Abstract
Microbiome assemblages of plants and animals often show a degree of correlation with host phylogeny; an eco-evolutionary pattern known as phylosymbiosis. Using 16S rRNA gene sequencing to profile the microbiome, paired with COI, 18S rRNA and ITS1 host phylogenies, phylosymbiosis was investigated in four groups of coral reef invertebrates (scleractinian corals, octocorals, sponges and ascidians). We tested three commonly used metrics to evaluate the extent of phylosymbiosis: (a) intraspecific versus interspecific microbiome variation, (b) topological comparisons between host phylogeny and hierarchical clustering (dendrogram) of host-associated microbial communities, and (c) correlation of host phylogenetic distance with microbial community dissimilarity. In all instances, intraspecific variation in microbiome composition was significantly lower than interspecific variation. Similarly, topological congruency between host phylogeny and the associated microbial dendrogram was more significant than would be expected by chance across all groups, except when using unweighted UniFrac distance (compared with weighted UniFrac and Bray–Curtis dissimilarity). Interestingly, all but the ascidians showed a significant positive correlation between host phylogenetic distance and associated microbial dissimilarity. Our findings provide new perspectives on the diverse nature of marine phylosymbioses and the complex roles of the microbiome in the evolution of marine invertebrates.
Subject terms: Marine microbiology, Phylogenetics, Symbiosis
Introduction
Phylosymbiosis occurs when microbial community relationships reflect the evolutionary history of the host [1–3]. The term was first coined to describe the impact of a host phylogenetic signal on gut microbial community relationships in Nasonia parasitoid wasps [2, 4], and the phenomenon has since been investigated in a diverse range of taxa and environments, e.g., the gut microbiomes of mammals and insects [1, 5, 6], the skin microbiome of ungulates [7], the endolithic microbiome of coral [8] and the root microbiome of plants [9]. These studies have confirmed that phylosymbiosis occurs in the simplest as well as the most diverse microbial communities and the discovery of virus/host phylosymbioses [10] demonstrates that the phenomenon is not limited to prokaryotes. As phylosymbiosis has become more frequently observed, the mechanisms underpinning these patterns are of increasing interest.
Evolutionary processes such as codivergence and coevolution are distinct from phylosymbiosis, establishing the need of an alternative term [1]. Namely, phylosymbiosis is a pattern observed at one moment in time and space, which does not assume a stable evolutionary association between a host and its microbiota or congruent ancestral splits, nor does it assume vertical transmission of microbial symbionts [11]. While it is possible that different evolutionary processes contribute to the mechanisms behind phylosymbiosis [8, 12], complex and dynamic systems that acquire high numbers of microbes from the environment are likely structured by other mechanisms. For example, horizontal transmission of microbes filtered through phylogenetically congruent host traits, biogeography of a host and the microbiota, and dispersal of microbes among conspecifics all potentially contribute to observed phylosymbiosis patterns [12–15]. These explanations are not necessarily mutually exclusive. Within a complex microbiome where both vertical and horizontal transmission occurs among obligate and facultative microbial members, phylosymbiosis is expected to rely on multiple mechanisms [3, 6].
Despite the extensive literature supporting phylosymbiotic relationships, host phylogeny does not always correlate with microbial community (dis)similarity. For example, in contrast to other mammals, no significant congruence was observed between skin microbiome composition and host phylogeny in the case of carnivores [7]. Similarly, no phylosymbiotic signal could be detected in the case of the intestinal microbiota of 59 Neotropical birds [16] and the gut microbiomes of bats are more similar to birds than other mammals [17]. There are multiple reasons why phylosymbiosis may not occur. First, factors such as environment and diet may obscure phylosymbiotic signals, which have been successfully controlled for in some studies [1, 4]. Second, in some cases, host genotype exerts strong effects on microbiome composition that are independent of host phylogeny [8, 18, 19]. Finally, host physiology can structure the microbiome [20], however, physiological traits may not always be consistent with host phylogeny [21]. Therefore, patterns of phylosymbiosis may be dependent on a certain host taxonomic level (i.e., host family), where host genotype effects are reduced and host physiological traits and phylogeny are congruent.
Reef invertebrates provide interesting opportunities for testing hypotheses of phylosymbiosis, as they often host diverse microbial communities acquired by combinations of vertical and horizontal transmission [22–25] that can be dynamic among different environments [26, 27]. Here, we first characterise the microbiomes of four groups of coral reef invertebrates: scleractinian corals, octocorals, sponges and ascidians. We then test three recommended analyses to investigate phylosymbiosis: (a) comparison of intraspecific and interspecific variation in microbiome composition, (b) comparison of the topology of host phylogeny and hierarchical clustering of its associated microbial community, and (c) correlation of host phylogenetic distance with microbial community dissimilarity [3, 14]. We hypothesise that a phylosymbiotic signal will be found across all four groups to show that host phylogeny is a dominant factor in microbiome structure of reef invertebrates. Through an improved understanding of microbial community dynamics using phylosymbiosis, our knowledge of how a microbiome is structured and maintained in complex marine holobionts will be enhanced [25].
