Abstract
Cyclic GMP-AMP synthase (cGAS) is an essential sensor of cytosolic DNA and critically mediates innate immune responses and autoimmunity. Modulating the activity and stability of cGAS provides potential strategies for treating viral or autoimmune diseases. Here, we report that ubiquitin-specific protease 29 (USP29) deubiquitinates and stabilizes cGAS and promotes cellular antiviral responses and autoimmunity. Knockdown or knockout of USP29 severely impairs Herpes simplex virus 1 (HSV-1)- or cytosolic DNA-induced expression of type I interferons (IFNs) and proinflammatory cytokines. Consistently, Usp29m/m mice produce decreased type I IFNs and proinflammatory cytokines after HSV-1 infection and are hypersensitive to HSV-1 infection compared to the wild-type littermates. In addition, genetic ablation of USP29 in Trex1−/− mice eliminated the detectable pathological and molecular autoimmune phenotypes. Mechanistically, USP29 constitutively interacts with cGAS, deconjugates K48-linked polyubiquitin chains from cGAS and stabilizes cGAS in uninfected cells or after HSV-1 infection. Reconstitution of cGAS into Usp29−/− cells fully rescues type I IFN induction and cellular antiviral responses after HSV-1 infection. Our findings thus reveal a critical role of USP29 in the innate antiviral responses against DNA viruses and autoimmune diseases and provide insight into the regulation of cGAS.
Subject terms: Pattern recognition receptors, Autoimmunity, Cell signalling, Ubiquitylation
Introduction
Innate immune responses to viral infection are initiated by the recognition of viral nucleic acid by pattern-recognition receptors (PRRs) including Toll-like receptors (TLRs), RIG-I-like receptors and cytosolic DNA sensors.1–3 While RNA polymerase III, IFI16, and DDX41 recognize cytosolic DNA in a sequence- and/or cell type-dependent manner, the nucleotidyltransferase cyclic GMP-AMP (cGAMP) synthase (cGAS) detects DNA of various sequences in most types of cells and is recognized as the major cytosolic DNA sensor.4–9 Structural and biochemical studies have demonstrated that a minimum length of 45 bp DNA is required to robustly activate human cGAS (hcGAS) in cells and longer DNAs at lower concentrations are sufficient for cGAS activation.10,11 The binding of DNA to cGAS induces the formation of stable lower molecular weight protein-DNA complexes or higher molecular weight protein-DNA ladders to activate the enzyme activity of cGAS that converts GTP and ATP into cGAMP in a manner of liquid phase condensation.11–14 Several co-sensors or coactivators including G3BP1, PQBP1, and ZCCHC3 facilitate the binding of DNA to cGAS and promote cGAS activation.15–17 Consequently, cGAMP binds to the adapter protein MITA (also known as STING and ERIS) to recruit downstream kinases and activate transcription factors such as IRF3 and NF-κB that collaborate to induce type I interferons (IFNs) and proinflammatory cytokines.18,19 Type I IFNs and proinflammatory cytokines further amplify the antiviral immune signaling by inducing the expression of a large array of genes whose products collaboratively elicit cellular antiviral responses.
In addition to viral DNA, the cGAS-STING pathway also recognizes apoptotic signal-induced mitochondrial DNA (mtDNA), stress- or enzyme-induced cytosolic accumulation of nuclear DNA and cytoplasmic leaked nuclear DNA from unstable genome or failed repair of DNA damage.20–23 For example, loss-of-function mutations of DNase three prime repair exonuclease 1 (TREX1), DNase II and RNase H2 result in hyper production of type I IFNs and proinflammatory cytokines and are linked to Aicardi-Goutieres Syndrome (AGS) and systemic lupus erythematosus (SLE) in humans. Trex1−/− or DNaseII−/− mice exhibit severe systemic inflammation and even lethal autoimmunity, whereas deletion of cGAS in Trex1−/− or DNaseII−/− mice completely rescues the autoimmune phenotypes, suggesting critical roles of cGAS in autoimmune diseases caused by self-DNA.24
Given the importance of the cGAS-STING pathway in antiviral immunity and autoimmunity, the activity and stability of cGAS must be tightly controlled to elicit protective immune responses and avoid harmful immune pathology. Various posttranslational modifications have been reported to control the activity and stability of cGAS. For example, Akt phosphorylates Ser305 on human or Ser291 on mouse cGAS to impair its enzyme activity.25 Tubulin tyrosine ligase-like 4 and 6 (TTLL4/6) and cytosolic carboxypeptidase 5 and 6 (CCP5/6) catalyze glutamylation and deglutamylation of cGAS at Glu302 and Glu272, thereby turning off or on the DNA binding and catalytic activity of cGAS, respectively.26 Acetylation of cGAS on Lys384, Lys394, or Lys414 inactivates cGAS and cytoplasmic DNA stimulation or HSV-1 infection leads to deacetylation of cGAS.27 TRIM56 and RINCK catalyze monoubiquitination of cGAS and RNF185 induces K27-linked polyubiquitination of cGAS,28–30 which facilitate the activation of cGAS after cytoplasmic DNA challenge. The autophagy inducer Beclin-1 interacts with cGAS and promotes autophagy-mediated degradation of DNA to prevent excessive activation of cGAS, whereas the autophagy receptor p62 recognizes K48-linked polyubiquitin modified cGAS to promote its degradation in the autophagosome after HSV-1 infection.31,32 TRIM14 recruits USP14 to cleave the K48-linked ubiquitin chains from Lys414 of cGAS after viral infection, thereby preventing the autophagy-dependent degradation of cGAS in a manner dependent on viral infection.31
The activity and stability of cGAS is also regulated by sumoylation in resting cells and after DNA virus infection. HSV-1-induced sumoylation at Lys335/372/382 of cGAS impairs its DNA binding ability and self-association and SENP7 desumoylates such a modification to promote cGAS activation.33 Another study demonstrates a step-wise sumoylation of cGAS controls its ubiquitination and degradation. A portion of cGAS is constitutively sumoylated at Lys217 in the absence of infection and HSV-1 infection at early phase triggers sumoylation at Lys464 of cGAS, which prevents the K48-linked ubiquitination and subsequent proteasomal degradation of cGAS.34 At late phase of infection, SENP2 mediates desumoylation of cGAS at Lys464 to allow the ubiquitin conjugation at the same residue and subsequent proteasome-dependent degradation of cGAS, thereby terminating the signaling to avoid excessive immune responses.34 Although the Lys217-unsumoylated and Lys464-desumoylated cGAS is unstable and prone to ubiquitination and degradation, it is noted that a large portion of unmodified cGAS is detectable in unstimulated or late-stage stimulated cells, indicating that additional regulatory mechanisms exist to control the stability of cGAS in resting or stimulated cells.
Since ubiquitination strictly controls stability and activity of cGAS, we hypothesize that deubiquitinating enzymes (DUBs) mediate deubiquitination and stability of cGAS and play essential roles in cGAS-mediated antiviral immunity and autoimmunity. By transient transfection and coimmunoprecipitation assays, we have identified USP29 as a cGAS-interacting DUB. USP29 constitutively interacts with cGAS and deconjugates K48-linked polyubiquitin chains from cGAS in the absence or presence of HSV-1 infection, which stabilizes cGAS and promotes cGAS-mediated cellular antiviral responses. Consistently, knockout of USP29 results in constitutive degradation of cGAS and eliminates the autoimmune phenotypes in Trex1−/− mice. These findings provide insight into the mechanisms that control cGAS stability, antiviral immunity and autoimmunity.
Results
Identification of USP29 as a cGAS-interacting DUB
It has been previously demonstrated that TRIM38 constitutively catalyzes sumoylation of cGAS at Lys217, which prevents cGAS from ubiquitination and protesomal degradation in unstimulated cells.34 Interestingly, we found that cGAS was rapidly degraded in THP-1 cells treated with cycloheximide (CHX), an inhibitor for eukaryotic translational elongation and protein synthesis, while the levels of TRIM38 remained stable in these cells (Supplementary information, Fig. S1a). In addition, CHX treatment-induced degradation of cGAS was completely rescued by the treatment of MG132, a proteasome inhibitor (Supplementary information, Fig. S1b), indicating that cGAS constitutively undergoes degradation in a proteasome-dependent manner. Consistent with this notion, we observed that MG132 treatment increased K48-linked ubiquitination of cGAS and led to the accumulation of cGAS (Supplementary information, Fig. S1c). These findings prompted us to hypothesize that E3 ligases and deubiquitinating enzymes (DUBs) might exist to regulate the stability of cGAS by modulating the ubiquitination and deubiquitination of cGAS (Supplementary information, Fig. S1d).
To test this hypothesis, we conducted an unbiased screening of cGAS-interacting DUBs by cotransfection and immunoprecipitation assays as previous established,35–37 and found that USP25 and USP29 interacted with cGAS in such a system (Fig. 1a). In a parallel luciferase reporter screening, we found that overexpression of USP29 but not USP25 potentiated cGAS and MITA-mediated activation of IFN-β promoter (Supplementary information, Fig. S2a). USP29 has been implicated in cell cycle and oxidative stress by deubiquitinating and stabilizing Claspin and p53, respectively.38,39 Whether and how USP29 regulated cGAS or cGAS-mediated antiviral signaling is unclear. We further found that overexpression of USP29 potentiated cGAS and MITA-mediated activation of IFN-β promoter in a dose-dependent manner (Supplementary information, Fig. S2b), whereas knockdown of USP29 had opposite effects (described below). Results from immunoblot analysis suggested that USP29 was ubiquitously expressed in various mouse organs, primary mouse cells and human cell lines (Supplementary information, Fig. S2c). In addition, USP29 selectively interacted with cGAS but not with other molecules including MITA, RIG-I, MAVS, TBK1 or IRF3 (Supplementary information, Fig. S2d). We thus focused on USP29 for further study.
