INTRODUCTION
Machine learning (ML) has the potential to transform oncology and, more broadly, medicine.1 The introduction of ML in health care has been enabled by the digitization of patient data, including the adoption of electronic medical records (EMRs). This transition provides an unprecedented opportunity to derive clinical insights from large-scale analysis of patient data.
CONTEXT
Key Objective
The objective of this review is to provide an overview of machine learning (ML) in oncology from a methods and applications perspective and to offer a framework for leveraging ML in clinical decision making.
Knowledge Generated
This review presents an overview of common ML algorithms and clinical data sources and discusses their relative merits. The data curation process is outlined, along with the technical challenges involved in working with large-scale health care data. Many aspects of oncology have benefited from these approaches, with applications ranging from early detection to treatment evaluation.
Relevance
ML presents an opportunity to transform cancer care through data-driven insights. This review provides practitioners with a practical view of the ML pipeline and its challenges.
Clinical decisions have traditionally been guided by medical guidelines and accumulated experience. ML methods add rigor to this process; algorithms can generate individualized predictions by synthesizing data across broad patient bases. On a policy level, these insights can be used to inform data-driven guidelines and risk cohorts. On a more granular level, these insights enable a personalized approach to medicine that accounts for a patient’s unique characteristics.
Despite its promise, there are numerous obstacles to the adoption of ML in medicine.2 The success of many methods depends on the availability of large-scale structured data. Variability in data capture across departments and health care systems leads to significant challenges in creating cohesive data sets for analysis. Furthermore, ML integration into clinical workflows presents its own set of challenges. Although this review focuses on the technical challenges of ML, we note that clinical decision support tools have implications on the treatment and subsequent outcomes of patients and thus must be handled with great care. There is well-placed scrutiny on ML methods in health care as a result of their potential consequences.3-5 ML models must gain the trust of clinicians through interpretability, collaboration between researchers and medical experts, and prospective validation in clinical settings.
In this article, we present an overview of ML in oncology. We introduce several classes of ML tasks and their interpretations. We discuss clinical data sources, the process of data curation, and challenges involved in creating useful data repositories for ML research. We conclude with a survey of ML applications in oncology, ranging across the continuum of care.
TOOLS AND TASKS
ML encompasses a broad range of tasks and methods.6 Supervised learning tasks have a known available outcome to predict, such as presence of a tumor, length of survival, or treatment response. Unsupervised learning identifies patterns and subgroups within data where there is no clear outcome to predict. It is often used for more exploratory analysis. Reinforcement learning is yet a third class of ML used for sequential decision making where a strategy must be learned from data; this has natural applications in determining optimal treatment regimens for patients with cancer.7,8 This review focuses on supervised and unsupervised learning settings.
Supervised Learning
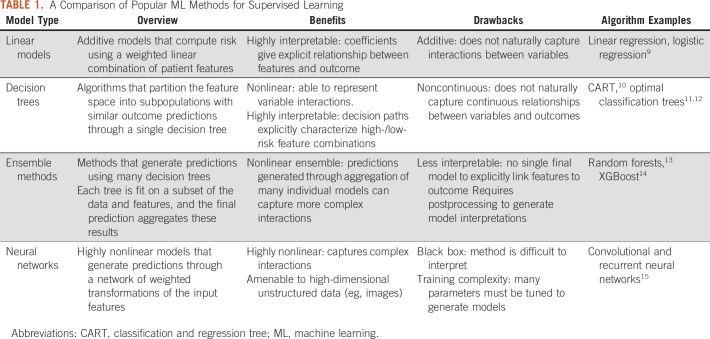
In this section, we introduce several common supervised learning approaches that appear throughout oncology applications. These algorithms take in a set of features and predict a chosen outcome, which could be either continuous (regression) or discrete (classification). Table 1 presents a summary and comparison of these methods.
TABLE 1.
A Comparison of Popular ML Methods for Supervised Learning
Linear models.
Linear models map the independent variables to the outcome of interest through a linear equation. A linear regression model finds coefficients β for each of the features. An observation’s prediction is then given by a weighted combination of these features (ie, β0 + β1x1+ …+ βpxp, where a patient’s features are given by variables x1, …, xp).
Linear regression assumes that the outcome linearly relates to the feature values and that there is an additive relationship between features. Other variants of regression models, such as logistic regression (for binary classification) and Cox regression (for survival analysis), similarly assume an additive relationship between features but involve a transformation of the linear function based on the prediction task.
Linear methods have been enduring popular choices for modeling as a result of their interpretability and straightforward methodology. Such models form the backbone of many existing risk scores and predictive models used throughout health care. However, outcomes are often inherently nonlinear in the features. For example, the effect of tumor size on cancer recurrence risk may be different for different age groups. A linear model does not naturally capture such interactions between variables. Interaction variables can be constructed to reflect nonlinearities; for example, one could create a derived feature that combines age and tumor size to model a joint effect. However, this is generally done on an ad hoc basis because it is impractical to consider all possible transformations of pairs, or larger groups, of variables. In the subsequent sections, we explore nonlinear methods that inherently account for variable interactions.
Decision tree models.
