Abstract
Programmed DNA recombination in mammalian cells occurs predominantly in a directional manner. While random DNA breaks are typically repaired both by deletion and by inversion at approximately equal proportions, V(D)J and class switch recombination (CSR) of immunoglobulin heavy chain gene overwhelmingly delete intervening sequences to yield productive rearrangement. What factors channel chromatin breaks to deletional CSR in lymphocytes is unknown. Integrating CRISPR knockout and chemical perturbation screening we here identify the Snf2-family helicase-like ERCC6L2 as one such factor. We show that ERCC6L2 promotes double-strand break end-joining and facilitates optimal CSR in mice. At the cellular levels, ERCC6L2 rapidly engages in DNA repair through its C-terminal domains. Mechanistically, ERCC6L2 interacts with other end-joining factors and plays a functionally redundant role with the XLF end-joining factor in V(D)J recombination. Strikingly, ERCC6L2 controls orientation-specific joining of broken ends during CSR, which relies on its helicase activity. Thus, ERCC6L2 facilitates programmed recombination through directional repair of distant breaks.
Subject terms: Immunology, Double-strand DNA breaks
Introduction
Programmed DNA recombination processes, including V(D)J and antibody class switch recombination (CSR), diversify lymphocyte antigen receptors for efficient adaptive immunity.1 RAG endonucleases assemble V, D and J gene segments to form variable region exons of B and T cell receptor genes,2 and activation-induced cytidine deaminase (AID) further introduces DNA lesions upstream of antibody constant region genes to switch the antibody class from IgM to other classes.3
General DNA repair pathways efficiently process immunoglobulin heavy chain gene (IgH) V(D)J and CSR lesions in B cells. Despite the many mechanistic differences, in both cases intermediate breaks are joined in an orientation-specific manner, i.e. rearrangements occur predominantly by deletion rather than by inversion.2,4 This feature increases the probability of productive rearrangements and consequently the number of peripheral lymphocytes available to fight infection. Conversely, random DNA breaks or designer endonuclease-cutting ends are typically joined both by deletion and by inversion at about equal proportions.5 RAG directional linear tracking and intrinsic properties of recombination signal sequences (RSSs) enforce orientation-specific joining of V(D)J breaks.2,6 How CSR ends are processed mostly by deletion is less understood.
In mammalian cells, double-strand breaks (DSBs) are sensed by the MRN complex, which activates the ATM-dependent DSB response (DSBR) pathway.7 ATM-substrate 53BP1 and its downstream effectors prevent excessive end resection to promote non-homologous end-joining (NHEJ) at the price of homologous recombination (HR).8 The mammalian NHEJ pathway is initiated by DSB end recognition through KU70-KU86 (KU80 in mouse) heterodimer, which further recruits nucleases and/or polymerases and finally Ligase4-XRCC4-XLF complex for repair.9 Besides the evolutionarily conserved core NHEJ factors found in all eukaryotes, several new NHEJ factors have involved in vertebrates and mammals, as exampled by the recently identified NHEJ factors PAXX and MRI/CYREN.10–12 NHEJ can function flexibly on a diverse range of DSB substrates in different chromatin contexts with its DNA-based factors (e.g. core NHEJ subunits) and chromatin factors, and specialized proteins could be involved in joining of a subset of breaks.
V(D)J recombination and CSR occur within the context of topological associated domains (TADs), where loop extrusion facilitates contacts between recombining elements as well as promoters and enhancers.13–15 During CSR, chromatin loop extrusion shapes the IgH chromatin architecture in a spatiotemporal manner.15 Upon antigen stimulation, the activation of I-promoters drives stepwise cohesin loading on the pre-assembled CSR center in naive B cells.15 The chromatin subdomains position the directional alignment of donor Sμ and acceptor S regions, which ensures deletional CSR in cis.15 However, the identity of the trans-acting factors behind orientation-specific end-joining remains unclear.16 DSBR factors, especially 53BP1, favor deletional CSR.4 The absence of 53BP1 is associated with excessive resection of DSB ends and near complete block of CSR,17 indicating that the loss of directional repair might be an indirect effect. Similarly, Lig4 deficient cells tend to have more inversional CSR joining during CSR, reflecting that the escaped broken ends from the joining complex are joined randomly in a diffusional manner at low levels.18
To search for additional end-joining factors, we combined CRISPR knockout screening and chemical perturbation screens, and functionally characterized the hit in the context of immune diversification. The strategy identified ERCC6L2 (Excision Repair Cross-Complementation Group 6 Like 2) as a new NHEJ factor that channels programmed CSR to directional repair.
Results
DNA repair genetic networks revealed by combined CRISPR-chemical screens
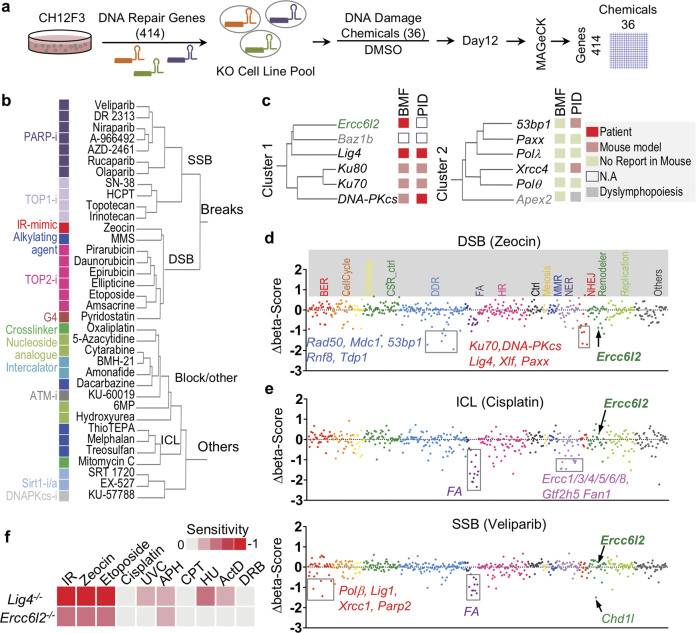
To identify potentially new NHEJ factors, we combined chemical perturbation screens on 36 compounds with focused CRISPR-knockout screens on 414 genes in the CH12 B cell line (Fig. 1a; Supplementary information, Table S1, see Materials and Methods for details). Genes were selected from known DNA repair factors and their homologs based on gene-ontology (GO) terms (Supplementary information, Table S1). DNA damage-inducing chemicals, including carcinogens, therapeutic agents, or DNA damage response (DDR) inhibitors, were selected based on their known functions and inhibitory concentrations (IC) in CH12 cells (Supplementary information, Table S1). Pools of knock-out cells were cultured with chemicals at IC20 (20% maximal inhibitory concentration) for 12 days. Hit genes were called with the MAGeCK algorithm and presented as a z-score,19 where a negative z-score indicates that the knockout of a gene renders the cell more sensitive to the chemical. Clustering of the chemicals by their CRISPR screen z-scores across all the 414 genes categorize the chemicals into two main groups (Fig. 1b; Supplementary information, Fig. S1a).
Fig. 1. CRISPR-chemical screening uncovers ERCC6L2 as a potential NHEJ factor.
a Schematic illustration of chemical genetic screening procedure. b Clustering tree of chemicals. Chemical names are listed and annotated by colored blocks. Similar chemicals based on their inhibitory targets or caused DNA damages types are marked with same color. c Tree-view of two NHEJ gene clusters. Gene names are listed. Roles in bone marrow failure (BMF) or primary immunodeficiency (PID) are annotated by colored blocks. d, e Sensitivity of gene knockouts to Zeocin (d) or Cisplatin/Veliparib (e) treatment. Genes are grouped by gene ontology and depicted with different colors based on the involved biological processes along the x-axis. The beta-score difference (chemical-DMSO) indicating positive or negative enrichment in the chemical treatment samples was calculated from two replicates and is plotted for each gene. Representative negatively enriched genes are labeled. FA indicated Fanconi anemia genes. f Sensitivity of ERCC6L2-deficient or LIG4-deficient B cells to different treatments. Cell viability curve was calculated and the area-under-the-curve (AUC) was computed. Heat map of sensitivity, which is indicated as “log2(AUCKO/AUCWT)”, is plotted. IR γ-irradiation, UVC ultraviolet wavelength 254 nm, APH aphidicolin, CPT camptothecin, HU hydroxyurea, ActD actinomycin D, DRB 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside.
The first group consists of reagents that generate single and double-strand breaks (SSBs and DSBs). The SSB subgroup included 7 poly-ADP ribose polymerase (PARP) and 4 DNA topoisomerase I (TOP1) inhibitors (Fig. 1b), consistent with the observation that these reagents produce similar DNA lesions.20,21 In the DSB subgroup, DNA topoisomerase II (TOP2) inhibitors clustered with γ-radiation mimicking reagent Zeocin, and G-quadruplex-interacting drug pyridostatin, all of which are known to induce DSBs.22–24 Although methyl methanesulfonate (MMS) does not directly cause breaks,25 the downstream lesions may be converted into DSBs at the dosage used in our screen.
