Abstract
PURPOSE
ASCO, through its wholly owned subsidiary, CancerLinQ LLC, developed CancerLinQ, a learning health system for oncology. A learning health system is important for oncology patients because less than 5% of patients with cancer enroll in clinical trials, leaving evidence gaps for patient populations not enrolled in trials. In addition, clinical trial populations often differ from the overall cancer population with respect to age, race, performance status, and other clinical parameters.
MATERIALS AND METHODS
Working with subscribing practices, CancerLinQ accepts data from electronic health records and transforms the local representation of a patient’s care into a standardized representation on the basis of the Quality Data Model from the National Quality Forum. CancerLinQ provides this information back to the subscribing practice through a series of tools that support quality improvement. CancerLinQ also creates de-identified data sets for secondary research use.
RESULTS
As of March 2020, CancerLinQ includes data from 63 organizations across the United States that use nine different electronic health records. The database includes 1,426,015 patients with a primary cancer diagnosis, of which 238,680 have had additional information abstracted from unstructured content.
CONCLUSION
As CancerLinQ continues to onboard subscribing practices, the breadth of potential applications for a learning health care system widen. Future practice-facing tools could include real-world data visualization, recommendations for treatment of patients with actionable genetic variations, and identification of patients who may be eligible for clinical trials. Feeding these insights back into oncology practice ensures that we learn how to treat patients with cancer not just on the basis of the selective experience of the 5% that enroll in clinical trials, but from the real-world experience of the entire spectrum of patients with cancer in the United States.
INTRODUCTION
In 2009, the Health Information Technology for Economic and Clinical Health Act was enacted as part of the American Recovery and Reinvestment Act (Public Law 111-5).1 Title VI of the Health Information Technology for Economic and Clinical Health Act provides financial incentives to medical practices for the adoption of electronic health records (EHRs). Since then, adoption of EHRs in US medical practices has increased significantly.2 Widespread adoption of EHRs permits the creation of learning health care systems in the United States. The Institute of Medicine defines a learning health care system “in which science and informatics, patient-clinician partnerships, incentives, and culture are aligned to promote and enable continuous and real-time improvement in both the effectiveness and efficiency of care.”3(p17) In short, a learning health care system uses real-world data (RWD) to improve patient care at the point of care.
CONTEXT
Key Objective
CancerLinQ is the only nonprofit, physician-led, big-data platform in cancer. In this article, we describe the process by which we ingest electronic health record (EHR) data. This information is relevant to entities that want to build learning health care systems and researchers interested in using the CancerLinQ Discovery databases.
Knowledge Generated
By adopting an EHR-agnostic approach to data ingestion, CancerLinQ created a scalable data pipeline that ingests clinically relevant structured and unstructured data from participating medical oncology organizations. CancerLinQ currently includes data from 63 organizations that use nine different EHRs. The database includes 1,426,015 patients with a primary cancer diagnosis, of which 238,680 have had additional information abstracted from unstructured content.
Relevance
As CancerLinQ grows, the number of potential applications for a learning health care system and real-world research grows. Expanding on the current data pipeline ensures that the oncology health care ecosystem learns from all patients, not just those enrolled in clinical trials.
The Institute of Medicine focused on oncology because it is ripe for the development of a learning health care system. Less than 5% of patients with cancer enroll in clinical trials,4 and clinical trial populations typically differ from the overall cancer population with respect to age, race, performance status, and other important clinical parameters. More than 1.7 million new patients with cancer are diagnosed in the United States annually, but as a result of diverse anatomic locations, driver mutations, and extent of disease spread, treatment is heterogenous. Consequently, many patients with cancer receive off-label treatments that have not been approved by the US Food and Drug Administration or may not be consistent with accepted guidelines.5
ASCO, through its wholly owned subsidiary, CancerLinQ LLC, created CancerLinQ6 in response to this need. Medical oncology organizations in the United States with at least one ASCO member are eligible to participate in CancerLinQ. CancerLinQ extracts data directly from EHRs or supporting data warehouses (the data from warehouses are native EHR data) of subscribing organizations, aggregates and harmonizes the data, and provides the data back to the subscriber for health care operations and quality improvement purposes. CancerLinQ also provides the practices with a dashboard of electronic clinical quality measures (and an electronic pathway toward achieving Quality Oncology Practice Initiative Certification), tools to assist physicians who have challenging or unusual cases, and reports to support business decisions. Most of the EHR systems used by CancerLinQ sites are not oncology specific; therefore, the processes described in this manuscript could be broadly applicable to learning health systems for other therapeutic areas.
As of March 2020, more than 100 oncology care delivery organizations have signed participation and business associate agreements with CancerLinQ. This publication summarizes the activation process for the first 63 oncology organizations that contribute data from nine EHR systems.