Materials and methods
Sample collection
Tissue samples from 3 to 5 replicates of 30 species of coral reef invertebrates (12 corals, 10 octocorals, 5 sponges and 3 ascidians) were collected on SCUBA from seven locations across the central and northern sectors of the Great Barrier Reef (GBR) (Table S1; Fig. S1). On sampling trips to Broadhurst Reef, Davies Reef and Orpheus Island, August 2017 (Table S1), adult colonies no larger than 30 × 30 cm were collected using hammer and chisel and returned to the reef after sampling. Alternatively, sampling of invertebrates was performed in situ. On the surface, colonies/samples were isolated and placed in running seawater (0–2 h) until processing. Each invertebrate was sampled for 3–5 fragments ~5 cm in length using either a hammer and chisel or dive knife (coral), or sterile razor blades (all other invertebrates). In addition, seawater samples were collected from the central GBR sites in August 2017 as an environmental control (Table S1). All samples were collected under the marine parks permits G12/35236.1 and G15/37574.1
Sample processing and preservation
Fragments were rinsed in autoclaved calcium- and magnesium-free seawater (CMFSW; NaCl: 26.2 g, KCl: 0.75 g, Na2SO4: 1 g, NaHCO3: 0.042 g, per 1 L) to remove any loosely attached microbes. For scleractinian coral, tissue was removed from the skeleton by pressurised air into ~30 ml of CMFSW. Coral blastate was homogenised by vortex for 1 min and 2 × 2 ml aliquots were kept for DNA extraction. Aliquots were centrifuged for 10 min at 10,000 × g, the supernatant was removed, and tissue pellet was either snap frozen in liquid nitrogen or preserved in 1 ml dimethyl sulfoxide-EDTA salt saturated solution (DESS) and kept at −80 °C (Table S1). For octocorals and sponges, fragments were cut into small pieces ~0.5 × 0.5 cm3 using a sterile razor blade, snap frozen in a 2 ml cryovial and stored at −80 °C until DNA extraction. Alternatively, a 15 ml falcon tube with ~7 ml DESS was filled with the dissected tissue until approximately a 1:1 ratio of tissue:DESS was reached. The ascidians Lissoclinum patella and Polycarpa aurata were dissected longitudinally and the tunic layer was removed and snap frozen as described above. Colonies of the remaining ascidian Didemnum molle were dissected into three equal parts as the tunic was too small to isolate and preserved in 1 ml DESS and kept at −80 °C. Seawater was collected from each site (excluding the Ribbon Reefs (RR) and Osprey Reef) ~1 m above the benthos at the area of sample collection using 4 × 5 L retractable water bottles (washed and sterilised with 10% hydrochloric acid). Approximately 2–3 L were then filtered through 0.22 µm sterivex filters and stored at −80 °C (where −80 °C was not available, samples were stored at −20 °C for 1–5 days before being transferred to −80 °C upon returning to the lab).
DNA extraction and sequencing
Approximately 0.05 g of tissue was used for DNA extraction using the DNeasy PowerBiofilm Kit (QIAGEN Pty Ltd, VIC Australia 3148). Extraction was performed following the manufacturers protocol with the BioSpec Mini-Beadbeater-96 used for mechanical lysis at 3–5 cycles of 30–60 s depending on the difficulty to break down the tissue. Genomic DNA was sent to the Ramaciotti Centre for Genomics (UNSW, Sydney Australia) for 16S rRNA amplicon sequencing on the Illumina MiSeq platform using the modified V4 region primer set, 515F (GTGYCAGCMGCCGCGGTAA) [28] and 806R (GGACTACNVGGGTWTCAAT) [29]. Samples were prepared for sequencing with the Earth Microbiome Project’s 16S Illumina Amplicon protocol and sequencing was performed following the standard Illumina protocol for 16S rRNA gene amplicon library prep. Sequencing of the host phylogenetic markers COI, 18S rRNA and ITS1 was performed at the Beijing Genome Institute following the BGISEQ-500 library prep protocol on the BGISEQ-SE400 module. COI (~712 bp), 18 S (~470 bp) and ITS1 (~288 bp) were amplified using the primer pairs, LCO1490 (GGTCAACAAATCATAAAGATATTGG) and HCO2198 (TAAACTTCAGGGTGACCAAAAAATCA) for COI [30] and V4_18S_Next.For (CCAGCASCYGCGGTAATTCC) and V4_18S_Next.Rev.B (ACTBTCGYTCTTGATYARNGA) were modified from Pirredda et al. [31] for 18S rRNA. For ITS1, the custom primers 18S-F1759 (GGTGAACCTGCGGAWGGATC) and 5.8S-R40 (CGCASYTDGCTGCGTTCTTC) were designed by retrieving all available sequences from our target species and aligning them using MAFFT [32]. Full length barcodes were assembled from single-end 400 bp reads using the HIFI-SE pipeline [33].
16S rRNA gene amplicon analysis
Sequences were analysed using QIIME2 (v 2018.4) [34] by first demultiplexing reads and denoising following the DADA2 pipeline [35]. Taxonomic assignment was performed using a Naive Bayes classifier pre-trained on the Silva 132 99% OTU database modified to the V4 region primer set 515F/806R. The resulting amplicon sequence variant (ASV) table was filtered for chloroplast, mitochondrial and eukaryotic sequences. A phylogenetic tree was reconstructed using the qiime fragment-insertion sepp command (QIIME2 v 2019.1), which places the ASVs into a larger, well-curated 16S rRNA reference phylogeny containing >200,000 representative tips (GreenGenes 13.8, 99% OTU) [36]. The resulting tree was then trimmed to the original reference sequences and used for subsequent UniFrac analyses. ASV and taxonomic tables were imported into R studio v.3.5.0 [37] for further analysis with extensive use of the packages ‘phyloseq’ [38], ‘vegan’ [39], ‘ggplot2’ [40], ‘ggtree’ [41], ‘ape’ [42], ‘phangorn’ [43] and ‘dplyr’ [44].
Characterisation of microbial diversity and composition
The following analyses were conducted at the ASV level, excluding visual representations of relative abundance. Relative abundance for each microbial phylum was calculated and grouped by invertebrate taxonomy to give a broad overview of microbial profiles of each invertebrate group. In addition, the top 25 most abundant microbial families across the entire dataset were shown to give an overview of the lower taxonomic levels. As the taxonomic profile of the blanks was sufficiently different from the marine invertebrate profiles, with only 0.4% of sequences present in the top 25 family level ASVs, these samples were removed from further analysis. Rarefaction curves were calculated and plotted to illustrate the total diversity of ASVs captured against the sampling effort. Alpha diversity was calculated using both species richness (total number of ASVs retrieved per sample) and Shannon–Wiener diversity index on a dataset rarefied to 3500 sequences (equal to the sample with the lowest number of sequences). Beta diversity was calculated on non-rarefied data using the Bray–Curtis dissimilarity measure by first standardising the data by the species maximum and then by the sample total (Wisconsin double standardisation). This method of normalisation was chosen for beta diversity as transforming data to proportions returns the most accurate Bray–Curtis dissimilarities [45]. The resulting dissimilarity scores were visualised using non-metric multidimensional scaling (NMDS) to observe overall patterns in microbial community structure among the different invertebrates. Analysis of variance (ANOVA) and a post-hoc Tukey’s test with unplanned comparisons and a Bonferroni correction were used for significance testing of alpha diversity, while permutational multivariate analysis of variance (PERMANOVA) was used for beta diversity using the pairwiseAdonis function for post-hoc analysis.
