Abstract
Evidence supports a possible role of BANK1 in innate immune signaling in B cells. In the present study, we investigated the interaction of BANK1 with two key mediators in interferon and inflammatory cytokine production, TRAF6 and MyD88. We revealed by coimmunoprecipitation (CoIP) analyses the binding of BANK1 with TRAF6 and MyD88, which were mediated by the BANK1 Toll/interleukin-1 receptor (TIR) domain. In addition, the natural BANK1–40C variant showed increased binding to MyD88. Next, we demonstrated in mouse splenic B cells that BANK1 colocalized with Toll-like receptor (TLR) 7 and TLR9 and that after stimulation with TLR7 and TLR9 agonists, the number of double-positive BANK1–TLR7, –TLR9, –TRAF6, and –MyD88 cells increased. Furthermore, we identified five TRAF6-binding motifs (BMs) in BANK1 and confirmed by point mutations and decoy peptide experiments that the C-terminal domain of BANK1-full-length (-FL) and the N-terminal domain of BANK1–Delta2 (-D2) are necessary for this binding. Functionally, we determined that the absence of the TIR domain in BANK1–D2 is important for its lysine (K)63-linked polyubiquitination and its ability to produce interleukin (IL)-8. Overall, our study describes a specific function of BANK1 in MyD88–TRAF6 innate immune signaling in B cells, clarifies functional differences between the two BANK1 isoforms and explains for the first time a functional link between autoimmune phenotypes including SLE and the naturally occurring BANK1–40C variant.
Keywords: autoimmunity, B cells, innate immune signaling, BANK1 isoforms, SLE
Subject terms: Mechanisms of disease, NF-kappaB, Systemic lupus erythematosus, Protein translocation, Interleukins
Introduction
Systemic lupus erythematosus (SLE) is a multisystemic and chronic disorder that is considered the prototype autoimmune disease. SLE is caused by a complex combination of genetic, epigenetic, and environmental factors that eventually result in abnormal pathologic immune responses.1 B cells are considered key components contributing to the complex pathophysiology of SLE.2
Under normal conditions, B cells are maintained in an anergic state by peripheral tolerance checkpoints.3 However, after chronic activation and/or in genetically predisposed individuals, self-reactive B cells may be released from anergy to produce autoantibodies and change their pattern of cytokine expression. Therefore, a better understanding of the molecular mechanisms in B cells that lead to autoimmunity is needed. Several candidate genes have been identified by genome-wide association studies (GWAS) to be associated with SLE and to predispose individuals to increased B-cell responsiveness.4,5 In addition, some B-cell depletion therapies have been developed, although with varying success.6–8 The exploration of new pathways and approaches to pursue more effective therapies is a main task for current research on autoimmunity.
Dysfunction of innate pathways has been increasingly associated with a breakdown in immune tolerance and the development of autoimmunity.9,10 Overactivation of nucleic acid-sensing Toll-like receptor (TLR) 7 and TLR9 by massive release of nuclear material in lupus patients is a clear example of this phenomenon.11 Both receptors are known to be extremely relevant for proinflammatory cytokine production in plasmacytoid dendritic cells; however, they are also expressed and play a key role in B cells.12,13 Tlr7 transgenic mice spontaneously develop a B cell-dependent SLE-like disease because of TLR7 overexpression.14 In addition, increased secretion of interleukin (IL)-6 was shown in another model to be orchestrated by TLR7, B-cell receptor (BCR) and interferon-type (IFN) I receptor signaling integration in B cells,15 highlighting once again the close relationship between B cells and innate pathways. Although more controversial, the role of TLR9 in autoimmunity has also been highlighted by a number of studies.16–19 Production of antidouble-stranded DNA and antichromatin autoantibodies in the MRL-lpr/lpr murine lupus model was specifically diminished by Tlr9 deficiency, although strikingly, no effect was observed on clinical outcomes or nephritis.20 Considering all of this, it is most likely that B cells and deregulation of their TLR7 and TLR9 pathways play a central role in the development of autoimmunity.
TLR7 and TLR9 signal propagation is strictly dependent on myeloid differentiation primary response 88 (MyD88),21 which is recruited upon activation via its Toll/interleukin-1 receptor (TIR) domain. MyD88 binds to IL-1R-associated kinase (IRAK) 4, thereby activating IRAK1 and IRAK2,22,23 which directly interact with tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6). As a result, TRAF6 undergoes lysine (K)63-linked polyubiquitination to further propagate the signal, leading to the activation of interferon regulatory factor 7 (IRF7), nuclear factor-kappa-B (NF-κB) and mitogen-activated protein kinases (MAPKs).24,25
The MyD88 adaptor has two homotypic interaction motifs: a death domain (DD)26 and a TIR domain.27 Although MyD88 has not been genetically associated with autoimmune diseases, some gain-of-function mutations in this gene were found in lymphomas or correlated with increased susceptibility to certain infections.28 Involvement in innate autoimmune processes is therefore not surprising because of its strategic position in the transduction cascade.29–31 TRAF6 in turn was first reported by GWAS analyses to be associated with rheumatoid arthritis (RA)32 and subsequently reported to be associated with SLE.33 TRAF6 is a member of the TRAF family and acts as an adaptor protein downstream of multiple receptor families.34,35 It is involved in the direct activation of NF-κB, MAPK, phosphoinositide 3-kinase (PI3K), the Akt/PKB pathway, and IRFs5 and 7. TRAF family members consist of an N-terminal Zn RING finger domain, a series of five Zn finger domains, a coiled-coil TRAF N-domain, and a member-specific C-terminal TRAF-C domain. In the case of TRAF6, the latter provides it with interaction specificity towards consensus P-X-E-X-X-(X)-aromatic/acidic motifs,36,37 which are found in interacting proteins such as IRAK1, IRAK2, and IRAK334,36,38; CD4039; or RANK.40 In addition to ubiquitinating targeted proteins, TRAF6 catalyzes its own site-specific polyubiquitination. This ubiquitination was found to be of the K63 type rather than the degradation-associated K48 type. This specific polyubiquitination is believed to promote protein complex formation and/or pathway activation and is therefore often used as readout of such signaling events in experimental setups.41 Finally, Traf6-deficient mice revealed that TRAF6 is required for proper B-cell maturation and functions at different levels.42
The B-cell scaffold protein with ankyrin repeats, BANK1, has been genetically associated through GWASs with SLE43 and other inflammatory diseases such as systemic sclerosis44 and RA.45,46 Four isoforms of BANK1 have been reported at the protein level, of which only isoforms 1, 3, and 4 were experimentally corroborated. Produced by alternative splicing, isoform 1 contains the canonical sequence, named full-length (FL) BANK1 (785 amino acids). The fourth isoform lacks exon 2, which encodes a putative conformational TIR domain, and is named Delta2 (D2) (652 amino acids).43,47 To our knowledge, no experimental data have confirmed the functionality of this homotypic interaction domain. Of note, higher expression of this shorter D2 isoform and lower expression of the FL form were linked to protection against SLE in a GWAS.43 In addition, the protective allele of one of the strongest disease-associated variants in BANK1 (rs10516487; A allele; 61H) correlated with poor splicing of exon 2, rendering lower amounts of the FL form.48
BANK1 was first proposed as a positive regulator of B cell signaling through the induction of calcium mobilization upon BCR stimulation of chicken B cells. Its expression was found to be confined to functional BCR-expressing B cells.49 In contrast, removal of Bank1 in knockout mice presented enhanced germinal center formation and IgM production in response to T-dependent antigens, supporting a regulatory role for BANK1 in B cell signaling. Further analyses in Bank1 and Cd40 double-knockout mice suggested that BANK1 negatively regulates CD40-induced Akt activation, thus preventing B cells from becoming hyperactive.50 Instead, a more recent study comparing BANK1 risk vs. nonrisk genotypes in human peripheral B cells has reported possible opposite findings. In this case, the decreased levels of Akt activation in subjects with BANK1 risk haplotypes presented an expanded memory B-cell compartment. Differences between mouse models and humans or between nonsynonymous variants and knockout systems may account for these inconsistencies across studies.51
The role of BANK1 in innate pathways has also been studied in mice and, unlike BCR-CD40 signaling, shows no controversy thus far.52,53 Experiments in Bank1−/− splenic B cells reported less IL-6 production after CpG induction and linked BANK1 to MAPK p38 phosphorylation and TLR9 signaling.52 BANK1 deficiency in the lupus Sle1.yaa mouse model, in which Tlr7 duplication plays a key role in disease development, also confirmed the role of BANK1 as an enhancer of TLR7 signaling. Purified splenic B cells from B6.Sle1.yaa.Bank1−/− mice showed reduced Ifnb, Ifna4, Irf7, Aicda, and Stat1 expression after TLR7 stimulation. These findings were accompanied by impaired STAT1-Tyr701 phosphorylation, reduced nuclear translocation of IRF7, and a significant reduction in serum autoantibodies in mice.53 However, the specific niche of BANK1 in the TLR7 and TLR9 pathways and the consequences of its autoimmunity-related genetic variability are still unclear.
The aim of this study was to explore candidate partners of BANK1 in innate immune pathways based on its interaction domains as well as the possible mechanisms of BANK1 signal transduction. We found that BANK1 isoforms harbor functional TRAF6-binding motifs (BM) and confirmed for the first time the functionality of the BANK1 TIR domain. BANK1 binding to the key components of TLR pathways, TRAF6 and MyD88, is mediated by these interaction domains. We further analyzed BANK1 isoforms and determined differences in their levels of interaction with MyD88, K63-linked polyubiquitination, and production of the cytokine IL-8, which is explained by the conformational TIR domain that is absent in BANK1–D2. In addition, we report new data on the naturally occurring rare variant BANK1–40C. Overall, our data support previous observations made in Bank1 knockout mice and provide new insights into the role of BANK1 in innate immune signaling in B cells, which is an essential task in the advance towards more effective and personalized therapies in patients suffering from autoimmune diseases such as SLE.
