Abstract
We determined the effect of sample size on performance of polygenic hazard score (PHS) models in prostate cancer. Age and genotypes were obtained for 40,861 men from the PRACTICAL consortium. The dataset included 201,590 SNPs per subject, and was split into training and testing sets. Established-SNP models considered 65 SNPs that had been previously associated with prostate cancer. Discovery-SNP models used stepwise selection to identify new SNPs. The performance of each PHS model was calculated for random sizes of the training set. The performance of a representative Established-SNP model was estimated for random sizes of the testing set. Mean HR98/50 (hazard ratio of top 2% to average in test set) of the Established-SNP model increased from 1.73 [95% CI: 1.69–1.77] to 2.41 [2.40–2.43] when the number of training samples was increased from 1 thousand to 30 thousand. Corresponding HR98/50 of the Discovery-SNP model increased from 1.05 [0.93–1.18] to 2.19 [2.16–2.23]. HR98/50 of a representative Established-SNP model using testing set sample sizes of 0.6 thousand and 6 thousand observations were 1.78 [1.70–1.85] and 1.73 [1.71–1.76], respectively. We estimate that a study population of 20 thousand men is required to develop Discovery-SNP PHS models while 10 thousand men should be sufficient for Established-SNP models.
Subject terms: Genetics research, Risk factors
Introduction
Polygenic risk models have been studied extensively for several diseases such as prostate cancer [1], breast cancer [2], type 2 diabetes [3], dementia [4], and atherosclerosis [5]. Polygenic scores in the context of survival models are a more recent advancement in the field, but have been garnering interest in the Alzheimer’s disease [6] and prostate cancer [7]. The steady increase in genetic testing [8, 9], both in public and clinical domains, suggests that survival models could be applied to new diseases. The largest obstacle to the development of these models is the large number of study subjects, often in the tens of thousands [8], which are required for robust training and testing.
Our aim was to quantify the effect of sample size on the performance of a polygenic survival model. This was explored through a specific disease condition that is expected to be representative, namely prostate cancer. We investigated two potential model development strategies. For the ‘Established-SNP’ model, we selected single-nucleotide polymorphisms (SNPs) that had previously been shown to be associated with prostate cancer, and estimated the coefficients for these SNPs in a Cox proportional hazards framework. For the ‘Discovery-SNP’ model, we implemented the SNP selection technique described by Seibert et al. [7] to identify SNPs in our genotyping data for inclusion in the Cox proportional hazards framework. The Established (EST) SNP and Discovery (DIS) SNP represent two strategies that researchers could employ to build a polygenic survival model. In order to simulate samples of different sizes, we randomly sampled our training and testing sets. The results of this work will help inform the design of future studies to develop polygenic survival models for other diseases.
Materials and methods
Training and testing set
As previously described [7], we obtained genotype and age data from 21 studies included in the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium. We analyzed data from 40,861 men consisting of 20,551 individuals with prostate cancer and 20,310 individuals without. For analysis, the age for each man was recorded as either their age at prostate cancer diagnosis (cases) or at interview (controls). Genotype data were restricted to SNPs with missing value rates <5%, resulting in 201,590 SNPs available for analysis. Missing calls were assigned the mean value for that SNP [7]. The genotype data had been assayed using a custom iCOGS chip (Illumina, San Diego, CA) the details for which are elaborated elsewhere [10]. The sample was split into training (34,444 men, consisting of 18,962 cases and 15,482 controls) and testing (6417 men consisting of 1589 cases and 4828 controls) sets. The testing set was selected using men who were enrolled in the Prostate testing for cancer and Treatment (ProtecT [11]) trial. ProtecT (ClinicalTrials.gov: NCT02044172) is a large, multicenter trial within the United Kingdom, which aims to investigate the effectiveness of treatments for localized prostate cancer. The ProtecT study group was chosen for testing as it represented a well-characterized group of individuals that had been used for measuring testing performance for our earlier work. The Data Availability Statement describing how readers can gain access to the PRACTICAL dataset is provided in the Supplementary information.
The present study used only de-identified data from the PRACTICAL consortium. All studies contributing data have the relevant Institutional Review Board approval in each country in accordance with the Declaration of Helsinki [12]. The details of each study set, including the consent and accrual process are previously published [12].
