In this study, Rickel et al. identified essential histone methylatransferase-independent functions of Trr, used a screen to identify a minimal Trr domain sufficient to rescue Trr-null lethality, and demonstrated that this domain binds and stabilizes Utx in vivo. Their findings show that COMPASS-dependent UTX stabilization is an essential noncatalytic function of Trr/MLL3/MLL4, suggesting that stabilizing UTX could be a therapeutic strategy for cancers with MLL3/4 loss-of-function mutations.
Keywords: COMPASS, chromatin, epigenetics, transcription
Abstract
Catalytic-inactivating mutations within the Drosophila enhancer H3K4 mono-methyltransferase Trr and its mammalian homologs, MLL3/4, cause only minor changes in gene expression compared with whole-gene deletions for these COMPASS members. To identify essential histone methyltransferase-independent functions of Trr, we screened to identify a minimal Trr domain sufficient to rescue Trr-null lethality and demonstrate that this domain binds and stabilizes Utx in vivo. Using the homologous MLL3/MLL4 human sequences, we mapped a short ∼80-amino-acid UTX stabilization domain (USD) that promotes UTX stability in the absence of the rest of MLL3/4. Nuclear UTX stability is enhanced when the USD is fused with the MLL4 HMG-box. Thus, COMPASS-dependent UTX stabilization is an essential noncatalytic function of Trr/MLL3/MLL4, suggesting that stabilizing UTX could be a therapeutic strategy for cancers with MLL3/4 loss-of-function mutations.
Trithorax-related (trr), MLL3 (KMT2C), and MLL4 (KMT2D) represent the “branch” of COMPASS family lysine methyltransferases responsible for catalyzing histone 3 lysine 4 monomethylation (H3K4me1) at enhancer chromatin in Drosophila and mammals, respectively (Herz et al. 2012; Hu et al. 2013). These enzymes form complexes with components common to all COMPASS members, as well as complex-specific subunits that enable unique functions, and this subunit composition is conserved from Drosophila to humans (Shilatifard 2012). In particular, the H3K27 demethylase, UTX (KDM6A), is specific to Trr/MLL3/MLL4 complexes and is believed to function in the process of enhancer activation by facilitating acetylation of H3K27 by CBP/p300 (Agger et al. 2007; Cho et al. 2007; Mohan et al. 2011; Herz et al. 2012). Indeed, depletion of either Utx or Trr in Drosophila results in decreased levels of H3K27ac, as well as H3K4me1, underscoring the functional interdependence between Trr and Utx in modifying chromatin structure in vivo (Herz et al. 2012). Trr is critical for maintaining the stability of Utx, as well as Lpt, a protein homologous to the N-terminal half of MLL3/4, which is expressed from a separate gene but still complexes with Trr/COMPASS in the nucleus (Mohan et al. 2011; Chauhan et al. 2012; Herz et al. 2012). Thus, the function and stability of these two enzymes are intimately entwined. Genetic links between MLL4 and UTX were initially identified in a rare pediatric congenital disorder called Kabuki Syndrome (Ng et al. 2010; Lederer et al. 2012; Miyake et al. 2013a,b), and are also frequently mutated across a broad swath of human malignancies, highlighting a connection between regulators of enhancer histone modification and cancer pathogenesis (van Haaften et al. 2009; Ntziachristos et al. 2012; Kandoth et al. 2013; The Cancer Genome Atlas Research Network 2014; Zhang et al. 2015; Wang and Shilatifard 2019).
We recently demonstrated loss of H3K4me1, or its conversion to H3K4me2/3 at enhancers is compatible with proper development and adult homeostasis in Drosophila melanogaster (Rickels et al. 2017). Using complementation assays in a lethal Trr-null background (trr[1]), we rescued viability by expressing Trr with either catalytic-defective (C2398A) or catalytic-hyperactive (Y2383F) point mutations, each producing unique yet subtle phenotypes only observable under temperature stress (Rickels et al. 2017). Consistent with data showing H3K4me1 is largely dispensable for enhancer function in vivo, our group and others also reported similar catalytic-inactivating mutations to MLL3/4 only minimally affect gene expression compared with MLL3/4-null deletions, which cause down-regulation of thousands of genes in mouse embryonic stem cells (mESCs) (Dorighi et al. 2017; Rickels et al. 2017). Enhancer chromatin accessibility in mESCs was reduced in the absence of MLL3/4, with no change in the catalytic-dead mutants. Furthermore, enhancer-associated RNA synthesis at MLL3/4-bound enhancers was diminished only in the double-knockout cell line, concomitant with increased RNA polymerase II pausing at associated gene promoters (Dorighi et al. 2017). This is in agreement with a growing body of research suggesting enhancer-mediated transcriptional activation functions by stimulating transcription elongation (Chen et al. 2018; Rickels and Shilatifard 2018). Thus, localization of MLL3/4, but not its catalytic activity, is required to maintain open chromatin at enhancers, which indicates a catalytic-independent function of Trr/MLL3/MLL4 is critical for its role in development.
