Abstract
Alterations in brain–gut–microbiome (BGM) interactions have been implicated in the pathogenesis of irritable bowel syndrome (IBS). Here, we apply a systems biology approach, leveraging neuroimaging and fecal metabolite data, to characterize BGM interactions that are driving IBS pathophysiology. Fecal samples and resting state fMRI images were obtained from 138 female subjects (99 IBS, 39 healthy controls (HCs)). Partial least-squares discriminant analysis (PLS-DA) was conducted to explore group differences, and partial correlation analysis explored significantly changed metabolites and neuroimaging data. All correlational tests were performed controlling for age, body mass index, and diet; results are reported after FDR correction, with q < 0.05 as significant. Compared to HCs, IBS showed increased connectivity of the putamen with regions of the default mode and somatosensory networks. Metabolite pathways involved in nucleic acid and amino acid metabolism differentiated the two groups. Only a subset of metabolites, primarily amino acids, were associated with IBS-specific brain changes, including tryptophan, glutamate, and histidine. Histidine was the only metabolite positively associated with both IBS-specific alterations in brain connectivity. Our findings suggest a role for several amino acid metabolites in modulating brain function in IBS. These metabolites may alter brain connectivity directly, by crossing the blood–brain-barrier, or indirectly through peripheral mechanisms. This is the first study to integrate both neuroimaging and fecal metabolite data supporting the BGM model of IBS, building the foundation for future mechanistic studies on the influence of gut microbial metabolites on brain function in IBS.
Subject terms: Physiology, Pathogenesis, Neuroscience
Introduction
Alterations in brain–gut–microbiome (BGM) interactions have been implicated in the pathogenesis of irritable bowel syndrome (IBS)1. However, despite a growing body of preclinical and clinical studies, the precise mechanisms by which these interactions contribute to IBS symptom generation remains incompletely understood.
Neuroimaging studies have previously demonstrated structural and functional differences between IBS patients and healthy controls (HCs), especially in the default mode (DMN), somatosensory, and salience networks2. One of the most consistent findings in IBS and other disorders of visceral hypersensitivity and chronic pain has been alterations in the structure and function of key regions of the somatosensory network, including the basal ganglia (composed of the caudate, putamen, and globus pallidus)2. The basal ganglia, due in part to the structure’s many cortical and thalamic afferent and cortical efferent connections, is responsible for central processing and modulation of both visceral and somatic nociception3,4. As females tend to experience more severe and frequent IBS symptoms than males, it is unsurprising that many of the described brain network alterations are sex-specific2,5.
We have previously provided evidence supporting a role for spore forming gut bacteria of the order Clostridiales in contributing to alterations in evoked visceral hypersensitivity through microbial modulation of both cortical and subcortical brain regions, including the basal ganglia6. Although this work and many others reveal insights into the complex relationship between gut microbes and IBS, there remains insufficient granularity for robust mechanistic studies to take place. Microbes may modulate BGM interactions by directly activating vagal afferents or by stimulating dendritic cells, thereby triggering an immune-mediated signaling cascade to the central nervous system (CNS) without the help of any small molecule intermediates7.
However, it is perhaps more likely that gut microbiota-derived or microbiota-modified metabolites play a role in driving clinically meaningful changes in the BGM axis8. Tryptophan and related metabolites have been the most extensively studied within this context9, with previously described alterations in levels of blood tryptophan10 and immediate downstream products11,12 in patients with IBS, compared with HCs. Recent evidence also suggests that histamine, derived from the amino acid histidine, may play an important role in IBS pathophysiology and symptomatology13. The H1R, H2R, and H4R gut histamine receptors mediate sensorineural signaling14, immune activation15, and nociception16, respectively—representing key avenues for gut–brain communication17. Histamine may also act centrally, given that the basal ganglia is densely populated by histamine receptors, with a large body of evidence to suggest that histamine can directly influence basal ganglia output18.
In this cross-sectional study, we integrate fecal metabolite and neuroimaging data to explore the relationship between BGM alterations in IBS females. Although results from cross-sectional studies do not allows us to make inferences about causality, we hypothesize that amino acid metabolites are playing a key role in driving some of the neuroimaging changes in IBS. These results will allow for novel hypothesis generation and will serve as the foundation for future, mechanistic studies.
Methods
Subjects
138 right-handed adult premenopausal females were recruited (99 female IBS patients and 39 female HCs) provided a stool sample and underwent multimodal brain-imaging studies at UCLA. All MRI scans were taken during the follicular phase of the menstrual cycle, with stool collections typically occurring within a week of the scan. Subjects were all premenopausal as determined by medical history. IBS subjects met Rome III symptom criteria for IBS19. A gastroenterologist or nurse practitioner with expertise in IBS obtained a medical history and physical exam to confirm the IBS diagnosis. IBS patients with any bowel habit were included.
