Abstract
Biomolecules form dynamic ensembles of many inter-converting conformations which are key for understanding how they fold and function. However, determining ensembles is challenging because the information required to specify atomic structures for thousands of conformations far exceeds that of experimental measurements. We addressed this data gap and dramatically simplified and accelerated RNA ensemble determination by using structure prediction tools that leverage the growing database of RNA structures to generate a conformation library. Refinement of this library with NMR residual dipolar couplings provided an atomistic ensemble model for HIV-1 TAR, and the model accuracy was independently supported by comparisons to quantum-mechanical calculations of NMR chemical shifts, comparison to a crystal structure of a substate, and through designed ensemble redistribution via atomic mutagenesis. Applications to TAR bulge variants and more complex tertiary RNAs support the generality of this approach and the potential to make the determination of atomic-resolution RNA ensembles routine.
Subject terms: RNA, Biophysics, RNA folding, Solution-state NMR
Determining dynamic ensembles of biomolecules is still challenging. Here the authors present an approach for rapid RNA ensemble determination that combines RNA structure prediction tools and NMR residual dipolar coupling data and use it to determine atomistic ensemble models for a variety of RNAs.
Introduction
The functions of many regulatory RNAs crucially depend on changes in three-dimensional (3D) structures that occur in response to a diverse array of cellular inputs, including the binding of small-molecule ligands1, proteins2, epitranscriptomic modifications3, mutations4, and even to nascent RNA elongation during transcription5. Initially described as changes from one 3D structure to another, these transitions are now better understood as changes in dynamic ensembles of many inter-converting conformations, from one conformational distribution to another6.
To deeply understand RNAs at a level that ultimately makes it possible to rationally manipulate their behavior in drug discovery and synthetic biology, we need the ability to determine their dynamic ensembles at atomic resolution. This however presents a challenge to current biophysical techniques: the information required to specify the position of all atoms in thousands of conformations in an ensemble far exceeds the information content of experimental measurements.
NMR spectroscopy is a rich source of ensemble-averaged measurements, and in combination with computational modeling, has been applied with success to determine ensembles of proteins at atomic resolution7–9. In contrast, fewer atomic-level experimental measurements are typically available for characterizing RNA ensembles6. Moreover, while computational modeling methods such as molecular dynamics (MD) simulations are needed to address the experimental data gap10, nucleic acid force fields remain underdeveloped relative to proteins, and MD simulations of RNAs often poorly predict experimental data even for simple motifs11,12. Shortcomings in RNA force field potentials can be addressed using experimental data to readjust the populations of conformers in MD-generated ensembles13. However, simulation times are typically short relative to the dynamic timescales measured experimentally, and force field limitations may prevent sampling of particular conformations; ensemble models refined using the experimental data therefore would not include conformations that were not sampled in the original MD simulation. Enhanced sampling approaches can be used, but can lead to an erroneous sampling of high energy conformations, thereby increasing the possibility of over-fitting the data14. Compounding these difficulties is the lack of methods for assessing the accuracy of various structural features in an RNA ensemble.
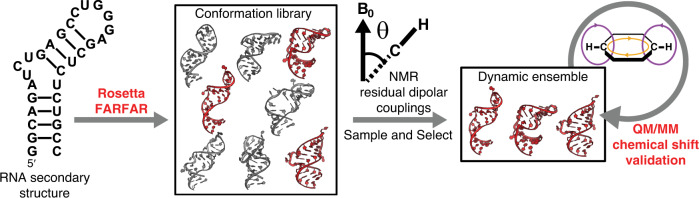
To address the data gap, as well as markedly simplify and accelerate RNA ensemble determination, we took advantage of structure prediction tools that leverage the growing database of RNA structures to directly generate a conformation library that broadly samples energetically favorable 3D conformations given a secondary structure (Fig. 1). We used Fragment Assembly of RNA with Full-Atom Refinement (FARFAR)15, given its high performance in extensive tests of blind prediction of 3D RNA structure16. We then determined RNA ensembles by using previously published NMR residual dipolar coupling (RDC) data17,18 to guide the selection of conformers from the FARFAR-library12,19,20. Finally, we used quantum-mechanical calculations of NMR chemical shifts21 and cross-validation analysis12 to test the accuracy of the generated ensembles (Fig. 1).
Fig. 1. The FARFAR-NMR pipeline.
An RNA conformation library is generated by Rosetta FARFAR structure prediction using an RNA secondary structure. The FARFAR-NMR ensemble can be refined from the conformation library using NMR RDCs and cross-validated by computing NMR chemical shifts using QM/MM calculations.
Results
FARFAR-library better predicts TAR RDCs compared to MD-generated library
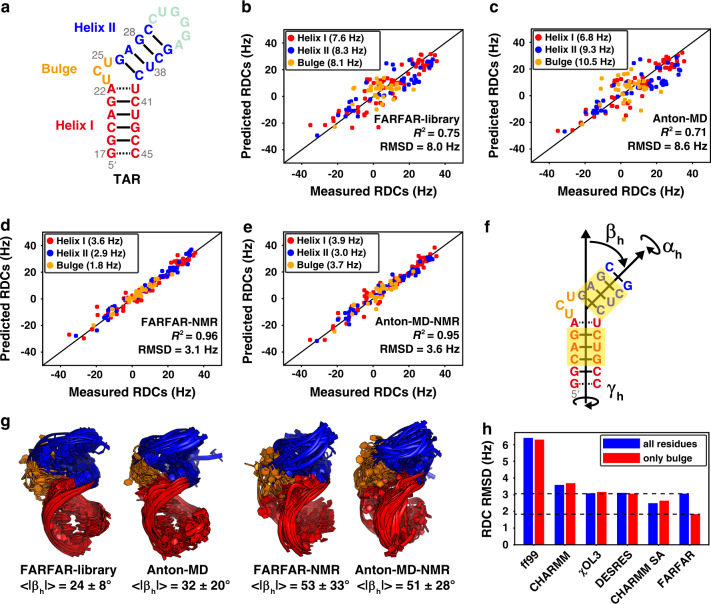
We tested our approach on the transactivation response element (TAR) (Fig. 2a) from HIV-122,23, which has served as a model system for bulge motifs. As one of the most common RNA secondary structural elements, bulges often serve as dynamic joints connecting helical elements, enabling their relative orientation to change adaptively during folding and function12,22–24. We used FARFAR to directly generate a conformation library (N = 10,000) from an input TAR secondary structure only constraining Watson–Crick base pairs (bps) observed by NMR and assuming an idealized A-form geometry25 for these bps while predicting the structure for all remaining nucleotides (“Methods”). We then tested and optimized the FARFAR-library using a rich data set of four independent RDCs (~8 RDCs per nucleotide) previously measured for four TAR molecules that were variably elongated to modulate alignment relative to the NMR magnetic field12,20.
Fig. 2. Using FARFAR-NMR to determine a TAR ensemble.
a Secondary structure of TAR. b–e Comparison between measured and predicted TAR RDCs for the b FARFAR-library (N = 10,000), c Anton-MD library (N = 10,000), d FARFAR-NMR (N = 20), and e Anton-MD-NMR (N = 20) ensembles. Values in parentheses denote the RDC RMSD. RDCs are color-coded according to the structural elements in a. f Definition of the three inter-helical Euler angles (αh, βh, γh) describing the relative orientation of the two helices (“Methods”). The bps of the upper and lower helices used for defining the Euler angles are highlighted in yellow. g Structural overlay of the TAR libraries and NMR refined ensembles (N = 20) (“Methods”). |βh| is the average absolute magnitude of the bend angle. h Comparison of RDC RMSD overall (blue) or bulge (red) residues for ensembles generated using different MD force fields (ff9950, CHARMM3626, ff99bsc0χOL351, and modified ff14 DESRES55), multiple simulations using a simulated annealing approach (CHARMM SA), and FARFAR.
Intriguingly, the FARFAR-library showed better agreement with the RDCs (Fig. 2b) as compared to a previously reported12 TAR library (Anton-MD) that was generated by subjecting an experimentally determined NOE-based NMR structure of TAR (PDBID 1ANR)22 to MD simulations with the CHARMM36 force field26 (Fig. 2c; RMSD 8.0 versus 8.6 Hz). An optimized ensemble with N = 20 conformers (Supplementary Fig. 1b) was generated (“Methods”) by using the agreement with the RDCs to guide selection of conformers from the FARFAR-library (Fig. 2d)12,19. The optimized FARFAR-NMR ensemble also better predicted the RDCs (RMSD 3.1 Hz) relative to the optimized Anton-MD-NMR ensemble (RMSD 3.6 Hz) obtained using a similar procedure and the Anton-MD library (Fig. 2e and Supplementary Fig. 1b). Cross validation12,14 showed that the improved RDC agreement was not due to over-fitting (Supplementary Fig. 1c, d).
The improved agreement observed with the FARFAR-NMR ensemble, while small, was surprising given that the global inter-helical distribution27 of the Anton-MD-NMR TAR ensemble had been independently validated via X-ray scattering interferometry (XSI)28. Indeed, the FARFAR-NMR and Anton-MD-NMR ensembles show comparable agreement with the RDCs measured for helical bps (Fig. 2d, e) and the two ensembles sample similar inter-helical orientational distributions (Fig. 2f, g and Supplementary Fig. 2a–d).