Results
Identification of an essential Trr domain sufficient to rescue Trr-null associated lethality
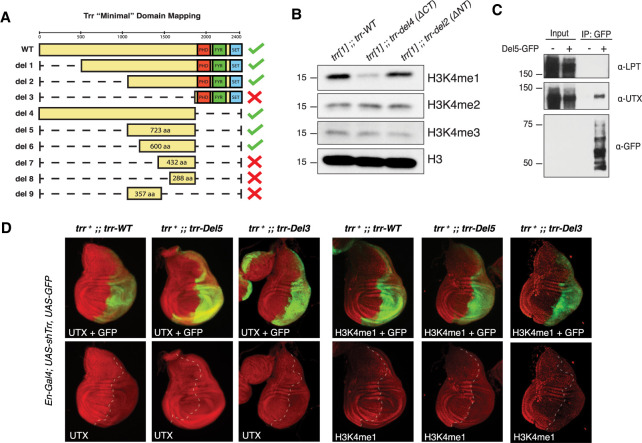
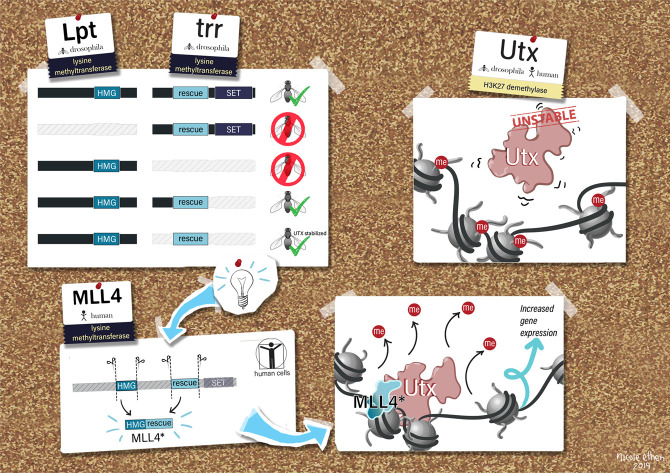
In search of the catalytic-independent function for Drosophila Trr and mammalian MLL3/4, we generated a suite of transgenic Drosophila lines that express different Trr domain deletions to assay for complementation with the trr[1]-null mutant. Males from each line were crossed with trr[1] females and their progeny were assessed to determine whether or not individual domain deletions could rescue viability (Supplemental Fig. S1A), as previously described (Rickels et al. 2017). While our previous study used a point mutation to disrupt Trr catalytic activity, we further found deletion of the entire C terminus (del4), which includes the SET domain, was also able to rescue lethality (Fig. 1A). Consistent with our previous report, bulk reductions in H3K4me1 (Fig. 1B) were not associated with obvious effects on Drosophila development (Rickels et al. 2017). Interestingly, when reared at higher temperatures (29°C), Trr-del4 flies phenocopy the catalytic-dead Trr-C2398A mutation by producing ectopic L3/L4 wing cross-veins, while N-terminal deletions do not (Supplemental Fig. S1B; data not shown). This supports our theory that H3K4me1 is important in fine-tuning enhancer-mediated gene expression, especially under temperature stress (Rickels et al. 2017). While N-terminal deletions, such as Trr-del2, readily rescue viability, we found this mutation dramatically reduces female fertility in the trr[1] background (data not shown). Ultimately, our genetic complementation strategy successfully identifies a 600-amino-acid region of unknown function, located roughly in the middle of Trr, sufficient to rescue Trr-null lethality (Fig. 1A).
Figure 1.
A UTX-interacting region of Trr is sufficient to rescue Trr-null viability. (A) The diagram depicts full-length Trr coding sequence and the various deletion constructs assayed for complementation with the trr[1] lethal allele. Green check marks designate which constructs rescue trr[1] viability according to the complementation assay illustrated in Supplemental Figure S1. (B) Western blots for H3K4-methylation confirm bulk H3K4me1 decrease in the del4 rescue line, which lack the catalytic SET domain. (C) Immunoprecipitation of the Del5-GFP fragment in Drosophila S2 cells copurifies UTX, but not LPT. (D) Immunofluorescence microscopy in larval wing imaginal discs confirms the “minimal” Del5 fragment is sufficient to stabilize UTX in the absence of full-length TRR. Endogenous TRR is depleted by RNAi in the posterior compartment (GFP-labeled), leading to reductions in H3K4me1. However, UTX levels are not affected upon expression of Del5, which is insensitive to Trr-RNAi.
To study the biochemical properties of this domain, we expressed a GFP-tagged peptide corresponding to the del5 rescue fragment in Drosophila S2 cells, and performed protein purification to identify interacting partners of this domain. The protein products of two essential genes, Lpt and Utx, are known to form a complex with Trr; however, only Utx was confirmed by Western blot to interact with the Trr-del5 fragment (Fig. 1C). This is consistent with a previous report showing UTX interacts with a 489-amino-acid portion near the C terminus of MLL4 (Kim et al. 2014). Our laboratory and others have previously reported Trr and MLL4 are responsible for stabilizing Utx protein in flies and mammals, respectively (Herz et al. 2012; Lee et al. 2013). To test whether this region of Trr is sufficient to stabilize Utx in vivo, we expressed either full-length Trr-WT, Trr-del3, or Trr-del5 in a trrWT;en-Gal4>Trr-RNAi background, and then assessed Utx stability in wing imaginal discs. The En-Gal4 system drives Trr-RNAi expression in the posterior half of imaginal discs, providing an internal control for comparison across all three genotypes. Importantly, both the endogenous trr and Trr-WT are sensitive to Trr-RNAi, while Trr-del3 and –del5 are insensitive. In all three genotypes, knockdown of endogenous trr is evident by reduced H3K4me1, as previously published (Herz et al. 2012). Likewise, Utx levels are diminished in the Trr-WT and Trr-del3, but unaffected in Trr-del5 (Fig. 1D). These findings suggest stabilization of Utx is an essential catalytic-independent function of Trr.