Exclusionary criteria for all subjects included (1) serious medical conditions or were taking medications which could compromise interpretation of the brain imaging; (2) ongoing major psychiatric diagnoses or use of psychotropic medications in the past 6 months (subjects were not excluded for lifetime incidence of psychiatric disorders or for intake of low-dose tricyclic antidepressants for non-psychiatric indications); (3) use of antibiotics in the past 3 months, selective serotonin reuptake inhibitors, opioids; and (4) excessive physical exercise (e.g., marathon runners).
Ethics and statement of informed consent
All procedures complied with the principles of the Declaration of Helsinki. Informed consent was obtained from all participants. This study was approved by the Institutional Review Board (12-001802, 16-000187, 15-001591) at the University of California, Los Angeles.
Diet questionnaire
Diet was assessed using through self-reported questionnaire, where participants were asked to select which diet is consumed on a regular basis. Options included: Standard American (characterized by high consumption of processed, frozen, and packaged foods, pasta and breads, and red meat; vegetables and fruits are not consumed in large quantities), Modified American (high consumption of whole grains including some processed, frozen, and packaged foods; red meat is consumed in limited quantities; vegetables and fruit are consumed in moderate to large quantities), Mediterranean (high consumption of fruits, vegetables, beans, nuts, and seeds; olive oil is the key monounsaturated fat source; dairy products, fish, and poultry are consumed in low to moderate amounts and little red meat is eaten), and all other diets that do not fit into the above categories.
Fecal metabolomics
Fecal samples were stored at −80 °C and shipped to Metabolon (Durham, NC) for processing and analysis as a single batch on their global metabolomics and bioinformatics platform using ultrahigh performance liquid chromatography and tandem mass spectrometry20. Raw data was curated by mass spectrometry using specialized software as previously described20. The number of missing data was low (<3%); missing values of raw data were filled up by median value, and ineffective peaks were removed through the interquartile range denoising method. In addition, the internal standard normalization method was employed in the data analysis. The dataset for the multiple classification analysis was compiled from the metabolite profiling results and a 3D matrix involving metabolite numbers, sample names, and normalized peak intensities were fed into the MetaboAnlyst web software 3.0 (http://www.metaboanalyst.ca)21.
Resting state brain connectivity
Magnetic resonance imaging acquisition
All patients underwent an imaging session in a 3 T Siemens Allegra MRI Scanner (Siemens, Erlangen, Germany) for a high-resolution T1 structural scan, and a resting-state functional scan. Whole-brain structural and functional (resting-state) imaging data was acquired using the following parameters: acquisition parameters for high-resolution T1-weighted images were as follows: echo time/repetition time (TE/TR) = 2.85/2200 ms, inversion time = 750 ms, field of view (FOV) = 256 mm, slice thickness = 1 mm, 176 slices, 256 × 240 acquisition matrix, voxel size = 1 mm3. Functional resting-state scans were acquired with eyes closed and an echo-planar sequence with the following parameters: TE/TR: 28/2000 ms, flip angle = 77°, scan duration = 10 min, FOV = 220 mm, slices = 300, slice thickness = 4.0 mm, and slices were obtained with whole brain coverage.
MRI processing
Structural preprocessing
Preprocessing and quality control of structural images were done using Statistical Parametric Mapping 12 (SPM12)4. All structural images were skull stripped, segmented, then normalized to the MNI T1 template. This created normalized T1 images for every subject along with segmented images (gray matter, white matter, and cerebrospinal fluid (CSF)) in normalized space.
Structural processing
Structural image parcellation
T1-image segmentation and cortical and subcortical regional parcellation were conducted using FreeSurfer v.6.05–7 following the nomenclature described in the Destrieux and Harvard-Oxford subcortical atlas8,9. The parcellation results in the labeling of 165 cortical regions, 74 bilateral cortical structures, 7 subcortical structures, the midbrain, and the cerebellum.
Resting-state fMRI preprocessing
Preprocessing and quality control of functional images was done using SPM-12 software (Welcome Department of Cognitive Neurology, London, UK) and involved slice–time correction and motion correction for the six realignment parameters. If any motion was detected above 2 mm translation or 2˚ rotation, the scan, along with the paired structural scan was discarded. In order to robustly take account the effects of motion, root mean squared (RMS) realignment estimates were calculated as robust measures of motion using publicly available MATLAB code from GitHub. The resting state images were then co-registered to their respective anatomical T1 images. Each T1 image was then segmented and normalized to a smoothed template brain in Montreal Neurological Institute (MNI) template space. Each subject’s T1 normalization parameters were then applied to that subject’s resting state image, resulting in an MNI space normalized resting state image. The resulting images were smoothed with 5 mm3 Gaussian kernel. For each subject, a sample of the volumes was inspected for any artifacts and anomalies. Levels of signal dropout were also visually inspected for excessive dropout in a priori regions of interest.