The second major group contains reagents that cause blockage on DNA, such as interstrand crosslink (ICL) reagents, nucleoside analogs, crosslinkers, and DNA intercalators as well as DDR inhibitors against ATM and DNA-PKcs (Fig. 1b). This clustering suggests that blockage of DNA replication/transcription may cause the cell proliferation defects observed in the group. We conclude that the combined CRISPR-chemical screens cluster similar chemicals (illustrated by the same color block in Fig. 1b) based on the kind of DNA damage they cause, demonstrating the effectiveness of our approach at dissecting the function of DNA repair genes and DNA damage chemicals.
ERCC6L2 clusters with other NHEJ factors
Next, we clustered all 414 DNA repair genes by their z-scores across the 36 chemicals used, which categorized genes into three major groups depending on their impact on cell growth (Supplementary information, Fig. S1a). Consequently, epistatic genes segregated together, such as those involved in Fanconi anemia and NHEJ factors (Supplementary information, Fig. S1a). NHEJ factors segregated in turn into two main clusters (Fig. 1c): cluster 1 contained core subunit genes (Ku70/80, Lig4, DNA-PKcs) and potentially new members (Baz1b, Ercc6l2). Cluster 2 comprised several other known NHEJ genes: Xrcc4, Paxx, 53bp1, Polλ, Polθ and the apurinic/apyrimidinic endonuclease gene Apex2. Mutations in ERCC6L2 have recently been identified in inherited bone marrow failure (BMF) patients.26–30 Several classic NHEJ gene mutants have been implicated in BMF,28 leading us to wonder whether ERCC6L2 contributes to NHEJ pathway.
Interestingly, ERCC6L2 deficient cells were depleted upon Zeocin treatment which induces DSBs (Fig. 1d), but not in the presence of Cisplatin or Veliparib treatment which creates ICLs and SSBs, respectively (Fig. 1e). This is consistent with results obtained from patient-derived lines carrying ERCC6L2 mutations.27 To confirm the screening results, we deleted Ercc6l2 in CH12 B cells with two sets of sgRNAs. Set 1 deleted the predicted catalytic domain on ERCC6L2, while set 2 created an out of frame mutation (Supplementary information, Fig. S1b, Table S2). We found that all resulting clones were hypersensitive to treatments that induce DSBs, such as γ-irradiation (IR), Zeocin and Etoposide (Fig. 1f; Supplementary information, Fig. S1c). This phenotype is similar to, but less severe than that observed in isogenic cells lacking the major NHEJ ligase Lig4 (Fig. 1f). Increased sensitivity to DSBs was also evident in ERCC6L2-deleted Abelson virus-transformed mouse pro-B cells and human osteosarcoma U2OS cells (Supplementary information, Fig. S1d, e). Altogether, these data demonstrate that ERCC6L2 promotes DSB repair.
ERCC6L2 is required for optimal CSR
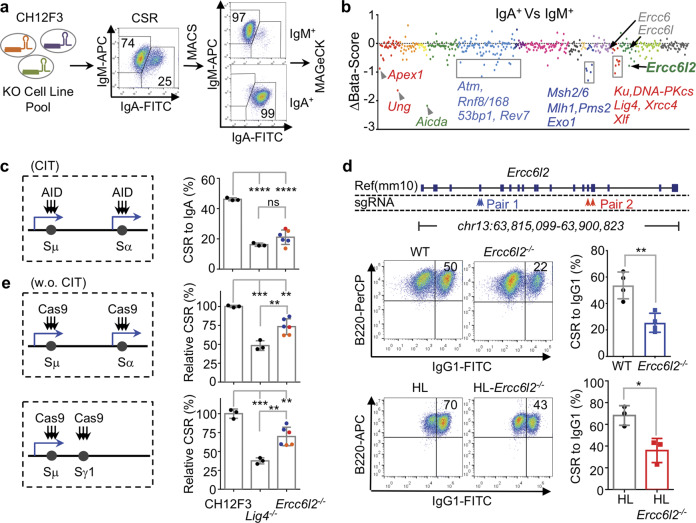
V(D)J recombination and CSR have been used to characterize the function of DSBR/NHEJ factors, and deleterious mutants of NHEJ genes frequently lead to primary immunodeficiencies (PID) (Fig. 1c). We therefore performed a focused CRISPR-knockout screen in CH12 B cells stimulated to undergo IgM to IgA CSR, and compared the enrichment of knocked-out genes between IgA+ and IgM+ populations (Fig. 2a). As controls, the screen included sgRNAs targeting known genes required for CSR (e.g. Aicda, Stat6). Consistent with previous reports,31 the assay confirmed the function of various base excision repair (BER), mismatch repair (MMR), DSBR and NHEJ factors in CSR (Fig. 2b; Supplementary information, Table S1). Among the potentially novel CSR genes, Ercc6l2 was ranked highest in the MAGeCK analysis. Conversely, other Ercc6 family members, Ercc6/CSB (functioning in transcription-coupled nucleotide excision repair) and Ercc6l/PICH (playing a role in spindle assembly checkpoint) were not required for CSR (Fig. 2b).
Fig. 2. ERCC6L2 is required for optimal CSR.
a Schematic illustration of CSR screening procedure. Representative flow cytometry plots are showed. b Enriched CSR genes. Genes are grouped and illustrated as Fig. 1d. The beta-score difference (IgA+–IgM+) is plotted and representative genes are labeled. c ERCC6L2 is required for optimal CSR in CH12F3 cells. AID-initiated CSR is illustrated at left, and CSR to IgA in presence of cytokines (CIT, α-CD40/IL4/TGFβ) of indicated cells are plotted at right. Blue arrows indicate transcription. Colored points indicate knockout clones obtained with different sets of sgRNAs. d ERCC6L2 is required for optimal CSR in ex vivo activated splenic B cells. Gene knockout strategies with two sets of sgRNAs are illustrated on top. Representative CSR flow cytometry plots are showed at left. Data from four pairs of Ercc6l2−/− (sgRNA Pair 1) and wild-type (WT) mice and three pairs of HL-Ercc6l2−/− (sgRNA Pair 2) and corresponding Ig heavy and light chain knockin (HL) mice are summarized. e ERCC6L2 is required for optimal Cas-CSR. CRISPR/Cas9-initiated CSR is schematically illustrated at left, and normalized CSR level of indicated cells are plotted at right. Data are represented as mean ± SD (standard deviation) in (c, d, e). Two-tail unpaired t-test was performed for (c, d, e). ****p < 0.0001, ***p < 0.001, **p < 0.01; *p < 0.05; ns: p > 0.05.
In cytokine activated CH12 cells, CSR was decreased ~50% in the absence of ERCC6L2, which is comparable to results in isogenic Lig4−/− cells32 (Fig. 2c). To study this phenotype under more physiological conditions, we deleted Ercc6l2 by CRISPR-Cas9 (Fig. 2d; Supplementary information, Fig. S2a) in mouse embryos, which were either wild type or carried preassembled heavy and light chain (HL) genes.33,34 Ercc6l2−/− mice were viable (Supplementary information, Fig. S2b). Splenic naive B cells were purified from KO and control mice and stimulated with LPS or LPS + IL4 to induce CSR to IgG3 or IgG1 respectively. Consistent with the CH12 results, ERCC6L2 deficiency resulted in a ~50% reduction in CSR (Fig. 2d; Supplementary information, Fig. S2c), with no obvious impact in naive splenic B cell number, proliferation, AID expression, Ch gene transcription, or genome-wide transcription (Supplementary information, Fig. S2d–k, and Table S3). Correspondingly, the serum IgG/IgA levels were significantly decreased in Ercc6l2−/− mice (Supplementary information, Fig. S2l). In germinal centers of immunized mice, AID-induced somatic hypermutation (SHM) was unaffected (Supplementary information, Fig. S2m). This is reminiscent to the phenotype reported for DSBR/NHEJ-deficient germinal center B cells.31 These results demonstrate that ERCC6L2 facilitates antibody isotype switching.
During CSR, AID deaminates cytosines at switch region DNA. BER and MMR enzymes convert the uracils into DSBs, which are then processed by NHEJ factors during the recombination step.31 To test whether ERCC6L2 functions upstream or downstream of DSB induction, we used Cas9 to create breaks at IgH switch regions in the absence of AID (Supplementary information, Table S2). Under these conditions, Cas9 can promote efficient CSR to IgG1 or IgA (a process dubbed Cas-CSR, Fig. 2e; Supplementary information, Fig. S3a). End-joining level of Cas9 breaks in the absence of ERCC6L2 was reduced to 70%–75% of that in isogenic control cells (Fig. 2e; Supplementary information, Fig. S3b). These results indicate that ERCC6L2 promotes general DSB end-joining. It is of note that the reduction of Cas-CSR in ERCC6L2 deficiency is less than that caused by Lig4 deficiency (Fig. 2e), while ERCC6L2 and Lig4 deficiencies had comparable effects on AID-initiated CSR (Fig. 2c). In the Cas-CSR assay, the expression of Cas9 was well controlled by the co-transfected mCherry+ control cells in the same transfection reaction (Supplementary information, Fig. S3). The levels of Cas9-generated break levels among difference genotypes cannot be quantitatively revealed by the current technology. However, our observations are reminiscent of the fact that ERCC6L2 deficient cells showed less sensitivity to IR- or chemical-induced DSBs comparing to isogenic Lig4 deficient cells (Fig. 1f). Thus, it is unlikely that ERCC6L2 affected Cas9 cutting efficiency. The comparison of AID-CSR and Cas-CSR suggests ERCC6L2 might have an additional role in physiological CSR besides end-joining.
