Abstract
Mammalian male meiosis requires homologous recombination between the X and Y chromosomes. In humans, such recombination occurs exclusively in the short arm pseudoautosomal region (PAR1) of 2.699 Mb in size. Although it is known that complete deletion of PAR1 causes spermatogenic arrest, no studies have addressed to what extent male meiosis tolerates PAR1 size reduction. Here, we report two families in which PAR1 partial deletions were transmitted from fathers to their offspring. Cytogenetic analyses revealed that a ∼400-kb segment at the centromeric end of PAR1, which accounts for only 14.8% of normal PAR1 and 0.26% and 0.68% of the X and Y chromosomes, respectively, is sufficient to mediate sex chromosomal recombination during spermatogenesis. These results highlight the extreme recombinogenic activity of human PAR1. Our data, in conjunction with previous findings from animal studies, indicate that the minimal size requirement of mammalian PARs to maintain male fertility is fairly small.
Keywords: crossover, homologous recombination, pseudoautosomal region, sex chromosome, SHOX
Significance
Mammalian X and Y chromosomes consist of different genetic components, except for small homologous chromosomal elements designated as pseudoautosomal regions (PARs). PARs are essential for spermatogenesis, because they serve as the sole platform for X–Y crossovers during meiosis. Here, we show that loss of 85% of human PAR does not necessarily result in male infertility, based on the observation of two men who carried chromosomal deletions within PAR. Our findings suggest that mammalian PARs have a potential to acquire gross structural changes during evolution without affecting male reproductive fitness.
Introduction
Homologous recombination between the X and Y chromosomes is indispensable for mammalian male meiosis (Koller and Darlington 1934; Simmler et al. 1985; Raudsepp and Chowdhary 2015). Such recombination occurs exclusively in the pseudoautosomal regions (PARs), in which the two sex chromosomes share homologous sequences (Simmler et al. 1985; Skaletsky et al. 2003; Ross et al. 2005). Although the human genome contains two PARs, only PAR1 spanning 2.699 Mb at the end of Xp/Yp (GRCh37/hg19) serves as the platform for sex chromosomal recombination (Charchar et al. 2003; Raudsepp and Chowdhary 2015).
The high frequency of meiotic recombination in PAR1 gives rise to various deletions and duplications (Bussell et al. 2006; Otto et al. 2011; Shima et al. 2016). Of note, Gabriel-Robez et al. (1990) and Mohandas et al. (1992) identified Xp terminal deletions encompassing the entire PAR1 in two men with spermatogenic arrest. These X chromosomal deletions contained no known spermatogenic genes, indicating that infertility of these two men results from the loss of PAR1. Consistent with this, animal studies revealed that the structural or sequence divergences of PARs between two murine subspecies affect the success rate of X–Y crossover (Dumont 2017). To date, however, no study has addressed to what extent human spermatogenesis tolerates PAR1 size reduction.
Results and Discussion
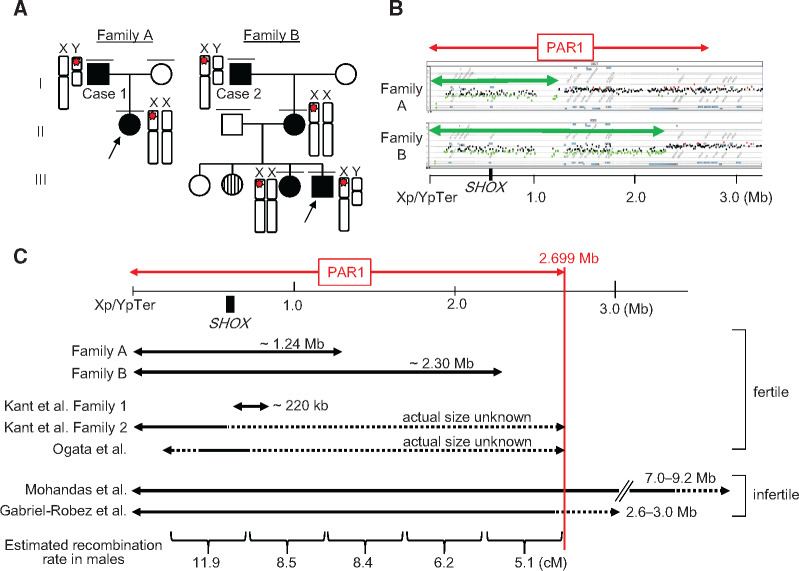
We identified two families (families A and B) with large terminal deletions of PAR1 (fig. 1A). These families were found through molecular analyses of SHOX, an osteogenic gene located in PAR1 at a position ∼600 kb from the end of Xp/Yp (Rao et al. 1997; the UCSC Genome Browser, http://genome.ucsc.edu/ [GRCh37/hg19]). The probands of these families (II-1 of family A and III-4 of family B) presented with mesomelic short stature and skeletal deformity indicative of SHOX haploinsufficiency (Belin et al. 1998). Multiplex ligation-dependent probe amplification showed decreased copy number of all SHOX exons and their flanking regions in both individuals. The same copy-number losses were also identified in their relatives (fig. 1A). Microarray-based comparative genomic hybridization (CGH) detected PAR1 terminal deletions of ∼1.24 Mb (maximum interval, chrXY:1–1,256,608; minimum interval, chrXY:1–1,235,344) in family A and of ∼2.30 Mb (maximum interval, chrXY:1–2,309,402; minimum interval, chrXY:1–2,297,925) in family B (fig. 1B). Fluorescent in situ hybridization revealed that the deletion in family A was located on the X chromosome of the proband and on the Y chromosome of her father (A-I-1) (supplementary fig. 1, Supplementary Material online), whereas the deletion in family B resided on the X chromosome of the proband, his elder sister (B-II-3), mother (B-II-2), and maternal grandfather (B-I-1) (fig. 1A). All deletion-positive individuals exhibited skeletal features indicative of SHOX haploinsufficiency (Belin et al. 1998), but no other congenital anomalies. Allegedly, another individual of family B (III-2) also had short stature, although genomic DNA samples and detailed clinical information of this individual were unavailable.
