In this study, Low et al. investigated the signal that triggers CMG unloading in Xenopus egg extracts. Using single-molecule and ensemble approaches, the authors show that DNA suppresses CMG ubiquitylation during elongation and that this suppression is relieved when CMGs converge, leading to CMG unloading.
Keywords: CMG, CRL2Lrr1, DNA replication, replication termination, single-molecule imaging, ubiquitylation
Abstract
When converging replication forks meet during replication termination, the CMG (Cdc45–MCM2–7–GINS) helicase is polyubiquitylated by CRL2Lrr1 and unloaded from chromatin by the p97 ATPase. Here, we investigate the signal that triggers CMG unloading in Xenopus egg extracts using single-molecule and ensemble approaches. We show that converging CMGs pass each other and keep translocating at the same speed as before convergence, whereafter they are rapidly and independently unloaded. When CMG unloading is blocked, diverging CMGs do not support DNA synthesis, indicating that after bypass CMGs encounter the nascent lagging strands of the converging fork and then translocate along double-stranded DNA (dsDNA). However, translocation on dsDNA is not required for CMG's removal from chromatin because in the absence of nascent strand synthesis, converging CMGs are still unloaded. Moreover, recombinant CMG added to nuclear extract undergoes ubiquitylation and disassembly in the absence of any DNA, and DNA digestion triggers CMG ubiquitylation at stalled replication forks. Our findings suggest that DNA suppresses CMG ubiquitylation during elongation and that this suppression is relieved when CMGs converge, leading to CMG unloading.
The faithful duplication of genomic DNA is a fundamental property of all living systems. In vertebrate cells, DNA replication initiates from ∼60,000 sites called origins of replication. The two replisomes assembled at each origin travel in opposite directions, copying DNA as they go, in a process called elongation. The enzymatic machinery of the replisome includes the CMG helicase, which unwinds DNA by translocating along the leading strand template; DNA polymerase (pol) ε, which binds tightly to CMG and synthesizes the leading strand; DNA pol α, which primes Okazaki fragments; DNA pol δ, which completes Okazaki fragments; and PCNA, which confers processivity to polymerases ε and δ. When converging replisomes meet, DNA synthesis is completed, all nascent strands are ligated, replisomes are disassembled, and daughter molecules are decatenated (“replication termination”) (Dewar and Walter 2017). Compared with initiation and elongation, replication termination is poorly understood.
In yeast and metazoans, replisome disassembly during termination involves ubiquitylation of the Mcm7 subunit of the CMG helicase, followed by CMG removal from chromatin by the p97 ATPase (Maric et al. 2014; Moreno et al. 2014). CMG ubiquitylation and removal leads to unloading of many CMG interacting partners (e.g., DNA pol ε, Timeless-Tipin, Claspin, Mcm10, TRAIP) effectively dismantling the replisome (Dewar et al. 2017). In yeast, Mcm7 is ubiquitylated by the E3 ubiquitin ligase SCFDia2 (Maric et al. 2014), whereas in metazoans, this function is carried out by the cullin 2 RING ligase, CRL2Lrr1 (Dewar et al. 2017; Sonneville et al. 2017). If CMG fails to be ubiquitylated by CRL2Lrr1 in interphase, it can be ubiquitylated and unloaded in mitosis by a back-up pathway that depends on the E3 ubiquitin ligase TRAIP (Sonneville et al. 2017; Deng et al. 2019; Moreno et al. 2019). A major unanswered question is which termination-specific event triggers CMG ubiquitylation and unloading upon fork convergence in interphase. This is a critical issue because premature unloading of CMG would lead to replication fork collapse and DNA underreplication, whereas failure to unload CMG might interfere with transcription or the next round of replication. Consistent with the importance of timely CMG removal, mutations in Dia2 cause genome instability in yeast (Blake et al. 2006), and ablating both interphase and mitotic CMG unloading pathways is lethal in worms (Sonneville et al. 2017).
To identify the signal that promotes CMG ubiquitylation and replisome disassembly after fork convergence, it is essential to consider in detail the events underlying termination. When two replisomes meet, their leading strands pass each other and are rapidly ligated to the last Okazaki fragment of the converging fork (Fig. 1A). CMG is unloaded after nascent strands are ligated, demonstrating that CMG removal is a late event that occurs only after converging replication forks have fully matured into double-stranded DNA (Dewar et al. 2015). To explain this observation, we previously proposed that CMGs pass each other during termination and that when they reach the 5′ end of the downstream Okazaki fragment, they pass over the single-stranded DNA (ssDNA)–double-stranded DNA (dsDNA) junction and keep translocating along dsDNA (Fig. 1A; Dewar et al. 2015). However, direct evidence for this hypothesis is lacking, and how CMG might behave on dsDNA is unclear. Finally, catenated daughter DNA molecules are resolved by topoisomerase II independently of CMG unloading. Importantly, CRL2Lrr1 binds chromatin de novo only after replisome convergence (Dewar et al. 2017; Sonneville et al. 2017), but how termination triggers CRL2Lrr1 recruitment remains unknown. Notably, SCFDia2 immunoprecipitates with components of the yeast replisome (Morohashi et al. 2009), but whether this interaction is constitutive during replication or forms during preparation of cell lysates is unclear.
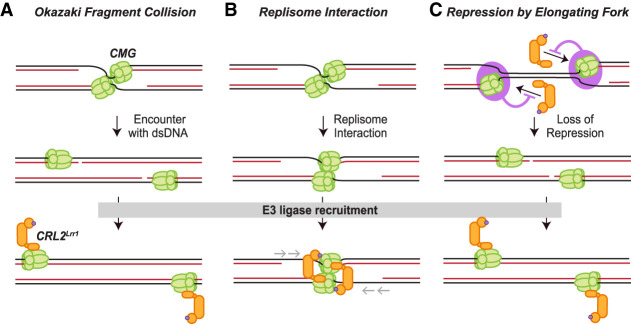
Figure 1.
Possible models of CRL2Lrr1 recruitment upon DNA replication termination. (A) Okazaki fragment collision model. Upon CMG encounter with the Okazaki fragment of the opposing fork, CMG undergoes a conformational change that triggers CRL2Lrr1 recruitment. (B) Replisome interaction model. When two replication forks meet, the two CMG helicases interact, creating a structure that recruits CRL2Lrr1. In this model, CMGs reel in the template strands to complete DNA synthesis (gray arrows). In an alternative version of this model (not depicted), CMGs pass each other and interact with other protein components of the converging fork (e.g., PCNA), leading to CRL2Lrr1 recruitment. (C) Repression model. A protein or DNA structure (pink circle) suppresses binding of CRL2Lrr1 to CMG at the elongating replication fork. When forks converge, the repressive protein or DNA structure is lost, leading to CRL2Lrr1 recruitment.
Three general models have been proposed to explain how CMG ubiquitylation is limited to termination (Dewar and Walter 2017; Mukherjee and Labib 2019). (1) CRL2Lrr1 recruitment is triggered when converging CMGs encounter a termination-specific nucleic acid structure. In this model, the collision of CMG with an Okazaki fragment from the oncoming replication fork triggers CMG ubiquitylation, possibly due to a conformational change in CMG that occurs when dsDNA enters CMG's central channel (Fig. 1A). (2) CMG's interaction with proteins of the converging replisome enables the recruitment of CRL2Lrr1. For example, the interaction of two CMGs (Fig. 1B) or the collision of CMG with lagging-strand processing enzymes of the converging fork might promote CRL2Lrr1 binding. (3) A protein or DNA structure associated with the elongating replisome suppresses CRL2Lrr1 binding; upon fork convergence, this structural feature disappears, allowing ligase recruitment (Fig. 1C). While the above discussion focuses on CRL2Lrr1, the same models can in principle also apply to SCFDia2.
To distinguish between these models and to better understand how CMGs behave after fork convergence, we used single-molecule imaging of CMG in frog egg extracts. We show that fluorescently labeled CMG complexes normally undergo unloading within minutes of fork convergence. However, when CMG unloading is blocked, most CMGs pass each other and keep moving without changing speed. Strikingly, CMGs that bypass each other no longer support DNA synthesis, probably reflecting CMG translocation along dsDNA, as previously proposed (Dewar et al. 2015). However, when CMGs converge in the absence of nascent strand synthesis, CMGs are still unloaded, indicating that the encounter of CMG with a ssDNA–dsDNA junction is not essential for unloading. Moreover, DNase treatment of replisomes that have not yet converged induces CMG ubiquitylation, and recombinant CMG added to egg extracts in the absence of any DNA undergoes CRL2Lrr1-dependent ubiquitylation and p97-dependent disassembly. Together, our data argue that CMG ubiquitylation is suppressed by an elongation-specific DNA structure that is lost when forks meet during termination (Fig. 1C).
