Synopsis
Given their remarkable phenotypic diversity, dogs present a unique opportunity for investigating the genetic bases of cognitive and behavioral traits. Our previous work demonstrated that genetic relatedness among breeds accounts for a substantial portion of variation in dog cognition. Here, we investigated the genetic architecture of breed differences in cognition, seeking to identify genes that contribute to variation in cognitive phenotypes. To do so, we combined cognitive data from the citizen science project Dognition.com with published breed-average genetic polymorphism data, resulting in a dataset of 1654 individuals with cognitive phenotypes representing 49 breeds. We conducted a breed-average genome-wide association study to identify specific polymorphisms associated with breed differences in inhibitory control, communication, memory, and physical reasoning. We found five single nucleotide polymorphisms (SNPs) that reached genome-wide significance after Bonferroni correction, located in EML1, OR52E2, HS3ST5, a U6 spliceosomal RNA, and a long noncoding RNA. When we combined results across multiple SNPs within the same gene, we identified 188 genes implicated in breed differences in cognition. This gene set included more genes than expected by chance that were (1) differentially expressed in brain tissue and (2) involved in nervous system functions including peripheral nervous system development, Wnt signaling, presynapse assembly, and synaptic vesicle exocytosis. These results advance our understanding of the genetic underpinnings of complex cognitive phenotypes and identify specific genetic variants for further research.
Introduction
Comparative phylogenetic studies of animal cognition have the potential to illuminate genetic mechanisms that contribute to variation in cognitive phenotypes (MacLean and Nunn 2017). However, our ability to identify genotype–phenotype associations in these contexts requires both cognitive assays that can be meaningfully employed in large and diverse comparative samples and high-quality genomic data on the taxa being studied. Due to these challenges, few studies have implemented genomic approaches in comparative studies of animal cognition (Chittka et al. 2012).
As a result of recent advances in the study of canine cognition and dog genomics, domestic dogs (Canis lupus familiaris) present rich opportunities for these types of studies. As the first domesticated species, and one which has undergone strong diversifying selection due to selective breeding by humans, dog breeds are characterized by an extraordinary degree of phenotypic diversity. Currently more than 400 dog breeds are recognized internationally, each representing a closed breeding population with distinct phenotypic features (Karlsson and Lindblad-Toh 2008). In the last two decades, scientists have made rapid progress toward characterizing the genetic underpinnings of phenotypic variation among breeds. In 2003, the first complete dog genome was published (Kirkness et al. 2003), which was shortly followed by a dense single nucleotide polymorphism (SNP) map (Lindblad-Toh et al. 2005), the development of several commercially-available genotyping arrays (Karlsson et al. 2007; Boyko et al. 2010; Vaysse et al. 2011), and phylogenetic analyses of dog breeds (VonHoldt et al. 2010; Parker et al. 2017). These tools have been used in genome-wide association studies (GWASs) to identify genetic contributions to phenotypic variability among breeds, including aspects of morphology (Boyko et al. 2010), disease susceptibility (Ahonen et al. 2013), and athleticism (Kim et al. 2018).
However, we know considerably less about genetic factors associated with variance in cognitive and behavioral phenotypes, despite growing evidence for breed differences in these domains (Scott and Fuller 1965; Wilsson and Sundgren 1997; Wobber et al. 2009; Jakovcevic et al. 2010; Serpell and Duffy 2014; Horschler et al. 2019). Most studies to date have focused on genetic correlates of behavioral differences between breeds. For example, Vaysse et al. (2011) performed an across-breed GWAS for breed boldness (using data from Jones et al. 2008) and five personality traits—sociability, curiosity, playfulness, chase-proneness, and aggressiveness—using data from the Swedish Kennel Club (Svartberg and Forkman 2002). These analyses identified a small set of loci associated with boldness and sociability, some of which occurred in the same genomic regions associated with ear morphology and body size. Using data from the Canine Behavioral Assessment and Research Questionnaire (C-BARQ), Zapata et al. (2016) assessed associations between allele frequencies across approximately 45,000 SNPs and breed differences in fear and aggression; they identified several loci associated with these traits, some of which were known to also control variation in body size. Most recently, MacLean et al. (2019) conducted an across-breed GWAS on traits measured by the C-BARQ, using a sample of 101 breeds in conjunction with genetic data from more than 100,000 loci in the dog genome. These analyses identified 131 SNPs implicated in breed differences in behavior, which were found disproportionally in genes that are highly expressed in the brain and which are implicated in nervous system functions.
Although breed differences in cognition are less well studied, we have recently analyzed patterns of variation across breeds using data from the citizen science project Dognition.com (Gnanadesikan et al. 2020). Analysis of this dataset identified four factors underlying individual differences in cognition, which were interpreted as reflecting variation in inhibitory control, communication, memory, and physical reasoning (Table 1 and Supplementary Fig. S1), although it should be noted that the inhibitory control factor is in a single social context and is therefore likely to be an incomplete measure. Gnanadesikan et al. (2020) investigated the extent to which breed-level variation in these traits covaried with genetic similarity among breeds—a modified form of narrow-sense heritability (Visscher et al. 2008) which we term “among-breed heritability”—finding that all four traits were moderately to highly heritable. In this study, we build on this work by conducting a GWAS on breed-average differences in cognitive phenotypes. Given the heritable variation in these traits, we hypothesized that by modeling breed-average cognitive phenotypes as a function of allele frequency across a large set of SNPs, we could identify molecular variants associated with these cognitive traits. We further hypothesized that if the genes implicated in these analyses contribute to breed differences in cognition, they should be highly expressed in brain tissue and enriched for biological processes related to nervous system functions.
Table 1.
Overview of the Dognition measures and their primary (>0.2) factor loadings, with positive (+) and negative (−) loadings noted accordingly
| Factor | (Loading direction) | Task | Description | Measure |
|---|---|---|---|---|
|
Inhibitory Control (+) |
Forbidden food: watching mean |
The participant places a treat on the floor in front of dog, verbally forbids the dog from taking it, and watches their dog | Mean latency to eat the treat, up to 90 s | |
|
Forbidden food: back turned |
Identical to the watching condition, but the participant turns their back to the dog after placing the food on the ground | Mean latency to eat the treat, up to 90 s | ||
|
Forbidden food: eyes covered |
Identical to the watching condition, but the participant covers their eyes after placing the food on the ground | Mean latency to eat the treat, up to 90 s | ||
|
Communication (+) |
Arm pointing | The participant places two treats on the ground—one to the left and one to the right—and gestures towards one of them using an extended arm and index finger | Proportion of first approaches to the indicated treat | |
| Foot pointing | Identical to arm pointing, except the gesture is performed with the participant’s foot | Proportion of first approaches to the indicated treat | ||
|
Memory (+) |
Delayed memory | The participant visibly places food under one of two cups, but waits for a delay (60, 90, 120, 180 s) before the dog is allowed to search | Proportion of first approaches to the correct (remembered) cup | |
|
Memory (+) / Communication (−) |
Memory vs. pointing |
The participant visibly places food under one of two cups and points with their arm and index finger to the other cup | Proportion of first approaches to the visibly baited (remembered) cup | |
|
Memory (+) / Physical reasoning (−) |
Memory vs. smell |
The participant visibly places food under one of two cups, but then blocks the dog’s view and moves the food to under the other cup | Proportion of first approaches to the visibly baited (remembered) cup | |
|
Physical reasoning (+) |
Inferential reasoning | Out of view of the dog, the participant hides food under one of two cups. The participant then lifts the empty cup, revealing that there is no food underneath | Proportion of first approaches to the correct (not shown) cup | |
| Physical reasoning | The participant places food under one of two folded sheets of paper, such that the food props up the one piece of paper, while the other lies flat | Proportion of first approaches to the correct (displaced) side |
Two tasks were excluded from the analysis: yawning, which as a binary measure was not well suited to factor analysis, and eye contact, which did not load significantly onto any factor in an initial analysis. For more information on the battery, including the order of trials and familiarizations, see Stewart et al. (2015). For a visual representation of the factor loadings, see Supplementary Fig. S1, and for a full description of the factor analysis see Gnanadesikan et al. (2020)
In previous studies, body and brain weight have been strongly associated with aspects of behavior and cognition in dogs (McGreevy et al. 2013; Horschler et al. 2019) as well as other species (Deaner et al. 2007; Sol et al. 2008; Kotrschal et al. 2013; MacLean et al. 2014; Benson-Amram et al. 2016). Whether body weight is included as a covariate in genomic analyses of behavioral traits in dogs has varied in prior research, with advantages and disadvantages to both approaches. While models that do not control for body mass tend to identify size-related genetic variants (e.g., Jones et al. 2008; Zapata et al. 2016), these same variants may be functionally linked to cognition or behavior through effects on brain architecture (Horschler and MacLean 2019; Horschler et al. 2019); in contrast, models that do control for body mass should reveal residual variation among breeds not explained by effects of body or brain mass (e.g., MacLean et al. 2019). Given the tradeoffs between these approaches, we conducted analyses both with and without controlling for breed-average weight.