Host phylogenetic reconstructions
Representative sequences for COI, 18S rRNA and ITS1 from each species in each taxonomic group were aligned separately using MUSCLE [46] and then concatenated using DAMBE [47]. Concatenated octocoral and sponge alignments were further curated using Gblocks [48] to remove poorly aligned, high gap regions. Evolutionary model selection was performed using JModelTest2 (Supplementary Table 2) [49] and phylogenetic analysis was conducted in Mr Bayes v3.2.7 [50] using the outgroups Carteriospongia foliascens for corals, octocorals and ascidians and Cladiella sp. for sponges. Outgroups were selected based on their low phylogenetic relatedness to the ingroup and low variability in microbiome composition among sample replicates. Evolutionary history was inferred using Bayesian inference with the Markov Chain Monte Carlo method using two independent runs of 5,000,000 generations and all models converged at <0.01.
Phylosymbiosis analysis
The 16S rRNA gene dataset was subsampled to each taxonomic group and analysed independently. Gorgonians did not contain enough species within our dataset to compare host phylogeny with microbial composition and were added to the soft coral dataset to create an octocoral group. Intraspecific against interspecific variability of microbiome composition was compared using pairwise comparisons of Bray–Curtis dissimilarity between each sample. Welches t test was used for significance testing following an arcsine transformation to normalise the 0–1 distribution, while an ANOVA and post-hoc Tukey’s test with unplanned comparisons and a Bonferroni correction were used to test for significant differences in intraspecific variation among invertebrate groups.
Microbial dendrograms were built in QIIME2 using the qiime diversity beta-rarefaction command. Within each invertebrate ASV table subset, all ASVs that appear two times or less and those that are present in only one sample were removed to reduce noise from potentially spurious and transient ASVs. Each sample was then pooled by host species and rarefied over 1000 iterations to the host species with the lowest number of reads following the method of Brooks et al. [1]. Hierarchical clustering of host species from the resulting table was performed using the UPGMA clustering method based on Bray–Curtis dissimilarity and both weighted and unweighted UniFrac distances. Microbial dendrograms along with phylogenetic trees and pooled ASV tables were imported into R studio for analysis.
To assess topological congruency, host phylogenetic tree topology was compared with the microbial dendrograms using the normalised Robinson–Foulds (nRF) metric, where 0 is complete congruence and 1 is no congruence. Branch lengths were removed in host phylogenetic trees for visualisation and a significance value was calculated using the RFmeasures function [14] with 9999 permutations. Correlation between host phylogenetic distance and microbial dissimilarity was analysed by first creating a distance matrix of pairwise phylogenetic distances between each host species and distance matrices of Bray–Curtis dissimilarity and weighted and unweighted UniFrac distances using the pooled ASV tables. A Mantel test was used to test for correlation between host and microbial distance matrices using Pearson correlation with 9999 permutations. A similarity percentages (SIMPER) analysis was used to identify which ASVs were contributing to dissimilarity between host species that showed incongruence.
Results
Sample collection and sequencing
Field collections resulted in a total of 161 samples across 30 species of reef invertebrates (Table S1). In addition, eight seawater samples, two blank extractions and two sequencing positive controls were sequenced. For 16S rRNA amplicon sequencing, this yielded a total of 10,415,183 reads in 173 samples, which was reduced to 8,611,147 high quality reads following quality control and denoising. For host phylogeny, successful COI sequences were obtained for all 30 species, however, 18S rRNA sequencing was unsuccessful for Acropora formosa, Acropora hyacinthus, Diploastrea heliopora, Heteroxenia sp. and Isis hippuris and ITS1 sequencing was unsuccessful for Lissoclinum patella and Didemnum mole. As a result, ITS1 was not used for ascidian phylogeny.
Characterisation of microbial diversity and composition
Rarefaction curves for each sample approached asymptotes, illustrating that total ASV richness for each sample was captured (Fig. S2). However, rarefaction to the sample with the lowest number of reads (Isis hippuris: 3323 reads; excluding blanks) resulted in a loss in diversity in some samples. Nonetheless, overall trends showed that both ASV richness and ASV diversity (Shannon–Wiener Index) were both significantly different across the broad taxonomic associations (richness; ANOVA; F(5, 163) = 7.01, p < 0.001; Fig. 1) (Shannon diversity; ANOVA; F(5, 163) = 4.64, p < 0.001; Fig. 1). Post-hoc comparisons revealed that seawater had a significantly higher ASV richness than the ascidians (p = 0.024), while coral had a significantly higher ASV richness than ascidians (p = 0.006), soft corals (p = 0.006) and sponges (p = 0.003). For ASV diversity, post-hoc comparisons revealed an increase in diversity in coral compared with the ascidians (p = 0.009) and soft corals (p = 0.046), and an increase in seawater compared with the ascidians (p = 0.014). However, unlike richness, no difference was seen in ASV diversity between corals and sponges (p = 1.0).
Fig. 1. ASV richness (top panel) and Shannon–Wiener diversity index (bottom panel) for each invertebrate group and seawater.
Letters indicate groups which are significantly different from each other.
A total of 62 microbial phyla were observed across the invertebrate groups and microbial profiles showed a high degree of uniformity at the phylum level. Microbial taxonomy mentioned here and herein are ASV sequences affiliated to that taxonomic classification, with Proteobacteria, Cyanobacteria and Bacteroidetes among the dominant phyla across all marine invertebrates (Fig. S3). However, differences were evident even at the broad taxonomic level, with the octocorals (soft coral and gorgonians) hosting a higher relative abundance of Tenericutes (mean = 4.71% ± 1.63 SE and 11.12% ± 6.28 SE, respectively) compared with other invertebrates, while sponges were associated with more Chloroflexi, Acidobacteria and Cyanobacteria (mean = 19.09% ± 2.29 SE, 9.86% ± 1.64 SE and 28.31% ± 3.67 SE, respectively).
