In this study, Ramamoorthy et al. investigate EBF1 and PAX5 combined haploinsufficiency in the development of a B-ALL phenotype in mice. Using transcriptional and metabolomic profiling, the authors report that EBF1 and Pax5 may safeguard early stage B cells from transformation to B-ALL by limiting IL-7 signaling, folate metabolism, and Myc expression.
Keywords: EBF1, Pax5, B-ALL, IL-7 signaling, cMyc, folate pathway
Abstract
EBF1 and PAX5 mutations are associated with the development of B progenitor acute lymphoblastic leukemia (B-ALL) in humans. To understand the molecular networks driving leukemia in the Ebf1+/−Pax5+/− (dHet) mouse model for B-ALL, we interrogated the transcriptional profiles and chromatin status of leukemic cells, preleukemic dHet pro-B, and wild-type pro-B cells with the corresponding EBF1 and Pax5 cistromes. In dHet B-ALL cells, many EBF1 and Pax5 target genes encoding pre-BCR signaling components and transcription factors were down-regulated, whereas Myc and genes downstream from IL-7 signaling or associated with the folate pathway were up-regulated. We show that blockade of IL-7 signaling in vivo and methotrexate treatment of leukemic cells in vitro attenuate the expansion of leukemic cells. Single-cell RNA-sequencing revealed heterogeneity of leukemic cells and identified a subset of wild-type pro-B cells with reduced Ebf1 and enhanced Myc expression that show hallmarks of dHet B-ALL cells. Thus, EBF1 and Pax5 may safeguard early stage B cells from transformation to B-ALL by limiting IL-7 signaling, folate metabolism and Myc expression.
Differentiation of hematopoietic stem cells via increasingly lineage-restricted progenitors generates highly specialized effector cells that carry out specific functions or synthesize specific products. Differentiation toward the B-cell fate involves a progressive loss of lineage potential and a stepwise acquisition of the B-cell phenotype (Nutt and Kee 2007). Rearrangement of the immunoglobulin (Ig) heavy chain (H) gene in pro-B cells leads to the expression of the pre-B-cell receptor (pre-BCR) and subsequent rearrangement of immunoglobulin light chain (IgL) genes in pre-B cells that eventually generates a functional B-cell receptor as the hallmark of immature and mature B cells (Hardy et al. 2007). Pre-BCR signaling allows the expansion and differentiation of precursor cells, whereas signaling via the interleukin 7 receptor (IL7R) mediates expansion of pro-B cells and early pre-B (large pre-B) cells prior to the rearrangement of IgL genes and transition to the more quiescent, small pre-B-cell stage (Hayashi et al. 1990; Herzog et al. 2009; Clark et al. 2014).
The establishment of B lymphoid lineage potential is governed by a complex regulatory network of transcription factors that include Ikaros (Ikzf1), E2A (Tcf3), EBF1, Pax5, and FoxO1 (for review, see Singh et al. 2007; Boller and Grosschedl 2014; Hu et al. 2017; Sigvardsson 2018). In this network, the transcription factors cross-regulate each other's expression and reinforce the acquisition of lineage identity (Roessler et al. 2007; Lin et al. 2010; Mansson et al. 2012). However, these transcription factors fulfill nonredundant functions in early B lymphopoiesis. Ikaros is required at the stage of lymphoid-primed multipotent progenitors and subsequently at the stage of large pre-B cells, in which Ikaros limits their adherence to stroma and proliferation (Thompson et al. 2007; Ng et al. 2009; Joshi et al. 2014; Arenzana et al. 2015; Hu et al. 2017). Early B-cell factor 1 is a key determinant of B-cell specification and can bind and activate B lineage genes in the context of naïve progenitor chromatin (Maier et al. 2004; Zandi et al. 2008; Treiber et al. 2010; Boller et al. 2016; Li et al. 2018). EBF1 is also involved in the repression of lineage-inappropriate genes and cooperates with Pax5 to safeguard pro-B cells from adopting an alternative cell fate (Nutt et al. 1999; Cobaleda et al. 2007; Pongubala et al. 2008; Nechanitzky et al. 2013). Pax5, a key determinant of B lineage commitment, regulates gene sets that overlap with and are distinct from those targeted by EBF1 (Revilla et al. 2012; Vilagos et al. 2012).