CancerLinQ DATA ARCHITECTURE
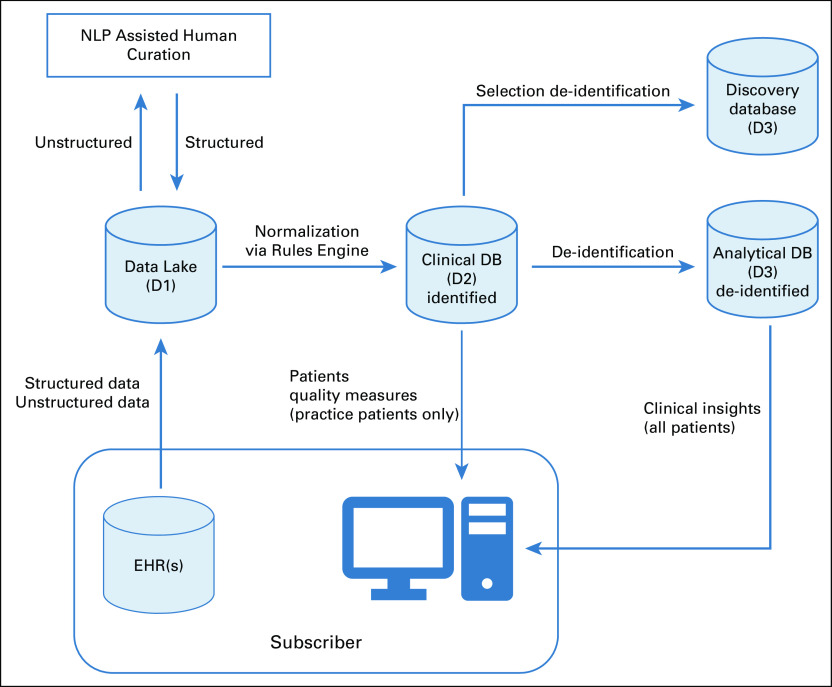
CancerLinQ uses a series of data repositories to support participating subscribers and secondary use researchers (Fig 1). Practice data are extracted via query from the underlying database (pull), or the practice puts the data into an agreed upon format and pushes it to CancerLinQ. CancerLinQ uses Jitterbit (Alameda, CA) to develop and maintain these connections and templates between CancerLinQ and each subscriber’s EHR. Practices that push data receive templates from CancerLinQ partner Jitterbit. Whereas CancerLinQ performs quality control checks on the inbound data, the organization is ultimately responsible for the queries that generate the data and for ensuring data completeness. Once extracted, data are converted to a JavaScript Object Notation format and then landed in a secure file transfer protocol site for additional processing. Pull practices provide nightly data feeds, whereas push practices set the schedule for data feeds.
FIG 1.
High-level data assets and flows within CancerLinQ. DB, database; EHR, electronic health record; NLP, natural language processing.
Data are then processed into the Data Lake or D1. Master D1 data are implemented as an Amazon S3 structure and also presented in Amazon Redshift (Amazon Web Services, Seattle, WA). D1 is not harmonized or standardized in any meaningful way from its initial structure and content in the local EHR or warehouse. This flexible design allows the data to convey the attribute names, detailed values, and its implicit and explicit relationships from the underlying EHR, supporting data provenance. Content varies greatly among EHR vendors. The complexity is compounded by practice customization. D1 contains protected health information (PHI) as defined by the Health Insurance Portability and Accountability Act of 1996 (HIPAA), such as names, addresses, medical record numbers, etc, as well as information about the specific practice that sent the data and the name(s) of treating physicians.
The Clinical Database (D2) is the point at which aggregated data are available to CancerLinQ subscribers. Data from D1 are processed into D2 via a set of proprietary rules into a common information model. This includes deduplication of patients via an electronic master patient index, transformation of the data structure, and conversion of local data values—for example, drug names, laboratory test names, specific clinical evaluation names, etc—into standard representations via a process termed codification. Data in D2 retain both the original value from the EHR—for example, total neutrophil count—and the harmonized value—for example, neutrophils—and contain PHI. Quality measures for participating organizations and their individual physicians are calculated using the D2 codified data. Because D2 data contain PHI, access is restricted to the respective participating practices that provided the data. D2 is implemented in Amazon S3 and Redshift. Figure 2 illustrates an example of a quality improvement report.
FIG 2.