Results
BANK1 interacts and colocalizes with TRAF6 and MyD88
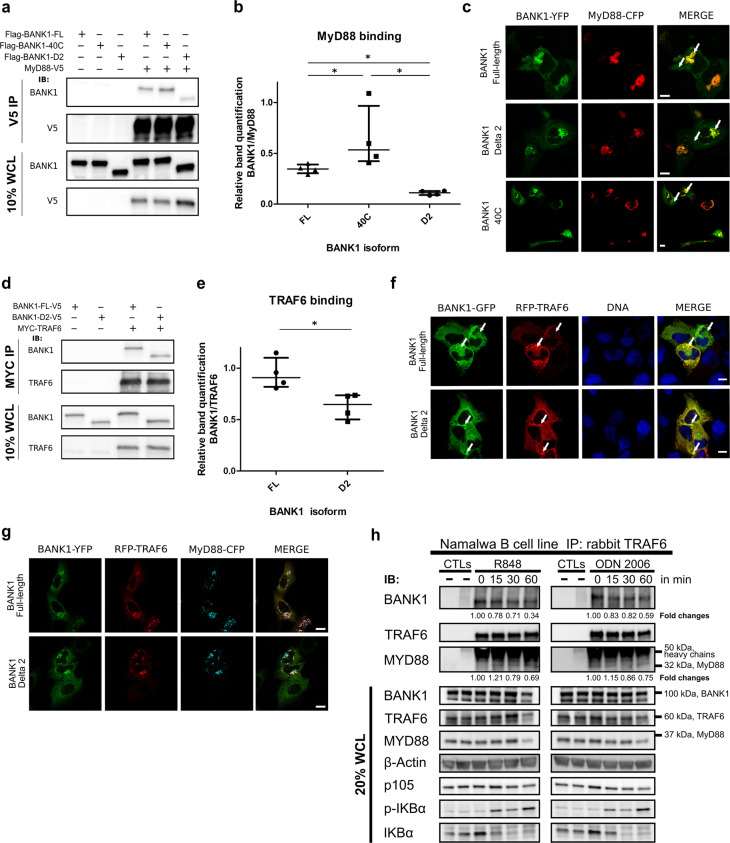
The TLR signaling adaptor MyD88 contains two interaction motifs: a death domain and a TIR domain, which are both involved in homotypic interactions.21 We therefore investigated by coimmunoprecipitation (CoIP) and colocalization studies the possible binding to BANK1 through its putative conformational TIR domain encoded by exon 2.47 Western blot analysis of immunoprecipitations conducted in nonstimulated HEK293 cells confirmed the interaction (Fig. 1a). Notably, the BANK1–D2 isoform lacking the TIR domain exhibited a sharp decrease in binding to MyD88 (mean decrease of approximately one-third) compared to BANK1–FL, suggesting that this protein motif considerably mediates the interaction (Fig. 1a, b). Consequently, we searched for BANK1 natural variants lying within exon 2 to test their possible effects on this interaction. SNP rs35978636 is the only described deleterious variant (as predicted by PolyPhen-2)54 with a frequency higher than 0.01 in the European population (dbSNP).55 It is represented by a substitution of a tryptophan for a cysteine in position 40 (40C) (Supplementary Fig. S1A, highlighted in red). Interestingly, this variant was found to be overrepresented within families showing several autoimmune phenotypes, including SLE.56 Notably, compared to BANK1–FL, the BANK1–40C mutant showed an average increase of one-third in the interaction with MyD88 (Fig. 1a, b). Studies by confocal microscopy in nonstimulated U2OS cells showed that expression of BANK1 alone resulted in a homogenous distribution throughout the cytoplasm with occasional small cytoplasmic aggregates, whereas MyD88 displayed various patterns (Supplementary Fig. S2A–D). However, when expressed together, MyD88 and all three BANK1 proteins predominantly colocalized in large aggregated structures (Fig. 1c, arrows).
Fig. 1.
BANK1 interacts and colocalizes with MyD88 and TRAF6. a Nonstimulated HEK293 cells transfected with V5-tagged MyD88 or empty vector (EV) together with Flag-tagged BANK1 constructs. Coimmunoprecipitations (CoIP) were carried out against the V5 epitope, and proteins were detected with the indicated antibodies. b Quantification with ImageJ of four independent experiments. Dots indicate BANK1 CoIP band intensity relative to the amount of MyD88 immunoprecipitated. Medians with interquartile ranges are represented. c BANK1-YFP vectors and MyD88-CFP were transfected into nonstimulated U2OS cells to study their subcellular localization and colocalization. Note that BANK1-YFP isoforms are shown in green and MyD88-CFP in red for a better appreciation of merged images. d Overexpression of Myc–TRAF6 in combination with V5-tagged BANK1–FL or BANK1–D2 in nonstimulated HEK293 cells and immunoprecipitation with an anti-MYC antibody. Immunoblotting with the indicated antibodies shows TRAF6 interaction with both isoforms of BANK1. e Quantification with ImageJ of four independent experiments as in b. Dots indicate BANK1 CoIP band intensity relative to the amount of TRAF6 immunoprecipitated. Medians with interquartile ranges are represented in the graph. f BANK1–GFP vectors for both isoforms and RFP-TRAF6 were transfected into nonstimulated U2OS cells. BANK1–FL and –D2 formed accumulations and defined small punctate structures in which both isoforms were highly colocalized with TRAF6 (arrows, MERGE). Nuclei were stained with DAPI. g BANK1–YFP isoforms were transfected with RFP-TRAF6 and MyD88-CFP vectors into nonstimulated U2OS cells. h Endogenous CoIP reactions in the B cell line Namalwa showed a TRAF6–BANK1–MyD88 interaction. TRAF6 was pulled down, and immunoblotting reactions were developed against BANK1, TRAF6, and MyD88. The image shown is from a single experiment that is representative of at least three separate experiments. Western blot bands from β-actin were densitometrically measured by ImageJ to determine the lane normalization factor for samples that were then used to normalize bands from CoIP reactions. Mean fold changes in the BANK1 and MyD88 interaction relative to TRAF6 over time of stimulation are shown, where time point 0 was taken as the control. See also Supplementary Fig. S4 for graphic presentations of the results and statistics. p105, p-IKBα, and IKBα immunoblots indicate successful stimulation of TLRs. The unpaired nonparametric Mann–Whitney t-test was applied *p < 0.05. All images were taken with a ×63 objective using a ZEISS confocal microscope. Scale bars = 10 µm
TRAF6 is known to form a complex with MyD88 and IRF7 after TLR7, TLR8, or TLR9 stimulation, triggering the production of IFN-alpha.57 This E3 ubiquitin ligase requires a specific consensus sequence in its interacting proteins represented by the motif P-X-E-(X)2-aromatic/acidic residue36 or P-X-E-(X)3-aromatic/acidic residue.37 We therefore searched the protein sequences of BANK1 isoforms for these BMs (Supplementary Fig. S1). Indeed, we found four putative TRAF6 BMs in the FL isoform (Supplementary Fig. S1A, underlined) and five TRAF6 BMs in the D2 short isoform of BANK1 (Supplementary Fig. S1B, underlined). A Wald statistical test revealed that these findings deviate significantly from those expected at random (p < 0.002 for both isoforms), highlighting the relevance of these motifs in BANK1. The fifth BM is formed by the absence of exon 2 in the D2 isoform (Supplementary Fig. S1A, exon 2 is highlighted in blue). This motif is not a full consensus TRAF6 BM because it contains aspartate (Asp) instead of glutamate (Glu) in the third position (Supplementary Fig. S1B). However, we considered it in our study for two reasons: (1) the high similarity between Asp and Glu in terms of protein structure conservation and (2) a mutation in CD40, a well-known target of TRAF6, consisting of a change from Asn to Asp, generated a motif with enhanced affinity for TRAF6.36 Moreover, this protein displays both TRAF6 BM consensus sequences (P-X-E-(X)2-aromatic/acidic residue or P-X-E-(X)3-aromatic/acidic residue). To determine whether TRAF6 can interact with both isoforms of BANK1, we performed transient transfections of Myc–TRAF6, BANK1–FL–V5, and BANK1–D2–V5 in nonstimulated HEK293 cells and immunoprecipitations with an anti-MYC antibody. We found that TRAF6 interacts with both isoforms of BANK1 (Fig. 1d), although we noticed a significant decrease of approximately one-third in binding to BANK1–D2 compared to BANK1–FL (Fig. 1d, e). The BANK1–40C mutant resulted in inconsistent binding to TRAF6 relative to BANK1–FL and was thus not quantified (data not shown). We then performed colocalization analyses of TRAF6 with the two BANK1 isoforms and the BANK1-40C mutant in nonstimulated U2OS cells. All three BANK1 proteins formed aggregate-like structures in which they sometimes colocalized with TRAF6 (Figs. 1f, arrows and S3A). Interestingly, we determined a significant decrease in colocalization between BANK1–D2 and TRAF6 compared to BANK1–FL, which corresponds to decreased binding to TRAF6 (Supplementary Fig. S3B).
The MyD88–TRAF6 interaction has been studied in the context of TLR signaling.57 We cotransfected both proteins into nonstimulated U2OS cells and observed that some cells displayed colocalization in punctate structures across the cytoplasm (Supplementary Fig. S3C, white arrows). Then, we studied the colocalization pattern of MyD88 and TRAF6 together with BANK1 proteins in nonstimulated U2OS cells. We found that all three proteins colocalized in most of the cells in large aggregated structures, suggesting that BANK1 participates as part of the MyD88–TRAF6-signaling complex (Figs. 1g and S3D).
Next, we tested whether BANK1, MyD88, and TRAF6 are also able to interact endogenously in the B cell line Namalwa by immunoprecipitating TRAF6 under nonstimulated conditions and after TLR7 (agonist R848) or TLR9 (agonist ODN 2006) engagement (Figs. 1h and S4). An interaction among TRAF6, BANK1, and MyD88 was already observed under nonstimulated conditions (Fig. 1h). We followed the interaction kinetics of BANK1 with TRAF6 upon R848 stimulation and determined that the interaction decreased gradually with time and was reduced by approximately two-thirds within an hour (0.34). The kinetics for cells following TLR9 induction also showed a decrease at 15 and 30 min and further declined by two-fifths (0.59) at 60 min. In contrast, TRAF6 interaction kinetics with MyD88 increased strongly upon stimulation with the TLR7 agonist R848 but then decreased slowly by one-third compared to the initial levels within an hour (0.69) (Fig. 1h). Namalwa cells stimulated with the TLR9 agonist ODN 2006 also showed a slight increase at 15 min of stimulation but then declined continually and reaching a value one-fourth lower than that in nonstimulated cells after an hour (0.75) (Fig. 1h). These results suggest that BANK1 is slowly released from the MyD88–TRAF6–BANK1 protein complex after stimulation. Immunoblots against p105, p-IκBα and IκBα, and ELISAs for IL-8 and TNF-α confirmed the activation of innate immune signaling (Figs. 1h and S5A).