Methods
Cognitive data
The cognitive dataset was collected through Dognition.com, a citizen science website that guides owners through experiments they can conduct at home with their own dogs. Previous analyses of data from Dognition.com have replicated findings from similar protocols implemented in traditional laboratory settings, supporting the validity of this citizen science approach (Stewart et al. 2015). The cognitive outcome measures used here are the results of a factor analysis reported in (Gnanadesikan et al. 2020). In brief, an exploratory factor analysis was conducted using the psych package (Revelle 2018) in R version 3.5.2 (R Core Team 2018), with data from all adult (>1-year old) purebred dogs in the Dognition dataset (n = 2044; nbreeds = 172). Factoring was conducted using the minimizing residuals method (minres) and an oblique rotation (oblimin). Two tasks were excluded from the analysis: yawning, which as a binary measure was not well suited to factor analysis, and eye contact, which did not load significantly onto any factor in initial analysis. Four factors were extracted, as indicated by a parallel analysis of simulated and resampled data. These factors were interpreted as reflecting latent cognitive variables related to inhibitory control, communication, memory, and physical reasoning (Table 1 and Supplementary Fig. S1), although as in any factor analysis, these names are simplifications of a more complex factor structure. For a more detailed discussion of these interpretations and their limitations, see the supplementary information, as well as Gnanadesikan et al. (2020).
Genetic data
Genetic data were obtained from a publicly available data set (Parker et al. 2017) that combined newly analyzed data with previously published data (Vaysse et al. 2011; Hayward et al. 2016), all of which was collected using the Illumina CanineHD bead array, supplemented with three publicly available genome sequences. The full genetic dataset includes 150,067 SNPs from 1346 dogs representing 161 breeds.
Breed average similarity was calculated in an identical manner to Gnanadesikan et al. (2020), as an identity-by-state (IBS) matrix: the proportion of SNPs that were identical by state for each pair of individuals was calculated using PLINK (Purcell et al. 2015; Purcell and Chang 2018). These values were then averaged for every pair of breeds to generate a breed-average IBS matrix. This breed-level IBS matrix was extrapolated to an individual-level IBS matrix by assuming breed-average similarity between each pair of individuals: for individuals of different breeds, the IBS value was set to the average similarity between those two breeds; for individuals of the same breed, the average similarity of individuals within that breed was used (supplementary information).
Combined dataset
Our combined dataset included cognitive data on 1654 individuals; these individuals represented 49 breeds for which we had both cognitive data from at least 10 individuals and breed-average genetic data aggregated from Parker et al. (2017).
Analysis
Genome-wide association
The associations between SNPs and cognitive measures were modeled using an efficient mixed model association approach (Kang et al. 2008), as implemented in the EMMREML package (Akdemir and Godfrey 2015) in R version 3.5.1 (R Core Team 2018). We modeled breed-average cognitive factor scores as a function of breed-average allele frequency while controlling for breed-average relatedness (model details in the supplementary information). We fit two models per factor: one while controlling for breed-average body weight—using data from the C-BARQ (Hsu and Serpell 2003; McGreevy et al. 2013)—and one without this covariate. We considered both models because (1) brain weight and volume covary strongly with body weight in dogs (Jardim-Messeder et al. 2017; Horschler et al. 2019) and brain size has repeatedly been suggested to affect various cognitive processes (Deaner et al. 2007; Sol et al. 2008; Kotrschal et al. 2013; MacLean et al. 2014; Benson-Amram et al. 2016); (2) Horschler et al. (2019) found estimated brain weight was positively associated with some measures in the Dognition.com test battery, namely those related to executive functions, including short-term memory and self-control; and (3) we previously found that controlling for breed-average body weight reduced the estimated among-breed heritability of the inhibitory control factor from 0.7 to 0.5 (Gnanadesikan et al. 2020).
To avoid models on rare variants, the SNPs used for GWAS modeling were further filtered to those with a median minor allele frequency across breeds of at least 0.05, resulting in 124,821 SNPs. Although we control for breed-average relatedness, cryptic relatedness and population structure can lead to an increased false-positive rate in genomic studies (Devlin and Roeder 1999). The distributions of P-values for the first three factors (inhibitory control, communication, and memory) were found to be inflated (λ > 1) both with breed-average weight as a covariate (λ1 = 1.077, λ2 = 1.208, λ3 = 1.117, λ4 = 0.941) and without (λ1 = 1.054, λ2 = 1.200, λ3 = 1.135, λ4 = 0.998). This inflation may be due in part to polygenicity (Yang et al. 2011), however, we took a conservative approach and corrected the inflated distributions using the genomic control method (Devlin and Roeder 1999; Amin et al. 2007).
Results are reported (1) at the SNP level, with a Bonferroni corrected threshold for genome-wide significance (<4 × 10−7; α/NSNP = 0.05/124,821) and (2) at the gene level, by aggregating across all SNPs in a given gene. In the latter, P-values were combined across SNPs within a gene using Fisher’s method and the Nyholt correction for multiple testing of SNPs in linkage disequilibrium (Nyholt 2004), as implemented in the R package poolr (Cinar and Viechtbauer 2020). The Nyholt correction was designed to correct for linkage disequilibrium and the resultant nonindependence of SNPs (Nyholt 2004), and simulations have supported its ability to control false positives under conditions of both moderate and high linkage disequilibrium (Nicodemus et al. 2005). We implemented a false discovery rate threshold for the number of genes included (n = 14,442) when determining gene-level significance (Benjamini and Hochberg 1995). Using BEDtools (Quinlan and Hall 2010), SNPs were mapped to their closest gene in the most recent dog genome (CanFam 3.1 assembly [Hoeppner et al. 2014], accessed [June 2020] through the UCSC Table Browser [Karolchik 2004]). We only considered SNPs that were found in genes (distance = 0), which includes both upstream (5′) and downstream (3′) untranslated regions.