Identification of a conserved MLL4-UTX-binding domain
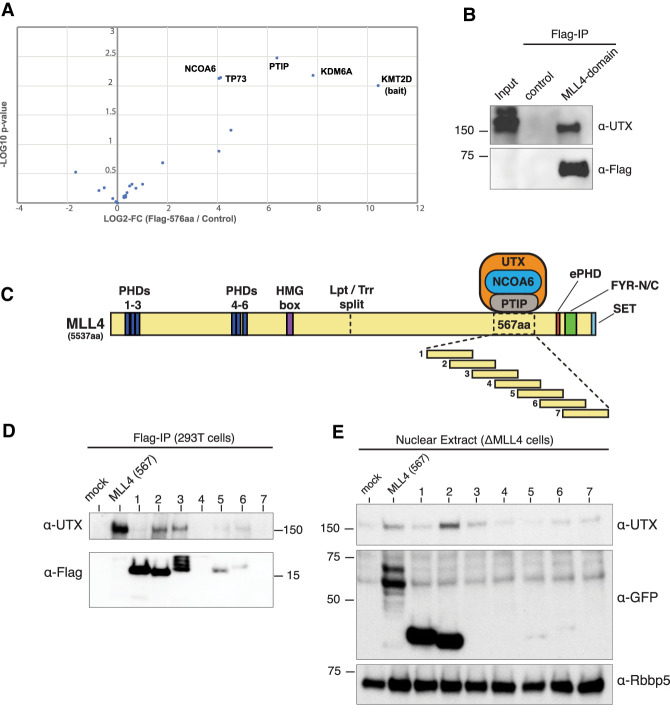
To apply our findings from Drosophila in a mammalian system, we expressed the homologous 567-amino-acid region of MLL4 in 293Trex cells as a FLAG fusion protein, followed by FLAG purification to identify interacting factors. Mass spectrometry from three independent experiments identified UTX (KDM6A) as the most significantly enriched protein interaction, followed by PTIP (PAXIP1) and NCOA6, which are also known components of MLL3/4 COMPASS-like complexes (Fig. 2A). TP73 was also enriched similarly to NCOA6, but, to our knowledge, has yet to be reported as a biochemical interactor of either UTX or MLL4. We confirmed the MLL4 567-amino-acid region coimmunoprecipitates with UTX by Western blot (Fig. 2B). It is important to note that, just as we did not detect Lpt in the Trr-del5 pull-down, we also did not detect endogenous MLL4 peptides in our mass spectrometry, indicating this domain is not sufficient for participating in a full COMPASS. We performed size exclusion chromatography with nuclear extracts from doxycycline-inducible 293Trex cells expressing a GFP-tagged MLL4 567-amino-acid fragment and found the vast majority of UTX coelutes with MLL4 in uninduced cells. However, in the presence of the MLL4 domain, UTX coelutes in later fractions where GFP-tag is detected (Supplemental Fig. S2A). These results indicate the MLL4-UTX-binding domain is not incorporated into the endogenous MLL4 complex, but instead competes for UTX binding.
Figure 2.
MLL4/UTX-binding interactions are important for UTX protein stability. (A) Volcano plot of mass spectrometry analysis reveals UTX is the most significantly enriched factor in the MLL4-FLAG (567 amino acids) purification, among other known MLL4/COMPASS subunits. Common contaminants (Mellacheruvu et al. 2013) were filtered out and remaining hits are plotted as –log10 P-value (t-test) versus log2 fold change (Supplemental Table 1). (B) Western blots confirm UTX interaction with MLL4-FLAG (567 amino acids). (C) Diagram of the full-length MLL4 protein structure including annotated domains. To fine-map the interacting region, we overexpress overlapping fragments tiled across the UTX interaction domain. (D). FLAG-tagged fragments #1–7 were transfected in 293T cells and IPs show UTX interaction is most evident with fragments #2 and #3. (E) Fragments #1–7 transfected in ΔMLL4 HCT116 cells in which UTX levels are diminished due to loss of stabilizing interaction with MLL4 (Supplemental Fig. S2B). Note that the full-length 567-amino-acid domain, as well as fragment #2, provide some stabilizing effect on UTX.
While an approximate location of the MLL4-UTX-binding domain was previously reported (Kim et al. 2014), we began narrowing down the interaction surface to better understand how MLL4 stabilizes UTX. We created a series of seven constructs tiled across the 567-amino-acid MLL4 domain, such that each fragment of ∼150 amino acids overlaps its neighbors by ∼50% (Fig. 2C). The FLAG-tagged series was expressed in 293T cells, and although we had difficulty achieving equal expression of each fragment, coimmunoprecipitation shows fragments #2 and #3 both pull down UTX (Fig. 2D), indicating the amino acid sequence shared between these fragments is sufficient to bind to UTX. Using CRISPR/Cas9 to delete the MLL4 promoter in HCT116 cells, we generated a ΔMLL4 cell line that displays significantly lower levels of UTX compared with parental HCT116 at the protein level (Supplemental Fig. S2B) but not at the mRNA level (Supplemental Fig. S2C).
Although we were unable to improve expression by replacing FLAG with GFP, after transfecting ΔMLL4 cells we observe fragment #2 does provide some stabilizing effect on endogenous UTX levels (Fig. 2E).
