Abstract
Background
Circular RNAs (circRNAs) play a crucial role in tumorigenesis. However, the effects of circRNAs on acute myeloid leukemia (AML) remain largely unexplored. We explored the function of circRAD18 in AML development.
Methods
QRT-PCR was performed for the levels of circRAD18, RAD18, microRNA-206 (miR-206) and protein kinase CAMP-activated catalytic subunit beta (PRKACB). Cell Counting Kit-8 (CCK-8) assay and colony formation assay were utilized for cell proliferation. Flow cytometry analysis was carried out to analyze cell apoptosis and cell cycle process. Transwell assay was manipulated for cell migration and invasion. Western blot assay was conducted for protein levels. Dual-luciferase reporter assay was adopted to verify the interaction between miR-206 and circRAD18 or PRKACB.
Results
CircRAD18 level was increased in AML patients’ blood specimens and AML cell lines compared to normal controls. CircRAD18 knockdown impeded the proliferation, migration and invasion and facilitated the apoptosis and cell cycle arrest in AML cells. Moreover, circRAD18 was identified as a sponge for miR-206, and circRAD18 knockdown-mediated effect on AML cell progression was reversed by miR-206 suppression. Additionally, PRKACB was the target gene of miR-206. MiR-206 overexpression suppressed the malignant behaviors of AML cells, while PRKACB elevation restored the effects.
Conclusion
CircRAD18 aggravated the malignancy of AML cells through reducing miR-206 expression and elevating PRKACB expression, indicating circRAD18 might be a therapeutic target for AML.
Keywords: AML, circRAD18, miR-206, PRKACB
Introduction
Acute myeloid leukemia (AML) is a malignant myeloid disease caused by uncontrolled proliferation, retarded apoptosis and abnormal differentiation of myeloid leukocytes.1,2 Although significant advancements in diagnostic and therapeutic strategies have been made, the prognosis of patients with AML remains unsatisfied.3,4 Moreover, the underlying pathogenic mechanisms of AML are elusive due to the heterogeneity of AML.5 Hence, understanding the pathogenesis of AML is of great significance for developing novel therapy methods.
Circular RNAs (circRNAs) are a sort of novel non-coding RNAs (ncRNAs), which are featured by covalently closed-loop structures.6 Presently, circRNAs have been demonstrated to sponge microRNAs (miRNAs) and then regulate their functions via serving as competitive endogenous RNAs (ceRNAs).7 Several of circRNAs are aberrantly expressed and play a vital role in leukemogenesis. For example, circ_100290 level was enhanced in AML and facilitated AML cell growth and impeded apoptosis through sponging miR-203.8 High level of circ_0009910 predicted a worse outcome of AML patients and circ_0009910 silencing restrained the malignancy of AML.9 As a member of circRNAs, circRAD18 (circ_0001264) was dysregulated in doxorubicin-resistant AML cells.10 However, the expression level of circRAD18 in AML and related functions are unclear.
As a class of small ncRNAs, miRNAs can recognize the 3ʹUTR of target mRNA for cleavage or translational repression.11 Multiple miRNAs have been reported to be closely linked to AML progression. For instance, miR-4792 was able to hinder AML cell growth and invasion and induce apoptosis by targeting Kindlin-3.12 MiR-34a repressed the autophagy and facilitated the apoptosis of AML cells by interacting with HMGB1.13 More importantly, Liu et al reported that miR-206 served as a tumor inhibitor in pediatric AML via targeting CyclinD1.14 Even so, the underlying mechanisms of miR-206 in AML progression still need to be further explored.
The protein kinase CAMP-activated catalytic subunit beta (PRKACB) is a key effector of cAMP/PKA-stimulated signal pathway and participates in the regulation of diverse cellular processes.15 Furthermore, Wu et al disclosed that PRKACB contributed to AML cell proliferation and hindered apoptosis via functioning as the target of miR-496.16 However, the interaction between miR-206 and PRKACB is still undefined.
In this paper, the expression profiles of circRAD18, miR-206 and PRKACB in AML patients and cells were determined. Furthermore, the functional roles and underlying mechanisms of circRAD18 were further assessed.
Materials and Methods
Blood Samples Collection
Fifty-five AML patients and 55 healthy donors were enrolled in our research. The blood specimens were collected from the participants at Ruijin Hospital, Shanghai Jiao Tong University School of Medicine and then preserved at −80°C before use. This work was approved by the Ethics Committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine and was carried out according to the guidelines of Declaration of Helsinki. Written informed consents were obtained from the participants.
Cell Culture
Human normal marrow stromal cell line (HS-5) and AML cell lines (THP-1 and HL-60) were obtained from the ATCC (ATCC, Manassas, VA, USA). These cells were maintained in RPMI 1640 medium (Gibco, Grand Island, NY, USA) added with 1% penicillin-streptomycin (Gibco) and 10% FBS (Gibco) at 37°C in a humid incubator containing 5% CO2.
