynthesis are disturbed in ipe2 (Table 1). Taken together, we inferred that defective IPE2 may result in severe disruptions of lipid metabolism, which leads to metabolic disorders of cuticle and sporopollenin in anther and pollen wall, and ultimately causes male sterility in ipe2 mutant.
Table 1. DEGs involved in anther cuticle and pollen exine development.
| Gene ID | Function | Log2 (ipe2/Wild Type) |
|---|---|---|
| Zm00001d048337 | Cutin, suberine, and wax biosynthesis | −3.353 |
| Zm00001d042814 | Cutin, suberine, and wax biosynthesis | −3.337 |
| Zm00001d012326 | Cutin, suberine, and wax biosynthesis | −3.499 |
| Zm00001d012274 | Cutin, suberine, and wax biosynthesis | −2.072 |
| Zm00001d008622 | Fatty acid metabolism, fatty acid elongation, and unsaturated fatty acids biosynthesis | −2.077 |
| Zm00001d008623 | Fatty acid metabolism, fatty acid elongation, and unsaturated fatty acids biosynthesis | −2.978 |
| Zm00001d044908 | α-linolenic acid metabolism | 3.158 |
| Zm00001d021297 | Glycerolipid metabolism | −2.023 |
| Zm00001d051043 | Glycerolipid metabolism | −1.396 |
| Zm00001d010078 | Glycerophospholipid metabolism | −2.835 |
| Zm00001d041962 | Glycerophospholipid metabolism | −1.204 |
| Zm00001d049960 | Glycerophospholipid metabolism | −2.212 |
| Zm00001d053107 | Phe metabolism | −2.187 |
| Zm00001d003702 | Phe metabolism and phenylpropanoid biosynthesis | −2.351 |
| Zm00001d014601 | Phenylpropanoid biosynthesis | −2.642 |
| Zm00001d002898 | Phenylpropanoid biosynthesis | 3.702 |
| Zm00001d037547 | Phenylpropanoid biosynthesis | −1.606 |
| Zm00001d051754 | β-Ala metabolism and pyruvate metabolism | −1.330 |
| Zm00001d033619 | Pantothenate and CoA biosynthesis | −1.652 |
| Zm00001d015445 | Pantothenate and CoA biosynthesis | 8.549 |
| Zm00001d015183 | Pantothenate and CoA biosynthesis | −2.525 |
| Zm00001d017077 | Flavone and flavonol biosynthesis | −1.873 |
| Zm00001d001960 | Flavonoid biosynthesis | 1.863 |
| Zm00001d033225 | Pyruvate metabolism | −1.663 |
| Zm00001d037332 | Phosphatidylinositol signaling system | −1.466 |
| Zm00001d016990 | GPI-anchor biosynthesis | 1.286 |
| Zm00001d044564 | ABC transporters | −1.997 |
DISCUSSION
IPE2 Is a GDSL Lipase Essential for Maize Male Fertility
The GDSL-lipase gene family, which belongs to the supergene family of SGNH esterases, is one of the largest gene families in plants. The GDSL family members exist in both prokaryotes and eukaryotes. Phylogenetic and evolutionary studies have demonstrated that the family has greatly expanded in land plants, indicating the important roles of the family in plant kingdom (Volokita et al., 2011). The evolution of GDSL-lipase gene family has been widely studied in model plant species, such as Arabidopsis (Lai et al., 2017), rice (Chepyshko et al., 2012), and Brassica rapa (Dong et al., 2016). However, the biological functions of the family members are largely unknown because GDSL lipases often have a wide range of substrates. To our knowledge, GDSL lipases mainly participate in biological processes including plant defense (Gao et al., 2017), hormonal metabolism (Cao et al., 2006), cutin and wax metabolism (Girard et al., 2012), and a secondary wall formation (Zhang et al., 2019). We predicted a total of 102 GDSL-lipase family members dispersed in the entire maize genome, and only AchE and MS30 were functionally studied. AchE was preferentially expressed in coleoptile nodes and seeds. It mainly contributes to cell-wall matrix development and plays a positive role in maize heat tolerance (Yamamoto et al., 2011). MS30 encodes a GDSL lipase crucial for the aliphatic metabolic pathway required for pollen exine formation and anther cuticle development (An et al., 2019). It seems that IPE2 and MS30 have similar functions in contributing to male fertility of maize. However, IPE2 was located on the long arm of chromosome 5 while MS30 was located on chromosome 4. In addition, the amino acid sequences of the two genes were highly diverged (24.69% identity; Supplemental