Resting-state fMRI processing
Functional network construction
Functional brain networks were constructed using the CONN 17 toolbox10 in MATLAB. Regions from the Destrieux and Harvard-Oxford Subcortical Atlases were entered as ROIs8,9. To summarize, all pre-processed, normalized images were first corrected for noise using the CompCor method to remove physiological noise without regressing out the global signal22. Confounds for the six motion parameters along with their first-order temporal derivatives, along with confounds emerging from white matter and CSF, and first-order temporal derivatives of motion, and RMS were removed using regression. The images were then band-passed filtered between 0.01/s < f < 0.1/s to reduce low-frequency and high-frequency noise that are not indicative of intrinsic brain activity. Linear measures of ROI-to-ROI functional connectivity were computed using Fisher-transformed correlations representing the association between average temporal BOLD time series signals across all voxels in a brain region. The final outputs for each subject consisted of a 165 × 165 matrix consisting of Fisher-transformed Z correlation values between each ROI.
Partial least-squares discriminant analysis (PLS-DA)
PLS-DA was conducted in R (Boston, MA) to explore the difference between groups by incorporating known classifications for the metabolites and for whole brain for the resting state connectivity. PLS-DA was ran separately for brain and metabolites. In order to prevent overfitting of the model with PLS-DA, we ran permutation tests as previously published23,24. The metabolites and brain connectivity regions with values of the first PLS-DA component of variable importance projection (VIP) in PLS-DA >1.0 were assessed by Student’s t-test controlling for age, body mass index (BMI), and diet. P-values were adjusted with the Benjamini–Hochberg false discovery rate (FDR) procedure25. FDR-corrected P values, which referred to as q values were reported. The metabolite and brain connectivity with VIPs > 1.0 and q values <0.05 was selected as significantly different between the two groups. For the metabolites the FC was also calculated to investigate the difference by comparing the mean value of the peak area obtained between the two groups. The KEGG pathway databases for Homo sapiens in the MetaboAnalyst 3.0 were used to explore metabolic impact pathways21. The pathway with a q value <0.05 and an impact value >0.05 was defined as a significant impact pathway.
Correlation analysis
The partial correlation analysis between significantly changed metabolites and resting-state connectivity controlling for age, BMI, and diet, was conducted using SPSS software version 21 (SPSS Inc., Chicago, IL, USA). The relations between metabolites and resting-state function with P value < 0.05 were considered as significantly correlated. Means and standard deviations (SD) are presented for normally distributed data. Medians and IQR are reported for non-normally distributed data.
Results
Clinical measures
In total, 138 female subjects participated in this study (99 IBS, 39 HCs). Ages ranged from 18 to 44 (IBS: median = 24, interquartile ranges [IQR] = [21, 29]); HC: median = 27, IQR = [20, 34]; p = 0.357). BMI ranged from 18.4 to 40.2 (IBS: mean = 23.5, SD = 3.94; HC: mean = 24.0, SD = 2.74; p = 0.415). With respect to bowel habit, 36 subjects reported constipation-predominant IBS, 28 reported diarrhea-predominant, 26 reported mixed, and 9 were unspecified.
For HCs, 64% of females reported a standard American diet, 5% a modified American diet, 5% a Mediteranian diet, and 26% a diet in the “other” category. For IBS, 53% of females reported a standard American diet, 12% a modified American diet, 1% a mediteranian diet, and 34% a diet in the “other” category.
Significant metabolites differences between IBS and HC females
A total of 1154 metabolites were identified by metabolomic profiling. Classifications were found in the PLS-DA (Fig. 1). A total of 125 significant differences in metabolites (VIP > 1.0 and q value < 0.05) were identified between premenopausal IBS and HCs, including 32 amino acid metabolites, 15 carbohydrate metabolites, 5 cofactors, and vitamins metabolites, 2 energy metabolites, 35 lipid metabolites, 17 nucleotide metabolites, 7 peptide metabolites, and 12 xenobiotic compounds (Table S1). In detail, compared with HC females, 43 metabolites were significantly lower in IBS, particularly lipids, such as N-palmitoyl-sphinganine (d18:0/16:0) (fold change [FC] = 0.14, q = 1.7E−09), palmitoyl-oleoyl-glycerol (16:0/18:1) (FC = 0.12, q = 1.1E−07), and oleoyl-oleoyl-glycerol (18:1/18:1) (FC = 0.19, q = 7.11167E−07), while 82 metabolites were significantly higher in IBS females, such as gamma-glutamylleucine (FC = 1.57, q = 8.44067E−11), 2-hydroxyglutarate (FC = 1.47, q = 1.2E−09), and N-acetylmethionine sulfoxide (FC = 1.73, q = 7.2E−08).
Fig. 1. Multivariate analysis in the fecal samples for human subjects.
