Abstract
Marfan syndrome (MFS) is a connective tissue disease caused by variants in the FBN1 gene. Nevertheless, other genes influence the manifestations of the disease, characterized by high clinical variability even within families. We mapped modifier loci for cardiovascular and skeletal manifestations in the mg∆loxPneo mouse model for MFS and the synthenic loci in the human genome. Corroborating our findings, one of those loci was identified also as a modifier locus in MFS patients. Here, we investigate the HSPG2 gene, located in this region, as a candidate modifier gene for MFS. We show a correlation between Fbn1 and Hspg2 expression in spinal column and aorta in non-isogenic mg∆loxPneo mice. Moreover, we show that mice with severe phenotypes present lower expression of Hspg2 than those mildly affected. Thus, we propose that HSPG2 is a strong candidate modifier gene for MFS and its role in modulating disease severity should be investigated in patients.
Subject terms: Gene expression, Animal breeding
Introduction
Marfan syndrome (MFS—MIM# 154700) is an autosomal dominant disorder of the connective tissue with high clinical variability both between and within families [1]. It is caused by variants in FBN1 gene encoding fibrilin-1, the major component of microfibrils [2]. Microfibrils are present in several tissues, which makes MFS a pleiotropic disease affecting mostly the ocular, cardiovascular and musculoskeletal systems [3].
Previous works have suggested that variations in FBN1 expression caused by polymorphisms in the gene could play a role as modifier of disease severity [4, 5]. However, giving the poor genotype–phenotype correlations and the large intrafamilial clinical variability of the disease, recent works focused on understanding how variants in other genes influence MFS phenotypes [6–8].
The effect of genetic background on phenotypic variability in MFS was demonstrated in mice by our group [9]. We showed that mg∆loxPneo mice in the 129/Sv (129) isogenic background presented earlier age of onset of the disease when compared with those in the C57BL/6 (B6) background. Subsequently, we identified loci modulating the phenotypic variability in B6/129 mixed mg∆loxPneo mice [8]. One locus on chromosome 4 was associated with variability of the cardiovascular phenotype [8].
One of the candidate genes we identified in this region was Hspg2, which encodes perlecan, a heparan-sulfate proteoglycan. Variants in Hspg2 are associated with Schwartz–Jampel Syndrome (SJS1; MIM# 255800), an autosomal recessive disease characterized by skeletal manifestations. Knockout mice for Hspg2 presented severe skeletal abnormalities and died around birth due to heart arrest, showing that Hspg2 plays a role in the formation of the skeletal and cardiac system [10]. Interestingly, in our mapping study the Hspg2 locus presented an almost suggestive association with the skeletal phenotype, indicating that it may modulate both cardiovascular and skeletal phenotypes of MFS [8].
Biochemical studies show that perlecan is also involved in maintenance of vascular homeostasis by its interaction with several extracellular matrix (ECM) components, including fibrillin-1 [11, 12]. This interaction is essential for positioning fibrilin-1 multimeres in the pericellular space and, consequently, for the assembly of microfibrils [12–14].
Given these findings, we propose that Hspg2 is a strong candidate modifier gene for MFS. Here, we used the MFS mouse model mg∆loxPneo on a mixed background (B6/129) to compare Hspg2 expression between mild and severely affected mice. We show a correlation between Fbn1 and Hspg2 expression, and an association between lower Hspg2 expression and more severe vascular and skeletal phenotypes, corroborating our hypothesis of Hspg2 as a modifier gene of MFS.
Material and methods
B6/129mg∆loxPneo mice tissues collection and phenotyping
B6/129mg∆loxPneo mice were generated as previously described [8], and the F2 generation was analyzed at 3 months of age. The project was approved by the Ethics Committee for Animal Experimentation of the Institute of Biosciences, University of São Paulo.
Full body digital radiographic images of euthanatized animals were obtained using In Vivo Imaging System FX PRO (Bruker, Germany). Hiperkyphosis was quantified using the kyphosis index (KI) as described [15, 16]. We selected ten mice (five males and five females) with the lowest ten mice (five males and five females) with the highest KI values as the severe and mild group, respectively. Thoracic spinal column fragments were collected for RNA extraction.