Fig. S11) and they were phylogenetically classified into distinct clades in the ML tree (Fig. 8). It seems that IPE2 and MS30 did not originate from simple gene duplication during maize evolution. Interestingly, the expression patterns of IPE2 and MS30 were very similar and both were specifically expressed in meiotic anthers. This was also indicated in the phylogenetic trees based on gene expression data of 79 tissues from B73 (Supplemental Fig. S10). In the tree, GDSL lipase gene family members were classified into two clades, with IPE2 and MS30 clustered into the same clade. We predicted that GDSL lipase gene family has gone through subfunctionalization in maize, and sequence identity as well as expression patterns should be considered when predicting biological functions of this gene family. Besides IPE2 and MS30, 10 additional GDSL genes were detected to be specifically or strongly expressed in immature tassel, meiotic tassel, or anthers (Stelpflug et al., 2016). They are Zm00001d038211 (Immature_Tassel_V13); Zm00001d045302, Zm00001d031855, Zm00001d035871, Zm00001d046136, Zm00001d044045, Zm00001d016130, and Zm00001d037556 (Meiotic_Tassel_V18); and Zm00001d014308 and Zm00001d046642 (Anthers_R1; Supplemental Fig. S10). These genes belong to diverse clades in the ML tree and may be potential genes contributing to maize male fertility, but further studies are needed to confirm their functions.
IPE2 Is Essential for Anther Cuticle and Pollen Exine Formation
A series of genes have been reported to be involved in anther cuticle and/or pollen exine development in maize, including MS26, MS45, MS7, ms44, IPE1, APV1, MS6021, MS30, and MS33 (Cigan et al., 2001; Djukanovic et al., 2013; Chen et al., 2017; Fox et al., 2017; Somaratne et al., 2017; Tian et al., 2017; Xie et al., 2018; Zhang et al., 2018; An et al., 2019). Notably, all the products are involved in the processes of lipid and/or fatty acid metabolism. We infer that the lipid metabolism plays crucial roles in anther or pollen wall development. In this study, we identified a completely male-sterile mutant ipe2 displaying impaired cuticle and exine. Through metabolomic study, we found that the amount of total cutin and wax in ipe2 anthers significantly decreased compared with wild type (Fig. 4). It is known that hydrophobic cutin is a polymer of hydroxylated and epoxylated fatty acids and their derivatives with chain lengths of C16 and C18 (Heredia, 2003). Cuticle wax is composed of long-chain fatty acids, alkanes, fatty alcohols, and other derivatives (Kunst and Samuels, 2003). The exine is composed of sporopollenins, which are highly resistant biopolymers derived from fatty acids, phenylpropanoids, and phenolic compounds. Consistently, the metabolome results showed that C16 and C18 fatty acids and other derivatives were severely reduced in ipe2. From the TEM observation, we could see the irregular sporopollenin deposition on the primexine of microspores in ipe2, which may result in the discontinuous dashed pollen nexine in ipe2 mutant. For the wax constituents, the contents of C22 hydroxyl alcohol and alkanes like C33 and C35 were significantly decreased, while the contents of C25 and C27 alkanes were significantly increased in ipe2. All the facts imply that IPE2 may contribute to the synthesis of soluble fatty acids and cutin monomers, and the elongation of long-chain fatty acids from C25/C27 to C33/C35. GO analysis confirms that DEGs between ipe2 and wild-type anthers are mainly involved in pollen exine formation and sporopollenin biosynthetic process (Fig. 9B). Additional Kyoto Encyclopedia of Genes and Genomes analysis demonstrates that a series of biological processes, such as cutin and wax biosynthesis, flavonoid biosynthesis, and phenylpropanoid biosynthesis, are disturbed in ipe2 (Table 1). Metabolome combined with transcriptome studies suggests that IPE2 participates in the lipid metabolism that is required for anther cuticle formation, sporopollenin biosynthesis, and pollen exine formation.