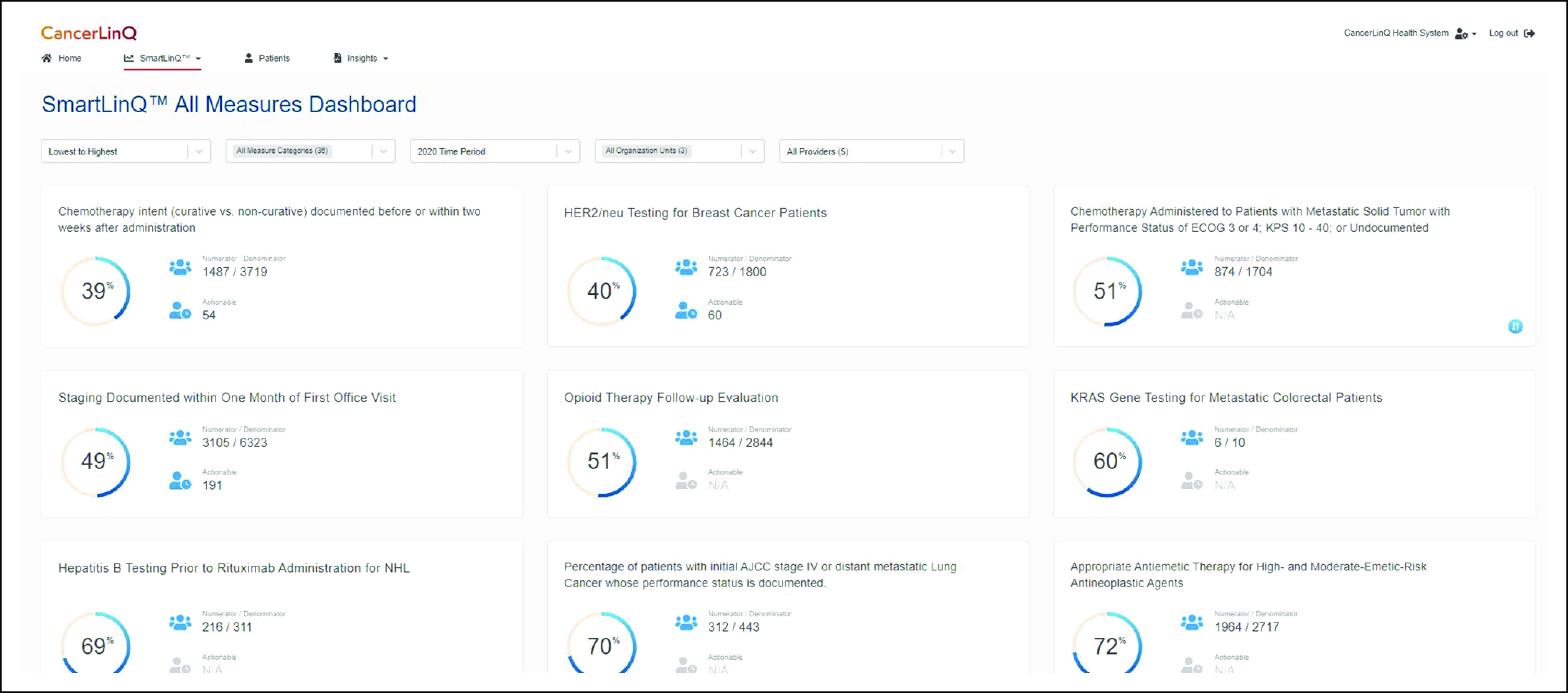
CancerLinQ quality measure dashboard.
The Analytical Databases (D3s) are de-identified representations of D2. Access to D3 is provided to subscribing organizations through tools embedded in the CancerLinQ application for health care operations and clinical quality improvement. Unlike D2, where practices can only see their own patients’ data, subscribers are granted access—via the application—to de-identified data from all practices so that they can benefit from collective experience. CancerLinQ primarily uses Expert Determination (HIPAA privacy rule § 164.514(b)(1)) as its de-identification method, although Safe Harbor (§ 164.514(b)(2)) is used for some data sets. Expert Determination requires that “a person with appropriate knowledge of and experience with generally accepted statistical and scientific principles and methods for rendering information not individually identifiable: (i) Applying such principles and methods, determines that the risk is very small that the information could be used, alone or in combination with other reasonably available information, by an anticipated recipient to identify an individual who is a subject of the information; and (ii) Documents the methods and results of the analysis that justify such determination.”7,8 CancerLinQ utilizes Privacy Analytics Eclipse software (Privacy Analytics, Ottawa, ON, Canada) to perform Expert Determination de-identification. Practice-facing D3s are implemented in Amazon Redshift, and access is via a data exploration tool.
The CancerLinQ Discovery (CLQD) program allows researchers to conduct real-world evidence studies on CancerLinQ’s de-identified data. CLQD data sets are provided as D3’s, typically as a tumor site–specific subset for a given research purpose. Researchers from CancerLinQ subscribing organizations, academic institutions, medical specialty societies, government entities, and others can access these de-identified data sets via CancerLinQ. For-profit entities, including life sciences companies and payers, must access CancerLinQ Discovery data through two coexclusive licensees: ConcertAI (Boston, MA) and Tempus Labs (Chicago, IL). Approved data sets are implemented in purpose-built Amazon Redshift instances in Amazon Web Services environments. CancerLinQ has migrated CLQD to a new environment that will provide additional functionality for researchers. Full up-to-date details about the CLQD program are available online.9 Identifiable data in CancerLinQ are collected from the contributing practices for quality improvement purposes, and uses of CancerLinQ data, once de-identified, do not themselves constitute human subjects research activities, a determination made by an independent institutional review board evaluating CancerLinQ’s regulatory framework in 2013.10 Although obtaining individual patient authorization for the use of de-identified data is not required under HIPAA, end users of CLQD data sets are responsible for managing any additional institutional review board requirements at their institutions.