Rather, the improved agreement was primarily driven by the RDCs measured in the locally more flexible bulge residues. For the FARFAR-NMR ensemble, the RDC RMSD (1.8 Hz) for bulge residues (Fig. 2d) was within experimental error (~2 Hz), but it was substantially higher (3.7 Hz) for the Anton-MD-NMR ensemble (Fig. 2e). The bulge RDC RMSD could not be improved by running MD simulations using different force fields or by running multiple simulations using the CHARMM36 force field with simulating annealing to more broadly sample conformational space (“Methods”) (Fig. 2h). It appears that improved conformational sampling in the FARFAR-library makes it possible to surpass the accuracy with which the TAR bulge can be described using conventional MD simulations. This improved performance is particularly noteworthy when considering that solving a high-resolution structure and then running MD can in totality consume several months and often years whereas the FARFAR-library is generated within 24 h running on 100 cores in parallel.
FARFAR-NMR ensembles better predict chemical shifts compared to MD
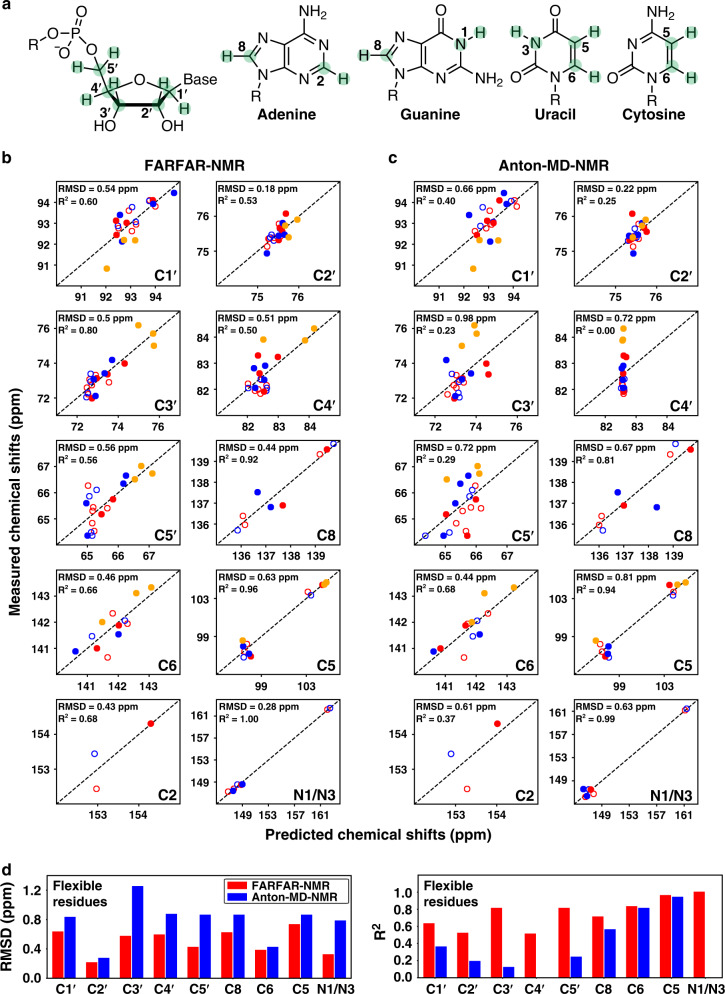
To further evaluate the accuracy of the FARFAR-NMR TAR ensemble, we substantially expanded the breadth and depth of atomic-level experimental data that can be brought to bear when evaluating the accuracy of RNA ensembles by predicting ensemble-averaged 1H, 13C, and 15N chemical shifts (~15 chemical shifts per nucleotide) using quantum-mechanical AF-QM/MM calculations21,29. Although rich in structural information that is complimentary to that obtained from RDCs, the agreement between measured chemical shifts and values predicted from crystal structures of nucleic acids has traditionally been poor21,30. We recently showed in studies of DNA duplexes that at least some of this disagreement originates from neglecting ensemble-averaging when predicting 13C chemical shifts29. This revelation and the improved FARFAR-NMR TAR ensemble led us to test this approach on flexible RNAs and to extend it by incorporating 1H and 15N shifts (Fig. 3a).
Fig. 3. Evaluating TAR ensembles using chemical shifts.
a Chemical structures of the sugar and base moieties with chemical shift probes used to test ensemble accuracy highlighted in green. b, c Comparison of measured and predicted 13C/15N chemical shifts for b FARFAR-NMR (N = 20) and c Anton-MD-NMR (N = 20) by AF-QM/MM (“Methods”). Values are color-coded according to the structural elements in Fig. 2a. Chemical shifts for central Watson–Crick bps within A-form helices (C19-G43, A20-U42, G21-C41, A27-U38, G28-C37) are denoted using open circles. A correction was applied to the predicted chemical shifts (“Methods”) as described previously29. For 1H chemical shifts see Supplementary Fig. 3. d Comparison of RMSD (left) and R2 (right) between measured and predicted 13C/15N chemical shifts for flexible residues (U23, C24, U25, A22-U40, G26-C39, C29-G36, G18-C44) for FARFAR-NMR (red) and Anton-MD-NMR (blue).
Remarkably, good agreement was observed between the measured 1H, 13C, and 15N base and sugar chemical shifts and values back-calculated for the FARFAR-library (Supplementary Fig. 3a). The agreement improved substantially for the FARFAR-NMR ensemble following RDC optimization for 48% of the atom-types analyzed, while the agreement was unaffected for the remaining atom-types (Fig. 3b and Supplementary Fig. 3b). The agreement was poor for any one conformer member of the ensemble, underscoring the critical importance of ensemble-averaging (Supplementary Fig. 4). The R2 values were >0.5 for ~70% of all the nuclei examined and the RMSD for 13C (0.18–0.63 ppm) was decreased relative to the value of 3.6 ppm obtained previously for a static representation of a simple RNA motif21 and that obtained for RDC-selected DNA duplex ensembles (0.66–1.27 ppm)29 using the same QM/MM approach. The agreement with an independent set of measurements that were not used in ensemble determination and that is highly sensitive to structural features of the base, sugar, and backbone strongly suggests that the FARFAR-NMR ensemble describes the solution behavior of TAR with atomic accuracy (<2 Å, “Methods”).
In sharp contrast, the agreement was much weaker for the Anton-MD-NMR ensemble relative to FARFAR-NMR for 90% of the atom-types, particularly for the sugar chemical shifts, some of which show no apparent correlation even following RDC optimization (Fig. 3c, d, Supplementary Fig. 3c–h and Supplementary Discussion). These results establish the utility of chemical shifts computed using quantum-mechanical calculations in RNA ensemble validation and also show that the improved accuracy of the FARFAR-NMR ensemble relative to Anton-MD-NMR is even greater than revealed by the RDC data alone (also see below).
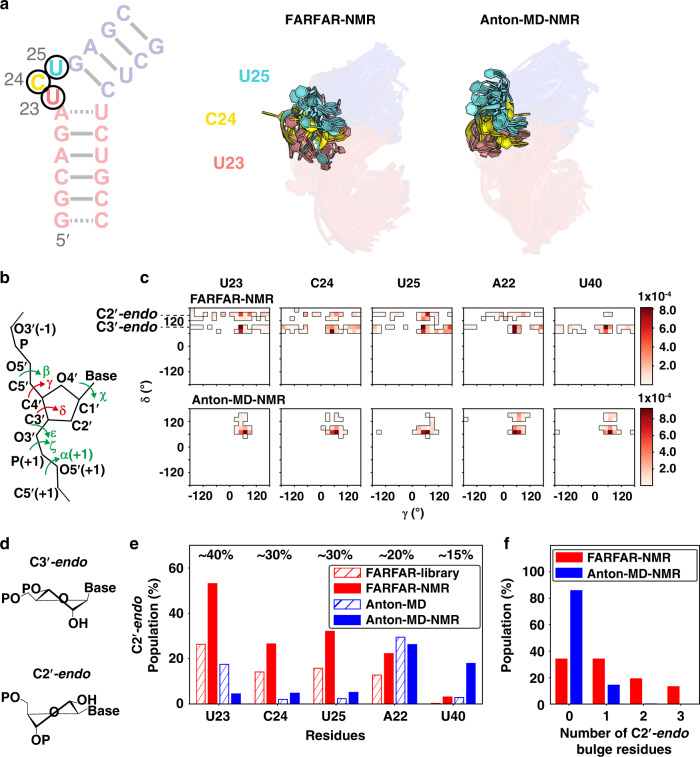
FARFAR ensemble broadly samples the sugar-backbone conformation at bulge nucleotides
We compared the FARFAR-NMR and Anton-MD-NMR ensembles (Fig. 4a) to better understand the features responsible for the more accurate ensemble description of the TAR bulge. Approximately 75% of the conformers in the FARFAR-NMR ensemble have the canonical UCU bulge, while the remaining 25% have a non-canonical AUC bulge that forms through a single nucleotide register shift (Supplementary Fig. 5a and Supplementary Discussion). In contrast, conformers in the Anton-MD-NMR ensemble sample a broader set of junction topologies some of which deviate from the NMR-derived Watson–Crick pairing (Supplementary Fig. 5b and Supplementary Discussion), potentially contributing to the poor agreement observed for the imino 15N/1H chemical shifts (Fig. 3c and Supplementary Fig. 3).
Fig. 4. FARFAR-NMR ensemble broadly samples non-canonical sugar-backbone torsion angles relative to Anton-MD-NMR.
a Overlay of the dynamic ensembles of the TAR bulge. b RNA backbone torsion angles exhibiting different and similar distributions between Anton-MD-NMR and FARFAR-NMR are colored red and green, respectively (“Methods”). c 2D density maps of δ versus γ comparing Anton-MD-NMR and FARFAR-NMR ensembles (N = 2000) for bulge residues as well as A22 and U40. The bin width is 20°. d Structure of the ribose moiety in C3′-endo and C2′-endo conformations. e Population of C2′-endo pucker at bulge residues as well as A22 and U40 in the FARFAR-library (N = 10,000, red open), FARFAR-NMR (N = 20 × 100 = 2000, red fill), Anton-MD library (N = 10,000, blue open) and Anton-MD-NMR (N = 20 × 100 = 2000, blue fill). Experimental estimates of the C2′-endo population based on 13C chemical shifts are indicated above the bars (“Methods”). f The population of conformers in the ensemble as a function of the number of C2′-endo bulge residues for FARFAR-NMR (red, N = 20 × 100 = 2000) and Anton-MD-NMR (blue, N = 20 × 100 = 2000).