Enrichment analyses
To examine the biological functions of genes identified in the gene-level analyses described above, we performed two types of gene set enrichment analysis (Subramanian et al. 2005): gene ontology (GO) and tissue-specific expression. GO analyses assessed whether the set of genes identified in the GWAS are disproportionately related to specific biological functions, as annotated in the Gene Ontology knowledgebase (The Gene Ontology Consortium 2000, 2019). GO analyses were conducted using the topGO R package (Alexa and Rahnenfuhrer 2018), using a Fisher exact test and the “weight01” algorithm. ENSEMBL gene identifiers were associated with GO terms using the biomaRt R package (Durinck et al. 2005, 2009). The gene sets used for enrichment analyses included all genes with an aggregated gene-level P ≤0.05 after false discovery rate correction (Benjamini and Hochberg 1995). Network plots for enriched GO terms were made by calculating term similarities (Resnik score) with all enriched terms using NaviGO (Wei et al. 2017) followed by network visualization using the igraph R package (Csardi and Nepusz 2006). For the no-covariates analysis only, due to the larger number of enriched terms, the plot was hand-curated to highlight terms related to nervous system functions and genetic regulation.
Tissue-specific enrichment analyses assessed whether the genes identified in our gene-level analysis are biased toward expression in specific tissues, controlling for background rates of tissue-specific expression across all genes included in the analysis. Statistical significance was assessed using hypergeometric tests. For these analyses, we did not control for multiple tests across different tissue types, as we were most interested in relative patterns across tissues and species-specific expression profiles (see below). We compared the gene sets produced by our analysis with data on tissue-specific gene expression in both dogs (Briggs et al. 2011) and humans (Uhlén et al. 2015) using the TissueEnrich R package (Jain and Tuteja 2019). The dog expression data come from a study across 10 tissues from 4 dogs using a microarray, from which the data has been averaged across dogs (two beagles and two mixed-breed dogs) (Briggs et al. 2011). Because tissue-specific expression is highly conserved for orthologous genes in dog and humans (Briggs et al. 2011; Li et al. 2013), we also conducted analyses using human gene-expression data, which have been measured in a greater diversity of tissue types (ntissue = 35). Enrichment analyses were performed separately for the results of models with and without breed-average body weight as a covariate.
Contextual analysis of candidate genes
In order to compare our results to previously identified candidate genes implicated in behavior and domestication, we cross-referenced the combined results of our gene-level analyses with a database compiled from 13 canid behavioral genomics studies (Saetre et al. 2004; Karlsson et al. 2007; Cadieu et al. 2009; Chase et al. 2009; VonHoldt et al. 2010; Vaysse et al. 2011; Axelsson et al. 2013; Wang et al. 2013, 2018; Freedman et al. 2016; Hekman et al. 2018; Kukekova et al. 2018; MacLean et al. 2019).
Results
Genome-wide association
Models not including body mass as a covariate
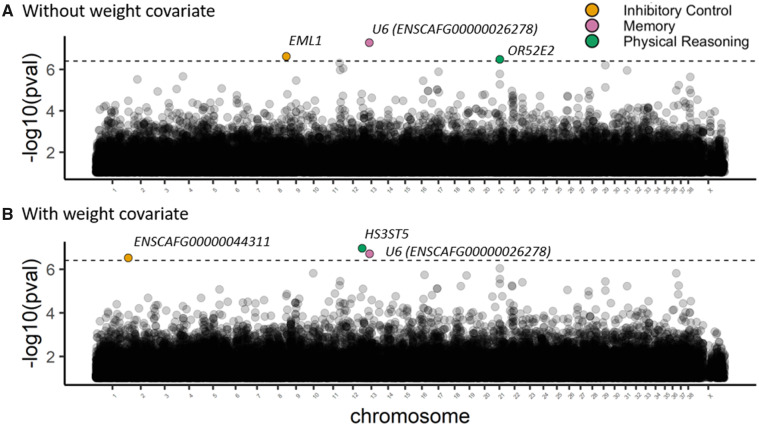
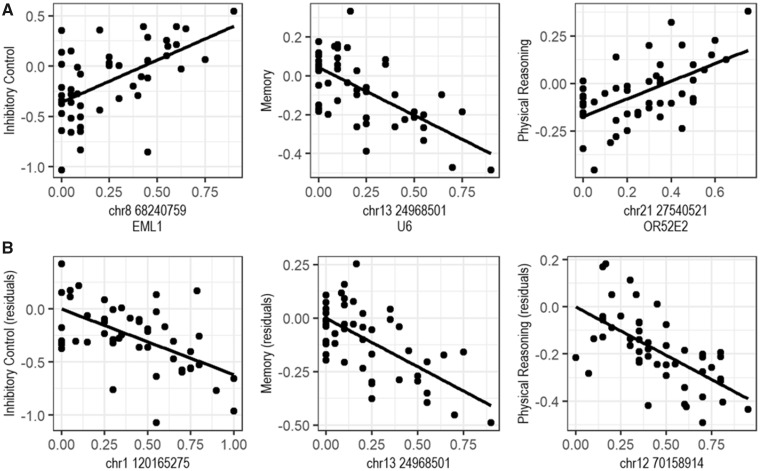
Without controlling for body mass, our breed-average GWAS of cognitive traits revealed three SNPs that reached genome-wide significance after a Bonferroni correction (Fig. 1). Scores on the inhibitory control factor were associated with a SNP (chromosome 8, position 68240759, and rs24514902) in EML1 (P = 0.030); scores on the memory factor were associated with a SNP (chromosome 13 and position 24968501) in ENSCAFG00000026278, a U6 spliceosomal RNA (P = 0.006); and scores on the physical reasoning factor were associated with a SNP (chromosome 21, position 27540521, and rs851264582) in OR52E2 (P = 0.043). No significant associations were found for the communication factor (see Supplementary Table S2 for all SNP-level results). Allele frequencies at these SNPs accounted for 7–34% of the variance in the inhibitory control (34.2%), memory (9.88%), and physical reasoning factor (7.31%) scores (Fig. 2).
Fig. 1.
Manhattan plots showing SNP-level associations with cognitive factors both (A) without covariates and (B) with weight as a covariate. The dashed line represents the threshold for genome-wide significance after Bonferroni correction, and SNPs achieving genome-wide statistical significance are color-coded for the associated cognitive factor.
Fig. 2.
Breed-average trait scores or residuals by allele frequency for the SNPs that reached genome-wide significance. The top panel (A) shows the no-covariates results, and the bottom panel (B) shows the weight-controlled results.
Models including body mass as a covariate
Controlling for breed-average weight, three SNPs were identified as significantly associated with a cognitive phenotype, one of which was the same as in the previous analysis (Fig. 1). As in the analyses without the body weight covariate, we found that breed-average allele frequency for one SNP in ENSCAFG00000026278 (chromosome 13 and position 24968501) was associated with breed differences in memory (P = 0.025). Additionally, a SNP in ENSCAFG00000044311 (chromosome 1 and position 120165275), a long noncoding RNA, was associated with breed differences in inhibitory control (P = 0.039). Lastly, breed differences on the physical reasoning factor were associated with allele frequency for a SNP in HS3ST5 (chromosome 12 and position 70158914; P = 0.014). Again, no significant associations were found for the communication factor. In analyses controlling for body weight, the variance explained by allele frequency at these SNPs was 28.9% for inhibitory control, 8.47% for memory, and 6.74% for physical reasoning (Fig. 2).
