Abstract
For children with neuroblastoma, the likelihood of cure varies widely according to age at diagnosis, disease stage, and tumor biology. Treatments are tailored for children with this clinically heterogeneous malignancy on the basis of a combination of markers that are predictive of risk of relapse and death. Sequential risk-based, cooperative-group clinical trials conducted during the past 4 decades have led to improved outcome for children with neuroblastoma. Increasingly accurate risk classification and refinements in treatment stratification strategies have been achieved with the more recent discovery of robust genomic and molecular biomarkers. In this review, we discuss the history of neuroblastoma risk classification in North America and Europe and highlight efforts by the International Neuroblastoma Risk Group (INRG) Task Force to develop a consensus approach for pretreatment stratification using seven risk criteria including an image-based staging system—the INRG Staging System. We also update readers on the current Children’s Oncology Group risk classifier and outline plans for the development of a revised 2021 Children’s Oncology Group classifier that will incorporate INRG Staging System criteria to facilitate harmonization of risk-based frontline treatment strategies conducted around the globe. In addition, we discuss new approaches to establish increasingly robust, future risk classification algorithms that will further refine treatment stratification using machine learning tools and expanded data from electronic health records and the INRG Data Commons.
INTRODUCTION
Neuroblastoma is notable for its broad spectrum of clinical behavior,1,2 and efforts to tailor treatment according to the predicted clinical aggressiveness of the tumor have been ongoing for decades.3 The first risk classification algorithm used age, stage of disease, and ferritin to subdivide patients into groups with good, intermediate, or poor prognosis4 (Fig 1). The prognostic strength of tumor histology,5 tumor cell ploidy,6 and MYCN status7 identified in the 1980s led to additional refinements in treatment stratification and improved survival.8-11 The North American cooperative groups, Children’s Cancer Group (CCG) and Pediatric Oncology Group (POG), developed classifiers using these prognostic markers, although the risk criteria differed12,13 (Fig 1).
FIG 1.
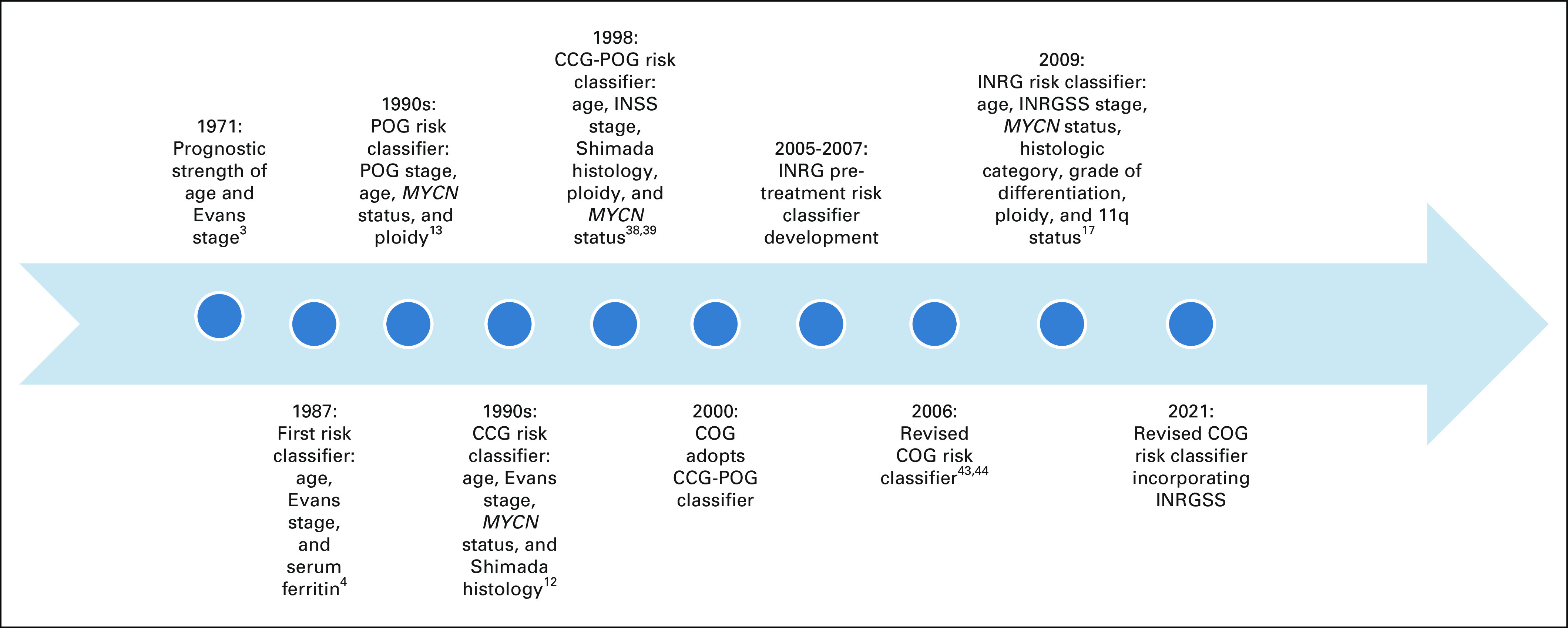
Timeline of neuroblastoma risk classification schemas. CCG, Children’s Cancer Group; COG, Children’s Oncology Group; INRG, International Neuroblastoma Risk Group; INRGSS, International Neuroblastoma Risk Group Staging System; POG, Pediatric Oncology Group.
CONTEXT
Key Objective
To provide an overview of how risk classification algorithms have advanced treatment of children with neuroblastoma and highlight new approaches for maximizing the scientific value of available data and accelerating the pace of developing refined risk classification algorithms.
Knowledge Generated
Outcomes have been improved for children with neuroblastoma using risk-based treatment approaches, although the criteria used to define risk differ around the world. Comparison of frontline clinical trials conducted in different geographic regions has therefore been difficult.
Relevance
Refinements of the current risk classification system are needed to more precisely tailor treatment and improve outcomes for patients with neuroblastoma. The International Neuroblastoma Risk Group Data Commons, electronic health records, and new machine learning methods may accelerate the development of future risk classifiers. To further advance treatment, international neuroblastoma community collaboration will be needed to develop harmonized approaches for risk classification.