In addition, despite having a narrower range of junction topologies, the sugar-pucker distributions (defined by angle δ) for bulge residues in the FARFAR-NMR ensemble are broader (Fig. 4b, c), substantially enriched in the non-canonical C2′-endo conformation (Fig. 4d) relative to the Anton-MD-NMR ensemble, and are in better agreement with the NMR-derived31 sugar puckers (Fig. 4e and Supplementary Fig. 6a). Moreover, ~15% of the conformers in the FARFAR-NMR ensemble had all three bulge residues simultaneously in the C2′-endo conformation whereas none did in the Anton-MD-NMR ensemble (Fig. 4f). Excluding all conformers containing C2′-endo sugar puckers in U23, C24, U25, A22 or U40 from the FARFAR-library diminished the RDC agreement to a level similar to the Anton-MD derived ensembles again confirming that the improved agreement is not due to over-fitting of the data (Supplementary Fig. 6b, c). Similar behavior was observed for the backbone torsion angle γ (Fig. 4b), which shows a greater sampling of non-gauche + angles in the FARFAR relative to the Anton-derived ensembles (Fig. 4c and Supplementary Fig. 6b, d). This difference can account for the better agreement observed for sugar chemical shifts31 in the FARFAR-NMR versus Anton-MD-NMR ensembles (Fig. 3b–d).
It should be noted that sugar repuckering observed in the FARFAR-NMR ensemble likely occurs on the fast nanosecond to microsecond timescales and is distinct from the slower microsecond to millisecond timescale repuckering modes reported recently for TAR using NMR chemical exchange measurements that are coupled to localized changes in base pairing and secondary structure when forming low-populated “excited states”31.
Atomic view of coaxial stacking and cooperative flipping of bulge nucleotides
What causes conformers in the FARFAR-NMR ensemble to more greatly sample non-canonical sugar-backbone conformations at the bulge relative to conformers in the Anton-MD-NMR ensemble? Unpaired pyrimidine RNA nucleotides unconstrained by other interactions are enriched in the C2′-endo conformation (C2′-endo:C3′-endo is 40:60)32; C3′-endo becomes the predominant sugar-pucker when the nucleotides form bps or stack intra-helically31. Consistent with these expectations, in the FARFAR-NMR ensemble, ~80% of the residues with C2′-endo sugar puckers were extra-helical (Fig. 5a and Supplementary Fig. 5a). Moreover, linear inter-helical conformers within the FARFAR-NMR ensemble had a strong preference to have all three bulge residues simultaneously flip out and be enriched in C2′-endo sugar puckers (Fig. 5b, c, Supplementary Fig. 5a and Supplementary Movie 1). Such coaxial inter-helical conformations have previously been hypothesized to exist within the TAR ensemble based on the Mg2+ dependence of the inter-helical ensemble24 and a crystal structure of Ca2+-bound TAR33.
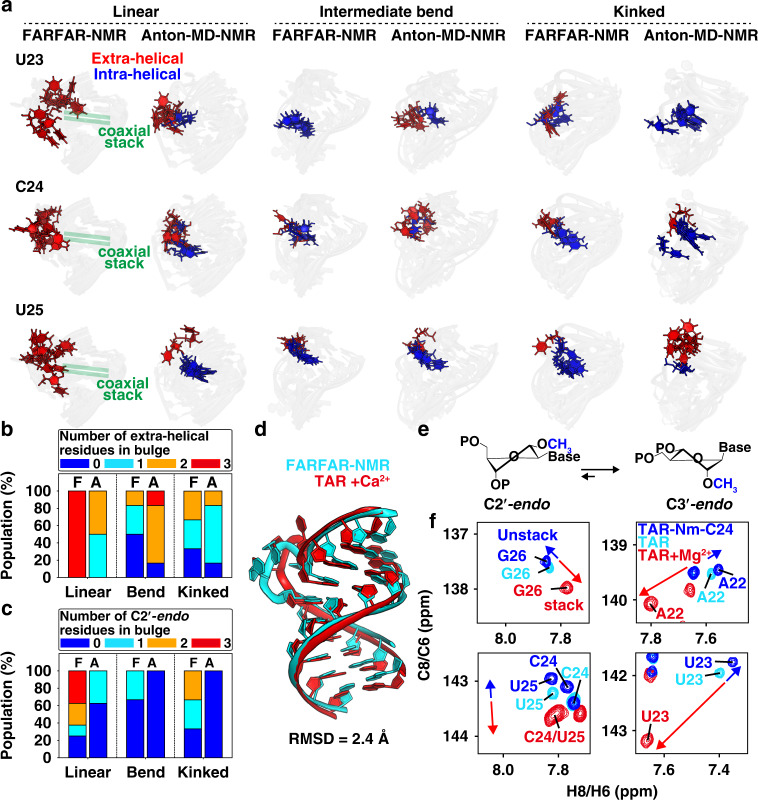
Fig. 5. Cooperative extra-helical flipping and sugar repuckering of bulge residues are coupled to coaxial stacking.
a Overlay of conformers showing motions in bulge residues for linear (|βh| < 45°), intermediate bend (45° < |βh| < 70°) and kinked (|βh| > 70°) inter-helical conformations in the FARFAR-NMR and Anton-MD-NMR ensembles. b, c The fractional populations of conformers with the number of b extra-helical and c C2′-endo residues (color-coded) in the bulge residues (U23, C24, and U25) as a function of bending angle in the FARFAR-NMR (“F”) and Anton-MD-NMR (“A”) ensembles (N = 20). d Comparison of one coaxially stacked FARFAR-NMR conformer with the crystal structure of Ca2+-bound TAR (PDBID: 397D)33. e Nm shifts sugar-pucker equilibrium towards C3′-endo. f Overlay of 2D [13C, 1H] HSQC NMR spectra of the aromatic spins for TAR-Nm-C24 without Mg2+ (blue) with blue arrows indicate unstacking, TAR without Mg2+ (cyan) and TAR with 3 mM Mg2+ (red) with red arrows indicating increased coaxial stacking.
Strikingly, one of the coaxial conformations in the FARFAR-NMR ensemble superimposes with the Ca2+-bound TAR crystal structure with a heavy-atom RMSD of 2.4 Å (Fig. 5d). Thus, the TAR crystal structure captures a substate of the ensemble in solution, providing support for the validity of the FARFAR ensemble and underscoring limitations of static structures as accurate representations of RNAs in solution. In sharp contrast, none of the conformations in the Anton-MD-NMR ensemble had the two helices coaxially stacked, and none had all three bulge residues simultaneously flipped out and with C2′-endo sugar pucker (Fig. 5a–c and Supplementary Fig. 5b). Based on the population of conformations in which all three bulge residues are simultaneously flipped out relative to the population in which individual bulge residues are flipped out in the FARFAR-NMR ensemble (Fig. 5a and Supplementary Movie 1), the three bulge residues simultaneously flip out with estimated cooperativity of ~2 kcal/mol (“Methods”). This cooperativity may arise because flipping of all three bulge residues permits favorable coaxial stacking of the helices (Fig. 5a, Supplementary Fig. 5a and Supplementary Movie 1). Indeed, changing the sequence identity of the Watson–Crick bps at the interface of the two TAR helices such to promote inter-helical stacking34 results in a predominantly coaxially stacked conformation in which all three bulge residues are simultaneously flipped out.
Further test of the TAR ensemble using atomic mutagenesis
We put key atomic features of the FARFAR-NMR ensemble to a test by rationally redistributing the conformer populations via atomic mutagenesis. The bulge residues in kinked, unstacked conformations are more likely to adopt the C3′-endo sugar pucker relative to these residues in the coaxially stacked conformations (Fig. 5c). This difference leads to a prediction: substitutions like 2′-O-Methyl (Nm) modifications (Fig. 5e) at U23 or C24 that bias the sugar pucker toward C3′-endo will favor the unstacked over stacked conformational states, although the effect will be small (~0.2 kcal/mol per substitution)35.
To test this feature of the ensemble, we incorporated Nm modifications at U23 and C24 and assessed the resultant ensemble using NMR chemical shift measurements. Indeed, methylating either U23 or C24 resulted in chemical shift perturbations in 2D NMR spectra of TAR (Fig. 5f and Supplementary Fig. 7) throughout the bulge and neighboring residues that are directed towards the chemical shifts of the unstacked conformation24, as expected for a cooperative redistribution in favor of the kinked conformation.
Applications to TAR bulge variants and in presence of Mg2+
To test the generality and limits of our approach, we used FARFAR-NMR to generate ensembles for three additional TAR mutants (Fig. 6a) containing one (U1-TAR), two (U2-TAR), and seven (U7-TAR) bulge nucleotides in the absence and presence of Mg2+. The conformational dynamics of these TAR bulge variants have recently been characterized using NMR24, though with only a single set of RDCs for each variant, other than for U2-TAR in absence of Mg2+ where two sets of RDC measurements were made20. With the exception of U7-TAR in the absence of Mg2+, the agreement observed between RDCs measured for these TAR mutants and values computed for the FARFAR-library or FARFAR-NMR ensembles was similar to that obtained for TAR (Fig. 6b and Supplementary Fig. 8). The lower agreement for U7-TAR in the absence of Mg2+ may reflect limited structural information for kinked RNAs with long bulges24 and/or a need to sample a much larger number of conformations.