Established-SNP model
A list of 65 SNPs [13] was chosen to represent those on the iCOGS array that had been published as associated with prostate cancer. The coefficients of the SNPs within the EST-SNP model were then estimated using the “coxphfit” function in MATLAB (Mathworks, Natwick, MA). It should be noted that the 65 SNPs used were discovered, in large part, using the data presently defined as the test set. The effect allele for all 65 SNPs was defined as “A” to simplify analysis.
Discovery-SNP model
For every SNP, a trend test was used to check for associations between SNP count and the binary classification of individuals with or without prostate cancer. The SNP selection pool was then reduced to those whose trend test p value was less 1 × 10−6. In order of increasing p value, each SNP was tested in a multiple logistic regression model for association with the binary classification of men as with or without prostate cancer, after adjusting for age, six principal components based upon genetic ancestry, and previously selected SNPs. If the p value of the coefficient of the tested SNP was <1 × 10−6, it was selected for the final Cox proportional hazard model estimation. The coefficients of the selected SNP pool within the DIS-SNP model were estimated as previously described [7].
Polygenic hazard score (PHS)
The PHS for each of the EST-SNP and DIS-SNP models was calculated as the linear product of the coefficients of the SNPs used in the model and the corresponding patient genotype counts [6, 7].
PHS performance metrics
Several performance metrics for PHS models were investigated, and are described in Table 1. In each case, the PHS for each test subject was calculated as the dot product of SNP coefficients, either EST or DIS, and SNP counts. A Cox proportional hazards model was then fit using PHS as the sole predictor of age in the test set. The z-score and beta of this Cox proportional hazards model relate to how well PHS was associated with age within the test set. The hazard ratios were calculated as the exponential of the differences in predicted log-relative hazards of different groups within the test set. The groups were defined using centile cut-points for those controls within the training set whose age was <70 years. This list of performance metrics expands on those (z-score and HR98/50) that were used in our earlier work [7]. In addition, sample-weight performance metrics were estimated using a weighted Cox proportional hazard model [7, 14, 15] with PHS as the sole predictor of age in the test set. The weighting factor for the cases and controls were estimated using published prevalence data from the ProtecT randomized phase 3 trial [11].
Table 1.
Performance metrics used in the evaluation of polygenic hazard scores.
| Performance metric | Description |
|---|---|
| HR98/50 | Hazard ratio of the top 2% to the average (30–70%) in the test set |
| HR20/50 | Hazard ratio of the bottom 20% to the average (30–70%) in the test set |
| HR98/20 | Hazard ratio of the top 2% to the bottom 20% in the test set |
| HR80/20 | Hazard ratio of the top 20% to the bottom 20% in the test set |
| z-score | z-score of Cox proportional hazards model using PHS as a sole predictor of age in the test set |
| beta | Coefficient of PHS in a Cox proportional hazards model using PHS as a sole predictor of age in the test set |
Random sampling of training set
Random sampling of the training set was performed with replacement while ensuring equal proportions of men with and without prostate cancer. The training set was randomly sampled to include 1, 5, 10, 15, 20, 25, and 30 thousand total observations. Performance of the EST- and DIS-SNP models using random samples of the training data was measured in the entire test set.
A sub-analysis investigating the effect of the percentage of cases in the training set was conducted using the EST-SNP model with 5000 and 25,000 random samples of the training set. The results are presented in Supplementary Fig. 5.
Random sampling of the testing set
Random sampling of the testing set was performed with replacement while ensuring equal proportion of men with and without prostate cancer. The testing set was randomly sampled to include 0.5, 1, 2, 3, 4, 5, and 6 thousand total observations. Performance in the randomly sampled testing sets was performed using a representative EST-SNP model. The representative model was chosen as that whose parameters were estimated using a training sample size of 30 thousand total observations, and whose performance metrics were the shortest Euclidean distance to the average performance across all EST-SNP models using a training sample size of 30 thousand.