C-terminal half of ERCC6L2 leads its catalytic activity to DNA damage sites
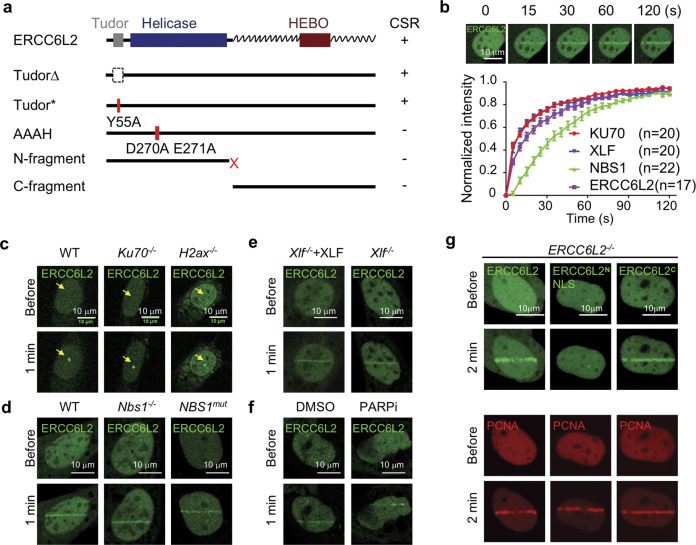
ERCC6L2 contains three conserved domains: a Tudor domain, an ATPase/Helicase domain and a conserved HEBO domain of unknown function (Fig. 3a). Bioinformatic analysis revealed that except for the HEBO domain, the C-terminal half of ERCC6L2 is less conserved and contains intrinsically disordered sequence by bioinformatic prediction (Supplementary information, Fig. S4a). We found that Tudor domain mutants can fully support CSR in Ercc6l2−/− cells (Fig. 3a; Supplementary information, Fig. S4b). However, ERCC6L2 N-terminal or C-terminal fragments failed to do this (Fig. 3a; Supplementary information, Fig. S4b). Furthermore, the helicase catalytic-dead (DEAH > AAAH) mutant did not promote CSR (Fig. 3a; Supplementary information, Fig. S4b), indicating that ERCC6L2’s predicted catalytic activity is required for DNA end-joining. Consistent with this idea, ERCC6L2 helicase catalytic-dead mutant and various frame-shift mutants (containing N-terminal fragments only) were also identified in BMF patients (Supplementary information, Fig. S4c).
Fig. 3. ERCC6L2 quickly engages in DNA damage repair.
a Summary of ERCC6L2 mutants in CSR. ERCC6L2 domain architecture is illustrated on top. The zigzag line indicates predicted internal disordered sequence. The CSR levels in ERCC6L2 mutant-complemented B cells are summarized at right. +: wild-type level, −: empty vector control level. b Recruitment of indicated proteins to microirradiation damaged sites in U2OS cells. Upper panel, a representative time-lapse view of GFP-ERCC6L2 recruitment. Lower panel, normalized fluorescent intensity of indicated proteins at DNA damage sites. The intensity was normalized to maximal intensity value at 120 s. Data are represented as mean ± SEM. Observed cell numbers (n) are listed. The curves of KU70 and XLF recruitment are highly overlapped. c Representative images of ERCC6L2 recruitment in MEFs of indicated genotypes. DNA damage sites (dot) are pointed. d Representative images of ERCC6L2 recruitment in MEFs (left and middle) or human fibroblast (right) of indicated genotypes. e, f Representative images of ERCC6L2 recruitment with/without XLF (e) or in presence of 10 μM PARP inhibitor Veliparib (f). g Representative images of ERCC6L2 fragments recruitment in ERCC6L2−/− U2OS cells. GFP-ERCC6L2, GFP-ERCC6L2N-NLS or GFP-ERCC6L2C fusion protein was expressed in ERCC6L2−/− U2OS cells. The recruitment of mCherry-PCNA was assayed at the same time as a control for DNA damage.
To shed further light on ERCC6L2 protein domains, we fused the full-length protein or its mutants to GFP and studies the dynamics in cells exposed to laser microirradiation. We found that GFP-ERCC6L2 is recruited to micro-irradiated sites within seconds of DNA damage, with similar kinetics to that of Ku70 and XLF (Fig. 3b; Supplementary information, Fig. S4d). This recruitment was observed in more than 90% of cells, and was independent of KU70, H2AX, NBS1, XLF or PARPs (Fig. 3c–f; Supplementary information, Fig. S4e). The ERCC6L2 C-terminal half fragment was sufficient to drive nuclear localization and recruitment to damaged sites (Fig. 3g), while the N terminal fragment could not be efficiently transported into the nucleus (Supplementary information, Fig. S4f). Remarkably, fusing ERCC6L2N to a nuclear localization signal peptide did not rescue DNA damage foci formation (Fig. 3g). These results indicate that the C-terminal domains recruits ERCC6L2 catalytic activity to damaged chromatin.
ERCC6L2 interacts with end-joining factors
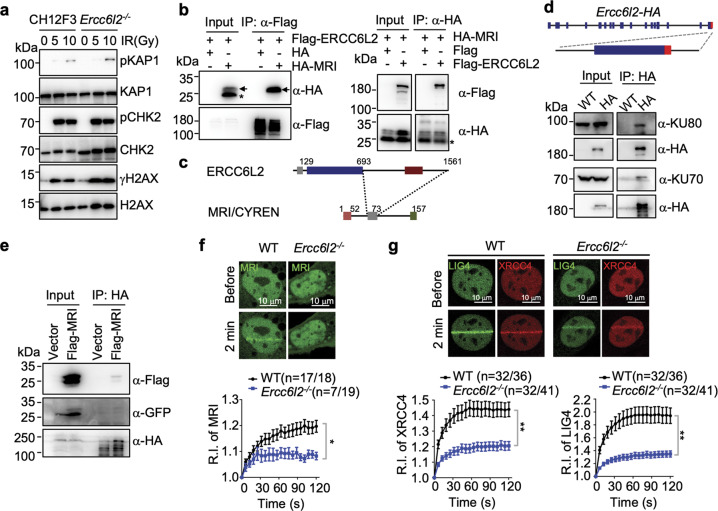
ERCC6L2 has been suggested to be an early DSB response factor.27 However, IR-induced phosphorylation of ATM substrates H2AX, CHK2 and KAP1 was unaffected in Ercc6l2−/− cells (Fig. 4a). Instead, we found that ectopically overexpressed ERCC6L2 co-immunoprecipitated with several NHEJ subunits in HEK293T cells in a DNA-independent manner (Supplementary information, Fig. S5a, b). This has been previously observed by Immunoprecipitation-Mass Spectrometry (IP-MS) analysis,35 although we cannot conclude whether it is a direct or indirect interaction. Interestingly, consistent with IP-MS analysis and genome-wide yeast two-hybrid report,11,36 we observed a DNA-independent interaction between ERCC6L2 and MRI/CYREN (Fig. 4b). This interaction was mediated by ERCC6L2 C-terminus and a conserved motif in the middle of MRI/CYREN protein (Fig. 4c; Supplementary information, Fig. S5c, d).
Fig. 4. ERCC6L2 is a component of end-joining machinery.
a Phosphorylation of ATM substrates was examined by western blot. Indicated CH12F3 cell lines were treated with IR at different dosages, and representative blots are showed from three repeats. b Co-immunoprecipitation (Co-IP) of ERCC6L2 and MRI/CYREN in HEK293T cells. The panels show western blots with anti-Flag or anti-HA antibodies of total extract (input) and IP samples. Arrows indicate bands of HA-MRI. Asterisk marks an unknown form of HA-MRI that does not interact with ERCC6L2. c Schematic illustration of ERCC6L2-MRI interaction domains. d Inputs and anti-HA-immunoprecipitated samples from CSR-activated B cells were assayed for NHEJ subunits. WT: B cells from wild-type mice; HA: B cells from Ercc6l2-HA knock-in mice. Knock-in strategy of Ercc6l2-HA mice is showed at top. e Inputs and anti-HA-immunoprecipitated samples from Ercc6l2-HA knock-in CSR-activated B cells with retroviral-expressed Flag-tagged MRI. Flag-MRI-IRES-GFP was ectopically expressed in CSR-activated B cells with an empty retroviral vector (IRES-GFP) as control. IRES internal ribosome entry site. f Recruitment of MRI to DNA damage site in WT and ERCC6L2-deficient MEFs. Upper, total numbers of micro-irradiated cells are listed alone with numbers of cells showed detectable MRI accumulation. Percentage of cells with MRI accumulation are showed in parenthesis. Exampled images of GFP-MRI recruitment are showed at lower. Relative intensity (R.I.) of GFP-MRI at DNA damages are calculated. g Recruitment of XRCC4/LIG4 to DNA damage site in WT and ERCC6L2-deficient MEFs. GFP-LIG4 and mCherry-XRCC4 were co-expressed in cells, and the panel is illustrated as in (f). Data are represented as mean ± SEM in (f) and (g). A t-test was applied as described in the “Materials and Methods” section. **p < 0.01; *p < 0.05.