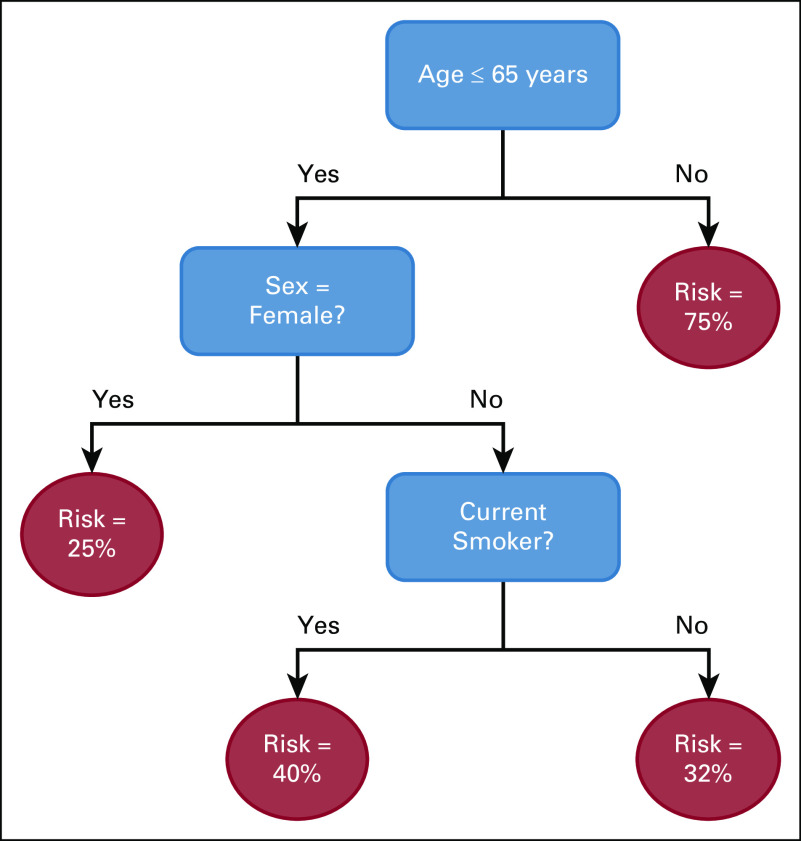
Classification and regression trees (CARTs) were initially proposed by Leo Breiman as an alternative to linear models.10 A decision tree consists of feature splits, which split observations into subgroups, and leaves, which contain the final subgroups of observations. The final tree partitions the population; every observation is assigned to a single leaf based on the feature splits. A single prediction is generated for each leaf. The prediction is a probability in the case of classification, generally calculated as the frequency of the most common outcome in the leaf, and a numerical value for continuous outcomes, generally the average value of the outcomes in the leaf. An example is shown in Figure 1.
FIG 1.
An example of a binary classification decision tree.
The tree-based structure of the model allows it to capture nonlinear relationships between features. It can identify cutoff thresholds, such as discretely differentiating risk levels between patients above or below a certain age. It can also reflect dependencies between variables, such as determining that certain comorbidities are only relevant for male patients.
The feature splits in decision trees are chosen to minimize a loss function, a measure of prediction error. In a classification task, this could be the misclassification rate (the proportion of observations incorrectly classified); in a regression task, this could be the mean absolute difference between the predicted and actual outcome values. CART forms decision trees by making greedy recursive splits. It first separates the data into two subsets based on the split that minimizes the error and then splits these subsets and continues to further levels without modifying earlier splits. A complexity parameter controls the splitting by only allowing splits that meet a certain error improvement threshold. Finally, the decision tree is pruned to remove splits that do not sufficiently improve the model error.
More recently, optimal classification trees (OCTs) were introduced as an alternative decision tree algorithm.12 OCTs use an optimization framework that considers the full structure of the tree when evaluating potential splits. A local search procedure enables the recovery of data partitions that are not identifiable from a greedy approach. The method also restricts tree depth through a complexity parameter. OCTs generally demonstrate stronger performance while maintaining the high interpretability of CART.
Ensemble models.
Ensemble methods, such as random forests13 and gradient boosted machines,16,17 extend the decision tree framework. These methods build many decision trees and generate predictions based on the resultant set of models. In random forests, each tree is trained using a random subset of features and data, resulting in varied models. The final prediction aggregates the predictions of the individual trees. Gradient boosted machines train individual trees iteratively: subsequent trees are built to place higher weight on observations that had high error in previous trees. This error-correcting approach often gives a performance advantage over random forests.
Because ensemble methods aggregate many individual trees, there is no single model that explicitly ties the input features to the final prediction. This makes the models more difficult to understand than linear models, which have coefficients, and decision trees, which have clear feature partitions. The lack of transparency poses a challenge in application spaces where interpretability is critical to adoption. Feature importance measures calculated from the models can offer more general insights,13,16 and frameworks such as SHapley Additive exPlanations18,19 have been proposed as alternative ways of extracting insights.
Neural networks.
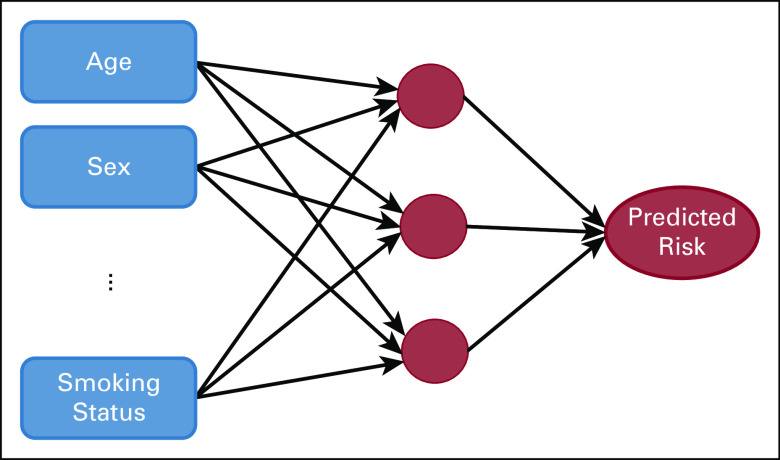
Neural networks map features to predicted outcomes through a layered network of mathematical transformations. Figure 2 displays a simple feedforward neural network with a single layer. The model maps the input features to nodes in a hidden layer through linear functions. These nodes then map to an outcome using a nonlinear activation function. These network dynamics allow neural networks to capture complex interactions between features and the outcome.