Results
Terminating CMGs can pass each other
To better understand the mechanism of replication termination, we studied the dynamics of converging CMGs using single-molecule imaging in replication-competent Xenopus egg extracts (Fig. 2A), as described previously (Sparks et al. 2019). Briefly, stretched λ DNA molecules immobilized in a microfluidic flow cell were first incubated in a high-speed supernatant (HSS) of total egg lysate that supports MCM2–7 recruitment (“licensing”). We subsequently replaced HSS in the flow cell with a concentrated nucleoplasmic extract (NPE) containing recombinant GINS, a component of CMG that was labeled with Alexa fluor 647 (GINSAF647). After a brief incubation to allow CMG assembly and initiation, we flowed in NPE lacking any GINS, followed by total internal reflection microscopy to visualize AF647-labeled CMG (Fig. 2A). While we previously intentionally limited origin firing during this procedure (Sparks et al. 2019), here we allowed multiple origins to fire per DNA molecule, enabling us to visualize CMG unloading during termination.
Figure 2.
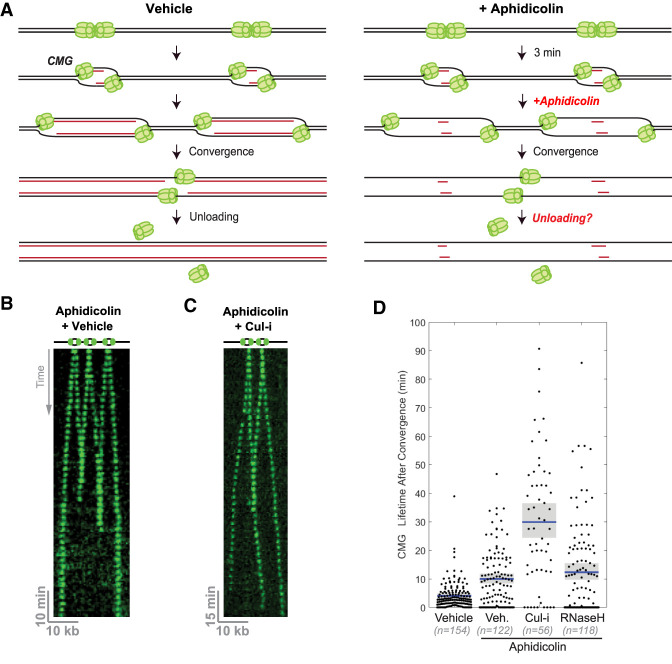
CMG complexes pass one another when unloading is prevented. (A) Scheme of the single-molecule imaging experiment. Green ovals represent GINSAF647. (B) CMGs converge and are rapidly unloaded. λ DNA was replicated in the presence of DMSO (vehicle) and imaged every 30 sec. Images of individual molecules were stacked to create kymograms. CMGs were false-colored in green. The signal in the region of interest (gray box) was integrated and is shown in the box at the right. The moment of CMG unloading was determined by the stepwise drop in signal intensity. Scale bars are shown at the bottom left. (C) Same as B but depicting an example in which CMGs are unloaded after divergence. (D) CMG lifetime after replisome convergence in the indicated conditions from C, E, and F. A schematic of the event being quantified is shown in green. (E) Same as B, but NPE contained p97-i. Note that rightward-moving CMG is brighter than the leftward-moving molecule, clearly showing the two helicases passing each other after convergence (as opposed to backtracking). (F) Same as B, but NPE contained Cul-i. (G) Percentage of total termination events where CMGs diverged after convergence. (H) Time interval between the moment of convergence and divergence. (I) Time interval between the unloading of the first and the second CMG molecule. (J) CMG speed during convergence (Conv.) or divergence (Div.) for termination events where the CMGs diverged. In D and H–J, the blue bar represents the mean, and the gray box indicates the 95% confidence interval estimated by bootstrapping. Values are reported in Supplemental Table S1.
Following their convergence, CMGs were rapidly unloaded (Fig. 2B), and they sometimes passed each other before being unloaded (Fig. 2C). For quantification, the trajectory of each CMG molecule prior to convergence was fitted to a straight line, and the moment of CMG convergence was estimated by determining when the two lines intersect (Fig. 2B). Similarly, the moment of CMG divergence was estimated using linear fits to CMG trajectories after divergence (Fig. 2C). CMG bypass was scored when the two helicases diverged by more than two pixels (∼300 nm), and therefore became well resolved according to the Rayleigh resolution limit criterion (Fig. 2C). Based on these criteria, CMGs were unloaded within 4.0 [3.4–5.0] min (mean [confidence interval {CI} of the mean]) after convergence (Fig. 2D), consistent with ensemble experiments (Dewar et al. 2017). When CMG unloading was prevented with NMS-873, an inhibitor of the p97 ATPase (“p97-i”), or MLN4924, which prevents neddylation and activation of cullin RING ligases (“Cul-i”), converged CMG molecules persisted on DNA much longer after convergence: 32.0 [27.9–36.7] min and 29.5 [25.9–33.4] min, respectively (mean [CI of the mean]) (Fig. 2D). In this setting, CMG lifetimes were likely an underestimate, as some CMG molecules disappear because of AF647 photobleaching (with an estimated half-time of ∼60 min) (Sparks et al. 2019), and others—due to DNA tethers rupturing during the experiment. In the presence of inhibitors, CMGs diverged, and travelled for several kilobases (Fig. 2E,F). Because the signal from individual fluorophores can vary substantially (Mutch et al. 2007), we occasionally observed converging CMGs whose brightness was clearly distinct, enabling us to unambiguously conclude that the two helicases passed each other (Fig. 2E). CMGs bypassed each other in 30% of unperturbed termination events, and almost 80% of the time in the presence of p97-i or Cul-i (Fig. 2G). Moreover, CMGs diverged within a few minutes of converging (mean [CI of the mean] = 1.0 [0.5–2.3] min), and in most CMG bypass events, there was no detectable pause between the estimated moment of helicase convergence and that of divergence (Fig. 2H). These results disfavor the model in which replisome dimerization triggers CMG unloading (Fig. 1B). To determine whether unloading of converging CMGs is concerted, we measured the time between the two CMG unloading events for each termination instance. In the vast majority of cases, the two CMGs were not unloaded simultaneously (Fig. 2I), indicating that converging CMGs are unloaded independently of each other. Finally, we found that the speed of the replicative helicase did not change significantly after CMGs diverged: 267 [237–302] nt/min before convergence versus 323 [252–455] nt/min (mean [CI of the mean]) after divergence for vehicle control (Fig. 2J). We conclude that converging CMGs normally pass each other and continue rapid translocation, but in the absence of inhibitors, they are usually unloaded before being optically resolved.
We next asked whether diverging CMGs support DNA synthesis. To monitor DNA synthesis in our single-molecule assay, we added catalytically inactive, fluorescently labeled flap endonuclease Fen1-D179AmKikGR to NPE (Supplemental Fig. S1A). Fen1-D179AmKikGR binds to PCNA and delays its dissociation from DNA, decorating the nascent lagging strands and thereby labeling most of the replication bubble (Loveland et al. 2012). Although CMGs diverged in the presence of Fen1-D179AmKikGR (hereafter referred to as Fen1mKikGR) and p97-i or Cul-i, they travelled very slowly (Supplemental Fig. S1B) suggesting that Fen1mKikGR impaired the ability of diverging CMG to translocate. Interestingly, the gap between the two diverging helicases was devoid of new Fen1mKikGR signal (Supplemental Fig. S1C). Although this observation could reflect abnormal DNA synthesis (e.g., leading strand only replication), the simplest interpretation is that diverging CMGs do not support DNA rereplication. We propose that advancing CMGs displace Fen1mKikGR of the converging fork, but cannot rule out that Fen1mKikGR naturally dissociates. In further agreement with a lack of rereplication, we did not detect additional DNA synthesis when replication termination was carried out in the presence of p97-i (Supplemental Fig. S1D,E), nor did this drug delay the appearance of fully ligated DNA replication products (Supplemental Fig. S1F; Dewar et al. 2017). Finally, during unperturbed replication, the leading strand from one replication fork stops when it reaches the lagging strand from the converging fork (Dewar et al. 2015), arguing against rereplication in the absence of any inhibitors. Taken together, these data indicate that during replication termination, CMGs pass each other and switch to a translocation mode that does not support DNA synthesis, even when CMG unloading is blocked. We propose that this distinct translocation regime involves CMG movement along dsDNA, which has been observed previously for recombinant CMG on model DNA templates (Langston and O'Donnell 2017). The data presented so far disfavor the replisome dimerization model (Fig. 1B) but are consistent with our previous hypothesis that CMG's encounter with a ssDNA–dsDNA junction triggers CMG unloading (Fig. 1A; Dewar et al. 2015).