During this era, therapy was also stratified in clinical trials conducted in Europe on the basis of a combination of prognostic markers. However, instead of designing risk-based clinical trials, treatment was tailored according to tumor genomic markers among clinical subsets of patients, including: (1) those with localized resectable and unresectable disease,9,14 (2) children > 1 year of age with disseminated neuroblastoma,15 and (3) infants with metastatic neuroblastoma.16 The unique eligibility criteria and nonstandardized methods of data collection used by North American, European, and other cooperative groups internationally impeded the comparison of upfront neuroblastoma clinical trials conducted around the world.
INTERNATIONAL NEUROBLASTOMA RISK GROUP CLASSIFICATION SYSTEM
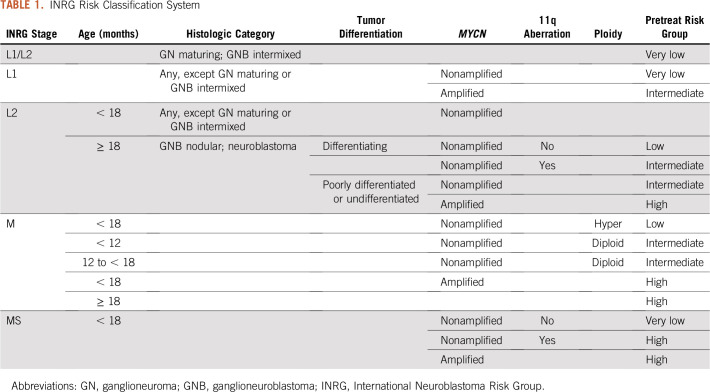
To establish a consensus approach for pretreatment risk stratification, an International Neuroblastoma Risk Group (INRG) Task Force representing the major pediatric cooperative groups around the world was formed in 2005.17 Risk criteria incorporated into the INRG Classification System were based on statistical analyses of 13 potential prognostic factors in a cohort of 8,800 children diagnosed with neuroblastoma between 1990 and 2002, including patients from North America and Australia (Children’s Oncology Group [COG]), Europe (International Society of Pediatric Oncology Europe Neuroblastoma Group [SIOPEN]), Germany (German Pediatric Oncology and Hematology Group), and Japan (Japanese Advanced Neuroblastoma Study Group and the Japanese Infantile Neuroblastoma Co-operative Study Group). Survival tree analysis was performed and the most highly statistically significant and clinically relevant factors were included in the INRG classifier. The task force also developed a new staging system defined by imaging, the INRG Staging System (INRGSS), for the pretreatment INRG Classification System.18 According to INRGSS criteria, locoregional tumors are categorized as L1 or L2 based on the absence or presence of tumor infiltration or invasion of nerves, vessels, and organs (ie, image-defined risk factors [IDRFs]).18,19 IDRFs have been shown to be associated with an increase in intraoperative complications, incomplete tumor resection, and worse survival in numerous studies.20-22
Although the INRG Classification System (Table 1) provides a platform for uniformly defining risk, the more recently discovered genomic and molecular biomarkers, including segmental chromosome aberrations (SCAs) and copy number changes,23 gene expression signatures,24 mutational profiles,25 and telomere maintenance mechanisms,26 were not available during the development of this risk algorithm. It is anticipated that incorporating these robust prognostic markers in the next-generation INRG Classification System will lead to refined treatment stratification and improved survival.
TABLE 1.
INRG Risk Classification System
HISTORY OF COG NEUROBLASTOMA RISK STRATIFICATION
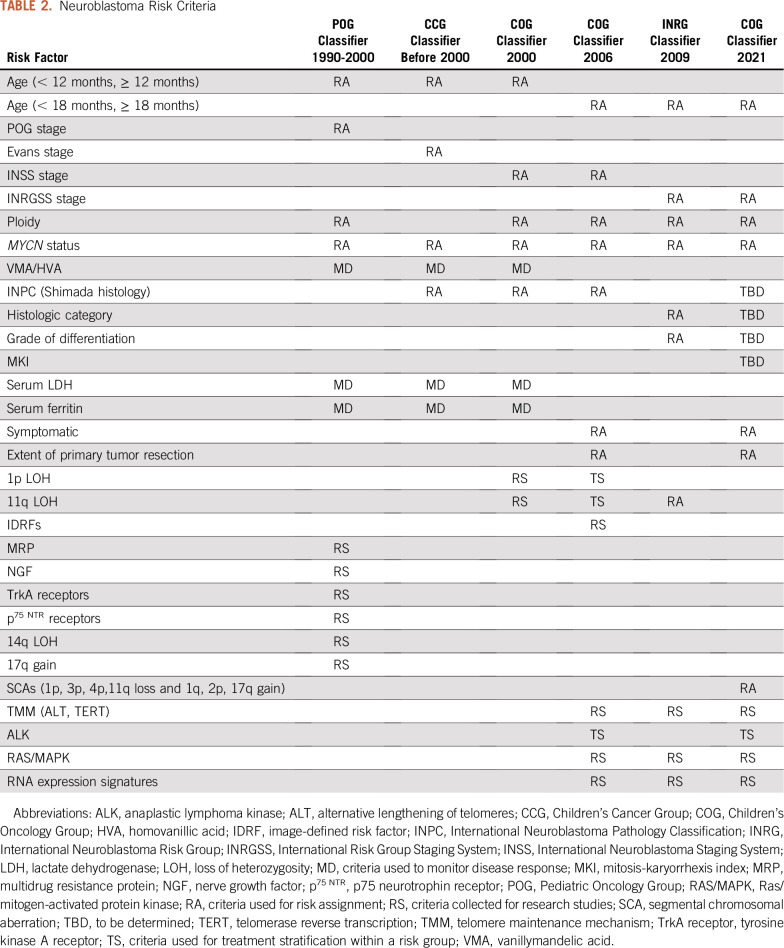
Criteria in the CCG classifier included Evans stage, age at diagnosis, MYCN status, Shimada tumor histology, and serum ferritin,12 whereas POG criteria included POG stage, age at diagnosis, MYCN status, and tumor cell ploidy.13 In parallel with the development of risk-based therapeutic trials, POG also established the first neuroblastoma biology protocol (9047), which was designed to collect tumor samples for prospective analyses of potential prognostic markers and was amended in 1998 to collect outcome data. Initial markers included DNA content (ploidy),6 N-myc (MYCN) copy number,7 chromosome 1p loss,27 and multidrug resistance–related protein expression.28 As new variables were identified, the study was amended to evaluate the prognostic strength of additional genetic29-31 and molecular markers32 (Table 2).