QRT-PCR Assay
Total RNA was extracted from blood samples and cells utilizing RNeasy Mini kit (Qiagen, Valencia, CA, USA). RNA treatment was executed at 37°C for 15 min utilizing RNase R (Epicenter Biotechnologies, Madison, WI, USA). Then, M-MLV Reverse Transcriptase Kit (Promega, Madison, WI, USA) or mirVanaTM qRT-PCR miRNA Detection Kit (Ambion, Austin, TX, USA) was used to transcribe RNAs into cDNAs. The qRT-PCR analysis was manipulated on a CFX96 Touch Real-time PCR detection system (Bio-Rad, Hercules, CA, USA) utilizing miScript SYBR Green PCR Kit (Qiagen) and indicated primers. The sequences of primers were shown: circRAD18: (F: 5ʹ-CAGCTCATTAAAAGGCACCA-3ʹ and R: 5ʹ-GGAAGAAGCAGGAGATTTGG-3ʹ); RAD18: (F: 5ʹ-CTGTTTGCGGGGTTAACATT-3ʹ and R: 5ʹ-TTCCCCCAAGTAAGCACAAG-3ʹ); miR-206: (F: 5ʹ-CGATGGAATGTAAGGAAGT-3ʹ and R: 5ʹ-GTGCAGGGTCCGAGGT-3ʹ); PRKACB: (F: 5ʹ-TGGCAGCTTATAGAGAACCACC-3ʹ and R: 5ʹ-CTCTTTCATTGATCTGTCCCA-3ʹ); GAPDH: ((F: 5ʹ-AAGGTGAAGGTCGGAGTCA-3ʹ and R: 5ʹ-GGAAGATGGTGATGGGATTT-3ʹ); U6: (F: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and R: 5ʹ-CGCTTCACGAATTTGCGTGTCAT −3ʹ). The expression was estimated via the 2−ΔΔCt strategy with GAPDH or U6 as an internal reference.
Cell Transfection
CircRAD18 small interfering RNA (si-circRAD18#1 and si-circRAD18#2) and scramble siRNA (si-NC), miR-206 mimics (miR-206) and miR-NC, miR-206 inhibitor (anti-miR-206) and anti-miR-NC, PRKACB overexpression vector (PRKACB) and its control (vector) were synthesized by GenePharma (Shanghai, China). Cell transfection was executed through the usage of Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Cell Counting Kit-8 (CCK-8) Assay
In each well of 96-well plates, 1×104 THP-1 and HL-60 cells were seeded after relevant transfection and inoculated overnight. After 0 h, 24 h, 48 h or 72 h, 10 μL CCK-8 (Dojindo, Kumamoto, Japan) was added and maintained for a further 4 h. The absorption at 450 nm was measured with a microplate reader (Potenov, Beijing, China).
Colony Formation Assay
The THP-1 and HL-60 cells after transfected for 48 h were plated into 6-well plates (300 cells/well) and cultured in the growth medium for about 14 days. The colonies were fixed in methanol and dyed with 0.1% crystal violet (Solarbio, Beijing, China) for 30 min at 37°C. Next, the colonies that had >50 cells were counted under an inverted microscope (Olympus, Tokyo, Japan) at the magnification of 40×.
Flow Cytometry Analysis
For cell apoptosis determination, the transfected THP-1 and HL-60 cells were collected and washed twice using PBS (Solarbio). Next, the cells were resuspended and then labeled with Annexin V-fluorescein isothiocyanate (FITC; Vazyme, Nanjing, China) and propidium iodide (PI; Vazyme) for 20 min. After washing with PBS (Solarbio), the apoptotic cells were examined by a FACScan® flow cytometry (BD Bioscience, Franklin Lakes, NJ, USA).
For cell cycle determination, the collected cells were fixed with 70% ethanol overnight at 4°C and then washed using PBS (Solarbio). Then, the cells were suspended and mixed with PI (Vazyme) for 30 min at 37°C. After that, cell cycle was estimated with a FACScan® flow cytometry (BD Bioscience).
Western Blot Assay
The extraction of total protein was finished through the usage of RIPA (CWBio, Beijing, China) and the concentrations of proteins were determined through the usage of BCA Protein Quantification Kit (Vazyme). Next, the proteins were loaded on SDS-PAGE gel (Solarbio) and blotted onto PVDF membranes (Amersham Biosciences, Chicago, IL, USA). After blocking in 5% defatted milk for 1 h, the membranes were cultivated overnight with primary antibodies at 4°C followed by incubation with secondary antibody (ab205719; Abcam, Cambridge, MA, USA) for 1 h at indoor temperature. The blots were examined with an ECL detection kit (Pierce Biotechnology, Rockford, IL, USA). The primary antibodies were: Bcl-2 (ab182858; Abcam), Bax (ab32503; Abcam), cleaved-caspase-3 (c-caspase-3; ab49822; Abcam), PRKACB (ab26322; Abcam) and GAPDH (ab9485; Abcam).
Transwell Assay
The invasion and migration capacities of THP-1 and HL-60 cells were, respectively, assessed utilizing transwell inserts (BD Bioscience) pre-coated with or without Matrigel (BD Bioscience). Briefly, 1×105 transfected AML cells in serum-free medium were plated into the top chamber of the inserts. The lower chamber was added with 600 μL culture medium. The cells were allowed to grow for 24 h and then treated with 70% ethanol and 0.1% crystal violet (Solarbio). After washing with PBS (Solarbio), the migrated and invaded cells were analyzed utilizing a microscope (Olympus, Tokyo, Japan) at the magnification of 100×.