Inhibition of nascent strand synthesis does not prevent CMG unloading
To investigate whether a ssDNA–dsDNA junction is essential to trigger CMG unloading, we allowed replication to initiate for 3 min, and then added high levels of aphidicolin, an inhibitor of replicative DNA polymerases that uncouples CMG from the point of synthesis (Sparks et al. 2019). Under these conditions, CMGs should encounter no ssDNA–dsDNA junctions upon convergence (Fig. 3A, +aphidicolin). Indeed, we used Fen1mKikGR imaging to confirm that polymerase activity was blocked by aphidicolin (Supplemental Fig. S2A). CMG helicases were then imaged as in Figure 2A. Aphidicolin slowed the rate of fork progression from 267 [237–302] nt/min to 71 (62–80) nt/min (mean [CI of the mean]) (Supplemental Fig. S2B), consistent with our previous observation of uncoupled CMGs in the absence of termination (Sparks et al. 2019). The uncoupled CMGs eventually converged and were unloaded (Fig. 3B), but apparently more slowly than in the absence of aphidicolin (Fig. 3D; see the Discussion). This effect was not caused by the activation of the DNA damage checkpoint (Supplemental Fig. S2C). We observed similar results when extracts were supplemented with aphidicolin and recombinant RNase H to degrade any RNA primers synthesized by pol α (Fig. 3D). Importantly, Cul-i addition delayed CMG unloading in the presence of aphidicolin, indicating that the unloading of uncoupled CMGs was still CRL2Lrr1-dependent (Fig. 3C,D). Thus, assuming that the trigger for CMG unloading is the same in the presence and absence of aphidicolin, these results suggest that a ssDNA–dsDNA junction (with or without an RNA primer) is not an essential trigger for CMG unloading by CRL2Lrr1, contrary to the model depicted in Figure 1A.
Figure 3.
CMG unloading does not require encounter with dsDNA. (A) Schematic of replication termination in the presence or absence of aphidicolin, which inhibits all replicative polymerases. Short red lines in the panel at the right represent limited nascent strand synthesis that occurs before addition of aphidicolin. Without DNA synthesis, replisomes converge without CMG encountering dsDNA, as shown at the right. (B) λ DNA was replicated as in Figure 2A, but in the presence of aphidicolin. (C) Same as B, but extracts were supplemented with Cul-i. (D) Quantification of CMG lifetime after convergence in the presence or absence of aphidicolin, Cul-i, or RNase H. (Blue bar) Mean, (gray boxes) 95% confidence interval estimated by bootstrapping. Values are reported in Supplemental Table S1.
Purified CMG is ubiquitylated by CRL2Lrr1 in the absence of DNA
Given that a ssDNA–dsDNA junction is dispensable for CMG ubiquitylation, we tested whether CMG can be ubiquitylated in the absence of any DNA. To test this possibility, we added recombinant Xenopus CMG (Fig. 4A, rCMGWT; Wu et al. 2019) containing an N-terminal FLAG tag on Mcm3 to NPE, which is largely devoid of DNA (Lebofsky et al. 2009). After incubation in NPE, CMG was isolated on anti-FLAG resin and immunoblotted for components of CMG and CRL2Lrr1. Strikingly, incubation of recombinant CMG in NPE was sufficient to trigger its disassembly, as seen from the loss of GINS from the FLAG-Mcm3 IP (Fig. 4B, cf. lanes 4 and 2). We speculate that the MCM2–7 subcomplex is also disassembled in NPE, but that once Mcm7 has been extracted, the complex rapidly reassembles, explaining the coimmunoprecipitation of Mcm7 and Mcm6 with FLAG-Mcm3 (Fig. 4B, lane 4). Whether the coimmunoprecipitated Mcm7 represents recombinant molecules that have been deubiquitylated upon extraction or endogenous Mcm7 that was never ubiquitylated is unclear. When NPE was supplemented with p97-i, CMG was not disassembled, and it associated with CRL2Lrr1 and accumulated ubiquitylated Mcm7 (Fig. 4B, lane 3). Ubiquitylation of rCMG in extract involved K48 linkages (Supplemental Fig. S3A,B), as seen for endogenous CMG during bona fide termination (Maric et al. 2014; Moreno et al. 2014; Dewar et al. 2017). Moreover, depletion of CRL2Lrr1 from NPE abolished rCMG ubiquitylation, and the defect was partially rescued by recombinant CRL2Lrr1 (Supplemental Fig. S3C–E), which was also active in CMG unloading during normal replication termination (Supplemental Fig. S3F–J). In contrast, TRAIP depletion had no effect on rCMG ubiquitylation (data not shown). Thus, in extracts lacking replication or exogenous DNA, rCMG complexes are ubiquitylated on Mcm7 by CRL2Lrr1 and disassembled in a p97-dependent manner.
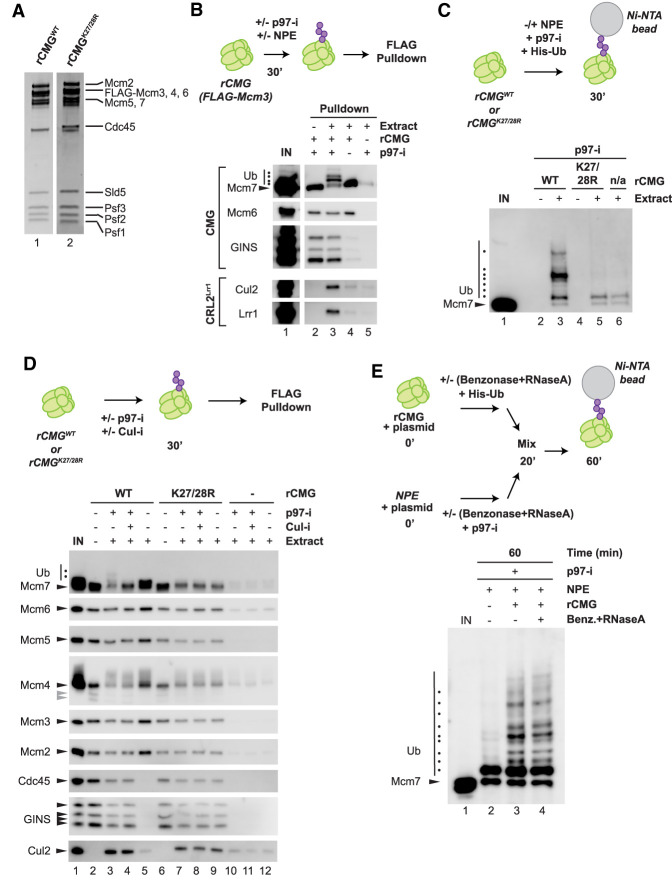
Figure 4.
Recombinant CMG undergoes Mcm7 ubiquitylation in extract in the absence of DNA. (A) rCMG and rCMGK27/28R visualized with SYPRO Ruby stain. (B) rCMG from A was incubated in NPE or buffer in the presence or absence of p97-i, and then recovered using anti-FLAG resin and blotted for the indicated proteins. (C) rCMG and rCMGK27/28R from A were incubated in buffer or NPE in the presence of p97-i and His-tagged ubiquitin (His-Ub). Ubiquitylated proteins were isolated with Ni-NTA resin and immunoblotted for Mcm7. (D) rCMG and rCMGK27/28R from A were incubated in buffer or NPE in the presence or absence of p97-i or Cul-i, CMG and associated proteins were isolated on anti-FLAG resin, and the samples were immunoblotted with the indicated antibodies. (Gray arrowheads) Degradation products of Mcm4. (E) rCMG supplemented with His-Ub was treated with 2 U/µL benzonase and 2.5 µg/µL RNase A for 20 min. Simultaneously, NPE supplemented with p97-i was treated with benzonase and RNase A for 20 min. Both NPE and rCMG samples were supplemented with a 5.4-kb plasmid to verify the efficiency of benzonase treatment. The two treated samples were mixed together and incubated for a further 40 min. At that point, His-ubiquitin pull-down was performed, followed by immunoblotting with Mcm7 antibodies. Samples were also removed at the 20- and 60-min time points for DNA analysis (Supplemental Fig. S5A).