TABLE 2.
Neuroblastoma Risk Criteria
POG 9047 also provided a mechanism for banking clinically annotated neuroblastoma samples, which were made available to the research community.33,34 This protocol served as a template for the COG biology study (ANBL00B1; ClinicalTrials.gov identifier: NCT00904241). The clinical, genomic, specimen, and outcome data collected in these biology studies were collated to generate the COG Neuroblastoma Virtual Tumor Bank, a resource that facilitated the discovery of new genomic and molecular biomarkers.35,36 SIOPEN is currently developing a bioportal to establish a similar virtual biobank.
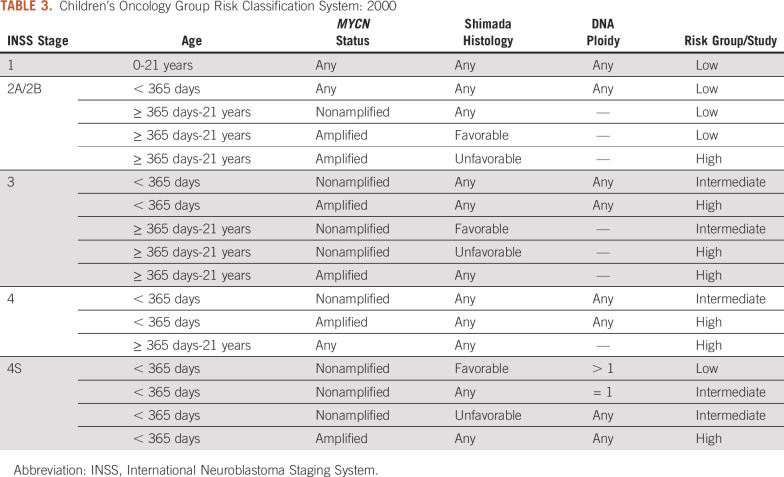
Because neuroblastoma is a rare cancer, leaders from POG, CCG, and the Cancer Therapy Evaluation Program at the National Cancer Institute recognized that significant efficiencies would be gained with the development of intergroup (POG-CCG) risk-based clinical trials. A new neuroblastoma risk classification schema was developed for these intergroup clinical trials that included age, International Neuroblastoma Staging System (INSS) stage,37 MYCN status, ploidy, and Shimada histology (Fig 1 and Table 3). It is important to note that the legacy staging systems cannot be directly mapped to INSS stage,38 and that the resulting neuroblastoma classification system was based on consensus rather than a statistical methods approach. Patient eligibility for the CCG-POG intergroup low-risk (P9641; ClinicalTrials.gov identifier: NCT00003119)),39 intermediate-risk (A3961; ClinicalTrials.gov identifier: NCT00003093),40 and high-risk (A3973; ClinicalTrials.gov identifier: NCT00004188)41 clinical trials were defined by this risk schema.
TABLE 3.
Children’s Oncology Group Risk Classification System: 2000
THE 2000 COG NEUROBLASTOMA RISK CLASSIFICATION SYSTEM AND 2006 REVISION
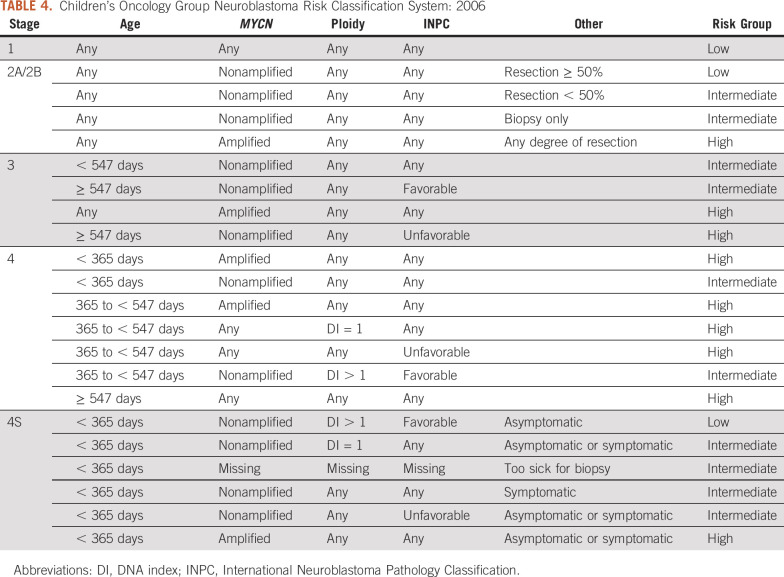
In 2000, COG adopted the POG-CCG Intergroup Neuroblastoma Risk Classification System (Fig 1). The COG biology study ANBL00B1 serves as the infrastructure for rapid and reliable acquisition of tumor prognostic markers used for risk classification and clinical trial eligibility, and approximately 500 to 600 patients per year are enrolled in this biology study. In 2006, the COG Neuroblastoma Risk Classification System was revised based on studies demonstrating the poor outcome of symptomatic infants with stage 4S disease42,43 and that 18 months is a more optimal age cutoff than 12 months for clinically relevant risk stratification44,45 (Fig 1 and Table 4). In addition, the COG 2006 classifier categorized histology according to International Neuroblastoma Pathology Classification System (INPC).46 The revised schema also classified patients with 2A/B tumors as low versus intermediate risk according to the degree of tumor resection based on the treatment approaches used in the COG P9641 clinical trial.39
TABLE 4.
Children’s Oncology Group Neuroblastoma Risk Classification System: 2006
SIOPEN CLINICAL TRIALS
Risk-based treatment strategies are also used in European clinical trials. Eligibility for the high-risk phase III (HR-NBL1) SIOPEN trial included children with INSS stage 2, 3, 4S or tumors with MYCN amplification, and patients age 12 months or older with INSS stage 4 disease.47 In the European Low- and Intermediate-Risk Neuroblastoma Protocol (ClinicalTrials.gov identifier: NCT01728155), risk is defined by age, INRGSS stage, MYCN status, SCAs, grade of differentiation, and the presence of life-threatening symptoms. This study assesses a reduction of therapy question in a subset of low-risk patients with L2 tumors without SCAs and no life-threatening symptoms. For infants with localized suprarenal masses discovered before birth or neonatally, the primary aim is to maintain a 3-year event-free survival of ≥ 80% with a nonoperative therapeutic approach. The study is also evaluating the efficacy of adding radiotherapy and 13-cis retinoic acid to the multiagent chemotherapy regimen used in the legacy SIOPEN study48 in a subset of intermediate-risk patients ≥ 18 months of age with L2 tumors with poorly differentiated or undifferentiated histology.