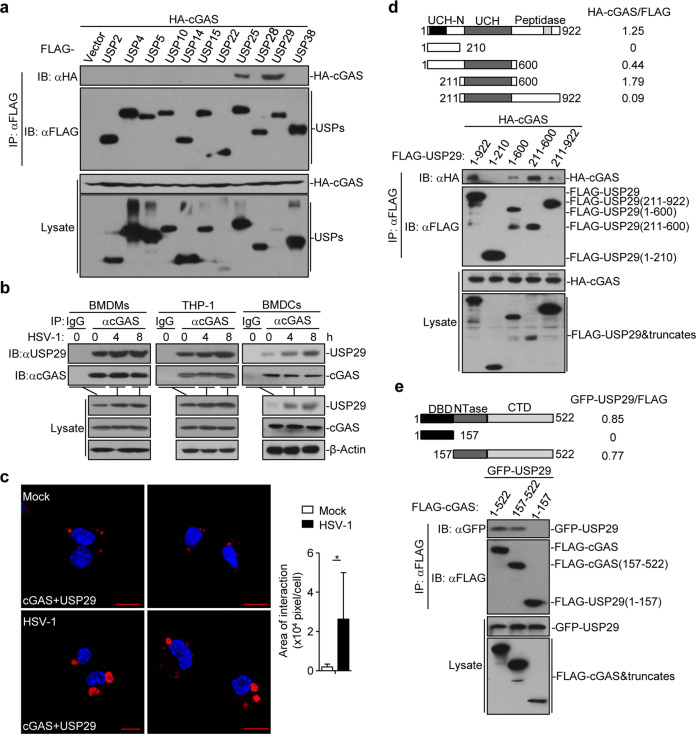
Fig. 1. USP29 interacts with cGAS.
a Immunoprecipitation (IP, with anti-FLAG) and immunoblot analysis (with anti-FLAG and anti-HA) of HEK293 cells transfected with plasmids encoding FLAG-tagged USPs and HA-cGAS for 24 h. b Immunoprecipitation (with anti-cGAS or control IgG) and immunoblot analysis (with anti-cGAS or anti-USP29) of BMDMs (left), THP-1(middle) and BMDCs (right) left uninfected or infected with HSV-1 for 4-8 h. c Proximity ligation assay (PLA) and confocal microscopy imaging of cGAS and USP29 interaction in MEFs that were left uninfected (Mock) or infected with HSV-1 for 4 h (left images). The association intensities of the red spots were quantified with Image-Pro Plus software and calculated as pixels per cell (right graph) (n = 8). d, e Immunoprecipitation (with anti-FLAG) and immunoblot analysis (with anti-FLAG or anti-HA) of HEK293 cells transfected with plasmids encoding HA-cGAS and FLAG-tagged USP29 or USP29 truncates for 24 h (d) or transfected with plasmids encoding GFP-USP29 and FLAG-cGAS or cGAS truncates for 24 h (e). *P < 0.05 (two-tailed Student’s t-test). Scale bars represent 10 μm. Data are representative of three independent experiments (means ± SD in c).
Results from endogenous immunoprecipitation assays suggested that USP29 constitutively interacted with cGAS in bone marrow-derived macrophages (BMDMs), bone marrow-derived dendritic cells (BMDCs), mouse embryonic fibroblasts (MEFs), mouse lung fibroblasts (MLFs) or human monocytic THP-1 cells and their association was potentiated after HSV-1 infection (Fig. 1b and Supplementary information, Fig. S2e). In contrast, USP25 did not interact with cGAS in MEFs before or after HSV-1 infection (Supplementary information, Fig. S2f). Results from proximity ligation assay and confocal microscopy imaging further confirmed that a portion of cGAS and USP29 was constitutively colocalized without infection and such a colocalization was significantly increased after HSV-1 infection (Fig. 1c). Domain mapping analysis showed that the ubiquitin carboxyl-terminal hydrolase (UCH) domain of USP29 and the C-terminal domain containing the nucleotidyltransferase (NTase) domain of cGAS were responsible for USP29-cGAS association (Fig. 1d, e). In addition, results from GST pulldown assay suggested that the enzymatic motif of USP29 directly associated with in vitro translated FLAG-cGAS (Supplementary information, Fig. S2g). Interestingly, we found that TRIM38 constitutively interacts with cGAS but not with USP29, whereas USP29 constitutively interacts with cGAS but not with TRIM38 (Supplementary information, Fig. S2h), indicating that TRIM38 and USP29 independently interacts with and regulates cGAS. Together, these data suggest that USP29 interacts with cGAS directly and constitutively in resting cells and after HSV-1 infection.
Interestingly, we found that USP29 protein was accumulated in various primary mouse cells and THP-1 cells after HSV-1 infection (Fig. 1b and Supplementary information, Fig. S2e). Quantitative reverse-transcription PCR (qRT-PCR) analysis suggested that HSV-1 infection but not Sendai virus (SeV) infection, cytoplasmic immunostimulatory DNA 45 bp (ISD45) challenge or IFN-β treatment upregulated the mRNA of USP29 gene in THP-1 cells (Supplementary information, Fig. S2i). Sequence analysis of USP29 promoter (−5000 bp ~ +100 bp) failed to identify any recognizable IRF, NF-κB, or STAT binding sites, indicating that the transcriptional regulation of USP29 by viral infection is different from that of OTUD4, USP25 or INKIT previously reported.37,40,41 In this context, it has been shown that transcription factors JTV1 and FBP co-activate USP29 transcription in response to oxidative stress and that one of the evolutionarily conserved sequence elements (CSE1) on Usp29 promoter suppresses its transcription.39,42
USP29 promotes HSV-1-triggered innate antiviral signaling in THP-1 cells
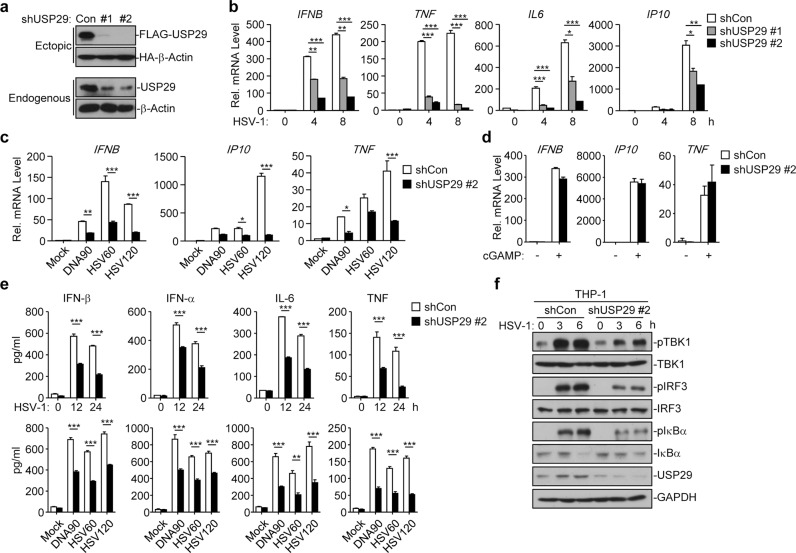
Because USP29 interacted with cGAS and was upregulated by HSV-1 infection, we investigated its role in regulating HSV-1- or cytoplasmic DNA-triggered innate immune signaling. We designed two shRNAs of USP29, both of which potently downregulated the protein levels of endogenous or ectopic expressed USP29 in THP-1 cells or HEK293 cells, respectively (Fig. 2a). Knockdown of USP29 by these two shRNAs significantly inhibited HSV-1-induced expression of IFNB, TNF, IL6, and IP10 in THP-1 cells (Fig. 2b) (The #2 shRNA was used for the following experiments and similar results were observed with #1 shRNA). Interestingly, we found that knockdown of USP29 significantly impaired various cytoplasmic DNA-induced expression of IFNB, TNF, and IP10 but had little effect on cGAMP-induced expression of IFNB, TNF, and IP10 (Fig. 2c, d). In addition, knockdown of USP29 significantly impaired the production of IFN-β, IFN-α, IL-6, and TNF after infection of HSV-1 or transfection of various DNA ligands (Fig. 2e). Consistent with the results from gene induction analysis, knockdown of USP29 substantially impaired HSV-1-induced phosphorylation of TBK1, IRF3, or IκBα (Fig. 2f). These data together suggest that USP29 positively regulates DNA virus- or cytoplasmic DNA-triggered innate immune signaling.
Fig. 2. Knockdown of USP29 impairs HSV-1-induced innate immune signaling.
a Immunoblot analysis (with anti-FLAG or anti-HA) of HEK293 cells transfected for 36 h with plasmids encoding FLAG-tagged USP29 and HA-β-Actin and either USP29-targeting shRNA (shUSP29#1 and shUSP29#2) or control shRNA (Con) to test the knockdown efficiency of shRNA (upper two panels). Immunoblot analysis (with anti-USP29 or anti-β-Actin) of THP-1 cells stably transfected with plasmids encoding control (Con) shUSP29 (#1 or #2) (lower two panels). b qRT-PCR analysis of IFNB, TNF, IL6, and IP10 in THP-1 cells stably transfected with control shRNA, shUSP29#1 or shUSP29#2 that were infected with HSV-1 for 0–8 h. c qRT-PCR analysis of IFNB, IP10, and TNF in THP-1 cells stably transfected with control shRNA, shUSP29#2 that were mock transfected or transfected with DNA90, HSV60 or HSV120 for 6 h. d qRT-PCR analysis of IFNB, TNF and IP10 mRNA in THP-1 cells stably transfected with control shRNA or shUSP29#2 that were left untreated (Mock) or treated with cGAMP for 4 h. e ELISA analysis of IFN-α, IFN-β, IL-6 and TNF in the supernatants of THP-1 cells stably transfected with control shRNA or shUSP29#2 that were infected with HSV-1 for 0–24 h, or mock transfected or transfected with DNA90, HSV60 or HSV120 for 12 h. f Immunoblot analysis of phosphorylated and total TBK1, IRF3 and IκBα, and USP29 and GAPDH in THP-1 cells stably transfected with control shRNA or shUSP29#2 that were infected with HSV-1 for 0–6 h. *P < 0.05; **P < 0.001; ***P < 0.001 (analysis of two-way ANOVA followed by Bonferroni post-test or two-tailed Student’s t-test). Data are representative of three independent experiments (means ± SD in b–e).