The role of MLL4 HMG-box domain in UTX stability
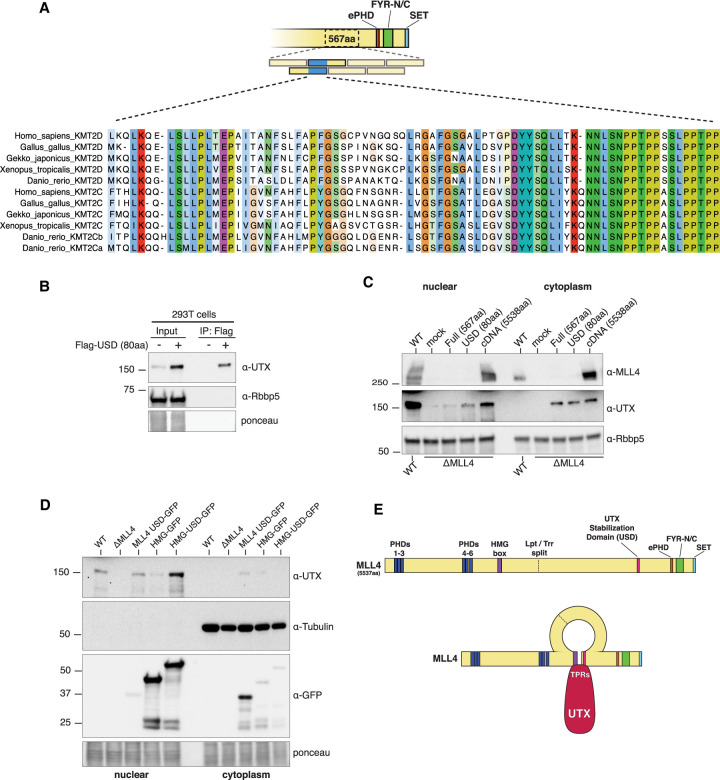
Based on the results above, we focused our attention on the overlapping region between fragments #2 and #3 (Fig. 3A) that were able to interact with UTX, and which is conserved in Drosophila (Supplemental Fig. S3). Since this ∼80-amino-acid peptide was sufficient to bind and stabilize UTX in 293T cells, we refer to it as the UTX stabilization domain (USD) (Fig. 3B). However, when transiently transfected into ΔMLL4 cells, neither the original 567-amino-acid nor the 80-amino-acid USD were able to fully stabilize UTX to the extent observed with a full-length MLL4 cDNA, which achieves UTX levels similarly to WT-HCT116 (Fig. 3C). This suggests an additional MLL4 domain is also required to provide full UTX stability in the nucleus and, based on combined genetic and biochemical data, we hypothesized that the MLL4 HMG-box could be important for this function. First, our genetic complementation assays in Drosophila were always conducted in an lptWT background, indicating the N-terminal half of MLL4 could also play a role in maintaining UTX stability. Second, a previous study in Drosophila demonstrated that deletion of the Lpt HMG-box disrupts Lpt's ability to pull down Trr, although direct binding was never demonstrated (Chauhan et al. 2012). Third, we used CRISPR in HCT116 cells to delete the N terminus from MLL4, which contains the PHD domains, while leaving the HMG-box intact. Bulk UTX levels are not disrupted in these cells, whereas deleting the entire C terminus, including the UTX-binding domain, results in strong depletion of UTX from the nucleus (Supplemental Fig. S4A). To test this hypothesis in ΔMLL4 cells, we transiently expressed a GFP-tagged MLL4 HMG-box domain alone, or fused it with the USD, and observed a dramatic increase in nuclear UTX stability with the “HMG-USD,” comparable with WT levels (Fig. 3D). Surprisingly, the HMG-box alone had a small effect on promoting UTX stability, suggesting that this domain might also contact UTX directly (Fig. 3D). A similar MLL3 HMG-USD fusion also stabilized UTX to near WT levels (Supplemental Fig. S4B). Thus far, all of the constructs used for protein expression in cell culture have necessarily included an SV40 nuclear localization signal (NLS); however, we found inclusion of the HMG-box in the fusion protein obviates the need for an NLS, indicating that this domain might be important for MLL4 nuclear localization (Supplemental Fig. S4B).
Figure 3.
Mapping the MLL4 domains required for UTX nuclear stability. (A) An 80-amino-acid “minimal” UTX interaction sequence was narrowed down to the overlapping region between fragments #2 and #3. Peptide alignment reveals a conserved acidic patch. (B) The 80-amino-acid “minimal” sequence is sufficient to bind UTX in 293T cells, and also has a stabilizing effect on endogenous UTX protein. (C) Transient transfections of the full-length (567-amino-acid) or minimal (80-amino-acid) domains into ΔMLL4 HCT116 cells stabilize UTX in nuclear and cytoplasmic fractions, but never restore wild-type UTX levels compared with WT-HCT116 or transfection with full-length MLL4 cDNA (5538 amino acids). (D) Western blots show transiently expressing the “minimal” 80-amino-acid domain fused with the MLL4 HMG-box domain (HMG-USD) fully restores nuclear UTX levels. Note the HMG-box alone has a minor stabilizing effect on UTX. (E) A diagram of the full-length MLL4 protein including annotated domains with approximate location of the Lpt/Trr gene split. Genetic and biochemical evidence suggests UTX is stabilized through intramolecular interactions between the MLL4 HMG-box and UTX-interacting domain.