The sites on Mcm7 that are ubiquitylated during termination remain uncharacterized in higher eukaryotes. Proteomic studies have identified lysine 28 (K28) on human Mcm7 as being ubiquitylated in a cullin-dependent manner (Emanuele et al. 2011; Kim et al. 2011). Additionally, ubiquitylation of budding yeast Mcm7 occurs on K29 in vitro, but other sites can be used in vivo (Maric et al. 2017). Xenopus laevis Mcm7 contains lysines at positions 27 and 28, and at least one of these sites is conserved in Drosophila, worms, mice, and humans (Supplemental Fig. S4). To determine whether K27 and/or K28 of Mcm7 are ubiquitylated in NPE, we expressed and purified CMG containing a FLAG tag on the N terminus of Mcm3 and K-to-R mutations at both of these sites in Mcm7 (rCMGK27/28R) (Fig. 4A). To recover ubiquitylated Mcm7, His-tagged ubiquitin (His-Ub) was included in the reaction, and the ubiquitylated species were stabilized by addition of p97-i. His-Ub-conjugated proteins isolated on Ni-NTA resin were blotted with Mcm7 antibodies. Whereas highly modified Mcm7 was recovered from extracts supplemented with wild-type rCMG, no polyubiquitylated Mcm7 species were detectable in extracts containing rCMGK27/28R (Fig. 4C, cf. lanes 3 and 5). Similar results were observed when rCMGK27/28R was immunoprecipitated with anti-FLAG resin (note that for rCMGWT, ubiquitin chains appear shorter than in the His-Ub pull-down because the latter method preferentially recovers long chains) (Fig. 4D, lanes 3,7). Furthermore, rCMGK27/28R incubated in NPE without p97-i retained its GINS and Cdc45 subunits, unlike wild-type rCMG (Fig. 4D, cf. lanes 5 and 9), but both complexes interacted with Cul2 (Fig. 4D, cf. lanes 3 and 7). Together, these data argue that Mcm7 is ubiquitylated at K27 and/or K28 in Xenopus egg extract and that ubiquitylation at these sites is important for CMG disassembly. Because this ubiquitylation was also dependent on CRL2Lrr1 and K48 linkages (Supplemental Fig. S3B,C), rCMG ubiquitylation and disassembly appear to involve the same mechanism as seen during bona fide termination.
We wanted to rule out that ubiquitylation of rCMG in extract was due to RNA or residual DNA. We therefore separately treated rCMG and NPE with benzonase, which digests both single-stranded and double-stranded DNA and RNA, and with RNase A, which digests RNA. The nuclease-treated rCMG preparation was mixed with the nuclease-treated NPE in the presence of His-Ub and p97-i, and ubiquitylated species were recovered with Ni-NTA resin and immunoblotted (Fig. 4E). Importantly, the ubiquitylation pattern after treatment with nucleases looked similar to the pattern in the untreated samples (Fig. 4E, cf. lanes 3 and 4). We verified that nucleic acids were efficiently removed from the extract by the nucleases (Supplemental Fig. S5A). Together, these data indicate that no termination-associated nucleic acid structures are critical to trigger CMG ubiquitylation.
DNA digestion induces CMG ubiquitylation before termination
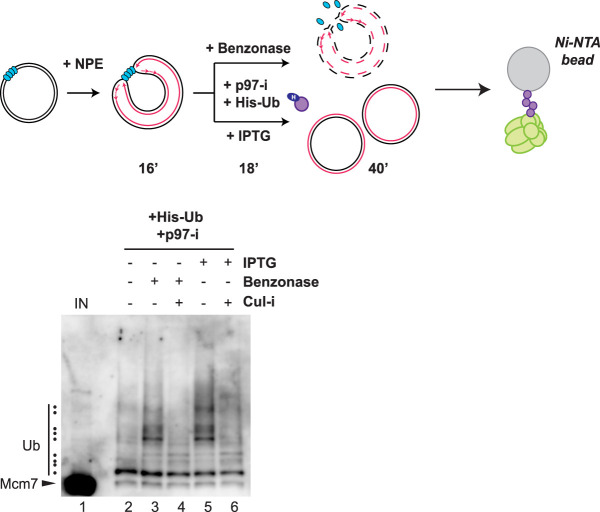
Combined with our evidence against the Okazaki fragment collision (Fig. 1A) and replisome dimerization models (Fig. 1B), the finding that rCMG incubated in extract lacking any nucleic acid undergoes ubiquitylation suggested that a DNA structure involved in replication elongation might suppress CRL2Lrr1 recruitment (Fig. 1C). To test this idea, we asked whether the elimination of DNA is sufficient to induce ubiquitylation of CMGs at replication forks that have not yet completed replication. Thus, forks were stalled at a LacR array, DNA was digested with benzonase, and the level of ubiquitylation was compared with that observed when termination was induced with IPTG (Fig. 5, top). To recover ubiquitylated Mcm7, His-Ub was included in the reaction. Benzonase, which efficiently digested the plasmid substrate in this experiment (Supplemental Fig. S5B), induced a similar extent of cullin-dependent Mcm7 ubiquitylation as IPTG addition (Fig. 5, lanes 3–6), and benzonase-induced ubiquitylation was replication-dependent (Supplemental Fig. S5C). However, given the limited amount of CMG loaded on chromatin, we were not able to address whether benzonase induces CMG disassembly. Taken together, these data suggest that the elimination of DNA from a stalled replisome is sufficient to trigger CRL2Lrr1-dependent CMG ubiquitylation.
Figure 5.
Release of CMG from DNA results in Mcm7 ubiquitylation. Replication forks were stalled at the outer edges of a LacR array via sequential incubation of a LacR array containing plasmid in HSS and NPE. Reactions were then treated with benzonase to digest the plasmid or incubated with IPTG to promote termination. His-Ub was added to the extracts at the same time as IPTG or benzonase. Proteins pulled down with Ni-NTA beads under denaturing conditions were blotted for Mcm7. Ubiquitylated species are indicated by the dots on the left of the image. Samples were also removed for DNA analysis (Supplemental Fig. S5B).
Discussion
In this report, we characterize the signal that triggers CMG unloading during replication termination. Using single-molecule imaging, we present evidence that converging CMGs pass each other, travel along dsDNA, and then undergo rapid unloading from dsDNA. The data argue against the idea that converging replisomes must dimerize to be unloaded. The replisome dimerization model is further disfavored by our recent observation that a single replisome is unloaded when it reaches a nick in the lagging strand template (Vrtis et al. 2020). Importantly, we also found that CMG is unloaded when DNA synthesis is inhibited by aphidicolin (Fig. 3B). Therefore, collision of CMG with a ssDNA–dsDNA junction cannot be an essential trigger for CMG removal. Instead, we provide two lines of evidence that CMG is ubiquitylated when it loses some or all of its interaction with DNA. First, recombinant CMG added to egg extracts undergoes ubiquitylation and disassembly in the absence of any DNA. Second, when replisomes stalled at a LacR array are treated with benzonase, CMG undergoes ubiquitylation. Collectively, our data strongly suggest that a DNA structure associated with elongating replisomes suppresses CMG ubiquitylation before termination.
How might DNA repress CMG ubiquitylation? During elongation, CMG encircles and travels along the leading strand template while excluding the lagging strand template from its central channel, a DNA unwinding mechanism called steric exclusion (Fu et al. 2011; Li and O'Donnell 2018). We speculate that before termination, the excluded strand blocks CRL2Lrr1 recruitment to CMG. This could occur in two ways. First, the excluded strand might compete with CRL2Lrr1 for binding to a specific surface on CMG (Fig. 6, pink patch). In possible agreement with this idea, in archaea, the excluded strand contacts charged residues on the outer surface of the MCM complex (Graham et al. 2011). In addition, a recent study identified a conformational state of the Drosophila CMG complex where the lagging strand template interacts with a channel between Mcm3 and Mcm5 (Eickhoff et al. 2019). Given that Mcm3 resides adjacent to Mcm7, it is not difficult to envision that ssDNA binding to Mcm3 could disrupt association of CRL2Lrr1 with CMG. On the other hand, Mcm7 appears to participate in the separation of dsDNA ahead of the fork (Baretić et al. 2020), suggesting that Mcm7 itself may be able to detect the presence of ssDNA. Alternatively, the excluded strand might dynamically sample many sites on the surface of CMG or associated replisome subunits and thereby prevent prolonged binding of CRL2Lrr1, inhibiting processive ubiquitylation. Whatever the precise mechanism, as soon as the last parental duplex between converging CMGs is unwound, the excluded strand loses its intimate contact with CMG or the replisome, relieving the repression of ligase binding (Fig. 6). This model is consistent with our observation that CMG is unloaded at a nick in the lagging strand template (Vrtis et al. 2020), which likely disengages the lagging strand from the replisome. The model also predicts that CMGs are disassembled when they slide off the end of a telomere.