USP29 deficiency impairs HSV-1-triggered expression of downstream genes
To further investigate the role of USP29 in regulating antiviral signaling in vivo, we generated USP29-deficient mice by CRISPR/Cas9-mediated genome editing (Supplementary information, Fig. S3a). Results from sequencing of tail DNA showed that a deoxylguanine and a deoxylthymine were inserted at +193 position on exon 8 of Usp29 gene, which caused a frame shift and led to an early translational termination of USP29 (aa1-62) (Supplementary information, Fig. S3b). The Usp29m/+ mice bred normally with the Mendelian inheritance ratio (Supplementary information, Fig. S3c). The numbers and percentages of various lymphocytes in thymus, spleen or peripheral lymph nodes were comparable between the wild-type and Usp29m/m mice (Supplementary information, Fig. S3d-f), indicating that USP29 is dispensable for the development and homeostasis of lymphocytes. The differentiation of Usp29m/m bone marrow cells into BMDCs or BMDMs was similar to that of wild-type counterparts in the presence of GM-CSF or M-CSF, respectively (Supplementary information, Fig. 3g), suggesting a dispensable role of USP29 for in vitro BMDM or BMDC differentiation.
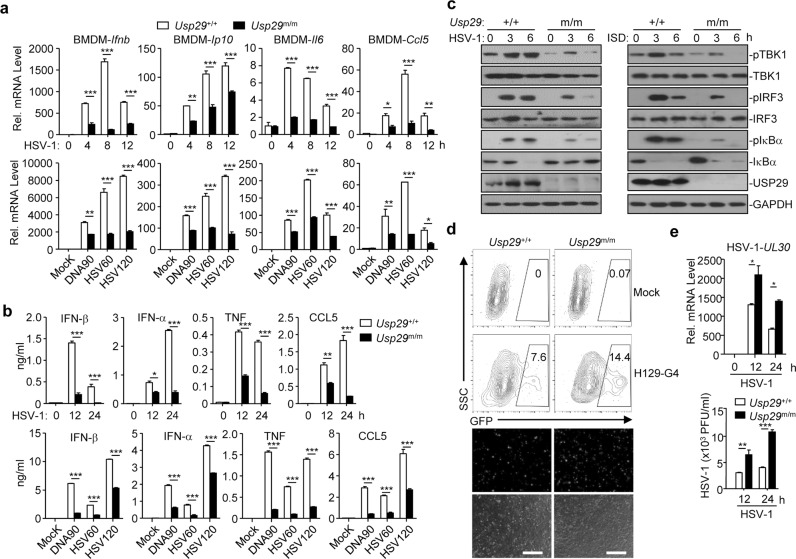
We next examined the effect of USP29 deficiency on HSV-1-triggered innate immune responses in primary murine cells and found that HSV-1- or cytoplasmic DNA-induced expression of Ifnb, Ip10, Il6, or Ccl5 and the production of IFN-β, IFN-α, TNF, or CCL5 were impaired in Usp29m/m BMDMs or MEFs compared to the Usp29+/+ counterparts (Fig. 3a, b and Supplementary information, Fig. S4a). In contrast, knockout of USP29 had little effect on cGAMP-, SeV- or transfected Poly(I:C)-induced expression of Ifnb, Ifna6, Ifna4, Tnf, and Il6 in MEFs or BMDMs (Supplementary information, Fig. S4b, c). In addition, LPS-, Poly(I:C)- or CpG-induced expression of Ifna, Ifnb and Tnf was comparable between Usp29+/+ and Usp29m/m BMDCs (Supplementary information, Fig. S4d). Consistent with these observations, HSV-1- but not SeV-induced phosphorylation of TBK1, IRF3, and IκBα was significantly compromised in Usp29m/m BMDMs or MEFs compared to the Usp29+/+ counterparts (Fig. 3c and Supplementary information, Fig. S4e), indicating that USP29 selectively regulates DNA virus- and cytoplasmic DNA-triggered cGAS-dependent innate antiviral signaling.
Fig. 3. USP29 deficiency impairs HSV-1-triggered expression of downstream genes.
a qRT-PCR analysis of Ifnb, Ip10, Il6, and Ccl5 mRNA in Usp29+/+ and Usp29m/m BMDMs infected with HSV-1 for 0–12 h, mock transfected (Mock) or transfected with DNA90, HSV60, or HSV120 for 6 h. b ELISA analysis of IFN-β, IFN-α, TNF, and CCL5 in the supernatants of Usp29+/+ and Usp29m/m BMDMs infected with HSV-1 for 0–24 h, or mock transfected (Mock) or transfected with DNA90, HSV60 or HSV120 for 12 h. c Immunoblot analysis of phosphorylated and total TBK1, IRF3 and IκBα, USP29 and GAPDH in Usp29+/+ and Usp29m/m BMDMs infected with HSV-1 for 0–6 h or transfected with ISD45 for 0–6 h. d Flow cytometry analysis (upper flow charts) and fluorescent microscopy imaging (lower images) of Usp29+/+ and Usp29m/m BMDMs infected with or without H129-G4 for 16 h. e qRT-PCR analysis of HSV-1 UL30 (upper) and plaque assays (bottom) of Usp29+/+ and Usp29m/m BMDMs infected with or without HSV-1 for 0–24 h. *P < 0.05; **P < 0.001; ***P < 0.001 (two-tailed Student’s t-test). Scale bars represent 300 μm. Data are representative of two (a, b, d, e) or three (c) independent experiments (means ± SD in a, b, e).
Viral infection-induced production of cytokines is critical for control of virus replication. We next examined the effect of USP29 deficiency on HSV-1 replication and found that the replication of H129-G4 (a GFP-tagged strain of HSV-1)43 was potentiated in Usp29m/m BMDMs or MEFs compared to the Usp29+/+ counterparts as monitored by the GFP percentages (Fig. 3d and Supplementary information, Fig. S4f). Results from qRT-PCR and plaque assays further confirmed that knockout of USP29 promoted HSV-1 replication in BMDMs and MEFs as monitored by the expression of HSV-1 UL30 gene in cells and the HSV-1 titers in the supernatants (Fig. 3e and Supplementary information, Fig. S4g). These data collectively suggest that USP29 deficiency restricts cellular antiviral responses against HSV-1.
Usp29m/m mice exhibit hypersensitivity to HSV-1 infection
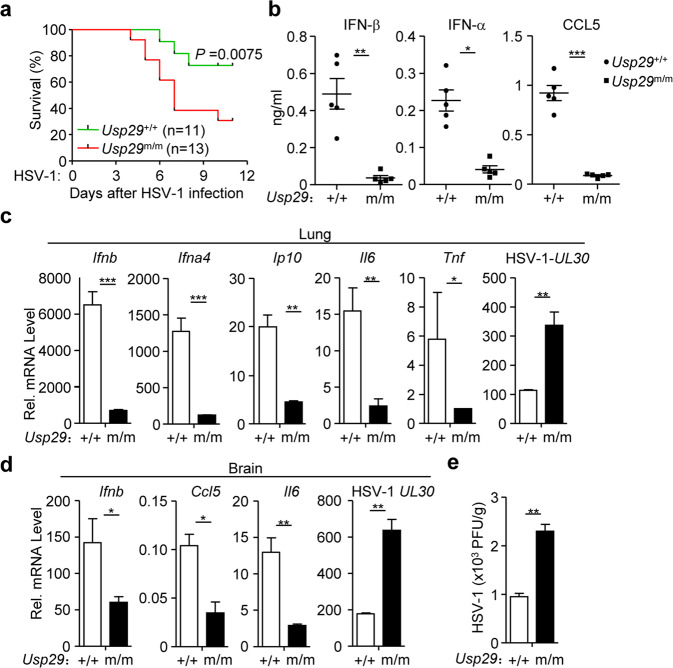
To investigate the physiological role of USP29 in response to HSV-1 infection, we challenged Usp29+/+ and Usp29m/m mice with HSV-1 through intravenous (i.v.) injection and monitored their survival. The Usp29m/m mice exhibited an earlier death onset and were more susceptible to HSV-1 infection than the wild-type littermates (Fig. 4a). The protein levels of IFN-β, IFN-α, and CCL5 were significantly compromised in the sera of Usp29m/m mice compared to those in sera from the wild-type littermates at 12 h after HSV-1 infection (Fig. 4b). The expression of Ifnb, Ifna4, Ip10, Il6, and Tnf was impaired and the expression of HSV-1 UL30 gene was increased in the lungs from Usp29m/m mice compared to Usp29+/+ mice at 24 h after HSV-1 infection (Fig. 4c). We also analyzed the expression of cytokines and viral titers in the brains 4 days after HSV-1 infection and observed that the expression of Ifnb, Ccl5, and Il6 was decreased and the titers of HSV-1 were increased in the brains from Usp29m/m mice compared to those from Usp29+/+ mice (Fig. 4d, e). Collectively, these data suggest that USP29 physiologically regulates HSV-1-induced production of type I IFNs and proinflammatory cytokines and innate immune responses against HSV-1 infection.
Fig. 4. Usp29m/m mice are hypersensitive to HSV-1 infection.
a Survival (Kaplan–Meier curve) of Usp29+/+ (n = 11) and Usp29m/m mice (n = 13) intravenously injected with HSV-1 (1.5 × 106 PFU per mouse) monitored survival for 12 days. b ELISA analysis of IFN-β, IFN-α, and CCL5 in the sera of Usp29+/+ and Usp29m/m mice (n = 5) intravenously injected with HSV-1 (2 × 106 PFU per mouse) for 12 h. c qRT-PCR analysis of Ifnb, Ifna4, Ip10, Il6, Tnf or HSV-1 UL30 mRNA in the lung from Usp29+/+ and Usp29m/m mice intravenously injected with HSV-1 (2 × 106) for 24 h. d, e qRT-PCR analysis of Ifnb, Ccl5, Il6 and HSV-1 UL30 (d) and plaque assays (e) of the brains from Usp29+/+ and Usp29m/m mice intraperitoneally injected with HSV-1 (2 × 106) for 4 days. *P < 0.05; **P < 0.01; ***P < 0.001 (two-tailed Student’s t-test, b–e). Data are of combined two independent experiments (a) or representative of two (b–e) independent experiments (means ± SD in b–e).