Despite extensive testing, we were unable to obtain a workable anti-ERCC6L2 antibody or serum and currently no commercial anti-ERCC6L2 antibody is available to detect the endogenous protein. To confirm the protein interaction in vivo, we generated Ercc6l2-HA knock-in mice (Fig. 4d, top), in which the HA-tag was fused to the last exon of Ercc6l2. We detected high levels of ERCC6L2-HA protein in CSR-activated B cells (Supplementary information, Fig. S5e), where it co-immunoprecipitated with KU (Fig. 4d) and an ectopically-expressed Flag-tagged MRI (Fig. 4e). Together, these findings indicate that ERCC6L2 is associated with other NHEJ subunits.
Next, we examined how the physical interaction might contribute to ERCC6L2’s function in end-joining. Similar to Ku70, MRI is not required for ERCC6L2 recruitment to DNA damage sites (Supplementary information, Fig. S6a). We then checked the recruitment kinetics of NHEJ factors in ERCC6L2−/− U2OS cells, and found KU70/80, PAXX and XLF to be unaffected while the recruitment of XRCC4 was slightly decreased (Supplementary information, Fig. S6b, c). To further investigate this, we generated Ercc6l2−/− MEFs. Compared to controls, there were fewer KO cells showing MRI or XRCC4/LIG4 recruitment to micro-irradiated sites, and those that did, showed a significant decrease in MRI or XRCC4/LIG4 signals at foci (Fig. 4f, g; Supplementary information, Fig. S6d). The overall expression levels of MRI, XRCC4 or LIG4 protein were unaffected by ERCC6L2 deficiency, based on total fluorescent intensity (Supplementary information, Fig. S6e). The recruitment of XLF was slightly affected in Ercc6l2−/− MEFs (Supplementary information, Fig. S6f, p = 0.07). The results indicate that the presence of ERCC6L2 at DSB sites could facilitate ligase complex loading.
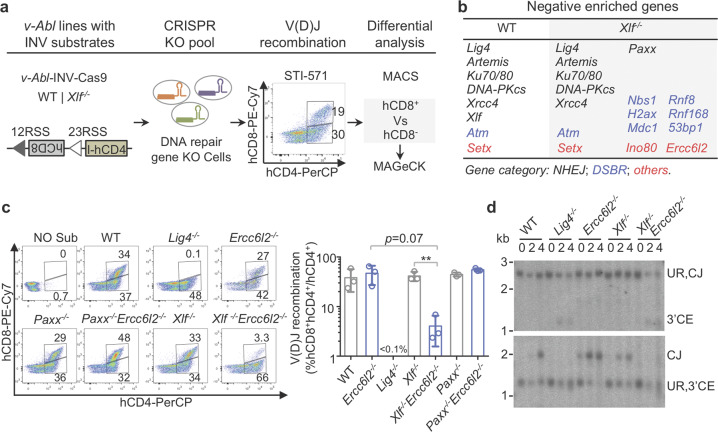
ERCC6L2 is required for V(D)J recombination in sensitized XLF-deficient cells
Core NHEJ factors are absolutely required for V(D)J recombination.37 Thus, we examined the lymphocyte development in ERCC6L2 deficient mice to check the V(D)J recombination efficiency (Supplementary information, Fig. S7a). Distribution of pro-B (CD43+B220+IgM−), pre-B (CD43−B220+IgM−), immature B (IgM+B220low) and recirculating B (IgM+B220hi) cells in Ercc6l2−/− bone marrow was similar to that of WT. Although the total cell numbers in the Ercc6l2−/− thymus were ~50% of that in WT, the distribution of CD4−CD8−, CD4+CD8+, CD4+CD8− and CD4−CD8+ Ercc6l2−/− T cells were comparable to that of WT (Supplementary information, Fig. S7a). The lymphocyte development in ERCC6L2 deficient mice is different from that in core NHEJ factor deficiency.37 Previous studies have uncovered a functional redundancy between XLF and ATM-dependent DSB response factors, or between XLF and other non-essential NHEJ factors including PAXX and MRI.11,38–44 Therefore, we hypothesized that ERCC6L2 may play a role in V(D)J recombination that is masked by other NHEJ factors, possibly XLF. To test this idea, we performed a focused CRISPR screen in WT and Xlf−/− v-Abl lines with a modified pMX-INV substrate (Fig. 5a; Supplementary information, Fig. S7b, c). DNA repair genes required for recombination were identified as negatively enriched in the recombined hCD8+ population (see Materials and Methods). As expected, core NHEJ factors and Artemis were identified in both genotypes (Fig. 5b). Also, as previously reported,39–44 XLF was functionally redundant with PAXX, 53BP1, and H2AX. The assay however identified new XLF redundant factors, including NBS1, MDC1, RNF8, RNF168, INO80, and importantly ERCC6L2 (Fig. 5b). Of note, Mri was not included in our original focused CRISPR sgRNA library (Supplementary information, Table S1), so Mri did not show up here.
Fig. 5. ERCC6L2 and XLF are functionally redundant in DNA recombination.
a Schematic illustration of V(D)J recombination screening procedure. Representative flow cytometry plot of chromosomal V(D)J recombination assay is showed. b Negatively enriched genes are showed for indicated genotypes. c Deficiencies of both ERCC6L2 and XLF cannot support efficient V(D)J recombination in a chromosomal V(D)J recombination assay. Representative flow cytometry plots of samples collected at Day 4 after STI-stimulation are showed at left and data from three replicates are summarized at right with mean ± SEM. Two-tail unpaired t-test was performed. **p < 0.01. d Southern blot analysis of INV substrate rearrangement products and intermediates. Genomic DNA was digested by Nde I/Nhe I (upper) or Nco I/Nhe I (lower), respectively. Unrearranged substrate (UR), coding end (CE) intermediates and coding join (CJ) are labeled.
The functional redundancy between ERCC6L2 and XLF was confirmed by chromosomal V(D)J recombination assays in Ercc6l2−/−Xlf−/− and isogenic v-Abl lines (Fig. 5c; Supplementary information, Fig. S7d). At the same time, there was no obvious redundancy in Ercc6l2−/−Paxx−/− cells. Similar to Lig4−/−, Ercc6l2−/−Xlf−/− cells showed a near complete block in V(D)J recombination (Fig. 5c; Supplementary information, Fig. S7d). This defect could be rescued by ectopic expression of ERCC6L2 but not ERCC6L2 N-terminal fragment (Supplementary information, Fig. S7e). Absence of coding joins and a smear signal of coding ends were also noticed in Ercc6l2−/−Xlf−/− cells (Fig. 5d), reminiscent of extensive resection of unprocessed DNA ends. In summary, ERCC6L2 and XLF play functional-redundant roles in the repair of DNA ends during V(D)J recombination.