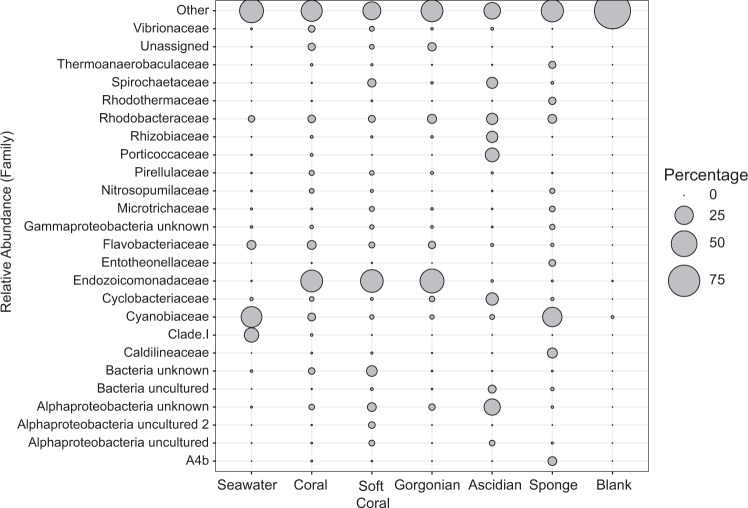
Relative abundance at the family level indicated far more variation in taxonomic profiles among the invertebrate groups (Fig. 2). The three groups of anthozoans (coral, soft coral and gorgonian) were clearly different to the other marine invertebrate classifications and mostly dominated by the common Endozoicomonadaceae (mean = 33.52% ± 4.19 SE, 38.41% ± 4.58 SE and 42.88% ± 10.74 SE, respectively). Sponges consisted of a high relative abundance of Cyanobiaceae (mean = 27.87% ± 3.74 SE), comprised of the commonly found cyanobacteria Prochlorococcus and Synechococcus (Silva database classification), as did seawater (mean = 32.26% ± 2.46 SE). Ascidians appeared more variable, with Rhodobacteraceae, Porticoccaceae, Cyclobacteriaceae and unclassified Alphaproteobacteria, all abundant within the top 25 bacteria at the family level.
Fig. 2. Relative abundance of the top 25 prokaryotic families found across each invertebrate group as well as seawater and blank extractions.
Bubble size is proportional to the relative abundance of each prokaryotic family (y-axis) within a host group (x-axis).
Between sample variability (beta diversity) showed there was an overall weak clustering of samples by their broad taxonomic classifications (Fig. 3). Particularly the three anthozoans (coral, soft coral and gorgonian) and ascidians had low homogeneity in microbial composition. Comparatively, sponge and seawater samples formed clusters that indicated consistent microbial composition across samples. Microbial composition was confirmed statistically to be associated with host taxonomy (PERMANOVA; F(5, 163) = 2.58, p < 0.001), however, only a small amount of variation in the data was explained by the broad taxonomic classification (R2 = 0.073). When samples were instead grouped by host species, the amount of variation explained increased dramatically (PERMANOVA; F(30, 138) = 2.01, R2 = 0.30, p < 0.001). Lastly, beta-diversity analysis showed there was a significant association to collection site (PERMANOVA; F(6, 162) = 1.90, R2 = 0.066, p < 0.001), however, only a small amount of variation could be explained by this variable, and since many species were collected from only one reef, it is likely the variation is due to species-specific microbiomes.
Fig. 3. Bray–Curtis dissimilarity based on microbial composition visualised using NMDS.

Each symbol represents a sample where colour is the associated host and shape is reef zone where sample was collected.
Assessment of phylosymbiosis among coral reef invertebrates
All four marine invertebrate groups showed lower intraspecific Bray–Curtis dissimilarity in microbial composition compared with interspecific Bray–Curtis dissimilarity (coral: t(364) = 13.53, p < 0.001; octocoral: t(302) = 18.84, p < 0.001; sponge: t(200) = 34.80, p < 0.001; ascidian: t(69) = 19.09, p < 0.001), confirming lower microbiome variability among conspecifics (Fig. 4). Furthermore, intraspecific variation was significantly different among the invertebrate groups (ANOVA; F(3, 818) = 231.15, p < 0.001), with the exception of the ascidians and octocorals (t = 1.85, p = 0.39), highlighting sponges and coral with the highest and lowest microbiome homogeneity, respectively.
Fig. 4. Intraspecific and interspecific Bray–Curtis dissimilarity scores for each invertebrate group.

Interspecific variation (red boxplots) in the microbiome was significantly greater than intraspecific variaion (blue boxplots) for each invertebrate group.
Comparing the topology of host phylogenetic trees with the corresponding microbial dendrograms (nRF test) and measuring the correlation of host phylogenetic distance with microbial dissimilarity (Mantel test) further revealed significant levels of phylosymbiosis across all four groups of invertebrates (Table 1). Patterns of phylosymbiosis were significant in sponges using all tests and metrics (Figs. 5a and S4), while Bray–Curtis and weighted UniFrac metrics found significant patterns of phylosymbiosis using the nRF and Mantel tests in corals (Figs. 5b and S5a) and octocorals (Figs. 5c and S6a). Using the unweighted UniFrac distance, phylosymbiosis patterns were significant only using the Mantel test but not the nRF test for coral (Fig. S5b) and octocoral (Fig. S6b) and no patterns were detected in the ascidians (Fig. S7b). Perfect congruency between host phylogeny and microbial dendrograms was observed in the ascidians using both the Bray–Curtis and weighted UniFrac metrics (Figs. 5d and S7a). Despite this, no significant phylosymbiosis was observed using the Mantel test. This opposing result is likely due to the low sample size combined with marked differences in microbial composition among the three ascidians (Fig. S8).
Table 1.