CancerLinQ DATA MODEL
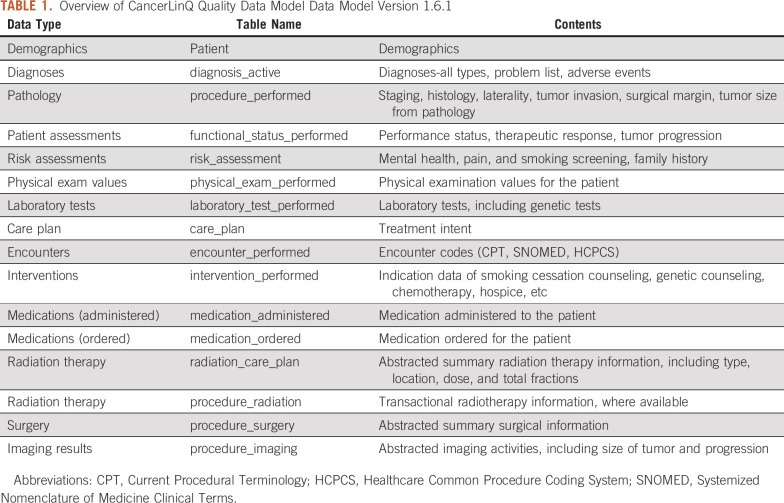
In 2009, the National Quality Forum established a quality data model (QDM) to describe clinical concepts and provide a common framework for electronic performance measurement. The QDM is now maintained by the Centers for Medicare & Medicaid Services and the Office of the National Coordinator for Health Information Technology.11 CancerLinQ adopted an expanded version of the QDM as its D2 and D3 clinical data model.
The CancerLinQ QDM (Table 1) schema is built around the patient. CancerLinQ collects PHI on patients for two purposes: to provide meaningful data back to treating physicians at the practice(s), and to identify patients who have been treated at multiple CancerLinQ participating facilities to create a longitudinal patient record for patient care. Provider information is collected and used to calculate quality measures for each practice and treating physician. Providers are only shown their subscribing organization–specific identified data in D2. They are also able to view the de-identified D3 data through a data exploration tool in the CancerLinQ platform. This D3 includes de-identified longitudinal data for all patients.
TABLE 1.
Overview of CancerLinQ Quality Data Model Data Model Version 1.6.1
QDM elements are described with three discrete units of information: data types, value sets, and code systems. In D2, this means that most key elements are represented as quadruplets: an original value, a coded value, a code system, and a code label. For example, stage group values are stored in the procedure_performed table as stagegroup (original value; eg, ‘IIA’), stagegroup_code (261614003), stagegroup_codesystem (SNOMED CT), and stagegroup_codelabel (‘Stage 2A’). We use established terminologies to represent value sets and coding systems, depending on the data type of interest—for instance, Logical Observation Identifiers Names and Codes for Laboratory Tests,12 RxNorm codes for Medications,13,14 and International Classification of Disease (ICD) codes for diagnoses.15 Where available, CancerLinQ has retained specific codes from the source data—for example, Logical Observation Identifiers Names and Codes in certain EHRs—and codified additional structured data elements as the data are transferred from D1 into D2. Data codification is of central importance as the quality measures require codified values to operate correctly, and de-identified research data sets are only be derived from codified values.
The QDM used by CancerLinQ is not fully congruent with the current version of the Centers for Medicare & Medicaid Services QDM. It has been modified to support a variety of clinically relevant oncology data elements that are not well supported by the base model. We have thus adapted several variant categories and data types, including Procedure, Imaging; Procedure, Surgery; Procedure, Radiation; and Radiation Care Plan tables. These tables were derived by making a more specific version of the Procedure Performed table and the Care Plan tables from the QDM. Given the high value of these data elements to patient care and quality improvement, the need to model additional curated attributes and the necessity of supporting a degree of backward compatibility for applications built on the previous CancerLinQ QDM, we determined that these changes added value to D1 and D2 without disrupting the core QDM.
Patients in CancerLinQ often have multiple diagnoses. The diagnosis table contains the complete problem list as structured data. The treating oncologist enters the data, including tumor types and noncancer diagnoses. The completeness of these noncancer diagnoses varies across practices and physicians as a result of differences in documentation practices. In addition, some patients have multiple cancer diagnoses. This may be because of the actual presence of multiple primary tumors in a single patient, but in some cases, metastases are initially or incorrectly coded as primary tumors. Specificity of a diagnosis code may also vary. For example, a patient with a lung tumor might have ICD-10 code C34 (malignant neoplasm of bronchus or lung), C34.1 (malignant neoplasm of upper lobe, bronchus, or lung), or C34.12 (malignant neoplasm of upper lobe, left bronchus, or lung), depending on the recording practices of the individual physician. This coding variability means that certain analyses—for example, patient counts by anatomic site—will yield a sum greater than the total number of patients, but it also provides both an accurate record of the data in the EHR and allows maximum flexibility to manage the data in ways that support specific uses for health care operations and/or research.