Results
Established- vs. Discovery-SNP model performance
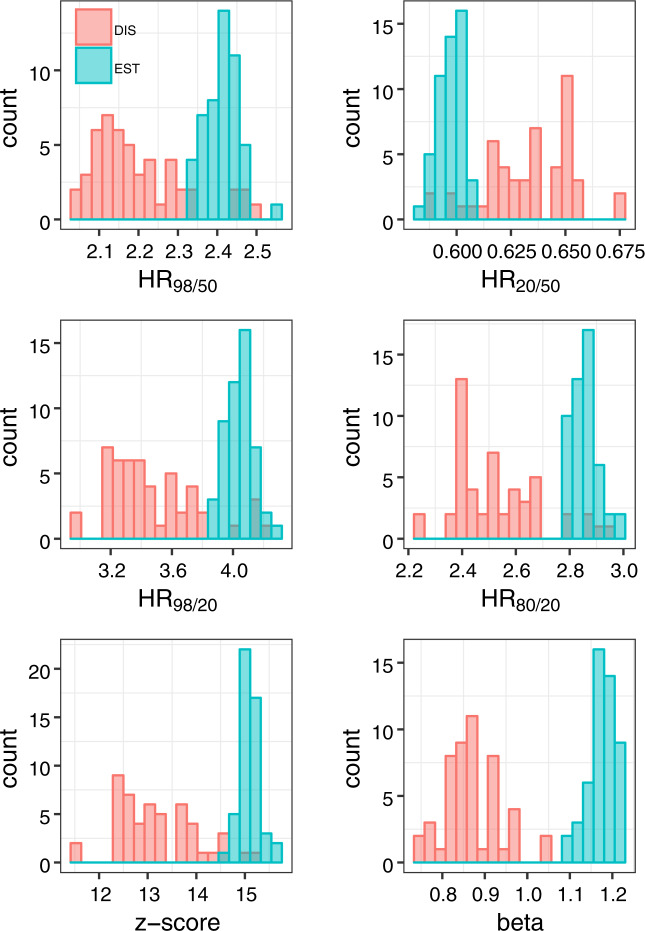
Histogram comparisons of performance metrics of EST- and DIS-SNP models are illustrated in Fig. 1. The performance metrics are shown for 50 random samplings of the training set using a sample size of 30 thousand total observations. Qualitatively, there appears to be more variability in performance metrics associated with the DIS process.
Fig. 1. Comparison of performance metrics between Established (EST) and Discovery (DIS) SNP models using 50 random samples of the training set using a sample size of 30 thousand.
There is more variability with the Discovery process. Established SNPs, though, were discovered using the data in the training set; this circularity is not accounted for in the present study, which focuses on sample size effects.
Coefficients of Established-SNP model
The mean coefficients for the 65 SNPs used in the EST-SNP model are plotted in Supplementary Fig. 1.
Effect of training set sample size on performance
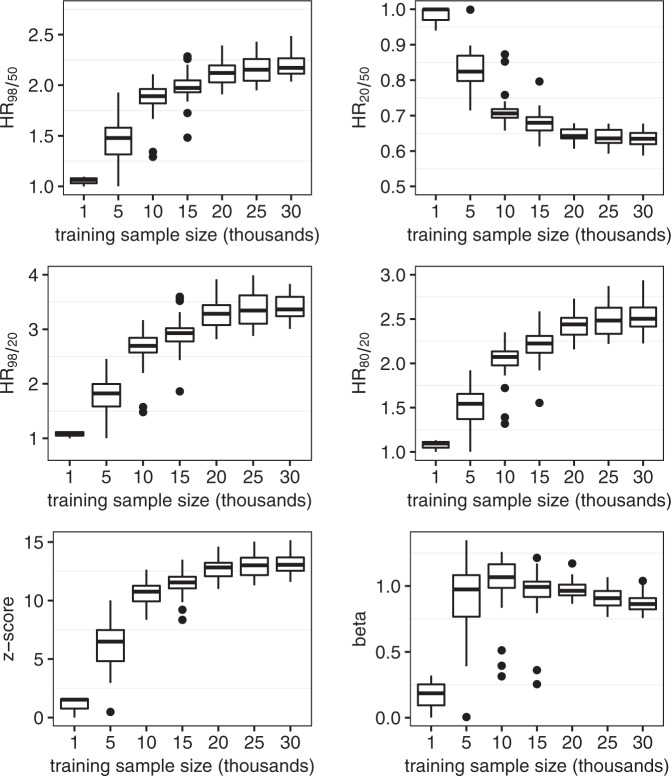
Box plots of the performance metrics of the EST-SNP and DIS-SNP models for random samples of the training set are shown in Figs. 2 and 3, respectively. The mean values of HR98/50, HR20/50, HR98/20, HR80/20, z-score, and beta using a random training sample of 1 thousand total observations in the EST-SNP model were 1.73 [95% CI: 1.69–1.76], 0.71 [0.71–0.73], 2.42 [2.35–2.50], 1.96 [1.92–2.01], 9.92 [9.57–10.28], and 0.45 [0.43–0.47], respectively. The corresponding values using a random training sample of 30 thousand total observations were 2.41 [95% CI: 2.40–2.43], 0.60 [0.60–0.60], 4.04 [4.02–4.07], 2.86 [2.84–2.87], 15.1 [15.04–15.16], and 1.18 [1.17–1.18], respectively.