Like most male-sterile mutants identified so far, ipe2 exhibits normal vegetative growth as well as female fertility. We infer that these male-sterile–related genes including IPE2 are specifically involved in the development of male reproductive organs. This was further verified by the temporal and spatial expression patterns of IPE2. It is known that tapetum is the innermost layer of anther wall, which directly contacts developing gametophytes. The tapetum provides enzymes for the release of microspores from tetrads and further provides nutrients for pollen development (Li et al., 2006a). During the late stage of pollen maturation, tapetum degeneration occurs through a PCD process (Wu and Cheun, 2000). Postmeiotic male-sterile mutants have defects in tapetum development, demonstrating the crucial roles of tapetum on male gametophyte development. These studies referred to model species including Arabidopsis (Zhang et al., 2007; Xu et al., 2010; Gu et al., 2014; Wang et al., 2018), rice (Xu et al., 2017; Chang et al., 2018; Uzair et al., 2020), as well as maize (Moon et al., 2013; Wang et al., 2013; Fox et al., 2017; Nan et al., 2017; Somaratne et al., 2017; Xie et al., 2018; Zhang et al., 2018; An et al., 2019). In ipe2 mutant, we also detected a delayed degeneration of tapetum and an undegraded middle layer at the vacuolated stage (Fig. 2P). In the late stage of anther development, the tapetum was invisible while the middle layer was still intact or even swelling (Fig. 2, R and T). We predicted that the delayed tapetum degeneration and undegraded middle layer may contribute to the impaired pollen wall formation and further affect starch accumulation in pollen. This was also confirmed by in situ hybridization, which demonstrates strong signals of IPE2 in microspores, tapetum, as well as the middle layer during microspore release stage (Fig. 6, B and C). TAPETUM DEGENERATION RETARDATION (TDR) in rice and its ortholog gene ABORTED MICROSPORES (AMS) in Arabidopsis were previously identified to encode a basic helix-loop-helix transcription factor, which plays key roles in tapetal PCD and pollen exine formation (Li et al., 2006a, 2006b; Xu et al., 2010). TDR/AMS has been commonly regarded as a landmark for tapetum degeneration during gametophyte development (Zhang et al., 2011). We thus identified the putative ortholog of TDR/AMS in maize (Gene number: Zm00001d053895) and checked the expression pattern of Zm00001d053895 through transcriptome analysis. As expected, compared with the wild type, the expression of Zm00001d053895 was significantly downregulated in ipe2, i.e. FPKM 64065.7 in wild type versus FPKM 20490.1 in ipe2 (Supplemental Table S3). The results help to strengthen our judgement that tapetum degradation is delayed or attenuated in ipe2.
Given that IPE2 encodes a GDSL lipase modulating lipid metabolism and homeostasis, IPE2 is believed to regulate the biosynthesis of anther cuticle and pollen exine in maize. The inference was verified by both metabolome and transcriptome data. IPE2 may achieve this by contributing to the synthesis of soluble fatty acids and cutin monomers, as well as by participating in the elongation of long-chain fatty acids from C25/C27 to C33/C35. Besides, IPE2 was also hypothesized to be involved in the degeneration of tapetum and middle layer during late stages of anther development. The delayed degeneration of tapetum and middle layer may largely affect enzymes and nutrients required for exine formation and pollen starch accumulation. Although PCD of tapetum is much emphasized during male gametophyte development, the underlying mechanism is still poorly understood in plants. The PCD of tapetum is supposed to be a complex regulatory network, which involves diverse enzymes, transcription factors, signaling proteins, transporters, etc. Compared with tapetum, the middle layer is often underestimated, and the function of this unique layer is less known so far. Further studies should be performed to decipher the pathway through which IPE2 regulates the development of tapetum and middle layer, and how these two layers contribute to male fertility in maize.
In conclusion, the identification of IPE2 will provide insight to the molecular mechanisms of male sterility and the biological functions of GDSL lipase gene family in maize, and will also facilitate its future application in hybrid production. Nevertheless, due to the universal substrates that GDSL lipase may have, its functional substrates have not been uncovered. Further genetic and biochemical studies are needed to decipher the function and regulatory network of IPE2 in lipid metabolism and male fertility.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
ipe2 and its allelic materials (ipe2-a, ipe2-b and ipe2-c) were obtained from the Maize Genetics Cooperation Stock Center (https://www.maizegdb.org/data_center/stock). The F2 and BC1F1 mapping populations were derived from crosses between ipe2 and B73/Chang7-2. All the plants were grown in a field in Beijing or Sanya, China. Tobacco (Nicotiana benthamiana) plants for subcellular localization experiment were grown in the light incubator with 16 h of light/8 h of dark at 22°C. Transgenic plants and populations for complementary experiment were grown in a greenhouse in Beijing, China.