COG NEUROBLASTOMA RISK CLASSIFICATION SYSTEM: 2021 REVISION
In an effort to harmonize risk-based frontline treatment strategies around the world, COG has replaced INSS stage with the INRGSS to determine eligibility in current high-risk and non–high-risk clinical trials (ClinicalTrials.gov identifiers: NCT03126916, NCT03786783, and NCT02176967); however, the 2006 COG neuroblastoma risk classification system has not yet been modified to include INRGSS. Recently, a comprehensive analysis of IDRF data collected on 4,569 patients enrolled in the ANBL00B1 biology study between 2006 and 2016 was performed to assess the prognostic significance of INRGSS stage.49 Results of these analyses provide a statistical framework to develop a revised COG risk classification incorporating the INRGSS (Table 2).
Additional analyses are ongoing to determine which prognostic variables should be included in the revised 2021 COG risk classifier. In addition to INRGSS stage, the current plan is to include the established risk criteria age, MYCN status, and ploidy. SCAs will also be incorporated on the basis of their prognostic strength,50 especially in subgroups of patients with locoregional disease and otherwise favorable features.51 INPC is another powerful prognostic marker,49 and tumor histology will be included in the revised classifier. However, age at diagnosis is used as a criterion to define favorable versus unfavorable INPC histology52; therefore, its inclusion within a classification schema that also includes age as a separate variable results in a duplication of the prognostic contribution (confounding) of age to define risk, obscuring the effect of age on the dependent variable, event-free survival.
In an effort to develop a neuroblastoma classifier that eliminates the confounding of age and facilitates more precise prognostication, a recent study analyzed INRG data from more than 18,000 patients with known tumor histology.53 The study demonstrated the independent prognostic significance of age and each histologic feature that underlies INPC, including histologic category, mitosis-karyorrhexis index (MKI), and grade of differentiation. Furthermore, survival tree regression analyses revealed a novel, unfavorable prognostic subgroup of patients ≥ 547 days old with stage 1 or 2, MYCN nonamplified, diploid tumors with intermediate/high MKI.53 Taken together, these results provide statistical methodologic support to incorporate histologic category, MKI, and grade instead of INPC category in a future COG classifier.
A revised COG neuroblastoma risk classification schema that includes risk criteria harmonized with those currently used by SIOPEN and other pediatric cooperative groups around the world is needed to compare clinical trial results. The proposed changes in the revised 2021 classifier will modify risk assignment, treatment stratification, and treatment intensity for specific subsets of patients compared with the 2006 risk algorithm. Future studies will be needed to assess whether the change in treatment stratification for these subsets of patients leads to improved outcomes. As new genomic and molecular biomarkers are identified, it will be important to test the independent prognostic strength of these factors. Candidate genomic biomarkers currently being prospectively studied that may be incorporated into risk classifiers in the next 5 to 10 years include specific mutations (eg, ALK, RAS-MAPK pathway)54 and alterations in telomere maintenance mechanisms (eg, high TERT expression, status of the alternative lengthening of telomeres).26
NEUROBLASTOMA RISK SCORES
The current neuroblastoma classification systems use a rules-based table to assign risk groups based on categorical prognostic variables.17,45 The use of categorical variables allows for ease of calculation but lacks precision; lost is the gradation of risk associated with continuous variables. The resulting risk groupings are clinically useful for initial treatment assignment, but lack precision in risk determination. For example, within the high-risk neuroblastoma grouping, there is evidence of an ultra–high-risk subgroup with worse prognosis that cannot be identified using current risk classification algorithms.55 A risk score, in addition to a risk group assignment, may provide more accurate information regarding the patient’s risk relative to others receiving the same therapy. As proof of principle, Moreno et al developed a nomogram that calculates a risk score using three markers prognostic of overall survival56. In a discovery and validation cohort of high-risk patients (≥ 18 months old with metastatic neuroblastoma), the risk score could be used to identify patients at greatest risk of death within 3 years. Similarly, in an analysis of data from the HR-NBL1/SIOPEN trial (ClinicalTrials.gov identifier: NCT01704716), Morgenstern et al, developed a risk score based on age, serum LDH and metastatic site index (MSI) that identified an ultra-high-risk cohort with 5-year event-free survival of < 10%.57 Future risk scores that include continuous risk variables and additional tumor biomarkers may provide more precise prognostic information.
REFINING FUTURE NEUROBLASTOMA RISK CLASSIFIERS: CHALLENGES AND OPPORTUNITIES
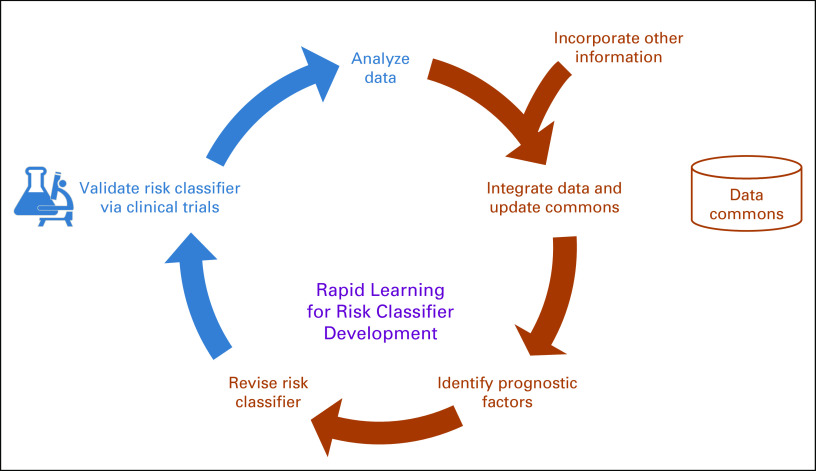
Refinements of neuroblastoma risk classification are made through an iterative process akin to the rapid-learning health care model, described more than 10 years ago.58 In the rapid-learning system for cancer care, data routinely generated through patient care and clinical research feed into an ever-growing set of coordinated databases and registries, driving the scientific discovery that subsequently leads to improved patient care (Fig 2). For risk algorithms, data from clinical trials, clinical registries, tumor biology studies, data commons, and other sources are analyzed to identify factors that are predictive of survival. An updated risk classification schema that incorporates new knowledge is then developed and the proposed risk classification schema is validated through clinical trials, leading to the beginning of the next cycle. Each iteration results in an increasingly `accurate and robust risk classification model.