Fig. 2. Performance metrics of Established-SNP model.
Box plots of performance metrics are shown for random samples of the training set using sample sizes of 1, 5, 10, 15, 20, 25, and 30 thousand total observations. Within each box plot, the horizontal line represents the median and the box extends from the 25th to 75th percentile.
Fig. 3. Performance metrics of the Discovery-SNP model.
Box plots of performance metrics are shown for random samples of the training set using sample sizes of 1, 5, 10, 15, 20, 25, and 30 thousand total observations. Within each box plot, the horizontal line represents the median and the box extends from the 25th to 75th percentile.
The mean values of HR98/50, HR20/50, HR98/20, HR80/20, z-score, and beta using a random training sample of 1 thousand total observations in the DIS-SNP model were 1.05 [0.93–1.18], 0.98 [0.89–1.07], 1.07 [0.91–1.24], 1.08 [0.91–1.24], 1.06 [−1.20 to 3.31], and 0.17 [−0.23 to 0.65], respectively. The corresponding performance values using a training sample size of 30 thousand observations were 2.20 [2.16–2.23], 1.60 [1.59–1.62], 3.47 [3.39–3.56], 2.53 [2.49–2.58], 13.19 [12.96–13.41], and 0.87 [0.85–0.89], respectively.
Box plots of the sample-weight corrected performance metrics for the EST-SNP and DIS-SNP model are shown in Supplementary Figs. 2 and 3, respectively. The trends observed in the sample-weight corrected performance metrics are identical to those observed in the raw, uncorrected metrics.
Effect of testing set sample size on performance
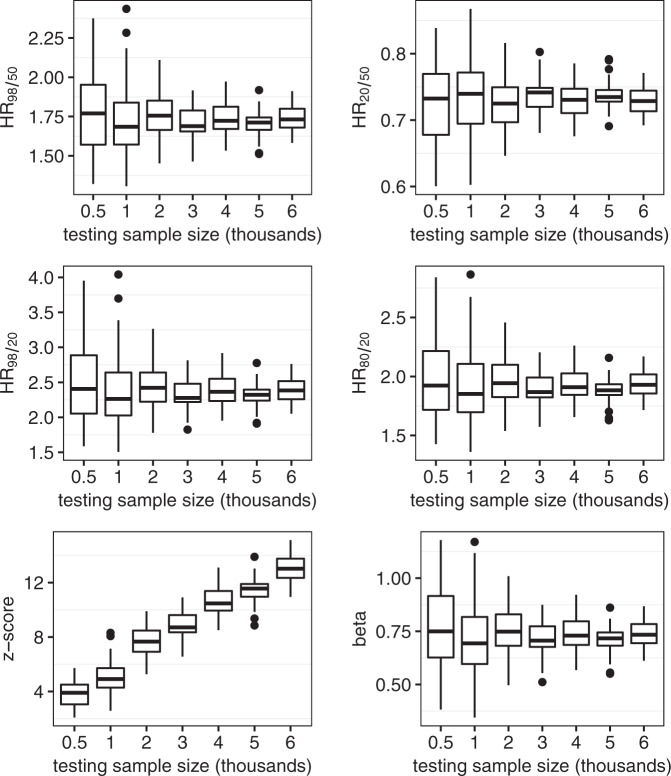
Box plots of the performance metrics of the representative EST-SNP model for random samples of the testing set are shown in Fig. 4. Box plots of the corresponding sample-weight corrected performance metrics are presented in Supplementary Fig. 4. The mean values of HR98/50, HR20/50, HR98/20, HR80/20, z-score, and beta using a random testing sample of 0.5 thousand total observations in the representative EST-SNP model were 1.78 [1.71–1.85], 0.73 [0.71–0.74], 2.50 [2.33–2.66], 1.99 [1.89–2.09], 3.82 [3.57–4.08], and 0.76 [0.70–0.82], respectively. The corresponding values using a testing sample of 6 thousand observations were: 1.73 [1.72–1.76], 0.73 [0.72–0.73], 2.39 [2.34–2.44], 1.93 [1.90–1.96], 13.07 [12.80–13.32], and 0.74 [0.72–0.76], respectively.