Fig. 1.
Molecular findings of families A and B. (A) The pedigrees of families A and B. Black boxes and circles indicate individuals with mesomelic short stature and/or skeletal deformities, whereas the white box and circles depict unaffected family members. The striped circle indicates an individual with short stature, whose genomic DNA sample and detailed clinical information were unavailable. Red stars on the X and Y chromosomes indicate SHOX-containing deletions in the pseudoautosomal region 1 (PAR1). (B) Representative results of microarray-based comparative genomic hybridization for the probands of families A and B. PAR1 is indicated by the red arrow. Black, green, and red dots denote signals indicative of the normal, decreased (<−0.8) and increased (>+0.4) copy numbers, respectively. Green arrows indicate the deleted regions in families A and B. Genomic positions refer to the human genome database (GRCh37/hg19). The position of SHOX is indicated by the black box. (C) Schematic representation of PAR1. The deleted regions in families A and B, together with those in the three previously reported cases with normal fertility (Ogata et al. 2002; Kant et al. 2011) and two cases with spermatogenic failure (Gabriel-Robez et al. 1990; Mohandas et al. 1992), are shown as black arrows. The broken lines depict dosage-unknown regions. The position of SHOX is indicated by the black box. The panel at the bottom shows the recombination rates of normal males (in cM) calculated by Hinch et al. (2014).
The most striking finding from these families was that two adult men, that is, the proband’s father in family A (A-I-1; hereafter referred to as case 1) and the proband’s grandfather in family B (B-I-1; case 2), were fertile and transmitted their PAR1 deletions to daughters (fig. 1A). Cases 1 and 2 retained only ∼1.44-Mb and ∼400-kb segments of PAR1, respectively (fig. 1B). In case 1, homologous recombination between the X and Y chromosomes must have occurred within the ∼1.44-Mb segment in the most centromeric part of PAR1, because during meiosis, the SHOX-containing deletion was translocated from the Y chromosome to the X chromosome (fig. 1A and supplementary fig. 1, Supplementary Material online). The normal female phenotype of the daughter of case 1 (A-II-1) provides evidence that the X–Y crossover in case 1 occurred telomeric to SRY, the sex-determining gene located in the Y-specific region only ∼5 kb from the PAR1 boundary (the UCSC Genome Browser). It is known that male meiotic homologous recombination occurs predominantly in the telomeric part of PAR1, with the hottest hotspot being at the SHOX locus (May et al. 2002; Flaquer et al. 2009; Hinch et al. 2014). Moreover, in several species, telomeric regions are predicted to play an important role in the meiotic chromosomal pairing (McKee 2004). However, the results of case 1 indicate that loss of the telomeric half of PAR1 does not necessarily lead to spermatogenic failure. Consistent with this, previous studies have identified three fertile men with PAR1 partial deletions, in whom meiotic homologous recombination occurred between SHOX and the centromeric end of PAR1 (fig. 1C) (Ogata et al. 2002; Kant et al. 2011). In case 2, furthermore, the site of meiotic recombination was restricted to a ∼400-kb region at the most centromeric part of PAR1. The SHOX-containing deletion in this individual resided on the X chromosome throughout meiosis, indicating that the recombination occurred between the Y chromosome and the nontransmitted sister chromatid of the X chromosome. We cannot completely exclude the possibility that the sex chromosomal recombination in case 2 occurred outside PAR1. For example, PAR2 on Xq/Yq also has the potential to mediate male meiotic recombination (Ciccodicola et al. 2000; Raudsepp and Chowdhary 2015). However, this probability is low, because 1) complete loss of X chromosomal PAR1 was observed in two men with spermatogenetic arrest (Gabriel-Robez et al. 1990; Mohandas et al. 1992), 2) the estimated genetic size of PAR1 in normal males is ∼50 cM (Flaquer et al. 2009; Evers et