Phenotypic Characterization of the ipe2 Mutant
A Canon digital camera (model no. EOS 550D) was used to take photographs for plants and flowers. Pollen grains were stained with 1% (m/v) I2-KI and photographed by the Olympus BX53 microscope. For SEM analysis, fresh anthers from both wild type and ipe2 mutants were ordinally fixed, graded-dehydrated, dried, and gold-coated as described by Tian et al. (2017). A HITACHI S-3400N scanning electron microscope was used to observe and take pictures of the samples.
For cytological observation, anthers from both wild type and ipe2 were ordinally fixed, graded-dehydrated, and embedded in spur resin for semithin sections, as described by Tian et al. (2017). A Leica RM2265 was used to obtain the sections, which were then stained with 0.05% (m/v) Toluidine Blue and observed with the Olympus BX-53 microscope.
For TEM analysis, fresh anthers were vacuum-infiltrated and fixed in 4% (v/v) glutaraldehyde (with 0.1 m of phosphate buffer, pH 7.4), followed by rinsing with 0.1 m of phosphate buffer. The tissues were then immersed in 1% (m/v) osmium tetraoxide and rinsed with 0.1 m of phosphate buffer. The samples were dehydrated using an acetone series from 30% to 100% (v/v) and embedded in epoxy resin. Leica EM-UC6 ultramicrotome was used to obtain ultra-thin sections, which were then stained with uranyl acetate and double-stained with lead citrate. Images were obtained with a HITACHI H-7500 transmission electron microscope.
Wax and Cutin Analysis of Mature Anthers
Fresh mature anthers were used for wax and cutin analysis. The anther surface area was calculated considering the anthers as cylinders, and then plotted against the corresponding fresh weight (Li et al., 2010). Wax, cutin constituents, and soluble fatty acids were extracted and analyzed using the methods in Bonaventure et al. (2004) and Franke et al. (2005).
Allelism Test and Map-Based Cloning of the IPE2 Gene
ipe2 was respectively crossed with ipe2-a, ipe2-b, and ipe2-c to test the allelism between ipe2 and ipe2-a, as well as ipe2-b and ipe2-c. F1 individuals were generated by crossing ipe2 with inbred line B73/Chang7-2, and F2 populations were then obtained from the self-pollination of F1 plants. The BC1F1 segregation populations were generated by backcrossing F1 with ipe2. The locus was initially mapped to the long arm of Chromosome 5. Additional insertion or deletion, simple sequence repeat, and SNP markers were developed for fine mapping. The primers for gene mapping are listed in Supplemental Table S4.
For functional complementation, a 7,524-bp genomic fragment, including a 2,869-bp upstream promoter, the entire IPE2 genomic region, and a 1,773-bp downstream segment, was amplified from the wild type and transferred into the vector pCAMBIA3300. This p3300::IPE2 expression vector was then transformed into the maize (Zea mays) inbred Zong31 with Agrobacterium tumefaciens. The positive transgenic lines were detected using primers listed in Supplemental Table S4 and then backcrossed to ipe2 to obtain T1. Pollen viability of T2 individuals was assayed with 1% (m/v) I2-KI.
RNA Extraction and RT-qPCR
To study the detailed expression pattern of IPE2, we collected anther samples of 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, and 5 mm (±0.03 mm) in length corresponding to St6, St7, St8b, St9, St10, St11, St12, St13, and St14, respectively. Furthermore, we included three biological replicates and three technical replicates for each stage group, and analyzed a total of 27 anther groups. At least 10 anthers from each group were double-checked for the cytological stages by semithin sections. For each group, only the anthers at correct developmental stages were selected for RT-qPCR study. Total RNA was extracted from root, stem, leaf, cob, silk, mature pollen, 12 d-after-pollination seed, 24 d-after-pollination seed, and different stages of anthers using TRIzol reagent (Invitrogen). The developmental stages of anthers were determined based on the semisection morphology. Complimentary DNA (cDNA) of the sample was synthesized from 1 μg of total RNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). A LightCycle480 system (Roche) was used to perform RT-qPCR, with a 20-μL reaction mixture containing 10 μL of SYBR Green Premix (TAKARA), 2 μL of cDNA, and 10 pmol of each primer. ZmGAPDH was used as the internal control for normalization. All reactions were conducted in triplicate and the expression data were analyzed using 2−ΔΔCt method. All primers for RT-qPCR are listed in Supplemental Table S4.