Fig. 4. Performance as a function of testing sample size.
Box plots of performance metrics of the representative Established-SNP model in random samples of the testing set from 0.5 to 6 thousand total observations.
Discussion
We identified several trends in the effect of training and testing sample size on the performance of PHS models in prostate cancer using SNP genetic variants. When using SNPs that had already been associated with prostate cancer risk, our analysis suggests that very little improvement in performance can be achieved once the training sets become larger than 10–15 thousand observations. When attempting to discover SNPs, a similar plateau in performance was observed from training sets larger than 20–25 thousand observations. Apart from z-scores, the performance metrics of the chosen Cox proportional hazards model did not vary with testing sample size. However, we did observe that the distribution of performance metrics narrows until a testing sample size of 3 to 4 thousand observations, after which the distribution remains relatively stable.
Our results may be used to inform researchers on the approximate number of subjects needed to develop PHS models using SNP counts. A dataset of 20 thousand observations may be the minimum needed to accurately estimate the PHS coefficients of SNPs that have been previously discovered in the setting of a logistic model. Such a dataset would allow for the accurate estimation of SNP coefficients as well as the testing of model performance in an independent holdout set. Based on our results, this number would have to be increased to roughly 30 thousand observations if the researchers intend on discovering the SNPs from scratch using the approach described here.
The PHS model developed by Desikan et al. [6] to estimate age-associated risk of Alzheimer’s disease used a training set with roughly 55,000 individuals. A similarly structured model developed by Seibert et al. [7] to guide screening for aggressive prostate cancer was developed with roughly 31,000 men. Studies such as these require large investments in time, money, and resources in order to acquire the genetic data needed for the analysis. The results of our analysis help elucidate that the minimum sample size needed to translate this technology to other diseases and processes may be lower than what has been used so far in previous studies. This seems to be particularly true if the researchers use SNPs that have already been discovered and validated as associated with the process of interest.
The results of this study must be considered in the context of its limitations. The list of EST-SNPs was previously selected from a larger dataset that included the sample patients used in the test set in the present study. As such, there is leakage of information from the test set to the development of the EST-SNP model. Therefore, the performance metrics of the EST-SNP model should not be directly compared with those of the DIS-SNP model, as the values of the former may be inflated.
In addition, we have chosen to focus on only two of countless possible model development schemes. The role of sample size in other development strategies—such as regularized Cox proportional models, parametric survival functions, or random survival forests—is yet to be explored. Finally, the analysis is limited to prostate cancer and to the SNPs available on the iCOGS array. Future studies to investigate the influence of additional SNPs, such as those on HapMap 3 or 1000 Genomes, on the performance of PHS models are underway at our institution.
In conclusion, we have studied the effect of sample size on the performance of PHS models to study the association between SNPs and the age at diagnosis of prostate cancer. We have determined that models require roughly 20 to 30 thousand samples before their performance would not be improved greatly by expansion of the training set. Using SNPs that have already been established in the literature may help reduce the number of training samples required to reach this performance plateau by almost 10 thousand samples.
Supplementary information
Acknowledgments
Funding
This study was funded in part by a grant from the United States National Institute of Health/National Institute of Biomedical Imaging and Bioengineering (#K08EB026503, TMS), United States Department of Defense (#W81XWH-13-1-0391, AMD, TMS), the Research Council of Norway (#223273, OAA), KG Jebsen Stiftelsen (OAA) and South East Norway Health Authority (OAA). Funding support for the PRACTICAL consortium, from which the data were obtained, is detailed in the Supporting information A2.