The DUB activity of USP29 is required for optimal cellular antiviral responses
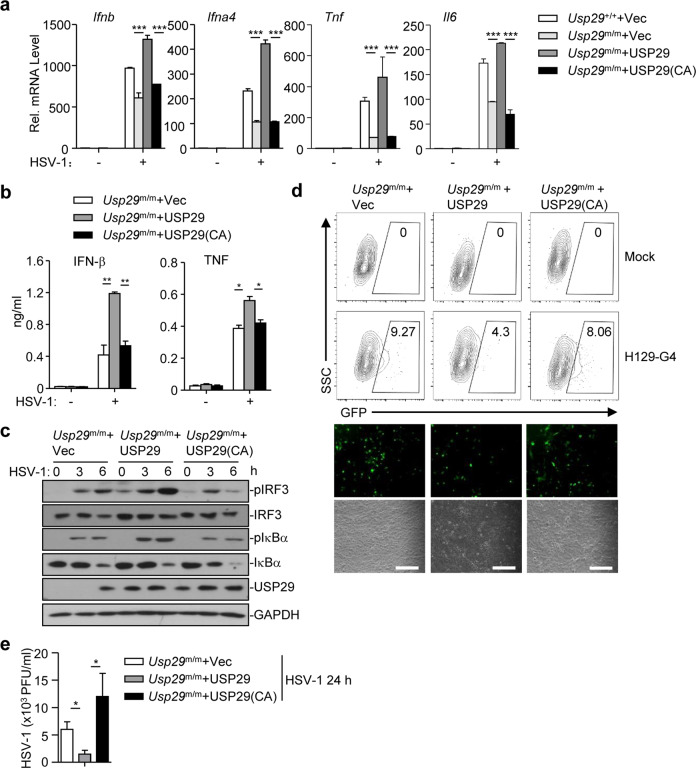
We next examined whether the enzymatic activity of USP29 was required for its regulation of cellular antiviral responses against HSV-1. As shown in Fig. 5a, reconstitution of wild-type USP29 but not the inactive mutant USP29(C298A) into Usp29m/m MLFs fully restored the expression of Ifnb, Ifna4, Tnf, or Il6. In addition, the production of IFN-β or TNF and phosphorylation of IRF3 and IκBα was rescued by reconstitution of USP29 but not USP29(C298A) into Usp29m/m MLFs (Fig. 5b, c). Consistently, the replication of H129-G4 or HSV-1 was substantially inhibited in Usp29m/m MLFs reconstituted with USP29 but not USP29(C298A) as determined by the GFP signals and the HSV-1 titers in the supernatants, respectively (Fig. 5d, e). These data suggest that USP29 promotes cellular antiviral responses in a manner dependent on its DUB activity.
Fig. 5. The DUB activity of USP29 is required for optimal cellular antiviral responses.
a qRT-PCR analysis of Ifnb, Ifna4, Tnf, or Il6 mRNA in Usp29+/+ MLFs reconstituted with the empty vector (Usp29+/++Vec) or in Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A) [Usp29m/m + USP29 (C298A)] infected with HSV-1 for 0–6 h. b, c ELISA analysis of IFN-β and TNF (b) and immunoblot analysis of phosphorylated and total IRF3 and IκBα, USP29, and GAPDH (c) in Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A)[Usp29m/m + USP29(C298A)] infected with HSV-1 for 0–6 h. d Flow cytometry analysis (upper flow charts), fluorescent microscopy imaging (lower images) of Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A) [Usp29m/m + USP29(C298A)] infected with or without H129-G4 for 24 h. e Plaque assays of Usp29m/m MLFs reconstituted with the empty vector (Usp29m/m + Vec), USP29 (Usp29m/m + USP29), or USP29 (C298A) [Usp29m/m + USP29(C298A)] infected with or without HSV-1 for 24 h. *P < 0.05; **P < 0.001; ***P < 0.001 (analysis of two-way ANOVA followed by Bonferroni post-test). Scale bars represent 300 μm. Data are representative of three (a, b, d, e) or two (c) independent experiments (means ± SD in a, b, e).
USP29 deubiquitinates cGAS in the presence or absence of HSV-1 infection
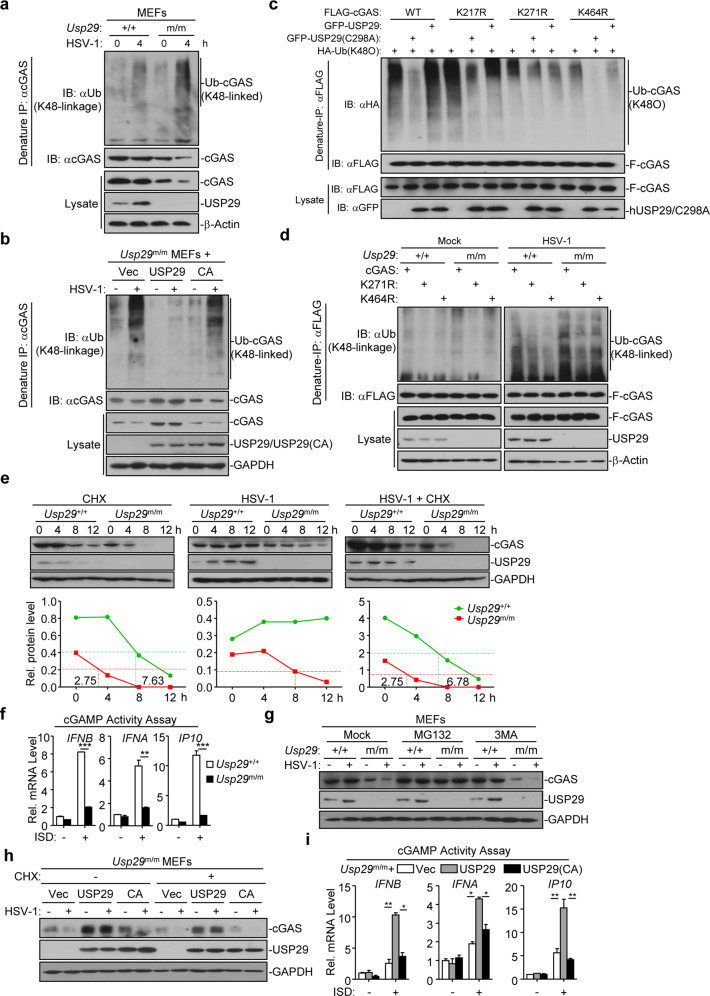
Because USP29 interacts with cGAS and its enzyme activity is required for regulating antiviral signaling, we investigated whether USP29 eliminated polyubiquitin chains from cGAS. Expectedly, hUSP29 but not hUSP29(C294I) which is the human analogous inactive form of murine USP29(C298A) catalyzed deubiquitination of hcGAS in cells or in vitro (Supplementary information, Fig. S5a, b). To examine whether hUSP29 removed K48- or K63-linked polyubiquitin chains from hcGAS, we transfected hcGAS together with ubiquitin, or ubiquitin mutants either retaining a single lysine residue (KO) or retaining all but one lysine residues (KR) in the presence or absence of hUSP29 followed by deubiquitination assays. The results showed that hUSP29 catalyzed removal of K48O-linked but not K48R-linked polyubiquitin chains from hcGAS in HEK293 cells (Supplementary information, Fig. S5c). We further found that the basal or HSV-1-induced K48-linked ubiquitination of cGAS was substantially potentiated in Usp29m/m MEFs compared to the Usp29+/+ counterparts (Fig. 6a), whereas reconstitution of USP29 but not USP29(C298A) into Usp29m/m MEFs eliminated basal or HSV-1-induced K48-linked ubiquitination of cGAS (Fig. 6b). Taken together, these data suggest that USP29 targets cGAS for deubiquitination in the presence or absence of HSV-1 infection.
Fig. 6. USP29 deubiquitinates and stabilizes cGAS.
a Denature-IP (with anti-cGAS) and immunoblot analysis [with anti-Ub (K48-linkage), anti-cGAS, anti-USP29 or anti-β-Actin] of Usp29+/+ and Usp29m/m MEFs left uninfected or infected with HSV-1 for 4 h. b Denature-IP (with anti-cGAS) and immunoblot analysis [with anti-Ub (K48-linkage), anti- cGAS, anti-USP29 or anti-GAPDH] of Usp29m/m MEFs reconstituted with empty vector, USP29 or USP29(C298A) left uninfected or infected with HSV-1 for 4 h. c Denature-IP (with anti-FLAG) and immunoblot analysis (with anti-FLAG, anti-HA or anti-GFP) of HEK293 cells transfected with plasmids encoding HA-Ub(K48O), FLAG-cGAS or FLAG-cGAS mutants together with an empty vector, GFP-USP29 or GFP-USP29(C298A) for 24 h. d Denature-IP (with anti-FLAG) and immunoblot analysis [with anti-Ub (K48-linkage), anti-FLAG, anti-USP29 or anti-β-Actin] of Usp29+/+ and Usp29m/m MLFs reconstituted with cGAS, cGAS(K271R) or cGAS(K464R) left uninfected or infected with HSV-1 for 6 h. e Immunoblot analysis of cGAS, USP29 and GAPDH (upper blots) and quantitation of the intensities of cGAS (relative to GAPDH) (lower graphs) in Usp29+/+ and Usp29m/m MLFs infected with or without HSV-1 in the presence or absence of CHX for 0–12 h. f Usp29+/+ and Usp29m/m MLFs were transfected with ISD45 for 4 h, and then the cell extracts containing cGAMP were delivered to digitonin-permeabilized HFFs for 4 h followed by qRT-PCR analysis. g Immunoblot analysis of cGAS, USP29 and GAPDH in Usp29+/+ and Usp29m/m MEFs that were treated with MG132 or 3MA, followed by HSV-1 infection for 0–4 h. h Immunoblot analysis of cGAS, USP29 and GAPDH in Usp29m/m MEFs reconstituted with the empty vector (Vec), USP29 (USP29), or USP29 (C298A) (CA) that were infected by HSV-1 in the presence or absence of CHX for 0–4 h. i Usp29m/m MEFs reconstituted with the empty vector (Vec), USP29 (USP29) or USP29 (C298A) (CA) were transfected with ISD45 for 4 h, and then cell extracts containing cGAMP were delivered to digitonin-permeabilized HFFs for 4 h followed by qRT-PCR analysis. *P < 0.05; **P < 0.001; ***P < 0.001 (analysis of two-way ANOVA followed by Bonferroni post-test or two-tailed Student’s t-test). Data are representative of three (f, i) or two (a, b, c, d, e, g, h) independent experiments (means ± SD in f, i).