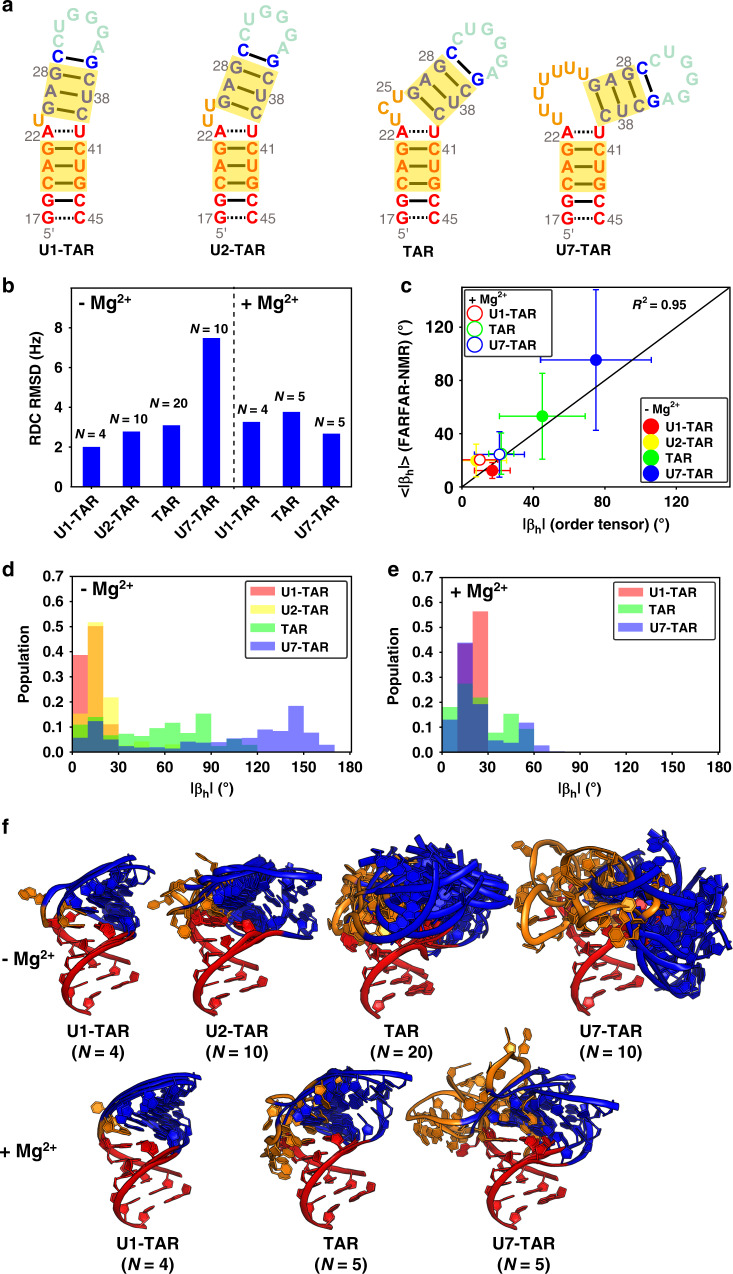
Fig. 6. Dynamic ensembles of TAR and its bulge variants in the absence and presence of 3 mM Mg2+.
a Secondary structure of TAR and its bulge variants (U1-TAR, U2-TAR, and U7-TAR). The bps at different helices used for defining the Euler angles are highlighted in yellow. b RDC RMSD for ensembles of TAR variants with ensemble size N obtained using FARFAR-NMR. c Comparison between the average bend angle (<|βh|>) and its standard deviation for the FARFAR-NMR (N = 2000, “Methods”) derived ensembles with the best-fit |βh| values obtained from an order tensor analysis of the RDCs24. The error bar in the order tensor analysis corresponds to half the cone radius angle assuming an isotropic model69. d, e Distributions of the inter-helical bend angle magnitude |βh| for the ensembles (N = 2000, “Methods”) in the absence d and presence e of Mg2+. f The FARFAR-NMR ensembles of TAR and its bulge variants in the absence (upper) and presence (lower) of 3 mM Mg2+. The ensemble size (N) is labeled below for each TAR bulge variant. Motifs in the ensembles are color-coded as in a.
The FARFAR-NMR ensembles reproduce trends in ensemble properties observed previously for the bulge variants based on an independent analysis of RDCs and chemical shifts24. The average and standard deviation of the inter-helical bend angle in the FARFAR-NMR ensembles are in good agreement with values reported based on an order tensor analysis of the RDCs24, which does not involve explicit ensemble modeling (Fig. 6c). In addition, the distributions of the bend angle have the expected bimodal character24 with a narrow stacked state and broader set of kinked conformations and with Mg2+ increasing the population of the stacked state (Fig. 6d–f). These results suggest that FARFAR-NMR can be used to generate ensembles for simple RNA motifs under a variety of conditions in solution.
Initial applications to other RNAs
To test the general applicability of FARFAR-NMR to larger and more complex RNAs, we used FARFAR to generate conformation libraries for four additional RNAs: human telomerase P2ab36, the fluoride riboswitch apo state37, the preQ1 Class I riboswitch holo state38 and the preQ1 Class II riboswitch holo state39. These RNAs range in size between 35 and 59 nucleotides and include three riboswitches with complex tertiary structures (Fig. 7a). The secondary structures of these RNAs were inferred from prior NMR studies36–39. In addition to the secondary structure, long-range non-Watson–Crick bps, which can be identified based on analysis of imino resonances were also specified as restraints during the FARFAR ensemble calculations (“Methods”). We determined preliminary ensembles for these RNAs using a single set of previously published RDCs and compared the results with the reported solution NMR structures determined using a combination of RDCs and NOE data.
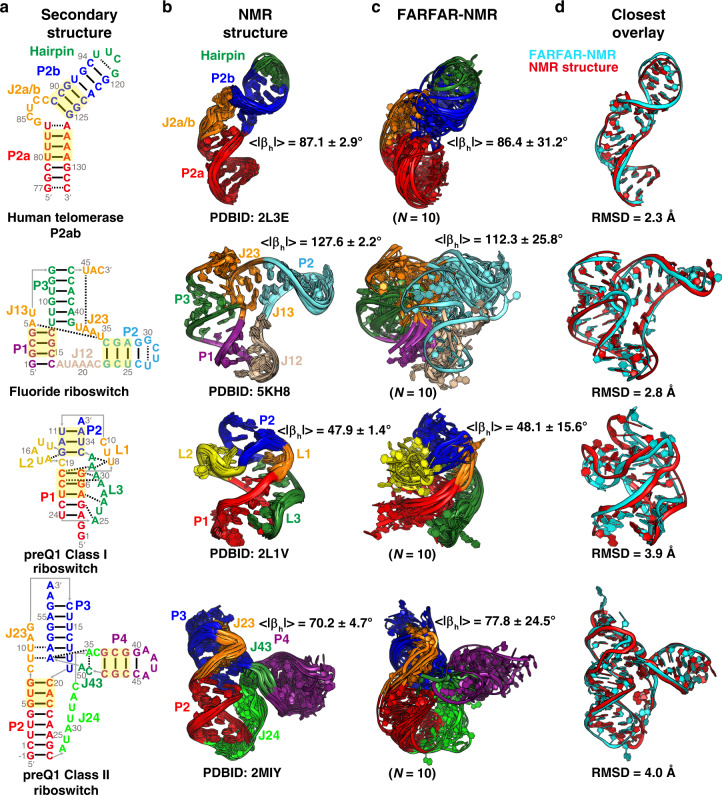
Fig. 7. Initial ensembles of various RNA.
a The RNA secondary structure for each RNA from top to bottom: human telomerase P2ab, fluoride riboswitch, preQ1 Class I riboswitch and preQ1 Class II riboswitch. The bps at different helices used for defining the Euler angles are highlighted in yellow. b The NMR structure bundles. c The FARFAR-NMR ensembles with ensemble size (N) labeled below. Different structural elements are color-coded according to a. The average bend angles < |βh|> (N = 10 × 200 = 2000) of the two helices are labeled along with the ensembles. d Comparison of the FARFAR-NMR conformer (cyan) with the closest heavy-atom RMSD to the NMR structure (red).
In all cases, the FARFAR-NMR ensembles generated using the FARFAR-library reproduced the RDCs (RDC RMSD = 2.7–3.3 Hz) within experimental precision (Supplementary Fig. 9). The ensembles in all cases contain substates that are similar to those in the NMR structures, superimposing with all-atom RMSDs of 2.3–4.0 Å (Fig. 7b–d). The inter-helical orientation in the FARFAR-NMR ensembles also agreed well with that from the NMR structures (Fig. 7b, c). These examples support the generality of the approach but are expected to have limited precision since they were determined based on a single set of RDCs. Further refinement of these ensembles will require measurements of additional RDC data sets followed by evaluation using our chemical shift approach.
Discussion
FARFAR-NMR lays the foundation for a new paradigm for ensemble determination by combining the most easily obtained and most reliably measured NMR data with 3D structure prediction. The approach can immediately be applied to render many existing NOE-based NMR structures of RNAs currently in the PDB into dynamic ensembles, without the need for additional measurements, and the accuracy of these ensembles can be evaluated using published chemical shift data. It should therefore be possible to rapidly and greatly expand the number of atomistic models of ensembles available for RNAs, and new RNA structures can now be reported as ensembles, not merely as measures of structural uncertainty but as more accurate and complete representations of the RNA’s structural properties.