Normalised Robinson–Foulds (nRF) and mantel statistics across Bray–Curtis, weighted and unweighted UniFrac beta-diversity metrics.
| Bray–Curtis | Weighted UF | Unweighted UF | |
|---|---|---|---|
| Sponge | |||
| nRF | RF = 0.02, p < 0.001 | RF = 0.4, p = 0.006 | RF = 0.4, p = 0.01 |
| Mantel | r = 0.71, p < 0.001 | r = 0.78, p = 0.006 | r = 0.75, p = 0.03 |
| Coral | |||
| nRF | RF = 0.69, p < 0.001 | RF = 0.69, p < 0.001 | RF = 0.92, p = 0.15 |
| Mantel | r = 0.37, p = 0.02 | r = 0.38, p = 0.01 | r = 0.42, p = 0.03 |
| Octocoral | |||
| nRF | RF = 0.64, p < 0.001 | rRF = 0.82, p = 0.02 | RF = 0.91, p = 0.24 |
| Mantel | r = 0.23, p < 0.001 | r = 0.36, p < 0.001 | r = 0.25, p < 0.001 |
| Ascidian | |||
| nRF | RF = 0, p < 0.001 | RF = 0, p < 0.001 | RF = 0.5, p = 0.34 |
| Mantel | r = −0.03, p = 0.63 | r = 0.46, p = 0.17 | r = 0.18, p = 0.46 |
Fig. 5. Host phylogeny and microbial dendrogram comparisons for each invertebrate group.
a–d Host phylogenies are inferred from COI, 18S rRNA and ITS1 sequences while microbial dendrograms are based on Bray–Curtis dissimilarity for microbial composition of each host species. Cladiella sp. was used as an outgroup for a sponges, while C. foliascens was used as an outgroup for b coral, c octocoral and d ascidians. Numbers at nodes reflect posterior probability for clade support in host trees and jackknife support values in dendrograms. Branch tips are coloured to reflect clades in host phylogeny. Initials in brackets next to species names refer to collection site. BR Broadhurst Reef, DR Davies Reef, OR Osprey Reef, PI Palm Islands (Orpheus and Pelorus), PR Pandora Reef, RB Rib Reef, RR Ribbon Reefs. P. massive refers to massive Porites sp.
A select few species were collected from multiple locations and showed contrasting results in relation to phylosymbiosis. The sponge Ircinia ramosa and octocoral Sarcophyton sp. were collected from two locations and both correctly formed a clade with their conspecifics (Figs. 5, S4 and S6), which was supported by uniform microbial profiles (Figs. S9 and S10). Conversely, the octocoral Sinularia sp. and the coral species Porites cylindrica and Seriatopora hystrix did not form clades with their conspecifics from different locations and there was a reduced overall phylosymbiotic signal (Figs. 5, S5 and S6). A SIMPER analysis revealed that shifts in the relative abundance of ASVs assigned to Endozoicomonadaceae were consistently the top contributors to the dissimilarity observed between species collected from two sites (Table S3; Fig. S11). For example, Porites cyclindrica collected from the Palm Islands (PI) had a dramatic reduction in Endozoicomonadaceae compared with those collected from the RR, where the mean relative abundance of Endozoicomonadaceae fell from 82.9% (±4.32 SE) to 3.31% (±1.69 SE). Similarly, the microbial profile of Sinularia collected from RR differed from the two Sinularia species collected from PI, with colonies from RR hosting a lower relative abundance of Endozoicomonadaceae and a higher relative abundance of unknown bacteria and Fusobacteriaceae (Fig. S11).
Additional incongruences were observed among the groups where sample location was not a factor. The overwhelming majority of extant corals fall into one of two major clades, the Robusta and Complexa. This split was only partially reflected in the Bray–Curtis and weighted UniFrac microbial dendrograms, although in most cases, species within a genus or family clustered together (Figs. 5b and S5). Similarly, host phylogeny was recapitulated in the microbiome of only certain clades of octocorals using Bray–Curtis and weighted UniFrac metrics, such as the microbiome of Briareum and species within the family Alcyoniidae (Sarcophyton, Sinularia and Cladiella), with the exception of Sinularia collected from the RR (Figs. 5c and S6). However, no congruence was seen between gorgonian phylogeny and microbial composition, which can again be attributed to ASVs assigned to Endozoicomonadaceae (Table S3; Fig. S10). Lastly, although the signal of phylosymbiosis in sponges was strong and robust across all analyses, the main incongruence was due to an unclassified Ircinia sp., which did not form a clade with its sister species in the host phylogeny (Fig. 5a), and highlights the unresolved phylogenetic relationships among the Ircinia [51].
Discussion
This study evaluates the signal of phylosymbiosis in diverse coral reef invertebrates, finding evidence that host evolutionary history helps shape the microbiome in sponges, corals, octocorals and ascidians. By testing three commonly used methods for phylosymbiosis analysis, we show that all groups have lower intraspecies microbiome variability compared with interspecies. This was combined with greater topological congruency between host phylogeny and the microbial dendrogram than would be expected by chance, except when using the unweighted UniFrac distance in corals, octocorals and ascidians. Interestingly, all invertebrate groups but the ascidians exhibited a significant correlation between host phylogenetic distance and microbial dissimilarity across all beta-diversity metrics.
Our results demonstrate that sponges have a strong signature of phylosymbiosis, which likely reflects the uniform microbiome structure in sponges compared with other coral reef invertebrates [18]. This was observed through low intraspecific variation and high homogeneity in the microbiome when the same species was collected from different reefs. Sponges are also known to have a relatively stable microbiome in response to temporal variation and environmental perturbations [52–54]. A stable microbiome may lead to a strong phylosymbiotic signal if there is less influence from the surrounding environment, leaving host factors to be the primary structuring element of the microbiome [53]. Importantly, while sister species were included in the analysis, overall the sponges sampled here span a larger phylogenetic diversity compared with the other groups, which may increase the chance to observe phylosymbiosis. Our results agree with previous conclusions of a significant correlation between host phylogeny and microbiome dissimilarity and validate a prominent role of host phylogeny in shaping the sponge microbiome [18, 55].