It has been shown that cGAS undergoes K48-linked ubiquitination at Lys271 in resting cells and at Lys464 after viral infection, respectively.34 We examined whether USP29 cleaved polyubiquitin chains from Lys271 or Lys464 of cGAS. Consistent with the previous report,34 mutation of Lys271 into Arg diminished basal K48-linked ubiquitination of cGAS (Fig. 6c and Supplementary information, Fig. S5d). Ectopic expression of USP29 but not USP29(C298A) impaired K48-linked ubiquitination of wild-type cGAS or cGAS(K464R) but not cGAS(K271R) (Fig. 6c and Supplementary information, Fig. S5d). In addition, we adjusted the lenti-viral titers of cGAS or cGAS mutants for reconstitution into Usp29+/+ or Usp29m/m MLFs to assure the equal expression of cGAS and the mutants in these cells followed by HSV-1 infection and ubiquitination assays. The results showed that the K48-linked ubiquitination of cGAS and cGAS(K464R) was increased in Usp29m/m MLFs compared to Usp29+/+ MLFs before or after HSV-1 infection (Fig. 6d, the Mock panel and the HSV-1 panel, lane 4 v.s. 1, and lane 6 v.s. 3). In contrast, the K48-linked ubiquitination of cGAS(K271R) was comparable between Usp29m/m and Usp29+/+ MLFs (Fig. 6d, the Mock panel and the HSV-1 panel, lane 5 v.s. 2), suggesting that USP29 regulates deubiquitination of cGAS at K271 in resting and HSV-1-infected cells. These data together indicate that USP29 removes K48-linked polyubiquitin chains at Lys271 of cGAS in the presence or absence of HSV-1 infection.
USP29 protects cGAS from proteasomal degradation
We next examined whether USP29-mediated deubiquitination of cGAS promoted its stability. In the absence of HSV-1 infection, the basal cGAS protein level was substantially lower in Usp29m/m MLFs than in Usp29+/+ MLFs, although the basal cGas mRNA was comparable between Usp29m/m and Usp29+/+ MLFs (Fig. 6e and Supplementary information, Fig. S5e). The half-life of basal cGAS was substantially shorter in Usp29m/m MLFs compared to Usp29+/+ MLFs in the presence or absence of HSV-1 infection (Fig. 6e), indicating that USP29 controls the stability of cGAS. Interestingly, however, HSV-1 infection alone did not downregulate cGAS protein in Usp29+/+ MLFs, which was a result of transcriptional induction of cGas by HSV-1 infection (Fig. 6e and Supplementary information, Fig. S5e). In contrast, cGAS was still degraded in Usp29m/m MLFs (half-life ~8 h) after HSV-1 infection, which was due to inefficient induction of cGas transcription and accelerated degradation of basal and newly synthesized cGAS caused by the absence of USP29 (Fig. 6e and Supplementary information, Fig. S5e). In support of this notion, we found that the cGAMP activity was significantly lower in Usp29m/m MLFs than in Usp29+/+ MLFs after transfection of ISD45 (Fig. 6f). These data suggest that USP29 is required for the stabilization of cGAS, which facilitates efficient cGAMP production after cytoplasmic DNA challenge.
Consistent with the notion that USP29 deficiency promoted K48-linked ubiquitination and degradation of cGAS, we found that treatment of MG132 but not 3MA fully prevented the degradation of cGAS in Usp29m/m MEFs in the presence or absence of HSV-1 infection (Fig. 6g). In addition, reconstitution of USP29 but not USP29(C298A) into Usp29m/m MEFs restored the protein levels of cGAS with or without HSV-1 infection and cGAMP production after cytoplasmic DNA challenge (Fig. 6h, i). These data together suggest that USP29 deconjugates K48-linked polyubiquitin chains from cGAS and thereby inhibits the proteasomal degradation of cGAS.
To further substantiate our conclusion that USP29 positively regulates antiviral immunity by stabilizing cGAS, we reconstituted an empty vector or cGAS into Usp29+/+ or Usp29m/m MLFs and examined HSV-1-triggered signaling in these cells. Results from qRT-PCR and ELISA analysis showed that HSV-1-induced expression of Ifnb, Ip10, Ccl5 or Il6 and production of IFN-β or TNF were rescued in Usp29m/m MLFs reconstituted with cGAS (Supplementary information, Fig. S6a, b). Consistently, reconstitution of cGAS into Usp29m/m MLFs restored cGAMP production by cytoplasmic DNA challenge and phosphorylation of TBK1, IRF3 and IκBα after HSV-1 infection (Supplementary information, Fig. S6c, d). In addition, the replication of H129-G4 or HSV-1 was substantially inhibited in Usp29m/m MLFs reconstituted with cGAS (Supplementary information, Fig. S6e, f). Taken together, these data suggest that USP29 targets cGAS for deubiquitination to stabilize cGAS and positively regulate cellular antiviral responses against HSV-1 infection.
USP29 deficiency alleviates autoimmunity in Trex1−/− mice
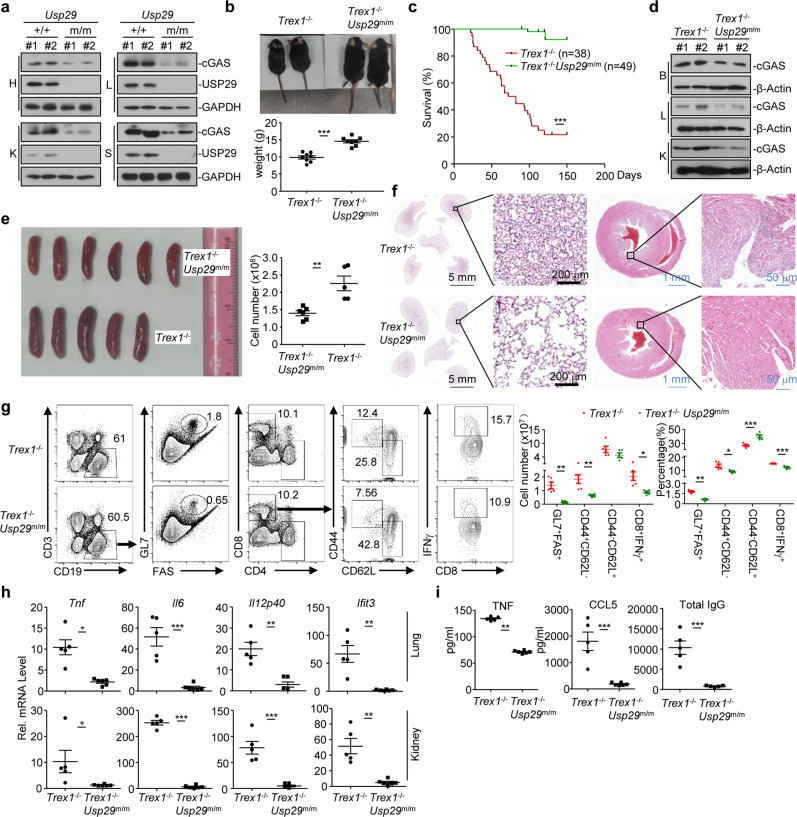
Consistent with the results obtained with USP29 knockout cells, we found that the protein levels of cGAS were substantially lower in various organs such as heart, liver, kidney and spleen from Usp29m/m mice than in those from Usp29+/+ mice (Fig. 7a). Considering that cGAS critically mediates autoimmune phenotypes in Trex1−/− mice,24 we investigated the role of USP29 in autoimmunity caused by Trex1 deficiency. We generated Trex1 knockout mice by CRIPSR/Cas9-mediated genome editing and obtained the Trex1−/−Usp29m/m mice (Supplementary information, Fig. S7a-c). Interestingly, the levels of cGAS and the expression of Tnf, Il6, Il12p40 and Ifit3 were significantly decreased in Trex1−/−Usp29m/m BMDCs compared to Trex1−/− BMDCs (Supplementary information, Fig. S7d). In addition, knockout of USP29 in Trex1−/− background rescued developmental retardation of Trex1−/−mice (Fig. 7b). Expectedly, ablation of USP29 in Trex1−/− mice rescued the autoimmune lethal phenotypes of Trex1−/− mice (Fig. 7c), consistent with the observations that the levels of cGAS in brain, liver, and kidney from Trex1−/−Usp29m/m mice were substantially lower than in those from Trex1−/− mice (Fig. 7d). These data suggest that ablation of USP29 results in decreased levels of cGAS and rescues the Trex1−/− mice from autoimmune lethality.