While we have focused on FAFAR, our approach can be extended to other RNA structure prediction programs40 and can also incorporate other sources of experimental data6. Given its higher throughput, FARFAR-NMR may enable broader and deeper explorations of ensemble behavior across multiple different RNA motifs under varying solution conditions, providing a broad view of the RNA ensemble landscape that is impossible to attain with current methods. The approach can also be extended to proteins by leveraging advances in protein structure prediction to determining protein ensembles41.
Our ability to extensively explore potential conformations of the TAR bulge, and to extensively test the accuracy of resulting ensembles using chemical shifts resulted in unexpected new insights about the TAR ensemble. We have obtained an unprecedented atomic view of the bulge ensemble and how coaxial stacking of helices cooperatively expels bulge residues into an extra-helical conformation with non-canonical sugar-backbone conformations.
Our results show that it is now possible to bring to bear the power of the NMR chemical shift to assess the accuracy of RNA ensembles. Information about motional averaging obtained from accurate ensembles allows accurate chemical shifts to be computed even for highly flexible residues. In the future, it may be possible to directly use chemical shifts in ensemble determination. Such an approach might obviate the need for multiple RDC data sets and further accelerate ensemble determination.
Our results also suggest that there may be biases in MD in at least some current RNA force fields11. These force fields favor intra-helical conformations with canonical sugar-backbone conformations for the flexible nucleotides in and around the bulge of TAR. By using chemical shifts and RDCs, it should be possible to greatly expand the data that is available for testing and guiding the optimization of MD-based methods.
While the conformational sampling in the FARFAR-NMR approach is limited by the current repertoire of high-resolution structures of RNA motifs, we can anticipate continual improvements as more structures of RNAs are determined. Future studies should explore other approaches for determining ensembles from the FARFAR generated library including Bayesian approaches13 and compare the generated ensembles with those generated using data-driven approaches such as simulated annealing42 and restrained MD43. Our preliminary application to complex tertiary RNAs underscores the importance of having information regarding long-range contacts when generating the FARFAR-library (Supplementary Fig. 9).
In conclusion, FARFAR-NMR lays the foundation for a new paradigm for RNA ensemble determination by combining reliably measurable NMR data with 3D structure prediction. The approach can immediately be applied to render many existing NMR structures of RNAs that were determined using conventional approaches into dynamic ensembles, and their accuracies can be tested using the available chemical shift data. The approach is general, rapid, and can also incorporate additional sources of experimental data. Given its ease of implementation and higher throughput, FARFAR-NMR has the potential to unleash the ensemble description of RNAs to all corners of biology, from which a deeper and broader understanding of folding and function will undoubtedly emerge.
Methods
Generating ensembles using FARFAR
FARFAR15 is implemented as the rna_denovo program in Rosetta Software Suite. FARFAR requires as input the RNA sequence, which can be constrained by an optional secondary structure. The input secondary structures for TAR and its bulge variants were derived based on the imino 1H resonances in NMR spectra. Therefore, Watson–Crick base pairing was imposed for G18-C44, C19-G43, A20-U42, G21-C41, G26-C39, A27-U38, G28-C37, and C29-G36, whereas nucleotides in the apical loop and bulge, and the terminal G17-C45 bp were unconstrained. Given the indirect NMR observation of the A22-U40 bp in U2-TAR by H6(C5)NN experiment as reported previously24, two sets of FARFAR simulations were conducted for each TAR variant with A22-U40 either paired or unpaired. Interestingly, with the exception of U2-TAR, constraining A22-U40 when generating FARFAR-libraries did not improve the agreement with the RDCs for TAR and other variants. This highlights the importance of differentiating between well-formed versus labile bps, an important advantage offered by NMR as compared to other methods for determining secondary structure such as chemical probing. Thus, all the FARFAR-libraries in this study correspond to those in which the A22-U40 bp is not constrained, except for the U2-TAR ensemble in the absence of Mg2+. The input secondary structures of TAR and its bulge variants were summarized in Supplementary Tables 2–5.
The secondary structure constraints of other RNAs including human telomerase P2ab36, fluoride37, and preQ1 Class I38 and Class II39 riboswitches were derived from prior NMR studies based on a combination of observed imino 1H resonances, HNN-COSY experiments, direct observation of resonances in slow exchange, and 15N chemical shifts, and are summarized in Supplementary Tables 6–9. For these RNAs, non-Watson–Crick base pairing was also imposed during the FARFAR calculations using the “-obligate_pair_explicit” flag with the specification of Leontis–Westhof (LW) classification (Supplementary Tables 6–9). Specifically, for the U-G bp in UUCG apical loop, the bp geometry was set to “tSW” and only G in the syn conformation (χ between 0 and 100°)44 was kept. Additional inspection of steric clashes was performed on these bps specified by the “-obligate_pair_explicit” flag to remove stereochemically unreasonable structures. To significantly reduce the run time, we modeled the helices of TAR and its bulge variants, human telomerase P2ab, and fluoride riboswitch as static idealized A-form helices shown previously to satisfy NMR RDC data in RNA24,25. For preQ1 Class I and II riboswitches, since the helices are involved in tertiary interactions, a better RDC RMSD was achieved while allowing the helices to deviate from the idealized A-form during the calculation.
rna_helix.py is a python wrapper for the Rosetta executable rna_helix. rna_helix.py is available in $ROSETTA/tools/rna_tools/bin, where $ROSETTA is the Rosetta installation path.
rna_helix.py -seq gcag cugc -resnum 18-21 41-44 -o helix1.pdb
rna_helix.py -seq gagc gcuc -resnum 26-29 36-39 -o helix2.pdb
in which RNA sequences of both strands as well as their residue indices are required as input. While generating FARFAR-library assuming idealized A-form helices, we then executed rna_denovo with the following command:
rna_denovo -nstruct 100 -s helix_1.pdb helix_2.pdb -fasta input.fasta -secstruct_file input.secstruct -minimize_rna true
where -nstruct is the number of models per run, which is 100 in the current study, -fasta is the path of a fasta file containing the RNA sequence, -secstruct is the path of a file (input.secstruct) containing the RNA secondary structure in dot-bracket notation, -minimize_rna true minimizes the RNA after fragment assembly, and -s specifies the path to the pdb files that contain static structures of our helices we do not wish to generate via fragment assembly to save computation (the fasta, secstruct files as well as example commands can be found in Supplementary Tables 2–9).
The entire procedure was repeated 100–200 times and 10,000 structures were randomly selected from the entire resulting output with Rosetta energy units < 0 (for TAR and its bulge variants) to remove models that potentially may have chain breaks and severe steric clashes, which do not satisfy the RDCs (Supplementary Fig. 1a), to generate the FARFAR-library (N = 10,000). The Rosetta energy cutoff for human telomerase P2ab, fluoride riboswitch, preQ1 Class I riboswitch and preQ1 Class II riboswitch was set to be 50, 200, 150, 100, respectively. The cutoff was determined as described for TAR (Supplementary Fig. 1a).
Molecular dynamics (MD) simulations
Simulations of HIV-1 TAR starting from the PDB structure 1ANR using the CHARMM36 force field26 (8.3 μs) were performed as described previously12. The structures from this simulation were then clustered based on the heavy-atom RMSD of TAR (excluding the apical loop and terminal bp) using the cluster command in the CPPTRAJ suite45 to give 16 structures, which served as starting points for the simulated annealing runs. The simulated annealing simulations using the CHARMMM 36 force field were performed with the GROMACS MD simulation package46. GROMACS force field files for CHARMM36 were obtained from (http://mackerell.umaryland.edu/charmm_ff.shtml#gromacs) as of March 2019. The structures were solvated with a rhombic dodecahedral box of TIP3P47 water molecules, with box size chosen such that the boundary was at least 10 Å away from any of the RNA atoms, and was then neutralized using Na+ ions. After energy minimization without restraints, the system was then gradually heated to a temperature of 300 K using a Nose–Hoover thermostat48 (τ = 0.1 ps), under constant volume conditions for 100 ps with harmonic restraints on the solute (20 kJ/mol/nm2). The restraints were gradually reduced in two 50 ps NVT equilibration steps (10 and 5 kJ/mol/nm2, respectively). This was followed by two 50 ps NVT and NPT equilibration steps at 300 K without restraints on the solute. NPT equilibration was performed using a modified Berendsen thermostat with a stochastic term49 with τ = 0.1 ps and at 1 bar using a Berendsen barostat with τ = 2.0 ps. Simulated annealing was then performed by subjecting the system to a pair of successive 50 ps NVT and NPT equilibration steps at 400 and 300 K. This was followed by a 5 ns NVT equilibration step at 300 K prior to the production run (500 ns). A non-bonded cutoff of 10 Å was used for treating short-range non-bonded interactions while the particle mesh Ewald method was used to treat long-range electrostatic interactions. Non-bonded van-der Waals forces were switched to 0 from 8 to 10 Å. Covalent bonds involving hydrogen were constrained using the LINCS algorithm to enable the use of a 2 fs timestep. A total of 625 equally spaced snapshots were obtained from each of the 16 production runs to get create a pool of 10,000 conformers for RDC selection.
MD simulations of TAR starting from the PDB structure 1ANR using the ff99 force field50 with and without χOL3 corrections for RNA51 were performed using periodic boundary conditions as implemented in the AMBER MD simulation package52. All starting structures were solvated using a truncated octahedral box of TIP3P47 water molecules with box size chosen such that the boundary was at least 11 Å away from any of the RNA atoms for all simulations with the ff99 force field with χOL3 corrections. For the TAR simulations using the ff99 force field without χOL3 corrections, the system was solvated in a truncated octahedral box of SPC/E water molecules such that the boundary was at least 15.4 Å away from any of the RNA atoms. Na+ ions treated using the Joung–Cheatham parameters53 was then added to neutralize the charge of the system in all cases. The system was then energy minimized in two stages with the solute being fixed (with restraint of 500 kcal/mol/Å2) during the first stage. Equilibration and production runs (1 μs) were then performed as described previously54.