FIG 2.
Rapid learning health care model.
Two major limitations to using this rapid-learning model to refine the neuroblastoma classifier are the length of time an iteration takes and the paucity of data available for modeling. It commonly takes 5 to 10 years to obtain results from a clinical trial in which survival is a primary end point, representing a major bottleneck. In addition, as neuroblastoma is a rare disease, the number of patients enrolled in these studies and the amount of data generated are limited. Methods that can maximize the scientific value of available data and accelerate the pace of risk classification development are needed to further refine treatment approaches that will ultimately improve outcomes for children with neuroblastoma.
INRG DATA COMMONS
One way to address the paucity of available data in neuroblastoma and other rare diseases is by combining data from multiple sources. There is a culture of international collaboration and data sharing within the pediatric oncology community and, in particular, within neuroblastoma, as demonstrated by the development of uniform staging systems,37 the INRG Risk Classification System,17 and the International Neuroblastoma Response Criteria.59 In addition, the international neuroblastoma community has pioneered emerging data-sharing models within pediatric oncology through the development of the INRG database.1,17 Clinical information from patients enrolled in the COG, SIOPEN, German Pediatric Oncology and Hematology Group, Japanese Advanced Neuroblastoma Study Group, and Japanese Infantile Neuroblastoma Co-operative Study Group studies included in this database are available to the research community and have enabled studies that were not previously possible with smaller patient cohorts.60 In 2015, phenotype data were uploaded into the INRG Data Commons, which was established at the University of Chicago as an international, centralized ecosystem for neuroblastoma data analysis.61 This cloud-based infrastructure colocates neuroblastoma-related data, analysis tools, and high-performance computational resources in a secure environmental that adheres to FAIR (Findable, Accessible, Interoperable, Reusable) principles for data sharing and is interoperable with the National Cancer Institute’s cancer research data commons infrastructure.61 The INRG Data Commons currently houses data from more than 20,000 individual patients with neuroblastoma, the largest such neuroblastoma data repository in the world, with a subset linked to biospecimen and genomic data through a common identifier62 . It serves as a model and integral data repository within the University of Chicago’s Pediatric Cancer Data Commons, a data commons for all pediatric oncology data63. Clinical data within the INRG Data Commons can be linked with tumor genomic data in the TARGET (Therapeutically Applicable Research to Generate Effective Treatments) database64 and the Gabriella Miller Kids First Data Resource,65 as well as data generated in neuroblastoma laboratories around the world. By providing qualified researchers easy access to both the data and the computational resources necessary to perform secondary analyses, the INRG Data Commons has the potential to accelerate risk classification development.
ELECTRONIC HEALTH RECORD DATA
Another way to address the paucity of neuroblastoma data is to expand available data sources, which currently include clinical trials, tumor biology studies, and clinical registries. These data are high quality and meticulously collected. However, data analysis is limited to the data variables that were defined and collected prospectively; uncollected variables are obviously not available. Electronic health record (EHR) data would allow investigators to explore all clinical data that were captured during routine clinical care, including detailed information about treatments received, toxicities, responses, and long-term health outcomes, expanding the spectrum of potential questions that can be asked.66 The EHR has been leveraged to analyze real-world performance of treatments in patients with adult malignancies.67 Whereas the lack of interoperability and standardization has traditionally hampered the widespread use of EHR data for research, new initiatives, such as the eMERGE consortium, demonstrate that large EHR data sets may be used successfully to uncover previously unknown associations between specific clinical variables and rare conditions, such as specific genomic variants.68 Enriching the INRG data repository with linked EHR data could lead to the discovery of previously unconsidered prognostic factors that are unavailable in the existing data set.
COMPUTER-ASSISTED IMAGE CLASSIFICATION
The prognostic value of current risk criteria may be improved using machine learning methods to analyze pathologic and radiographic images. Classification of image-based findings is a task for which machine learning is well suited,69 and classification algorithms have been applied to radiographic and histopathologic images to assist with classifying prognostic image-based findings.70 Computer-assisted image interpretation allows for the development of more precise prognostic variables. In addition, when radiographic and histopathologic data are linked to clinical data, machine learning may be used to identify novel image-based predictive factors. Recent studies have demonstrated that machine learning techniques that quantitatively analyze radiologic or pathologic image features provide valuable prognostic information in adults with various malignancies, including lung and breast cancer,71 and in children with neuroblastoma.72 The INRG Data Commons has recently linked patient data with meta-iodo-benzylguanidine imaging studies and computed tomography, and the prognostic value of imaging data patterns is currently being analyzed using machine learning data processing methods.
CONCLUSION
Although risk-based treatment approaches have led to improvements in outcomes for children with neuroblastoma,1 comparison of frontline clinical trials conducted in different regions around the world has been difficult as a result of the lack of uniform eligibility criteria. The INRG Risk Classification System was established to provide a consensus approach for pretreatment risk stratification17; however, the classifier, which was developed more than 10 years ago, does not include more recently identified genomic and molecular biomarkers that are currently used to tailor treatments.73-75 To further advance treatment, nimble approaches must be developed to identify the most robust prognostic markers for future neuroblastoma risk classifiers, testing biomarkers, such as telomere lengthening/maintenance pathways, the composition of the tumor microenvironment, and host germline aberrations.2 More rapid refinements in risk classification may be achieved using machine learning tools and expanded data from EHRs and the INRG Data Commons. To optimize the treatment of individual patients, close collaboration between the international neuroblastoma community and data scientists will be needed to ensure that harmonized criteria are used to define risk and guide therapy.
EQUAL CONTRIBUTION
W.H.L. and S.M.F. are co-first authors.
SUPPORT
Supported in part by the National Institutes of Health, National Cancer Institute Grant No. U10-CA180899 to Children’s Oncology Group Statistics and Data Center, National Clinical Trials Network Operations Center Grant No. U10-CA180886, and St Baldrick's Foundation.