Dual-Luciferase Reporter Assay
The sequences of circRAD18 or PRKACB 3'UTR, which consisted of the wild-type (WT) or mutant (MUT) miR-206 binding sites, were cloned into pmirGLO plasmid (Promega), constructing circRAD18 WT, circRAD18 MUT, PRKACB 3ʹUTR WT and PRKACB 3ʹUTR MUT, respectively. THP-1 and HL-60 cells were cultivated in 6-well plates and co-transfected the constructs and miR-NC or miR-206. The luciferase activity was examined through the Dual-Luciferase Reporter Assay System (Promega) after 48 h.
Statistical Analysis
The experiments were executed triple times and the data were analyzed by GraphPad Prism 7 and represented as mean ± SD. The statistical significance was estimated via Student’s t-test or one-way analysis of variance (ANOVA). P<0.05 was deemed as statistically significant.
Results
CircRAD18 Was Overexpressed in AML Patients and AML Cell Lines
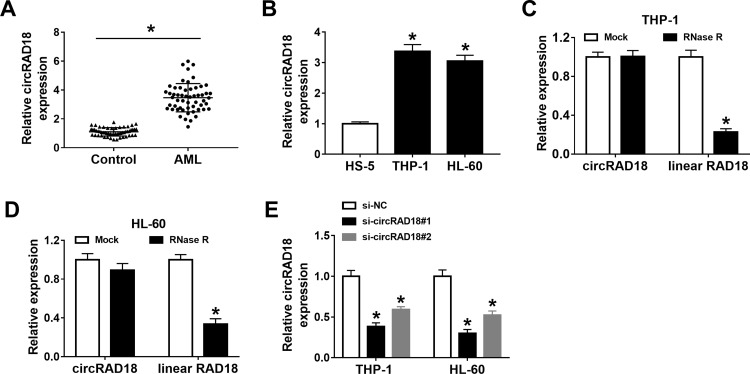
To explore the potential roles of circRAD18 in AML, we initially determined the expression level of circRAD18 in the blood samples from AML patients and healthy volunteers. The results exhibited that circRAD18 level was evidently raised in AML patients compared to healthy controls (Figure 1A). We also determined the expression level of circRAD18 in AML cell lines and normal cell lines. The results showed that circRAD18 was highly expressed in THP-1 and HL-60 cells in comparison with normal cells (Figure 1B). The characteristic of circRAD18 was then verified by RNase R assay, which showed that linear RAD18 could be digested by RNase R, but circRAD18 level was not affected (Figure 1C and D). In addition, si-circRAD18#1 or si-circRAD18#2 was transfected into THP-1 and HL-60 cells to knock down the expression of circRAD18. As demonstrated by qRT-PCR analysis, the transfection of si-circRAD18#1 or si-circRAD18#2 markedly decreased circRAD18 level in THP-1 and HL-60 cells; moreover, the knockdown efficiency of si-circRAD18#1 was more significant than si-circRAD18#2 (Figure 1E). Thus, si-circRAD18#1 was selected for the subsequent experiments. These results indicated that circRAD18 might play a role in AML.
Figure 1.
High level of circRAD18 in AML patients and cells. (A) The level of circRAD18 in the blood samples from 55 AML patients and 55 healthy donors was examined with qRT-PCR analysis. (B) The level of circRAD18 in HS-5, THP-1 and HL-60 cells was determined by qRT-PCR assay. (C and D) After total RNA in THP-1 and HL-60 cells was treated with or without RNase R, circRAD18 and linear RAD18 levels were detected via qRT-PCR assay. (E) The expression level of circRAD18 in THP-1 and HL-60 cells transfected with si-NC, si-circRAD18#1 or si-circRAD18#2 was determined with qRT-PCR assay. *P<0.05.