BANK1, TRAF6, and MyD88 colocalize in mouse splenic B cells
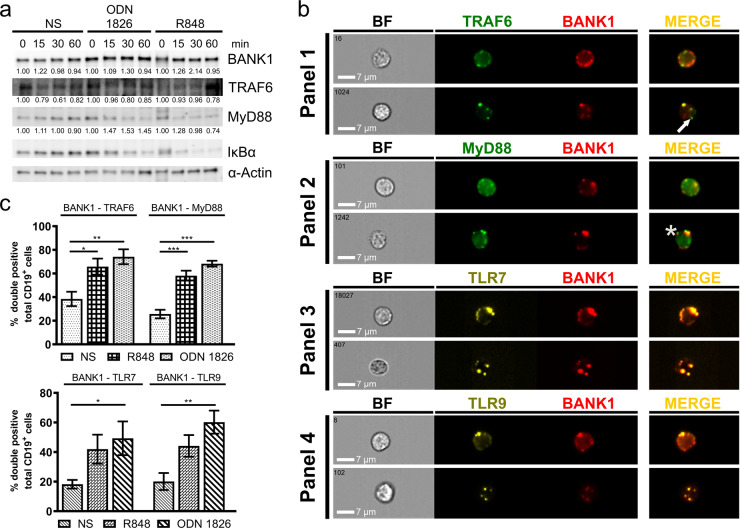
Previous studies in Bank1−/− mice suggest a regulatory role for BANK1 in B-cell signaling,50 and a study in purified splenic B cells from B6.Sle1.yaa.Bank1−/− lupus mice showed impaired activation of the pathway after TLR7 stimulation.53 Thus, we asked whether our results obtained in vitro recapitulate the in vivo situation in mouse B cells, if BANK1 colocalizes with TLRs 7 and 9 and if so, whether colocalization is changed upon B cell stimulation by TLR7 and 9 engagement. First, we measured the expression levels of the BANK1, TRAF6, and MyD88 proteins in purified CD19+ mouse cells (Fig. 2a) and found no substantial changes in the expression of BANK1 and TRAF6 in ODN 1826- and R848-stimulated cells compared to that in nonstimulated cells. Compared to those in nonstimulated cells, MyD88 expression levels increased by approximately one-third in CD19+ cells stimulated with ODN 1826 for all three time points and increased by approximately one-tenth after 15 min of TLR7 engagement but then gradually decreased (Fig. 2a, statistics Table S1 Excel file). To determine the activation of B cells by different TLR agonists at 24 and 48 h, we measured IκBα degradation (Fig. 2a) and IL-6 production (Supplementary Fig. S5B). Cells responded to the stimuli most effectively within 30 min, and thus, we used this time for imaging flow cytometry analyses. First, we conducted single immunofluorescence (IF) reactions for MyD88, BANK1, TRAF6, and TLR9 in purified mouse B cells to optimize the reactions by confocal microscopy (data not shown) and imaging flow cytometry (Supplementary Fig. S6A). Then, we performed triple IF reactions for BANK1 with either TRAF6 or MyD88 in combination with TLR7 or 9. We determined that BANK1 was localized in the cytoplasm and formed small single-protein accumulations alone (Fig. 2b, panels 1, arrow and 2, asterisk) or colocalized with either TRAF6 or MyD88 (Fig. 2b, panels 1 and 2). Our IF analysis also showed that BANK1 colocalized with TLR7 and 9, indicating the possible involvement of BANK1 in innate immune signaling (Fig. 2b, panels 3 and 4). Colocalization was measured by the bright detail similarity coefficient, R3, which was designed to quantify the colocalization of two probes in a defined region. We observed similar colocalization in stimulated and nonstimulated cells for all reactions analyzed (Supplementary Fig. S6B). BANK1 colocalized with TRAF6 and MyD88 even in the absence of stimuli, confirming our results from endogenous CoIPs conducted in human B cells (Fig. 1h) and from confocal microscopy (Supplementary Fig. S7A).
Fig. 2.
In vivo analyses of purified splenic mouse B cells. a Western blot of BANK1, TRAF6, MyD88, and IκBα upon stimulation of purified mouse B cells with TLR9 and TLR7 agonists ODN 1826 and R848 for 0, 15, 30, and 60 min. Densitometric measurements were performed on western blot bands using ImageJ. Samples were normalized using β-actin to calculate fold changes in protein expression relative to time point 0. Mean fold changes are shown. The results are from a single experiment that is representative of at least four separate experiments. See also Supplementary material Table S1 in the Excel file for statistics. b Panels 1–4 show imaging flow cytometry images of immunofluorescence reactions from nonstimulated purified mouse splenic B cells, with staining for BANK1 with TRAF6 (Panel 1), MyD88 (Panel 2), TLR7 (Panel 3), and TLR9 (Panel 4). Strong colocalization was observed for all reactions according to the bright detail similarity feature R3 (R3 > 1.0). c Percentages of cells double positive for BANK1–TRAF6, BANK1–MyD88, and BANK1 with TLRs 7 and 9 were determined in nonstimulated cells and after 30 min of R848 and ODN 1826 stimulation. One-way ANOVA was applied for statistical analyses with *p < 0.033, **p < 0.002, and ***p < 0.001 indicating significance. NS, nonstimulated; BF, brightfield. Scale bars = 7 µm
In the next step, we quantified the percentage of colocalization of the studied proteins to determine the degree of change in double-positive cells upon stimulation (Figs. 2c and S7B, C). In nonstimulated cells, ~40% of cells were BANK1 and TRAF6 double positive; however, we observed a significant increase of ~25% in R848-stimulated cells and 35% in ODN 1826-stimulated cells (Fig. 2c). Interestingly, the changes in the number of cells double positive for BANK1 and MyD88 were even stronger. We observed that 20% of nonstimulated cells were already positive for both factors, while this value increased to 60% in cells stimulated with R848 and to 70% in cells stimulated with ODN 1826 (Fig. 2c). We also investigated the changes in cells double positive for BANK1–TLR7 and BANK1–TLR9 and observed a similar pattern for both receptors, namely, that 20% of nonstimulated cells were already double positive. The percentage increased 20% following R848 stimulation for both receptors and 30% and 40%, respectively, following ODN 1826 stimulation (Fig. 2c). These results indicate that BANK1 is indeed involved in the TRAF6 and MyD88 interaction complex together with TLR7 and 9. As expected, we observed an increase in cells double positive for TLR7 with MyD88 or TRAF6 and TLR9 with MyD88 or TRAF6, respectively, upon TLR7 and 9 agonist stimulation (Supplementary Fig. S7B, C).
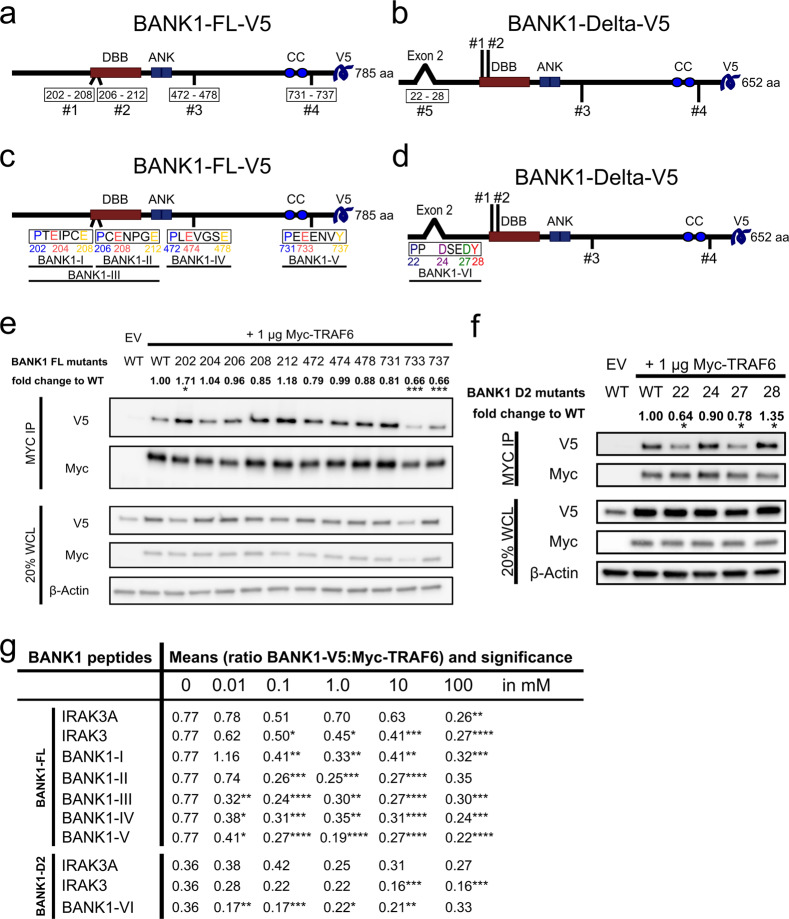
BANK1 binds to TRAF6 via the C-terminal domain in the FL isoform and the N-terminal domain in the D2 isoform
The BANK1–FL protein sequence consists of 785 amino acids (aa). In this sequence, we can distinguish the Dof/BCAP/BANK (DBB) domain (200–327 aa), followed by two ankyrin repeats (ANK1 342–371 aa; ANK2 378–408 aa) and a coiled-coil region close to the C-terminus (677–705 aa)58 (Fig. 3a). In the BANK1–FL protein sequence, there are four TRAF6 BMs, numbered and located within the sequence as follows: #1 (202–208 aa), #2 (206–212 aa), #3 (472–478 aa), and #4 (731–737 aa) (Fig. 3a). BANK1–D2 distinguishes itself from BANK1–FL by the lack of exon 2, which leads to the creation of another TRAF6 BM in BANK1–D2 at residues 22–28 in the aa sequence that was numbered #5 (Fig. 3b). To determine which of the four TRAF6 BMs in BANK1–FL and the fifth motif in BANK1–D2 are responsible for the interaction with TRAF6, mutants harboring point mutations in each of the three core amino acids of each TRAF6 consensus motif were constructed (Figs. 3c, d and S8, statistics in Tables S2 and S3 in the Excel file). We determined in our analyses that the BANK1–FL–202–V5 mutant located in TRAF6 BM #1 in the FL protein showed a significant increase of three-quarters in the protein interaction with Myc–TRAF6 (1.71) (Fig. 3e). The point mutant BANK1–FL–212–V5 in motif #2 also showed an increase; however, this change was insignificant. The other mutants showed decreased interaction with Myc–TRAF6, with decreases ranging between one-tenth and one-fifth. However, mutants BANK1–FL–733–V5 and -737–V5 located in TRAF6 BM #4 close to the BANK1 C-terminus showed a significant reduction by one-third (0.66) in their interactions with Myc–TRAF6 (Fig. 3e). We determined significantly reduced interactions with the Myc–TRAF6 protein of more than one-third for BANK1–D2-22–V5 and of one-fifth for BANK1-D2-27-V5 (0.64 and 0.78) (Fig. 3f), while the mutant BANK1–D2-24–V5 showed a slight insignificant reduction. In contrast, the BANK1–D2-28 mutant resulted in an increase in its interaction by one-third (1.35) compared to the control. These results suggest that the new TRAF6 BM in BANK1–D2 is functional and that the core motif P-X-D-X-X-D bearing an Asp in the third position might be responsible for mediating TRAF6 binding rather than the motif P-X-D-X-X-X-Y (Fig. 3d–f).