Simulations of TAR starting from PDB 1ANR using the DESRES force field55 were performed using the GROMACS MD simulation package46. DESRES force field files for GROMACS were obtained from a port by Giovanni Bussi (https://github.com/srnas/ff/tree/desres). The starting structure (PDB 1ANR) was solvated using a rhombic dodecahedral box of TIP4P-D56 water molecules, with box size chosen such that the boundary was at least 10 Å away from any of the RNA atoms. Na+ ions treated using the parameters from MacKerell et al. 57 were then added to neutralize the charge of the system. After energy minimization without restraints, the system was then gradually heated to a temperature of to 298 K using a modified Berendsen thermostat with a stochastic term49 (τ = 0.1 ps), under constant volume conditions for 100 ps with harmonic restraints on the solute (1000 kJ/mol/nm2). The system was then allowed to equilibrate for 100 ps under constant pressure (1 bar), using the Parinello–Rahman barostat58 (τ = 2 ps) and temperature (at 298 K, using a modified Berensen thermostat49, τ = 0.1 ps) conditions, with harmonic restraints on the solute (1000 kJ/mol/nm2). This was followed by NPT equilibration for 30 ns without harmonic restraints, following by a production run of 1 μs. A non-bonded cutoff of 9 Å was used for treating short-range non-bonded interactions while the particle mesh Ewald method was used to treat long-range electrostatic interactions. All bonds were constrained using the LINCS algorithm to enable the use of a 2 fs timestep. A set of evenly (5 ps) spaced snapshots were used for subsequent analysis of all simulations using the CPPTRAJ suite of programs45.
Additional MD simulations using the ff99 force field with χOL3 corrections were also performed to assess the extent to which sampling of sugar puckers in the bulge could be influenced by changes in the starting structures used for the simulations (Supplementary Fig. 10). Two starting structures were derived from PDB 1ANR with the sugar puckers of U23 (TARU23 C2′-endo) and U25 (TARU25 C2′-endo) individually switched from C3′-endo to C2′-endo, and another was a conformer from the FARFAR-NMR ensemble (TARFARFAR) in which the sugar puckers of U23, C24 and U25 were all C2′-endo. The starting structures for the TARU23 C2′-endo and TARU25 C2′-endo simulations were generated by the superposition of a C2′-endo uridine nucleotide onto U23/U25 in 1ANR using the uridine base atoms, and replacing the sugar-backbone atoms with those of the superimposed C2′-endo uridine. For TARU23 C2′-endo and TARU25 C2′-endo, only the backbone of the C2′-endo uridine was fixed during an energy minimization, while for TARFARFAR, all the heavy atoms were fixed during minimization.
NMR residual dipolar coupling (RDC) data
The RDC data (sugar C1′-H1′/C2′-H2′/C3′-H3′/C4′-H4′ and base C8-H8/C6-H6/C2-H2/C5-H5/N1-H1/N3-H3) used in the RNA ensemble determination were reported previously12,36–39 and are summarized in Supplementary Table 1. The raw data can also be downloaded from https://github.com/alhashimilab/RDC.
RDC calculations
Ensemble-averaged RDCs were calculated by computing the RDCs for each conformer in an ensemble using the program PALES59. PALES computes RDCs based on global molecular shape. The RDC was computed using a cylindrical wall model using the following command:
pales -pdb input.pdb -inD input_rdc.tab -outD output_rdc.tab -H -pf1 -wv 0.022
where -pdb is the path of an input PDB file (input.pdb), -inD is the path of an input data file indicating the bond vectors for which RDC should be computed (input_rdc.tab), -outD is the path of the output file containing all the calculated RDCs, -H means selecting all atoms including proton and -pf1 -wv specifies the pf1 effective concentration (0.022 g/mL) assuming a rod liquid crystal model.
The RDC values were then averaged over all conformers in an ensemble assuming that they are equiprobable. Individual scaling factors were applied to the predicted RDCs of each construct to account for the difference of alignment magnitude in part arising due to differences in phage concentrations in the experiments, as described previously12. The elongated constructs were elongated in silico using an idealized A-form geometry prior to RDC calculation as described previously12.
The TAR apical loop was modeled using the wild-type CUGGGA loop12. As reported previously for the Anton-MD-NMR ensemble12, replacing the loop with the UUCG loop44 used to measure RDCs minimally impacted the RDC agreement for the FARFAR ensembles (Supplementary Fig. 1e, f). This is consistent with prior NMR studies showing that the apical loop replacement minimally impacts the dynamics of the bulge12,60.
Sample and select (SAS)
We used the SAS approach19 to generate ensembles from a structured pool that best satisfied the measured RDCs. Briefly, a simulated annealing Monte Carlo sampling scheme was used to select an ensemble that minimizes the cost function depicting the differences between the measured and predicted RDCs:
| 1 |
and are the calculated and measured RDCs, respectively, of the ith bond vector measured on the jth TAR construct, is the overall scaling factor of alignment magnitude for construct j, and N is the total number of bond vectors. The initial effective temperature for simulation annealing was 100 and decreased by a factor of 0.9 in every step for a total of 5 × 105 steps. A series of SAS runs were performed varying the ensemble size from N = 1 to an ensemble size in which the RDC RMSD reaches a plateau (Supplementary Figs. 1b, 8, and 9). The resultant ensemble size for different RNA ensembles were: N = 4 (U1-TAR, no Mg2+), N = 10 (U2-TAR, no Mg2+), N = 20 (TAR, no Mg2+), N = 10 (U7-TAR, no Mg2+), N = 4 (U1-TAR, with Mg2+), N = 5 (TAR, with Mg2+), N = 5 (U7-TAR, with Mg2+), N = 10 (human telomerase P2ab), N = 10 (fluoride riboswitch), N = 10 (preQ1 Class I riboswitch) and N = 10 (preQ1 Class II riboswitch). To analyze distributions of structural parameters (Figs. 4, 6, 7 and Supplementary Fig. 2), we also generated larger sized ensembles by running SAS multiple times to ensure the total size N = 2000 for all systems.
The SAS analysis of TAR and its bulge variants excluded RDCs from the flexible terminal G17-C45 bp as well as those of the C29-G36 bp flanking the apical loop, given differences between the apical loop sequences used to measure RDCs (wild-type or UUCG) and to model (wild-type) TAR. Note that RDCs from G17-C45 and C29-G36 bp were included in the prior study12 and this explains the small differences in RDC RMSD for the two Anton-MD-derived ensembles relative to that reported earlier.
Cross-validation analysis
Cross-validation was performed using two approaches as described previously12. In one approach (inactive random, Supplementary Figs. 1c, d, 8, 9), 10% of the RDC data was randomly removed and SAS was used to generate an ensemble. The RMSD between measured and predicted RDCs was then computed for the left out RDC data. This procedure was repeated 10 times and the final RDC RMSD was averaged over all ten independent runs. For TAR (absence of Mg2+) where we have RDCs measured on four constructs, a second mode (inactive media, Supplementary Fig. 1c, d) of cross-validation was also performed, in which the RDC data set of each construct was left out individually before running SAS. The RMSD between measured and predicted RDCs was then computed for the left out RDC data set. The final RDC RMSD is averaged over the four iterations corresponding to leaving out each RDC data set.
NMR chemical shift data
The 1H, 13C, and 15N chemical shift assignments of TAR have been published previously31,60 and were compared to quantum-mechanical chemical shift predictions. The numerical populations for C2′-endo shown in Fig. 4e were obtained based on the C1′ chemical shift assuming a linear dependence between the range 86 ppm (100 % C2′-endo)54 and 94 ppm (100 % C3′-endo)31.
Automated fragmentation quantum mechanics/molecular mechanics (AF-QM/MM) chemical shift calculations
Chemical shift calculations were performed using a previously described fragmentation procedure21. Each RNA structure was subjected to five steps of conjugate gradient minimization with harmonic restraints of 2 kcal/mol Å2 on all heavy atoms; this regularizes bond lengths and angles to minimize the noise in the results that can arise from very small changes in these geometric parameters. Next, each structure was broken into “quantum” fragments centered on each nucleotide, containing 2–6 neighboring nucleotides, using a heavy-atom distance cutoff of 3.4 Å. The effects of RNA atoms outside the quantum region, and of water and ions in the solvent, were represented as point charges uniformly distributed on the molecular surface of the quantum region and resolved by fitting to Poisson−Boltzmann calculations using the “solinprot” program from the MEAD package61,62. The quantum region was assigned a local dielectric ε of 1 (vacuum); the remaining RNA region had an ε of 4, and the solvent region an ε of 80. GIAO chemical shift calculations were carried out for each fragment, using version 5.0 of the demon-2k program63 using the OLYP functional64 with the pcSseg-1 (triple-ς plus polarization) basis set optimized for chemical shifts65 for the central nucleotide (whose results are reported here), and a DZVP basis for the remaining atoms. Reference shieldings were computed for tetramethylsilane (TMS) using the same functional and basis set. The ensembles of Anton-MD-NMR, FARFAR-NMR as well as a randomly selected Anton-MD and FARFAR-library of size N = 20 were examined. A linear correction was applied to the predicted chemical shifts for each nucleus type individually as described previously29. Note that R2 can be artificially low for spins with small ranges of chemical shifts (e.g., ~1 ppm for C3′/C4′/C6 for central Watson–Crick bps).