Silencing of circRAD18 Suppressed AML Cell Malignant Behaviors
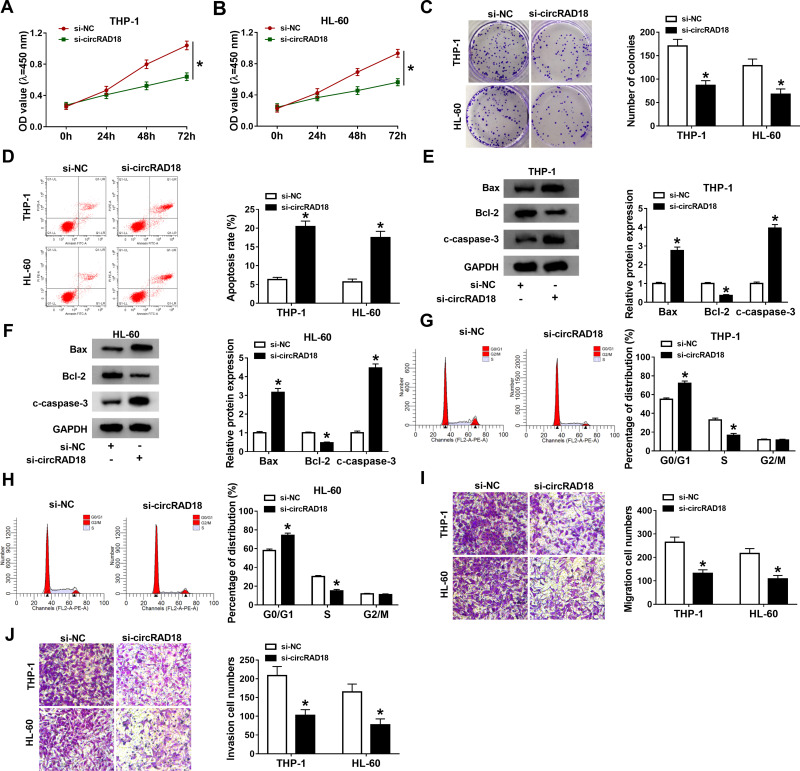
Subsequently, the exact roles of circRAD18 in AML were investigated. As illustrated by CCK-8 assay, circRAD18 knockdown conspicuously restrained the proliferation of THP-1 and HL-60 cells relative to si-NC control groups (Figure 2A and B). The results of colony formation assay indicated that the colony formation ability of THP-1 and HL-60 cells was apparently inhibited by circRAD18 knockdown (Figure 2C). Flow cytometry analysis exhibited that circRAD18 silencing markedly facilitated cell apoptosis in THP-1 and HL-60 cells in comparison with si-NC groups (Figure 2D). Then, we detected the levels of pro-apoptotic proteins Bax and c-caspase-3 and anti-apoptotic protein Bcl-2 through Western blot assay, showing that Bax and c-caspase-3 levels were enhanced and Bcl-2 level was declined in both THP-1 and HL-60 cells transfected with si-circRAD18 compared to control groups (Figure 2E and F). We also analyzed cell cycle process using flow cytometry analysis. The results exhibited that the percentage of THP-1 and HL-60 cells in G0/G1 phase was increased and the percentage of THP-1 and HL-60 cells in S phase was reduced, indicating that cell cycle process was arrested (Figure 2G and H). In addition, circRAD18 deficiency repressed the migration and invasion of THP-1 and HL-60 cells in comparison with control groups (Figure 2I and J). Collectively, circRAD18 knockdown inhibited the progression of AML.
Figure 2.
Effects of circRAD18 knockdown on AML cell biological behaviors. (A and B) The proliferation, (C) colony formation and (D) apoptosis of THP-1 and HL-60 cells were assessed by CCK-8 assay, colony formation assay and flow cytometry analysis, respectively. (E and F) The protein levels of Bax, Bcl-2 and c-caspase-3 in THP-1 and HL-60 cells were measured by Western blot assay. (G and H) The cell cycle process in THP-1 and HL-60 cells was analyzed by flow cytometry analysis. (I and J) The migration and invasion of THP-1 and HL-60 cells were explored by transwell assay. *P<0.05.
CircRAD18 Directly Interacted with miR-206 in AML Cells
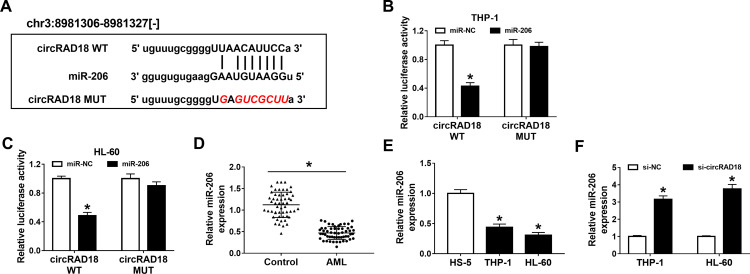
To clarify the underlying molecular mechanism of circRAD18 in AML, online software starBase v2.0 was employed to analyze the potential target of circRAD18. As a result, miR-206 contained the complementary sequences of circRAD18, indicating that miR-206 might be a target of circRAD18 (Figure 3A). Dual-luciferase reporter assay presented that miR-206 transfection notably decreased the luciferase activity of circRAD18 WT, while no effect was observed in the luciferase activity of circRAD18 MUT in both THP-1 and HL-60 cells, further suggesting the interaction between circRAD18 and miR-206 (Figure 3B and C). Furthermore, we found that miR-206 was lowly expressed in AML patients’ blood samples and AML cell lines when compared to normal controls (Figure 3D and E). Additionally, our results showed that circRAD18 knockdown prominently elevated the level of miR-206 in AML cells (Figure 3F). These findings suggested that circRAD18 directly bound to miR-206 and negatively regulated miR-206 expression in AML cells.
Figure 3.
CircRAD18 directly sponged miR-206. (A) The potential binding sites between circRAD18 and miR-206. (B and C) The association between circRAD18 and miR-206 was estimated by dual-luciferase reporter assay. (D and E) The expression level of miR-206 in AML patients’ blood samples and AML cells was determined by qRT-PCR assay. (F) The relative expression of miR-206 in THP-1 and HL-60 cells transfected with si-circRAD18 or si-NC was detected using qRT-PCR assay. *P<0.05.