Fig. 3.
TRAF6-binding motifs (BMs) are necessary for the BANK1–TRAF6 interaction. a Drawings of BANK1–FL–V5 showing the localization of the Dof/BCAP/BANK (DBB) motif (200–327, amino acids, aa, in red), the double ankyrin repeat-like (ANK) motifs (342–408 aa, in blue) and the putative coiled-coil (CC) region (677–705 aa, blue circles). Below, the protein with its four TRAF6 BMs numbered #1 (202–208 aa), #2 (206–212 aa), #3 (472–478 aa), and #4 (731–737 aa). b Drawing of BANK1–D2–V5 that differs from BANK1–FL by the lack of exon 2, creating a new TRAF6 BM #5. c Drawing of the four TRAF6 BMs in BANK1–FL and their aa sequences. Core aa that were exchanged with alanine to create BANK1 mutants are colored. Peptides used for decoy peptide assays are shown. d Drawing explicitly shows TRAF6 BM #5 in BANK1–Delta2 with core aa P(22), D(24), D(27), and Y(28) in color. The location of peptide BANK1-VI is shown underneath. e Analysis of the binding of BANK1–FL–V5 mutants by CoIP in nonstimulated HEK293 cells. Pull-down was carried out with an anti-Myc antibody, and band levels for Myc–TRAF6 were equalized by ImageJ. The figure is from a single experiment that is representative of at least ten independent experiments. Densitometric measurements were performed. Signals of bands from lysates of BANK1–V5 proteins and Myc–TRAF6 were normalized by using β-actin as a housekeeping protein. From those normalized V5/Myc signals, lane normalization factors were calculated to remove variations in transfection efficiencies among samples. The factors were used to normalize signals from CoIP bands for V5 and Myc. Fold changes in BANK1 interaction relative to TRAF6 were calculated. Interaction of BANK1–WT–V5 with Myc–TRAF6 was used as a control. See also Supplementary Fig. S8A for a graphic presentation of the results and Table S2 in the Excel file for statistical analyses. f Analyses of CoIP reactions of BANK1–D2–V5 mutants with Myc–TRAF6 were performed as described for BANK1–FL in nonstimulated HEK293 cells. See also Supplementary Fig. S8B for a graphic presentation of the results and Table S3 in the Excel file for statistical analyses. g Table summarizes the results from decoy peptide experiments. Nonstimulated HEK293 cells were transiently transfected with 1 µg Myc–TRAF6 and 4 µg BANK1–FL/D2–V5, harvested, and subjected to pull-down with an anti-Myc antibody, and the cleared lysates were incubated either without any peptide added (0 mM) or with 0.01 mM, 0.1 mM, 1 mM, 10 mM, and 100 mM peptide. For BANK1–FL, peptides BANK1-I to -V were tested, while for BANK1-D2, peptide BANK1-VI was tested. Peptide IRAK3A was applied as a negative control, while peptide IRAK3 was used as a positive control for the BANK1–V5:Myc–TRAF6 interaction. Experiments were performed for each concentration at least three times. Western blot bands of CoIP reactions were measured by ImageJ and BANK1–V5:Myc–TRAF6 ratios were calculated. The results are shown as the means of these ratios and their significance. See also Supplementary Tables S4 and S5 in the Excel file for statistical analyses. The Wilcoxon signed-rank test was used for statistical analyses of BANK1 mutants with p < 0.12 (ns), *p < 0.033, **p < 0.002, and ***p < .001 and the unpaired Mann–Whitney t-test for decoy peptides with p < 0.1234 (ns), *p < 0.0332, **p < 0.0021, ***p < 0.0002, and ****p < 0.0001. ns, not significant
Next, to verify our results obtained by BANK1 point mutant analyses, we designed decoy peptides for each TRAF6 BM in BANK1–FL and BANK1–D2 and tested the effects of these decoy peptides at different concentrations on the interaction of BANK1 with TRAF6. These peptides might be useful as potential treatments for human autoimmune diseases such as SLE by modulating TRAF6 signaling and its associated biological functions. Thus, we designed five decoy peptides for BANK1–FL, named BANK1-I, -II, -III, -IV, and -V. Their locations are shown in Fig. 3c. Each decoy peptide was designed to cover one entire TRAF6 BM. We also designed peptide BANK1-III, which covers TRAF6 BMs 1 and 2 entirely (Fig. 3c). For BANK1–D2, we designed the decoy peptide BANK1-VI (Fig. 3d). We added peptide IRAK3A as a negative control and IRAK3, a well-described TRAF6 target, as a positive control.36 As expected, the negative control, IRAK3A, resulted in both BANK1 isoforms in an unchanged BANK1–TRAF6 interaction, and only at high concentrations was an effect detectable (Figs. 3g and S9A, B and Tables S4 and S5 in the Excel file for statistics). We detected a dose-dependent decrease in the BANK1–TRAF6 interaction for the positive control peptide IRAK3 for both BANK1 isoforms, which demonstrated the functionality of the peptide assay. When we tested our BANK1 peptides, we noticed that all peptides could interfere with the BANK1–TRAF6 interaction but with different affinities. In addition, the peptide assays reflected the results of BANK1 point mutant analyses. Peptides BANK1-I and -II failed to block the BANK1–TRAF6 interaction at the lowest concentration of 0.01 mM but did block this interaction at 0.1 mM. Both peptides stopped inhibiting the interaction again at higher concentrations. Interestingly, the BANK1-III peptide, which covers TRAF6 BMs 1 and 2 for peptides I and II, reduced the BANK1–TRAF6 interaction by approximately half at 0.01 mM compared to the reaction without peptide, which was even stronger than the positive control, and continued inhibiting at 0.1 mM but then declined its action at high concentrations. This may reflect a synergistic effect of the simultaneous blockade of BM 1 and 2 in BANK1. Peptide BANK1-IV gave results similar to the double peptide BANK1-III. However, peptide BANK1-V was the only peptide tested that inhibited the BANK1–TRAF6 interaction in a dose-dependent manner with consistent and significant results. The interaction of BANK1–D2–V5:Myc–TRAF6 was significantly reduced by approximately half when we added peptide BANK1-VI at 0.01 mM but then stagnated (Fig. 3g), indicating the relevance of the N-terminal domain of the D2 isoform in the interaction with TRAF6, which is in agreement with our point mutation results (Fig. 3f). In combination, our mutational and decoy peptide assays point to BM 4 in the C-terminus as the more relevant mediator of the BANK1–TRAF6 interaction in BANK1–FL. In contrast, the N-terminus with the newly formed BM 5 seems to be important in the BANK1–D2 isoform.
The TIR domain in BANK1 is important for its K63-linked polyubiquitination and TLR pathway activation
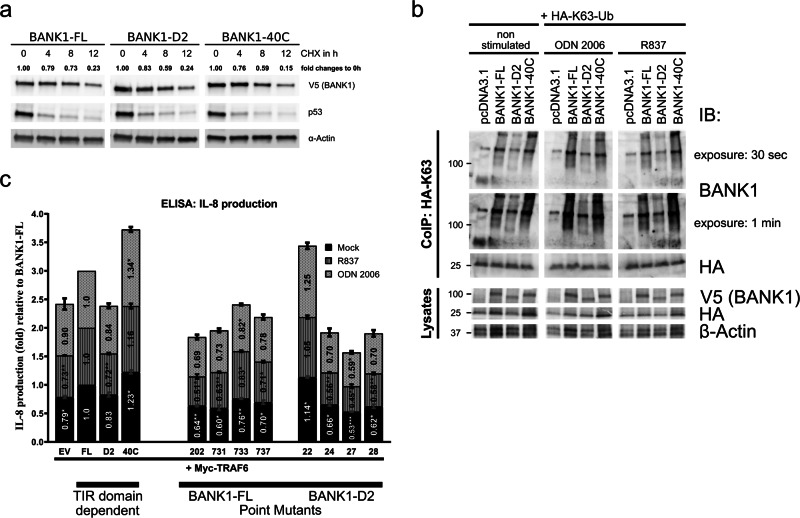
We performed functional analyses to study possible signaling differences between BANK1 isoforms and BANK1–40C. One reason might be related to altered protein half-lives due to different protein sequences, which would translate into different protein abundances. We conducted cycloheximide (CHX) chase assays in nonstimulated U2OS cells for 0, 4, 8, and up to 12 h (Fig. 4a). We used p53, a well-known short-lived protein, as an internal control. We tested the BANK1–V5 proteins and observed that their stability was higher than that of p53 and that the protein levels decreased gradually within 12 h (Fig. 4a). BANK1–D2 and BANK1–40C showed a reduction of two-fifths after 8 h of CHX treatment (0.59), while BANK1–FL was reduced approximately by one-fourth (0.73). At 12 h of treatment, BANK1 proteins showed similar reductions (0.23, 0.24, and 0.15) (Fig. 4a). In conclusion, BANK1 proteins have similar protein half-lives and display a medium- to long-term stability of more than 6 h, as reported previously by Yen et al.59 To confirm this result, we performed ubiquitination assays on BANK1 proteins, showing that BANK1 ubiquitination is generally low with very little K48-linked polyubiquitination (Supplementary Fig. S10). All these data suggest that the possible signaling properties of the different BANK1 proteins are not defined by their protein stability.