A signal of phylosymbiosis was demonstrated in coral, which was characterised by a tendency of corals of the same genus or family to cluster together. However, incongruences were observed where the same species was collected from two different locations, primarily due to a shift in the relative abundance of Endozoicomonadaceae. Shifts in Endozoicomonadaceae have been documented previously, normally in response to host stress [26, 56]. As shifts in the microbial community can often precede visual signs of an unhealthy holobiont [57, 58], it is plausible the decrease in Endozoicomonadaceae is linked to an unknown event. Second, coral tissue samples are often contaminated by the coral mucus, which is known to have a dynamic microbial community shifting in composition between new and aged mucus [59]. However, bacteria within the tissues of corals are housed within coral-associated microbial aggregates and these communities likely have a more stable association with the host [60, 61]. Therefore, developing approaches to target tissue-specific microbes could be beneficial to understanding phylosymbiosis and other questions related to microbial symbiosis in corals.
Similar clustering of coral microbiomes has been observed in Caribbean corals. This partially reflected coral phylogeny, as congenerics showed comparatively low microbial dissimilarity and the two major coral clades tended to cluster together, however, inconsistencies were seen when looking at the species level [62], and reflect the results seen here on the GBR. Further evidence of phylosymbiosis in coral was found in an analysis of 691 coral samples collected Australia wide [8]. The endolithic microbial community showed the strongest signal and was the best predictor of the deep phylogeny between the Robusta and Complexa clades. Tissue microbiomes also illustrated evidence of phylosymbiosis, however, the signal was absent in the coral’s surface mucus layer. This emphasises an increasing strength of phylosymbiosis where direct environmental factors are reduced. In addition, a small number of microbial lineages, including those within Endozoicomonadaceae, demonstrated co-phylogeny with their host, while other clades had a more generalist host distribution. It is possible that host-specialist clades play a minor role in phylosymbiosis through codivergence and future work should aim to untangle the mechanisms behind phylosymbiosis [14].
Research on the microbiome structure of octocorals is limited compared with corals, and we show for the first-time direct evidence for phylosymbiosis. The phylosymbiotic signal in octocorals was similar to corals and incongruences also occurred when there was a shift in the relative abundance of Endozoicomonadaceae. Octocorals are known to have a more stable and less diverse microbial community than hard corals [63], consistent with our finding that overall microbial diversity was lower and microbiome uniformity higher in octocorals compared with hard corals. While this likely influences the phylosymbiotic signal, a direct comparison between octocorals and corals (and other invertebrate groups) cannot be drawn due to the differences in phylogenetic relatedness between host species. Furthermore, the phylogenetic markers used in this study were chosen to capture both mitochondrial and nuclear evolution across a broad range of diverse species. However, octocorals have poorly understood phylogenetic relationships, with little concordance between morphological, nuclear and mitochondrial data [64]. The incorporation of alternative phylogenetic markers optimised for each taxonomic group may further improve the analyses of phylosymbiosis and comparisons among groups. Finally, octocoral identification in the field is extremely challenging especially when trying to resolve to species level [65]. Despite these limitations, we still observe a significant signal of phylosymbiosis, which is likely to strengthen with improved phylogenetic relationships and species identification.
Ascidians showed complete congruence between the host phylogeny and microbial dendrogram for both Bray–Curtis and weighted UniFrac metrics, yet no correlation existed between host phylogenetic distance and microbial dissimilarity. Our results therefore do not provide strong support for phylosymbiosis in the group, yet they highlight the need for multiple lines of evidence when evaluating phylosymbiosis [3]. For example, we find that when sample numbers are low, particularly when marked changes are observed among the microbiomes of host species, the dendrogram approach was more sensitive to patterns of phylosymbiosis compared with the Mantel test. Furthermore, unweighted UniFrac methods were unable to identify a phylosymbiotic signal in the ascidians and had the least power to identify a signal across all invertebrate groups, which agrees with previous conclusions on weighted and unweighted beta-diversity metrics [14]. As this method does not account for the abundance of ASVs, it is less likely to identify beta-diversity patterns in highly diverse microbiomes that are dominated by a relatively small number of bacteria
Our study overwhelmingly found that host phylogeny is reflected in the microbiome of marine invertebrates, particularly notable when considering several confounding factors. Sampling of the reef invertebrates occurred over four field trips that spanned a 1-year timeframe, potentially obscuring phylosymbiosis patterns due to seasonal influences on the microbiome [66]. Furthermore, our samples are from wild colonies collected from multiple locations on the GBR which introduces local environmental differences including water quality and the pelagic communities that serve as host diet. Preservation methods also varied across organisms, including snap freezing and the use of salt saturated dimethyl sulfoxide-EDTA. While these preservation approaches have been shown to have little effect on the microbial composition of coral, it could have influenced alpha diversity [67]. Finally, sample representation differed among the four groups and likely has an important impact on the strength of the phylosymbiosis signal. For example, only three ascidian species (and one outgroup) were used, whereas four related species are recommended [3]. Had more species been included in the analysis, with a larger number of taxonomic sister species, a more reliable representation of phylosymbiosis would likely have been achieved.
This is the first study to systematically assess phylosymbiosis among diverse groups of marine invertebrates. We identified a phylosymbiotic signal across all invertebrate groups with multiple methods, of which sponges consistently showed a significant signal using all beta-diversity metrics. Increased intraspecific variability of the microbiome in both scleractinian corals and octocorals was often associated with a change in the relative abundance of Endozoicomonadaceae. This microbial family is characterised by host-specialist and host-generalist clades and is assumed to be a dynamic member of the coral holobiont [8]. Host-specialist clades may contribute to phylosymbiosis in corals and octocorals through codivergence, while host-generalist clades obscure the signal through host infidelity. Here, we provide a foundation to begin exploring the mechanisms behind phylosymbiosis and further our understanding on host-microbe symbiosis and coevolution in marine invertebrates.