Inhibition of miR-206 Reversed the Effects of circRAD18 Silencing on AML Cell Malignant Phenotypes
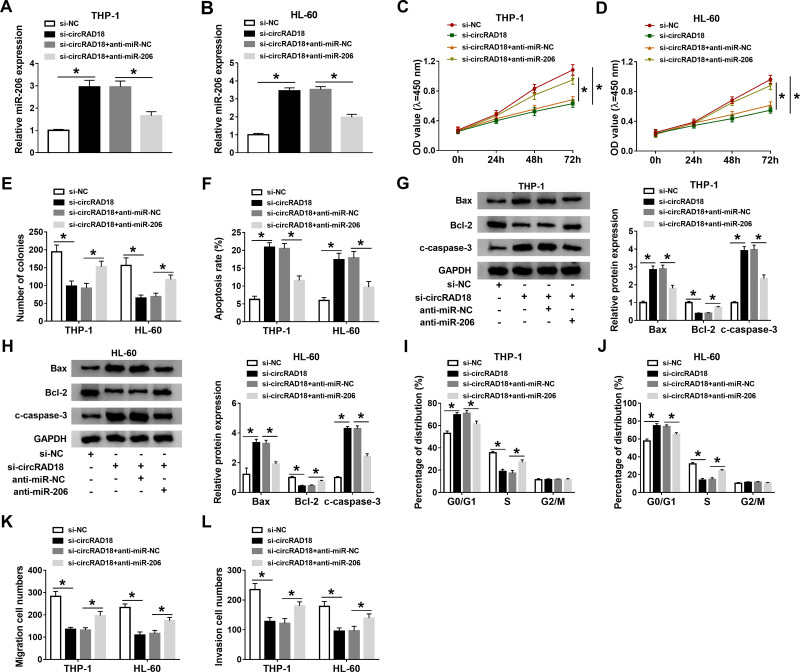
To further explore the effects of circRAD18 and miR-206 in AML, THP-1 and HL-60 cells were introduced with si-NC, si-circRAD18, si-circRAD18+anti-miR-NC or si-circRAD18+anti-miR-206. As exhibited in Figure 4A and B, anti-miR-206 transfection effectively overturned the upregulation of miR-206 in THP-1 and HL-60 cells caused by circRAD18 knockdown. The results of CCK-8 assay and colony formation assay indicated that miR-206 inhibition ameliorated the inhibitory roles of circRAD18 silencing in cell proliferation and colony formation in THP-1 and HL-60 cells (Figure 4C-E). Flow cytometry analysis showed that circRAD18 silencing induced the apoptosis of AML cells, while miR-206 suppression ameliorated the effect (Figure 4F). Western blot assay exhibited that the effects of circRAD18 knockdown on Bax, Bcl-2 and c-caspase-3 protein levels were also abated by the inhibition of miR-206 in AML cells (Figure 4G and H). Flow cytometry analysis demonstrated that the promotional effect of circRAD18 knockdown on cell cycle arrest in THP-1 and HL-60 cells was also abrogated by decreasing miR-206 (Figure 4I and J). Additionally, circRAD18 deficiency markedly restrained the migration and invasion of THP-1 and HL-60 cells, whereas miR-206 inhibition reversed the impacts (Figure 4K and L). In summary, circRAD18 knockdown repressed AML cell progression by targeting miR-206.
Figure 4.
CircRAD18 knockdown regulated AML cell progression by sponging miR-206. THP-1 and HL-60 cells were transfected with si-NC, si-circRAD18, si-circRAD18+anti-miR-NC or si-circRAD18+anti-miR-206. (A and B) The expression of miR-206 in THP-1 and HL-60 cells was tested by qRT-PCR assay. (C–F) The proliferation, colony formation and apoptosis of THP-1 and HL-60 cells were investigated by CCK-8 assay, colony formation assay and flow cytometry analysis, respectively. (G and H) The protein levels of Bax, Bcl-2 and c-caspase-3 in THP-1 and HL-60 cells were measured through Western blot assay. (I and J) The cell cycle process was analyzed by flow cytometry analysis. (K and L) The migration and invasion of THP-1 and HL-60 cells were assessed by transwell assay. *P<0.05.