FIG 2.
An example of a feedforward neural network.
In recent years, significant advancements have been made in neural networks, including the introduction of recurrent neural networks, convolutional neural networks, and generative adversarial networks. We refer the reader to Schmidhuber15 for a comprehensive review of these innovations. These methods form the foundation of deep learning, a subfield of ML built on neural networks.
Neural networks have become especially popular as a result of their ability to synthesize raw images and free text. They are amenable to unstructured data formats and can scale to high-dimensional settings, namely cases where the number of input features greatly exceeds the number of observations. However, the modeling power and complexity come at the expense of interpretability. Neural networks have been coined as black box methods as a result of the difficulty in extracting insights. As with ensemble methods, the lack of interpretability limits its utility in certain clinical settings.
Unsupervised Learning
Although the methods described earlier predict a specific outcome, unsupervised learning is less targeted; it seeks to identify underlying structures within data. The outputs of these methods are not task specific (ie, not based on a specific predicted outcome such as survival) and provide general insight. There are several variants of unsupervised learning, which are listed in Table 2.
TABLE 2.
Overview of Unsupervised Learning Tasks
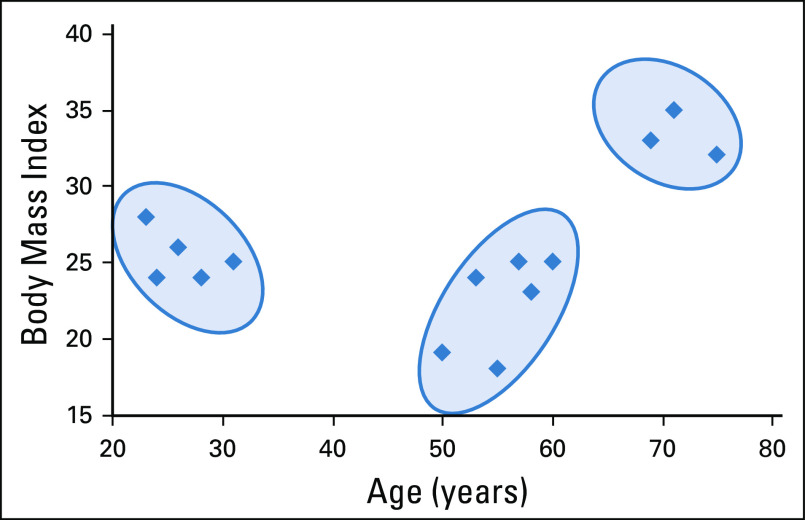
We focus on clustering in this review because it has the most natural interpretation in a health care setting. For example, clustering EMR data for patients with a certain disease type could offer insight into different patient profiles within the disease. Figure 3 illustrates a simple example of clusters where there are only two features, age and body mass index (BMI). In general, clustering algorithms partition the data into K clusters with a goal of maximizing similarity within clusters and separation between clusters. In other words, a good cluster assignment would have homogenous clusters that are highly distinct from each other. Similarity is measured by the difference between two observations; in this example, patients are more similar the closer they are in age and BMI.
FIG 3.
An example of clusters in two dimensions with K = 3.
K-means and hierarchical clustering are two of the most popular clustering methods. K-means clustering uses a heuristic to find the best assignment for a fixed K. Hierarchical clustering begins with each observation in a separate cluster and aggregates them incrementally by increasing distance. This results in a tree-like structure that allows the user to find the corresponding cluster assignment for any choice of K clusters.
Cluster interpretation poses a central challenge in unsupervised learning, particularly given the relevance of clustering in exploratory data analysis. Users often want to understand the distinguishing features of chosen clusters. For example, a given disease may have a cluster composed of older patients and another of younger patients with a high number of comorbidities. A simple approach is to look at the mean and variance of each feature for all clusters to identify which features differ most between groups, although this can be challenging with large feature spaces.26 Alternatively, one can fit a multiclass classification model such as CART or OCT to the data, where the outcome is the assigned cluster.27,28 The output tree would then give paths that are predictive of cluster membership, providing insight into the distinctive features of each group. However, this is a postprocessing step; the clustering algorithms do not inherently consider interpretation. More recent clustering methods have been proposed to construct interpretable clusters directly.29-31
CLINICAL DATA SOURCES
ML models offer a scalable and objective way of gleaning insights from data. Health care data sources vary widely, both in their structure and the information that they capture. Increased computing power, algorithmic developments, and data encoding schemes have introduced rich new data sources to leverage in ML. We focus on clinical data but note that there are additional sources that can provide valuable information, such as financial claims records or national registry data.
EMR
EMR data are composed of patient-level data recorded in patient encounters with a health care system. These data includes basic patient demographics, medical and social history, and the record of the patient’s care, including medications, diagnoses, procedures, vitals, and laboratory results. This is an extremely valuable asset for large-scale analysis because it provides structured data that are relatively standardized across patients within a health care system and somewhat consistent across systems using the same EMR vendor. In addition, for patients who stay within a single health care system, the EMR offers a comprehensive and longitudinal view of their health trajectories. The EMR data structure makes it naturally amenable to ML algorithms because many clinical features can be used directly as model inputs.
Although EMR data are appealing as a result of their structure, there are significant components that are only available in unstructured form, including free text notes and reports. For example, tumor descriptors might be indicated on a radiology report but not entered into a coded field within the EMR. In such cases, the information is inaccessible when restricted to structured data. The field of natural language processing (NLP) addresses this issue by converting raw text into discrete features that can be used as inputs into ML algorithms.32 NLP methods range from simple approaches of counting word frequencies within a note to more advanced methods such as GloVe33 that represent words as vectors based on their contextual meaning.