Fig. 7. Ablation of USP29 alleviates autoimmunity in Trex1−/− mice.
a Immunoblot analysis(with anti-cGAS, anti-USP29, or anti-GAPDH) in heart (H), liver (L), kidney (K), or spleen (S) of Usp29+/+ and Usp29m/m mice (6-week old). b Representative image (upper image) and body weight (lower graph) of Trex1−/− and Trex1−/−Usp29m/m mice (5-week old). c Survival (Kaplan–Meier curve) of Trex1−/− (n = 38) and Trex1−/−Usp29m/m (n = 49) mice. d Immunoblot analysis (with anti-cGAS or anti-β-Actin) in brain (B), liver (L) or kidney (K) of Trex1−/− and Trex1−/−Usp29m/m mice (5-week old). e, f Representative image (e, left) and cell number (e, right) of spleens and HE staining of lungs and hearts (f) from Trex1−/− and Trex1−/−Usp29m/m mice (8-week old). g Flow cytometry analysis of spleenocytes from Trex1−/− and Trex1−/−Usp29m/m mice (8-week old) strained with fluorescence-conjugated antibodies against the indicated surface or intracellular molecules. h qRT-PCR analysis of Tnf, Il6, Il12p40 and Ifit3 mRNA in lungs and kidneys of Trex1−/− and Trex1−/−Usp29m/m mice (8-week old). i ELISA analysis of TNF, CCL5 and total IgG in the sera of Trex1−/− and Usp29m/m Trex1−/− mice (8-week old). *P < 0.05; **P < 0.001; ***P < 0.001 (log-rank analysis or two-tailed Student’s t-test). Data are representative of two independent experiments (means ± SD in b, e, g–i).
We next analyzed the autoimmune phenotypes of Trex1−/−Usp29m/m mice and Trex1−/− mice. Firstly, the sizes of and total cell numbers in spleens from Trex1−/−Usp29m/m mice were significantly smaller than in spleens from Trex1−/− mice (Fig. 7e), and the infiltration of immune cells into lung or heart was impaired in Trex1−/−Usp29m/m mice compared to Trex1−/− mice (Fig. 7f), which was consistent with the observation that cGAS levels were decreased in the USP29-deficient organs and suggested that knockout of USP29 alleviated the inflammation in various organs caused by Trex1 deficiency. Secondly, the percentages and numbers of GL7+FAS+ B cells, CD8+CD44+CD62L− T cells and CD8+IFNγ+ T cells were significantly less in the spleens from Trex1−/−Usp29m/m mice than in those from Trex1−/− mice (Fig. 7g), indicating that the spontaneous activation of lymphocytes in Trex1−/−Usp29m/m mice is inhibited compared to Trex1−/− mice. Thirdly, the expression of proinflammatory cytokines and ISGs were significantly impaired in lungs and kidneys from Trex1−/−Usp29m/m mice than in those from Trex1−/− mice (Fig. 7h). Lastly, the levels of TNF, CCL5 and total IgG were significantly lower in the sera from Trex1−/−Usp29m/m mice than in those from Trex1−/− mice (Fig. 7i), suggesting less systemic inflammation in Trex1−/−Usp29m/m mice than in Trex1−/− mice. Collectively, these data together suggest that knockout of USP29 impairs Trex1 deficiency-related autoimmunity, indicating that targeting USP29 could provide a potential strategy for autoimmune diseases.
Discussion
The cytoplasmic DNA sensor cGAS critically mediates innate immune responses against DNA viruses or retroviruses and inflammation-related autoimmune diseases.44,45 The stability of cGAS is strictly regulated by various posttranscriptional modifications such as sumoylation and ubiquitination.46 In this study, we demonstrated that USP29 deconjugated K48-linked polyubiquitin chains from K271 of cGAS in resting cells or after HSV-1 infection, which stabilized cGAS to produce cGAMP and activate MITA-mediated antiviral immunity as well as autoimmunity (Supplementary information, Fig S7e). In support of this notion, we observed that (i) USP29-deficient cells retained less cGAS in steady conditions or after HSV-1 infection and produced less cGAMP in response to cytoplasmic DNA challenge than did wild-type cells; (ii) USP29 deficiency in mouse cells or knockdown of USP29 in human cell lines impaired HSV-1-triggered activation of TBK1, IRF3 and NF-κB and subsequent induction of type I IFNs and proinflammatory cytokines, and potentiated HSV-1 replication; (iii) USP29-deficient mice were more susceptible to HSV-1 infection compared with the wild-type littermates and ablation of USP29 in Trex1−/− mice rescued lethal autoimmune phenotypes of Trex1−/− mice; (iv) hUSP29 removed K48O- but not K48R-linked deubiquitination of hcGAS and regulated HSV-1-triggered signaling in a manner dependent on its DUB activity; and (v) USP29 deficiency promoted K48-linked ubituitination of and proteasome-dependent degradation of cGAS. These findings thus uncover a new regulatory role of USP29 in innate antiviral signaling and autoimmunity by deubiquitinating and stabilizing cGAS.
Our results suggest that HSV-1 infection upregulated USP29, which might be responsible for increased USP29-cGAS association after HSV-1 infection. Previous studies have demonstrated that accumulation of dsDNA in the cytoplasm induces micronuclei-localization of cGAS.14,22,23 In our study, we found that a portion of USP29-cGAS associating spots were localized at perinuclear compartments after HSV-1 infection. Although the nature of such subcellular compartments is unclear, the localization of cGAS might also play a role for increased USP29-cGAS association after HSV-1 infection. Interestingly, SeV infection, transfection of dsDNA or Poly(I:C) or treatment of type I IFNs did not induce upregulation of USP29. In this context, a previous study has shown that oxidative stress induces JTV1-FBP association to co-activate transcription of USP29.39 Whether and how HSV-1 infection promotes oxidative stress to activate transcription of USP29 requires further investigations.
It has been proposed that three pools of cGAS exist in uninfected cells: unmodified, Lys217-sumoylated and constitutively Lys271-ubiquitinated cGAS.34 Sumoylation at Lys217 antagonizes K48-linked ubiquitination of Lys271 of cGAS, thereby inhibiting its proteasomal degradation.34 In our study, we found that USP29 constitutively interacted with cGAS and USP29 deficiency substantially potentiated basal K48-linked ubiquitination of cGAS and downregulated cGAS protein level without affecting its mRNA expression in uninfected cells, indicating that USP29 constitutively removes K48-linked ubiquitination of cGAS to maintain a relatively high level of cGAS under steady conditions. In support of this notion, we found that USP29 deubiquitinated cGAS(K464R) but did not affect the ubiquitination of cGAS(K271R). We also observed that K48-linked ubiquitination of cGAS was potentiated in Usp29m/m cells after HSV-1 infection, which might be due to constitutive ubiquitination of newly synthesized cGAS at Lys271. In this context, we noted that USP29 inhibited ubiquitination of cGAS(K464R) but not cGAS(K271R) after HSV-1 infection, and that knockout of USP29 potentiated ubiquitination of cGAS(K464R) but not cGAS(K271R) before or after HSV-1 infection. In addition, we found that TRIM38 and USP29 interacts with cGAS independently, suggesting that TRIM38-mediated sumoylation and USP29-mediated deubiquitination of cGAS cooperatively regulate the stability of cGAS. Results from the CHX treatment and MG132 rescue experiments strongly indicated that cGAS was constitutively ubiquitinated and degraded through the proteasome pathway in resting cells. Further investigations are required to characterize the E3 ligase(s) that constitutively ubiquitinate and induce the degradation of cGAS.
Consistent with the notion that USP29 regulates the stability of cGAS, we observed that the protein levels of cGAS were substantially decreased in USP29 knockout organs and cells. It has been demonstrated that activation of cGAS by self-DNA causes autoimmunity and deletion of cGAS significantly alleviates the autoimmune symptoms in Trex1−/− mice and rescues the lethality of those mice.24 We found that Trex1−/−Usp29m/m mice did not show any developmental retardation or autoimmune phenotypes and ablation of USP29 rescued the lethality caused by Trex1 deficiency, suggesting that USP29 serves as a promoter of autoimmune diseases. In addition, emerging clinical observations suggest that various autoimmune disorders including antibody-mediated autoimmune encephalitis, anti-N-methyl-D-aspartate (NMDA) receptor encephalitis and neuromyelitis optica spectrum disorders (NMOSD) are triggered by virus infection such as HSV-1 or Epstein-Barr virus (EBV).47–50 It would be of great interest to develop molecules targeting USP29 on the transcriptional level or on the enzymatic activity for effective treatment of infection-related autoimmunity.
In addition to mediate antiviral immune responses and autoimmunity, cGAS also mediates autophagy to protect the liver from ischemia-reperfusion injury and promotes senescence and immortalization of cells.51–54 Because USP29 dubiquitinates and stabilizes cGAS, it is worthy to investigate whether USP29 regulates the cGAS-related autophagy or senescence. Indeed, we observed that Usp29m/m MEFs were easier to be spontaneously immortalized (~10 passages) than the Usp29+/+ counterparts (~15 passages) in in vitro cultures, which is consistent with the report that deletion of cGAS accelerates the spontaneous immortalization of MEFs.53 This phenomenon indicates that USP29 is involved in the senescence of cells, which awaits further studies.
USP29 has been shown to deubiquitinate and stabilize p53 and Claspin that control cellular response to DNA damage caused by oxidative stress and DNA replication, respectively.38,39 Interestingly, a recent study has shown that oxidative stress induces translocation of cGAS from cytoplasm to nucleus, which inhibits homologous recombinant (HR) repair of DNA damage.51 Upon virus infection, cells activate antiviral signaling, shut down cell cycle and are prone to apoptosis. USP29 may simultaneously play dual roles in such a process, i.e. stabilizing cGAS to promote antiviral signaling and inhibit HR repair of DNA damage and targeting p53 and Claspin to induce apoptosis and cell cycle arrest. Therefore, USP29 might be involved in cell malignancy as a result of stabilized cGAS-caused-HR inhibition and genome instability. Collectively, our findings not only provide insight into the complex regulatory mechanisms of innate immune signaling triggered by cytosolic DNA and DNA virus, but also point to potential targets for drug development against infectious, autoimmune diseases and cancer.