Descending thoracic aorta fragments were collected from each mouse for RNA extraction and histology. For each animal, three transversal slices of the aorta were analyzed for elastic fibers fragmentation by optical microscopy, where number of fragmentations (N) was counted (Supplementary Fig. 1B). Elastic fibers integrity index (EFI) was calculated as following:
Mice with an EFI in the highest quartile were classified as mild (five males and 4 females) and those with an EFI in the lowest quartile as severe (four males and three females). Transversal sections of descending thoracic aorta were also screened for aneurysms. Mice with saccular aneurysms (five males; two females) were considered as a separated group.
RNA extraction and gene expression assay
Spinal column and thoracic aorta fragments were macerated in liquid nitrogen in Trizol® reagent (ThermoFisher). RNA was isolated by RNeasy MiniSpin using RNeasy MiniKit (Qiagen). Complementar DNA (cDNA) was obtained from 500 ng of total RNA using SuperScript™ III Reverse Transcriptase (Thermofisher). For Fbn1 and Hspg2 expression analysis we used TaqMan® Gene Expression Assay (Thermofisher—Mm01334119_m1 and Mm01181173_g1, respectively). The gene Actb (TaqMan® Gene Expression Assay—Mm00607939_s1) was used as endogenous control to calculate fold change based on the 2−[ΔΔCt] method [17].
Statistical analysis
Wilcoxon nonparametric test was used for statistical analysis of comparative gene expression. Pearson correlation test was used for gene expression correlation analysis. Expression values in the aorta were adjusted for sex effects by linear model. Tests with p value < 0.05 were considered statistically significant. Computation was performed using R software (version 3.6.1). All data are available in the Supplementary data file.
Results
Hspg2 expression and phenotypes severity
Heterozygous mg∆loxPneo (F2 B6/129) animals were phenotyped for skeletal and cardiovascular systems and separated in groups according to severity (Supplementary Fig. 1A/B). Mice with aneurysm had EFI similar to those in the severe group (Supplementary Fig. 1B).
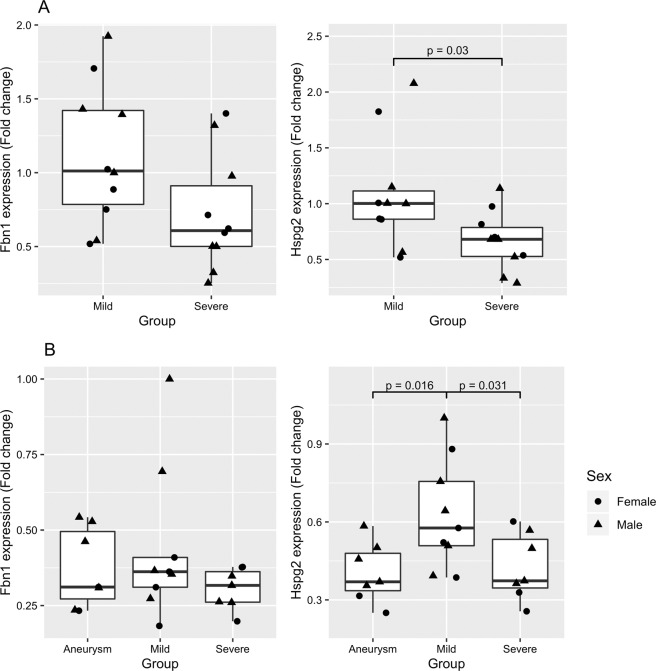
Expression of Fbn1 and Hspg2 was quantified in aorta and spinal column (Fig. 1). No difference of Fbn1 expression in the spinal column and aorta was observed between severe and mild mice for each phenotype (Fig. 1). In contrast, mice with severe hyperkyphosis had lower expression of Hspg2 in comparison with the mild group (p < 0.05) (Fig. 1a). Similarly, expression of Hspg2 in aorta was lower in the severely affected and the aneurysm group when compared with mildly affected animals (p < 0.05) (Fig. 1b). Similar results were obtained when correcting for differences in sex distribution among groups (Supplementary Fig. 1C). No differences in Hsgp2 and Fbn1 expression between the different isogenic wild type and mutant mice were detected (data not shown).
Fig. 1. Expression of Hspg2 and Fbn1 in mg∆loxPneo mice.
Box plots with expression of Fbn1 (left) and Hspg2 (right) in animals with mild and severe phenotypes in a spinal column; and b aorta.