al. 2011; Otto et al. 2011), suggesting that virtually all spermatocytes leading to live births undergo homologous recombination in this region, and 3) in midpachytene spermatocytes, chiasmata were observed exclusively in PAR1 (Sarbajna et al. 2012). Of note, the ∼400-kb PAR1 segment retained in case 2 accounts for only 14.8% of normal PAR1 and corresponds to 0.26% and 0.68% of the length of the X and Y chromosomes, respectively (the UCSC Genome Browser). The estimated genetic size of this segment in normal males is <5 cM (fig. 1C; Hinch et al. 2014), indicating that during normal spermatogenesis, this short segment is rarely involved in sex chromosomal recombination. Nevertheless, in case 2, this segment is likely to have hosted homologous recombination in most spermatocytes, because animal studies have shown that X–Y pairing in 50% of germ cells, but not in 30% of cells, permits sperm production (Faisal and Kauppi 2016).
The aforementioned results indicate that the minimal size requirement of human PAR1 to maintain spermatogenesis is fairly small. In this regard, it is noteworthy that the size of PARs is highly variable among mammalian species (Graves et al. 1998; Raudsepp and Chowdhary 2015). PARs are believed to be under the constant evolutionary pressure to shrink, yet such PAR attrition can be counteracted by the insertion of DNA fragments through chromosomal translocation (Graves et al. 1998; Mensah et al. 2014). Indeed, recent studies have shown that a small percentage of healthy men carry a ∼110-kb insertion polymorphism in PAR1 that expands the size of the recombination platform to some extent (Mensah et al. 2014; Poriswanish et al. 2018). Thus, human PAR1 is still evolving. The present study provides evidence that human PAR1 is highly tolerant to size reduction. These data are consistent with the prior observation that the size of murine PARs is only 700 kb or less (Perry et al. 2001; Raudsepp and Chowdhary 2015). The high recombinogenic activity of mammalian PARs is likely to reflect their long chromosome axes, which leads to the frequent occurrence of double-strand DNA breaks (Kauppi et al. 2011; Acquaviva et al. 2020).
In summary, the results indicate that a ∼400-kb segment at the centromeric end of PAR1 is sufficient to produce homologous recombination during human spermatogenesis. This study highlights the extreme recombinogenic activity of PARs in the maintenance of male fertility.
Materials and Methods
This study was approved by the Institutional Review Board Committee and performed after obtaining written informed consent to participate. The probands of families A and B were identified through molecular analyses of SHOX for patients with short stature and/or skeletal deformity. The other members of these families were ascertained by familial studies of the probands.
Genomic DNA samples were obtained from peripheral leukocytes of the participants. Multiplex ligation-dependent probe amplification for SHOX and its flanking regions was performed using the commercially available kit (SALSA P018-G1, MRC-Holland, the Netherlands). CGH was performed using a human catalog array (4× 180 k format; Agilent Technologies, California, USA) according to the manufacturer’s instructions. The results of CGH were assessed using the Genomic Workbench (version 7.0, Agilent Technologies) with the default settings of the aberration detection algorithm.
Metaphase spreads were generated from peripheral leukocytes. Fluorescent in situ hybridization was carried out using a standard procedure (LSI Medience, Tokyo, Japan). We utilized a ∼14-kb probe containing SHOX exons 3–5 and a part of exon 6a and a probe for the Xq/Yq telomere region (TelVysion VYS33-260023; Abbott Laboratories, Illinois, USA). The cells were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (ThermoFisher Scientific, Massachusetts, USA).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (17H06428), the Japan Agency for Medical Research and Development (20ek0109464h0001), the National Center for Child Health and Development (2019A-1), and the Takeda Science Foundation.