Genomics
The past decades have seen rapid growth in the availability of genomic data as sequencing has become increasingly cost-effective and data storage capabilities have increased.34 Genomic data have natural structure—gene expression, mutations, and copy number variation data can be used directly as features in an algorithm. However, the high dimensionality and noisy measurement of genomic data can make it difficult to extract meaningful signals. There are various techniques for synthesizing genomic data, including matrix factorization (Table 2) and feature subset selection, which are used in combination with biologic expertise.
Imaging
Imaging, such as radiology results or pathology slides, provides critical data about a patient’s condition and is particularly relevant in cancer treatment, diagnosis, and continued tumor evaluation. These data are entirely unstructured and have no natural features for an algorithm. However, image digitization has led to the emergence of the field of computer vision, which applies algorithms to images. Radiomics translates digital images into high-dimensional feature spaces by dividing the image into small segments and encoding each segment’s characteristics.35 More recently, deep learning has been applied to directly leverage raw imaging data. Neural networks are able to ingest images directly without explicit feature conversion. This has greatly expanded the utility of imaging in predictive algorithms and has emerged as one of the most popular areas of ML in health care.36
DATA CURATION CHALLENGES
ML depends on the availability of high-quality data and its translation into meaningful clinical features. EMR data present unique challenges in extraction and cleaning. EMR systems vary significantly in data capture, making it difficult to merge data sets across organizations. Even within a single EMR, there are challenges to processing patient records into a single, complete data set. These challenges can be summarized in the following broad categories: data extraction and transfer, data imputation, and clinical validation.
Data Extraction and Transfer
Data curation begins with the extraction of raw EMR data. Oncology projects also require information about the cancer itself, such as staging and treatment information. These data are often hosted across several sources including hospital EMRs, cancer-specific software, and registries. Moreover, data capture often differs between hospital departments.
For example, a researcher may be interested in extracting absolute neutrophil count (ANC). There is often not a single standardized field that captures a desired feature. Rather, there may be several laboratory entries that measture ANC, some in different units, using different equipment, or being taken in different parts of the hospital. To accurately retrieve this value, one must merge together all alternative definitions into a single unified field.
There is a fundamental trade-off between physician flexibility and data standardization. Although it is often beneficial to combine (approximately) equivalent fields from a research perspective, there are many considerations that lead physicians to pursue customized workflows. Flexibility ensures minimal loss in the physician’s ability to convey information, but it leads to difficulties in data aggregation. This challenge highlights the need for clinical ontologies that map data elements to their clinically meaningful fields. This would allow researchers or clinicians to easily filter and group together clinical features. Although coding schemes such as Logical Observation Identifier Names and Codes37 or Anatomic Therapeutic Chemical Classification Scheme38 exist for various data elements, they are often incomplete and not consistently applied throughout EMRs.
Data Imputation
Inevitably, some clinical data will be missing, even after successful extraction. It is likely that not all laboratory values are recorded at every patient visit or that patients have only a partial available medical history. There are several approaches to handling missing data. Observations with any missing values are often excluded, called complete case analysis, but this can be misleading. Some fields may be systematically missing. For example, smokers may be hesitant to report their smoking status, so excluding patients with missing values would systematically bias the population toward nonsmokers. Missing data can be easily imputed by taking the average of the columns—the mean or median in the case of continuous variables and the mode in the case of categoric variables. However, this can also lead to bias in the imputation.
As large-scale data analysis has become more prevalent, more nuanced methods to imputation have been introduced. Multiple imputation by chained equations learns from the full feature space in imputation rather than considering each variable independently.39 Optimal imputation (OptImpute) takes an optimization approach to imputation that leverages the global structure of the data.40 This has more recently been extended to a medical setting (MedImpute), which further accounts for temporal data sets in which the same patient appears in multiple instances over time.41
Clinical Validation
Once data have been extracted and organized into an initial feature space, they must be reviewed for clinical validity. Although small cohorts can be chart reviewed, ML thrives on large data sets where it is necessary to standardize validation checks in a way that is reproducible and scalable. Data cleaning and verification are primary obstacles to ML,42,43 and medical applications introduce additional domain-specific complexities. There is also a challenge in distinguishing between errant values and true exceptional cases. Although data should be cleaned to minimize value errors or inconsistencies that are attributable to data entry issues, it is simultaneously important to ensure that extreme cases are not mistakenly altered.
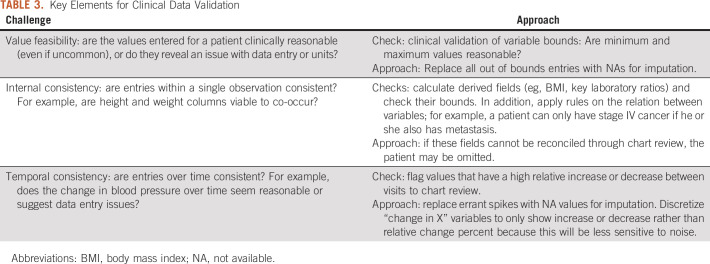
Data cleaning has been tackled algorithmically through conditional functional dependencies,44 statistical estimation methods,45 and crowdsourcing,46 among others. In Table 3, we present three challenge areas—value feasibility, internal consistency, and temporal consistency—and propose basic checks and approaches to data validation.
TABLE 3.