Fig. 4.
Functional analyses of BANK1 proteins. a Protein stability of BANK1–FL, BANK1–D2, and BANK1–40C was studied in nonstimulated U2OS cells by treating cells with 100 µg/ml cycloheximide (CHX) for a time frame of 12 h. β-Actin was used as a housekeeping protein to normalize samples by ImageJ. See also Supplementary Fig. S10. b K63-linked polyubiquitination of BANK1–FL and -D2 and BANK1–40C was tested in nonstimulated HepG2 cells and in cells stimulated O/N with 2 µM ODN 2006 and 8 µg/ml R837. Pull-downs were performed with an anti-HA antibody O/N at 4 °C, and eluates of immunoprecipitates were run on a western blot and interrogated with an anti-BANK1 antibody. c Proinflammatory IL-8 cytokine production was measured in supernatants derived from HepG2 cells transfected with 0.1 µg Myc–TRAF6 only and with 0.1 µg Myc–TRAF6 together with 0.4 µg BANK1–FL–V5, -D2–V5, and -40C–V5 and treated for 72 h with R837 and ODN 2006. The results are shown as the mean fold changes in IL-8 production relative to BANK1–FL from three independent experiments. The results are presented in bars for mock (black), R837-stimulated (dark gray) and ODN 2006-stimulated (light gray) HepG2 cells. Samples are categorized as TIR domain-dependent and BANK1 point mutants. See also Supplementary Fig. S11 for transient transfection efficiency in HepG2 cells and Table S6 in the Excel file for statistical analyses. Two-way ANOVA was applied with p < 0.12 (ns), *p < 0.033, **p < 0.002, and ***p < 0.001. ns, not significant
It is generally known that ubiquitination is important for the activation, fine-tuning, and termination of immune system signaling. We know from our study that BANK1 is a part of the so-called Myddosome signaling complex involving MyD88 and TRAF6. K63-linked polyubiquitination of TRAF6 is crucial to activate the TLR pathway.41 Therefore, we tested K63-linked polyubiquitination of BANK1 proteins. We observed in nonstimulated HepG2 cells that BANK1–FL–V5 and −40C–V5 showed a similar strong signal for K63-linked polyubiquitination (Fig. 4b). We also determined similar results in response to ODN 2006 and R848 (Fig. 4b).
Then, we asked whether BANK1 isoforms, the −40C mutant and point mutants of BANK1–FL–V5 and BANK1–D2–V5 had effects on proinflammatory cytokine production in the presence of TRAF6 in either nonstimulated or stimulated HepG2 cells (Figs. 4c and S11 and Table S6 in the Excel file for statistics). We determined that IL-8 cytokine secretion was independent of the stimulus and that the presence of the BANK1–V5 proteins increased IL-8 production compared to HepG2 cells expressing Myc–TRAF6 only; however, this difference was only marginal for BANK1–D2–V5 (Fig. 4c).
In addition, the lack of the TIR domain in BANK1–D2–V5 resulted in reduced IL-8 cytokine levels compared to BANK1–FL–V5 and BANK1–40C–V5. Next, we studied whether the intensity of TRAF6 binding to BANK1 influences IL-8 production by testing our BANK1 mutants. Interestingly, we determined that point mutants in the C-terminus of BANK1–FL–V5, −731, −733, and −737 produced less IL-8 than BANK1–FL–V5 (Fig. 4c). This result was in line with the reduced interaction with Myc–TRAF6 (Fig. 3e). However, BANK1–FL-202–V5 showed lower IL-8 production than BANK1–FL–V5, although its binding to TRAF6 was stronger (Fig. 3e). BANK1–D2–V5 mutants −24, −27 and −28 showed lower IL-8 production than BANK1–D2–V5, but the mutant −22 showed higher levels of IL-8 production. In general, the binding of BANK1 point mutants to TRAF6 correlated only partially with IL-8 production. Thus, we cannot exclude the possibility that the altered interaction between BANK1 mutant proteins and TRAF6 might impact different signaling outcomes. These results demonstrate that the absence or modification of the BANK1 TIR domain influences proinflammatory cytokine production.
Discussion
In this study, we placed for the first time the SLE-linked B-cell adaptor BANK1 within the TLR pathway. We found that in B cells, BANK1 interacts, and colocalizes with two key TLR signaling molecules: the master adaptor MyD88 and the E3 ubiquitin ligase TRAF6. Specific modifications of the BANK1 protein sequence can alter these interactions with important implications in the context of disease-related genetic variability of this gene. The BANK1 exon 2-encoded domain was recently proposed to be a TIR domain because of its high homology to the TIR domain of the BCAP adaptor (~40%).60 To our understanding, our data are the first to experimentally show the functionality of the BANK1 TIR domain. This homotypic motif was crucial for (a) the interaction with the other TIR-containing adaptor, MyD88; (b) K63-linked polyubiquitination; and (c) induction of the proinflammatory cytokine IL-8 production as a readout. We have also described five previously unidentified TRAF6 BMs within the BANK1 isoforms that indicated a link between TRAF6 binding and BANK1.
TRAF6 requires the presence of consensus BMs within targeted proteins to allow their interaction.36,37 Of the two types of TRAF6 BMs described to date, we found four of the type P-X-E-(X)3-aromatic/acidic residues within the BANK1–FL isoform, with two of them overlapping (BM #1 and #2).36,37 In BANK1–D2, we found an extra BM formed following alternative splicing of exon 2 and the subsequent removal of the TIR domain (BM #5). This N-terminal BM 5 presented an Asp instead of a Glu in position 3, but these two amino acids are considered highly similar in terms of protein structure conservation. Indeed, our mutational analyses showed that a TRAF6 BM with an Asp in position 3 was able to mediate the interaction with TRAF6. This fact should be considered when searching for new putative TRAF6 targets.
Mutational analysis of the BANK1–FL isoform showed that BM 4 at the C-terminus was crucial to maintain its interaction with TRAF6 at normal levels. However, we could only partially link TRAF6-binding ability in the C-terminus of BANK1–FL with IL-8 proinflammatory cytokine production. Our results connecting TRAF6-binding affinity to BANK1 and IL-8 production require caution. Further studies are needed to demonstrate the importance of the BANK1–TRAF6 interaction in innate immune signaling. In the case of the BANK1–D2 modification, two of the core amino acids of the newly formed BM 5 indeed decreased the binding to the E3 ubiquitin ligase, supporting its functional role as well. Additional decoy peptide assays confirmed the functionality of the TRAF6 BMs in BANK1. Moreover, we showed that the BANK1 TIR domain is also important for the TRAF6 interaction since the absence of this domain in the D2 isoform decreased binding by one-third. The BANK1–TRAF6 interaction might be indirectly and/or partially mediated by MyD88 and/or other TIR-containing proteins, such as MAL/TIRAP or TICAM-1/TRIF. Another explanation is that the D2 isoform may present differential folding that could affect the exposure of the rest of the TRAF6 BMs in BANK1, decreasing their direct interaction. These hypotheses are not mutually exclusive, and they could both occur given the complex nature of signaling platform formation. Our CoIP experiment in the Namalwa B cell line shows that BANK1 interacts with MyD88 and TRAF6 endogenously. This complex is formed naturally under nonstimulated conditions, and we observed that the amount of interacting BANK1 and TRAF6 decreased with time after stimulation with TLR7 or TLR9 agonists, suggesting a release of BANK1 from the MYD88–TRAF6-signaling complex after pathway initiation. We further determined in mouse B cells that BANK1 colocalized with MyD88 and TRAF6 as expected but also with TLR7 and TLR9 under nonstimulated conditions and after stimulation. However, upon B cell stimulation with the TLR7 agonist R848 and the TLR9 agonist ODN 1826, we determined similar expression levels of BANK1, TRAF6, and MyD88 but significantly increased levels of B cells double positive for BANK1 in combination with MyD88, TRAF6, TLR7, and TLR9. These results are interesting in the context of endogenous BANK1 interactions and need further experiments on the kinetics of binding and the proximity of MyD88–TRAF6–BANK1 as a signaling platform.
Previous work by Kozyrev et al.43 showed that the risk allele (G; R61) of the SLE-associated SNP rs10516487 in BANK1 correlates with increased levels of BANK1–FL mRNA and BANK1 protein in lymphoblastoid cell lines. Conversely, the mRNA of the FL isoform was decreased in PBMCs from individuals with the protective allele of the same SNP (A; H61), which in turn presented increased levels of the BANK1–D2 transcript.43 We recently performed a high-density genetic analysis of BANK1 in which our genetic and epigenetic observations suggested a link between the BANK1 exon 2-encoded TIR domain and the development of autoimmunity.61 BANK1 has been previously related to TLR pathways,52,53 so we decided to investigate the interaction of BANK1 with the key player in TLR activation, MyD88. Our CoIP experiments confirmed the interaction, which was primarily mediated by the BANK1 TIR domain. Other regions of BANK1 may account for the residual binding observed after removal of the TIR domain. MyD88 aggregation occurs after activation of TLR pathways in a process related to K63-linked polyubiquitination.62 Similarly, we have shown that BANK1 and MyD88 form large aggregated structures where they colocalize together with the TRAF6 ubiquitin ligase. Interestingly, the BANK1–D2 isoform displayed an inability to undergo K63-linked polyubiquitination in vitro. Since this type of ubiquitination is associated with increased TLR pathway activity63 and enhanced cell activation,24 our results could be interpreted as a mechanism by which the D2 isoform confers protection against SLE by reducing activation. In fact, the decreased K63-linked polyubiquitination of the D2 isoform was accompanied by impaired IL-8 production in HepG2 cells, especially upon TLR7 engagement. It is tempting to hypothesize that B cells may be less activated in individuals with higher levels of D2 than FL, given the signaling properties of the TIR-expressing BANK1 isoform. Interestingly, the BANK1–40C mutant showed even higher IL-8 production than BANK1–FL, which was accompanied by stronger binding to MyD88. All these data suggest that the appearance of autoimmune phenotypes in carriers of the mutation56 might be linked to altered innate signaling through the Myddosome signaling platform.