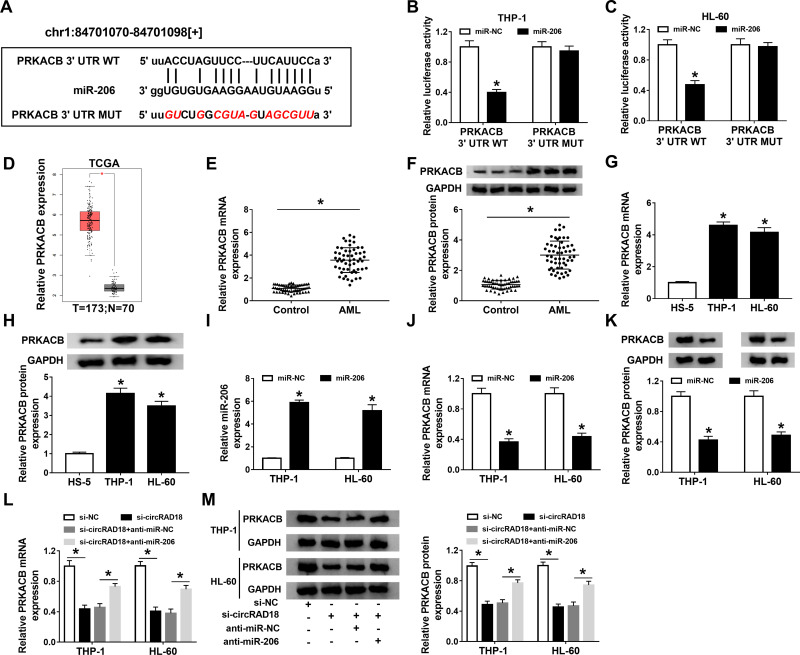
PRKACB Acted as the Direct Target Gene of miR-206 in AML Cells
Through further analyzing starBase v2.0, PRKACB was found to be a target gene of miR-206 and their complementary sequences are presented in Figure 5A. Then, dual-luciferase reporter assay was performed to verify this prediction. The data showed that miR-206 transfection led to a noteworthy suppression in the luciferase activity of PRKACB 3ʹUTR WT in both THP-1 and HL-60 cells instead of PRKACB 3ʹUTR MUT (Figure 5B and C). Through analyzing database TCGA (http://gepia.cancer-pku.cn/detail.php?gene=PRKACB###), PRKACB level was found to be elevated in AML patients compared to normal controls (Figure 5D). Moreover, our results showed that PRKACB mRNA and protein levels were all drastically increased in AML patients’ blood samples and AML cells compared to normal controls (Figure 5E-H). Next, we transfected miR-NC or miR-206 into THP-1 and HL-60 cells, and found that miR-206 transfection apparently increased the level of miR-206, indicating that miR-206 was successfully transfected into AML cells (Figure 5I). We also observed that miR-206 transfection remarkably reduced PRKACB mRNA and protein levels in THP-1 and HL-60 cells compared to miR-NC groups (Figure 5J and K). Additionally, our results exhibited that circRAD18 knockdown conspicuously decreased the mRNA and protein levels of PRKACB in THP-1 and HL-60 cells, while miR-206 suppression rescued the effects (Figure 5L and M). To sum up, circRAD18 positively modulated PRKACB expression by targeting miR-206 in AML cells.
Figure 5.
MiR-206 directly targeted PRKACB in AML cells. (A) The predicted complementary sequences between miR-206 and PRKACB. (B and C) The interaction between miR-206 and PRKACB in THP-1 and HL-60 cells was analyzed by dual-luciferase reporter assay. (D) The database TCGA showed PRKACB expression was upregulated in AML patients. (E–H) The mRNA and protein levels of PRKACB in AML patients’ blood samples and AML cells were measured by qRT-PCR assay and Western blot assay, respectively. (I–K) After THP-1 and HL-60 cells were transfected with miR-NC or miR-206, the levels of miR-206, PRKACB mRNA and PRKACB protein were determined by qRT-PCR assay or Western blot assay. (L and M) The mRNA and protein levels of PRKACB in THP-1 and HL-60 cells transfected with si-NC, si-circRAD18, si-circRAD18+anti-miR-NC or si-circRAD18+anti-miR-206 were detected using qRT-PCR assay and Western blot assay, respectively. *P<0.05.
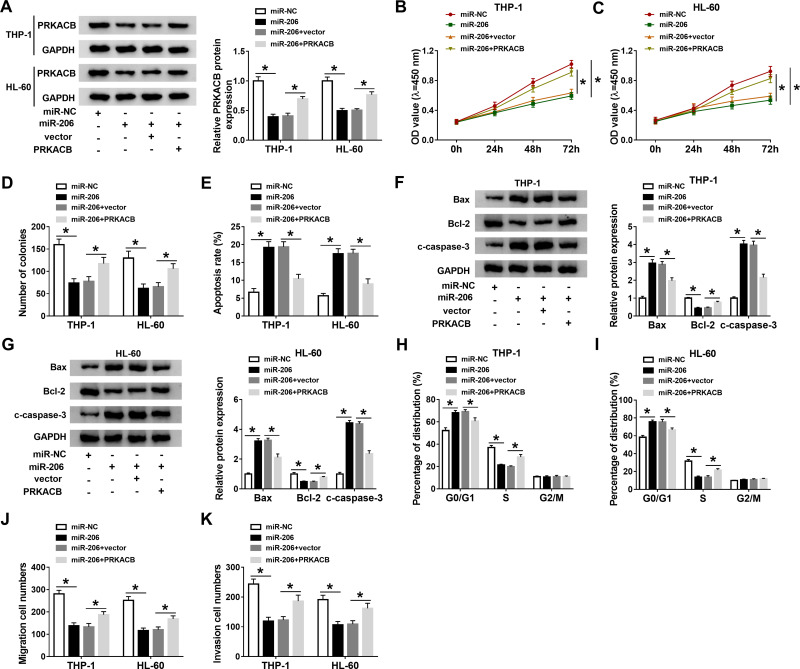
Overexpression of miR-206 Repressed AML Cell Progression by Binding to PRKACB
To further elucidate the relationship between miR-206 and PRKACB in AML development, THP-1 and HL-60 cells were treated with miR-NC, miR-206, miR-206+vector or miR-206+PRKACB. As shown in Figure 6A, miR-206 transfection markedly reduced PRKACB protein level in THP-1 and HL-60 cells, while PRKACB transfection overturned the reduction. As illustrated by CCK-8 assay, colony formation assay and flow cytometry assay, miR-206 upregulation evidently repressed cell proliferation and colony formation and accelerated apoptosis in AML cells, while these effects were abolished following PRKACB elevation (Figure 6B-E). Moreover, miR-206 elevation increased Bax and c-caspase-3 levels and decreased Bcl-2 level in THP-1 and HL-60 cells, while PRKACB overexpression abrogated the effects (Figure 6F and G). We also found that miR-206 overexpression inhibited cell cycle process in THP-1 and HL-60 cells, while this effect was abolished by increasing PRKACB (Figure 6H and I). Furthermore, the migration and invasion of THP-1 and HL-60 cells were suppressed by miR-206 overexpression, while PRKACB upregulation reversed the impacts (Figure 6J and K). All these outcomes illustrated that miR-206 overexpression relieved the malignant phenotypes of AML cells by interacting with PRKACB.