Key Elements for Clinical Data Validation
APPLICATIONS
Diagnosis and Early Detection
Cancer diagnosis requires synthesis of detailed clinical data, whether gene expression, radiology images, histopathology, or a combination of these data. Since the early 2000s, ML has been used to detect cancer biomarkers through gene expression profiles.47-50 With advances in computer vision, focus has shifted toward diagnosis from raw images. Breast cancer has been a natural pioneer in this domain given the importance of mammograms in cancer diagnosis. Related work dates back to 199551 with great progress in mammography-based diagnosis more recently.52,53 Similar approaches have been taken to diagnose lung cancer through computed tomography (CT) scans.54 Hu et al55 provide a detailed review of imaging-based diagnosis applications. Histology has also been explored as an application of image-based diagnosis.56 Convolutional neural networks have been applied to a range of diagnostic tasks using pathology results, including diagnosing prostate cancer57 and bladder cancer,58 as well as identifying breast cancer lymph node metastasis.57,59
ML also offers potential for early detection of cancers by scalably synthesizing trends across patients over a potentially distant time horizon. The value of early cancer detection is widely recognized60 yet challenging, because characteristics that are predictive of cancer emergence are often subtle and varied across patients. Computer vision methods have been used to predict future breast cancer diagnosis using breast density in mammography61 or lung cancer using CT scans.62 Other work has focused on identifying cancer susceptibility using gene expression data,63 and yet other investigators have used EMR data to predict pancreatic cancer risk within a high-risk cohort.64 These early warning systems could potentially inform cancer screening policies and methods. Most importantly, they offer potential for earlier intervention and improved patient outcomes.
Cancer Classification and Staging
Cancer staging forms the basis of much cancer classification. It often defines eligibility criteria for clinical trials and prognosis estimates and thus has broad implications for treatment guidelines and patient care. The American Joint Committee on Cancer (AJCC) guidelines have represented the gold standard of cancer staging in current practice since their introduction in 1977.65,66 The TNM classification in particular allows for stage classification on a sparse set of features—primary tumor size (T), affected lymph nodes (N), and the presence of metastasis (M). Cancer classification schemes benefit from this simplicity because they require minimal data collection of a consistent and well-accepted set of features. However, these schemes ignore potentially important clinical features and rely on clinically derived cutoff values. The limits of the existing system have been recognized and have prompted investigation into alternative approaches.
Cancer staging can be viewed more broadly as an attempt to stratify patients into well-differentiated risk cohorts. ML presents an opportunity to designate staging criteria directly from data, potentially providing better prognostic differentiation between stages. For example, a model that predicts disease-free survival can be used to stratify patients into prognostic groups. In a sense, this then becomes a de facto cancer classification system.
Researchers have applied this approach to pancreatic cancer67 and intrahepatic cholangiocarcinoma,68 among others. Although the individual methods vary, they all leverage large-scale data to derive insights into novel predictors and demonstrate better patient stratification than the AJCC scheme. These works take advantage of the ability to simultaneously parse many features with potentially thousands of patients, in a way that was infeasible when the AJCC initially formed.
Cancer staging is generally approached through a supervised learning framework to predict survival; the predictors are then analyzed to define staging criteria. However, unsupervised learning has also proved useful in identifying distinct cohorts within cancer types. Researchers have applied clustering to lung cancer69,70 and breast cancer71 and found the resultant subgroups to be prognostically distinct even though the algorithm does not consider survival directly. It can be advantageous to derive subgroups without an explicit outcome, particularly given the noise and difficulty in measuring survival. Clustering offers a new perspective in cancer classification to more generally partition patients into clinical subgroups.
Unsupervised learning has also been used to identify gene signatures for cancers.72 Researchers have applied this to learn about distinct profiles within cancer. The identification of such cohorts presents an opportunity for better disease understanding and a more tailored approach to treatment decisions.
Predicting and Evaluating Treatment Response
ML also provides prescriptive insights. Personalized predictions for treatment response to alternative therapies, as well as their potential adverse effects, can inform treatment decisions and patient monitoring. Genomic data have played an important role in this effort; the growing availability of cell line data has enabled large-scale drug sensitivity prediction based on genomic profiles.73-76 Genomic information has also been leveraged to predict clinical response metrics, both in pancancer analysis77 and for more targeted interactions such as leucovorin, fluorouracil, and oxaliplatin response in patients with colorectal cancer.78 ML has been used to predict treatment response for patients receiving neoadjuvant chemotherapy; radiomics has been leveraged for non–small-cell lung cancer (NSCLC),79 and a combination of clinical and imaging features has been used for breast cancer.80 Other work has been done to identify adverse effects of treatments, either at the drug level81 or at the patient level.82
ML can also be used to evaluate tumor response, which has traditionally relied on two-dimensional tumor measurements assessed using RECIST.83,84 The reliance on two-dimensional measurements came from necessity, namely the need to use features that could feasibly be measured by radiologists. There are shortcomings to this approach, and researchers have found that RECIST may not accurately track with outcome improvements.85 Just as ML has proved useful in diagnostic imaging, it has been applied to automatically detect the RECIST criteria in patients with NSCLC.86 Other studies have introduced RECIST alternatives for response evaluation with sequences of CT scans for NSCLC87 and volumetric measurements from magnetic resonance imaging for brain tumors.88
In conclusion, ML offers great promise in oncology. It can be used to derive risk cohorts, predict prognosis, inform treatment plans, and aid with diagnosis and early interventions. Given the proliferation of patient data that are available, data-driven approaches can enhance our understanding of cancer and its effect on individuals. Although ML presents numerous technical and organizational challenges, it is a worthwhile endeavor that will transform cancer care.
SUPPORT
Supported by the National Science Foundation Graduate Research Fellowship under Grant No. 174530.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Dimitris Bertsimas
Stock and Other Ownership Interests: Reclaim, Alexandria Health
No other potential conflicts of interest were reported.