ERCC6L2 controls the orientation of DNA recombination
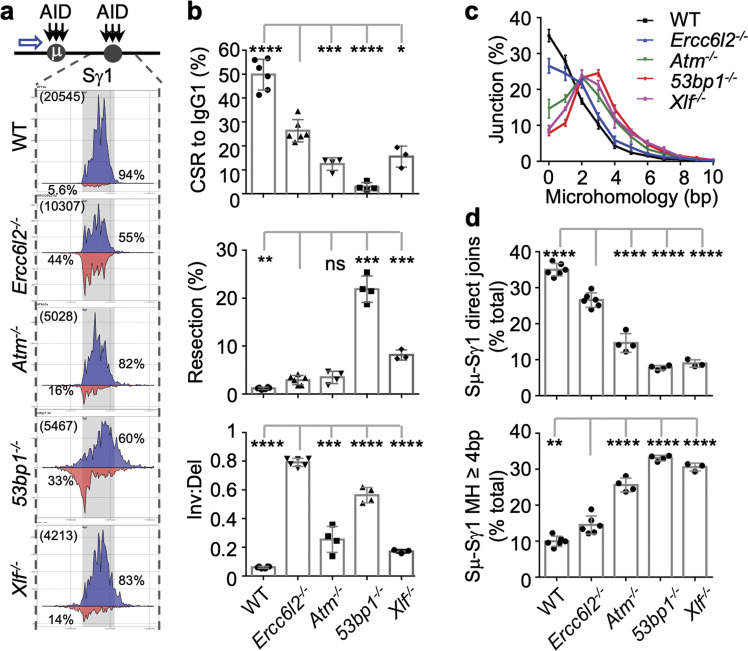
Our experiments have so far conclusively established ERCC6L2 promoting DSB end-joining in conjunction with XLF, primarily during programmed recombination in lymphocytes. The precise molecular mechanism however is unclear. We thus applied high-throughput genome-wide translocation sequencing (HTGTS), which can simultaneously quantify end resection, microhomology (MH) usage and orientation of DNA end joining,4 to explore whether CSR junctions are improperly joined in Ercc6l2−/− cells. As bait, we used AID-mediated breaks at Sμ (AIDSμ), which are joined to Sγ1/Sε in B cells undergoing IgM to IgG1/IgE switching (Fig. 6a; Supplementary information, Fig. S8a). Remarkably, Ercc6l2−/− B cells displayed a profound defect in orientation-specific joining (Fig. 6a, b; Supplementary information, Fig. S8a, b). Compared to controls, where inter-S recombination occurs predominantly by deletion (>90%), in Ercc6l2−/− cells deletions and inversions were present at nearly equal frequencies (55% vs 44%, Fig. 6a). In 53bp1−/− cells, which were previously found defective in directional CSR repair,4 the ratio was 60% vs 33%, whereas in both Atm−/− and Xlf−/−cells this ratio was around 80% vs 15% (Fig. 6a).
Fig. 6. ERCC6L2 determines orientation-specific recombination.
a Distribution of S junctions in CSR-activated B cells. Top, AID targeting (black arrow) in Sμ and Sγ1, and HTGTS bait site (blue arrow) are depicted. Bottom, linear distribution of pooled junctions along Sγ1 and flanking regions recovered from LPS/IL4-stimulated B cells. Gray boxes indicate Sγ1. Blue, junctions orientated from left to right; Red, Junctions orientated from right to left. Total Sμ–Sγ1 junction number (bracketed) and percentages of inversional/deletional junctions in the region are labeled. b CSR to IgG1, percentage of resection, and inversional/deletional joining ratio are plotted from top to bottom for the indicated B cells. Data were from 3 to 6 mice. c Microhomology usage of Sμ–Sγ1 DNA rearrangement junctions in the indicated activated B cells. Percentage of junctions with different length of microhomology are plotted. d Percentages of direct joins of 5′ Sμ to Sγ1 (upper panel) and percentage of junctions with 4 bp or longer MH in Sγ1 (lower panel) in activated B cells with indicated genotypes. Data are represented as mean ± SD in (b, c, d). Two-tail unpaired t-test was performed for (b) and (d). Data from Ercc6l2 knockout are compared with those from other genotypes. ****p < 0.0001, ***p < 0.001, **p < 0.01; *p < 0.05, ns: p > 0.05.
In ERCC6L2 deficiency, the resection levels at S regions were slightly higher than wild type and similar to those in ATM deficiency, but significantly lower than that in 53BP1 or XLF deficiency (Fig. 6b). The much mild increase of resection levels observed in ERCC6L2 deficiency distinguished ERCC6L2 from 53BP1 in DSB repair or directional DSB repair. Similarly, the MH usage analysis also revealed a mild but significantly decreased direct joining and increased joining with longer MH in CSR junctions of ERCC6L2 deficiency comparing to WT (Fig. 6c). However, the MH usage bias in ERCC6L2 deficiency was less pronounced than in ATM, 53BP1 or XLF deficiencies18 (Fig. 6d). Thus, although ERCC6L2 deficiency shows similar trends of more resection and increased longer MH usage in CSR-junctions as NHEJ/DSBR deficiencies, orientation-specific joining did not correlate with the extend of end-resection or MH usage. This suggests that ERCC6L2 plays a much more robust and unique role in directional end joining during CSR.
To bypass the B cell development defect of core-NHEJ factor deficiencies and quickly access the directional CSR, we deleted the non-productive IgH allele in CH12F3 cells as previous described18 (Supplementary information, Fig. S8c, named CH12-NCDel cells) and perform HTGTS assay with endogenous AIDSμ baits. Similar results were obtained in this system, and moreover, we found MRI did not affect the inversion/direction ratio (Supplementary information, Fig. S8d, e). These results therefore demonstrate that ERCC6L2 has a unique role in orientation-specific class switch recombination of antibody genes.
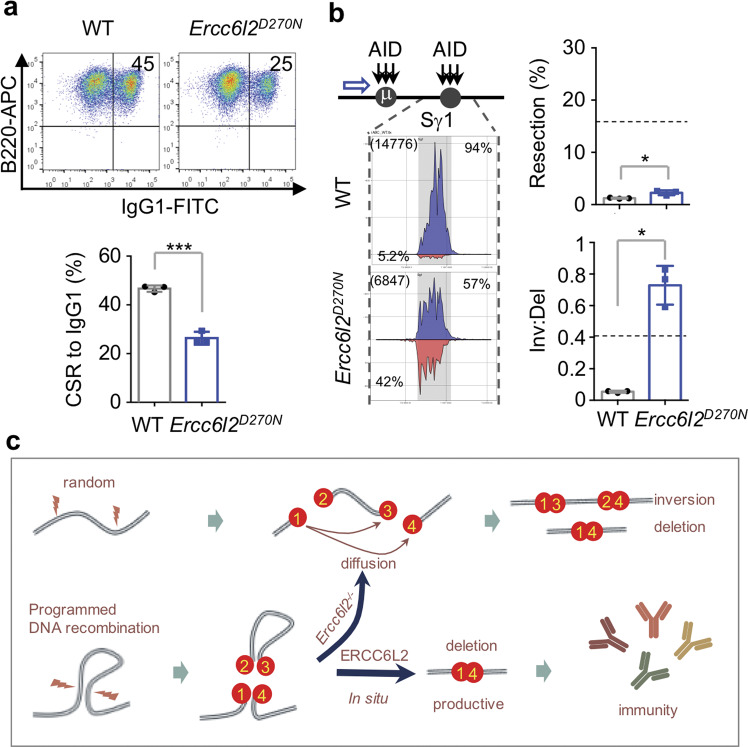
ERCC6L2 activity is required for directional end-joining
The catalytic activity of ERCC6L2 is required for optimal CSR (Fig. 3a), suggesting the catalytic activity is required for its functions in DSB end-joining. To further examine ERCC6L2 catalytic activity in mediating directional end-joining during CSR, we generated an Ercc6l2D270N mouse line with a BMF patient-derived mutation at DEAH-box helicase catalytic site (Supplementary information, Fig. S9a). Normal numbers of splenic mature B cells were obtained from the knockin mice (Supplementary information, Fig. S9b). Upon cytokine stimulation, Ercc6l2D270N B cells switched at a lower frequency comparing to WT B cells (Fig. 7a and Supplementary information, Fig. S9c). The ~50% reduction of CSR level was comparable to the CSR reduction in Ercc6l2 KO B cells (Fig. 2d). We then performed HTGTS to access the CSR junctions, and found that catalytic-dead ERCC6L2 phenocopied the Ercc6l2 knockout in end-resection and directional end-joining (Fig. 7b). In Ercc6l2D270N B cells, deletions and inversions were present at nearly equal frequencies (57% vs 42%, Fig. 7b) during CSR. We conclude that ERCC6L2’s catalytic activity is required for deletional repair during CSR.
Fig. 7. ERCC6L2 activity is required for directional end-joining during CSR.
a CSR levels to IgG1 in CSR-activated WT and Ercc6l2D270N B cells at Day 4 upon LPS/IL4 stimulation. b Distribution of Sμ–Sγ1 junctions at Sγ1, end-resection and inversion/deletion ratio of CSR junctions in WT and Ercc6l2D270N B cells. Dashed lines indicate the levels in 53bp1−/− B cells, which were assayed at the same time. c A working model to explain roles of ERCC6L2, see “Discussion” for details. Data are represented as mean ± SD and two-tail unpaired t-test was performed in (a, b). ***p < 0.001, *p < 0.05.
Discussion
CRISPR-chemical screens dissect the function of DNA repair factors
In this study, we used a comprehensive CRISPR and chemical screening approach to define a core DNA repair genetic network. In our 414X36 screen matrix, functionally-related genes or chemicals clustered together, offering new insights into gene function and how DNA damage-causing agents impact cells. Even though our focused CRISPR screening could not cover the whole genome, it has the advantage of better signal-to-noise ratios which can easily pinpoint regulators involved in subtle mechanisms. Our focused approach successfully identified factors that impact recombination by ~50%. Focused CRISPR screens can also be used as a reverse genetic tool to dissect genetic interactions, as we demonstrated with V(D)J recombination. Although our studies focused on the role of ERCC6L2 in NHEJ, it is worth noting that the assay revealed new functions of many other factors, including BAZ1B, a bromodomain-containing protein involved in H2AX phosphorylation.45 Moreover, our data provided important hints on combinatorial cancer treatments, since many of the DNA damage reagents tested are widely used in the clinic.