This study shows for the first time the importance of the TIR domain of BANK1, thereby functionally linking previous genetic analyses and studies in mice with the risk of developing autoimmune diseases, including SLE.
Materials and methods
Mice and cell culture
C57BL/6 wild type mice were purchased from the Jackson Laboratory. All mice were bred and maintained in a pathogen-free animal facility at the Biomedical Research Center of the University of Granada. Mouse housing, handling, and experimental protocols were approved by the Ethics Committee for Animal Experimentation of the University of Granada and the Ministry of Agriculture of Spain. Human embryonic kidney HEK293, osteosarcoma U2OS, and hepatocellular carcinoma HepG2 cells (all kindly provided by Daniel Krappmann (Helmholtz Zentrum, Munich)) were maintained in Dulbecco’s modified Eagle’s medium with GlutaMAX (Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FCS (Biowest). The B cell lymphoma cell line Namalwa, obtained from the American Type Culture Collection, and purified splenic mouse B cells were maintained in RPMI medium 1640 with GlutaMAX (Thermo Scientific) supplemented with 10% FCS.
Primary and secondary antibodies
The following primary antibodies were used: rat anti-HA (clone 3F10, Roche), rabbit anti-MyD88 (#3699S), rabbit anti-p105 (#4717), rabbit anti-IκBα (44D4) (#4812S), rabbit anti-p-IκBα (Ser32) (#2859) (all from Cell Signaling), mouse anti-V5 (Thermo Scientific, #R96025), rabbit anti-BANK1 (HPA037002), mouse anti β-actin (clone AC-15) (A5441) (both from Sigma-Aldrich), mouse anti-BANK1 (F-8) (sc-393611), rabbit anti-TRAF6 (H-274) (sc-7221), mouse anti-TRAF6 (D-10) (sc-8409), goat anti-TLR7 (V-20) (sc-16245), goat anti-TLR9 (N-15) (sc-13215), and mouse anti-Myc (9E10) (sc-40) antibodies; as a nonspecific control, the immunoglobulins rabbit IgG (sc-2027) and mouse IgG (sc-2025) (all from Santa Cruz) were used. The HRP-linked secondary antibodies for western blotting were anti-rabbit IgG (sc-2004) (#7074), anti-mouse IgG (sc-2005) (#7076) (both from Santa Cruz and Cell Signaling), anti-goat IgG (sc-2354) or anti-rat IgG (sc-2032) (both from Santa Cruz). The secondary antibodies for IF reactions were Alexa Fluor 555 donkey anti-goat IgG (H + L) (#A-21432), Alexa Fluor 647 goat anti-mouse/rabbit IgG (H + L) (#A-21235; #A-31634) and Alexa Fluor 488 goat anti-rabbit/mouse IgG (H + L) (#A-11034; #A-11001) (all from Thermo Scientific).
Plasmids and transient transfection
For interaction analyses in HEK293 cells, the vectors pRK5 (kindly provided by Daniel Krappmann, Helmholtz Zentrum, Munich) and pcDNA3.1D/V5-His (#K490001, Thermo Fisher Scientific) were used either as empty vectors or containing pRK5–Myc–TRAF6 and pcDNA3.1D–BANK1–FL–V5, -BANK1–D2–V5,58 -BANK1–40C–V5, -BANK1 point mutants-V5, and -MyD88–V5, respectively. In addition, pIRES S2-EGFP was used as an empty vector or containing BANK1–FL-EGFP, -D2-EGFP, and -BANK1-40C-EGFP. For ubiquitination assays in HepG2 cells, vectors pcDNA3.1(+)-HA-Ub (kindly provided by Prof. Yukiko Gotoh), pRK5-HA-Ubiquitin-K63, and pRK5-HA-Ubiquitin-K48 were used (kindly provided by Ted Dawson64; Addgene plasmid 17606 and 17605). For confocal microscopy, U2OS cells were transfected either alone or in combination with the expression vectors pRK5-RFP-TRAF6 (kindly provided by Daniel Krappmann, Helmholtz Zentrum, Munich), pcDNA3.1D/V5–BANK1–FL/D2/40C-YFP, and pcDNA3.1D/V5–MyD88-CFP. jetPRIME transfection reagent was used to perform transient transfections according to the manufacturer’s protocol (Polyplus transfection).
Cytokine production
Stimulation of Namalwa and mouse B cells was tested by the ELISA for either human IL-8 and TNFα or mouse IL-6 (both BD Biosciences), respectively, following the manufacturer’s instructions. Briefly, cells were seeded at a concentration of 3 × 106 cells in 12-well plates and were left nonstimulated or stimulated with 1 μM CpG B (human ODN 2006, mouse ODN 1826) (#10336022, Thermo Fisher Scientific; tlrl-1826, Invivogen) or 5 μg/ml Resiquimod (R848) (tlrl-r848, Invivogen) for 24 and 48 h; supernatants were collected. IL-8 cytokine production in HepG2 cells was measured by ELISA 72 h upon stimulation of cells with 8 µg/ml Imiquimod (human R837, tlrl-imqs, Invivogen) and 2 µM ODN 2006. Cells were seeded in 24-well plates, transfected separately with 0.1 µg pRK5–Myc–TRAF6 and 0.4 µg pcDNA3.1D/V5–BANK1–FL/D2/40C–V5, and stimulated the next day.
Immunoprecipitation reactions
Nonstimulated HEK293 cells were seeded in 100-mm dishes (2 × 106) and transiently transfected with a total of 5 μg pDNA. Twenty-four hours post transfection, the cells were lysed in 900 μl Triton-X lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and protease/phosphatase inhibitor cocktail tablets (#04693159001, #04906837001, Roche)). Thirty microliters of cleared lysate was used for immunoblotting, while the remaining lysates were used to perform IP reactions with 1 µg primary antibody overnight (O/N) at 4 °C. Protein G Sepharose (#101241, Thermo Fisher Scientific) was added for 1 h at 4 °C, and IPs were washed, boiled in 30 μl 2× LDS sample buffer (#NP0007, Thermo Fisher Scientific) and analyzed by western blotting. For endogenous IP reactions, Namalwa B cells were seeded in 100-mm dishes (5 × 107) and left nonstimulated or stimulated for 5, 15, 30, and 60 min with 5 μg/ml R848 and 1 µM ODN 2006. Cells were lysed in 1 ml TRITON-X lysis buffer for 60 min at 4 °C while rotating, and the cleared lysates were incubated O/N with 2 µg primary or rabbit control IgG antibody. Immune complexes were eluted in 20 µl 2× LDS buffer and analyzed by western blotting. The detection solution for the western blot membranes with strong signals was Clarity Western ECL Substrate (#170-5061, BioRad), and that the for membranes with weak signals was the SignalFire Elite ECL Reagent (#12757S, Cell Signaling).
Cloning of BANK1 mutants by PCR-mediated deletion of plasmid DNA
Primer pairs to introduce an exchange to alanine in the desired position of each TRAF6 BM in BANK1 were designed so that there were nonoverlapping sequences at the 3′ end and primer–primer complementary (overlapping) sequences at the 5′ end containing the change in the codon of interest. The KAPA HiFi PCR kit with dNTPs (#KK2101, KAPA Biosystems) was used to introduce point mutations in 50 ng of the template vectors pcDNA3.1D–BANK1–FL–V5 and –D2–V5 according to the manufacturer’s instructions. O/N digestion at 37 °C with 10 units DpnI (#1701, Thermo Scientific) followed. Clones were verified for the introduced point mutations by DNA sequencing (StabVida, Lisbon, Portugal).
Decoy peptides
Four peptides were designed for the four TRAF6 BMs in BANK1–FL, plus an additional fifth peptide that included overlapping TRAF6 BMs 1 and 2 of BANK1–FL, called BANK1-III. For BANK1-D2, one peptide was designed that reflected the BM created by the deletion of exon 2 (BANK1-VI). The BANK1–FL peptides (with TRAF6 BM underlined) BANK1-I (N-YVLPTEIPCE-C), BANK1-II (N-YEIPCENPGE-C), BANK1-III (N-YVLPTEIPCENPGE-C), BANK1-IV (N-YHSPLEVGSE-C), and BANK1-V (N-YKRPEEENVY-C) and the BANK1–D2 peptide BANK1-VI (N-YPAPPDSEDY-C) were chemically synthesized and purified by HPLC (Gene Cust, Luxembourg). In addition, a peptide called IRAK3 (N-YRQGPEESDEF-C) that is known to interact with TRAF636 was used as a positive control to interfere in the interaction of TRAF6 with BANK1, while the IRAK3A peptide, of which the canonical TRAF6 BM was synthetically interrupted by substitutions with alanine, was used as a negative control (N-YRQGAEASDEA-C). Three 100-mm dishes of HEK293 cells per BANK1 peptide were prepared, transfected with 1 µg pRK5–Myc–TRAF6 and 4 µg pcDNA3.1-BANK1–FL–V5/-BANK1–D2–V5 each, and incubated O/N at 37 °C. Cells were harvested in 1 ml Triton-X buffer and pooled in a 15 ml Falcon tube, lysed for 30 min at 4 °C, and then 300 µl of cleared lysate was mixed with 300 µl of various peptide concentrations (0.01, 0.1, 1.0, 10, and 100 mM). Cleared lysates of BANK1–FL/D2–V5 with Myc–TRAF6 without any peptide added were used as control reactions and were instead filled with 300 µl Triton-X buffer. The reactions were left rotating for 1 h at RT, and then 1 µg anti-Myc primary antibody was added for an O/N incubation at 4 °C. Immune complexes were eluted in 30 µl 4× LDS buffer and analyzed by western blotting. The signals were normalized, and the ratios of BANK1–V5 isoforms with Myc–TRAF6 were calculated.