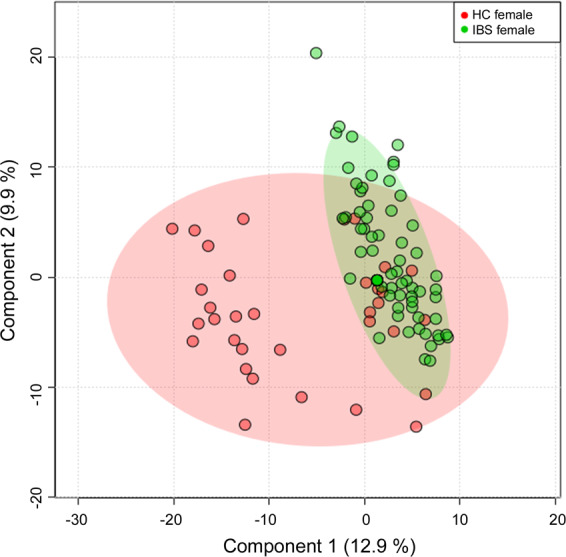
The PLS-DA score map in the fecal samples between HC and IBS females.
Significantly impacted pathways between IBS and HC females
A total of 51 pathways were identified that differed between IBS and HC females in KEGG database for H. sapiens (Supplementary information Table S2). In detail, four pathways including aminoacyl-tRNA biosynthesis (q = 0.002, impact value = 0.11268), pyrimidine metabolism (q = 0.005, impact value = 0.22819), alanine, aspartate, and glutamate metabolism (q = 0.02, impact value = 0.23362), and beta-alanine metabolism (q = 0.03, impact value = 0.07744) were identified as significant impact pathways (Table 1).
Table 1.
Significantly differentially abundant pathways between HC and IBS females.
| Pathways | Total | Expected | Hits | p value | −Log(p) | q valuea | Impact |
|---|---|---|---|---|---|---|---|
| Aminoacyl-tRNA biosynthesis | 75 | 2.6485 | 11 | 4.28E−05 | 10.058 | 0.002184 | 0.11268 |
| Pyrimidine metabolism | 60 | 2.1188 | 9 | 0.0001886 | 8.5759 | 0.004810 | 0.22819 |
| Alanine, aspartate, and glutamate metabolism | 24 | 0.84753 | 5 | 0.001221 | 6.7081 | 0.020757 | 0.23362 |
| Beta-alanine metabolism | 28 | 0.98878 | 5 | 0.002526 | 5.9812 | 0.032203 | 0.07744 |
| Glutathione metabolism | 38 | 1.3419 | 5 | 0.009777 | 4.6278 | 0.092727 | 0.04893 |
| Nitrogen metabolism | 39 | 1.3772 | 5 | 0.010909 | 4.5182 | 0.092727 | 0.00000 |
| Valine, leucine, and isoleucine biosynthesis | 27 | 0.95347 | 4 | 0.013666 | 4.2928 | 0.099567 | 0.11553 |
| Histidine metabolism | 44 | 1.5538 | 5 | 0.017928 | 4.0214 | 0.111191 | 0.14053 |
| Phenylalanine metabolism | 45 | 1.5891 | 5 | 0.019622 | 3.9311 | 0.111191 | 0.05939 |
| Glycine, serine, and threonine metabolism | 48 | 1.6951 | 5 | 0.025326 | 3.6759 | 0.129163 | 0.32425 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 36 | 1.2713 | 4 | 0.036090 | 3.3217 | 0.167326 | 0.06488 |
| Purine metabolism | 92 | 3.2489 | 7 | 0.041258 | 3.1879 | 0.175347 | 0.06907 |
ap value was calculated with FDR correction.
Significant differences in resting-state connectivity between IBS and HC females
The differences in resting-state connectivity between IBS and HCs were evaluated (Table 2). Compared with HCs, IBS had significantly higher resting-state connectivity, including left DMN (dorsal part of the posterior cingulate gyrus) to left basal ganglia (putamen) and right somatosensory network (superior frontal gyrus) to right basal ganglia (putamen), both q < 0.05.
Table 2.
Significantly changed resting state connectivity between IBS and HC females.
| Resting-state connectivity | Beta | SE | p value | t | q valuea | Change |
|---|---|---|---|---|---|---|
| Left PosDCgG to Left Pu | 0.129509684 | 0.029287569 | 2.01405E−05 | 4.422001763 | 0.049184611 | Increase in IBS |
| Right SupFG to Right Pu | 0.133066023 | 0.030322822 | 2.30643E−05 | 4.388312563 | 0.049184611 | Increase in IBS |
PosDCgG dorsal part of the posterior cingulate gyrus, SupFG superior frontal gyrus, Pu putamen.
aDifferences in resting-state connectivity between two groups were assessed by Student’s t test, controlling for age, BMI, and diet. q value was calculated with FDR correction.
Significant correlation of metabolites and resting-state connectivity between IBS and HC females
The partial correlations of significantly changed metabolites and RS connectivity between IBS and HCs, corrected for age, BMI, and diet are shown in Table 3 and Fig. 2. In particular, the differences in histidine, cysteine, glycine, glutamate, spermidine, and anserine were significantly associated with the alteration in left dorsal part of the posterior cingulate gyrus to the left putamen (p < 0.05). In addition, the changes in histidine, tryptophan, uracil, 2-deoxyuridine, thymidine, and succinate were differentially associated with the alteration in the right superior frontal gyrus to right putamen (p < 0.05).