TRANSFORMATION OF D1 TO D2
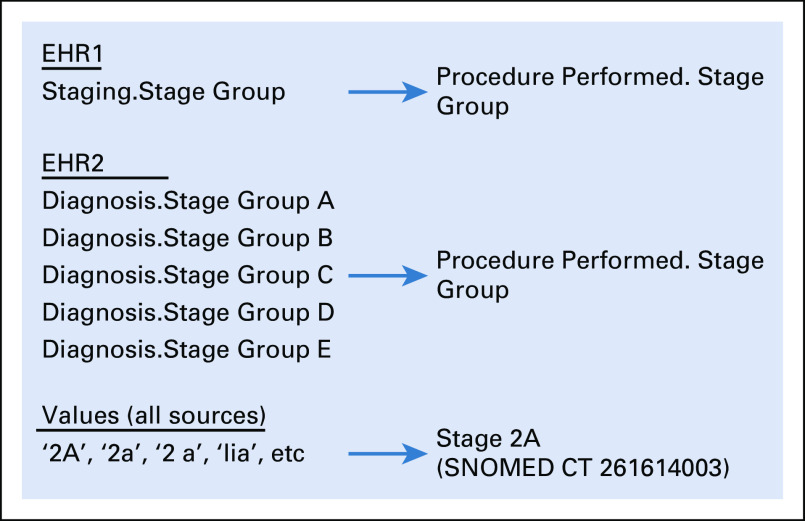
Transformation of D1 data into D2 is carried out via a proprietary rules engine. This engine takes input data types from the EHR and transforms them into output data types that are consistent with the QDM model, as described above. Two major transformations occur: conversion of the input data element type and transformation of the input data element value where it can be matched to a standard term. Rules can be generic for all subscribers, specific to an EHR, or specific to a specific implementation of an EHR. An example related to cancer staging is shown in Figure 3. In some EHR systems, the stage group data element is maintained as a single value in a single element in the underlying database, represented as Staging.Stage Group in the example. The rules engine takes all of these input values and maps them to the same QDM data element (procedure_performed.stagegroup). These transformations are of the first category—for example, mapping the data element type. In addition, the representation of the values requires modification. Figure 3 shows examples of mapping of these values, converting a variety of representations, such as 2A, 2a, 2 a, II A, etc, to the standard SNOMED CT representation of Stage 2A.
FIG 3.
Transformations within the rules engine. EHR, electronic health record.
PRACTICE ACTIVATION PROCESS
Practice activation is the process of integrating EHR data and/or data from other source systems—for example, secondary systems or enterprise data warehouses—in its native state into CancerLinQ. This is a cooperative effort between the subscribing practice and CancerLinQ staff. The complexity of the task varies depending on the number of EHRs, end points, and the degree of customization of each system. Success depends on a practice staff member, typically a physician champion, who serves as the liaison between the practice and CancerLinQ, the practice IT support group (when applicable), CancerLinQ staff, and the Jitterbit team that installs the data access connector. Activation time varies depending on the amount of EHR customization at each site and the level of local support available to develop queries to extract information. The process is shown in Figure 4. There are several key checkpoints where the data and transformations are validated. The final step is a review of the practice’s clinical quality measures with staff at the site. The rate of incremental updates is generally determined by the needs of the subscriber and the method—push versus pull—and varies from daily to monthly.
FIG 4.
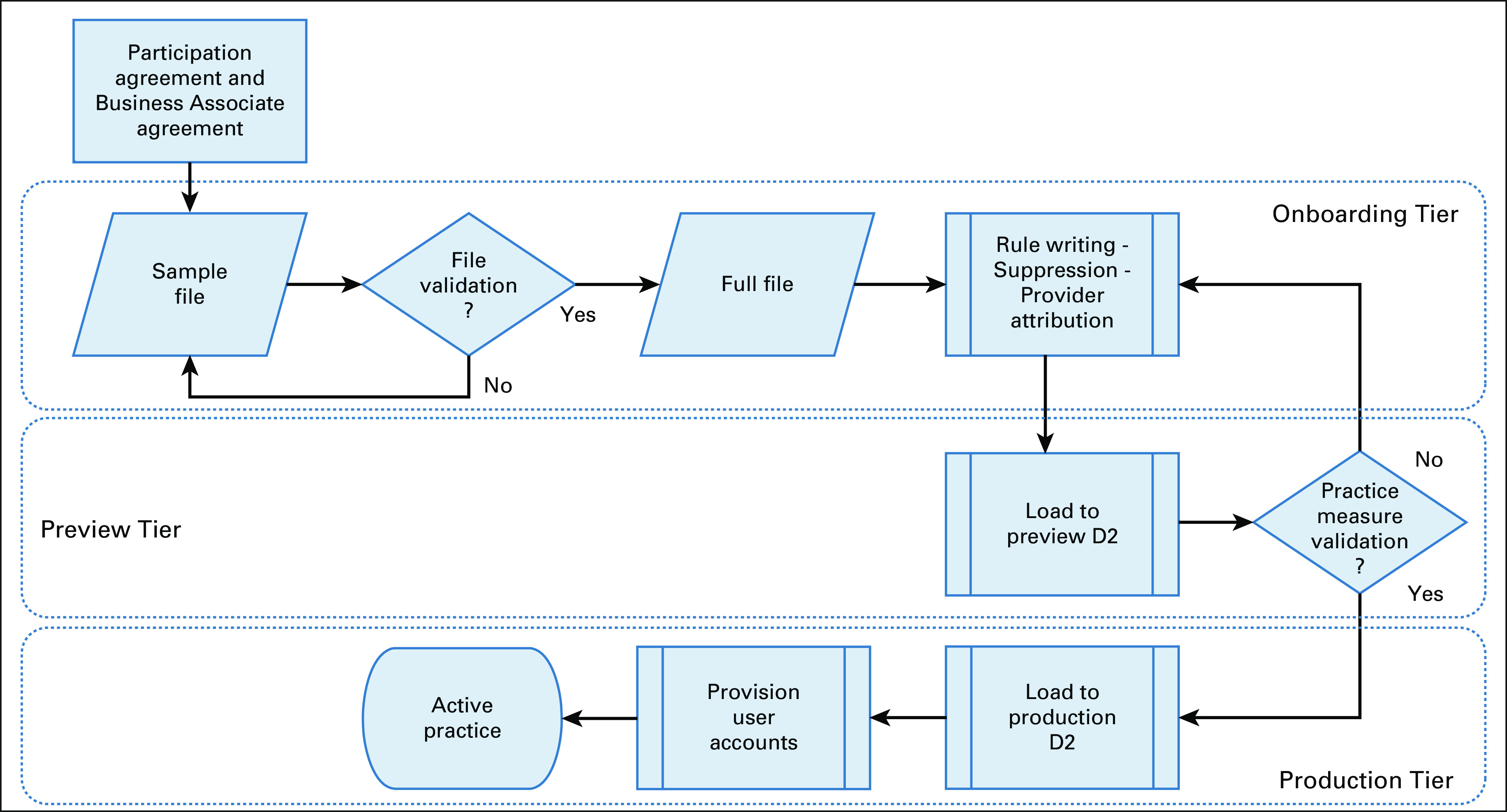
CancerLinQ activation process. Practices begin by signing a participation agreement and business associate agreement. Note that some electronic health records do not require a sample file and validation is completed on the full file.
NATURAL LANGUAGE PROCESSING–ASSISTED HUMAN CHART ABSTRACTION