Literature Cited
- Acquaviva L, et al. 2020. Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature 582(7812):426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin V, et al. 1998. SHOX mutations in dyschondrosteosis (Leri–Weill syndrome). Nat Genet. 19(1):67–69. [DOI] [PubMed] [Google Scholar]
- Bussell JJ, Pearson NM, Kanda R, Filatov DA, Lahn BT. 2006. Human polymorphism and human–chimpanzee divergence in pseudoautosomal region correlate with local recombination rate. Gene 368:94–100. [DOI] [PubMed] [Google Scholar]
- Charchar FJ, et al. 2003. Complex events in the evolution of the human pseudoautosomal region 2 (PAR2). Genome Res. 13(2):281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccodicola A, et al. 2000. Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Hum Mol Genet. 9(3):395–401. [DOI] [PubMed] [Google Scholar]
- Dumont BL. 2017. Meiotic consequences of genetic divergence across the murine pseudoautosomal region. Genetics 205(3):1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers C, et al. 2011. Pseudoautosomal inheritance of Léri–Weill syndrome: what does it mean? Clin Genet. 79(5):489–494. [DOI] [PubMed] [Google Scholar]
- Faisal I, Kauppi L. 2016. Sex chromosome recombination failure, apoptosis, and fertility in male mice. Chromosoma 125(2):227–235. [DOI] [PubMed] [Google Scholar]
- Flaquer A, Fischer C, Wienker TF. 2009. A new sex-specific genetic map of the human pseudoautosomal regions (PAR1 and PAR2). Hum Hered. 68(3):192–200. [DOI] [PubMed] [Google Scholar]
- Gabriel-Robez O, et al. 1990. Deletion of the pseudoautosomal region and lack of sex-chromosome pairing at pachytene in two infertile men carrying an X; Y translocation. Cytogenet Cell Genet. 54(1–2):38–42. [DOI] [PubMed] [Google Scholar]
- Graves JA, Wakefield MJ, Toder R. 1998. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum Mol Genet. 7(13):1991–1996. [DOI] [PubMed] [Google Scholar]
- Hinch AG, Altemose N, Noor N, Donnelly P, Myers SR. 2014. Recombination in the human pseudoautosomal region PAR1. PLoS Genet. 10(7):e1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant SG, et al. 2011. The jumping SHOX gene-crossover in the pseudoautosomal region resulting in unusual inheritance of Leri–Weill dyschondrosteosis. J Clin Endocrinol Metab. 96(2):E356–E359. [DOI] [PubMed] [Google Scholar]
- Kauppi L, et al. 2011. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331(6019):916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller PC, Darlington CD. 1934. The genetical and mechanical properties of the sex-chromosomes. J Genet. 29(2):159–173. [Google Scholar]
- May CA, Shone AC, Kalaydjieva L, Sajantila A, Jeffreys AJ. 2002. Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nat Genet. 31(3):272–275. [DOI] [PubMed] [Google Scholar]
- McKee BD. 2004. Homologous pairing and chromosome dynamics in meiosis and mitosis. Biochim Biophys Acta 1677(1–3):165–180. [DOI] [PubMed] [Google Scholar]
- Mensah MA, et al. 2014. Pseudoautosomal region 1 length polymorphism in the human population. PLoS Genet. 10(11):e1004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas TK, et al. 1992. Role of the pseudoautosomal region in sex-chromosome pairing during male meiosis: meiotic studies in a man with a deletion of distal Xp. Am J Hum Genet. 51(3):526–533. [PMC free article] [PubMed] [Google Scholar]
- Ogata T, et al. 2002. SHOX nullizygosity and haploinsufficiency in a Japanese family: implication for the development of Turner skeletal features. J Clin Endocrinol Metab. 87(3):1390–1394. [DOI] [PubMed] [Google Scholar]
- Otto SP, et al. 2011. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 27(9):358–367. [DOI] [PubMed] [Google Scholar]
- Perry J, Palmer S, Gabriel A, Ashworth A. 2001. A short pseudoautosomal region in laboratory mice. Genome Res. 11(11):1826–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poriswanish N, et al. 2018. Recombination hotspots in an extended human pseudoautosomal domain predicted from double-strand break maps and characterized by sperm-based crossover analysis. PLoS Genet. 14(10):e1007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao E, et al. 1997. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet. 16(1):54–63. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP. 2015. The eutherian pseudoautosomal region. Cytogenet Genome Res. 147(2–3):81–94. [DOI] [PubMed] [Google Scholar]
- Ross MT, et al. 2005. The DNA sequence of the human X chromosome. Nature 434(7031):325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbajna S, et al. 2012. A major recombination hotspot in the XqYq pseudoautosomal region gives new insight into processing of human gene conversion events. Hum Mol Genet. 21(9):2029–2038. [DOI] [PubMed] [Google Scholar]
- Shima H, et al. 2016. Systematic molecular analyses of SHOX in Japanese patients with idiopathic short stature and Leri–Weill dyschondrosteosis. J Hum Genet. 61(7):585–591. [DOI] [PubMed] [Google Scholar]
- Simmler MC, et al. 1985. Pseudoautosomal DNA sequences in the pairing region of the human sex chromosomes. Nature 317(6039):692–697. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423(6942):825–837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.