Enrichment analyses
Models not including body mass as a covariate
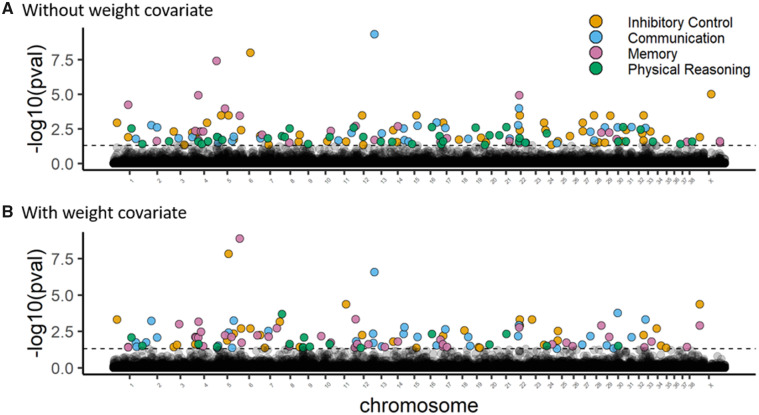
Aggregating across SNPs within each gene, we identified 52 genes associated with inhibitory control, 32 genes associated with communication, 27 genes associated with memory, and 39 genes associated with physical reasoning (Fig. 3 and Supplementary Table S3). In total, across these 4 phenotypes, 140 unique genes were identified. Three of these genes were associated with two traits: KLHL1 (communication and physical reasoning), WDR27 (inhibitory control and memory), and KCNQ5 (inhibitory control and physical reasoning).
Fig. 3.
Manhattan plots showing gene-level associations with cognitive factors both (A) without covariates and (B) with breed-average body weight as a covariate. Gene-level associations were produced by aggregation of SNPs within each gene using Fisher’s method, corrected for linkage disequilibrium by Nyholt’s method. The dashed line represents the threshold for genome-wide significance after false discovery rate correction, and genes achieving genome-wide statistical significance are color-coded for the associated cognitive factor.
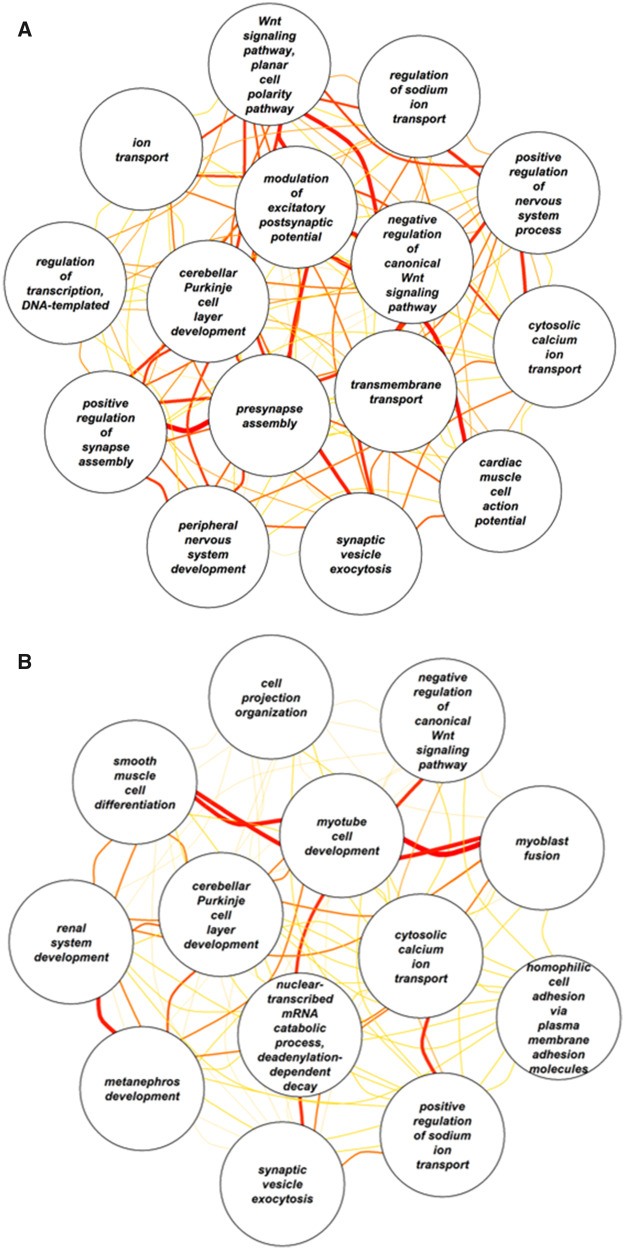
We conducted a GO analysis to assess whether this combined geneset was enriched for specific biological functions. This analysis identified 28 over-represented GO terms, including neurological functions such as peripheral nervous system development, presynapse assembly, cerebellar Purkinje cell layer development, negative regulation of canonical Wnt signaling pathway, and synaptic vesicle exocytosis; regulation of transcription, DNA templated was also enriched (Fig. 4). A full list of enriched GO terms is provided in Supplementary Table S4.
Fig. 4.
Network plot of significantly enriched GO terms for the analyses (A) without covariates, limited to nervous system and genetic regulation terms, and (B) with breed-average body weight as a covariate, showing all enriched terms. Line colors and widths represent Resnik’s similarity scores between GO terms, with wider and redder lines reflecting greater similarity between terms. A complete list of enriched GO terms is shown in Supplementary Tables S4 and S7, respectively.
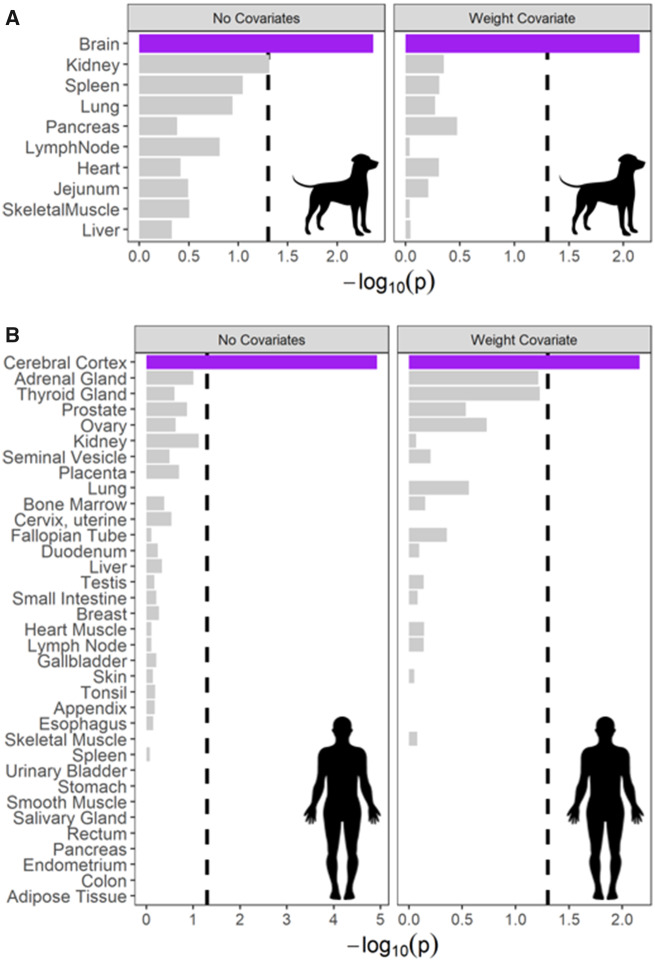
To explore the expression profiles of the collective set of genes identified in our gene-level analysis, we conducted tissue enrichment analyses using tissue-specific gene expression data from published sources. Using expression data from dogs (Briggs et al. 2011), we found that this gene set included more genes that are highly expressed in the brain (cerebrum) than are expected by chance (P = 0.004), as well as in the kidney (P = 0.048; Fig. 5A). Similarly, using expression data from humans (Uhlén et al. 2015), this gene set included more genes that are highly expressed in the cerebral cortex (P = 1.19 × 10−5; Fig. 5B) than would be expected by chance.
Fig. 5.
Results of a tissue-enrichment analysis based on tissue expression data from (A) dogs and (B)humans, both with weight as a covariate (right panels) and without (left panels). In all cases, a plurality of genes are more highly expressed in the brain or cerebral cortex (purple bars). This was more than expected by chance using a hypergeometric test (dashed line, P = 0.05, without correcting for multiple comparisons) in all cases.