To better understand the biochemical interactions between MLL3/4 and UTX, we determined the approximate location of UTX's interaction with MLL4. FLAG-tagged UTX was divided into three overlapping segments: NT, mid, CT, and IP/Western revealed the NT portion pulled down endogenous MLL4 in 293T cells (Supplemental Fig. S4C). This region of UTX contains at least six TPR domains, a structural motif known to mediate protein–protein interactions. Likewise, FLAG-IP after transfection with FLAG-UTX-NT and MLL4-HMG-USD-GFP shows these regions also interact (Supplemental Fig. S4D). Taken together, we propose a model in which the MLL4 HMG-box and USD come together to stabilize UTX (Fig. 3E), which is consistent with published genetic and biochemical data from Drosophila (Chauhan et al. 2012; Herz et al. 2012).
The MLL4-HMG-USD partially rescues gene expression via UTX recruitment to chromatin
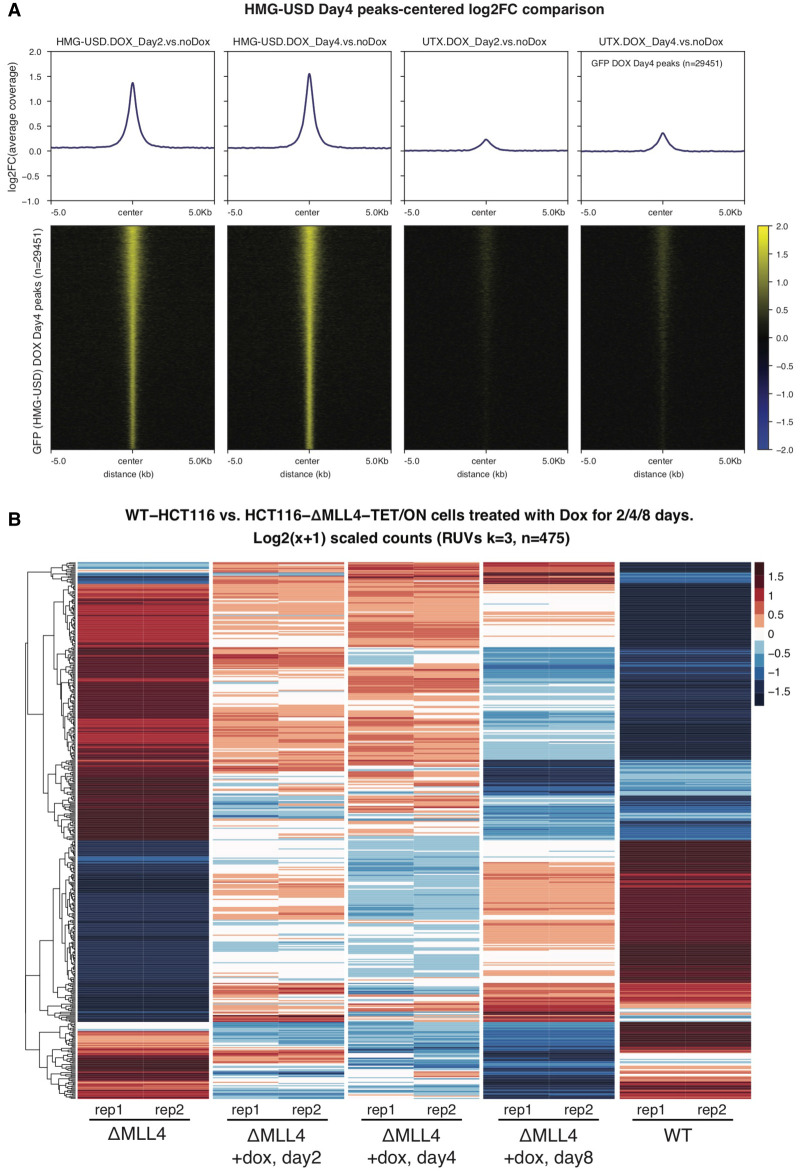
To investigate what effect, if any, the MLL4-HMG-USD has on gene expression, we modified our ΔMLL4 HCT116 cell line (already mutated for MLL3) to include a doxycycline-inducible (TET-ON, T/O) GFP-MLL4-HMG-USD transgene, and observed robust UTX nuclear stabilization ∼3 d following doxycycline treatment (Supplemental Fig. S4E). ΔMLL4-T/O GFP-MLL4-HMG-USD cells were treated with doxycycline for 2 and 4 d and chromatin occupancy of both MLL4 HMG-USD and UTX were determined by ChIP-seq. As shown by log2 fold change heat maps in Figure 4A, the HMG-USD readily binds chromatin after dox induction and this coincides with increased UTX binding at the same locations as HMG-USD. We also performed RNA-seq on samples collected after 2, 4, or 8 d of dox induction and, following a normalization step to remove unwanted batch effects (see the Materials and Methods), compared their gene expression profiles with that of WT-HCT116 and ΔMLL4 cells (no dox). Compared with WT-HCT116, our ΔMLL4 cells have 2112 down-regulated genes (adj.P < 0.05, absolute fold change >1.5), and after 8 d of dox induction we identified 177 genes up-regulated compared with uninduced ΔMLL4 cells. Interestingly, ∼83% (148/177) of those overlap with genes statistically down-regulated in the ΔMLL4 versus WT, indicating our synthetic HMG-USD transgene can at least partially rescue a subset of MLL4-dependent genes, presumably through UTX recruitment (Supplemental Fig. S5A). Likewise, roughly 66% (199/298) of dox-induced down-regulated genes overlap with those up-regulated in ΔMLL4 versus WT. This is visualized in Figure 4B where we hierarchically cluster all 475 differentially expressed genes (from dox day8) and compare their Z-scores across time points and treatments. Qualitatively, replicates from day8 induction bear a closer resemblance to WT than either day2/4 time points or uninduced ΔMLL4, suggesting sustained expression of HMG-USD can restore gene expression in certain contexts (Fig. 4B). We expect that these transcriptional effects are most likely independent of UTX's histone demethylase activity, as ChIP-seq reveals that the new HMG-USD-induced binding sites are largely devoid of H3K27me3 and existing H3K27me3 domains are little changed before and after induction (Supplemental Fig. S5B).