A working model of ERCC6L2 function
During immune diversification, programmed DNA breaks are processed into productive rearrangements within constrained topological microenvironments or TADs. We have shown that ERCC6L2 regulates how DNA ends are joined in a spatially oriented manner during repair (Fig. 7c). During CSR, transcribed S regions are synapsed and recruit AID deamination activity, which in turn engages BER and MMR enzymes that convert uracils into staggered DNA breaks with G-rich repetitive sequences.3 Several chromatin features appear to facilitate this activity, including R-loops,46 nascent G-rich RNA,47 and paused transcription complexes.48 These features, along with tightly packed nucleosomes, might slow down or interfere with end-joining.
In this context, ERCC6 family members are believed to function as nucleosome remodelers or DNA translocase.49–51 It is therefore tantalizing to speculate that ERCC6L2 removes nucleosomes or other protein/nucleotides near DNA lesions, probably to facilitate XRCC4/Lig4 sliding towards DNA ends.52 This function might ensure rapid in situ ligation of DSBs in the pre-assembled chromatin subdomains. Another non-mutually exclusive model is that AID-generated stagger ends within S regions could be mis-aligned and ERCC6L2 activity could detach the mis-aligned ends, leading to the dissociation of the intervening sequence from the CSR centre.15 Thus, ERCC6L2 could be directly involved in the spatiotemporal formation of IgH chromatin architecture,15 which is of great interest for further investigation. These possibilities suggest a model (Fig. 7c) on how ERCC6L2 facilitates deletional CSR of antibody genes. Considering that ERCC6L2 is the latest evolved member of the ERCC6 family, we speculate that it might have evolved to promote efficient end-joining under more complicated and specialized settings, such as in lymphocytes of higher organisms.
DNA damage in ERCC6L2-linked pathologies
Mutations of ERCC6L2 and LIG4 were previously identified in BMF patients, where DNA damage may originate from the same source. In Fanconi anemia, a subtype of BMF, ICLs generated by reactive aldehydes appear to be the major source of DNA damage in HSCs and other progenitor cells.53 However, the DNA lesions underlying ERCC6L2- and NHEJ-pathologies might be of different origin, as cells isolated from such patients are typically not hyper-sensitive to ICL-inducing agents.27 Our DNA repair genetic screen provided some hints in this regard, in which it showed that NHEJ and ERCC6L2 knockouts are markedly sensitive to TOP2 inhibitors. TOP2B-induced lesions are known to accumulate genome-wide at chromosome loop anchors,14,54 and at promoters of key genes during neuronal stimulation.55 Notably, some ERCC6L2-mutated BMF patients also show neurological dysfunctions.26,27,30 Whether TOP2B-induced breaks are causal to ERCC6L2- and NHEJ-pathologies represents an interesting future line of research.
Materials and methods
Mice
Ercc6l2 and Xlf knockout mouse lines were constructed through zygote injection of CRISPR/Cas9 constructs. Ercc6l2-HA knock-in mouse line was constructed by oocyte injection of androgenetic haploid embryonic stem cells harboring an Ercc6l2-HA allele obtained with homologous recombination (HR). Ercc6l2D270N mouse line was constructed by zygote injection of CRISPR/Cas9 constructs and a single-strand HR template. Guide RNA sequences are listed in Supplementary information, Table S2. 53bp1−/−56 and Atm−/−57 mouse lines have been described previously. All animal experiments were performed under protocols approved by the Institutional Animal Care and Use Committee of Shanghai Institute of Biochemistry and Cell Biology.
Cell lines
Cell lines used in this study were listed in the Supplementary information, Table S2. Parental B-lineaged CH12F3 cell line and isogenic Lig4−/−32 cell line, parental Abelson virus-transformed (v-Abl) mouse pro-B cell line containing the Eμ-Bcl2 transgene43 have been described previously. CH12F3 and its derived isogenic cells were cultured with RPIM1640 (10-040-CV; Corning), β-Mercaptoethanol (M6250-500ML; Sigma-Aldrich), Penicillin-Streptomycin-Glutamine (10378016; Thermo Fisher Scientific), plus 10% FBS (FCS500; ExCell Bio), and v-Abl cells were cultured with RPIM1640 (10-040-CV, Corning), β-Mercaptoethanol (M6250-500ML; Sigma-Aldrich), Penicillin-Streptomycin-Glutamine (10378016; Thermo Fisher Scientific), Sodium Pyruvate (11360670; Thermo Fisher Scientific), MEM Non-Essential Amino Acids Solution (11140050; Thermo Fisher Scientific), HEPES(15630080; Thermo Fisher Scientific), plus 10% FBS (35-053-CM; Corning). MEF, HEK293T and U2OS cells were cultured with DMEM (10-013-CV, Corning), Penicillin-Streptomycin-Glutamine (10378016; Thermo Fisher Scientific), plus 10% FBS (FSP500, ExCell Bio).
All cell lines are negative for mycoplasma contamination.
Primers, plasmids and antibodies
Primers, plasmids and antibodies used in this study were listed in the Supplementary information, Table S2.
Focused CRISPR screenings and data analysis
DNA repair-related CRISPR library design
DNA repair genes were picked based on gene ontology within AmiGO 2 database (http://amigo.geneontology.org/amigo). We also chose several CSR related genes from literature serving as potential CSR positive controls, and genes affecting cell viability (essential genes) and non-essential genes serving as CRISPR screening controls. Genes are listed in Supplementary information, Table S1. Guide RNAs were designed with CRISPR-FOCUS.58 Oligo pool was synthesized by Synbio Technologies (Suzhou) and cloned into lentiGuide-Puro by using EasyGeno Assembly kit (VI201; TIANGEN). Pooled sgRNA viruses were used to infect CH12F3 or v-Abl cell lines stably-expressing Cas9 protein. Resulting cell pools were selected with Puromycin for 7 days (Day −6 to Day 0) allowing efficient KO of targeted genes, before subjected to chemical treatment, CSR or V(D)J recombination assay.
Chemical treatment in CH12F3 cells
Inhibitory concentration (IC) was determined for each chemical in B-lineaged CH12F3 cells (Supplementary information, Table S1). Gene knockout cells were cultured with DNA damage chemicals at IC20 for 12 days (from Day 0 to Day 12) before harvested for genomic DNA purification. DMSO at a concentration of 0.1% was included as a control.
Enrichment of B cell sub-populations
For CSR screening, IgA+ cells were labeled with Biotin-conjugated anti-IgA antibody (407003; Biolegend), which were further enriched with anti-biotin microbeads (130-090-485; Miltenyi). Three replicates were performed for CSR screening of each genotype. For V(D)J recombination screening, v-Abl cells were stimulated by STI-571 for 4 days, hCD8+hCD4+ and hCD8−hCD4+ populations were enriched with anti-hCD4 (130-045-101; Miltenyi) and anti-hCD8 microbeads (130-045-201; Miltenyi). Two repeats were performed for V(D)J recombination screening of each genotype.
Library preparation and data analysis
The sgRNA sequences were PCR-amplified according to previous reported method,59 and subjected to Illumina sequencing. The raw reads were trimmed with fastx_trimmer in FASTX-Toolkit, and sgRNA sequences were counted by MAGeCK.19 Clonal batch difference was controlled by using a batch-removal sub-module in MAGeCK-Flute, and enriched genes were retrieved with MAGeCK test subcommand.
Cluster of chemical treatment result
We performed hierarchical clustering for both drugs and genes by R programming language. First, we used dist function to compute Euclidean distances for drugs or genes by using z-score computed by MAGeCK-MLE submodule. Then the distance matrix was passed to hclust function for clustering using ward.D method.
Exclude cell fitness genes and identify CSR/V(D)J recombination factors
We found that cell fitness genes which affect cell viability or growth rate often were retrieved in different CRISPR screening assays as false positive hits. Thus, we identified the cell fitness genes in DNA repair gene list by comparing recovered sgRNAs at day 7 after puromycin selection and sgRNAs in viral vector. The list was named as “Genes_Affecting_Cellular_Fitness” in Supplementary information, Table S1. After retrieving the enriched genes from CSR or V(D)J recombination screenings, cell fitness genes were first removed from the result. A false discovery rate (FDR) < 0.2 was further applied.
Chemical sensitivity assay
Cells were plated at a concentration of 6 × 10E4 cells/ml with indicated chemicals or treated with indicated doses of X-ray or UV. After 48 h, cell viability was measured with a Cell Counting Kit-8 assay (K1018; APExBio). The survival data were fit to a mixed-effects model using lmer function from the R package ‘lme4’, with “dosage” and “genotype” as fixed-effects parameters, and “cell viability data from each repeat” as the random effect parameter. The significance of the fixed effects parameters is obtained by the t-test.