Models including body mass as a covariate
Controlling for breed-average body weight, aggregation at the gene level identified 32 genes implicated in inhibitory control, 33 genes implicated in communication, 39 genes implicated in memory, and 17 genes implicated in physical reasoning (Fig. 3 and Supplementary Table S6). In total across these four phenotypes, 117 unique genes were identified. Three of these genes were associated with two traits: CDH13, NFIA (inhibitory control and communication) and MCU (memory and physical reasoning). This gene set was enriched for 13 GO terms, again including synaptic vesicle exocytosis, negative regulation of canonical Wnt signaling pathway, and cerebellar Purkinje cell layer development (Fig. 4 and Supplementary Table S7). Using gene expression data from dogs, we found that the genes identified in our analysis include more genes that are highly expressed in the brain than would be expected by chance (P = 0.007; Fig. 5A). Similarly, using expression data from humans, we found an enrichment for genes that are highly expressed in cerebral cortex (P = 0.007; Fig. 5B).
Out of these 117 genes identified after controlling for breed-average body weight, 69 genes overlapped with the gene set in our no-covariates analysis. This overlap between the two analyses is significantly more than expected by chance (hypergeometric test, P = 2.66 × 10−115). In total, 188 unique genes were identified across both analyses. Of these, at least 69 genes have also been previously identified in previous association studies of canid behavior and domestication (Supplementary Table S8), including 44 identified in a similar breed-average approach with behavioral data from the C-BARQ (MacLean et al. 2019) and 22 identified through both genomic and transcriptomic work in tame and aggressive foxes (Hekman et al. 2018; Kukekova et al. 2018; Wang et al. 2018).
Discussion
We used breed-average cognitive data from the citizen science project Dognition.com and breed-average allele frequencies across 124,821 SNPs from Parker et al. (2017) to perform a GWAS on breed differences in dog cognition. We identified three SNPs that reached genome-wide significance when not controlling for breed-average weight, and three SNPs—one of which was the same across analyses—when we included breed-average weight as a covariate. Using a meta-analytic approach to aggregate results at the gene level (while controlling for linkage disequilibrium), the genes identified in our analysis tended to be highly expressed in brain tissue and enriched for biological processes that include nervous system functions and genetic regulation, suggesting plausible mechanisms through which these genes may influence breed differences in performance on cognitive tasks.
The specific SNPs that reached genome-wide significance are found in genes that also have known roles in both neural functions and genetic regulation. Specifically, a variant in EML1, also known as echinoderm microtubule-associated protein-like 1, was associated with breed differences in inhibitory control. This gene is known to be involved in neurogenesis and neural organization, with variants in this gene associated with neuronal heterotopia and congenital hydrocephalus (Kielar et al. 2014; Shaheen et al. 2017). Intriguingly, in human studies, several variants in this gene have been associated with brain volume (Zhao et al. 2019). Given that comparative studies across dog breeds (Horschler et al. 2019) and other vertebrate taxa (MacLean et al. 2014) have identified positive associations between total brain volume and inhibitory control, EML1 is, therefore, a particularly promising candidate gene for further research in studies of both brain and cognitive evolution.
The SNP associated with memory in both analyses is located in a spliceosomal RNA in the U6 family, which is evolutionarily highly conserved (Brow and Guthrie 1988); variants in this gene could thus affect splicing of pre-mRNA transcripts. Similarly, the SNP associated with inhibitory control in the weight-controlled analysis is in a long noncoding RNA and therefore is likely to play a regulatory role (Yao et al. 2019). The SNP associated with physical reasoning in the no-covariate analysis is in OR52E2, which codes for an olfactory receptor. This is particularly interesting since the physical reasoning factor is positively loaded by a reliance on memory over smell when the two cues are pitted against each other (Table 1 and Supplementary Fig. S1); it is possible that this reflects a genetic contribution to olfactory salience that affects the ability of olfactory information to compete with visual memory. Lastly, the SNP associated with physical reasoning in the weight-controlled analysis is in HS3ST5, heparan sulfate-glucosamine 3-sulfotransferase 5, which is involved in the synthesis of heparan sulfate (Duncan et al. 2004). In mice, both heparan sulfate itself and heparan sulfate sulfotransferases generally have been shown to affect neural development (Inatani et al. 2003; Yabe et al. 2005).
It may initially seem surprising that we identified no SNPs that reached genome-wide significance for the communication factor, especially given that this was our second most heritable factor, with approximately 35% of the observed variation in this trait explained by breed-average relatedness (Gnanadesikan et al. 2020). However, this may be primarily due to the conservative nature of our correction for multiple comparisons, which yielded a small number of significant associations at the SNP level. It should be noted that in the gene-level aggregation analysis, we find a similar number of genes implicated in communication as we do for the other traits, although inhibitory control—our most heritable factor—was associated with the largest number of genes in our no-covariates analysis. It is also possible that the communication factor reflects a particularly polygenic trait, with many variants of small additive effect contributing to the phenotype.
Although we identified a limited set of SNPs that reached genome-wide significance in this study, it is important to note that the variance explained by allele frequency at these loci was generally <10%, except for the contribution to inhibitory control, which was considerably higher (34% no covariates; 29% controlling for weight). This finding is in stark contrast to the results of morphological trait-mapping studies in dogs which often find that one, or a few loci account for the majority of phenotypic variance across breeds (Sutter et al. 2007; Parker et al. 2009; Boyko et al. 2010). Nevertheless, compared to GWASs of cognitive traits in humans (Davies et al. 2011), the associated SNPs in our analyses explain significantly more variance, likely due to the effects of artificial selection on the dog genome (Lindblad-Toh et al. 2005; Ostrander and Wayne 2005; VonHoldt et al. 2010; Parker et al. 2017). Our findings echo those of MacLean et al. (2019) who conducted a similar GWAS on behavioral traits among breeds. For both cognitive and behavioral phenotypes, among-breed heritability of these traits (variance attributable to additive variation across the genome) can be high, yet there are only a few loci of possibly large effect, suggesting that breed differences in cognitive and behavioral traits are highly polygenic. Additionally, despite investigating only four cognitive traits, we found repeated association for a handful of genes across multiple traits. This finding emphasizes the importance of pleiotropic effects for complex traits (Visscher and Yang 2016), the limitations of simple one-to-one genotype–phenotype associations and frameworks, and the need for more integrative approaches (Solovieff et al. 2013).
Because our study took a breed-average approach and analyzed a limited set of SNPs across the genome, we did not intend to identify causal variants or to fine map any of the cognitive traits being studied. Rather, our principal aims were to identify a set of genes associated with breed differences in cognition and to assess whether these genes could plausibly influence breed differences in cognition through known actions in the nervous system. Our gene-level analyses identified 188 genes that were associated with variation in at least one of the four cognitive outcome measures, and enrichment analyses confirmed that these genes play important roles in diverse nervous system functions. First, the gene sets from each analysis were enriched for a variety of GO terms, many of which relate to neural functions, including peripheral nervous system development, presynapse assembly, cerebellar Purkinje cell layer development, negative regulation of canonical Wnt signaling pathway, and synaptic vesicle exocytosis. It should be noted that the Wnt signaling pathway—which was identified in both analyses—is known to be involved in neural crest cell development (Makoto et al. 1997; Dorsky et al. 1998), which may have been important in domestication (Wilkins et al. 2014). Second, through tissue-specific enrichment analyses, we found that the collective set of genes identified across our four phenotypes contained more genes that are highly expressed in brain tissue than would be expected by chance, controlling for background rates of genomic expression. This pattern held across analyses using tissue-specific gene expression data from both dogs and humans, and regardless of whether or not we controlled for breed-average body weight in the GWAS. Third, our results overlap considerably with previous association studies of canid behavior, including a recent study that took a similar breed-average approach to studying dog behavior using the C-BARQ (MacLean et al. 2019) and a variety of studies on experimentally bred tame and aggressive fox strains (Hekman et al. 2018; Kukekova et al. 2018; Wang et al. 2018). Thus, while we still know little about the specific mechanisms through which variants in these genes may influence cognition, the genes implicated in our analyses have strong potential to influence developmental and neurobiological functions with relevance for cognitive phenotypes. Future functional molecular work exploring variation in these genes could prove fruitful in illuminating specific mechanisms through which they may influence performance on cognitive tasks. We also note that while the results of our analyses did change when controlling for breed-average weight, there was also considerable overlap in the gene sets identified by each analysis (69 overlap/188 total unique genes). Combined with previous findings that certain cognitive and behavioral traits (McGreevy et al. 2013; Horschler et al. 2019) and their heritability estimates (Gnanadesikan et al. 2020) are more dependent on body weight than others, this suggests that body or brain size might contribute differentially to certain cognitive processes (MacLean et al. 2012).