Correlation between Fbn1 and Hspg2 expression
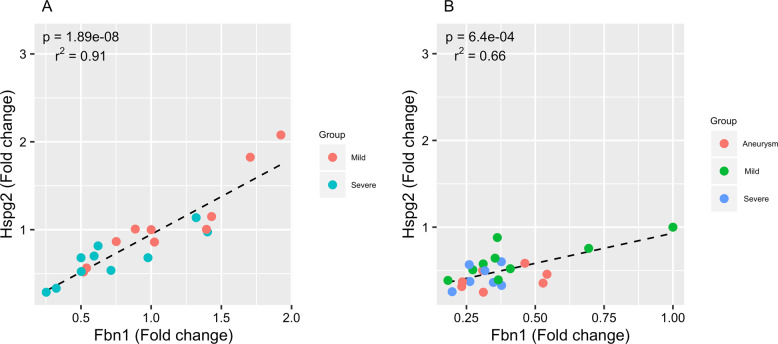
Although we did not find any differences in Fbn1 expression between groups in the two phenotypes analyzed, we tested for correlation between Fbn1 and Hspg2 expression which could suggest co-function of the corresponding proteins. We observed a significant positive correlation between Fbn1 and Hspg2 expression on both spinal column and aorta in mgΔloxPneo mice (p < 0.01 | r2 = 0.66 and p < 0.01 | r2 = 0.91, respectively) (Fig. 2).
Fig. 2. Correlation between Fbn1 and Hspg2 expression in mg∆loxPneo mice.
Scatter plots showing positive correlation between Fbn1 and Hspg2 expression in a spinal column from mg∆loxPneo mice with severe and mild skeletal phenotype; b in aorta from the three mg∆loxPneo groups of vascular phenotype.
Discussion
The mg∆loxPneo mouse model of the clinical variability of MFS allowed us to identify the Awtq1 locus in mouse chromosome 4/human chromosome 1 as a modifier of the cardiovascular phenotype. Within that locus we highlighted Hspg2 as a candidate modifier gene based on its involvement in skeletal and cardiovascular function, and its direct interaction with fibrillin-1 [10, 12, 18].
More recently, a study with 1070 MFS patients identified a smaller modifier locus for the cardiovascular phenotype named gMod-M1 which overlaps with Awtq1 [7]. The only candidate gene identified within that locus was ECE1, highly expressed in the aortic wall and involved in regulation of endothelial-to-mesenchymal transition [7]. However, the region also contains the HSPG2 gene which, interestingly, has a 2.5-fold higher expression in aorta than ECE1 (227.2 TPM vs. 93.88 TPM, n = 432; GTEx Portal). Thus, by the expression and function criteria, HSPG2 should also be considered a candidate modifier gene.
Our expression data from mg∆loxPneo mice show a positive correlation between Hspg2 and Fbn1, strengthening the hypothesis of co-function of the corresponding proteins [12–14, 19]. Moreover, we showed that lower expression of Hspg2 is associated with more severe hyperkyphosis, and with lower integrity of elastic fibers and presence of aneurysms. Thus, although the Hspg2/HSPG2 locus has been originally identified as a modifier of the cardiovascular phenotype, our findings indicate that Hspg2 expression may influence the severity of both skeletal and vascular phenotypes in MFS mice.
The identification of modifier genes of monogenic diseases gives clues about the molecular mechanism of pathogenesis and novel therapeutic strategies and contributes to the prediction of disease severity. Here, we build a case for HSPG2 as a modifier gene for MFS by reviewing the literature and presenting corroborating evidence in our mouse model of MFS clinical variability. We propose that the role of HSPG2 in modulating the severity of skeletal and cardiovascular manifestations should be investigated in large cohorts of MFS patients.
Supplementary information
Acknowledgments
Funding
This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2016/16077-0; 2016/18255-3 and 2018/11708-8) This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES)—Finance Code 001.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-020-0666-0) contains supplementary material, which is available to authorized users.