Compliance with ethical standards
Conflict of interest
TMS reports honoraria from Multimodal Imaging Services Corporation for imaging segmentation, honoraria from WebMD, Inc. for educational content, as well as a past research grant from Varian Medical Systems unrelated to this work. OAA reports speaker honoraria from Lundbeck and consultant for HealthLytix, Inc. OAA has a patent application # U.S. 20150356243 pending; AMD also applied for this patent application and assigned it to UC San Diego. AMD has additional disclosures outside the present work: founder, equity holder, and advisory board member for CorTechs Labs, Inc.; founder and equity holder in HealthLytix, Inc., advisory board member of Human Longevity, Inc.; recipient of nonfinancial research support from General Electric Healthcare.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Membership of The PRACTICAL Consortium is provided in the Supporting information.
Contributor Information
Roshan A. Karunamuni, Email: rakarunamuni@health.ucsd.edu
Tyler M. Seibert, Email: tseibert@health.ucsd.edu
Supplementary information
The online version of this article (10.1038/s41431-020-0664-2) contains supplementary material, which is available to authorized users.
References
- 1.Aly M, Wiklund F, Xu J, Isaacs WB, Eklund M, D’Amato M, et al. Polygenic risk score improves prostate cancer risk prediction: results from the Stockholm-1 cohort study. Eur Urol. 2011;60:21–8. doi: 10.1016/j.eururo.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machiela MJ, Chen C, Chanock SJ, Hunter DJ, Kraft P. Evaluation of polygenic risk scores for predicting breast and prostate cancer risk. Genet Epidemiol. 2011;35:506–14. doi: 10.1002/gepi.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vassy JL, Hivert MF, Porneala B, Dauriz M, Florez JC, Dupuis J, et al. Polygenic type 2 diabetes prediction at the limit of common variant detection. Diabetes. 2014;63:2172–82. doi: 10.2337/db13-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marden JR, Walter S, Tchetgen Tchetgen EJ, Kawachi I, Glymour MM. Validation of a polygenic risk score for dementia in black and white individuals. Brain Behav. 2014;4:687–97. doi: 10.1002/brb3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desikan RS, Fan CC, Wang Y, Schork AJ, Cabral HJ, Cupples LA, et al. Genetic assessment of age-associated Alzheimer disease risk: development and validation of a polygenic hazard score. PLoS Med. 2017;14:1–17.. doi: 10.1371/journal.pmed.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seibert TM, Fan CC, Wang Y, Zuber V, Karunamuni R, Parsons JK, et al. Polygenic hazard score to guide screening for aggressive prostate cancer: development and validation in large scale cohorts. BMJ. 2018;360:1–7. doi: 10.1136/bmj.j5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee N, Shi J, García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat Rev Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–90. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 10.Eeles RA, Olama AAAl, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–91. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane JA, Donovan JL, Davis M, Walsh E, Dedman D, Down L, et al. Active monitoring, radical prostatectomy, or radiotherapy for localised prostate cancer: study design and diagnostic and baseline results of the ProtecT randomised phase 3 trial. Lancet Oncol. 2014;15:1109–18. doi: 10.1016/S1470-2045(14)70361-4. [DOI] [PubMed] [Google Scholar]
- 12.Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, Ingles SA, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–61. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szulkin R, Whitington T, Eklund M, Aly M, Eeles RA, Easton D, et al. Prediction of individual genetic risk to prostate cancer using a polygenic score. Prostate. 2015;75:1467–74. doi: 10.1002/pros.23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minh-Phuong H-L, Chieh Fan C, Karunamuni R, Martinez ME, Eeles RA, Kote-Jarai Z, et al. Polygenic hazard score predicts aggressive and fatal prostate cancer in multi-ethnic populations. medRxiv. 2019. 10.1101/19012237.
- 15.Therneau TM, Li H. Computing the Cox Model for case cohort designs. Lifetime Data Anal. 1999;5:99–112. doi: 10.1023/A:1009691327335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.