Supplementary information
Acknowledgements
The authors would like to thank Katharina Fabricius, Georgina Torras and Bettina Glasl for their assistance in the field. We also thank Orpheus Island Research Station and the Molecular Ecology and Evolution Laboratory for facilitating field and laboratory work. We thank Zhenyu Peng, Guohai Hu, Bo Wang, Xudan Li, Wei Zhou, Sha Liao and Junqiang Xu for providing SE400 sequencing. Guohai Hu and Bo Wang are affiliated with Guangdong Provincial Key Laboratory of Genome Read and Write (No. 2017B030301011). We also thank Long Zhou and Qiye Li for coordinating the project. This work was funded by the Beijing Genome Institute, Earthwatch Institute and Mitsubishi Corporation. PAO is supported by an AIMS@JCU postgraduate research scholarship.
Author contributions
PAO, DGB, NSW, DJM and GZ conceived and developed the study. PAO, DGB, NSW, PRF and HAS contributed to field work. PAO, ST, CY and HAS contributed to molecular lab work. PAO analysed the microbial data and generated figures and HAS finalised the figures. PAO, ST, CY and NA analysed the phylogenetic data. PAO drafted the paper and all authors revised the paper and approved the final version.
Data availability
All microbial data have been made available at the NCBI Sequence Read Archive under the BioProject accession number PRJNA577361 and host sequence data are available at the CNGB Sequence Archive under the accession numbers N_000000252.1–N_000000348.1.
Code availability
Code used for the analysis is available at https://github.com/paobrien.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guojie Zhang, Email: zhanggj@genomics.cn.
David G. Bourne, Email: david.bourne@jcu.edu.au
Supplementary information
The online version of this article (10.1038/s41396-020-0671-x) contains supplementary material, which is available to authorised users.
References
- 1.Brooks AW, Kohl KD, Brucker RM, van Opstal EJ, Bordenstein SR. Phylosymbiosis: relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016;14:1–29. doi: 10.1371/journal.pbio.2000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brucker RM, Bordenstein SR. The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus nasonia. Science. 2013;341:667–9. doi: 10.1126/science.1240659. [DOI] [PubMed] [Google Scholar]
- 3.Lim SJ, Bordenstein SR. An introduction to phylosymbiosis. Proc Biol Sci. 2020;287:20192900. doi: 10.1098/rspb.2019.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brucker RM, Bordenstein SR. The roles of host evolutionary relationships (genus: Nasonia) and development in sturcturing microbial communities. Evolution. 2011;66:349–62. doi: 10.1111/j.1558-5646.2011.01454.x. [DOI] [PubMed] [Google Scholar]
- 5.Ochman H, Worobey M, Kuo CH, Ndjango JBN, Peeters M, Hahn BH, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:3–10. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohl KD, Dearing MD, Bordenstein SR. Microbial communities exhibit host species distinguishability and phylosymbiosis along the length of the gastrointestinal tract. Mol Ecol. 2018;27:1874–83. doi: 10.1111/mec.14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross AA, Müller KM, Weese JS, Neufeld JD. Comprehensive skin microbiome analysis reveals the uniqueness of human skin and evidence for phylosymbiosis within the class Mammalia. Proc Natl Acad Sci. 2018;115:1–10. doi: 10.1073/pnas.1801302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, et al. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun. 2018;9:1–13. doi: 10.1038/s41467-018-07275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeoh YK, Dennis PG, Paungfoo-Lonhienne C, Weber L, Brackin R, Ragan MA, et al. Evolutionary conservation of a core root microbiome across plant phyla along a tropical soil chronosequence. Nat Commun. 2017;8:1–9. doi: 10.1038/s41467-017-00262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leigh BA, Bordenstein SR, Brooks AW, Mikaelyan A, Bordenstein SR. Finer-scale phylosymbiosis: insights from insect viromes. mSystems. 2018;3:e00131–18. doi: 10.1128/mSystems.00131-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Opstal EJ, Bordenstein SR. Phylosymbiosis impacts adaptive traits in Nasonia wasps. MBio. 2019;10:1–11. doi: 10.1128/mBio.00887-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TCG. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci. 2013;110:E3730–8. doi: 10.1073/pnas.1304960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas AE, Werren JH. Holes in the hologenome: why host-microbe symbioses are not holobionts. MBio. 2016;7:e02099–15. doi: 10.1128/mBio.02099-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazel F, Davis KM, Loudon A, Kwong WK, Groussin M, Parfrey LW. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems. 2018;3:e00097–18. doi: 10.1128/mSystems.00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Groussin M, Mazel F, Sanders JG, Smillie CS, Lavergne S, Thuiller W, et al. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat Commun. 2017;8:1–12. doi: 10.1038/ncomms14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hird SM, Sánchez C, Carstens BC, Brumfield RT. Comparative gut microbiota of 59 Neotropical bird species. Front Microbiol. 2015;6:1–16. doi: 10.3389/fmicb.2015.01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song SJ, Sanders JG, Delsuc F, Metcalf J, Amato K, Taylor MW, et al. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio. 2020;11:1–14. doi: 10.1128/mBio.02901-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas T, Moitinho-Silva L, Lurgi M, Björk JR, Easson C, Astudillo-García C, et al. Diversity, structure and convergent evolution of the global sponge microbiome. Nat Commun. 2016;7:1–12. doi: 10.1038/ncomms11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glasl B, Smith CE, Bourne DG, Webster NS. Disentangling the effect of host-genotype and environment on the microbiome of the coral Acropora tenuis. PeerJ. 2019;7:1–18. doi: 10.7717/peerj.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amato KR, G. Sanders J, Song SJ, Nute M, Metcalf JL, Thompson LR, et al. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 2019;13:576–87. doi: 10.1038/s41396-018-0175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amato KR, Mallott EK, McDonald D, Dominy NJ, Goldberg T, Lambert JE, et al. Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 2019;20:1–12. doi: 10.1186/s13059-019-1807-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharp KH, Distel D, Paul VJ. Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 2012;6:790–801. doi: 10.1038/ismej.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apprill A, Marlow HQ, Martindale MQ, Rappé MS. The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 2009;3:685–99. doi: 10.1038/ismej.2009.3. [DOI] [PubMed] [Google Scholar]
- 24.Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12:2070–82. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Brien PA, Webster NS, Miller DJ, Bourne DG. Host-microbe coevolution: applying evidence from model systems to complex marine invertebrate holobionts. MBio. 2019;10:1–14. doi: 10.1128/mBio.02241-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrow KM, Bourne DG, Humphrey C, Botté ES, Laffy P, Zaneveld J, et al. Natural volcanic CO2seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 2014;9:1–15. doi: 10.1038/ismej.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun. 2017;8:1–8. doi: 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–14. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 29.Apprill A, Mcnally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Micro Ecol. 2015;75:129–37. [Google Scholar]
- 30.Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–9. [PubMed] [Google Scholar]
- 31.Piredda R, Tomasino MP, D’Erchia AM, Manzari C, Pesole G, Montresor M, et al. Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean Long Term Ecological Research site. FEMS Microbiol Ecol. 2017;93:fiw200. doi: 10.1093/femsec/fiw200. [DOI] [PubMed] [Google Scholar]
- 32.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interact sequence choice and visualization. Brief Bioinform. 2019;20:1160–6. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C, Tan S, Meng G, Bourne DG, O’Brien PA, Xu J, et al. Access COI barcode efficiently using high throughput Single-End 400 bp sequencing. bioRxiv. 2018:498618.