REFERENCES
- 1. doi: 10.1056/NEJMra1814259. Rajkomar A, Dean J, Kohane I: Machine learning in medicine. N Engl J Med 380:1347-1358, 2019. [DOI] [PubMed] [Google Scholar]
- 2. doi: 10.1001/jama.2019.4914. Emanuel EJ, Wachter RM: Artificial intelligence in health care: Will the value match the hype? JAMA 321:2281–2282, 2019. [DOI] [PubMed] [Google Scholar]
- 3. doi: 10.1186/s12916-019-1426-2. Kelly CJ, Karthikesalingam A, Suleyman M, et al: Key challenges for delivering clinical impact with artificial intelligence. BMC Med 17:195, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. doi: 10.1371/journal.pmed.1002689. Vayena E, Blasimme A, Cohen IG: Machine learning in medicine: Addressing ethical challenges. PLoS Med 15 e1002689, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Price WN: Artificial intelligence in health care: Applications and legal issues. SciTech Lawyer 14:10-13, 2017. [Google Scholar]
- 6. Hastie T, Tibshirani R, Friedman J: The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York, NY, Springer, 2013. [Google Scholar]
- 7. Yu C, Liu J, Nemati S: Reinforcement learning in healthcare: A survey. http://arxiv.org/abs/1908.08796.
- 8.Padmanabhan R, Meskin N, Haddad WM. Reinforcement learning-based control of drug dosing for cancer chemotherapy treatment. Math Biosci. 2017;293:11–20. doi: 10.1016/j.mbs.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 9. doi: 10.3389/fninf.2014.00014. Pedregosa F, Varoquaux G, Gramfort A, et al: Scikit-learn: Machine learning in Python. http://scikit-learn.sourceforge.net. [DOI] [PMC free article] [PubMed]
- 10. Breiman L, Friedman JH, Olshen RA, et al: Classification and Regression Trees. New York, NY, Routledge, 1984. [Google Scholar]
- 11. Interpretable AI: Interpretable AI documentation. https://docs.interpretable.ai/stable/
- 12. Bertsimas D, Dunn J: Optimal classification trees. Mach Learn 106:1039-1082, 2017. [Google Scholar]
- 13.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 14. Chen T, He T, Benesty M, et al: xgboost: Extreme gradient boosting. https://cran.r-project.org/package=xgboost.
- 15. doi: 10.1016/j.neunet.2014.09.003. Schmidhuber J: Deep learning in neural networks: An overview. Neural Netw 61:85-117, 2015. [DOI] [PubMed] [Google Scholar]
- 16. Friedman JH: Greedy function approximation: A gradient boosting machine. https://projecteuclid.org/download/pdf_1/euclid.aos/1013203451.
- 17. Chen T, Guestrin C: XGBoost: A scalable tree boosting system. https://arxiv.org/pdf/1603.02754.pdf.
- 18. Lundberg SM, Lee SI: A unified approach to interpreting model predictions. https://github.com/slundberg/shap.
- 19.Lundberg SM, Erion G, Chen H, et al. From local explanations to global understanding with explainable AI for trees. Nat Mach Intell. 2020;2:56–67. doi: 10.1038/s42256-019-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. MacQueen J: Some methods for classification and analysis of multivariate observations. https://www.cs.cmu.edu/~bhiksha/courses/mlsp.fall2010/class14/macqueen.pdf.
- 21. Sneath PHA, Sokal RR: Numerical Taxonomy: The Principles and Practice of Numerical Classification. San Francisco, CA, W.H. Freeman and Company, 1973. [Google Scholar]
- 22. Ester M, Kriegel H-P, Sander J, et al: A density-based algorithm for discovering clusters in large spatial databases with noise. https://www.aaai.org/Papers/KDD/1996/KDD96-037.pdf.
- 23. Rao CR: The use and interpretation of principal component analysis in applied research. Sankhyā Indian J Stat Ser A 26:329-358, 1964. [Google Scholar]
- 24. Golub GH, Reinsch C: Handbook series linear algebra: Singular value decomposition and least squares solutions. http://people.duke.edu/~hpgavin/SystemID/References/Golub+Reinsch-NM-1970.pdf.
- 25. Agrawal R, Srikant R: Fast algorithms for mining association rules in large databases, in: Proceedings of the 20th International Conference on Very Large Data Bases. San Francisco, CA, Morgan Kaufmann Publishers, 1994, pp 487-499. [Google Scholar]
- 26.Radev DR, Jing H, Styś M, et al. Centroid-based summarization of multiple documents. Inf Process Manage. 2004;40:919–938. [Google Scholar]
- 27.Jain AK, Murty MN, Flynn PJ. Data clustering: A review. ACM Comput Surv. 1999;31:264–323. [Google Scholar]
- 28. Hancock TP, Coomans DH, Everingham YL: Supervised hierarchical clustering using CART. https://www.mssanz.org.au/MODSIM03/Volume_04/C07/06_Hancock.pdf.
- 29. Bertsimas D, Orfanoudaki A, Wiberg H: Interpretable clustering: An optimization approach. Mach Learn 1-50, 2020.
- 30. Fraiman R, Ghattas B, Svarc M: Interpretable clustering using unsupervised binary trees. Adv Data Anal Classif 7:125-145, 2013. [Google Scholar]
- 31. Blockeel H, De Raedt L, Ramon J: Top-down induction of clustering trees. https://arxiv.org/pdf/cs/0011032.pdf.
- 32. Manning C, Schutze H: Foundations of Statistical Natural Language Processing. Cambridge, MA, MIT Press, 1999. [Google Scholar]
- 33. Pennington J, Socher R, Manning CD: GloVe: Global vectors for word representation. https://nlp.stanford.edu/pubs/glove.pdf.