Figure 4.
The MLL4-HMG-USD domain rescues some gene expression via recruiting UTX to chromatin. (A) In ΔMLL4-T/O cells, ChIP-seq was performed for GFP-HMG-USD and UTX using antibodies recognizing GFP and UTX, respectively, after 0, 2, and 4 d of doxycycline induction. HMG-USD and UTX ChIP-seq average peaks (top) and heat maps (bottom) are centered at HMG-USD dox day 4 peaks and presented as log2FC. (B) After 8 d of dox induction, 475 genes are differentially expressed compared with uninduced ΔMLL4. Relative expression levels are hierarchically clustered and presented as Z-score heat maps for the same 475 genes across all five conditions. Note that the day8 induction time point bears closer resemblance to WT, indicating that a fraction of MLL4-dependent genes are “rescued” by sustained HMG-USD expression.
Figure 5.
Model for trr/MLL3/4-mediated UTX stabilization and function at chromatin. Lpt and Trr represent a gene split, corresponding to the N-terminal and C-terminal halves of MLL4, respectively, and both are required for Drosophila viability. We mapped an ∼600-amino-acid region of Trr, corresponding to ∼25% of the full-length protein, which is able to rescue Trr-null lethality. While this region does not overlap any known domains, we demonstrate its ability to bind and stabilize Utx both in flies and human cells. The HMG domain of Lpt was previously reported to associate with Trr by coimmunoprecipitation, suggesting that Lpt may also play a role in mediating Utx stability in Drosophila. We found that in the absence of MLL3/4, UTX stabilization is achieved by expressing a minimal UTX-interacting peptide fused to MLL4's HMG domain.
Discussion
In this study, we took advantage of the Lpt/trr gene split in Drosophila to investigate essential noncatalytic functions of Trr. We found a 600-amino-acid region of Trr rescued trr mutant flies and resulted in stabilization of UTX. Using mammalian cells, an ∼80-amino-acid portion of this domain from human MLL3 or MLL4, when combined with an HMG box from their N termini was sufficient to stabilize UTX in MLL3-MLL4 double mutant cells (Fig. 5). This stabilized UTX associates on chromatin and positively affected gene expression resulting from MLL4 knockout in the MLL3-mutant HCT116 cells. The discrepancy between the partial gene expression rescue of ΔMLL4 HCT116 cells with the MLL4 USD-HMG fusion and the ability of the 600-amino-acid region of Trr to rescue development of trr-null flies could be due to a number of factors. For example, it is possible that domains of the essential LPT protein, in addition to the HMG domain, could contribute to fly viability by providing additional chromatin interaction domains, and these are absent in ΔMLL4 HCT116 cells. In addition, PTIP, which is essential for fly development (Fang et al. 2009), has been shown to interact with MLL4 in the absence of UTX (Wang et al. 2017). We found that the 600-amino-acid rescue domain interacts with PTIP and NCOA6, which may contribute to the rescue of trr-null flies. Nonetheless, our findings indicate a major histone methylatransferase-independent function of Trr/MLL3/MLL4 branch of the COMPASS family is through UTX stabilization.
Loss-of-function mutations of KMT2C, KMT2D, and KDM6A are increasingly recognized as driving events in a variety of cancers and developmental disorders (Morgan and Shilatifard 2015; Wang and Shilatifard 2019). Similar to Trr/MLL3/MLL4, catalytic-dependent and -independent biological functions have been demonstrated for UTX in controlling various biological processes, both in Drosophila and mammals (Copur and Müller 2018). However, three homologs—UTX, UTY, and JMJD3—exist in mammals, and it remains to be determined which of these functions are redundant among those factors. Furthermore, it is unclear how UTX is recruited to enhancers, as this protein does not have a recognizable DNA-binding domain. One recent study demonstrates MLL3/UTX recruitment occurs through interactions between MLL3 and BAP1, and disrupting this interaction abrogates UTX enhancer binding (Wang et al. 2018). BAP1-dependent recruitment appears to be specific to MLL3, as MLL4 interactions were not detected.
Finally, our study demonstrates a conserved function of Drosophila Trr is to stabilize Utx, and raises the prospect that a pharmacologic strategy to stabilize UTX could be therapeutically beneficial for pathologies, such as bladder cancer, in which MLL3/4 and UTX mutations tend to be mutually exclusive (The Cancer Genome Atlas Research Network 2014; Fantini et al. 2018).