In Situ Hybridization
Wild-type anthers at different developmental stages were fixed in RNase free formaldehyde-acetic acid-ethanol fixative solution and then dehydrated in a gradient ethanol series, i.e. 50%, 70%, 85%, 95%, and 100% (v/v) ethanol series. After embedding in paraffin, 8-μm–thick sections were obtained using a Leica RM2265 rotary microtome. A 400-bp fragment from 627 to 1,026 bp of IPE2 cDNA was generated by PCR amplification to make antisense and sense probes. RNA in situ hybridization was performed according to Tian et al. (2017). The sequences of the probes are listed in Supplemental Table S4.
Subcellular Localization of IPE2
For the subcellular localization analysis, we first used SignalP 4.0 Server (http://www.cbs.dtu.dk/services/SignalP/) to predict the putative signal peptide. Then, IPE2 CDS, which contains a 1,179-bp coding sequence without the stop codon and IPE2ΔNCDS (i.e. CDS without the putative signal peptide), were cloned into pCAMBIA1300-GFP vector to obtain p35S::IPE2-GFP and p35S::IPE2ΔNCDS-GFP, respectively. These constructs, as well as empty construct, were cotransformed with an ER marker (RFP-KDEL) into the tobacco mesophyll cells. The fluorescence signals were observed by a laser scanning confocal microscope (LSM 710 NLO; Zeiss). IPE2-GFP and RFP-KDEL were excited at 488 and 561 nm, and emissions were collected at 500 to 540 nm and 600 to 650 nm, respectively. Data were produced from at least three independent biological replicates.
Phylogenetic Analysis of IPE2 and GDSL Gene Family
To elucidate the evolutionary relationship between IPE2 and its homologs, we searched the Phytozome V12 genome databases (https://phytozome.jgi.doe.gov/pz/portal.html) for homolog sequences using IPE2 protein sequence as the query. In total, 16 homologs from both monocot and eudicot species were retrieved and the alignment was performed with the tool ClustalW (https://www.genome.jp/tools-bin/clustalw) using default parameters. An unrooted neighbor-joining tree was constructed using the program MEGA 6.0 (https://www.megasoftware.net/) with the following parameters: Poisson model, partial deletion, and 1,000 bootstrap replicates (Tamura et al., 2013). The protein sequences of IPE2 homologs from the four model plants (i.e. sorghum [Sorghum bicolor], rice [Oryza sativa], Arabidopsis [Arabidopsis thaliana], and maize) were aligned using the program Geneious v5.1 (Drummond et al., 2010).
For maize GDSL gene family analysis, we first searched the maize genome database (B73_RefGen_v4) in EnsemblPlants (http://plants.ensembl.org/index.html) for annotated GDSL genes. The maize GDSL candidates were then tested against the HMM model profile, numbered PF00657 in the Pfam 32.0 HMM library (http://pfam.xfam.org/). Only 102 sequences with the complete GDSL domain and a length above 200 amino acids were selected for further study. The online program SMART (http://smart.embl.de/) was also used to further confirm the existence of the GDSL core sequences in the 102 candidate genes. The exon–intron organization of maize GDSL genes was determined by comparing predicted coding sequences with their corresponding full-length sequences using the online program CDD from the National Center for Biotechnology (https://www.ncbi.nlm.nih.gov/). The online MEME Suite 5.1.0 (http://meme-suite.org/) for protein sequence analysis was used to identify conserved motifs in maize GDSL genes. The optimized parameters were employed with the maximum number of motifs as 10. The alignment of GDSL genes was performed as described above. An unrooted ML tree was constructed with the program MEGA 6.0 using the best model WAG with frequencies (+F), partial deletion, and the 1,000 Bootstrap method. The phylogeny tree, motif pattern, and gene structure were finally combined by using the program TBtools v0.66831 (https://github.com/CJ-Chen/TBtools).