- 34.Stephens ZD, Lee SY, Faghri F, et al. Big data: Astronomical or genomical? PLoS Biol. 2015;13:e1002195. doi: 10.1371/journal.pbio.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. doi: 10.1038/ncomms5006. Aerts HJWL, Velazquez ER, Leijenaar RTH, et al: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miotto R, Wang F, Wang S, et al. Deep learning for healthcare: Review, opportunities and challenges. Brief Bioinform. 2018;19:1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. doi: 10.1373/49.4.624. McDonald CJ, Huff SM, Suico JG, et al: LOINC, a universal standard for identifying laboratory observations: A 5-year update. Clin Chem 49:624-633, 2003. [DOI] [PubMed] [Google Scholar]
- 38. WHO: The Anatomical Therapeutic Chemical Classification System with Defined Daily Doses (ATC/DDD). https://www.who.int/classifications/atcddd/en/
- 39.van Buuren S, Groothuis-Oudshoorn K. mice: Multivariate imputation by chained equations in R. J Stat Softw. 2011;45:2–20. [Google Scholar]
- 40. Bertsimas D, Pawlowski C, Zhuo YD: From predictive methods to missing data imputation: An optimization approach. http://jmlr.org/papers/v18/17-073.html.
- 41. Bertsimas D, Orfanoudaki A, Pawlowski C: Imputation of clinical covariates in time series. http://arxiv.org/abs/1812.00418.
- 42.Rahm E, Do H. Data cleaning: Problems and current approaches. IEEE Data Eng Bull. 2000;23:3–13. [Google Scholar]
- 43. Chu X, Ilyas IF, Krishnan S, et al: Data cleaning: Overview and emerging challenges. Proceedings of the ACM SIGMOD International Conference on Management of Data, San Francisco, CA, June 26-July 1, 2016, pp 2201-2206. [Google Scholar]
- 44. Bohannon P, Fan W, Geerts F, et al: Conditional functional dependencies for data cleaning. https://ieeexplore.ieee.org/document/4221723/
- 45. Krishnan S, Wang J, Franklin MJ, et al: SampleClean: Fast and reliable analytics on dirty data. http://sites.computer.org/debull/A15sept/p59.pdf.
- 46. Chu X, Morcos J, Ilyas IF, et al: Katara: A data cleaning system powered by knowledge bases and crowdsourcing. Proceedings of the ACM SIGMOD International Conference on Management of Data, Melbourne, Victoria, Australia, May 31-June 4, 2015, pp 1247-1261 . [Google Scholar]
- 47.Tan AC, Gilbert D. Ensemble machine learning on gene expression data for cancer classification. Appl Bioinformatics. 2003;2(suppl 3):S75–S83. http://europepmc.org/abstract/MED/15130820 [PubMed] [Google Scholar]
- 48. Hwang K-B, Cho D-Y, Park S-W, et al: Applying machine learning techniques to analysis of gene expression data: Cancer diagnosis, in Lin SM, Johnson KF (eds): Methods of Microarray Data Analysis: Papers from CAMDA ’00. Boston, MA, Springer, 2002, pp 167-182. [Google Scholar]
- 49. doi: 10.1142/9789813207813_0022. Danaee P, Ghaeini R, Hendrix DA: A deep learning approach for cancer detection and relevant gene identification. Pac Symp Biocomput 22:219-229, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. doi: 10.1038/nm843. Ye QH, Qin LX, Forgues M, et al: Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med 9:416-423, 2003. [DOI] [PubMed] [Google Scholar]
- 51.Wolberg WH, Street WN, Mangasarian OL. Image analysis and machine learning applied to breast cancer diagnosis and prognosis. Anal Quant Cytol Histol. 1995;17:77–87. [PubMed] [Google Scholar]
- 52. doi: 10.1038/s41598-019-48995-4. Shen L, Margolies LR, Rothstein JH, et al: Deep learning to improve breast cancer detection on screening mammography. Sci Rep 9:12495, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramos-Pollán R, Guevara-López MA, Suárez-Ortega C, et al. Discovering mammography-based machine learning classifiers for breast cancer diagnosis. J Med Syst. 2012;36:2259–2269. doi: 10.1007/s10916-011-9693-2. [DOI] [PubMed] [Google Scholar]
- 54.Sun W, Zheng B, Qian W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med. 2017;89:530–539. doi: 10.1016/j.compbiomed.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Hu Z, Tang J, Wang Z, et al. Deep learning for image-based cancer detection and diagnosis: A survey. Pattern Recognit. 2018;83:134–149. [Google Scholar]
- 56.Madabhushi A. Digital pathology image analysis: Opportunities and challenges. Imaging Med. 2009;1:7–10. doi: 10.2217/IIM.09.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. doi: 10.1038/srep26286. Litjens G, Sánchez CI, Timofeeva N, et al: Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 6:26286, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Z, Chen P, McGough M, et al: Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat Mach Intell 1:236-245, 2019. [Google Scholar]
- 59. Liu Y, Gadepalli K, Norouzi M, et al: Detecting cancer metastases on gigapixel pathology images. http://arxiv.org/abs/1703.02442.