Our current design benefited from a large sample size made possible by integrating data from the citizen science project Dognition.com, with publicly available genomic data on the breeds in the sample. However, this design is also subject to a number of important limitations that should be addressed in future work. Most notably, all analyses were conducted at the breed-average level, without paired genetic and cognitive data on the same individuals. Therefore, it will be critical for future association studies to build on this work through designs that integrate genotypes and phenotypes from the same individuals. Similarly, given that we used microarray data with a limited number of SNPs across the genome, we did not perform an exhaustive search for causal variants associated with cognitive phenotypes. Thus, future studies may benefit from incorporating greater coverage across the dog genome, for example through low coverage whole genome sequencing and imputation (Pasaniuc et al. 2012). Although the citizen science approach used here has been validated and implemented in several other studies of dog cognition (Stewart et al. 2015; Horschler et al. 2019; Watowich et al. 2020), it will be important for future work to explore a range of additional cognitive measures. With the recent establishment of several neuropsychological canine cognition test batteries (Wallis et al. 2014; Bray et al. 2017, 2020; MacLean et al. 2017), researchers will be well positioned to pursue these steps in the future. Lastly, although there is increasing evidence that dogs experience cognitive decline in middle to old age (Studzinski et al. 2006; Szabó et al. 2016; Watowich et al. 2020), the specific effects and progression—including variation across breeds—are not well understood, and we did not account for possible effects of age in our analyses. Despite these limitations, our study highlights how the remarkable phenotypic variation among dogs can be used to gain insights into the genetic factors contributing to cognitive variation among taxa and identifies an initial set of genes and biological processes to be considered in future research.
Data availability
Genetic data used in these analyses are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.266k4 (Hayward et al. 2016) and GEO accession nos. GSE90441, GSE83160, GSE70454, and GSE96736. The Dognition data used in these analyses are available from Brian Hare at b.hare@duke.edu.
Supplementary Material
Acknowledgments
We thank David Ivy, Eliot Cohen, Kip Frey, and everyone else who helped create Dognition.com, as well as Laurie R. Santos and Richard Wrangham, members of the scientific advisory board. We also thank all the dogs and people who participated in Dognition and made this work possible. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No (DGE-1746060). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Funding
This work was supported by the National Science Foundation Graduate Research Fellowship Program [DGE-1746060 to G.E.G.]; the National Institutes of Health [Grant 1R01HD097732-01 to B.H.]; MTA-ELTE Comparative Ethology Research Group [MTA01 031 to Á.M.], and the National Brain Research Program [2017-1.2.1-NKP-2017-00002 to Á.M.].
Author contributions
The data were collected by citizen scientists through Dognition, with tasks designed by B.H. The analysis was primarily designed and conducted by G.E.G and E.L.M, with B.H. and N.S-M. consulting. The paper was written by G.E.G. and E.L.M with significant contributions and revisions from B.H., N. S-M, J.C., J.K, and Á.M.
Conflict of Interest
B.H. is a founder of Dognition.com, and B.H., J.K., J.C., and Á.M. are members of its ScientificAdvisory Board. The authors declare no other competing interests.
Supplementary data
Supplementary data available at ICB online.
Contributor Information
Gitanjali E Gnanadesikan, School of Anthropology, University of Arizona, Tucson, AZ, USA; Cognitive Science Program, University of Arizona, Tucson, AZ, USA.
Brian Hare, Department of Evolutionary Anthropology, Duke University, Durham, NC, USA; Center for Cognitive Neuroscience, Duke University, Durham, NC, USA.
Noah Snyder-Mackler, Department of Psychology, University of Washington, Seattle, WA, USA; Center for Evolution and Medicine, Arizona State University, Tempe, AZ, USA; School of Life Sciences, Arizona State University, Tempe, AZ, USA.
Josep Call, School of Psychology and Neuroscience, University of St Andrews, St Andrews, UK.
Juliane Kaminski, Department of Psychology, University of Portsmouth, Portsmouth, UK.
Ádám Miklósi, Department of Ethology, Eötvös Loránd University, Budapest, Hungary; MTA-ELTE Comparative Ethology Research Group, Budapest, Hungary.
Evan L MacLean, School of Anthropology, University of Arizona, Tucson, AZ, USA; Cognitive Science Program, University of Arizona, Tucson, AZ, USA; Psychology Department, University of Arizona, Tucson, AZ, USA; College of Veterinary Medicine, University of Arizona, Tucson, AZ, USA.
From the symposium “Integrative Comparative Cognition: Can Neurobiology and Neurogenomics Inform Comparative Analyses of Cognitive Phenotype?” presented at the annual meeting of the Society for Integrative and Comparative Biology January 3–7, 2020 at Austin, Texas.
References
- Ahonen SJ, Pietilä E, Mellersh CS, Tiira K, Hansen L, Johnson GS, Lohi H. 2013. Genome-wide association study identifies a novel canine glaucoma locus. PLoS One 8:e70903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdemir D. Godfrey OU. 2015. EMMREML: fitting mixed models with known covariance structures (https://CRAN.R-project.org/package=EMMREML). [Google Scholar]
- Alexa A, Rahnenfuhrer J. 2018. topGO: enrichment analysis for gene ontology.
- Amin N, van Duijn CM, Aulchenko YS. 2007. A genomic background based method for association analysis in related individuals. PLoS One 2:e1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E, Ratnakumar A, Arendt ML, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar Å, Lindblad-Toh K. 2013. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495:360–4. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
- Benson-Amram S, Dantzer B, Stricker G, Swanson EM, Holekamp KE. 2016. Brain size predicts problem-solving ability in mammalian carnivores. Proc Natl Acad Sci U S A 113:2532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao KY, Brisbin A, Parker HG, VonHoldt BM, et al. 2010. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol 8:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EE, Gruen ME, Gnanadesikan GE, Horschler DJM, Levy KM, Kennedy BS, MacLean EL. 2020. Cognitive characteristics of 8-to-10-week-old assistance dog puppies. Anim Behav 166:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray EE, Sammel MD, Seyfarth RM, Serpell JA, Cheney DL. 2017. Temperament and problem solving in a population of adolescent guide dogs. Anim Cogn 20:923–39. [DOI] [PubMed] [Google Scholar]
- Briggs J, Paoloni M, Chen Q-RR, Wen X, Khan J, Khanna C. 2011. A compendium of canine normal tissue gene expression. PLoS One 6:e17107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. 1988. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334:213–8. [DOI] [PubMed] [Google Scholar]
- Cadieu E, Neff MW, Quignon P, Walsh K, Chase K, Parker HG, Vonholdt BM, Rhue A, Boyko AB, Byers A, et al. 2009. Coat variation in the domestic dog is governed by variants in three genes. Science 326:150–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Jones P, Martin A, Ostrander EA, Lark KG. 2009. Genetic mapping of fixed phenotypes: disease frequency as a breed characteristic. J Hered 100:S37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittka L, Rossiter SJ, Skorupski P, Fernando C. 2012. What is comparable in comparative cognition? Philos Trans R Soc Lond B Biol Sci 367:2677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinar O, Viechtbauer W. 2020. poolr: methods for pooling P-values from (dependent) tests (https://CRAN.R-project.org/package=poolr).