EHRs contain unstructured information, such as free text notes created within the EHR and scanned documents obtained from entities outside of the clinical practice—for example, surgical reports and molecular pathology reports. CancerLinQ has access to this content in varying degrees, depending on the EHR vendor and the willingness/ability of the site to make external scanned documents accessible. Trained health care professionals extract unstructured data by applying predefined chart abstraction business rules via user interfaces that use natural language processing to help identify important clinical facts. The abstracted content, which is now structured, is deposited into D1 and then processed like any other structured data.
Natural language processing–assisted abstraction is performed by CancerLinQ business associate subcontractors. All work performed on PHI, including curation work, is firewalled from work related to research. The limiting factors for high-volume, high-fidelity data abstraction are cost and the need for a sufficiently trained workforce to manage this task at scale. CancerLinQ, therefore, has focused curation efforts on important oncology data elements that are sourced at low rates from the structured content. CancerLinQ and its curation contractors focus on specific tumor types for which abstraction will more accurately reflect quality measure scores or for which there are significant opportunities for quality improvement as a result of active research. As of March 2020, the CancerLinQ database includes abstracted data for cancers of the lung, breast, ovary, pancreas, prostate, colon, rectum, liver, kidney, bladder, skin (melanoma), and chronic lymphocytic leukemia/small lymphocytic lymphoma.
The first tumor type selected for abstraction was non–small-cell lung cancer (NSCLC), chosen because of the many new targeted agents and immunotherapies approved in recent years and under investigation for this entity. Assessing NSCLC treatment requires many data elements that are not well captured by structured data in EHRs, particularly imaging, radiation therapy, surgery, and molecular tests. Even identifying the diagnosis of NSCLC requires the examination of histology data not typically present in structured content as ICD-9/10 codes are focused on anatomic location and not tumor morphology.
Identifying and characterizing adverse events (AEs) is crucial for quality improvement and research, but AEs pose a particular challenge for data abstraction. Most patients will experience at least one AE during treatment, and an occurrence of an AE in a given patient may be recorded in a discrete field in the EHR (eg, as a diagnosis in the problem list), in an unstructured text field or document, in both, or not at all. Limiting the capture of AEs to a list of expected toxicities is a simplistic solution, but would greatly restrict the value of the data for identifying newly described and emerging toxicities. Conversely, attempting to abstract all AEs, especially commonly occurring ones, such as nausea or alopecia, from RWD is not a judicious use of resources. CancerLinQ, therefore, abstracts AEs that are associated with a sentinel event. Sentinel events include change of therapy, discontinuation of therapy, emergency room visits, hospitalization, and death. Discontinuation of an individual drug, whether it is administered as a single agent or as part of a multidrug regimen, is considered a sentinel event.