Figure 6.
MiR-206 overexpression suppressed AML cell progression through targeting PRKACB. MiR-NC, miR-206, miR-206+vector or miR-206+PRKACB was transfected into THP-1 and HL-60 cells. (A) PRKACB protein level in THP-1 and HL-60 cells was measured by Western blot assay. (B–E) The proliferation, colony formation and apoptosis of THP-1 and HL-60 cells were assessed through CCK-8 assay, colony formation assay and flow cytometry analysis, respectively. (F and G) The protein levels of Bax, Bcl-2 and c-caspase-3 in THP-1 and HL-60 cells were measured by Western blot assay. (H and I) The cell cycle process was analyzed by flow cytometry analysis. (J and K) The migration and invasion of THP-1 and HL-60 cells were evaluated by transwell assay. *P<0.05.
Discussion
CircRNAs are differentially expressed in AML patients and act as potential diagnostic markers in AML.17–19 Nevertheless, the studies on the biological functions and molecular mechanisms of circRNAs in AML are still insufficient. Our work focused on the effects and related mechanisms of circRAD18 in AML progression.
Previous studies have reported that circRAD18 plays a tumor-promotional role in the carcinogenesis of human cancers. For instance, Zang et al uncovered that circRAD18 level was enhanced in breast cancer (BC) cells and its deletion hampered BC cell viability, colony formation, glycolysis and metastasis and induced apoptosis by altering miR-613/HK2 axis.20 Zou et al declared that circRAD18 knockdown hampered triple-negative breast cancer (TNBC) cell growth and migration and accelerated apoptosis by increasing miR-208a/miR-3164 and decreasing IGF1/FGF2.21 Herein, we, for the first time, investigated the roles of circRAD18 in AML. Compared to corresponding controls, circRAD18 was conspicuously upregulated in AML patients’ blood specimens and ALM cell lines. CircRAD18 silencing evidently hampered cell proliferation, migration and invasion in AML cells. CircRAD18 deficiency also facilitated the apoptosis of AML cells, along with the elevation of Bax and c-caspase-3 and the reduction of Bcl-2. All these findings indicated that circRAD18 aggravated the malignancy of AML cells.
Subsequently, the underlying mechanism of circRAD18 in AML was further explored. As a result, circRAD18 was able to positively modulate PRKACB expression through functioning as the sponge of miR-206 in AML cells. Li et al suggested that miR-206 participated in regulating the tumorigenesis of gastric cancer through the circ_0056618/miR-206/CXCR4 axis.22 Zheng et al manifested that circ_0056618 could aggravate colorectal cancer cell growth, migration and angiogenesis by modulation of miR-206/CXCR4 or miR-206/VEGF-A.23 Moreover, Li et al verified that miR-206 level was declined in pediatric AML and restrained cell growth and cell cycle process by binding to CyclinD1, indicating that miR-206 played an essential role in AML.14 In addition, PRKACB has been claimed to act as a vital mediator in several types of cancer, such as papillary thyroid cancer,24 non-small cell lung cancer,25 BC,26 as well as AML.16 Thus, we wondered whether circRAD18 knockdown mitigated the malignancy of AML cells through regulating miR-206 and PRKACB. Our results indicated that miR-206 suppression abrogated the impacts of circRAD18 knockdown on the malignant behaviors of AML cells. Furthermore, we found that miR-206 overexpression repressed AML cell progression by targeting PRKACB.
In conclusion, circRAD18 was abnormally expressed in AML and aggravated the malignant cell phenotypes of AML cells by regulating miR-206/PRKACB axis. The findings facilitated our understandings of AML pathogenesis and might offer a new sight for the therapy of AML.