Ensemble analysis
All the ensemble structure visualization was performed in PyMOL (https://pymol.org/). The local backbone and sugar torsion angles (Fig. 4b) were calculated using X3DNA-DSSR66. The inter-helical Euler angles (αh, βh, γh) were computed as described previously27 (Figs. 2, 5, 6, 7 and Supplementary Figs. 2, 5). Briefly, the two RNA helices connected by a junction were aligned to two idealized A-form RNA helices (upper helix and lower helix), respectively, and the relative orientation was specified by the inter-helical Euler angles between the upper and lower helix. For TAR and its bulge variants, helix II from G26 to G28 was aligned to the upper helix and helix I from C19 to G21 was aligned to the lower helix, respectively. For human telomerase P2ab, the P2b helix from C89 to G91 was aligned to the upper helix and the P2a helix from U80 to U82 was aligned to the lower helix, respectively. For fluoride riboswitch, the P2 helix from G23 to U25 was aligned to the upper helix and the P1 helix from G2 to G4 was aligned to the lower helix, respectively. For preQ1 Class I riboswitch, the P2 helix from C33 to A35 was aligned to the upper helix and the P1 helix from A5 to G7 was aligned to the lower helix, respectively. For the preQ1 Class II riboswitch, the P4 helix from G37 to G39 was aligned to the upper helix and the P2 helix from G5 to G7 was aligned to the lower helix, respectively. The sign of αh and γh is inverted relative to previously reported values12 such that a positive and negative inter-helical twist angle (αh + γh) corresponds to over- and under-twisting, respectively67.
Junctional topology (Supplementary Fig. 5) was defined as the base-pairing mode, which is detected by X3DNA-DSSR66. If two bases are forming a bp with Leontis–Westhof (LW) classification as “cWW” (e.g., Watson–Crick bp, Wobble bp), a solid line is indicated between the two bases, whereas other LW classifications are indicated as a dashed line (Supplementary Fig. 5).
TAR conformers were considered to be coaxially stacked when the bases comprising A22-U40 and G26-C39 or G21-C41 and U25-U40, were stacked with each other, as defined by X3DNA-DSSR66.
The lower bound estimate of pairwise RMSD between two ensembles was defined as the following: Consider two ensembles A and B with size NA and NB. For every conformer in ensemble A, we found the corresponding conformer in ensemble B that has the lowest pairwise RMSD. This procedure was repeated for every conformer in A to obtain a total of NA RMSD values. We then took the root mean square of all these NA RMSD values. The procedure was also repeated considering ensemble B. Then the minimum of the two root mean square values were selected as a lower bound of the similarity between the two ensembles. By this definition, Anton-MD-NMR and FARFAR-NMR ensembles differ by 3.7 Å while ensembles obtained from multiple independent FARFAR-NMR runs typically differ by 1.4 Å on average.
The cooperativity of flipping bulge nucleotides out was computed from the FARFAR ensemble as follows—for U23, C24, and U25, the probability of independently flipping out was computed as:
| 2 |
where N(nt out | other nt in) is the number of FARFAR-NMR conformers with the nucleotide (nt) of interest being flipped out with the other nucleotides (among U23, C24, and U25) being flipped in, and N(nt in | other nt in) is the number of FARFAR-NMR conformers with U23, C24, and U25 flipped in. Flipping in and flipping out of U23, C24, and U25 were gauged by visual examination of the conformers (Supplementary Fig. 5). The probability of simultaneously flipping out U23, C24, and U25 without cooperativity as then computed as the product of the probabilities of independently flipping out each nucleotide as defined above, i.e.,
| 3 |
This was then compared to the observed probability of U23, C24, and U25 being flipped out P(U23, C24, U25 out obs), which was computed as the fraction of FARFAR-NMR conformers with U23, C24 and U25 flipped out (8/20). Cooperativity was defined as:
| 4 |
where R is the universal gas constant, T is the temperature in Kelvin (298 K).
NMR sample preparation
TAR, TAR-Nm-U23, and TAR-Nm-C24 RNA samples were synthesized using a MerMade 6 Oligo Synthesizer (BioAutomation) via solid-phase synthesis using standard phosphoramidite chemistry and deprotection protocols. 2′-TBDMS protected phosphoramidites (ChemGenes) and 1 μmol standard synthesis columns (1000 Å) were used. The final 5′-DMT (4,4′-dimethoxytrityl) was removed during the synthesis for DMT-off deprotection and PAGE purification. Removal of nucleobase and phosphate protecting groups and cleavage from the 1 μmol columns was achieved using 1 ml of 30% ammonium hydroxide and 30% methylamine (1:1) followed by a 2-h incubation at room temperature. The solution was then air-dried and dissolved in 100 μL DMSO and 125 μL TEA-3HF, followed by 2.5 h incubation at 65 °C following Glen Research protocols (https://www.glenresearch.com/reports/gr19-22) for 2′-O deprotection. The sample was then ethanol precipitated overnight, air-dried, then dissolved in water for gel purification using a 20% (w/v) polyacrylamide gel with 8 M urea and 1× Tris/borate/EDTA. The RNA was removed from the excised gel by electro-elution in 1× Tris/acetic acid/EDTA followed by ethanol precipitation. The RNA was annealed in the water at a concentration of 50 µM by heating at 95 °C for 5 min followed by cooling on ice for 60 min. It was then buffer exchanged using an Amicon Ultra-15 centrifugal filter (EMD Millipore) with a 3 kDa cutoff into NMR buffer (15 mM sodium phosphate, 25 mM NaCl, 0.1 mM EDTA) at pH 6.4. The final concentrations were: ~0.8 mM for TAR, 2.5 mM TAR-Nm-U23, and 1.4 mM TAR-Nm-C24. TAR was also buffer exchanged into NMR buffer containing 3 mM Mg2+.
NMR spectroscopy
All the NMR 2D HSQC experiments in this study were carried out on Bruker Avance III 600-MHz NMR spectrometer equipped with a triple-resonance cryogenic probed at 25 °C. NMR Data were processed using NMRpipe68 and analyzed using SPARKY (T.D. Goddard and D.G. Kneller, SPARKY 3, University of California, San Francisco), respectively. The resonance assignments for Nm-modified TAR were obtained based on the previously reported assignments of TAR31,60 and further confirmed using 2D NOESY experiments.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank members of the Al-Hashimi laboratory and Dr. Dawn Merriman (University of Dayton) for assistance and critical comments on the manuscript. We would like to thank Dr. Andrew Watkins (Stanford University) for advice about FARFAR and Prof. Qi Zhang (University of North Carolina, Chapel Hill) for assistance with regard to secondary structures of complex RNAs. This work was supported by US National Institute for General Medical Sciences (1R01GM132899 to H.M.A. and D.H.), and US National Institute of Health (U54 AI150470 to H.M.A. and D.A.C.), and by Tobacco Settlement Fund (21-5734-0010 to J.D.Y.).
Author contributions
H.S., D.H., J.D.Y., and H.M.A. conceived the project and experimental design. H.S. and J.D.Y. performed FARFAR calculations. A.R. performed MD simulations. H.S. performed sample and select and other ensemble analysis, with assistance from R.R. H.A.A. prepared NMR samples, performed NMR experiments, and analyzed NMR data. D.A.C. performed QM/MM chemical shift calculations. H.M.A., H.S., A.R., and D.H. wrote the paper with input from D.A.C. and J.D.Y.
Data availability
The data that support this study are available from the corresponding authors upon reasonable request. Rosetta FARFAR commands as well as its required input files are included in Supplementary Information. The raw RDC data used in this study can be downloaded from https://github.com/alhashimilab/RDC. The FARFAR-NMR and Anton-MD-NMR ensembles of TAR (N = 20) as well as the idealized A-form helix used for in silico elongation can be downloaded from https://github.com/alhashimilab/Ensemble. The FARFAR-NMR ensemble of TAR (N = 20) with the chemical shifts and RDCs data has been deposited to the Biological Magnetic Resonance Data Bank under the accession code 30788 and the Protein Data Bank under the accession code 7JU1.
Code availability
The Rosetta software suite is available at https://www.rosettacommons.org/software/academic. The AFNMR programs are available at https://github.com/dacase/afnmr. Custom in-house Python scripts for sample and selection are available at https://github.com/alhashimilab/PySAS. Custom in-house Python scrips for calculation of inter-helical Euler angles are available at https://github.com/alhashimilab/ABG_calc.
Competing interests
H.M.A. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug discovery company. The remaining authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
David A. Case, Email: david.case@rutgers.edu
Daniel Herschlag, Email: herschla@stanford.edu.
Joseph D. Yesselman, Email: jyesselm@unl.edu
Hashim M. Al-Hashimi, Email: hashim.al.hashimi@duke.edu
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-19371-y.