Figure 6.
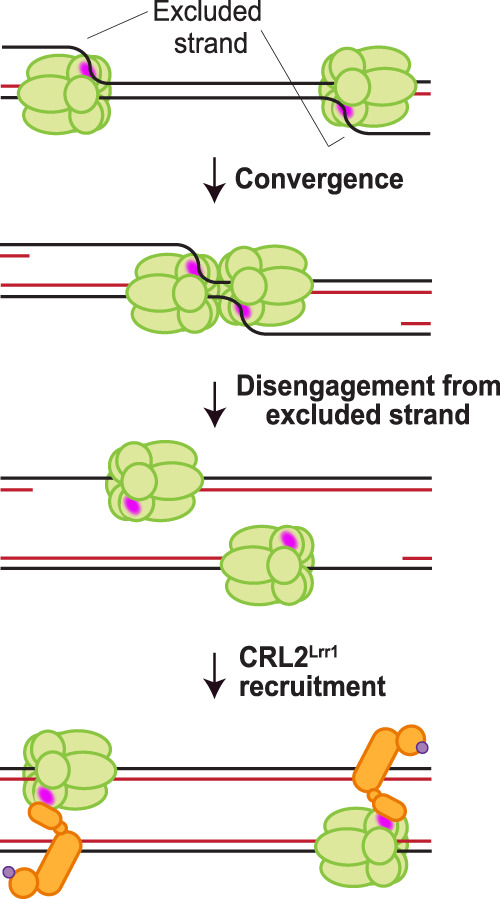
A model for the regulation of CRL2Lrr1 recruitment. During elongation, the excluded strand interacts with a surface of CMG (pink patch) or an associated replisome subunit that normally interacts with CRL2Lrr1, precluding binding of the ligase. Upon convergence of two CMGs during termination, the last parental duplex is unwound, and the excluded DNA strand disengages from the outer surface of CMG, allowing CRL2Lrr1 recruitment.
Several considerations suggest that the regulation of CMG ubiquitylation might be more complex than envisioned by this “excluded strand model.” For example, the time between CMG convergence and unloading is longer in the presence of aphidicolin than in control reactions (Fig. 3D). This observation may be due to uncoupled CMGs taking longer to meet than might be suggested by their colocalization in microscopy. This scenario could arise if uncoupled CMGs are less adept at unwinding the last stretch of parental DNA. Alternatively, uncoupled CMGs may not stably associate with one or more accessory factors that are required for CMG ubiquitylation. A potential candidate for such a factor is DNA pol ε (Pacek et al. 2006). Finally, though not essential, collision of CMG with nascent strands may in fact stimulate CMG unloading, perhaps by enhancing CMG's affinity for CRL2Lrr1. The steric exclusion model also raises the question of how ubiquitylation is avoided during initiation, since CMG is assembled at origins before expulsion of the excluded strand from its central channel (Douglas et al. 2018). Premature ubiquitylation could be prevented if initiation factors (e.g. TopBP1, Treslin/Ticrr, MTBP, or RecQL4) block CRL2Lrr1 binding until an excluded strand is present. Additionally, premature CMG ubiquitylation could be prevented by post-translational modifications that occur during initiation, such as DDK-dependent phosphorylation of MCM2–7 (Sheu and Stillman 2010). Assuming an efficient mechanism to protect preinitiated CMGs from ubiquitylation, the excluded strand model provides a robust means to ensure that CMGs are not unloaded before termination.
Formally, we cannot rule out that the repression of CMG ubiquitylation during elongation is exerted by a protein rather than a nucleic acid structure. In this view, termination, benzonase treatment, and use of rCMG all preclude binding of the relevant repressor(s). Mass spectrometry indicates that all replisome proteins that are intimately associated with CMG during elongation (Claspin, Timeless-Tipin, Traip, DNA pol ε, Ctf18, and Mcm10) are also bound to CMGs upon termination (Dewar et al. 2017; Sonneville et al. 2017). In contrast, lagging strand factors, including DNA pol δ, Fen1, and RPA, are not associated with terminated replisomes (Dewar et al. 2017, Sonneville et al. 2017), and their absence might enable CRL2Lrr1 recruitment. However, a mechanism relying on a protein repressor is potentially unreliable, as its premature dissociation during elongation would lead to CMG ubiquitylation and possible fork collapse. In contrast, the excluded strand is permanently linked to the replisome before termination and therefore unlikely to dissociate from CMG for prolonged periods. Future biochemical and structural analyses will be required to further test the excluded strand model. However, so far, we have not been able to reconstitute CMG ubiquitylation with recombinant CRL2Lrr1, likely because other cofactors are required.
Experiments in yeast suggested that SCFDia2 interacts constitutively with the elongating replisome (Morohashi et al. 2009). Dia2 contains a TPR motif that interacts with Mrc1 and Ctf4 (Morohashi et al. 2009), which is not present in Lrr1, implying a different regulatory mechanism in which prebound E3 ligase is activated upon fork convergence. However, in these experiments, the binding of SCFDia2 to CMG was invariably examined after digestion of chromatin with DNase. Given our results, it will be important to monitor the CMG–SCFDia2 interaction in the presence and absence of nuclease treatment.
Our data suggest that after converging CMGs pass each other, they subsequently translocate along dsDNA. This interpretation is based on prior biochemical experiments (Langston and O'Donnell 2017) and our observation that when CMG unloading is blocked, diverging CMGs translocate without promoting further DNA synthesis (Supplemental Fig. S1C), as expected for translocation along dsDNA. The fact that CMG's translocation rate is undiminished after bypass raises the interesting possibility that when CMG translocates along dsDNA, it still interacts primarily with one strand as seen during ssDNA translocation. We speculate that processive movement along dsDNA causes CMG to vacate the ssDNA–dsDNA junction for rapid Okazaki fragment processing. Interestingly, purified yeast CMG can undergo random diffusion in vitro on naked dsDNA (Wasserman et al. 2019), raising the possibility that an extract-derived factor imposes directional CMG movement on duplex DNA. Whether this still occurs on DNA that is not stretched and therefore contains a full complement of nucleosomes remains to be determined. Interestingly, although the E3 ubiquitin ligase TRAIP travels with CMG, TRAIP does not promote CMG unloading during interphase termination (Wu et al. 2019). In contrast, TRAIP acts on CMGs stalled on either side of an interstrand cross-link (ICL) (Wu et al. 2019). Together with the fact that TRAIP supports replication-coupled ubiquitylation of DNA protein cross-links (Larsen et al. 2019), it appears that TRAIP only ubiquitylates proteins that stably abut the replisome, and CMG ubiquitylation at ICLs likely occurs in trans. Thus, we propose that TRAIP does not function in CMG ubiquitylation during interphase termination because converging CMGs rapidly pass each other.
It was speculated previously that during replication stress, CMG may reversibly transition from encircling ssDNA to encircling dsDNA, in a process aided by Mcm10 (Wasserman et al. 2019). This would enable CMG to bypass DNA damage, or to remain on DNA in an inactive state during replication fork reversal. Upon fork restoration, CMGs would then revert to encircling ssDNA and resume their canonical DNA unwinding activity in DNA replication. Contrary to this proposal, our data show that CMG molecules encircling dsDNA are rapidly unloaded from chromatin. Therefore, the data presented here disfavor the idea of “mothballing” of CMG on dsDNA while replication stress is resolved.
In summary, our results provide strong evidence that converging CMGs pass each other and then translocate along dsDNA. However, we also show that specific DNA structures, including dsDNA, are not required to trigger CMG ubiquitylation, suggesting that the primary regulation of CMG unloading involves its repression during elongation.
Materials and methods
Cell lines
Sf9 cells and Tni cells used for recombinant protein expression were obtained from Expression Systems and cultured in ESF-921 medium according to the supplier's instructions.