Table 3.
Significant correlations of metabolites and RS connectivity between IBS and HC females.
| Metabolites | RS connectivity | r | p valuea |
|---|---|---|---|
| Histidine | Left PosDCgG to Left Pu | 0.21 | 0.016 |
| Cysteine | Left PosDCgG to Left Pu | 0.19 | 0.030 |
| Glycine | Left PosDCgG to Left Pu | 0.17 | 0.043 |
| Glutamate | Left PosDCgG to Left Pu | 0.18 | 0.042 |
| Spermidine | Left PosDCgG to Left Pu | 0.17 | 0.050 |
| Anserine | Left PosDCgG to Left Pu | 0.18 | 0.040 |
| Histidine | Right SupFG to Right Pu | 0.20 | 0.022 |
| Tryptophan | Right SupFG to Right Pu | 0.17 | 0.050 |
| Uracil | Right SupFG to Right Pu | 0.22 | 0.010 |
| 2-Deoxyuridine | Right SupFG to Right Pu | 0.19 | 0.027 |
| Thymidine | Right SupFG to Right Pu | 0.20 | 0.022 |
| Succinate | Right SupFG to Right Pu | −0.19 | 0.029 |
PosDCgG dorsal part of the posterior cingulate gyrus, SupFG superior frontal gyrus, Pu putamen.
aPartial correlation analysis between metabolites and RS connectivity controlling for age, BMI, and diet was performed.
Fig. 2. The partial correlation of significantly changed metabolites and resting-state connectivity controlling for age, BMI, and diet between IBS and HC females.
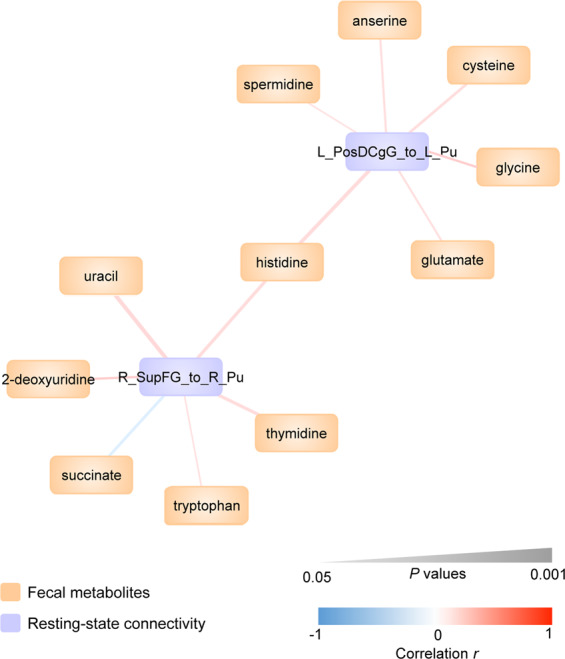
L_PosDCgG dorsal part of the left posterior cingulate gyrus, L_Pu left putamen, R_Pu right putamen, R_SupFG right superior frontal gyrus.
Discussion
In this study, we demonstrate key differences in brain connectivity and fecal metabolites between premenauposal IBS and HC subjects and these findings were integrated to create a brain–gut interactome map. This interactome likely represents the result of both brain to gut and gut to brain processes that are, at least in part, influenced by the gut microbiome. While not providing information about causality, our findings are an important foundation for future mechanistic studies.
IBS females showed higher connectivity between the putamen (basal ganglia) and a region in the DMN (dorsal part of the posterior cingulate gyrus) and somatosensory network (superior frontal gyrus). This is consistent with previous work, which demonstrated topological reorganization of the DMN26 and widespread microstructural white matter changes of the somatosensory network27 in IBS. Additionally, past studies using experimental rectal stimulation suggest that the basal ganglia play a key role in modulating IBS-related alterations in pain processing28,29.
Existing evidence from deep brain stimulation reveals that the basal ganglia are involved in IBS symptom generation30. We have also shown that the putamen is a key brain region through which the gut microbiota may impact to increase visceral hypersensitivity in IBS. In a recently published study integrating experimental rectal balloon distension, 16S rRNA gene sequencing, and functional neuroimaging, Clostridium XIVa demonstrated an association between rectal discomfort, rectal pain, and oral–anal-transit time, which was linked to altered connectivity of the putamen6. Furthermore, a previous study which investigated the relationship between the gut microbiome and brain volumes also showed an association between Firmicutes-associated Clostridia, including Clostridium XIVa, and increased gray matter volume of the putamen in IBS patients31. When viewed together, the current findings confirm an important role of the putamen in altered visceral perception in IBS.