Antibody class switch recombination assays
AID-initiated CSR assay
CH12F3 cells were stimulated with α-CD40 (16-0402-86; eBioscience), IL4 (CK15; Novoprotein) plus TGF-β (CA59; Novoprotein), and CSR to IgA were monitored at day 2 and 3. Splenic naive B cells were purified and cultured as previous described.60 CSR to IgG1 or IgG3 was monitored at day 3 and 4.
Cas9-initiated CSR assay
For CRISPR/Cas9-initiated CSR (Cas-CSR) in CH12F3 cells, test cells were mixed with control CH12F3-mCherry cells at a ratio of 3:1, sgRNAs targeting up- and down-stream S regions were transfected into the mixture via electroporation. The CH12F3-mCherry controls for electroporation decreased technical variations among each transfection. CSR level to other Ig in KO cells was first normalized to the CSR level of CH12F3-mCherry cells which was transfected in the same cuvette, and the relative CSR level was defined as a ratio of the CSR levels in KO and parental CH12F3 cells as below:
KO CSR level at transfection #1:
a = (KOIgG+/KOAll) / (mCherry#1IgG+/mCherry#1All)
WT CSR level at transfection #2:
b = (WTIgG+/WTAll) / (mCherry#2IgG+/mCherry#2All)
Relative Cas-CSR level = a/b.
It is of note that in the Cas-CSR assay, the Cas9-generated break levels among difference genotypes cannot be quantitatively revealed by the current technology. Thus, decreased Cas-CSR level could also be resulted from the low Cas9 cutting efficiency in a specific genotype.
Somatic hypermutation assay
Peyer’s Patch GC B cells (B220+PNAhi) were sorted from indicated mice. JH4 and Jκ5 introns were PCR amplified as previously reported61,62 and the PCR products were further tagged with illumine P5 and P7 index primers and subjected to Illumina HiSeq. Data were analyzed as previously described.63
Chromosomal V(D)J recombination assay
The v-Abl cells were infected with INV-invert-hCD8a cassette and enriched by using anti-hCD4 microbeads (130-045-101; Miltenyi). Cells which already underwent V(D)J recombination were removed with hCD8 microbeads (130-045-201; Miltenyi) at each experiment. Cells were stimulated with STI-571 for 48 h and 96 h, and then subjected to flow cytometry and/or southern blot. Southern blot was performed as previously described.39 For each sample, 10 μg genomic DNA was digested by Nde I/Nhe I and Nco I/Nhe I, respectively. A 900 bp Hind III/Nhe I fragment of hCD4 sequence was used as the probe.
Lymphocyte development
Flow cytometry was applied to assay lymphocyte development. Bone marrow and lymphocyte cellularity were calculated by using a hemocytometer. In brief, bone marrow cells were isolated by flushing or by crushing the long bones with a mortar and pestle in Ca2+ and Mg2+ free HBSS with 2% heat-inactivated serum. Spleen and thymus cells were obtained by crushing the tissue between two glass slides. The cells were filtered through a 70 μm nylon mesh before staining.
Gene deletion and complementation in cell line
For gene deletion in cell lines, pairs of sgRNAs were designed with SSC programe64 and selected based on the published gene knockout strategy or gene function domains. A GFP-expressing plasmid and pX330-based CRISPR/Cas9 plasmids were co-transfected into cells. Twenty-four hours after transfection, GFP-high cells were sorted with BD FCAS Aria III and plated into single clones in 96-well plates. Individual clones were genotyped by PCR (Supplementary information, Table S2) and positive clones were further confirmed by western blot or RT-qPCR. Despite many attempting, we were unable to get an anti-ERCC6L2 antibody or serum to detect the endogenous ERCC6L2 protein. Thus, the knockout of Ercc6l2 was confirmed by PCR-genotyping and RNA-Seq. ERCC6L2 plasmids were constructed by PCR with total cDNA templates from either mouse activated-B cells or human HEK293T cells. The gene was cloned into a lentiviral vector. The resulting lentiviral particles were used to generate Ercc6l2 or Ercc6l2-mutant cell lines. Empty vector (EV) was used as control. The Ercc6l2 mutant genes express at similar level as the WT gene in the experimental conditions.
Laser micro-irradiation
Laser microirradiation was performed as described.39 Briefly, U2OS cells were plated on 35 mm diameter glass-bottom plates (D35-20-1-N, Cellvis) at 1 × 10E5 cells/ml. On the next day, 1 μg GFP-GENE fusion plasmid with 0.05 μg mCherry-PCNA plasmid were transfected into cells. For MEFs, 10 μg GFP-GENE fusion plasmid was transfected into cells by electroporation (VCA-1001, Lonza Nucleofector 2b) following the manufacture’s instruction. The next day after transfection, cells were incubated with 10 μM 5-BrdU (HY-15910; MedChemExpress) overnight and exposed to 10 μg/ml Hoechst 33342 (C1022; Beyotime Technology) for 10 min just before irradiation. Laser microirradiation was performed with Nikon A1 confocal microscope and a 405 nm laser with 100% energy. The fluorescence data were fit to a mixed-effects model using lmer function from the R package ‘lme4’, with “acquired time” and “genotype” as fixed-effects parameters, and “data from each observed cell” as the random effect parameter. The significance of the fixed effects parameters is obtained by the t-test.
Immunoprecipitation
Co-transfection of GFP tagged NHEJ subunits and 3xFlag-ERCC6L2 or HA-ERCC6L2 in HEK293T cells was performed using Lipofectamine 2000 according to the manufacturer’s instruction. After 48 h, cells were washed, scraped and lysed for 30 min in lysis buffer (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 2.5 mM MgCl2, 10% Glycerol, 0.5% NP-40), supplemented with Complete protease inhibitor mix (4693132001, Roche), 25 U/ml Benzonase (E8263, Sigma-Aldrich) and 20 μg/ml Ethidium Bromide. Lysates were centrifuged at 16,000 g for 10 min at 4 °C, and supernatants were incubated with 15 μl anti-GFP nanobody beads (KTSM1301, Shenzhen KT Life technology), anti-Flag conjugated agarose beads (M20018, Abmart) or anti-HA conjugated agarose beads (M20013, Abmart) for 1 h at 4 °C. Beads were washed 3 times with wash buffer (10 mM Tris-HCl pH 7.4, 150 mM NaCl, 2.5 mM MgCl2, 10% Glycerol, 0.1% NP-40) and boiled in 20 μl 2 × SDS buffer. Proteins were analyzed by immunoblotting. For immunoprecipitation from activated splenic B cells, cells were stimulated with LPS/IL4 for 3 days, collected, washed and processed as above.
HTGTS
HTGTS was performed as previously reported.65 In primary B cell CSR assay, S region rearrangements were cloned from endogenous AID-initiated Sγ1 breaks with 5′-RED-Iμ primer.4 The HTGTS cloning primers are listed in Supplementary information, Table S2. S junction and resection ratios were plotted and calculated as previously described.4
Transcription analyses
RNA-seq and data analysis
Prepare RNA-Seq library
Total RNA was isolated using Trizol reagents (15596018; Life Technologies) and was treated with DNase I (M6101; Promega). RNA-seq libraries were prepared using KAPA stranded RNA-Seq kits with RiboErase (HMR) (KR1142; KAPA) following the manufacturer’s instructions. Then these libraries were sequenced with Illumina HiSeq at Geneseeq company. About ~25 million of 2 × 150 bp paired reads were retrieved for each library. Two biological replicates of mouse naive and CSR-activated B cells were performed for each genotype. Two repeats were performed in parental CH12F3 cell line along with two Ercc6l2−/− clones. One repeat was performed in parental U2OS cell line along with its isogenic Ercc6l2−/− cell line.
Genome-wide analysis
The raw reads were first cleaned with cutadapt (v. 1.14) cut adapter sequences and mapped to mouse or human rRNA reference using bowtie2 (v. 2.2.6) to filter out rRNA reads. Then the filtered reads were mapped to mouse (mm10) or human (hg38) reference genome with STAR (v. 2.5.3a) using default parameters. HTseq (v. 0.9.1) and DESeq2 (v. 1.14.1) were used for differential gene expression analysis. Package bamCoverage from deeptools (v. 2.5.3) was used to generate bigwig files from bam files and IGV was used for downstream visualization.
PRO-seq and data analysis
Prepare PRO-Seq library
PRO-Seq was performed according to previously published protocol.66 For primary B cells, WT and Ercc6l2−/− B cells were harvested at 60 h after CSR-activation, and three biological replicates of WT B cells and four biological replicates of Ercc6l2−/− B cells were subjected to PRO-Seq.
Data analysis
PRO-Seq data were first cut to remove adapter sequences with cutadapt (v. 1.14), then mapped to mouse reference genome (mm10) by bowtie2 (v. 2.2.6). Transcribed regions were defined as previously described. Only coordinate of the last base at 3′ end of each read was exacted for pausing index calculation. Pausing index was defined as previously described. Briefly, we calculate the ratio of mean counts between −30 bp to 300 bp relative to the TSS and the remaining length of the gene for NCBI RefSeq genes longer than 1 kb.