In vitro ubiquitination assays
HepG2 cells were seeded in 100-mm dishes (2 × 106), transiently transfected with 1 µg pRK5-HA-empty vector or pcDNA3.1(+)-HA-Ub, pRK5-HA-Ubiquitin-K63, and pRK5-HA-Ubiquitin-K48 with 4 µg each pcDNA3.1D-empty vector or pcDNA3.1D–BANK1–FL–V5, -BANK1–D2–V5, and -BANK1–40C–V5, and cells at 24 h post transfection were stimulated O/N with 2 µM ODN 2006 and 8 µg/ml R837 and lysed on the next day in 400 μl Triton-X buffer containing 1% SDS. To remove precipitated genomic DNA from the lysates, samples were first passed through 21G needles and then through 26G needles. Later, 30 µl of cleared lysate was collected to check the lysates for transfection efficiency by western blotting. To perform CoIP reactions, the remaining cleared lysates were diluted tenfold with TRITON-X buffer to a final concentration of 0.1% SDS and incubated with 1 µg anti-HA followed by O/N incubation at 4 °C. The precipitated immune complexes were washed three times in lysis buffer and eluted in 30 µl 4× LDS buffer, boiled for 5 min and analyzed by western blotting.
Protein stability by CHX chase assays
U2OS cells were seeded in 6-well plates and transiently transfected with 0.8 µg each pcDNA3.1D–BANK1–FL–V5, -BANK1–D2–V5, and -BANK1–40C–V5. Twenty-four hours post transfection, 100 µg/ml CHX (C7698, Sigma-Aldrich) was added to cells in a time course from 0 to a maximum of 12 h. The cells were lysed in 100 µl TRITON-X buffer and analyzed by western blotting. The short-lived protein p53 was used as a positive control for CHX treatment. The intensities of the protein bands were measured with ImageJ using β-actin to normalize the samples. BANK1 stability was expressed as the mean fold change from three independent experiments, where time point 0 h was used as the control time point and set as 1.0.
Confocal microscopy
U2OS cells were seeded on eight-well Nunc™ Lab-Tek™ chamber slides (Thermo Fisher Scientific, Waltham, MA) and transfected with 0.25 µg of plasmid DNA the next day. In the case of single and double transfections, the plasmid DNA was replaced with empty vector. Twenty-four hours post transfection, the cells were washed three times in ice-cold PBT, fixed in 4% PFA for 15 min and mounted with SlowFade Diamond Antifade Medium either with or without DAPI (Thermo Fisher Scientific, Waltham, MA, #S36964, #S36963) depending on the combination of expressed fluorescent proteins. Confocal microscopy was performed using a Zeiss 710 Laser Scanning Microscope, a Zeiss Plan-Apochromat 63X/1.40 NA oil-immersion DIC M27 objective (aperture Pinhole = 1.0 Airy Unit) and the Zeiss ZEN 2010 software. Fluorescence was acquired sequentially utilizing different laser excitation lines for excitation and different photomultipliers for the detection of cyan, yellow, and red fluorescent protein signals. The cross-channel effect was ruled out by measuring the emitted signal of each fluorescent protein in all channels in single transfection controls. For quantification of the colocalization of BANK1 isoforms and TRAF6, cells were individually selected as ROIs, and Pearson's correlation coefficient was measured inside these regions with the Zeiss ZEN 2010 software.
Imaging flow cytometry
Immunohistochemical analyses for imaging flow cytometry were performed three times on a pool of purified splenic B cells derived from four 8- to 12-week-old C57BL/6 females. Splenic B cells were purified on magnetic columns by negative selection using a mouse B-cell isolation kit (MACS Miltenyi Biotec, #130-090-862). Purity was assessed by flow cytometry with a FITC anti-mouse CD19 antibody (clone 1D3, Immunostep) and was ≥98% in all cases. Cells were stimulated with 1 µM ODN 1826 and 5 µg/ml R848 for 30 min or left nonstimulated, washed in 1 × MACS buffer (PBS, 2% BSA, 1 mM EDTA), fixed in 4% PFA, permeabilized for 10 min at RT (MACS, 0.01% Tween-20, 0.02% saponin), blocked first for 30 min at RT in Image-iT FX Fix signal enhancer (#I31933, Thermo Fisher Scientific) and then for 1 h at RT in MACS buffer containing 10% NGS, and incubated with primary antibodies O/N at 4 °C (mouse BANK1 (1:100), rabbit MyD88 (1:250), TLR7/9 (1:100), rabbit BANK1 (1:300), and mouse TRAF6 (1:100)). The cells were washed twice and incubated with secondary antibodies at a 1:1000 dilution, nuclei were stained with Hoechst 33342 (#H1399, Thermo Fisher Scientific), and the samples were washed before resuspension in 30 µl MACS buffer. Images were recorded (20,000 events per condition) using the INSPIRE™ software on the ImageStreamX Mark II Imaging Flow Cytometer (Amnis Corporation, Seattle, WA) at ×60 magnification with 405 nm (10.00 mW), 488 nm (150.00 mW), 560 nm (200.00 mW), 658 nm (15.00 mW), and side scatter (785 nm) (4.06 mW) lasers. Single color controls for each specific marker and Hoechst 33342 were also acquired to correct images for fluorescence that leaked into nearby channels and were used to create a compensation matrix via the compensation wizard. Any changes occurring upon stimulation in the cells gated as double positive with protein colocalization for two probes of interest were studied using the colocalization wizard included in the IDEAS™ (v6.2) software. The principle of the wizard is to first gate for cells in best focus and for single cells and then gate for cells double positive for two probes of interest with punctate staining and express the result as % of double-positive gated cells. Next, the program creates a histogram that provides the bright detail similarity feature R3 to gate on colocalized events of two probes of interest in a defined region. R3 is the log transformed Pearson’s correlation coefficient of the localized bright spots with a radius of max 3 pixels and can thus vary between 0 (uncorrelated, different spatially) and 1 (perfect correlation, same spatially) (IDEAS User’s Manual). The results were obtained and analyzed from three independent experiments.
Statistical analysis
The unpaired nonparametric Mann–Whitney t-test (CoIP quantification in HEK293 cells, ELISA assays of Namalwa and mouse B cells, decoy peptide assays), the Wilcoxon signed-rank test (BANK1 point mutants), one-way ANOVA (Amnis analyses), and two-way ANOVA (ELISA assays in HepG2 cells for BANK1 proteins) were applied using GraphPad Prism 8.1.1. The Wald statistical test was carried out to assess the significance of finding TRAF6 BMs within the BANK1 protein sequence. P values are indicated in the figure legends. Error bars indicate the standard error of the mean or the median with interquartile range. All experiments were repeated at least three times.
Supplementary information
Acknowledgements
We wish to thank the Unit of Microscopy and Flow Cytometry of GENYO for their constant help and support during experiments. This work was supported by the MINECO and cofinanced by the FEDER funds of the European Union [SAF2016-78631-P entitled “The Role of BANK1 in B Cell signaling through TLRs and autoimmunity”]; by the Swedish Research Council of Medicine to M.E.A.R.; by the Instituto de Salud Carlos III grant to I.G. [CD11/00277]; by the Proyecto de Excelencia, Consejería de Economía, Innovación y Ciencia of Andalucía [CTS-2548]; and by the Fundación Ramón Areces.
Author contributions
I.G. designed, analyzed, and provided overall guidance for the experiments and wrote the manuscript. A.D.B. designed, performed, and analyzed the experiments and wrote the manuscript. M.M. assisted with and performed experiments that involved mice. A.L.P. helped design the decoy peptide experiments. M.E.A.R. proposed the project, supervised the performance of the experiments and their analyses, revised the manuscript, and led the project.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ina Georg, Alejandro Díaz-Barreiro
Contributor Information
Ina Georg, Email: ina.georg@genyo.es.
Alejandro Díaz-Barreiro, Email: alediazbarreiro@gmail.com.
Supplementary information
The online version of this article (10.1038/s41423-019-0254-9) contains supplementary material.