Analysis of fecal metabolites revealed a diverse group of metabolites and metabolic pathways that successfully differentiate IBS from HCs. Notable pathways include those involved in nucleic acid (tRNA biosynthesis, pyrimidine metabolism) and amino acid (alanine, aspartate, and glutamate metabolism) metabolism. In contrast to previous human investigations32, which employed a very targeted approach and analyzed, for example, 19 discrete fecal metabolites, we interrogated 51 pathways and over 1000 metabolites, to allow for novel hypothesis generation.
In generating the brain–gut interactome map, we see that only a subset of metabolites—primarily those involving in amino acid metabolism—were associated with IBS-specific brain changes. The selected metabolites that emerged in the interactome map may represent key drivers of IBS brain–gut pathophysiology. In contrast, metabolites that differentiate IBS from HC females but that did not appear in the interactome might represent the result of secondary, brain to gut influences on the gut microenvironment that do not impact the brain or play a key role in IBS development.
The essential amino acid histidine emerged as the only metabolite that was positively associated with all IBS-specific changes in functional connectivity. Histidine may interact with the brain directly, by crossing the blood–brain-barrier33 or indirectly through conversion into histamine by either host or bacterial histidine decarboxylase enzymes34. The gut microbiome may also modulate these interactions, as one in vitro study demonstrated that histidine is extensively modified and digested by gut microbes of the small intestine35. This host–microbe cross-talk, however, extends beyond modifying gut metabolites directly related to histidine/histamine, as some bacteria such as Lactobacillus rhamnosus can modulate the immune system through direct activation of histamine receptors36. Changes in histidine/histamine may contribute to IBS symptomatology, as one randomized controlled trial comparing the effects of a diet low in fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAPs) and high in FODMAPs demonstrated reduction in IBS symptom severity scores and a greater than eightfold reduction in urine histamine levels in the low FODMAP group37. Furthermore, the association between this histamine metabolite and connectivity involving the putamen has not previously been described in IBS, but it is perhaps unsurprising, given that high expression of the histamine G-protein-coupled H1R, H2R, and H3R receptors has been identified in the basal ganglia38. When viewed together, the current results are consistent with a growing literature implicating histidine or its metabolites in IBS symptoms.
The amino acid tryptophan showed a positive association with increased connectivity between a region in the DMN (dorsal part of the posterior cingulate gyrus) to the basal ganglia (putamen). Tryptophan and its metabolites have been extensively studied within the context of the BGM axis and especially in IBS39. We have previously shown that spore forming microorganisms known to modulate gut motility can stimulate the biosynthesis and release of serotonin from intestinal enterochromaffin cells40. These microbes also demonstrate positive associations with putamen connectivity in IBS patients, but negative associations with HCs6. Our results further support the hypothesis that this interaction may be mediated by aberrant tryptophan signaling in IBS patients.
Our results also demonstrate an association between metabolites involved in pyrimidine metabolism (i.e. thymidine) and aberrant brain connectivity. Relatively few studies have explored this pathway within the context of IBS; however, one study of 23 IBS children and 22 HCs demonstrated an association between IBS pain intensity and pain frequency with thymine, the precursor to thymidine41. Data from a longitudinal human gut microbiota study suggests that enhanced pyrimidine metabolism may be indicative of gut microbiota dysbiosis42, further implicating the gut microbiome as playing a causative role in IBS. Exploring the role of this previously unstudied pathway in IBS may reveal new insights in how we understand this disease.
This study focused on fecal metabolites and functional connectivity of brain regions that have previously been implicated in IBS pathophysiology. Future work may benefit from incorporating clinical measures of IBS severity and alternative neuroimaging modalities, such as diffusion tensor imaging to also characterize differences in structural connectivity between IBS and HCs. Female patients with IBS are known to feel more fatigue, symptoms of depression, and report lower quality of life than men with IBS5, with high-quality data showing sex-differences in the neuroimaging findings of IBS2. Future studies would benefit from performing similar investigations in males with IBS. Although our discussion included the role of the gut microbiome in the interaction network between the described fecal metabolites and brain regions, we did not explicitly examine 16S rRNA gene-sequencing data in this study. Incorporating microbiota data into future analyses may provide important context for future investigations.
The directionality and causality between fecal metabolites and alterations in brain connectivity cannot be parsed, though previous work has suggested a bidirectional model for BGM communication in IBS43. In the absence of a truly valid IBS animal model and the lack of effective treatments targeting specific targets within the BGM axis which could be used as probes, and the challenges of doing studies in humans to address the bidirectional BGM interactions, cross-sectional studies such as the one we present here are a crucial step in identifying BGM interactions that may be driving IBS pathophysiology.
To our knowledge, this is the first study to integrate functional neuroimaging and fecal metabolite data to create an integrated BGM model of IBS. While supporting previous preclinical and clinical work in this patient population, the study revealed novel insights, which are essential to provide the foundation for previously unexplored avenues in understanding IBS pathophysiology, particularly with respect to how the gut microbiota participates in the complex cross-talk between gut metabolites and aberrant brain connectivity.