Methods
Mice
The Usp29m/m and Trex1−/−mice were generated by the CRISPR/Cas9-mediated genome editing (GemPharmatech Co. Ltd). In brief, the vectors encoding Cas9 (44758, Addgene) and guide RNA were in vitro transcribed into mRNA and gRNA followed by injection into the fertilized eggs that were transplanted into pseudopregnant mice. The targeted genome of F0 mice was amplified with PCR and sequenced and the chimeras were crossed with wild-type C57BL/6 mice (GemPharmatech Co. Ltd) to obtain the Usp29+/m or Trex1+/− mice. The F1 Usp29+/m or Trex1+/− mice were further crossed with wild-type C57BL/6 mice for at least three generations. Mice were genotyped by PCR analysis of tail DNA followed by sequencing and the resulted Usp29+/m or Trex1+/− mice were crossed to generate Usp29+/+ and Usp29m/m mice or Trex1+/+ and Trex1−/− mice, respectively. Trex1+/−mice were crossed with Usp29m/m mice to generate Trex1+/−Usp29+/m mice. Trex1+/−Usp29+/m mice were crossed to generate Trex1−/−Usp29m/m mice. Age- and sex-matched Usp29+/+ and Usp29m/m littermates or Trex1−/− mice and Trex1−/−Usp29m/m mice were randamonized into groups for animal studies. The sequences of genotyping primers were 5′-AATGGTTTGGTCCAGATCCGC-3′ and 5′-GGCATCTTGCGGGACAGTGG-3′ for Usp29m/m mice, and 5′-AGGCAAATAAGTAGTGGA-3′ and 5′-TCTCACTGGCCCCAGGGCTAC-3′ for Trex1−/− mice, respectively. All mice were housed in the specific pathogen-free animal facility at Wuhan University and all animal experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of Wuhan University.
Co-immunoprecipitation and immunoblot assays
These experiments were performed as previously described.37,55 Cells were harvested and lysed for 15 min with 800 μL lysis buffer (20 mM Tris·HCl, pH 7.4–7.5, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40) containing inhibitors for protease and phosphatases (Biotool). Cell lysates (700 μL) were incubated with a control IgG or specific antibodies and protein G agarose for 2–4 h. The immunoprecipitates were washed for 3 times by 1 mL lysis buffer and subject to immunoblot analysis. The rest of lysates (100 μL) were subject to immunoblot analysis to detect the expression of target proteins.
Deubiquitination assay
These experiments were performed as previously described.37,55 Cells were lysed in regular lysis buffer (100 μL) and the cell lysates were denatured at 95 °C for 5 min in the presence of 1% SDS. A portion of cell lysates (20 μL) were saved for immunoblot analysis to detect the expression of target proteins. The rest of cell lysates (80 μL) were diluted with 1 mL lysis buffer and immunoprecipitated (Denature-IP) with specific antibodies. The immunoprecipitates were washed by three times and subject to immunoblot analysis. For in vitro deubiquitination assays, denature-IP was performed to obtain ubiquitin-modified cGAS from HEK293 cells cotransfected with FLAG-tagged cGAS and HA-Ubiquitin. The immunoprecipitates were eluted by the FLAG peptide (0.5 mg/mL, 60 μL). USP29 or USP29(C294I) protein was obtained by an in vitro transcription and translation kit (Promega). The ubiquitin-modified FLAG-cGAS was incubated with hUSP29 or hUSP29(C294I) at 37 °C for 2 h followed overnight incubation at 16 °C in the presence of ATP (1 μM). The mixtures were analyzed by immunoblot with the indicated antibodies.
Transfection and reporter assay
HEK293 cells were transiently transfected with firefly luciferase reporter (100 ng) and TK-Renilla luciferase reporter (20 ng) and indicated plasmids or empty vector (100 ng) using standard calcium phosphate precipitation. Twenty-four hours later, luciferase assays were performed with a dual-specific luciferase reporter kit (Promega). The activity of firefly luciferase was normalized by that of Renilla luciferase to obtain relative luciferase activity.
Quantitative real time PCR and ELISA
Total RNA was isolated from cells or tissues with TRIzol reagent (Life Technologies) and then was reserve-transcribed with All-in-One cDNA Synthesis SuperMix (Biotool). The cDNA was amplified by a fast two-step amplification program using 2 × SYBR Green Fast qPCR Master Mix and specific primers which were previously described. Data were normalized to the expression of the gene encoding β-Actin. The sequences of primers for qRT-PCR analysis were previously described[36, 37, 40, 41]. The sequences of primers for human USP29 gene and mouse Usp29 gene were 5′-ATTATCCTACAGAGATGC-3′ and 5′-TTCACATGTGTTGGTGAT-3′, and 5′-CGAGAGTTCAGATCCCTCCA-3′ and 5′-CCTCCTTAGATGTGCTAAGG-3′, respectively. The IFN-α (PBL), IFN-β, IL-6, TNF and CCl5 (Biolegend) protein in the sera or cell supernatants were determined by ELISA kits from the indicated manufacturers.
Cell culture
Bone marrow cells were isolated from femurs of Usp29+/+ and Usp29m/m mice. Primary MEFs were prepared from E14.5 embryos. Primary MLFs were prepared from lung of mice at 8-week old. The cells were cultured in DMEM containing 10% (vol/vol) FBS, 1% streptomycin-penicillin and 10 μM β-mercaptoethanol. GM-CSF and M-CSF (20 ng/mL) were added to the bone marrow culture for differentiation of BMDCs and BMDMs, respectively. THP-1, Human foreskin fibroblasts (HFFs) and HEK293 cells were previously described.56
Viral infection
Cells were seeded into 24-well plates (2 × 105 cells per well) or 6-well plates (106 to 107 cells per well). Twenty-four hours later, cells were infected with HSV-1-GFP or HSV-1 for the indicated time points. The cells were collected for qRT-PCR or immunoblot assays. For mice infection, 8 to 10-week old and sex-matched Usp29+/+ and Usp29m/m littermates were intravenously or intraperitoneally injected with HSV-1 (1.5–2 × 106 pfu per mouse) and the survival of animals was monitored every day. The sera or lungs from mice infected with HSV-1 were collected for ELISA or qRT-PCR at 12 or 24 h after infection, respectively. For viral replication assays, viruses were removed at 1 h after infection and cells were washed with 1 mL prewarmed PBS twice followed by culture with full medium for the indicated time points.
Constructs
Various reporter plasmids and mammalian expression plasmids for cGAS, cGAS truncations were kindly provided by Drs. Ming-Ming Hu and Hong-Bing Shu.34 MITA, MAVS, TBK1, IRF3, RIG-I, ubiquitin and ubiquitin mutants were previously described.35–37 Mammalian expression plasmids for USP29, USP29 mutants and truncations were constructed by standard molecular biology techniques. Oligo DNAs targeting USP29 were synthesized, annealed and inserted into the pLenti-GFP vector. The sequences of USP29 shRNA are as following: #1: 5′-GTGATTGAGATGGAGAATA-3′; #2: 5’-GGGAACATGTTGAAAGACA-3′.
Lentivirus-mediated gene transfer
HEK293 cells (China Center for Type Culture Collection, Wuhan University, tested for mycoplasma negative) were transfected with phage-USP29-FLAG, phage-USP29(C298A)-FLAG or the empty vector along with the packaging vectors pSPAX2 and pMD2G. The shRNA lentivurses were packaged by cotransfection of the control or USP29 shRNA vector together with pSPAX2 and pMD2G into HEK293 cells. Eight hours later, the medium was changed with fresh full medium (10% FBS, 1% streptomycin-penicillin and 10 μM β-mercaptoethanol). Forty hours later, the supernatants were harvested to infect MLFs or THP-1 cells.
Antibodies and regents
Mouse control IgG (Santa Cruz Biotechnology, sc-2025) and rabbit control IgG (Millipore, 12–370), HRP-conjugated goat-anti mouse or rabbit IgG (Thermo Scientific, PA1-86717 and SA1-9510) (1:3000), mouse anti-GFP (Sungene Biotech, KM8009) (1:2000), mouse anti-FLAG (KM8002) (1:2000), mouse anti-β-Actin (KM9001) (1:2000), mouse anti-HA (COVANCE, MMS-101R) (1:2000), anti-pIκBα (9246 L)(1:1000), anti-Ubiquitin (sc-8017) (1:500), anti-Ubiquitin, K48 specific (Merck, 05-1307), anti-IRF3 (sc-9082) (1:1000), anti-IκBα (sc-371) (1:1000), anti-p-IRF3 (4947 S) (1:1000), anti-TBK1 (GR96328-11) (1:2000) and anti-cGAS (sc-515777) (1:1000)were purchased from the indicated manufactures. LPS (Sigma, L2630) and CpG-B (Invivogen, tlr-1826) were purchased from the indicated manufactures. Poly(I:C), ISD45, DNA90, and HSV120 were previously described.35 ISD45: 5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′; DNA90: 5′-TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACATACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA-3′; HSV120: 5′-AGACGGTATATTTTTGCGTTATCACTGTCCCGGATTGGACACGGTCTTGTGGGATAGGCATGCCCAGAAGGCATATTGGGTTAACCCCTTTTTATTTGTGGCGGGTTTTTTGGAGGACTT-3′.
Plaque assay
The supernatants of BMDMs or MEFs cultures and the homogenates of brains from infected mice (or the serial dilutions) were used to infect monolayers of Vero cells. One hour later, the supernatants or homogenates were removed and the infected Vero cells were washed with pre-warmed PBS twice followed by incubation with DMEM containing 2% methylcellulose for 48 h. The cells were fixed with 4% paraformaldehyde for 15 min and stained with 1% crystal violet for 30 min before counting the plaques.