Transcriptome Analysis
Total RNA was extracted with TRIzol reagent (Invitrogen) from anthers of wild type and ipe2 mutant, with three replicates for each genotype at the early uninucleate stage. The library was constructed according to Illumina TruSeq instructions, and sequenced with a HiSeq 2500 genome analyzer (Illumina) to obtain 101-nt long paired-end reads. Clean data after adaptor and quality trimming were mapped to the B73 reference genome with the program STAR (Dobin et al., 2013). Read counts per gene were used for differential expression analysis with the tool DESeq2 (Love et al., 2014) with 1% false discovery rate as the threshold to declare significant DEGs between ipe2 and wild type (Benjamini and Hochberg, 1995). GO analysis on the agriGO platform (Alexa et al., 2006) was performed for functional categorization of identified DEGs. A corrected P < = 0.01 was set as the threshold to identify GO significant enrichment of gene, while q < = 0.01 was selected as the threshold of pathway significant enrichment (Xie et al., 2016). KOBAS 2.0 software (http://kobas.cbi.pku.edu.cn/index.php) was used for pathway enrichment analysis. RT-qPCR of six randomly selected genes was performed to validate the RNA-seq data (Supplemental Fig. S12, A–G). The sequences of the primers are listed in Supplemental Table S4.
Accession Numbers
Sequence data of IPE2 can be found in GenBank/EMBL (https://www.ncbi.nlm.nih.gov/nuccore/NC_050100.1?report=genbank&from=133746326&to=133749208&strand=true) data libraries under the accession number AQK69811.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic comparison between wild type and ipe2 mutants.
Supplemental Figure S2. Weight/surface area ratio of wild type and ipe2 anthers.
Supplemental Figure S3. The content of total fatty acid and fatty acid constituents in wild-type and ipe2 anthers.
Supplemental Figure S4. Map-based cloning of IPE2.
Supplemental Figure S5. Coding sequence alignment of IPE2 in wild type and ipe2 mutants.
Supplemental Figure S6. Alignment of IPE2 amino acid sequences in wild type and ipe2 mutants.
Supplemental Figure S7. Signal peptide prediction of IPE2.
Supplemental Figure S8. Map of the constructs for IPE2 localization.
Supplemental Figure S9. The neighbor-joining tree of IPE2 homologs.
Supplemental Figure S10. The expression pattern of GDSL gene family in maize.
Supplemental Figure S11. Amino acid sequence alignment of maize IPE2 and MS30.
Supplemental Figure S12. RT-qPCR of six randomly selected DEGs and the correlation between RNA-seq and RT-qPCR data.
Supplemental Table S1. List of the cloned male-sterile genes in maize.
Supplemental Table S2. Results of the allelic test between ipe2 mutants.
Supplemental Table S3. RNA-seq data of selected DEGs and male-sterile genes.
Supplemental Table S4. List of primers used in the study.
Acknowledgments
We thank Caixia Gao (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for providing the GFP construct, Yanbao Tian (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the guidance of the use of SEM and TEM equipment, Fengxia Zhang (Metabolome platform of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for the wax, cutin, and fatty acid analysis, and Jenny Xiang (Department of Plant Biology, North Carolina State University) for valuable comments in the revision of this article. We acknowledge TopEdit LLC for the linguistic editing and proofreading during the preparation of this article.
Footnotes
This work was supported by the National Key Research and Development Programs of China (grant nos. 2017YFD0300301 and 2016YFD0101803) and the National Natural Science Foundation of China (grant no. 31601376).
References
- Albertsen MC, Fox T, Leonard A, Li B, Loveland B, Trimnell M, inventors (January 28, 2016) Cloning and use of the ms9 gene from maize. US Patent Application No. US20160024520A1 [Google Scholar]
- Albertsen MC, Fox T, Trimnell M, Wu Y, inventors (November 20, 2011) Msca1 nucleotide sequences impacting plant male fertility and method of using same. US Patent Application No. US007915478B2 [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T(2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- An X, Dong Z, Tian Y, Xie K, Wu S, Zhu T, Zhang D, Zhou Y, Niu C, Ma B, et al. (2019) ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol Plant 12: 343–359 [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K(2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M(2004) Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J 40: 920–930 [DOI] [PubMed] [Google Scholar]