Supplementary information
Acknowledgements
This research was supported by grants from the National Institutes of Health including K23 DK106528 (A.G.), ULTR001881/DK041301 (UCLA CURE/CTSI Pilot and Feasibility Study; A.G.), P50 DK064539 (E.A.M.), R01 DK048351 (E.A.M.), VA Career Development Award IK2CX001717 (J.P.J.), and pilot funds provided for brain scanning by the Ahmanson-Lovelace Brain Mapping Center. These funders played no role in study design, or the collection, analysis, and interpretation of the data.
Author contributions
V.O.: study concept and design, drafting of the manuscript, critical revision of the manuscript for important intellectual content. E.A.M.: funding, study concept and design, critical revision of the manuscript for important intellectual content, study supervision. K.G.: analysis and interpretation of data, statistical analysis, drafting of the manuscript. J.S.L., B.N., L.C., E.Y.H.: critical revision of the manuscript for important intellectual content. K.T., J.P.J.: funding, critical revision of the manuscript for important intellectual content. A.G.: funding, study concept and design, analysis, and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis, technical support. All authors approved the final draft for submission.
Conflict of interest
E.A.M. is a scientific advisory board member of Danone, Axial Biotherapeutics, Viome, Amare, Mahana Therapeutics, UBiome, Pendulum, Bloom Biosciences, APC Microbiome Ireland. V.O., K.G., J.S.L., B.N., K.T., L.C., J.P.J., E.Y.H., and A.G. have nothing to disclose.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41398-020-01071-2).
References
- 1.Osadchiy V, Martin CR, Mayer EA. The gut–brain axis and the microbiome: mechanisms and clinical implications. Clin. Gastroenterol. Hepatol. 2019;17:322–332. doi: 10.1016/j.cgh.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer EA, et al. Role of brain imaging in disorders of brain–gut interaction: a Rome Working Team Report. Gut. 2019;68:1701–1715. doi: 10.1136/gutjnl-2019-318308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker RA. The basal ganglia and pain. Int. J. Neurosci. 1988;41:29–34. doi: 10.3109/00207458808985739. [DOI] [PubMed] [Google Scholar]
- 4.Chudler EH, Dong WK. The role of the basal ganglia in nociception and pain. Pain. 1995;60:3–38. doi: 10.1016/0304-3959(94)00172-B. [DOI] [PubMed] [Google Scholar]
- 5.Kim YS, Kim N. Sex-gender differences in irritable bowel syndrome. J. Neurogastroenterol. Motil. 2018;24:544–558. doi: 10.5056/jnm18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labus JS, et al. Evidence for an association of gut microbial Clostridia with brain functional connectivity and gastrointestinal sensorimotor function in patients with irritable bowel syndrome, based on tripartite network analysis. Microbiome. 2019;7:45. doi: 10.1186/s40168-019-0656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Q, et al. Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J. Neuroinflamm. 2019;16:53. doi: 10.1186/s12974-019-1434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osadchiy V, et al. Correlation of tryptophan metabolites with connectivity of extended central reward network in healthy subjects. PLoS ONE. 2018;13:e0201772. doi: 10.1371/journal.pone.0201772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gheorghe CE, et al. Focus on the essentials: tryptophan metabolism and the microbiome–gut–brain axis. Curr. Opin. Pharm. 2019;48:137–145. doi: 10.1016/j.coph.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Christmas DM, et al. Increased serum free tryptophan in patients with diarrhea-predominant irritable bowel syndrome. Nutr. Res. 2010;30:678–688. doi: 10.1016/j.nutres.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Clarke G, et al. A distinct profile of tryptophan metabolism along the kynurenine pathway downstream of toll-like receptor activation in irritable bowel syndrome. Front. Pharm. 2012;3:90. doi: 10.3389/fphar.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thijssen AY, et al. Alterations in serotonin metabolism in the irritable bowel syndrome. Aliment. Pharm. Ther. 2016;43:272–282. doi: 10.1111/apt.13459. [DOI] [PubMed] [Google Scholar]
- 13.Fabisiak A, Wlodarczyk J, Fabisiak N, Storr M, Fichna J. Targeting histamine receptors in irritable bowel syndrome: a critical appraisal. J. Neurogastroenterol. Motil. 2017;23:341–348. doi: 10.5056/jnm16203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Togias A. H1-receptors: localization and role in airway physiology and in immune functions. J. Allergy Clin. Immunol. 2003;112:S60–S68. doi: 10.1016/S0091-6749(03)01878-5. [DOI] [PubMed] [Google Scholar]
- 15.Jutel M, Watanabe T, Akdis M, Blaser K, Akdis CA. Immune regulation by histamine. Curr. Opin. Immunol. 2002;14:735–740. doi: 10.1016/S0952-7915(02)00395-3. [DOI] [PubMed] [Google Scholar]
- 16.Sanna MD, et al. Histamine H4 receptor activation alleviates neuropathic pain through differential regulation of ERK, JNK, and P38 MAPK phosphorylation. Pain. 2015;156:2492–2504. doi: 10.1097/j.pain.0000000000000319. [DOI] [PubMed] [Google Scholar]