AUTHOR CONTRIBUTIONS
Conception and design: Wayne H. Liang, Wendy B. London, Meredith S. Irwin, Samuel L. Volchenboum, Susan L. Cohn
Administrative support: Sara M. Federico, Wendy B. London, Susan L. Cohn
Provision of study materials or patients: Susan L. Cohn
Collection and assembly of data: Sara M. Federico, Wendy B. London, Samuel L. Volchenboum, Susan L. Cohn
Data analysis and interpretation: Wayne H. Liang, Sara M. Federico, Wendy B. London, Arlene Naranjo, Meredith S. Irwin, Samuel L. Volchenboum
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Sara M. Federico
Honoraria: EUSA Pharma
Consulting or Advisory Role: EUSA Pharma
Wendy B. London
Consulting or Advisory Role: ArQule, Jubilant Draximage
Research Funding: Agios, Bristol Myers Squibb, Novartis, Aileron Therapeutics, Bluebird Bio
Travel, Accommodations, Expenses: ArQule
Arlene Naranjo
Consulting or Advisory Role: Novartis
Meredith S. Irwin
Honoraria: Bayer
Samuel L. Volchenboum
Stock and Other Ownership Interests: Litmus Health
Honoraria: Sanford Health
Consulting or Advisory Role: CVS Accordant
Research Funding: AbbVie (Inst)
Susan L. Cohn
Stock and Other Ownership Interests: United Therapeutics (I), United Therapeutics, Merck, Stryker (I), Stryker, Amgen (I), Pfizer (I), AbbVie, Amgen, Jazz Pharmaceuticals, Eli Lilly, Sanofi, Varex Imaging, Pfizer, Accelerated Medical Diagnostics, Anthem, Cardinal Health, Novo Nordisk, Regeneron, Zimmer BioMet
Research Funding: United Therapeutics (Inst), Merck (Inst)
Open Payments Link: https://openpaymentsdata.cms.gov/physician/46569/summary
No other potential conflicts of interest were reported.
REFERENCES
- 1.Pinto NR, Applebaum MA, Volchenboum SL, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33:3008–3017. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 3.Evans AE, D’Angio GJ, Randolph J. A proposed staging for children with neuroblastoma. Children’s Cancer Study Group A. Cancer. 1971;27:374–378. doi: 10.1002/1097-0142(197102)27:2<374::aid-cncr2820270221>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 4.Evans AE, D’Angio GJ, Propert K, et al. Prognostic factor in neuroblastoma. Cancer. 1987;59:1853–1859. doi: 10.1002/1097-0142(19870601)59:11<1853::aid-cncr2820591102>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 5.Shimada H, Chatten J, Newton WA, Jr, et al. Histopathologic prognostic factors in neuroblastic tumors: Definition of subtypes of ganglioneuroblastoma and an age-linked classification of neuroblastomas. J Natl Cancer Inst. 1984;73:405–416. doi: 10.1093/jnci/73.2.405. [DOI] [PubMed] [Google Scholar]
- 6.Look AT, Hayes FA, Nitschke R, et al. Cellular DNA content as a predictor of response to chemotherapy in infants with unresectable neuroblastoma. N Engl J Med. 1984;311:231–235. doi: 10.1056/NEJM198407263110405. [DOI] [PubMed] [Google Scholar]
- 7.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 8.Bowman LC, Castleberry RP, Cantor A, et al. Genetic staging of unresectable or metastatic neuroblastoma in infants: A Pediatric Oncology Group study. J Natl Cancer Inst. 1997;89:373–380. doi: 10.1093/jnci/89.5.373. [DOI] [PubMed] [Google Scholar]
- 9.Rubie H, Hartmann O, Michon J, et al. N-Myc gene amplification is a major prognostic factor in localized neuroblastoma: Results of the French NBL 90 study. Neuroblastoma Study Group of the Société Francaise d’Oncologie Pédiatrique. J Clin Oncol. 1997;15:1171–1182. doi: 10.1200/JCO.1997.15.3.1171. [DOI] [PubMed] [Google Scholar]
- 10.De Bernardi B, Gerrard M, Boni L, et al. Excellent outcome with reduced treatment for infants with disseminated neuroblastoma without MYCN gene amplification. J Clin Oncol. 2009;27:1034–1040. doi: 10.1200/JCO.2008.17.5877. [DOI] [PubMed] [Google Scholar]
- 11.Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. N Engl J Med. 1999;341:1165–1173. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 12.Matthay KK. Neuroblastoma: A clinical challenge and biologic puzzle. CA Cancer J Clin. 1995;45:179–192. doi: 10.3322/canjclin.45.3.179. [DOI] [PubMed] [Google Scholar]
- 13.Castleberry RP. Clinical and biologic features in the prognosis and treatment of neuroblastoma. Curr Opin Oncol. 1992;4:116–123. doi: 10.1097/00001622-199202000-00015. [DOI] [PubMed] [Google Scholar]
- 14.De Bernardi B, Conte M, Mancini A, et al. Localized resectable neuroblastoma: Results of the second study of the Italian Cooperative Group for Neuroblastoma. J Clin Oncol. 1995;13:884–893. doi: 10.1200/JCO.1995.13.4.884. [DOI] [PubMed] [Google Scholar]
- 15.De Bernardi B, Nicolas B, Boni L, et al. Disseminated neuroblastoma in children older than one year at diagnosis: Comparable results with three consecutive high-dose protocols adopted by the Italian Co-Operative Group for Neuroblastoma. J Clin Oncol. 2003;21:1592–1601. doi: 10.1200/JCO.2003.05.191. [DOI] [PubMed] [Google Scholar]
- 16.Canete A, Gerrard M, Rubie H, et al. Poor survival for infants with MYCN-amplified metastatic neuroblastoma despite intensified treatment: The International Society of Paediatric Oncology European Neuroblastoma Experience. J Clin Oncol. 2009;27:1014–1019. doi: 10.1200/JCO.2007.14.5839. [DOI] [PubMed] [Google Scholar]
- 17.Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monclair T, Brodeur GM, Ambros PF, et al. The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brisse HJ, McCarville MB, Granata C, et al. Guidelines for imaging and staging of neuroblastic tumors: Consensus report from the International Neuroblastoma Risk Group Project. Radiology. 2011;261:243–257. doi: 10.1148/radiol.11101352. [DOI] [PubMed] [Google Scholar]
- 20.Cecchetto G, Mosseri V, De Bernardi B, et al. Surgical risk factors in primary surgery for localized neuroblastoma: The LNESG1 study of the European International Society of Pediatric Oncology Neuroblastoma Group. J Clin Oncol. 2005;23:8483–8489. doi: 10.1200/JCO.2005.02.4661. [DOI] [PubMed] [Google Scholar]