- Cao D, Cheng H, Wu W, Soo HM, Peng J(2006) Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol 142: 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Jin M, Yan W, Chen H, Qiu S, Fu S, Xia J, Liu Y, Chen Z, Wu J, et al. (2018) The ATP-binding cassette (ABC) transporter OsABCG3 is essential for pollen development in rice. Rice (N Y) 11: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang H, Sun H, Luo H, Zhao L, Dong Z, Yan S, Zhao C, Liu R, Xu C, et al. (2017) IRREGULAR POLLEN EXINE1 is a novel factor in anther cuticle and pollen exine formation. Plant Physiol 173: 307–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepyshko H, Lai CP, Huang LM, Liu JH, Shaw JF(2012) Multifunctionality and diversity of GDSL esterase/lipase gene family in rice (Oryza sativa L. japonica) genome: New insights from bioinformatics analysis. BMC Genomics 13: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan AM, Unger E, Xu R, Kendall T, Fox TW(2001) Phenotypic complementation of ms45 maize requires tapetal expression of MS45. Sex Plant Reprod 14: 135–142 [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ(2009) A novel fatty Acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djukanovic V, Smith J, Lowe K, Yang M, Gao H, Jones S, Nicholson MG, West A, Lape J, Bidney D, et al. (2013) Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J 76: 888–899 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR(2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Lei Z, Nishikawa S, Urbanczyk-Wochniak E, Huhman DV, Preuss D, Sumner LW(2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol 153: 937–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Yi H, Han CT, Nou IS, Hur Y(2016) GDSL esterase/lipase genes in Brassica rapa L.: Genome-wide identification and expression analysis. Mol Genet Genomics 291: 531–542 [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, et al. (2010) Geneious v5.1. http://www.geneious.com
- Evans MM.(2007) The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falasca G, D’Angeli S, Biasi R, Fattorini L, Matteucci M, Canini A, Altamura MM(2013) Tapetum and middle layer control male fertility in Actinidia deliciosa. Ann Bot 112: 1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T, DeBruin J, Haug Collet K, Trimnell M, Clapp J, Leonard A, Li B, Scolaro E, Collinson S, Glassman K, et al. (2017) A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol J 15: 942–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L(2005) Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Gao M, Yin X, Yang W, Lam SM, Tong X, Liu J, Wang X, Li Q, Shui G, He Z(2017) GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog 13: e1006724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A-L, Mounet F, Lemaire-Chamley M, Gaillard C, Elmorjani K, Vivancos J, Runavot J-L, Quemener B, Petit J, Germain V, et al. (2012) Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24: 3119–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JN, Zhu J, Yu Y, Teng XD, Lou Y, Xu XF, Liu JL, Yang ZN(2014) DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J 80: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Heredia A.(2003) Biophysical and biochemical characteristics of cutin, a plant barrier biopolymer. Biochim Biophys Acta 1620: 1–7 [DOI] [PubMed] [Google Scholar]
- Kunst L, Samuels AL(2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Lai CP, Huang LM, Chen LO, Chan MT, Shaw JF(2017) Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol Biol 95: 181–197 [DOI] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006a) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, et al. (2006b) Genome-wide analysis of basic/helix–loop–helix transcription factor family in rice and Arabidopsis. Plant Physiol 141: 1167–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Skibbe D, Timofejeva L, Wang CJ, Kelliher T, Kremling K, Walbot V, Cande WZ(2013) Regulation of cell divisions and differentiation by MALE STERILITY32 is required for anther development in maize. Plant J 76: 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan GL, Zhai J, Arikit S, Morrow D, Fernandes J, Mai L, Nguyen N, Meyers BC, Walbot V(2017) MS23, a master basic helix–loop–helix factor, regulates the specification and development of the tapetum in maize. Development 144: 163–172 [DOI] [PubMed] [Google Scholar]
- Quilichini TD, Samuels AL, Douglas CJ(2014) ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell 26: 4483–4498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN, Rhoads DM, Ward GC, Levings CS III(1995) The relationship between the mitochondrial gene T-urf13 and fungal pathotoxin sensitivity in maize. Biochim Biophys Acta 1271: 235–240 [DOI] [PubMed] [Google Scholar]
- Skibbe DS, Schnable PS(2005) Male sterility in maize. Maydica 50: 367–376 [Google Scholar]
- Somaratne Y, Tian Y, Zhang H, Wang M, Huo Y, Cao F, Zhao L, Chen H(2017) ABNORMAL POLLEN VACUOLATION1 (APV1) is required for male fertility by contributing to anther cuticle and pollen exine formation in maize. Plant J 90: 96–110 [DOI] [PubMed] [Google Scholar]