- 34.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–7. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssen S, Mcdonald D, Gonzalez A, Navas-Molina JA, Jiang L, Xu ZZ, et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems. 2018;3:1–14. doi: 10.1128/mSystems.00021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 38.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P. McGlinn D, et al. vegan: community ecology package. R package version. 2019;2:5–4.
- 40.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 41.Yu G, Smith D, Zhu H, Guan Y, Tsan-Yuk Lam T. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 42.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2018;35:526–8. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 43.Schliep K. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–3. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham H, François R, Henry L, Müller K. dplyr: a grammar of data manipulation. R package version 0.8.0.1. 2019. https://CRAN.R-project.org/package=dplyr.
- 45.McKnight DT, Huerlimann R, Bower DS, Schwarzkopf L, Alford RA, Zenger KR. Methods for normalizing microbiome data: an ecological perspective. Methods Ecol Evol. 2019;10:389–400. [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. J Hered. 2001;92:371–3. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 48.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 49.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pöppe J, Sutcliffe P, Hooper JNA, Wörheide G, Erpenbeck D. CO I barcoding reveals new clades and radiation patterns of indo-pacific sponges of the family irciniidae (Demospongiae: Dictyoceratida) PLoS One. 2010;5:1–6. doi: 10.1371/journal.pone.0009950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erwin PM, Coma R, Paula L, Serrano E, Ribes M, Mar P. Stable symbionts across the HMA-LMA dichotomy: low seasonal and interannual variation in sponge-associated bacteria from taxonomically. FEMS Microbiol Ecol. 2015;91:1–11. doi: 10.1093/femsec/fiv115. [DOI] [PubMed] [Google Scholar]
- 53.Glasl B, Smith CE, Bourne DG, Webster NS. Exploring the diversity-stability paradigm using sponge microbial communities. Sci Rep. 2018;8:1–9. doi: 10.1038/s41598-018-26641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luter HM, Whalan S, Webster NS. Thermal and sedimentation stress are unlikely causes of brown spot syndrome in the coral reef sponge, Ianthella basta. PLoS One. 2012;7:e39779. doi: 10.1371/journal.pone.0039779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Easson CG, Thacker RW. Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front Microbiol. 2014;5:1–11. doi: 10.3389/fmicb.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer JL, Paul VJ, Teplitski M. Community shifts in the surface microbiomes of the coral porites astreoides with unusual lesions. PLoS One. 2014;9:e100316. doi: 10.1371/journal.pone.0100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bourne D, Iida Y, Uthicke S, Smith-keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–63. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- 58.Pollock FJ, Lamb JB, Van De Water JAJM, Smith HA, Schaffelke B, Willis BL, et al. Reduced diversity and stability of coral-associated bacterial communities and suppressed immune function precedes disease onset in corals. R Soc Open Sci. 2019;6:190355. doi: 10.1098/rsos.190355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glasl B, Herndl GJ, Frade PR. The microbiome of coral surface mucus has a key role in mediating holobiont health and survival upon disturbance. ISME J. 2016;10:2280–92. doi: 10.1038/ismej.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Work TM, Aeby GS. Microbial aggregates within tissues infect a diversity of corals throughout the Indo-Pacific. Mar Ecol Prog Ser. 2014;500:1–9. [Google Scholar]
- 61.Wada N, Ishimochi M, Matsui T, Pollock FJ, Tang S, Ainsworth TD, et al. Characterization of coral-associated microbial aggregates (CAMAs) within tissues of the coral Acropora hyacinthus. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-49651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLoS One. 2010;5:1–7. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van De Water JAJM, Allemand D, Ferrier-Pagès C. Host-microbe interactions in octocoral holobionts—recent advances and perspectives. Microbiome. 2018;6:1–28. doi: 10.1186/s40168-018-0431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McFadden CS, Sánchez JA, France SC. Molecular phylogenetic insights into the evolution of octocorallia: a review. Integr Comp Biol. 2010;50:389–410. doi: 10.1093/icb/icq056. [DOI] [PubMed] [Google Scholar]
- 65.Quattrini AM, Wu T, Soong K, Jeng M, Benayahu Y, Mcfadden CS. A next generation approach to species delimitation reveals the role of hybridization in a cryptic species complex of corals. BMC Evol Biol. 2019;19:1–19. doi: 10.1186/s12862-019-1427-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J, Chen Q, Long LJ, Dong JD, Yang J, Zhang S. Bacterial dynamics within the mucus, tissue and skeleton of the coral Porites lutea during different seasons. Sci Rep. 2014;4:1–8. doi: 10.1038/srep07320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hernandez-Agreda A, Leggat W, Ainsworth TD, Hernandez-Agreda A. A comparative analysisof microbial DNA preparation methods for use with massive and branching coralgrowth forms. Front Microbiol. 2018;9:1–14. doi: 10.3389/fmicb.2018.02146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microbial data have been made available at the NCBI Sequence Read Archive under the BioProject accession number PRJNA577361 and host sequence data are available at the CNGB Sequence Archive under the accession numbers N_000000252.1–N_000000348.1.
Code used for the analysis is available at https://github.com/paobrien.