- 21.Monclair T, Mosseri V, Cecchetto G, et al. Influence of image-defined risk factors on the outcome of patients with localised neuroblastoma. A report from the LNESG1 study of the European International Society of Paediatric Oncology Neuroblastoma Group. Pediatr Blood Cancer. 2015;62:1536–1542. doi: 10.1002/pbc.25460. [DOI] [PubMed] [Google Scholar]
- 22.Günther P, Holland-Cunz S, Schupp CJ, et al. Significance of image-defined risk factors for surgical complications in patients with abdominal neuroblastoma. Eur J Pediatr Surg. 2011;21:314–317. doi: 10.1055/s-0031-1280824. [DOI] [PubMed] [Google Scholar]
- 23.Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;28:3122–3130. doi: 10.1200/JCO.2009.26.7955. [DOI] [PubMed] [Google Scholar]
- 24.Oberthuer A, Hero B, Berthold F, et al. Prognostic impact of gene expression-based classification for neuroblastoma. J Clin Oncol. 2010;28:3506–3515. doi: 10.1200/JCO.2009.27.3367. [DOI] [PubMed] [Google Scholar]
- 25.Schramm A, Köster J, Assenov Y, et al. Mutational dynamics between primary and relapse neuroblastomas. Nat Genet. 2015;47:872–877. doi: 10.1038/ng.3349. [DOI] [PubMed] [Google Scholar]
- 26.Ackermann S, Cartolano M, Hero B, et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science. 2018;362:1165–1170. doi: 10.1126/science.aat6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maris JM, Guo C, Blake D, et al. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med Pediatr Oncol. 2001;36:32–36. doi: 10.1002/1096-911X(20010101)36:1<32::AID-MPO1009>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 28.Bordow SB, Haber M, Madafiglio J, et al. Expression of the multidrug resistance-associated protein (MRP) gene correlates with amplification and overexpression of the N-myc oncogene in childhood neuroblastoma. Cancer Res. 1994;54:5036–5040. [PubMed] [Google Scholar]
- 29.Maris JM, Guo C, White PS, et al. Allelic deletion at chromosome bands 11q14-23 is common in neuroblastoma. Med Pediatr Oncol. 2001;36:24–27. doi: 10.1002/1096-911X(20010101)36:1<24::AID-MPO1007>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 30.Fong CT, White PS, Peterson K, et al. Loss of heterozygosity for chromosomes 1 or 14 defines subsets of advanced neuroblastomas. Cancer Res. 1992;52:1780–1785. [PubMed] [Google Scholar]
- 31.Bown N, Cotterill S, Lastowska M, et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 32.Brodeur GM, Nakagawara A, Yamashiro DJ, et al. Expression of TrkA, TrkB and TrkC in human neuroblastomas. J Neurooncol. 1997;31:49–55. doi: 10.1023/a:1005729329526. [DOI] [PubMed] [Google Scholar]
- 33.Reale MA, Reyes-Mugica M, Pierceall WE, et al. Loss of DCC expression in neuroblastoma is associated with disease dissemination. Clin Cancer Res. 1996;2:1097–1102. [PubMed] [Google Scholar]
- 34.Norris MD, Bordow SB, Marshall GM, et al. Expression of the gene for multidrug-resistance-associated protein and outcome in patients with neuroblastoma. N Engl J Med. 1996;334:231–238. doi: 10.1056/NEJM199601253340405. [DOI] [PubMed] [Google Scholar]
- 35.Attiyeh EF, London WB, Mossé YP, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 36.Wang Q, Diskin S, Rappaport E, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–6062. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 37.Brodeur GM, Pritchard J, Berthold F, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 38.Castleberry RP, Shuster JJ, Smith EI. The Pediatric Oncology Group experience with the international staging system criteria for neuroblastoma. J Clin Oncol. 1994;12:2378–2381. doi: 10.1200/JCO.1994.12.11.2378. [DOI] [PubMed] [Google Scholar]
- 39.Strother DR, London WB, Schmidt ML, et al. Outcome after surgery alone or with restricted use of chemotherapy for patients with low-risk neuroblastoma: Results of Children’s Oncology Group study P9641. J Clin Oncol. 2012;30:1842–1848. doi: 10.1200/JCO.2011.37.9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baker DL, Schmidt ML, Cohn SL, et al. Outcome after reduced chemotherapy for intermediate-risk neuroblastoma. N Engl J Med. 2010;363:1313–1323. doi: 10.1056/NEJMoa1001527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreissman SG, Seeger RC, Matthay KK, et al. Purged versus non-purged peripheral blood stem-cell transplantation for high-risk neuroblastoma (COG A3973): A randomised phase 3 trial. Lancet Oncol. 2013;14:999–1008. doi: 10.1016/S1470-2045(13)70309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nickerson HJ, Matthay KK, Seeger RC, et al. Favorable biology and outcome of stage IV-S neuroblastoma with supportive care or minimal therapy: A Children’s Cancer Group study. J Clin Oncol. 2000;18:477–486. doi: 10.1200/JCO.2000.18.3.477. [DOI] [PubMed] [Google Scholar]
- 43.Katzenstein HM, Bowman LC, Brodeur GM, et al. Prognostic significance of age, MYCN oncogene amplification, tumor cell ploidy, and histology in 110 infants with stage D(S) neuroblastoma: The Pediatric Oncology Group experience—A Pediatric Oncology Group study. J Clin Oncol. 1998;16:2007–2017. doi: 10.1200/JCO.1998.16.6.2007. [DOI] [PubMed] [Google Scholar]
- 44.London WB, Castleberry RP, Matthay KK, et al. Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children’s Oncology Group. J Clin Oncol. 2005;23:6459–6465. doi: 10.1200/JCO.2005.05.571. [DOI] [PubMed] [Google Scholar]
- 45.Maris JM, Hogarty MD, Bagatell R, et al. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 46.Shimada H, Umehara S, Monobe Y, et al. International neuroblastoma pathology classification for prognostic evaluation of patients with peripheral neuroblastic tumors: A report from the Children’s Cancer Group. Cancer. 2001;92:2451–2461. doi: 10.1002/1097-0142(20011101)92:9<2451::aid-cncr1595>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 47.Ladenstein R, Pötschger U, Pearson ADJ, et al. Busulfan and melphalan versus carboplatin, etoposide, and melphalan as high-dose chemotherapy for high-risk neuroblastoma (HR-NBL1/SIOPEN): An international, randomised, multi-arm, open-label, phase 3 trial. Lancet Oncol. 2017;18:500–514. doi: 10.1016/S1470-2045(17)30070-0. [DOI] [PubMed] [Google Scholar]