Abstractors record AEs that occur within 30 days of sentinel events, but do not make specific assertions about causality. AEs occurring around sentinel events are more likely to correspond to grade 3 or higher toxicity as defined by the National Cancer Institute’s Common Terminology Criteria for Adverse Events.16
SUMMARY OF THE CancerLinQ D2 DATABASE
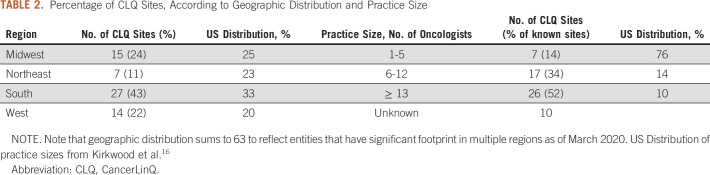
As of March 2020, the CancerLinQ D2 includes 1,426,015 patients with a primary cancer diagnosis, 733,107 patients with a primary diagnosis of a benign or in situ neoplasm, and 893,977 patients with a benign hematology diagnosis. The total patient population is 2,546,350, and 238,680 patients have chart-abstracted data. Note that patients with multiple diagnoses will be counted more than once in these totals. Table 2 summarizes the geographic distribution and practice size of active CancerLinQ sites.
TABLE 2.
Percentage of CLQ Sites, According to Geographic Distribution and Practice Size
DISCUSSION AND FUTURE DIRECTIONS
The CancerLinQ learning platform provides feedback to practices via e-measures that support practice improvements in health care operations and quality of care. Although this is a critical first step to building an oncology learning health system, it is certainly not the last. Over the next several years, we intend to increase the depth and breadth of quality improvement services that are available to subscribers, synthesize the data into more usable forms, and develop tools to support our subscribers’ clinical needs.
CancerLinQ prioritizes the codification and curation of data elements on the basis of its current health care operations and quality improvement services. As we gain experience transforming data from more EHR types, we anticipate integrating a more robust set of data elements. For example, with more integrated health systems joining, we will have the ability to bring in additional content, especially surgical and radiation oncology data. Integrating radiation oncology data is technically feasible, and because the market for such systems is less fragmented than that for transactional EHRs in medical oncology practices, and much radiation oncology data already exist in structured form, there may be fewer barriers to its integration. Genomics data are usually available in structured form within the bioinformatics pipelines at the testing laboratories, but typically are delivered to the EHR in the form of text—that is, PDF reports or scanned images. Integration with the source laboratory systems would reduce the amount of curation required and provide better value back to the practice.18 Future work will focus on the integration of these data sources into CancerLinQ.
CancerLinQ data are transactional in nature. It is a temporally organized set of medical observations and interventions relating to a single patient. Whereas we can transform any locally defined set of such transactional data into a standardized, standards-based representation of that data, realizing the full potential of CancerLinQ requires that we transform these data into knowledge. The first step is synthesizing the transactional data into a form more relevant to physicians. To accomplish this goal, we have begun to convert transactional drug order/administration events into regimens and lines of therapy. For example, physicians treating patients with advanced lung cancer are likely to want to identify patients who have received the specific standardized regimen of platinum doublet therapy rather than look for patients who have received certain numbers of either carboplatin or cisplatin concurrent with pemetrexed, etoposide, paclitaxel, etc, within certain defined time periods.
ASCO, CancerLinQ, the MITRE Corporation, the Alliance for Clinical Trials in Oncology Foundation, and the American Society for Radiation Oncology have developed a specification known as mCODE, Minimal Common Oncology Data Elements, based in part on the experience of CancerLinQ.19 The mCODE initiative envisions that EHRs would implement this standards-based model and be capable of delivering relevant information across systems using the Fast Healthcare Interoperability Resources standard.20 If fully implemented in EHR systems, and if physicians and other health care providers consistently record the data in structured form, it will substantially improve cancer data interoperability for all participants in the oncology ecosystem, especially patients, and it will enable CancerLinQ to leverage data more easily for quality of care.21
As CancerLinQ grows, the breadth of potential applications for a learning health care system widens. Future practice-facing tools could include RWD visualizations, treatment of patients with actionable genetic variations, and supporting treatment of unique or complex cases. Feeding these insights back into oncology practice ensures that we learn how to treat patients with cancer not just on the basis of the selective experience of the 5% who enroll in clinical trials, but from the real-world experience of the entire spectrum of patients with cancer in the United States.
ACKNOWLEDGMENT
The authors thank Yared Aragaw, Jeff Brown, Eric Chen, Meron Dantew, Dev Haberman, Anum Habib, Sirisha Kakamada, Manoj Kumar, Linh Mekuria, Jose Mena, MD, Shiblee Noonan, Trang Pham, Kruthi Rao, Kopila Sharma, Philip Stoeber, Arpitha Thakkalapally, Vamsidhar Meda Venkata, Alpna Wayal, Peter Wachira, Clyde Wentling, and Michael Zhao. In addition, the authors thank the CancerLinQ Development, DevOps, Implementation, and Product teams for their support.
SUPPORT
Supported by the direct expenditure of the American Society for Clinical Oncology.
W.S.R. participated in this work before joining the US Food and Drug Administration (FDA). This work and related conclusions reflect the independent work of study authors and do not necessarily represent the views of the FDA or US government.