Quantification and statistical analysis
Statistical analyses were performed by using the R (Version 3.5.1, R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org) or GraphPad Prism (version 8.0.1). The number of replicates, statistical test procedures are indicated in the figure legends.
Supplementary information
Acknowledgements
We thank Drs Frederick Alt, Klaus Rajewsky, Tasuku Honjo, Kefei Yu, Guoliang Xu, Ronggui Hu, Yin Jing, Lan Wang, Jumin Zhou, Matthew Weitzman, Ming-Han Tong, Jun Huang and Yu Zhang for providing reagents, and Drs Jin-Qiu Zhou and Bin-Bing Zhou for critical reading of the manuscript, and Dr Daniel Durocher for communicating unpublished observations. We thank the Genome Tagging Project (GTP) Center, SIBCB, CAS for generating Ercc6l2-HA mouse model. This work was supported by National Key R&D Program of China (2017YFA0506700, 2017YFC0908500), Strategic Priority Research Program of Chinese Academy of Sciences (XDB19030000), and National Natural Science Foundation of China (81622022, 31670929, 31800766). X.L. is supported by China Postdoctoral Science Foundation Grants (2018M632180, 2019T120363). GTP center is supported by a Shanghai Municipal Commission for Science and Technology Grant (17411954900).
Author contributions
F.L.M. conceived of the experiments. F.L.M., X.L., T.L., Y.S. and P.D. analyzed the data and prepared figures, with contributions from W.Z., B.J.L., X.Z. and S.Z. Experiments were performed by X.L., T.L., Y.S., P.D., M.H., B.J.L., D.Y. and L.D.L. W.Z., Q.W., T.X. and X.S.L. contributed to CRISPR screen design and data analyses. B.O.Z., J.D., L.S.Y. and J.H. provided essential materials and/or analysis tools. F.L.M., R.C., X.S.L., T.L., and Y.S. wrote the paper.
Data availability
All sequencing data generated in this study, including CRISPR screening, RNA-Seq, Pro-Seq, SHM amplicon-seq and HTGTS data, are deposited in the NCBI Sequence Read Archive (SRA accession: PRJNA624711). All other data are available from the authors on request.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Xiaojing Liu, Tingting Liu, Yafang Shang, Pengfei Dai
Senior Authors: X Shirley Liu and Fei-Long Meng
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0328-3.
References
- 1.Alt FW, Zhang Y, Meng FL, Guo C, Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu. Rev. Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 3.Casellas R, et al. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 2016;16:164–176. doi: 10.1038/nri.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong J, et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature. 2015;525:134–139. doi: 10.1038/nature14970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, et al. Chromosomal loop domains direct the recombination of antigen receptor genes. Cell. 2015;163:947–959. doi: 10.1016/j.cell.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 9.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnoult N, et al. Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature. 2017;549:548–552. doi: 10.1038/nature24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung PJ, et al. MRI is a DNA damage response adaptor during classical non-homologous end joining. Mol. Cell. 2018;71:332–342. doi: 10.1016/j.molcel.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochi T, et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature. 2019;573:600–604. doi: 10.1038/s41586-019-1547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vian L, et al. The energetics and physiological impact of cohesin extrusion. Cell. 2018;175:292–294. doi: 10.1016/j.cell.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature. 2019;575:385–389. doi: 10.1038/s41586-019-1723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeap LS, Meng FL. Cis- and trans-factors affecting AID targeting and mutagenic outcomes in antibody diversification. Adv. Immunol. 2019;141:51–103. doi: 10.1016/bs.ai.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panchakshari RA, et al. DNA double-strand break response factors influence end-joining features of IgH class switch and general translocation junctions. Proc. Natl. Acad. Sci. USA. 2018;115:762–767. doi: 10.1073/pnas.1719988115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murai J, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez R, et al. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012;8:301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benitez-Bribiesca L, Sanchez-Suarez P. Oxidative damage, bleomycin, and gamma radiation induce different types of DNA strand breaks in normal lymphocytes and thymocytes. A comet assay study. Ann. N. Y. Acad. Sci. 1999;887:133–149. doi: 10.1111/j.1749-6632.1999.tb07928.x. [DOI] [PubMed] [Google Scholar]
- 25.Lundin C, et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tummala H, et al. ERCC6L2 mutations link a distinct bone-marrow-failure syndrome to DNA repair and mitochondrial function. Am. J. Hum. Genet. 2014;94:246–256. doi: 10.1016/j.ajhg.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang S, et al. A nonsense mutation in the DNA repair factor Hebo causes mild bone marrow failure and microcephaly. J. Exp. Med. 2016;213:1011–1028. doi: 10.1084/jem.20151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bluteau O, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131:717–732. doi: 10.1182/blood-2017-09-806489. [DOI] [PubMed] [Google Scholar]
- 29.Jarviaho T, et al. Bone marrow failure syndrome caused by homozygous frameshift mutation in the ERCC6L2 gene. Clin. Genet. 2018;93:392–395. doi: 10.1111/cge.13125. [DOI] [PubMed] [Google Scholar]
- 30.Shabanova I, et al. ERCC6L2-associated inherited bone marrow failure syndrome. Mol. Genet. Genom. Med. 2018;6:463–468. doi: 10.1002/mgg3.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Methot SP, Di Noia JM. Molecular mechanisms of somatic hypermutation and class switch recombination. Adv. Immunol. 2017;133:37–87. doi: 10.1016/bs.ai.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells. J. Exp. Med. 2008;205:2745–2753. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonoda E, et al. B cell development under the condition of allelic inclusion. Immunity. 1997;6:225–233. doi: 10.1016/s1074-7613(00)80325-8. [DOI] [PubMed] [Google Scholar]
- 34.Pelanda R, et al. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- 35.Tummala H, et al. Genome instability is a consequence of transcription deficiency in patients with bone marrow failure harboring biallelic ERCC6L2 variants. Proc. Natl. Acad. Sci. USA. 2018;115:7777–7782. doi: 10.1073/pnas.1803275115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolland T, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu. Rev. Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar V, Alt FW, Oksenych V. Functional overlaps between XLF and the ATM-dependent DNA double strand break response. DNA Repair. 2014;16:11–22. doi: 10.1016/j.dnarep.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, Shao Z, Jiang W, Lee BJ, Zha S. PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nat. Commun. 2017;8:13816. doi: 10.1038/ncomms13816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hung PJ, et al. Deficiency of XLF and PAXX prevents DNA double-strand break repair by non-homologous end joining in lymphocytes. Cell Cycle. 2017;16:286–295. doi: 10.1080/15384101.2016.1253640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tadi SK, et al. PAXX is an accessory c-NHEJ factor that associates with Ku70 and has overlapping functions with XLF. Cell Rep. 2016;17:541–555. doi: 10.1016/j.celrep.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 42.Lescale C, et al. Specific roles of XRCC4 paralogs PAXX and XLF during V(D)J recombination. Cell Rep. 2016;16:2967–2979. doi: 10.1016/j.celrep.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar V, Alt FW, Frock RL. PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line. Proc. Natl Acad. Sci. USA. 2016;113:10619–10624. doi: 10.1073/pnas.1611882113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Balmus G, et al. Synthetic lethality between PAXX and XLF in mammalian development. Genes Dev. 2016;30:2152–2157. doi: 10.1101/gad.290510.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao A, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 47.Zheng S, et al. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biebricher A, et al. PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol. Cell. 2013;51:691–701. doi: 10.1016/j.molcel.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ke Y, et al. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J. 2011;30:3309–3321. doi: 10.1038/emboj.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu J, et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017;551:653–657. doi: 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brouwer I, et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature. 2016;535:566–569. doi: 10.1038/nature18643. [DOI] [PubMed] [Google Scholar]
- 53.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 54.Canela A, et al. Genome organization drives chromosome fragility. Cell. 2017;170:507–521. doi: 10.1016/j.cell.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madabhushi R, et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward IM, et al. 53BP1 is required for class switch recombination. J. Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zha S, et al. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cao Q, et al. CRISPR-FOCUS: a web server for designing focused CRISPR screening experiments. PLoS One. 2017;12:e0184281. doi: 10.1371/journal.pone.0184281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meng FL, et al. Convergent transcription atintragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jolly CJ, Klix N, Neuberger MS. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiang Y, Garrard WT. The downstream transcriptional enhancer, Ed, positively regulates mouse Ig kappa gene expression and somatic hypermutation. J. Immunol. 2008;180:6725–6732. doi: 10.4049/jimmunol.180.10.6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yeap LS, et al. Sequence-intrinsic mechanisms that target AID mutational outcomes on antibody genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu H, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu J, et al. Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc. 2016;11:853–871. doi: 10.1038/nprot.2016.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mahat DB, et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat. Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated in this study, including CRISPR screening, RNA-Seq, Pro-Seq, SHM amplicon-seq and HTGTS data, are deposited in the NCBI Sequence Read Archive (SRA accession: PRJNA624711). All other data are available from the authors on request.