Materials and methods
Fly (Drosophila melanogaster) stocks
Genomic trr rescue flies were generated using pattB plasmid for site-specific integration on 3R (89E11) and injections performed by BestGene (strain 9744). Transgenic flies were crossed to trr[1] flies and then made homozygous for the trr[1] allele and trr rescue construct.
Cell fractionation and immunoprecipitation
Cytosolic extracts were made using hypotonic buffer (10 mM HEPES at pH 7.9, 1.5 mM MgCl2, 10 mM KCl) with 0.2% Triton X-100. Nuclear extracts were made with high-salt buffer (20 mM HEPES at pH 7.9, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 350 mM NaCl). Anti-FLAG M2 agarose (Sigma) was used for all FLAG-IP, and nonspecific proteins were removed with wash buffer (10 mM HEPES at pH 7.9, 1.5 mM MgCl2, 300 mM NaCl, 10 mM KCl, 0.2% Triton X-100). SDS-PAGE was performed with 4%–20% mini-Protean TGX precast gels (Biorad) and transferred to nitrocellulose membrane for Western blotting.
Mass spectrometry
Sample preparation and mass spectrometry of immunoprecipitated protein complexes were performed as described (Hickox et al. 2017). See the Supplemental Material for details.
CRISPR–Cas9 gene editing
Plasmids expressing Cas9 and guide RNAs (gRNAs) were constructed in pX330 (Cong et al. 2013). Guide RNAs are listed in Supplemental Material. HCT116 cells were electroporated and single clones selected as described (Morgan et al. 2017). Cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Guide RNAs are listed in the Supplemental Material.
Protein sequence alignments
Full-length sequences were aligned with Muscle v3.8.31(Edgar 2004), trimmed and manually adjusted in AliView (Larsson 2014), and displayed with Jalview (Waterhouse et al. 2009) with Clustal colors with a coloring by conservation score of 40 for vertebrates and 20 for the combined vertebrate and invertebrate alignment.
NGS sample preparation
ChIP sequencing libraries were prepared using the KAPA HTP library preparation kit complemented with NEXTflex DNA Barcodes from Bioo Scientific. Ten nanograms of DNA was used as starting material for input and IP samples. Libraries were amplified using 13 cycles on the thermocycler. Postamplification libraries were size-selected at 250–450 bp in length using Agencourt AMPure XP beads from Beckman Coulter. Libraries were validated using the Agilent high-sensitivity DNA kit. RNA sequencing libraries were prepared using the NEBNext Ultra II directional RNA library preparation kits, used in conjunction with the NEBNext rRNA depletion kit. Input RNA quality was validated using the Agilent RNA 6000 nano kit. One microgram of total RNA was used as starting material. Libraries were validated using the Agilent DNA 1000 kit.
Chromatin immunoprecipitation analysis
The ChIP-seq was performed as described previously (Wang et al. 2018) and is detailed further in the Supplemental Material. For ChIP-seq analysis, H3K27me3 peaks were called with the MACS v2.1.0 software (Zhang et al. 2008) using default parameters (–broad, q = 0.01), while narrow peaks were called with MACS v1.4.2 with default parameters. Metaplots for called peaks signals were generated using R library SeqPlots v1.26.0 (Stempor and Ahringer 2016), while heat maps were generated using deeptools v3.1.1 (Ramírez et al. 2016).
RNA sequencing analysis
Gene counts were computed by HTSeq (Anders et al. 2015) and used as input for DESeq2 v1.26.0 (Love et al. 2014). As we were comparing RNA-seq samples from two separate experiments with different cell lines (wild-type HCT116 and MLL-KO HCT116 cells) we used RUVSeq v1.20.0 (Risso et al. 2014) p2013ackage to correct for unwanted batch effects. To estimate the factors of unwanted variation we applied RUVs protocol, using all expressed genes grouped by sample replicates, with parameter k = 3, resulting weights were passed as parameters to the design of DESeq2 log-ratio test model. Genes with adjusted P-values <0.05 and absolute fold change >1.5 (|log2FC|≥0.584) were considered as differentially expressed unless otherwise specified. RNA-seq heat maps adjacent to ChIP-seq heat maps display z-scored log2 fold change values of genes corresponding to TSSs nearest to ChIP-seq peaks and were created using R package pheatmap v1.0.12 (https://cran.r-project.org/web/packages/pheatmap/index.html). Functional analysis was carried out using R package clusterProfiler v.3.16.0 (Yu et al. 2012).
Accession number
ChIP-seq and RNA-seq data are available at GEO with accession number GSE158672.
Supplementary Material
Acknowledgments
We thank Nicole Ethen for assistance with illustrations. R.R. was supported by National Institutes of Health (NIH)/National Cancer Institute (NCI) F99CA222988. A.P. was supported by NIH/NCI K99CA234434. E.R.S. was supported by NIH/NCI R50CA211428. This study was supported in part by Outstanding Investigator Award R35CA197569 from the NCI to A.S.
Author contributions: R.R. and A.S. conceived the project. R.R., L.W., M.A.M., A.P., and S.H.A.S. performed experiments and interpreted the data. N.K. performed mass spectrometry analysis. E.J.R. performed Illumina sequencing. M.I., P.A.O., and E.R.S. analyzed data. J.N.S. and A.S. provided supervision. R.R. wrote the manuscript with input from L.W., M.I., E.R.S., and A.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.339762.120.