- Stelpflug SC, Sekhon RS, Vaillancourt B, Hirsch CN, Buell CR, de Leon N, Kaeppler SM(2016) An expanded maize gene expression atlas based on RNA sequencing and its use to explore root development. Plant Genome 9: 1–15 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S(2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LK, Chu H, Yip WK, Yeung EC, Lo C(2009) An anther-specific dihydroflavonol 4-reductase-like gene (DRL1) is essential for male fertility in Arabidopsis. New Phytol 181: 576–587 [DOI] [PubMed] [Google Scholar]
- Tian Y, Xiao S, Liu J, Somaratne Y, Zhang H, Wang M, Zhang H, Zhao L, Chen H(2017) MALE STERILE6021 (MS6021) is required for the development of anther cuticle and pollen exine in maize. Sci Rep 7: 16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton C, Buckley JT(1995) A new family of lipolytic enzymes? Trends Biochem Sci 20: 178–179 [DOI] [PubMed] [Google Scholar]
- Uzair M, Xu D, Schreiber L, Shi J, Liang W, Jung KH, Chen M, Luo Z, Zhang Y, Yu J, et al. (2020) PERSISTENT TAPETAL CELL2 is required for normal tapetal programmed cell death and pollen wall patterning. Plant Physiol 182: 962–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Laigle G, Rozier F, Meeley RB, Perez P, Rogowsky PM(2009) The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J 59: 883–894 [DOI] [PubMed] [Google Scholar]
- Volokita M, Rosilio-Brami T, Rivkin N, Zik M(2011) Combining comparative sequence and genomic data to ascertain phylogenetic relationships and explore the evolution of the large GDSL-lipase family in land plants. Mol Biol Evol 28: 551–565 [DOI] [PubMed] [Google Scholar]
- Wang CJ, Nan GL, Kelliher T, Timofejeva L, Vernoud V, Golubovskaya IN, Harper L, Egger R, Walbot V, Cande WZ(2012) Maize multiple archesporial cells 1 (mac1), an ortholog of rice TDL1A, modulates cell proliferation and identity in early anther development. Development 139: 2594–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Skibbe DS, Walbot V(2013) Maize male sterile 8 (Ms8), a putative β-1,3-galactosyltransferase, modulates cell division, expansion, and differentiation during early maize anther development. Plant Reprod 26: 329–338 [DOI] [PubMed] [Google Scholar]
- Wang K, Guo ZL, Zhou WT, Zhang C, Zhang ZY, Lou Y, Xiong SX, Yao XZ, Fan JJ, Zhu J, et al. (2018) The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol 178: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu D, Tian Y, Wu S, An X, Dong Z, Zhang S, Bao J, Li Z, Li J, et al. (2019) Map-based cloning, phylogenetic, and microsynteny analyses of ZmMs20 gene regulating male fertility in maize. Int J Mol Sci 20: 1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HM, Cheun AY(2000) Programmed cell death in plant reproduction. Plant Mol Biol 44: 267–281 [DOI] [PubMed] [Google Scholar]
- Xie K, Wu S, Li Z, Zhou Y, Zhang D, Dong Z, An X, Zhu T, Zhang S, Liu S, et al. (2018) Map-based cloning and characterization of Zea mays male sterility33 (ZmMs33) gene, encoding a glycerol-3-phosphate acyltransferase. Theor Appl Genet 131: 1363–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Zhang W, Wang Y, Xu L, Zhu X, Muleke EM, Liu L(2016) Comprehensive transcriptome-based characterization of differentially expressed genes involved in microsporogenesis of radish CMS line and its maintainer. Funct Integr Genomics 16: 529–543 [DOI] [PubMed] [Google Scholar]
- Xu D, Shi J, Rautengarten C, Yang L, Qian X, Uzair M, Zhu L, Luo Q, An G, Waßmann F, et al. (2017) Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiol 173: 240–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA(2010) The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 22: 91–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sakamoto H, Momonoki YS(2011) Maize acetylcholinesterase is a positive regulator of heat tolerance in plants. J Plant Physiol 168: 1987–1992 [DOI] [PubMed] [Google Scholar]
- Yeats TH, Rose JK(2013) The formation and function of plant cuticles. Plant Physiol 163: 5–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L(2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang D, Wu S, An X, Xie K, Dong Z, Zhou Y, Xu L, Fang W, Liu S, Liu S, et al. (2018) Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol J 16: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gao C, Mentink-Vigier F, Tang L, Zhang D, Wang S, Cao S, Xu Z, Liu X, Wang T, et al. (2019) Arabinosyl deacetylase modulates the arabinoxylan acetylation profile and secondary wall formation. Plant Cell 31: 1113–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, et al. (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J 52: 528–538 [DOI] [PubMed] [Google Scholar]