- Csardi G, Nepusz T. 2006. The igraph software package for complex network research. InterJ Complex Syst 1695:1–9. [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, et al. 2011. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry 16:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaner RO, Isler K, Burkart J, Van Schaik C. 2007. Overall brain size, and not encephalization quotient, best predicts cognitive ability across non-human primates. Brain Behav Evol 70:115–24. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. 1999. Genomic control for association study. Biometrics 55:997–1004. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Moon RT, Raible DW. 1998. Control of neural crest cell fate by the Wnt signalling pathway. Nature 396:370–2. [DOI] [PubMed] [Google Scholar]
- Duncan MB, Chen J, Krise JP, Liu J. 2004. The biosynthesis of anticoagulant heparan sulfate by the heparan sulfate 3-O-sulfotransferase isoform 5. Biochim Biophys Acta Gen Subj 1671:34–43. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. 2005. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21:3439–40. [DOI] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, Huber W. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman AH, Schweizer RM, Ortega-Del Vecchyo D, Han E, Davis BW, Gronau I, Silva PM, Galaverni M, Fan Z, Marx P, et al. 2016. Demographically-based evaluation of genomic regions under selection in domestic dogs. PLoS Genet 12:e1005851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadesikan GE, Hare B, Snyder-Mackler N, MacLean EL. 2020. Estimating the heritability of cognitive traits across dog breeds reveals highly heritable inhibitory control and communication factors. Anim Cogn 23:953–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward JJ, Castelhano MG, Oliveira KC, Corey E, Balkman C, Baxter TL, Casal ML, Center SA, Fang M, Garrison SJ, et al. 2016. Complex disease and phenotype mapping in the domestic dog. Nat Commun 7:10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekman JP, Johnson JL, Edwards W, Vladimirova AV, Gulevich RG, Ford AL, Kharlamova AV, Herbeck Y, Acland GM, Raetzman LT, et al. 2018. Anterior pituitary transcriptome suggests differences in ACTH release in tame and aggressive foxes. G3 Genes Genom Genet 8:859–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner MP, Lundquist A, Pirun M, Meadows JRS, Zamani N, Johnson J, Sundström G, Cook A, FitzGerald MG, Swofford R, et al. 2014. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS One 9:e91172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horschler DJ, Hare B, Call J, Kaminski J, Miklósi Á, MacLean EL. 2019. Absolute brain size predicts dog breed differences in executive function. Anim Cogn 22:187–98. [DOI] [PubMed] [Google Scholar]
- Horschler DJ, MacLean EL. 2019. Leveraging brain–body scaling relationships for comparative studies. Anim Cogn 22:1197–202. [DOI] [PubMed] [Google Scholar]
- Hsu Y, Serpell JA. 2003. Development and validation of a questionnaire for measuring behavior and temperament traits in pet dogs. J Am Vet Med Assoc 223:1293–300. [DOI] [PubMed] [Google Scholar]
- Inatani M, Irie F, Plump AS, Tessier-Lavigne M, Yamaguchi Y. 2003. Mammalian brain morphogenesis and midline axon guidance require heparan sulfate. Science 302:1044–6. [DOI] [PubMed] [Google Scholar]
- Jain A, Tuteja G. 2019. TissueEnrich: tissue-specific gene enrichment analysis. Bioinformatics 35:1966–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevic A, Elgier AM, Mustaca AE, Bentosela M. 2010. Breed differences in dogs’ (Canis familiaris) gaze to the human face. Behav Proc 84:602–7. [DOI] [PubMed] [Google Scholar]
- Jardim-Messeder D, Lambert K, Noctor S, Pestana FM, de Castro Leal ME, Bertelsen MF, Alagaili AN, Mohammad OB, Manger PR, Herculano-Houzel S. 2017. Dogs have the most neurons, though not the largest brain: trade-off between body mass and number of neurons in the cerebral cortex of large carnivoran species. Front Neuroanat 11:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Chase K, Martin A, Davern P, Ostrander EA, Lark KG. 2008. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics 179:1033–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. 2008. Efficient control of population structure in model organism association mapping. Genetics 178:1709–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NHC, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, et al. 2007. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet 39:1321–8. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Lindblad-Toh K. 2008. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet 9:713–25. [DOI] [PubMed] [Google Scholar]
- Karolchik D. 2004. The UCSC table browser data retrieval tool. Nucleic Acids Res 32:493D–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielar M, Tuy FPD, Bizzotto S, Lebrand C, De Juan Romero C, Poirier K, Oegema R, Mancini GM, Bahi-Buisson N, Olaso R, et al. 2014. Mutations in Eml1 lead to ectopic progenitors and neuronal heterotopia in mouse and human. Nat Neurosci 17:923–33. [DOI] [PubMed] [Google Scholar]
- Kim J, Williams FJ, Dreger DL, Plassais J, Davis BW, Parker HG, Ostrander EA. 2018. Genetic selection of athletic success in sport-hunting dogs. Proc Natl Acad Sci U S A 115:E7212–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, Delcher AL, Pop M, Wang W, Fraser CM. 2003. The dog genome: survey sequencing and comparative analysis. Science 301:1898–903. [DOI] [PubMed] [Google Scholar]
- Kotrschal A, Rogell B, Bundsen A, Svensson B, Zajitschek S, Brännström I, Immler S, Maklakov AA, Kolm N. 2013. Artificial selection on relative brain size in the guppy reveals costs and benefits of evolving a larger brain. Curr Biol 23:168–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukekova AV, Johnson JL, Xiang XXY, Feng SSH, Liu S, Rando HM, Kharlamova AV, Herbeck Y, Serdyukova NA, Xiong Z, et al. 2018. Red fox genome assembly identifies genomic regions associated with tame and aggressive behaviours. Nat Ecol Evol 2:1479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, VonHoldt BM, Reynolds A, Boyko AR, Wayne RK, Wu DDD, Zhang YPP, Von Holdt BM, Reynolds A, Boyko AR, et al. 2013. Artificial selection on brain-expressed genes during the domestication of dog. Mol Biol Evol 30:1867–76. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, Zody MC, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–19. [DOI] [PubMed] [Google Scholar]
- MacLean EL, Hare B, Nunn CL, Addessi E, Amici F, Anderson RC, Aureli F, Baker JM, Bania AE, Barnard AM, et al. 2014. The evolution of self-control. Proc Natl Acad Sci U S A 111:E2140–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean EL, Herrmann E, Suchindran S, Hare B. 2017. Individual differences in cooperative communicative skills are more similar between dogs and humans than chimpanzees. Anim Behav 126:41–51. [Google Scholar]
- MacLean EL, Matthews LJ, Hare BA, Nunn CL, Anderson RC, Aureli F, Brannon EM, Call J, Drea CM, Emery NJ, et al. 2012. How does cognition evolve? Phylogenetic comparative psychology. Anim Cogn 15:223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean EL, Nunn CL. 2017. Phylogenetic approaches for research in comparative cognition In: Call J, Burghardt GM, Pepperberg IM, Snowdon C, Zentall T, editors. APA handbook of comparative psychology: basic concepts, methods, neural substrate, and behavior Washington (DC: ): American Psychological Association; p. 201–16. [Google Scholar]
- MacLean EL, Snyder-Mackler N, vonHoldt BM, Serpell JA. 2019. Highly heritable and functionally relevant breed differences in dog behaviour. Proc R Soc B Biol Sci 286:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makoto I, Lee SMK, Johnson JE, Mc Mahon AP, Takada S. 1997. Wnt signalling required for expansion of neural crest and cns progenitors. Nature 389:966–70. [DOI] [PubMed] [Google Scholar]
- McGreevy PD, Georgevsky D, Carrasco J, Valenzuela M, Duffy DL, Serpell JA. 2013. Dog behavior co-varies with height, bodyweight and skull shape. PLoS One 8:e80529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemus KK, Liu W, Chase GA, Tsai YY, Fallin MD. 2005. Comparison of type 1 error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet 6:S78–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholt DR. 2004. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74:765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander E. a, Wayne RK. 2005. The canine genome. Genome Res 15:1706–16. [DOI] [PubMed] [Google Scholar]
- Parker HG, Dreger DL, Rimbault M, Davis BW, Mullen AB, Carpintero-Ramirez G, Ostrander EA. 2017. Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep 19:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, et al. 2009. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325:995–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasaniuc B, Rohland N, McLaren PJ, Garimella K, Zaitlen N, Li H, Gupta N, Neale BM, Daly MJ, Sklar P, et al. 2012. Extremely low-coverage sequencing and imputation increases power for genome-wide association studies. Nat Genet 44:631–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Chang C. 2018. PLINK [1.90] (https://www.cog-genomics.org/plink/).