- 48.Kohler JA, Rubie H, Castel V, et al. Treatment of children over the age of one year with unresectable localised neuroblastoma without MYCN amplification: Results of the SIOPEN study. Eur J Cancer. 2013;49:3671–3679. doi: 10.1016/j.ejca.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Naranjo A, Irwin MS, Hogarty MD, et al. Statistical framework in support of a revised Children’s Oncology Group neuroblastoma risk classification system. JCO Clin Cancer Inform. 2018;2:1–15. doi: 10.1200/CCI.17.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janoueix-Lerosey I, Schleiermacher G, Michels E, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 51.Schleiermacher G, Michon J, Ribeiro A, et al. Segmental chromosomal alterations lead to a higher risk of relapse in infants with MYCN-non-amplified localised unresectable/disseminated neuroblastoma (a SIOPEN collaborative study) Br J Cancer. 2011;105:1940–1948. doi: 10.1038/bjc.2011.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shimada H, Ambros IM, Dehner LP, et al. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 53.Sokol E, Desai AV, Applebaum MA, et al. Age, diagnostic category, tumor grade, and mitosis-karyorrhexis index are independtly prognostic in neuroblastoma: An INRG project. J Clin Oncol. 2020;38:1906–1918. doi: 10.1200/JCO.19.03285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eleveld TF, Oldridge DA, Bernard V, et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015;47:864–871. doi: 10.1038/ng.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morgenstern DA, Bagatell R, Cohn SL, et al. The challenge of defining “ultra-high-risk” neuroblastoma. Pediatr Blood Cancer. 2019;66:e27556. doi: 10.1002/pbc.27556. [DOI] [PubMed] [Google Scholar]
- 56.Moreno L, Dongjing G, Irwin M, et al. A nomogram of clinical and biologic factors to predict survival in children newly diagnosed with high-risk neuroblastoma: An International Neuroblastoma Risk Group project. Pediatr Blood Cancer. doi: 10.1002/pbc.28794. (in press. [DOI] [PubMed] [Google Scholar]
- 57.Morgenstern DA, Pötschger U, Moreno L, et al. Risk stratification of high-risk metastatic neuroblastoma: A report from the HR-NBL-1/SIOPEN study. Pediatr Blood Cancer. 2018;65:e27363. doi: 10.1002/pbc.27363. [DOI] [PubMed] [Google Scholar]
- 58.Abernethy AP, Etheredge LM, Ganz PA, et al. Rapid-learning system for cancer care. J Clin Oncol. 2010;28:4268–4274. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park JR, Bagatell R, Cohn SL, et al. Revisions to the International Neuroblastoma Response Criteria: A consensus statement from the National Cancer Institute clinical trials planning meeting. J Clin Oncol. 2017;35:2580–2587. doi: 10.1200/JCO.2016.72.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgenstern DA, London WB, Stephens D, et al. Metastatic neuroblastoma confined to distant lymph nodes (stage 4N) predicts outcome in patients with stage 4 disease: A study from the International Neuroblastoma Risk Group Database. J Clin Oncol. 2014;32:1228–1235. doi: 10.1200/JCO.2013.53.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Volchenboum SL, Cox SM, Heath A, et al. Data commons to support pediatric cancer research. Am Soc Clin Oncol Educ Book. 2017;37:746–752. doi: 10.1200/EDBK_175029. [DOI] [PubMed] [Google Scholar]
- 62.International Neuroblastoma Risk Group http://inrgdb.org/
- 63.Volchenboum Lab https://commons.cri.uchicago.edu/
- 64.National Cancer Institute Office of Cancer Genomics Therapeutically Applicable Research to Generate Effective Treatment (TARGET) https://ocg.cancer.gov/programs/target
- 65.Gabriella Miller Kids First Data Resource Center doi: 10.1093/jncics/pkad079. https://kidsfirstdrc.org/ [DOI] [PMC free article] [PubMed]
- 66.Safran C, Bloomrosen M, Hammond WE, et al. Toward a national framework for the secondary use of health data: An American Medical Informatics Association White Paper. J Am Med Inform Assoc. 2007;14:1–9. doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khozin S, Miksad RA, Adami J, et al. Real-world progression, treatment, and survival outcomes during rapid adoption of immunotherapy for advanced non-small cell lung cancer. Cancer. 2019;125:4019–4032. doi: 10.1002/cncr.32383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu J, Xue K, Zhang K. Current status and future trends of clinical diagnoses via image-based deep learning. Theranostics. 2019;9:7556–7565. doi: 10.7150/thno.38065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hosny A, Parmar C, Quackenbush J, et al. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kong J, Sertel O, Shimada H, et al. Computer-aided evaluation of neuroblastoma on whole-slide histology images: Classifying grade of neuroblastic differentiation. Pattern Recognit. 2009;42:1080–1092. doi: 10.1016/j.patcog.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oberthuer A, Juraeva D, Hero B, et al. Revised risk estimation and treatment stratification of low- and intermediate-risk neuroblastoma patients by integrating clinical and molecular prognostic markers. Clin Cancer Res. 2015;21:1904–1915. doi: 10.1158/1078-0432.CCR-14-0817. [DOI] [PubMed] [Google Scholar]
- 74.Defferrari R, Mazzocco K, Ambros IM, et al. Influence of segmental chromosome abnormalities on survival in children over the age of 12 months with unresectable localised peripheral neuroblastic tumours without MYCN amplification. Br J Cancer. 2015;112:290–295. doi: 10.1038/bjc.2014.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Twist CJ, Schmidt ML, Naranjo A, et al. Maintaining outstanding outcomes using response- and biology-based therapy for intermediate-risk neuroblastoma: A report from the Children’s Oncology Group study ANBL0531. J Clin Oncol. 2019;37:3243–3255. doi: 10.1200/JCO.19.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]