References
- 1.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarmoskaite I, et al. A quantitative and predictive model for RNA binding by human pumilio proteins. Mol. Cell. 2019;74:966–981. doi: 10.1016/j.molcel.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harcourt EM, Kietrys AM, Kool ET. Chemical and structural effects of base modifications in messenger RNA. Nature. 2017;541:339–346. doi: 10.1038/nature21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinert, H. et al. Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. Elife6, e21297 (2017). [DOI] [PMC free article] [PubMed]
- 6.Ganser LR, Kelly ML, Herschlag D, Al-Hashimi HM. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019;20:474–489. doi: 10.1038/s41580-019-0136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J. Mol. Biol. 2001;308:1011–1032. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- 8.Ozenne V, et al. Mapping the potential energy landscape of intrinsically disordered proteins at amino acid resolution. J. Am. Chem. Soc. 2012;134:15138–15148. doi: 10.1021/ja306905s. [DOI] [PubMed] [Google Scholar]
- 9.Lange OF, et al. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 10.Showalter SA, Brüschweiler R. Quantitative molecular ensemble interpretation of NMR dipolar couplings without restraints. J. Am. Chem. Soc. 2007;129:4158–4159. doi: 10.1021/ja070658d. [DOI] [PubMed] [Google Scholar]
- 11.Sponer J, et al. RNA structural dynamics as captured by molecular simulations: a comprehensive overview. Chem. Rev. 2018;118:4177–4338. doi: 10.1021/acs.chemrev.7b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salmon L, Bascom G, Andricioaei I, Al-Hashimi HM. A general method for constructing atomic-resolution RNA ensembles using NMR residual dipolar couplings: the basis for interhelical motions revealed. J. Am. Chem. Soc. 2013;135:5457–5466. doi: 10.1021/ja400920w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottaro S, Bussi G, Kennedy SD, Turner DH, Lindorff-Larsen K. Conformational ensembles of RNA oligonucleotides from integrating NMR and molecular simulations. Sci. Adv. 2018;4:eaar8521. doi: 10.1126/sciadv.aar8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salmon L, Yang S, Al-Hashimi HM. Advances in the determination of nucleic acid conformational ensembles. Annu Rev. Phys. Chem. 2014;65:293–316. doi: 10.1146/annurev-physchem-040412-110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watkins AM, Rangan R, Das R. FARFAR2: improved de novo rosetta prediction of complex global RNA folds. Structure. 2020;28:963–976. doi: 10.1016/j.str.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao Z, et al. RNA-puzzles round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA. 2017;23:655–672. doi: 10.1261/rna.060368.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 18.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins - information for structure determination in solution. Proc. Natl Acad. Sci. USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Campbell SL, Dokholyan NV. Deciphering protein dynamics from NMR data using explicit structure sampling and selection. Biophys. J. 2007;93:2300–2306. doi: 10.1529/biophysj.107.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 21.Swails J, Zhu T, He X, Case DA. AFNMR: automated fragmentation quantum mechanical calculation of NMR chemical shifts for biomolecules. J. Biomol. NMR. 2015;63:125–139. doi: 10.1007/s10858-015-9970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 24.Merriman DK, et al. Increasing the length of poly-pyrimidine bulges broadens RNA conformational ensembles with minimal impact on stacking energetics. RNA. 2018;24:1363–1376. doi: 10.1261/rna.066258.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musselman C, et al. Impact of static and dynamic A-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J. Biomol. NMR. 2006;36:235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 26.Denning EJ, Priyakumar UD, Nilsson L, Mackerell AD., Jr Impact of 2’-hydroxyl sampling on the conformational properties of RNA: update of the CHARMM all-atom additive force field for RNA. J. Comput. Chem. 2011;32:1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailor MH, Mustoe AM, Brooks CL, III, Al-Hashimi HM. 3D maps of RNA interhelical junctions. Nat. Protoc. 2011;6:1536–1545. doi: 10.1038/nprot.2011.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi X, Walker P, Harbury PB, Herschlag D. Determination of the conformational ensemble of the TAR RNA by X-ray scattering interferometry. Nucleic Acids Res. 2017;45:e64. doi: 10.1093/nar/gkx289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H, et al. Atomic structures of excited state A-T Hoogsteen base pairs in duplex DNA by combining NMR relaxation dispersion, mutagenesis, and chemical shift calculations. J. Biomol. NMR. 2018;70:229–244. doi: 10.1007/s10858-018-0177-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Case DA. Chemical shifts in biomolecules. Curr. Opin. Struct. Biol. 2013;23:172–176. doi: 10.1016/j.sbi.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clay MC, Ganser LR, Merriman DK, Al-Hashimi HM. Resolving sugar puckers in RNA excited states exposes slow modes of repuckering dynamics. Nucleic Acids Res. 2017;45:e134. doi: 10.1093/nar/gkx525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies DB. Conformations of nucleosides and nucleotides. Prog. Nucl. Magn. Reson. Spectrosc. 1978;12:135–225. doi: 10.1016/0079-6565(78)80006-5. [DOI] [Google Scholar]
- 33.Ippolito JA, Steitz TA. A 1.3-angstrom resolution crystal structure of the HIV-1 trans- activation response region RNA stem reveals a metal ion- dependent bulge conformation. Proc. Natl Acad. Sci. USA. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM. RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew. Chem. Int. Ed. Engl. 2010;49:5731–5733. doi: 10.1002/anie.201000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawai G, et al. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc. Natl Acad. Sci. USA. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao B, Guffy SL, Williams B, Zhang Q. An excited state underlies gene regulation of a transcriptional riboswitch. Nat. Chem. Biol. 2017;13:968–974. doi: 10.1038/nchembio.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Kang M, Peterson RD, Feigon J. Comparison of solution and crystal structures of preQ1 riboswitch reveals calcium-induced changes in conformation and dynamics. J. Am. Chem. Soc. 2011;133:5190–5193. doi: 10.1021/ja111769g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang M, Eichhorn CD, Feigon J. Structural determinants for ligand capture by a class II preQ1 riboswitch. Proc. Natl Acad. Sci. USA. 2014;111:E663–E671. doi: 10.1073/pnas.1400126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Popenda M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40:e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leman JK, et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods. 2020;17:665–680. doi: 10.1038/s41592-020-0848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nilges M, Gronenborn AM, Brunger AT, Clore GM. Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints. Application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng. 1988;2:27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- 43.Camilloni C, Vendruscolo M. Using pseudocontact shifts and residual dipolar couplings as exact NMR restraints for the determination of protein structural ensembles. Biochemistry. 2015;54:7470–7476. doi: 10.1021/acs.biochem.5b01138. [DOI] [PubMed] [Google Scholar]
- 44.Salmon L, et al. Modulating RNA alignment using directional dynamic kinks: application in determining an atomic-resolution ensemble for a hairpin using NMR residual dipolar couplings. J. Am. Chem. Soc. 2015;137:12954–12965. doi: 10.1021/jacs.5b07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 46.Abraham MJ, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 47.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 48.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/PhysRevA.31.1695. [DOI] [PubMed] [Google Scholar]
- 49.Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys.126, 014101 (2007). [DOI] [PubMed]
- 50.Cheatham TE, III, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 51.Zgarbova M, et al. Refinement of the Cornell et al. Nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 53.Joung IS, Cheatham TE., III Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rangadurai A, et al. Why are Hoogsteen base pairs energetically disfavored in A-RNA compared to B-DNA? Nucleic Acids Res. 2018;46:11099–11114. doi: 10.1093/nar/gky885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan D, Piana S, Dirks RM, Shaw DE. RNA force field with accuracy comparable to state-of-the-art protein force fields. Proc. Natl Acad. Sci. USA. 2018;115:E1346–e1355. doi: 10.1073/pnas.1713027115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piana S, Donchev AG, Robustelli P, Shaw DE. Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B. 2015;119:5113–5123. doi: 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- 57.MacKerell ADJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 58.Parrinello M, Rahman A. Polymorphic transitions in single-crystals - a new molecular-dynamics method. J. Appl Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 59.Zweckstetter M, Bax A. Predicition of sterically induced alignment in a dilute liquid crystalline phase; aid to protein structure determination by NMR. J. Am. Chem. Soc. 2000;122:3791–3792. doi: 10.1021/ja0000908. [DOI] [Google Scholar]
- 60.Dethoff EA, et al. Characterizing complex dynamics in the transactivation response element apical loop and motional correlations with the bulge by NMR, molecular dynamics, and mutagenesis. Biophys. J. 2008;95:3906–3915. doi: 10.1529/biophysj.108.140285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bashford D, Karplus M. pKa’s of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990;29:10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- 62.Richardson WH, Peng C, Bashford D, Noodleman L, Case DA. Incorporating solvation effects into density functional theory: calculation of absolute acidities. Int J. Quantum Chem. 1998;61:207–217. doi: 10.1002/(SICI)1097-461X(1997)61:2<207::AID-QUA3>3.0.CO;2-#. [DOI] [Google Scholar]
- 63.Koster, A. M. et al. deMon2k, version 5 (Cinvestav, Mexico City, 2011).
- 64.Handy NC, Cohen AJ. Left-right correlation energy. Mol. Phys. 2001;99:403–412. doi: 10.1080/00268970010018431. [DOI] [Google Scholar]
- 65.Jensen F. Segmented contracted basis sets optimized for nuclear magnetic shielding. J. Chem. Theory Comput. 2015;11:132–138. doi: 10.1021/ct5009526. [DOI] [PubMed] [Google Scholar]
- 66.Lu, X. J., Bussemaker, H. J. & Olson, W. K. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res.43, e142 (2015). [DOI] [PMC free article] [PubMed]
- 67.Zhou H, et al. New insights into Hoogsteen base pairs in DNA duplexes from a structure-based survey. Nucleic Acids Res. 2015;43:3420–3433. doi: 10.1093/nar/gkv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delaglio F, et al. Nmrpipe - a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 69.Al-Hashimi HM, et al. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that support this study are available from the corresponding authors upon reasonable request. Rosetta FARFAR commands as well as its required input files are included in Supplementary Information. The raw RDC data used in this study can be downloaded from https://github.com/alhashimilab/RDC. The FARFAR-NMR and Anton-MD-NMR ensembles of TAR (N = 20) as well as the idealized A-form helix used for in silico elongation can be downloaded from https://github.com/alhashimilab/Ensemble. The FARFAR-NMR ensemble of TAR (N = 20) with the chemical shifts and RDCs data has been deposited to the Biological Magnetic Resonance Data Bank under the accession code 30788 and the Protein Data Bank under the accession code 7JU1.
The Rosetta software suite is available at https://www.rosettacommons.org/software/academic. The AFNMR programs are available at https://github.com/dacase/afnmr. Custom in-house Python scripts for sample and selection are available at https://github.com/alhashimilab/PySAS. Custom in-house Python scrips for calculation of inter-helical Euler angles are available at https://github.com/alhashimilab/ABG_calc.