Funding Statement
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Khasawneh MK, Abdel-Wahab O. Recent discoveries in molecular characterization of acute myeloid leukemia. Curr Hematol Malig Rep. 2014;9(2):93–99. [DOI] [PubMed] [Google Scholar]
- 2.Rubnitz JE, Gibson B, Smith FO. Acute myeloid leukemia. Hematol Oncol Clin North Am. 2010;24(1):35–63. doi: 10.1016/j.hoc.2009.11.008 [DOI] [PubMed] [Google Scholar]
- 3.Webster JA, Pratz KW. Acute myeloid leukemia in the elderly: therapeutic options and choice. Leuk Lymphoma. 2018;59(2):274–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medinger M, Lengerke C, Passweg J. Novel Prognostic and Therapeutic Mutations in Acute Myeloid Leukemia. Cancer Genomics Proteomics. 2016;13(5):317–329. [PMC free article] [PubMed] [Google Scholar]
- 5.Shlush LI, Mitchell A, Heisler L, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547(7661):104–108. [DOI] [PubMed] [Google Scholar]
- 6.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–211. [DOI] [PubMed] [Google Scholar]
- 7.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- 8.Fan H, Li Y, Liu C, Liu Y, Bai J, Circular LW. RNA-100290 promotes cell proliferation and inhibits apoptosis in acute myeloid leukemia cells via sponging miR-203. Biochem Biophys Res Commun. 2018;507(1–4):178–184. [DOI] [PubMed] [Google Scholar]
- 9.Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis. 2019;75:41–47. [DOI] [PubMed] [Google Scholar]
- 10.Shang J, Chen W-M, Wang Z-H, Wei T-N, Chen -Z-Z, Wu W-B. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p–XIAP axis. Exp Hematol. 2019;70:e43. doi: 10.1016/j.exphem.2018.10.011 [DOI] [PubMed] [Google Scholar]
- 11.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 12.Qin Y, Wang Y, Liu D. miR-4792 Inhibits Acute Myeloid Leukemia Cell Proliferation and Invasion and Promotes Cell Apoptosis by Targeting Kindlin-3. Oncol Res. 2020;28(4):357–369. doi: 10.3727/096504020X15844389264424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L, Ren W, Chen K. MiR-34a Promotes Apoptosis and Inhibits Autophagy by Targeting HMGB1 in Acute Myeloid Leukemia Cells. Cell Physiol Biochem. 2017;41(5):1981–1992. doi: 10.1159/000475277 [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Wu H, Qin X. MicroRNA-206 serves as a tumor suppressor in pediatric acute myeloid leukemia by targeting Cyclin D1. Pathol Res Pract. 2019;215(10):152554. doi: 10.1016/j.prp.2019.152554 [DOI] [PubMed] [Google Scholar]
- 15.Skalhegg BS, Tasken K. Specificity in the cAMP/PKA signaling pathway. Differential expression, regulation, and subcellular localization of subunits of PKA. Front Biosci. 2000;5:D678–693. [DOI] [PubMed] [Google Scholar]
- 16.Wu D-M, Wen X, Han X-R, et al. Role of Circular RNA DLEU2 in Human Acute Myeloid Leukemia. Mol Cell Biol. 2018;38(20):e00259–18. doi: 10.1128/MCB.00259-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Zhong C, Jiao J, et al. Characterization of hsa_circ_0004277 as a New Biomarker for Acute Myeloid Leukemia via Circular RNA Profile and Bioinformatics Analysis. Int J Mol Sci. 2017;18(3):597. doi: 10.3390/ijms18030597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H, Liu T, Liu J, et al. Circ-ANAPC7 is Upregulated in Acute Myeloid Leukemia and Appears to Target the MiR-181 Family. Cell Physiol Biochem. 2018;47(5):1998–2007. doi: 10.1159/000491468 [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Zhou L-Y, Tang X, et al. Circ-Foxo3 is positively associated with the Foxo3 gene and leads to better prognosis of acute myeloid leukemia patients. BMC Cancer. 2019;19(1):930. doi: 10.1186/s12885-019-5967-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zang H, Li Y, Zhang X, Huang G. <p>Knockdown of circRAD18 Mitigates Breast Cancer Progression through the Regulation of miR-613/HK2 Axis. Cancer Manag Res. 2020;12:3661–3672. doi: 10.2147/CMAR.S243300 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Zou Y, Zheng S, Xiao W, et al. circRAD18 sponges miR-208a/3164 to promote triple-negative breast cancer progression through regulating IGF1 and FGF2 expression. Carcinogenesis. 2019;40(12):1469–1479. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Yao G, Feng B, Lu X, Fan Y. Circ_0056618 and CXCR4 act as competing endogenous in gastric cancer by regulating miR-206. J Cell Biochem. 2018;119(11):9543–9551. [DOI] [PubMed] [Google Scholar]
- 23.Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH, Han SG. Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-A in colorectal cancer. Eur Rev Med Pharmacol Sci. 2020;24(8):4190–4202. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Wang B, Zhou H, Zhang X, Qian X, Cui J. MicroRNA-384 Inhibits the Progression of Papillary Thyroid Cancer by Targeting PRKACB. Biomed Res Int. 2020;2020:4983420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Gao Y, Tian Y, Tian DL. PRKACB is downregulated in non-small cell lung cancer and exogenous PRKACB inhibits proliferation and invasion of LTEP-A2 cells. Oncol Lett. 2013;5(6):1803–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sigloch FC, Burk UC, Biniossek ML, Brabletz T, Schilling O. miR-200c dampens cancer cell migration via regulation of protein kinase A subunits. Oncotarget. 2015;6(27):23874–23889. [DOI] [PMC free article] [PubMed] [Google Scholar]