- Purcell SM, Chang CC, Chow CC, Tellier LC, Lee JJ, Vattikuti S. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna, Austria: R Core Team. [Google Scholar]
- Revelle W. 2018. psych: procedures for psychological, psychometric, and personality research (https://CRAN.R-project.org/package=psych).
- Saetre P, Lindberg J, Leonard JA, Olsson K, Pettersson U, Ellegren H, Bergström TF, Vilà C, Jazin E. 2004. From wild wolf to domestic dog: gene expression changes in the brain. Brain Res Mol Brain Res 126:198–206. [DOI] [PubMed] [Google Scholar]
- Scott JP, Fuller JL. 1965. Genetics and the social behavior of the dog. Vol. 111. Chicago (IL: ): University of Chicago Press. [Google Scholar]
- Serpell JA, Duffy DL. 2014. Dog breeds and their behavior Domestic dog cognition and behavior Springer. Heidelberg, Germany: Springer-Verlag Berlin Heidelberg; p. 31–57. [Google Scholar]
- Shaheen R, Sebai MA, Patel N, Ewida N, Kurdi W, Altweijri I, Sogaty S, Almardawi E, Seidahmed MZ, Alnemri A, et al. 2017. The genetic landscape of familial congenital hydrocephalus. Ann Neurol 81:890–7. [DOI] [PubMed] [Google Scholar]
- Sol D, Bacher S, Reader SM, Lefebvre L. 2008. Brain size predicts the success of mammal species introduced into novel environments. Am Nat 172:S63–71. [DOI] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. 2013. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet 14:483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart L, MacLean EL, Ivy D, Woods V, Cohen E, Rodriguez K, McIntyre M, Mukherjee S, Call J, Kaminski J, et al. 2015. Citizen science as a new tool in dog cognition research. PLoS One 10:e0135176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studzinski CM, Christie LA, Araujo JA, Burnham WMI, Head E, Cotman CW, Milgram NW. 2006. Visuospatial function in the beagle dog: an early marker of cognitive decline in a model of human aging and dementia. Neurobiol Learn Mem 86:197–204. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, et al. 2007. Ostrander E a. 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316:112–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svartberg K, Forkman B. 2002. Personality traits in the domestic dog (Canis familiaris). Appl Anim Behav Sci 79:133–55. [Google Scholar]
- Szabó D, Gee NR, Miklósi Á. 2016. Natural or pathologic? Discrepancies in the study of behavioral and cognitive signs in aging family dogs. J Vet Behav 11:86–98. [Google Scholar]
- The Gene Ontology Consortium. 2000. Gene ontology: tool for the unification of biology. Nat Genet 25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2019. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res 47:D330–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Tissue-based map of the human proteome. Science 347:1260419–347. [DOI] [PubMed] [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Pielberg GR, Sigurdsson S, Fall T, Seppälä EH, Hansen MSTT, Lawley CT, et al. 2011. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet 7:e1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. 2008. Heritability in the genomics era - concepts and misconceptions. Nat Rev Genet 9:255–66. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Yang J. 2016. A plethora of pleiotropy across complex traits. Nat Genet 48:707–8. [DOI] [PubMed] [Google Scholar]
- VonHoldt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, et al. 2010. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464:898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis L, Range F, Müller C, Serisier S, Huber L, Viranyi Z. 2014. The Vienna canine cognitive battery: assessment of cognitive functioning during development and aging in pet dogs. J Vet Behav 9:e18–19–e19. [Google Scholar]
- Wang GD, Zhai W, Yang HC, Fan RX, Cao X, Zhong L, Wang L, Liu F, Wu H, Cheng LG, et al. 2013. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat Commun 4:1860. [DOI] [PubMed] [Google Scholar]
- Wang X, Pipes L, Trut LN, Herbeck Y, Vladimirova AV, Gulevich RG, Kharlamova AV, Johnson JL, Acland GM, Kukekova AV, et al. 2018. Genomic responses to selection for tame/aggressive behaviors in the silver fox (Vulpes vulpes). Proc Natl Acad Sci U S A 115:10398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watowich MM, MacLean EL, Hare B, Call J, Kaminski J, Miklósi Á, Snyder-Mackler N. 2020. Age influences domestic dog cognitive performance independent of average breed lifespan. Anim Cogn 23:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Khan IK, Ding Z, Yerneni S, Kihara D. 2017. NaviGO: interactive tool for visualization and functional similarity and coherence analysis with gene ontology. BMC Bioinformatics 18:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins AS, Wrangham RW, Fitch WT. 2014. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197:795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsson E, Sundgren PE. 1997. The use of a behaviour test for the selection of dogs for service and breeding, I: method of testing and evaluating test results in the adult dog, demands on different kinds of service dogs, sex and breed differences. Appl Anim Behav Sci 53:279–95. [Google Scholar]
- Wobber V, Hare B, Koler-Matznick J, Wrangham R, Tomasello M. 2009. Breed differences in domestic dogs’ (Canis familiaris) comprehension of human communicative signals. Interact Stud 10:206–24. [Google Scholar]
- Yabe T, Hata T, He J, Maeda N. 2005. Developmental and regional expression of heparan sulfate sulfotransferase genes in the mouse brain. Glycobiology 15:982–93. [DOI] [PubMed] [Google Scholar]
- Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, Smith AV, Ingelsson E, O'Connell JR, Mangino M, et al. 2011. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet 19:807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao RW, Wang Y, Chen LL. 2019. Cellular functions of long noncoding RNAs. Nat Cell Biol 21:542––51. [DOI] [PubMed] [Google Scholar]
- Zapata I, Serpell JA, Alvarez CE. 2016. Genetic mapping of canine fear and aggression. BMC Genomics 17:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Luo T, Li T, Li Y, Zhang J, Shan Y, Wang X, Yang L, Zhou F, Zhu Z, et al. 2019. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet 51:1637–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genetic data used in these analyses are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.266k4 (Hayward et al. 2016) and GEO accession nos. GSE90441, GSE83160, GSE70454, and GSE96736. The Dognition data used in these analyses are available from Brian Hare at b.hare@duke.edu.