Protein expression and purification
The plasmid encoding CMG containing N-terminally FLAG-tagged Mcm3 (pGC187) was described previously (Wu et al. 2019). Mcm7 with K27/28R mutations was made by round-the-horn mutagenesis of Mcm7, followed by assembly into the complete CMG plasmid using the MultiBac system (Trowitzsch et al. 2010), containing N-terminally FLAG-tagged Mcm3. The bacmid encoding the full CMG complex was obtained by electroporation of the CMG plasmid into electrocompetent DH10EMBacY cells (Geneva Biotech) and isolated with the ZR BAC DNA minipreparation kit (Zymo Research). Sf9 insect cells (1 × 106) were infected with 3 μg of bacmid DNA using the FuGENE transfection protocol (Promega Corporation), and the baculovirus was amplified three times using Sf9 cells at a density of ∼2 × 106/mL. Cultures (250 mL) of Tni cells (∼2 × 106/mL to 3 × 106/mL) were infected with 40 mL of amplified baculovirus for 72 h at 27°C, and then harvested by spinning down at 500g, and snap-frozen in liquid nitrogen and stored at −80°C. The cell pellets were thawed in 40 mL of lysis buffer (20 mM Tris-HCl at pH 7.5, 5 mM magnesium acetate, 300 mM potassium acetate, 0.1% Tween-20, 10% glycerol, 1 mM PMSF [Roche] containing one tablet of EDTA-free Complete protease inhibitor cocktail [Roche]) by end-over-end rotation, and then sonicated at 40% amplitude for 30 sec (1 sec on, 6 sec off) on ice. The sonicated lysates were spun in a Beckman Optima L90-K ultracentrifuge in a Ti45 rotor at 25,000 rpm for 1 h at 4°C. The cleared lysates were collected and incubated with 450 μL of FLAG resin (Sigma) for 1.5 h at 4°C with end-over-end rotation. The beads were pelleted at 1000g for 2 min, and the supernatant was collected. The beads were placed in a 5-mL polypropylene column (Qiagen) and washed five times with 4 mL of lysis buffer and EDTA-free Complete protease inhibitor cocktail. To elute, the beads were incubated with 450 μL of lysis buffer containing 0.2 mg/mL 3xFLAG peptide (Sigma) for 1 h on ice with occasional resuspension by flicking. Furthermore, the supernatant from the first FLAG-binding reaction was incubated for a second round of binding with an additional 250 μL of FLAG resin for 1 h, and then washed as above and eluted with 250 μL of lysis buffer containing 0.2 mg/mL 3× FLAG peptide. The collected elutions were pooled, diluted threefold in start buffer (25 mM HEPES at pH 7.6, 0.02% Tween-20, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 0.4 mM PMSF, 1 mM DTT), and loaded onto a MonoS 5/50 GL column (GE) on the AKTA pure 25 M1 system (GE) (final salt concentration was 100 mM potassium chloride). The flowthrough from the MonoS column was loaded onto a MonoQ 5/50 GL column (GE). The CMG complexes bound to the MonoQ column were eluted over a gradient of 100–600 mM KCl. Fractions corresponding to the CMG peak were collected and concentrated by binding to 20 μL of NuviaQ anion exchange resin (Bio-Rad) for 1 h at 4°C and then eluting with two 10-min incubations with 20 μL of 600 mM KCl elution buffer (start buffer containing 600 mM KCl) followed by two 10-min incubations with 20 μL of 1 M KCl elution buffer (start buffer containing 1 M KCl). The eluates were flash-frozen in 3-μL aliquots in liquid nitrogen, and then stored at −80°C.
To generate the plasmid encoding for CRL2Lrr1, the open reading frames encoding Cul2, Lrr1, Elongin b, Elongin c, and Rbx1 were amplified from Xenopus cDNA, and a 3xFLAG tag was inserted at the 3′ end of the Lrr1 sequence. The amplified products were assembled into a single plasmid using the MultiBac system. The bacmid encoding the full complex was obtained by electroporation of the CRL2Lrr1 plasmid into electrocompetent DH10EMBacY cells (Geneva Biotech) and isolated with the ZR BAC DNA minipreparation kit (Zymo Research). Sf9 insect cells (1 × 106) were infected with 3 μg of bacmid DNA using the FuGENE transfection protocol (Promega Corporation), and the baculovirus was amplified three times using Sf9 cells that were at ∼2 × 106/mL. For expression of CRL2Lrr1, 500 mL of cultures of Sf9 cells (∼2 × 106/mL to 3 × 106/mL) were infected with 10 mL of amplified baculovirus for 72 h and then spun down, and the pellet was snap-frozen in liquid nitrogen and stored at −80°C. Cells were thawed in 45 mL of lysis buffer (50 mM Tris at pH 8.0, 300 mM NaCl, 10% glycerol, 0.1% NP-40) containing one tablet of EDTA-free Complete protease inhibitor (Roche). The resuspended cells were freeze–thawed once in liquid nitrogen, and then passed twice through a 21-gauge needle (BD Biosciences). The sample was sonicated at 40% amplitude for 30 sec (1 sec on, 6 sec off) on ice, and then spun down at 25,000 rpm in a Ti45 rotor in a Beckman Optima L-90K ultracentrifuge for 1 h at 4°C. The cleared lysate was incubated with 180 μL of anti-FLAG resin (Sigma), and then the resin was loaded onto a 5-mL polypropylene column (Qiagen) and washed five times with 10 resin volumes of lysis buffer. To elute the complex, the beads were incubated with one resin volume of lysis buffer + 0.2 mg/mL FLAG peptide, and this procedure was repeated four times. The elutions containing the complex were pooled and dialyzed twice against 1 L of dialysis buffer (20 mM HEPES at pH 7.5, 150 mM NaCl, 1 mM DTT, 10% glycerol, 0.05% NP-40, 0.5 μg/μL aprotinin/leupeptin) and then flash-frozen in liquid nitrogen and stored at −80°C. Lac repressor protein (LacR) was purified as described (Duxin et al. 2014).
SDS-PAGE analysis and immunoblotting
Samples were prepared with 2× SDS sample buffer for a final composition of 50 mM Tris (pH 6.8), 2% SDS, 0.1% Bromophenol blue, 10% glycerol, and 5% β-mercaptoethanol. All proteins were separated on precast 4%–15% polyacrylamide Mini-Protean gels (Bio-Rad) except for Mcm7, which was separated on homemade 6% polyacrylamide gels. Gels were run on the Bio-Rad Mini-Protean system at 200 V for 33 min with precast gels and 45 min with homemade gels. Protein gels were visualized with Instant Blue stain (Expedeon) or SYPRO Ruby stain (Invitrogen) according to the manufacturers’ protocols.
For immunoblots, gels were run as above and transferred to PVDF membranes (Perkin Elmer) at 300 mA for 75 min. Membranes were blocked with 5% dry milk made up in 1× PBST, and then incubated overnight at 4°C in primary antibody at a 1:2500–1:20,000 dilution in 1× PBST. All antibodies used for immunoblotting were described previously (Dewar et al. 2017). Membranes were rinsed with 1× PBST, and then incubated for 1 h at room temperature in secondary goat antirabbit HRP antibody (Jackson ImmunoResearch) at 1:30,000 dilution, made up in 5% dry milk in 1× PBST. Membranes were washed with 1× PBST, and then developed with either HyGLO chemiluminescent antibody detection reagent (Denville) or SuperSignal West Femto maximum sensitivity substrate (Thermo Fisher), and imaged using the chemiluminescence function on the Amersham Imager 600 (GE).
Immunoprecipitation of FLAG-CMG
FLAG-tagged CMG (5 nM final concentration) was incubated in 31 μL of nucleoplasmic extract (NPE) or IP buffer (1× ELB salts [2.5 mM MgCl2, 50 mM KCl, 10 mM HEPES at pH 7.7], 0.1% NP-40, 0.1 mg/mL BSA) in the presence of 200 μM p97-i (Sigma NMS-873) for 30 min at room temperature. Fifteen microliters of each reaction was added to 5 μL of anti-FLAG resin (Sigma) and incubated with end-over-end rotation for 1.5 h at 4°C. The beads were washed five times with 80 vol of IP buffer. Samples were eluted from anti-FLAG resin with 3 vol of IP buffer + 0.2 mg/mL 3xFLAG peptide prior to mixing with 1 vol of 2× SDS sample buffer. Samples were resolved by SDS-PAGE and immunoblotted as described above. All immunoprecipitation experiments were performed at least twice, with the exception of Figure 4B, which was performed once.