AUTHOR CONTRIBUTIONS
Conception and design: Danielle Potter, Raven Brothers, Jacob E. Koskimaki, Amy McNutt, Robert S. Miller, Jatin Nagda, Anil Nair, Wendy S. Rubinstein, Andrew K. Stewart, George A. Komatsoulis
Financial support: Danielle Potter
Provision of study materials or patients: George A. Komatsoulis
Collection and assembly of data: Danielle Potter, Andrej Kolacevski, Jacob E. Koskimaki, Amy McNutt, Robert S. Miller, Jatin Nagda, Wendy S. Rubinstein, Andrew K. Stewart, George A. Komatsoulis
Data analysis and interpretation: Danielle Potter, Jacob E. Koskimaki, Robert S. Miller, Jatin Nagda, Wendy S. Rubinstein, Andrew K. Stewart, Iris J. Trieb, George A. Komatsoulis
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Danielle Potter
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Research Funding: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Other Relationship: Tempus, Concerto Health AI
Jacob E. Koskimaki
Patents, Royalties, Other Intellectual Property: Coauthor on a patent filed in 2018 for work as a graduate student: Mimetic Peptides Derived From Collagen Type IV and Their Use for Treating Angiogenesis- and Lymphangiogenesis- Dependent Disease, Patent number: 10106597
Jatin Nagda
Stock and Other Ownership Interests: InVitae
Iris J. Trieb
Stock and Other Ownership Interests: Johnson & Johnson
No other potential conflicts of interest were reported.
REFERENCES
- 1.111th US Congress American Recovery and Reinvestment Act of 2009. https://www.govinfo.gov/content/pkg/PLAW-111publ5/pdf/PLAW-111publ5.pdf
- 2.Health IT Dashboard. https://dashboard.healthit.gov/quickstats/pages/physician-ehr-adoption-trends.php
- 3.Institute of Medicine . Best Care at Lower Cost: The Path to Continuously Learning Health Care in America. Washington, DC: The National Academies Press; 2013. [PubMed] [Google Scholar]
- 4.Unger JM, Cook E, Tai E, et al. The role of clinical trial participation in cancer research: Barriers, evidence, and strategies. Am Soc Clin Oncol Educ Book. 2016;35:185–198. doi: 10.14694/EDBK_156686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saiyed MM, Ong PS, Chew L. Off-label drug use in oncology: A systematic review of literature. J Clin Pharm Ther. 2017;42:251–258. doi: 10.1111/jcpt.12507. [DOI] [PubMed] [Google Scholar]
- 6.Miller RS, Wong JL. Using oncology real-world evidence for quality improvement and discovery: The case for ASCO’s CancerLinQ. Future Oncol. 2018;14:5–8. doi: 10.2217/fon-2017-0521. [DOI] [PubMed] [Google Scholar]
- 7.United States Department of Health and Human Services Guidance regarding methods for de-identification of protected health information in Accordance with the Health Insurance Portability and Accountability Act (HIPAA) privacy rule. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html
- 8.US Government The Health Insurance Portability and Accountability Act (HIPAA) http://purl.fdlp.gov/GPO/gpo10291
- 9.American Society of Clinical Oncology ASCO CancerLinQ Discovery. https://discovery.cancerlinq.org/
- 10.Schilsky RL, Michels DL, Kearbey AH, et al. Building a rapid learning health care system for oncology: The regulatory framework of CancerLinQ. J Clin Oncol. 2014;32:2373–2379. doi: 10.1200/JCO.2014.56.2124. [DOI] [PubMed] [Google Scholar]
- 11.Office of the National Coordinator for Health IT Quality data model, version 4.3. https://ecqi.healthit.gov/system/files/qdm_4_3_508_compliant.pdf
- 12.Regenstrief Institute Home--LOINC. https://loinc.org
- 13.Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18:441–448. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Library of Medicine RxNORM. doi: 10.1080/15360280801989377. https://www.nlm.nih.gov/research/umls/rxnorm/index.html [DOI] [PubMed]
- 15.World Health Organization The ICD-10 classification of mental and behavioural disorders: Clinical descriptions and diagnostic guidelines. https://apps.who.int/iris/handle/10665/37958
- 16.National Cancer Institute Common Terminology Criteria for Adverse Events version 5.0. https://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/CTCAE_v5.0.xlsx
- 17.Kirkwood MK, Hanley A, Bruinooge SS, et al. The state of oncology practice in America, 2018: Results of the ASCO Practice Census Survey. J Oncol Pract. 2018;14:e412–e420. doi: 10.1200/JOP.18.00149. [DOI] [PubMed] [Google Scholar]
- 18.Conway JR, Warner JL, Rubinstein WS, et al. Next-generation sequencing and the clinical oncology workflow: Data challenges, proposed solutions, and a call to action. JCO Precis Oncol. doi: 10.1200/PO.19.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.American Society of Clinical Oncology mCODE: Minimal Common Oncology Data Elements. https://mcodeinitiative.org/
- 20.Health Level 7 FHIR: Release 4. http://hl7.org/fhir/
- 21.Rubinstein WS. CancerLinQ: Cutting the Gordian knot of interoperability. J Oncol Pract. 2019;15:3–6. doi: 10.1200/JOP.18.00612. [DOI] [PubMed] [Google Scholar]