Proximity ligation assay (PLA)
The PLA reagents were purchased from Sigma and the experiments were performed as previously described.57 Briefly, cells were seeded on Teflon coted glasses and cultured for 20 h. After HSV-1 infection for 4 h, cells were f fixed in 4% paraformaldehyde for 10 min and washed with PBS for three times. After that, cells were fixed with 0.1% TritonX-100 on ice for 5 min, washed in PBS and blocked in 5% BSA in PBS for 1 h at 37 °C and incubated in primary antibodies overnight at 4 °C. On the next day, cells were washed with the wash buffer (DUO82049) for three times and incubated with the secondary antibodies with PLA probes (DUO92001-100RXN, DUO92005-100RXN) for 2 h at 37 °C followed by three times wash with the wash buffer. Cells were incubated with the Duolink in situ detection reagents red (DUO92008) for 2 h at 37 °C. Finally, cells were stained with the In Situ microplate nuclear stain and anti-fade (DUO82064-1KT) and covered onto slides. The images were acquired by using a confocal microscope (LSM 880, Carl Zeiss Microscopy Plan-Apochromat 40×/0.95 Korr M27) and the positive signals in the acquired images were quantified with Image-Pro Plus software and calculated as pixels per cell.
cGAMP assay
MEFs (1–3 × 107) transfected with dsDNA were harvested and homogenized in 1 mL sterile water by a 1 mL syringe pipetting up and down for 20 times. The homogenates were heated at 95 °C for 10 min followed by centrifuge at 37,000 rpm (Hitachi CP100NX, P90NT-1022) for 2 h. The supernatants (900 μL) were mixed with 10× digitonin (100 μL) and incubated with the single-layered HFF cells (2 × 105). Half an hour later, the supernatants were removed and full medium were added to treat HFFs for 4 h followed by qRT-PCR assays.
Flow cytometry analysis
Single-cell suspension was re-suspended in FACS buffer (PBS, 1%BSA) and blocked with anti-mouse CD16/32 antibodies for 10 min prior to staining with antibodies against surface markers. For intracellular cytokine staining, cells were fixed and permeabilized by using a fixation and permeabilization solution followed by intracellular staining (Biolgend). Flow cytometry data were acquired on a FACSCelesta or Fortesa flow cytometer and analyzed with FlowJo software (TreeStar).
Hematoxylin-eosin staining analysis
Tissues from mice were fixed in 4% araformaldehyde and embedded in paraffin blocks. The paraffin blocks were sectioned (5 μm) for HE staining (Beyotime Biotech) followed by coverslipped. Images were acquired using an Aperio VERSA 8 (Leica) multifunctional scanner.
Protein expression and purification
Competent BL21 cells (from Dr. Lei Yin, Wuhan University) were transformed with the desired recombinant DNA overnight (ON). An ON culture was inoculated in 20 ml LB (+ampicillin) and cultured at 37 °C with 180–200 rpm overnight. Dilute the ON culture 1:100 in 1 L LB-media (+ampicillin) shaking at 37 °C for 4–5 h. The culture was cooled down to 16 °C within 1 h, and was induced by addition of 500 µM IPTG. The cells were cultured at 16 °C at 180–200 rpm for about 16–20 h. The cells were harvested (10 min at 5500–6000 × g), re-suspended in 100 ml cold binding buffer (+protease inhibitors), and lysed by French Press followed by centrifugation for 30 min at 45000 × g. The supernatants were incubated with Glutathione Sepharose 4B resin for 2 h at 4 °C and the Resins were washed extensively with high salt buffer [250 mM Tris (pH 7.5), 500 mM NaCl, and 5 mM DTT] and low salt buffer [25 mM Tris (pH7.5), 150 mM NaCl, 10% glycerol, and 1 mM DTT]. Recombinant proteins were eluted from the resins and saved at −80 °C until use.
Statistical analysis
Differences between experimental and control groups were determined by Student’s t-test (where two groups of data were compared) or by two-way ANOVA analysis (where more than two groups of data were compared). P values less than 0.05 were considered statistically significant. For animal survival analysis, the Kaplan–Meier method was adopted to generate graphs, and the survival curves were analyzed with log-rank analysis.
Supplementary information
Acknowledgements
We thank Drs. Min-Hua Luo (Wuhan Institute of Virology, CAS), Lei Yin, Ming-Ming Hu and Hong-Bing Shu (Wuhan University) for reagents, members of Zhong lab and the core facilities of Medical Research Institute for technical help. This study was supported by grants from National Key Research and Development Program of China (2018YFE0204500 and 2018YFC1004601), Natural Science Foundation of China (31671454 and 31930040), Natural Science Foundation of Hubei Province (2018CFA016), and Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (TFJC2018004).
Author contributions
B.Z. designed and supervised the study; Q.Z., Z.T., R.A., and L.Y. performed the major experiments and analysis; Q.Z. and B.Z. wrote the paper; all the authors analyzed data.
Competing interests
The authors declare no competing interest.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-020-0341-6.
References
- 1.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 2.Tan X, Sun L, Chen J, Chen ZJ. Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 2018;72:447–478. doi: 10.1146/annurev-micro-102215-095605. [DOI] [PubMed] [Google Scholar]
- 3.Hu MM, Shu HB. Cytoplasmic mechanisms of recognition and defense of microbial nucleic acids. Annu. Rev. Cell Dev. Biol. 2018;34:357–379. doi: 10.1146/annurev-cellbio-100617-062903. [DOI] [PubMed] [Google Scholar]
- 4.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parvatiyar K, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X-D, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luecke S, et al. cGAS is activated by DNA in a length-dependent manner. EMBO Rep. 2017;18:1707–1715. doi: 10.15252/embr.201744017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, et al. Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell. 2018;174:300–311. doi: 10.1016/j.cell.2018.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andreeva L, et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature. 2017;549:394–398. doi: 10.1038/nature23890. [DOI] [PubMed] [Google Scholar]
- 14.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoh SM, et al. PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell. 2015;161:1293–1305. doi: 10.1016/j.cell.2015.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian H, et al. ZCCHC3 is a co-sensor of cGAS for dsDNA recognition in innate immune response. Nat. Commun. 2018;9:3349. doi: 10.1038/s41467-018-05559-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZS, et al. G3BP1 promotes DNA binding and activation of cGAS. Nat. Immunol. 2019;20:18–28. doi: 10.1038/s41590-018-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rongvaux A, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White MJ, et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding SM, et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548:466–470. doi: 10.1038/nature23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao D, et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo GJ, et al. Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 2015;13:440–449. doi: 10.1016/j.celrep.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia P, et al. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 2016;17:369–378. doi: 10.1038/ni.3356. [DOI] [PubMed] [Google Scholar]
- 27.Dai J, et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 2019;176:1447–1460. doi: 10.1016/j.cell.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo GJ, et al. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 2018;9:613. doi: 10.1038/s41467-018-02936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu ZS, et al. RINCK-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell Biosci. 2018;8:35. doi: 10.1186/s13578-018-0233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 2017;13:e1006264. doi: 10.1371/journal.ppat.1006264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen M, et al. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell. 2016;64:105–119. doi: 10.1016/j.molcel.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 32.Liang Q, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui Y, et al. SENP7 potentiates cGAS activation by relieving SUMO-mediated inhibition of cytosolic DNA sensing. PLoS Pathog. 2017;13:e1006156. doi: 10.1371/journal.ppat.1006156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu MM, et al. Sumoylation promotes the stability of the DNA Sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016;45:555–569. doi: 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Sun H, et al. USP13 negatively regulates antiviral responses by deubiquitinating STING. Nat. Commun. 2017;8:15534. doi: 10.1038/ncomms15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, et al. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 2016;26:1302–1319. doi: 10.1038/cr.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liuyu T, et al. Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res. 2019;29:67–79. doi: 10.1038/s41422-018-0107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martín Y, et al. USP29 controls the stability of checkpoint adaptor Claspin by deubiquitination. Oncogene. 2015;34:1058–1063. doi: 10.1038/onc.2014.38. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, et al. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO. J. 2011;30:846–858. doi: 10.1038/emboj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin D, et al. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc. Natl. Acad. Sci. USA. 2015;112:11324–11329. doi: 10.1073/pnas.1509968112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu B, et al. Induction of INKIT by viral infection negatively regulates antiviral responses through inhibiting phosphorylation of p65 and IRF3. Cell Host Microbe. 2017;22:86–98. doi: 10.1016/j.chom.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Kim JD, Yu S, Choo JH, Kim J. Two evolutionarily conserved sequence elements for Peg3/Usp29 transcription. BMC Mol. Biol. 2008;9:108. doi: 10.1186/1471-2199-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng, W.-B. et al. Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol. Neurodegener.12, 38 (2017). [DOI] [PMC free article] [PubMed]
- 44.Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018;215:1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- 46.Xiong M, Wang S, Wang Y-Y, Ran Y. The Regulation of cGAS. Virologica Sin. 2018;33:117–124. doi: 10.1007/s12250-018-0005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Getts DR, Chastain EM, Terry RL, Miller SD. Virus infection, antiviral immunity, and autoimmunity. Immunol. Rev. 2013;255:197–209. doi: 10.1111/imr.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venkatesan A, Benavides DR. Autoimmune encephalitis and its relation to infection. Curr. Neurol. Neurosci. Rep. 2015;15:3. doi: 10.1007/s11910-015-0529-1. [DOI] [PubMed] [Google Scholar]
- 49.Hellesen A, Bratland E. The potential role for infections in the pathogenesis of autoimmune Addison’s disease. Clin. Exp. Immunol. 2019;195:52–63. doi: 10.1111/cei.13207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levinson, J. B., Alvarez, M. R., Koci, K., Feoktistov, A. & McFarlane, I. M. Epstein - Barr virus infection in a patient with neuromyelitis optica spectrum disorder and Sjögren’s syndrome: A case report and review of literature. Clin. Case Rep. Rev.4, 1–5 (2018).
- 51.Liu H, et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature. 2018;563:131–136. doi: 10.1038/s41586-018-0629-6. [DOI] [PubMed] [Google Scholar]
- 52.Lei Z, et al. cGAS-mediated autophagy protects the liver from ischemia-reperfusion injury independently of STING. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;314:G655–g667. doi: 10.1152/ajpgi.00326.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glück S, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017;19:1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai, Z. et al. USP22 promotes IRF3 nuclear translocation and antiviral responses by deubiquitinating the importin protein KPNA2. J. Exp. Med.217, e20191174 (2020). [DOI] [PMC free article] [PubMed]
- 56.Ye L, et al. USP49 negatively regulates cellular antiviral responses via deconjugating K63-linked ubiquitination of MITA. PLoS Pathog. 2019;15:e1007680. doi: 10.1371/journal.ppat.1007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, et al. USP2a supports metastasis by tuning TGF-beta signaling. Cell Rep. 2018;22:2442–2454. doi: 10.1016/j.celrep.2018.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.