References
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734. 10.1038/nature06145 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507: 315–322. 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan C, Zraly CB, Parilla M, Diaz MO, Dingwall AK. 2012. Histone recognition and nuclear receptor co-activator functions of Drosophila cara mitad, a homolog of the N-terminal portion of mammalian MLL2 and MLL3. Development 139: 1997–2008. 10.1242/dev.076687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Smith ER, Shilatifard A. 2018. Born to run: control of transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 19: 464–478. 10.1038/s41580-018-0010-5 [DOI] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406. 10.1074/jbc.M701574200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copur O, Müller J. 2018. Histone demethylase activity of Utx Is essential for viability and regulation of HOX gene expression in Drosophila. Genetics 208: 633–637. 10.1534/genetics.117.300421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, Still CD II, Garcia BA, Adelman K, Wysocka J. 2017. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell 66: 568–576.e4. 10.1016/j.molcel.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Ren H, Liu J, Cadigan KM, Patel SR, Dressler GR. 2009. Drosophila ptip is essential for anterior/posterior patterning in development and interacts with the PcG and trxG pathways. Development 136: 1929–1938. 10.1242/dev.026559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini D, Glaser AP, Rimar KJ, Wang Y, Schipma M, Varghese N, Rademaker A, Behdad A, Yellapa A, Yu Y, et al. 2018. A carcinogen-induced mouse model recapitulates the molecular alterations of human muscle invasive bladder cancer. Oncogene 37: 1911–1925. 10.1038/s41388-017-0099-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi Yh, Mickey K, Voets O, Verrijzer CP, Shilatifard A. 2012. Enhancer-associated H3K4 monomethylation by trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 26: 2604–2620. 10.1101/gad.201327.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickox AE, Wong AC, Pak K, Strojny C, Ramirez M, Yates JR 3rd, Ryan AF, Savas JN. 2017. Global analysis of protein expression of inner Ear hair cells. J Neurosci 37: 1320–1339. 10.1523/JNEUROSCI.2267-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. 2013. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 33: 4745–4754. 10.1128/MCB.01181-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. 2013. Mutational landscape and significance across 12 major cancer types. Nature 502: 333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Sharma A, Dhar SS, Lee SH, Gu B, Chan CH, Lin HK, Lee MG. 2014. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res 74: 1705–1717. 10.1158/0008-5472.CAN-13-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. 2014. Aliview: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30: 3276–3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, Maystadt I, Dallapiccola B, Verellen-Dumoulin C. 2012. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am J Hum Genet 90: 119–124. 10.1016/j.ajhg.2011.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, et al. 2013. H3k4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2: e01503 10.7554/eLife.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. 2013. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods 10: 730–736. 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake N, Koshimizu E, Okamoto N, Mizuno S, Ogata T, Nagai T, Kosho T, Ohashi H, Kato M, Sasaki G, et al. 2013a. MLL2 and KDM6A mutations in patients with Kabuki syndrome. Am J Med Genet A 161A: 2234–2243. 10.1002/ajmg.a.36072 [DOI] [PubMed] [Google Scholar]
- Miyake N, Mizuno S, Okamoto N, Ohashi H, Shiina M, Ogata K, Tsurusaki Y, Nakashima M, Saitsu H, Niikawa N, et al. 2013b. KDM6A point mutations cause Kabuki syndrome. Hum Mutat 34: 108–110. 10.1002/humu.22229 [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. 2011. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 31: 4310–4318. 10.1128/MCB.06092-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MA, Shilatifard A. 2015. Chromatin signatures of cancer. Genes Dev 29: 238–249. 10.1101/gad.255182.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MAJ, Rickels RA, Collings CK, He X, Cao K, Herz HM, Cozzolino KA, Abshiru NA, Marshall SA, Rendleman EJ, et al. 2017. A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev 31: 2003–2014. 10.1101/gad.305201.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, et al. 2010. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42: 790–793. 10.1038/ng.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. 2012. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18: 298–302. 10.1038/nm.2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickels R, Shilatifard A. 2018. Enhancer logic and mechanics in development and disease. Trends Cell Biol 28: 608–630. 10.1016/j.tcb.2018.04.003 [DOI] [PubMed] [Google Scholar]
- Rickels R, Herz HM, Sze CC, Cao K, Morgan MA, Collings CK, Gause M, Takahashi YH, Wang L, Rendleman EJ, et al. 2017. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat Genet 49: 1647–1653. 10.1038/ng.3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. 2014. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32: 896–902. 10.1038/nbt.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. 2012. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempor P, Ahringer J. 2016. SeqPlots—interactive software for exploratory data analyses, pattern discovery and visualization in genomics. Wellcome Open Res 1: 14 10.12688/wellcomeopenres.10004.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523. 10.1038/ng.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shilatifard A. 2019. UTX mutations in human cancer. Cancer Cell 35: 168–176. 10.1016/j.ccell.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SP, Tang Z, Chen CW, Shimada M, Koche RP, Wang LH, Nakadai T, Chramiec A, Krivtsov AV, Armstrong SA, et al. 2017. A UTX–MLL4–p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol Cell 67: 308–321.e6. 10.1016/j.molcel.2017.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhao Z, Ozark PA, Fantini D, Marshall SA, Rendleman EJ, Cozzolino KA, Louis N, He X, Morgan MA, et al. 2018. Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nat Med 24: 758–769. 10.1038/s41591-018-0034-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, Vlasevska S, Mo T, Tang H, Basso K, et al. 2015. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 21: 1190–1198. 10.1038/nm.3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.