Egg extracts and replication reactions
High-speed supernatant (HSS) of egg lysate and nucleoplasmic extract (NPE) were prepared as described (Sparks and Walter 2019). Replication reactions were carried out as described (Lebofsky et al. 2009). Briefly, plasmids were incubated at a final concentration of 7 ng/µL for 30 min in HSS to carry out replication licensing. Subsequently, NPE containing an ATP regeneration system and [α-32P]dATP was added to initiate DNA replication. Replication of LacR array plasmids and induction of termination with IPTG were performed as described (Dewar et al. 2015). Inhibitors were added to replication reactions at the following final concentrations: p97-i (Sigma NMS-873) at 200 μM, and Cul-i (Active Biochem MLN4924) at 200 μM. At the specified time points, replication reactions were quenched with replication stop buffer (8 mM EGTA, 0.13% phosphoric acid, 10% Ficoll, 5% SDS, 0.2% Bromophenol blue, 80 mM Tris at pH 8.0) and treated with 1 μg/μL proteinase K. Samples were separated via agarose gel electrophoresis, and the resulting gels were exposed to phosphorscreens and imaged on the Typhoon FLA 7000 PhosphorImager (GE Healthcare). Band and lane intensities were measured using MultiGauge software (Fujifilm).
Immunodepletions
Lrr1 immunodepletions were performed as described previously (Dewar et al. 2017). For reactions containing recombinant CRL2Lrr1, the protein complex was added to a final concentration of ∼100 nM.
Single-molecule experiments and analysis
Protocols for immunodepletion of GINS, flow cell assembly, DNA tethering, DNA replication, imaging, and image analysis were performed as described previously (Sparks et al. 2019), with one exception: To maximize the number of termination events, the DNA was licensed for 8–10 min (instead of 2–5 min), and replication was initiated in the presence of fluorescently labeled GINS for 5–8 min. A fresh preparation of GINS-AF647 was performed following the protocol described previously (Sparks et al. 2019). Briefly, GINS was fluorescently labeled site-specifically at the C terminus of Psf3 via sortase-mediated conjugation of a small peptide, GGGGKYCAF647K, where the cysteine residue was labeled with Alexa fluor 647 maleimide. This protocol ensured that each GINS molecule bears either one or zero fluorescent dyes. The labeling efficiency was increased by removing unlabeled GINS from the final preparation via Ni-NTA affinity chromatography. UV-Vis spectrometer measurements indicated that 80%–90% of the GINS complex was conjugated to AF647. In all experiments where Fen1mKikGR was added to the single-molecule reaction, a catalytically dead variant of the protein was used: Fen1D179A. Imaging and data analysis were performed as described previously (Sparks et al. 2019), with the following modifications: (1) Bona fide termination events were manually selected on kymograms where the converging CMGs could be clearly resolved for at least three time points before they merged into a diffraction-limited spot (we used the Rayleigh resolution limit definition of 0.61 × λ/NA where NA = 1.4 and λ = 650–700 nm; i.e., the centroids of the two CMGs must be at least 300 nm apart to be resolved, corresponding to approximately two pixels at our magnification of 160 nm/pixel). (2) An automated MATLAB script was used to measure the speed of CMG convergence and estimate when the two helicases converged (as illustrated in Fig. 2B). Briefly, the position of each CMG molecule was determined to subpixel precision by fitting the diffraction-limited image of the molecule to a Gaussian approximation of the point spread function. The speed of each CMG was determined by fitting the trajectory of the molecule over two or more consecutive frames to a straight line. The moment of convergence or divergence for two helicases was determined by computing the intersection of the two linear fits for converging CMG trajectories (Fig. 2B,C). (3) An automated MATLAB script was used to determine the time interval between fork convergence and the disappearance of the CMG fluorescent signal. The sudden drop in integrated fluorescent signal was used to determine the moment when a CMG molecule was unloaded (Fig. 2B). The CMG signal was integrated within the region of interest, and the raw signal was fitted to a stepwise drop using a model-independent step-fitting algorithm (Kalafut and Visscher 2008), and the results were verified via manual inspection. The default MATLAB implementation of bootstrapping was used to estimate the 95% confidence intervals (CIs) for the mean or median values of CMG speed, the CMG lifetime after fork convergence, and the probability of CMG divergence. In experiments containing inhibitors, DNA was licensed for 8–10 min, and then replication was initiated in the absence of the drug for 5–8 min, after which the inhibitor was added to the reaction until the end of the experiment. Aphidicolin was used at a final concentration of 50 ng/μL, p97-i (NMS-873) was used at a final concentration of 200 μM, and Cul-i (MLN4924) was used at a final concentration of 200 μM. Mean, median, and 95% confidence interval values for all single-molecule experiments are shown in Supplemental Table S1.
Benzonase treatment of replication forks
Plasmids containing a 1.5-kb 48xlacO array were preincubated with LacR protein for 40 min at room temperature. LacR-bound plasmid (1.5 ng/μL) was replicated as above. Where indicated, ∼0.04 vol of [α-32P]dATP was added to label the nascent strands. After 18 min, when replication forks had stalled at the edges of the array, 15 mM IPTG was added to the reactions to release the array, or 0.5 U/μL benzonase was added to digest the plasmid DNA. At the same time, the reactions were supplemented with 200 μM NMS-873 (p97-i) and 25 μM His6-tagged ubiquitin (Boston Biochem). Twenty-two minutes after release of the array (40 min after initiation of replication), radioactive samples were collected by addition into replication stop buffer (8 mM EGTA, 0.13% phosphoric acid, 10% Ficoll, 5% SDS, 0.2% Bromophenol blue, 80 mM Tris-HCl at pH 8.0), and 1 μg/μL proteinase K was added for digestion of proteins for a minimum of 1 h at 37°C. Samples were separated on native 1% agarose gels and exposed to phosphorscreens before imaging on the Typhoon FLA 7000 PhosphorImager (GE Healthcare). Nonradioactive reactions were processed according to the protocol for His-ubiquitin pull-downs (see below). For Supplemental Figure 5C, replication licensing was inhibited by preincubating HSS with geminin for 10 min at room temperature. Next, replication of a plasmid containing 32xlacO array prebound with LacR protein was initiated as described above. DNA samples were processed as above and protein samples were processed as described below for His-ubiquitin pull-downs.
RNase A and benzonase treatment of recombinant CMG
A 5.4-kb plasmid (final concentration of 15 ng/μL) was added to NPE and rCMG prepared separately, in order to gauge efficiency of DNA digestion. Benzonase (Millipore) and RNase A (Sigma) were added to final concentrations of 2 U/μL and 2.5 μg/μL, respectively, and the reactions were incubated for 20 min at room temperature. Treated NPE (0.8 vol) was mixed with 0.2 vol of treated rCMG (final concentration of 10 nM) and incubated for an additional 40 min at room temperature. Samples were then split in half for processing of DNA or processing of protein. RNase A and benzonase experiments were performed at least twice.
For Supplemental Figure S5A, 1.5 μL of DNA sample was mixed with 10 μL of replication stop buffer (8 mM EDTA, 0.13% phosphoric acid, 10% Ficoll, 5% SDS, 80 mM Tris-HCl at pH 8.0) and treated with 1 μg/μL proteinase K for at least 1.5 h at 37°C to digest the proteins. The DNA was separated on a native 1% agarose gel for 2 h at 100 V. The gel was stained with 10 μL of SYBR Gold stain (Life Technologies) in 150 mL of 1× TBE for 1 h, and then destained twice with 1× TBE for 10 min before being imaged with the UV setting on an Amersham Imager 600 (GE).
His6-ubiquitin (His-Ub) pull-downs
Five microliters of NPE containing 25 μM His-tagged ubiquitin (Boston Biochem) was incubated with 10 μL of Ni-NTA resin in 200 μL of denaturing IP solution (100 mM NaH2PO4, 10 mM Tris-HCl at pH 8.0, 8 M urea, 10 mM imidazole, 0.2% NP-40) for 1 h at room temperature. The resin was washed three times with 50 vol of denaturing IP solution, and then boiled in 15 μL of 2× SDS sample buffer + β-mercaptoethanol. Fifteen microliters of 0.5 M imidazole was added to the bead mixture, and then shaken at 1500 rpm for 1 h to elute the His-tagged proteins, which were analyzed by SDS-PAGE and immunoblotting. All His6-ubiquitin pull-down experiments were performed at least twice.
Code and data availability
The custom-written MATLAB analysis code and raw data are available on request.