- 60. doi: 10.1038/nrc1041. Etzioni R, Urban N, Ramsey S, et al: The case for early detection. Nat Rev Cancer 3:243-252, 2003. [DOI] [PubMed] [Google Scholar]
- 61. doi: 10.1148/radiol.2019182716. Yala A, Lehman C, Schuster T, et al: A deep learning mammography-based model for improved breast cancer risk prediction. Radiology 292:60-66, 2019. [DOI] [PubMed] [Google Scholar]
- 62. doi: 10.1016/S2589-7500(19)30159-1. Huang P, Lin CT, Li Y, et al: Prediction of lung cancer risk at follow-up screening with low-dose CT: A training and validation study of a deep learning method. Lancet Digit Health 1:e353-e362, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim BJ, Kim SH. Prediction of inherited genomic susceptibility to 20 common cancer types by a supervised machine-learning method. Proc Natl Acad Sci USA. 2018;115:1322–1327. doi: 10.1073/pnas.1717960115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boursi B, Finkelman B, Giantonio BJ, et al. A clinical prediction model to assess risk for pancreatic cancer among patients with new-onset diabetes. Gastroenterology. 2017;152:840–850.e3. doi: 10.1053/j.gastro.2016.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. American Joint Committee on Cancer: Cancer Staging Manual. Chicago, IL, American Joint Committee on Cancer, 1977. [Google Scholar]
- 66. doi: 10.3322/caac.21388. Amin MB, Greene FL, Edge SB, et al: The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin 67:93-99, 2017. [DOI] [PubMed] [Google Scholar]
- 67.Das A, Ngamruengphong S. Machine learning based predictive models are more accurate than TNM staging in predicting survival in patients with pancreatic cancer. Am J Gastroenterol. 2019;114:S48. [Google Scholar]
- 68. doi: 10.1245/s10434-019-08067-3. Tsilimigras DI, Mehta R, Moris D, et al: A machine-based approach to preoperatively identify patients with the most and least benefit associated with resection for intrahepatic cholangiocarcinoma: An international multi-institutional analysis of 1146 patients. Ann Surg Oncol 27:1110-1119, 2020. [DOI] [PubMed] [Google Scholar]
- 69.Chen D, Xing K, Henson D, et al. Developing prognostic systems of cancer patients by ensemble clustering. J Biomed Biotechnol. 2009;2009:632786. doi: 10.1155/2009/632786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. doi: 10.1371/journal.pone.0184370. Lynch CM, Van Berkel VH, Frieboes HB: Application of unsupervised analysis techniques to lung cancer patient data. PLoS One 12:e0184370, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aure MR, Vitelli V, Jernström S, et al. Integrative clustering reveals a novel split in the luminal A subtype of breast cancer with impact on outcome. Breast Cancer Res. 2017;19:44. doi: 10.1186/s13058-017-0812-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. doi: 10.1016/j.bdq.2017.07.001. Kakushadze Z, Yu W: *K-means and cluster models for cancer signatures. Biomol Detect Quantif 13:7-31, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. doi: 10.1371/journal.pone.0061318. Menden MP, Iorio F, Garnett M, et al: Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS One 8:61318, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. doi: 10.1038/nature11005. Garnett MJ, Edelman EJ, Heidorn SJ, et al: Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483:570-575, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. doi: 10.1371/journal.pone.0186906. Huang C, Mezencev R, McDonald JF, et al: Open source machine-learning algorithms for the prediction of optimal cancer drug therapies. PLoS One 12:e0186906, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. doi: 10.1158/1541-7786.MCR-17-0378. Ding MQ, Chen L, Cooper GF, et al: Precision oncology beyond targeted therapy: Combining omics data with machine learning matches the majority of cancer cells to effective therapeutics. Mol Cancer Res 16:269-278, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. doi: 10.1038/s41598-018-34753-5. Huang C, Clayton EA, Matyunina LV, et al: Machine learning predicts individual cancer patient responses to therapeutic drugs with high accuracy. Sci Rep 8:16444, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. doi: 10.1002/cam4.2786. Lu W, Fu D, Kong X, et al: FOLFOX treatment response prediction in metastatic or recurrent colorectal cancer patients via machine learning algorithms. Cancer Med 9:1419-1429, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119:480–486. doi: 10.1016/j.radonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mani S, Chen Y, Arlinghaus LR, et al. Early prediction of the response of breast tumors to neoadjuvant chemotherapy using quantitative MRI and machine learning. AMIA Annu Symp Proc. 2011;2011:868–877. [PMC free article] [PubMed] [Google Scholar]
- 81. doi: 10.1007/s11095-018-2562-7. Bloomingdale P, Mager DE: Machine learning models for the prediction of chemotherapy-induced peripheral neuropathy. Pharm Res 36:35, 2019. [DOI] [PubMed] [Google Scholar]
- 82. doi: 10.1016/j.jvir.2018.01.769. Abajian A, Murali N, Savic LJ, et al: Predicting treatment response to intra-arterial therapies for hepatocellular carcinoma with the use of supervised machine learning: An artificial intelligence concept. J Vasc Interv Radiol 29:850-857.e1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 84.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 85.Villaruz LC, Socinski MA. The clinical viewpoint: definitions, limitations of RECIST, practical considerations of measurement. Clin Cancer Res. 2013;19:2629–2636. doi: 10.1158/1078-0432.CCR-12-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arbour KC, Anh Tuan L, Rizvi H, et al. ml-RECIST: Machine learning to estimate RECIST in patients with NSCLC treated with PD-(L)1 blockade. J Clin Oncol. 2019;37:9052–9052. (suppl; abst 9052) [Google Scholar]
- 87.Xu Y, Hosny A, Zeleznik R, et al. Deep learning predicts lung cancer treatment response from serial medical imaging. Clin Cancer Res. 2019;25:3266–3275. doi: 10.1158/1078-0432.CCR-18-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kickingereder P, Isensee F, Tursunova I, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: A multicentre, retrospective study. Lancet Oncol. 2019;20:728–740. doi: 10.1016/S1470-2045(19)30098-1. [DOI] [PubMed] [Google Scholar]