- 17.Martin CR, Osadchiy V, Kalani A, Mayer EA. The brain–gut–microbiome axis. Cell. Mol. Gastroenterol. Hepatol. 2018;6:133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolam JP, Ellender TJ. Histamine and the striatum. Neuropharmacology. 2016;106:74–84. doi: 10.1016/j.neuropharm.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drossman DA. The functional gastrointestinal disorders and the Rome III process. Gastroenterology. 2006;130:1377–1390. doi: 10.1053/j.gastro.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 21.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitfield-Gabrieli S, Nieto-Castanon A. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2012;2:125–141. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- 23.Carrola J, et al. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J. Proteome Res. 2011;10:221–230. doi: 10.1021/pr100899x. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J. Proteome Res. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 26.Qi R, et al. Topological reorganization of the default mode network in irritable bowel syndrome. Mol. Neurobiol. 2016;53:6585–6593. doi: 10.1007/s12035-015-9558-7. [DOI] [PubMed] [Google Scholar]
- 27.Ellingson BM, et al. Diffusion tensor imaging detects microstructural reorganization in the brain associated with chronic irritable bowel syndrome. Pain. 2013;154:1528–1541. doi: 10.1016/j.pain.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tillisch K, Mayer EA, Labus JS. Quantitative meta-analysis identifies brain regions activated during rectal distension in irritable bowel syndrome. Gastroenterology. 2011;140:91–100. doi: 10.1053/j.gastro.2010.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song GH, et al. Cortical effects of anticipation and endogenous modulation of visceral pain assessed by functional brain MRI in irritable bowel syndrome patients and healthy controls. Pain. 2006;126:79–90. doi: 10.1016/j.pain.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 30.Langguth B, et al. Deep brain stimulation for obsessive compulsive disorder reduces symptoms of irritable bowel syndrome in a single patient. Clin. Gastroenterol. Hepatol. 2015;13:1371–1374 e1373. doi: 10.1016/j.cgh.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labus JS, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome. 2017;5:49. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shankar V, et al. The networks of human gut microbe–metabolite associations are different between health and irritable bowel syndrome. ISME J. 2015;9:1899–1903. doi: 10.1038/ismej.2014.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamakami J, Sakurai E, Sakurada T, Maeda K, Hikichi N. Stereoselective blood–brain barrier transport of histidine in rats. Brain Res. 1998;812:105–112. doi: 10.1016/S0006-8993(98)00958-5. [DOI] [PubMed] [Google Scholar]
- 34.Lucas PM, Claisse O, Lonvaud-Funel A. High frequency of histamine-producing bacteria in the enological environment and instability of the histidine decarboxylase production phenotype. Appl. Environ. Microbiol. 2008;74:811–817. doi: 10.1128/AEM.01496-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai ZL, et al. Metabolism of select amino acids in bacteria from the pig small intestine. Amino Acids. 2012;42:1597–1608. doi: 10.1007/s00726-011-0846-x. [DOI] [PubMed] [Google Scholar]
- 36.Frei R, et al. Histamine receptor 2 modifies dendritic cell responses to microbial ligands. J. Allergy Clin. Immunol. 2013;132:194–204. doi: 10.1016/j.jaci.2013.01.013. [DOI] [PubMed] [Google Scholar]
- 37.McIntosh K, et al. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut. 2017;66:1241–1251. doi: 10.1136/gutjnl-2015-311339. [DOI] [PubMed] [Google Scholar]
- 38.Haas H, Panula P. The role of histamine and the tuberomamillary nucleus in the nervous system. Nat. Rev. Neurosci. 2003;4:121–130. doi: 10.1038/nrn1034. [DOI] [PubMed] [Google Scholar]
- 39.Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Irritable bowel syndrome: a microbiome–gut–brain axis disorder? World J. Gastroenterol. 2014;20:14105–14125. doi: 10.3748/wjg.v20.i39.14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yano JM, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hollister EB, et al. Leveraging human microbiome features to diagnose and stratify children with irritable bowel syndrome. J. Mol. Diagn. 2019;21:449–461. doi: 10.1016/j.jmoldx.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsen PE, Dai Y. Metabolome of human gut microbiome is predictive of host dysbiosis. Gigascience. 2015;4:42. doi: 10.1186/s13742-015-0084-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayer EA, Knight R, Mazmanian SK, Cryan JF, Tillisch K. Gut microbes and the brain: paradigm shift in neuroscience. J. Neurosci. 2014;34:15490–15496. doi: 10.1523/JNEUROSCI.3299-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