References
- 1.Ramirez F, Dietz HC. Marfan syndrome: from molecular pathogenesis to clinical treatment; 2007. www.sciencedirect.com. [DOI] [PubMed]
- 2.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–9. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 3.Bitterman AD, Sponseller PD. Marfan syndrome: a clinical update. J Am Acad Orthop Surg. 2017;25:603–9. doi: 10.5435/JAAOS-D-16-00143. [DOI] [PubMed] [Google Scholar]
- 4.Hutchinson S, Furger A, Halliday D, Judge DP, Jefferson A, Dietz HC, et al. Allelic variation in normal human FBN1 expression in a family with Marfan syndrome: a potential modifier of phenotype? Hum Mol Genet. 2003;12:2269–76. doi: 10.1093/hmg/ddg241. [DOI] [PubMed] [Google Scholar]
- 5.Díaz de Bustamante A, Ruiz-Casares E, Darnaude MT, Perucho T, Martínez-Quesada G. Phenotypic variability in Marfan syndrome in a family with a novel nonsense FBN1 gene mutation. Rev Española Cardiol (English Ed) 2012;65:380–1. doi: 10.1016/j.recesp.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Gentilini D, Oliveri A, Fazia T, Pini A, Marelli S, Bernardinelli L, et al. NGS analysis in Marfan syndrome spectrum: combination of rare and common genetic variants to improve genotype-phenotype correlation analysis; 2019. p. 1–20. 10.1371/journal.pone.0222506. [DOI] [PMC free article] [PubMed]
- 7.Aubart M, Gazal S, Arnaud P, Benarroch L, Gross MS, Buratti J, et al. Association of modifiers and other genetic factors explain Marfan syndrome clinical variability. Eur J Hum Genet. 2018;26:1759–72. doi: 10.1038/s41431-018-0164-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes GR, Massironi SM, Pereira LV. Identification of loci modulating the cardiovascular and skeletal phenotypes of Marfan syndrome in mice. Sci Rep. 2016;6:22426. doi: 10.1038/srep22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lima BL, Santos EJC, Fernandes GR, Merkel C, Mello MRB, Gomes JPA, et al. A new mouse model for marfan syndrome presents phenotypic variability associated with the genetic background and overall levels of Fbn1 expression. PLoS ONE. 2010;5:1–9. doi: 10.1371/journal.pone.0014136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costell M, Gustafsson E, Aszódi A, Mörgelin M, Bloch W, Hunziker E, et al. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–22. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo TA, Stoll D, Nader HB, Dreyfuss JL. Mechanical stretch implications for vascular endothelial cells: Altered extracellular matrix synthesis and remodeling in pathological conditions. Life Sci. 2018;213:214–25. doi: 10.1016/j.lfs.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 12.Tiedemann K, Sasaki T, Gustafsson E, Göhring W, Bätge B, Notbohm H, et al. Microfibrils at basement membrane zones interact with perlecan via fibrillin-1. J Biol Chem. 2005;280:11404–12. doi: 10.1074/jbc.M409882200. [DOI] [PubMed] [Google Scholar]
- 13.Sabatier L, Djokic J, Hubmacher D, Dzafik D, Nelea V, Reinhardt DP. Heparin/heparan sulfate controls fibrillin-1, -2 and -3 self-interactions in microfibril assembly. FEBS Lett. 2014;588:2890–7. doi: 10.1016/j.febslet.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 14.Ritty TM, Broekelmann TJ, Werneck CC, Mecham RP. Fibrillin-1 and -2 contain heparin-binding sites important for matrix deposition and that support cell attachment. Biochem J. 2003;375:425–32. doi: 10.1042/bj20030649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laws N, Hoey A. Progression of kyphosis in mdx mice. J Appl Physiol. 2004;97:1970–7. http://www.ncbi.nlm.nih.gov/pubmed/15234960. [DOI] [PubMed]
- 16.De Souza RB, Farinha-arcieri LE, Catroxo MHB, Martins AMCRP da F, Tedesco RC, Alonso LG, et al. Association of thoracic spine deformity and cardiovascular disease in a mouse model for Marfan syndrome. PLoS ONE. 2019;14:1–11. doi: 10.1371/journal.pone.0224581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winer J, Jung CKS, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–9. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 18.Zoeller JJ, McQuillan A, Whitelock J, Ho SY, Iozzo RV. A central function for perlecan in skeletal muscle and cardiovascular development. J Cell Biol. 2008;181:381–94. doi: 10.1083/jcb.200708022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes AJ, Smith SM, Melrose J. Comparative immunolocalisation of fibrillin-1 and perlecan in the human foetal, and HS-deficient hspg2 exon 3 null mutant mouse intervertebral disc. Histochem Cell Biol. 2013;139:1–11. doi: 10.1007/s00418-012-1041-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.