Competing interest statement
J.C.W. is a cofounder of MoMa therapeutics, in which he has a financial interest.
Supplementary Material
Acknowledgments
We thank Alan Brown, James Dewar, Ben Stinson, Kyle Vrtis, Alex Wu, and Maksym Shyian for comments on the manuscript. E.L. was supported by National Institutes of Health (NIH) grant F31 GM122277. G.C. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund. J.C.W. is supported by NIH grant GM80676. J.C.W. is a Howard Hughes Medical Institute Investigator and an American Cancer Society Research Professor.
Author contributions: E.L. and G.C. designed and executed protein expression and purification schemes. E.L. performed coimmunoprecipitations and His-ubiquitin pull-downs. G.C. performed single-molecule experiments and analyzed the resulting data. M.S.Z. performed the benzonase and RNase A experiments in Figure 4E and Supplemental Figure S5. O.V.K. performed the replication assay shown in Supplemental Figure S1, D–F. J.C.W. supervised all research. J.C.W., E.L., and G.C. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.339739.120.
References
- Baretić D, Jenkyn-Bedford M, Aria V, Cannone G, Skehel M, Yeeles JTP. 2020. Cryo-EM structure of the fork protection complex bound to CMG at a replication fork. Mol Cell 78: 926–940.e13. 10.1016/j.molcel.2020.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake D, Luke B, Kanellis P, Jorgensen P, Goh T, Penfold S, Breitkreutz BJ, Durocher D, Peter M, Tyers M. 2006. The F-box protein Dia2 overcomes replication impedance to promote genome stability in Saccharomyces cerevisiae. Genetics 174: 1709–1727. 10.1534/genetics.106.057836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wu RA, Sonneville R, Kochenova OV, Labib K, Pellman D, Walter JC. 2019. Mitotic CDK promotes replisome disassembly, fork breakage, and complex DNA rearrangements. Mol Cell 73: 915–929.e6. 10.1016/j.molcel.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Walter JC. 2017. Mechanisms of DNA replication termination. Nat Rev Mol Cell Biol 18: 507–516. 10.1038/nrm.2017.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Budzowska M, Walter JC. 2015. The mechanism of DNA replication termination in vertebrates. Nature 525: 345–350. 10.1038/nature14887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar JM, Low E, Mann M, Räschle M, Walter JC. 2017. CRL2Lrr1 promotes unloading of the vertebrate replisome from chromatin during replication termination. Genes Dev 31: 275–290. 10.1101/gad.291799.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Ali FA, Costa A, Diffley JFX. 2018. The mechanism of eukaryotic CMG helicase activation. Nature 555: 265–268. 10.1038/nature25787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dewar JM, Yardimci H, Walter JC. 2014. Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159: 346–357. 10.1016/j.cell.2014.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff P, Kose HB, Martino F, Petojevic T, Abid Ali F, Locke J, Tamberg N, Nans A, Berger JM, Botchan MR, et al. 2019. Molecular basis for ATP-hydrolysis-driven DNA translocation by the CMG helicase of the eukaryotic replisome. Cell Rep 28: 2673–2688.e8. 10.1016/j.celrep.2019.07.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. 2011. Global identification of modular cullin-RING ligase substrates. Cell 147: 459–474. 10.1016/j.cell.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Schärer OD, Walter JC. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146: 931–941. 10.1016/j.cell.2011.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham BW, Schauer GD, Leuba SH, Trakselis MA. 2011. Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res 39: 6585–6595. 10.1093/nar/gkr345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafut B, Visscher K. 2008. An objective, model-independent method for detection of non-uniform steps in noisy signals. Comput Phys Commun 179: 716–723. 10.1016/j.cpc.2008.06.008 [DOI] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. 2011. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44: 325–340. 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langston L, O'Donnell M. 2017. Action of CMG with strand-specific DNA blocks supports an internal unwinding mode for the eukaryotic replicative helicase. Elife 6: e23449 10.7554/eLife.23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Gao AO, Sparks JL, Gallina I, Wu RA, Mann M, Räschle M, Walter JC, Duxin JP. 2019. Replication-coupled DNA–protein crosslink repair by SPRTN and the proteasome in Xenopus egg extracts. Mol Cell 73: 574–588.e7. 10.1016/j.molcel.2018.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Takahashi T, Walter JC. 2009. DNA replication in nucleus-free Xenopus egg extracts. Methods Mol Biol 521: 229–252. 10.1007/978-1-60327-815-7_13 [DOI] [PubMed] [Google Scholar]
- Li H, O'Donnell ME. 2018. The eukaryotic CMG helicase at the replication fork: emerging architecture reveals an unexpected mechanism. Bioessays 40: 1700208 10.1002/bies.201700208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Habuchi S, Walter JC, van Oijen AM. 2012. A general approach to break the concentration barrier in single-molecule imaging. Nat Methods 9: 987–992. 10.1038/nmeth.2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric M, Maculins T, De Piccoli G, Labib K. 2014. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science 346: 1253596 10.1126/science.1253596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric M, Mukherjee P, Tatham MH, Hay R, Labib K. 2017. Ufd1–Npl4 recruit Cdc48 for disassembly of ubiquitylated CMG helicase at the End of chromosome replication. Cell Rep 18: 3033–3042. 10.1016/j.celrep.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SP, Bailey R, Campion N, Herron S, Gambus A. 2014. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science 346: 477–481. 10.1126/science.1253585 [DOI] [PubMed] [Google Scholar]
- Moreno SP, Jones RM, Poovathumkadavil D, Scaramuzza S, Gambus A. 2019. Mitotic replisome disassembly depends on TRAIP ubiquitin ligase activity. Life Science Alliance 2: e201900390 10.26508/lsa.201900390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi H, Maculins T, Labib K. 2009. The amino-terminal TPR domain of Dia2 tethers SCFDia2 to the replisome progression complex. Curr Biol 19: 1943–1949. 10.1016/j.cub.2009.09.062 [DOI] [PubMed] [Google Scholar]
- Mukherjee PP, Labib KPM. 2019. In vitro reconstitution defines the minimal requirements for Cdc48-dependent disassembly of the CMG helicase in budding yeast. Cell Rep 28: 2777–2783.e4. 10.1016/j.celrep.2019.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutch SA, Fujimoto BS, Kuyper CL, Kuo JS, Bajjalieh SM, Chiu DT. 2007. Deconvolving single-molecule intensity distributions for quantitative microscopy measurements. Biophys J 92: 2926–2943. 10.1529/biophysj.106.101428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. 2006. Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587. 10.1016/j.molcel.2006.01.030 [DOI] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. 2010. The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117. 10.1038/nature08647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Priego-Moreno S, Knebel A, Johnson C, Hastie J, Gartner A, Gambus A, Labib K. 2017. CUL-2LRR-1 and UBXN-3/FAF1 drive replisome disassembly during DNA replication termination and mitosis. Nat Cell Biol 19: 468–479. 10.1038/ncb3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks J, Walter JC. 2019. Extracts for analysis of DNA replication in a nucleus-free system. Cold Spring Harb Protoc 2019: pdb.prot097154 10.1101/pdb.prot097154 [DOI] [PubMed] [Google Scholar]
- Sparks JL, Chistol G, Gao AO, Räschle M, Larsen NB, Mann M, Duxin JP, Walter JC. 2019. The CMG helicase bypasses DNA–protein cross-links to facilitate their repair. Cell 176: 167–181.e21. 10.1016/j.cell.2018.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowitzsch S, Bieniossek C, Nie Y, Garzoni F, Berger I. 2010. New baculovirus expression tools for recombinant protein complex production. J Struct Biol 172: 45–54. 10.1016/j.jsb.2010.02.010 [DOI] [PubMed] [Google Scholar]
- Vrtis K, Dewar JM, Chistol G, Wu RA, Graham TG, Walter JC. 2020. Single-strand DNA breaks cause replisome disassembly. bioRxiv 10.1101/2020.08.17.254235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman MR, Schauer GD, O'Donnell ME, Liu S. 2019. Replication fork activation is enabled by a single-stranded DNA gate in CMG helicase. Cell 178: 600–611.e16. 10.1016/j.cell.2019.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Semlow DR, Kamimae-Lanning AN, Kochenova OV, Chistol G, Hodskinson MR, Amunugama R, Sparks JL, Wang M, Deng L, et al. 2019. TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 567: 267–272. 10.1038/s41586-019-1002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The custom-written MATLAB analysis code and raw data are available on request.