References
- 1.Teruel M, Alarcón-Riquelme ME. The genetic basis of systemic lupus erythematosus:what are the risk factors and what have we learned. J. Autoimmun. 2016;74:161–175. doi: 10.1016/j.jaut.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Ramos-Casals M, Sanz I, Bosch X, Stone JH, Khamashta MA. B-cell-depleting therapy in systemic lupus erythematosus. Am. J. Med. 2012;125:327–336. doi: 10.1016/j.amjmed.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crampton SP, Morawski PA, Bolland S. Linking susceptibility genes and pathogenesis mechanisms using mouse models of systemic lupus erythematosus. Dis. Model Mech. 2014;7:1033–1046. doi: 10.1242/dmm.016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H-S, Bae S-C. What can we learn from genetic studies of systemic lupus erythematosus? Implications of genetic heterogeneity among populations in SLE. Lupus. 2010;19:1452–1459. doi: 10.1177/0961203310370350. [DOI] [PubMed] [Google Scholar]
- 5.Taylor KE, et al. Risk alleles for systemic lupus erythematosus in a large case-control collection and associations with clinical subphenotypes. PLoS Genet. 2011;7:e1001311. doi: 10.1371/journal.pgen.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckwith H, Lightstone L. Rituximab in systemic lupus erythematosus and lupus nephritis. Nephron Clin. Pract. 2014;128:250–254. doi: 10.1159/000368585. [DOI] [PubMed] [Google Scholar]
- 7.Liossis S-NC, Melissaropoulos K. Molecular abnormalities of the B cell in systemic lupus erythematosus are candidates for functional inhibition treatments. Expert Opin. Pharm. 2014;15:833–840. doi: 10.1517/14656566.2014.894976. [DOI] [PubMed] [Google Scholar]
- 8.Vincent FB, Morand EF, Schneider P, Mackay F. The BAFF/APRIL system in SLE pathogenesis. Nat. Rev. Rheumatol. 2014;10:365–373. doi: 10.1038/nrrheum.2014.33. [DOI] [PubMed] [Google Scholar]
- 9.Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol. Rev. 2007;220:251–269. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 10.Theofilopoulos AN, et al. Sensors of the innate immune system: their link to rheumatic diseases. Nat. Rev. Rheumatol. 2010;6:146–156. doi: 10.1038/nrrheum.2009.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green NM, Marshak-Rothstein A. Toll-like receptor driven B cell activation in the induction of systemic autoimmunity. Semin. Immunol. 2011;23:106–112. doi: 10.1016/j.smim.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornung V, et al. Quantitative expression of Toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 2002;168:4531–4537. doi: 10.4049/jimmunol.168.9.4531. [DOI] [PubMed] [Google Scholar]
- 13.Hennessy EJ, Parker AE, O’Neill LAJ. Targeting Toll-like receptors: emerging therapeutics? Nat. Rev. Drug Discov. 2010;9:293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- 14.Walsh ER, et al. Dual signaling by innate and adaptive immune receptors is required for TLR7-induced B-cell-mediated autoimmunity. Proc. Natl Acad. Sci. USA. 2012;109:16276–16281. doi: 10.1073/pnas.1209372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain S, et al. Interleukin 6 accelerates mortality by promoting the progression of the systemic lupus erythematosus-like disease of BXSB.Yaa mice. PLoS One. 2016;11:e0153059. doi: 10.1371/journal.pone.0153059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadimitraki ED, et al. Expansion of Toll-like receptor 9-expressing B cells in active systemic lupus erythematosus: implications for the induction and maintenance of the autoimmune process. Arthritis Rheumatol. 2006;54:3601–3611. doi: 10.1002/art.22197. [DOI] [PubMed] [Google Scholar]
- 17.Komatsuda A, et al. Up-regulated expression of Toll-like receptors mRNAs in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin. Exp. Immunol. 2008;152:482–487. doi: 10.1111/j.1365-2249.2008.03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chauhan SK, Singh VV, Rai R, Rai M, Rai G. Distinct autoantibody profiles in systemic lupus erythematosus patients are selectively associated with TLR7 and TLR9 upregulation. J. Clin. Immunol. 2013;33:954–964. doi: 10.1007/s10875-013-9887-0. [DOI] [PubMed] [Google Scholar]
- 19.Lyn-Cook BD, et al. Increased expression of Toll-like receptors (TLRs) 7 and 9 and other cytokines in systemic lupus erythematosus (SLE) patients: ethnic differences and potential new targets for therapeutic drugs. Mol. Immunol. 2014;61:38–43. doi: 10.1016/j.molimm.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Christensen SR, et al. Toll-like receptor 9 controls anti-DNA autoantibody production in murine lupus. J. Exp. Med. 2005;202:321–331. doi: 10.1084/jem.20050338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janssens S, Beyaert R. A universal role for MyD88 in TLR/IL-1R-mediated signaling. Trends Biochem. Sci. 2002;27:474–482. doi: 10.1016/s0968-0004(02)02145-x. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Cousart S, Hu J, McCall CE. Characterization of interleukin-1 receptor-associated kinase in normal and endotoxin-tolerant cells. J. Biol. Chem. 2000;275:23340–23345. doi: 10.1074/jbc.M001950200. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Strelow A, Fontana EJ, Wesche H. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc. Natl Acad. Sci. USA. 2002;99:5567–5572. doi: 10.1073/pnas.082100399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walsh MC, Lee J, Choi Y. Tumor necrosis factor receptor- associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol. Rev. 2015;266:72–92. doi: 10.1111/imr.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann K, Tschopp J. The death domain motif found in Fas (Apo-1) and TNF receptor is present in proteins involved in apoptosis and axonal guidance. FEBS Lett. 1995;371:321–323. doi: 10.1016/0014-5793(95)00931-x. [DOI] [PubMed] [Google Scholar]
- 27.Hultmark D. Macrophage differentiation marker MyD88 is a member of the Toll/IL-1 receptor family. Biochem. Biophys. Res. Commun. 1994;199:144–146. doi: 10.1006/bbrc.1994.1206. [DOI] [PubMed] [Google Scholar]
- 28.O’Neill LAJ, Golenbock D, Bowie AG. The history of Toll-like receptors—redefining innate immunity. Nat. Rev. Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- 29.Isnardi I, et al. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity. 2008;29:746–757. doi: 10.1016/j.immuni.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivas MN, et al. MyD88 is critically involved in immune tolerance breakdown at environmental interfaces of Foxp3-deficient mice. J. Clin. Investig. 2012;122:1933–1947. doi: 10.1172/JCI40591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua Z, et al. Requirement for MyD88 signaling in B cells and dendritic cells for germinal center anti-nuclear antibody production in Lyn-deficient mice. J. Immunol. 2014;192:875–885. doi: 10.4049/jimmunol.1300683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amos CI, et al. High-density SNP analysis of 642 Caucasian families with rheumatoid arthritis identifies two new linkage regions on 11p12 and 2q33. Genes Immun. 2006;7:277–286. doi: 10.1038/sj.gene.6364295. [DOI] [PubMed] [Google Scholar]
- 33.Namjou B, et al. Evaluation of TRAF6 in a large multiancestral lupus cohort. Arthritis Rheumatol. 2012;64:1960–1969. doi: 10.1002/art.34361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 35.Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 36.Ye H, et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature. 2002;418:443–447. doi: 10.1038/nature00888. [DOI] [PubMed] [Google Scholar]
- 37.Meads MB, Li Z-W, Dalton WS. A novel TNF receptor-associated factor 6 binding domain mediates NF-kappa B signaling by the common cytokine receptor beta subunit. J. Immunol. 2010;185:1606–1615. doi: 10.4049/jimmunol.0902026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muzio M. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 39.Inoue Ji, et al. Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp. Cell Res. 2000;254:14–24. doi: 10.1006/excr.1999.4733. [DOI] [PubMed] [Google Scholar]
- 40.Darnay BG, Ni J, Moore PA, Aggarwal BB. Activation of NF-kappaB by RANK requires tumor necrosis factor receptor-associated factor (TRAF) 6 and NF-kappaB-inducing kinase. Identification of a novel TRAF6 interaction motif. J. Biol. Chem. 1999;274:7724–7731. doi: 10.1074/jbc.274.12.7724. [DOI] [PubMed] [Google Scholar]
- 41.Lamothe B, et al. The RING domain and first zinc finger of TRAF6 coordinate signaling by interleukin-1, lipopolysaccharide, and RANKL. J. Biol. Chem. 2008;283:24871–24880. doi: 10.1074/jbc.M802749200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi T, et al. TRAF6 is required for generation of the B-1a B cell compartment as well as T cell-dependent and -independent humoral immune responses. PLoS One. 2009;4:e4736. doi: 10.1371/journal.pone.0004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kozyrev SV, et al. Corrigendum: functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:484. doi: 10.1038/ng0408-484. [DOI] [PubMed] [Google Scholar]
- 44.Dieudé P, et al. BANK1 is a genetic risk factor for diffuse cutaneous systemic sclerosis and has additive effects with IRF5 and STAT4. Arthritis Rheumatol. 2009;60:3447–3454. doi: 10.1002/art.24885. [DOI] [PubMed] [Google Scholar]
- 45.Orozco G, et al. Study of functional variants of the BANK1 gene in rheumatoid arthritis. Arthritis Rheumatol. 2009;60:372–379. doi: 10.1002/art.24244. [DOI] [PubMed] [Google Scholar]
- 46.Suarez-Gestal M, et al. Rheumatoid arthritis does not share most of the newly identified systemic lupus erythematosus genetic factors. Arthritis Rheumatol. 2009;60:2558–2564. doi: 10.1002/art.24748. [DOI] [PubMed] [Google Scholar]
- 47.Troutman TD, et al. Role for B-cell adapter for PI3K (BCAP) as a signaling adapter linking Toll-like receptors (TLRs) to serine/threonine kinases PI3K/Akt. Proc. Natl Acad. Sci. USA. 2012;109:273–278. doi: 10.1073/pnas.1118579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozyrev SV, Bernal-Quirós M, Alarcón-Riquelme ME, Castillejo-López C. The dual effect of the lupus-associated polymorphism rs10516487 on BANK1 gene expression and protein localization. Genes Immun. 2012;13:129–138. doi: 10.1038/gene.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yokoyama K, et al. BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP(3) receptor. EMBO J. 2002;21:83–92. doi: 10.1093/emboj/21.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aiba Y, et al. BANK negatively regulates akt activation and subsequent B cell responses. Immunity. 2006;24:259–268. doi: 10.1016/j.immuni.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Dam EM, et al. The BANK1 SLE-risk variants are associated with alterations in peripheral B cell signaling and development in humans. Clin. Immunol. 2016;173:171–180. doi: 10.1016/j.clim.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y-Y, Kumar R, Haque MS, Castillejo-Lopez C, Alarcon-Riquelme ME. BANK1 controls CpG-induced IL-6 secretion via a p38 and MNK1/2/eIF4E translation initiation pathway. J. Immunol. 2013;191:6110–6116. doi: 10.4049/jimmunol.1301203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu Y-Y, Kumar R, Iida R, Bagavant H, Alarcón-Riquelme ME. BANK1 regulates IgG production in a lupus model by controlling TLR7-dependent STAT1 activation. PLoS One. 2016;11:e0156302. doi: 10.1371/journal.pone.0156302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang, S. H. et al. Functional rare and low frequency variants in BLK and BANK1 contribute to human lupus. Nat Commun10, 2201 (2019). [DOI] [PMC free article] [PubMed]
- 57.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 58.Bernal-Quirós M, Wu Y-Y, Alarcón-Riquelme ME, Castillejo-López C. BANK1 and BLK act through phospholipase C gamma 2 in B-cell signaling. PLoS One. 2013;8:e59842. doi: 10.1371/journal.pone.0059842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yen H-CS, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 60.Halabi S, Sekine E, Verstak B, Gay NJ, Moncrieffe MC. Structure of the Toll/Interleukin-1 receptor (TIR) domain of the B-cell adaptor that links phosphoinositide metabolism with the negative regulation of the Toll-like receptor (TLR) signalosome. J. Biol. Chem. 2017;292:652–660. doi: 10.1074/jbc.M116.761528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martínez-Bueno, M. et al. Trans-ethnic mapping of BANK1 identifies two independent SLE-risk linkage groups enriched for co-transcriptional splicing marks. Int. J. Mol. Sci.19, 10.3390/ijms19082331 (2018). [DOI] [PMC free article] [PubMed]
- 62.Into T, Inomata M, Niida S, Murakami Y, Shibata K. Regulation of MyD88 aggregation and the MyD88-dependent signaling pathway by sequestosome 1 and histone deacetylase 6. J. Biol. Chem. 2010;285:35759–35769. doi: 10.1074/jbc.M110.126904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13:816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 64.Lim KL, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.