In humans, genetic lesions of IKFZ1, EBF1, or PAX5 alleles are often associated with B-cell acute lymphoblastic leukemia (B-ALL), suggesting that the dosage of these transcription factors are important for preventing malignancy (Mullighan et al. 2007, 2008; Shah et al. 2013; Roberts and Mullighan 2019). A dose dependency of EBF1 function was further shown in mice in which Ebf1 heterozygosity results in a diminished B lineage potential that is enhanced by combined heterozygosity with Tcf3 or Runx2 (Lin and Grosschedl 1995; O'Riordan and Grosschedl 1999; Lukin et al. 2010; Åhsberg et al. 2013). Moreover, a combined heterozygosity of Ebf1 and Pax5 results in a B-ALL-like phenotype that includes cellular expansion, increased DNA damage and enhanced lineage infidelity (Prasad et al. 2015; Ungerbäck et al. 2015; Somasundaram et al. 2016). In addition, other B-cell-related transcription factors, such as Irf4 and Irf8, suppress pre-B-cell acute lymphoblastic leukemia in mice by cooperating with PU.1 (Pang et al. 2016). Recently, PAX5 and IKZF1 were shown to prevent pre-B-cell leukemia by limiting excess glucose metabolism (Chan and Müschen 2017). Although these studies indicated that altered expression of lineage-specific transcription factors results in cell transformation during B lymphopoiesis, the insight into the underlying molecular mechanisms remains limited.
Here, we report that EBF1 and Pax5 collaborate in a dose-dependent manner to regulate the IL-7-STAT5 signaling pathway and one-carbon metabolism, whereby we found both diminished and enhanced binding of EBF1 and Pax5 to target genes in compound heterozygous mutant mice. Moreover, single-cell RNA sequencing analysis identified a small subset of wild-type pro-B cells on the trajectory to pre-B cells that share gene expression signatures with leukemic Ebf1+/−Pax5+/− pro-B cells. Thus, a normal expression level of EBF1 and Pax5 is required for safeguarding a potentially vulnerable pro-B-cell subset from a transformation to B-ALL.
Results
Malignant expansion of CD19+AA4.1hi cells in Ebf1+/− Pax5+/− mice
EBF1 and PAX5 genes are frequently deleted or mutated in human B-progenitor acute lymphoblastic leukemia (B-ALL) and B-cell lymphoma (Mullighan et al. 2007; Shah et al. 2013; Okosun et al. 2014; Chan and Müschen 2017). Although heterozygous null mutations of Ebf1 or Pax5 in the mouse do not cause any obvious malignancy, the combined loss of single alleles of Ebf1 and Pax5 results in the development of a B-ALL-like malignancy (Prasad et al. 2015). To gain insight into the mechanism of this B-cell malignancy, we generated Ebf1+/−Pax5+/− mice and analyzed leukemic (dHet B-ALL) and preleukemic (dHet pro-B) relative to wild-type (wt) pro-B cells in terms of cell proliferation, metabolism, gene expression, and transcription factor binding. Consistent with previous studies (Prasad et al. 2015; Ungerbäck et al. 2015), flow cytometric analysis of Ebf1+/−Pax5+/− mice at 30–45 wk of age showed an accumulation of AA4.1+CD19+ B cells in primary and secondary lymphoid organs (Supplemental Fig. S1A,B, bottom panels). In most 20- to 35-wk-old Ebf1+/−Pax5+/− mice, we did not detect AA4.1+CD19+ B cells in the spleen (Supplemental Fig. S1A, middle panels). In the bone marrow, however, we detected reduced frequencies of pre-B and immature B cells and increased frequencies of pro-B cells relative to wild-type mice, suggesting a developmental block and/or expansion of cells representing the pro-B-cell stage (Supplemental Fig. S1B, top and middle panels). Analysis of surface markers and the rearrangement status of immunoglobulin heavy chain genes indicated that the accumulated cells represent late stage pro-B/early stage pre-B cells with rearrangements of proximal immunoglobulin (Ig) heavy chain variable (VH) gene segments (Supplemental Fig. S1B,C). Moreover, the small numbers of Ig gene rearrangements in AA4.1+CD19+ B cells from each analyzed mouse suggested that the accumulated cells are of oligo-clonal origin. We also examined the survival and proliferation status of splenic and bone marrow-resident AA4.1+CD19+ cells by flow cytometric analysis of 7-AAD and EdU incorporation, respectively. This analysis indicated that 34% of AA4.1+CD19+ cells in the bone marrow and spleen are proliferating and show a low frequency of apoptotic cells relative to splenic B cells from wild-type mice (Supplemental Fig. S1D,E). Moreover, immunoblot analysis indicated that dHet AA4.1+CD19+ cells expressed abundant levels of both cyclin D1 and cyclin D3 (Supplemental Fig. S1F). Finally, we confirmed the malignancy of the accumulated AA4.1+CD19+ cells by their rapid expansion in adoptive transfer experiments into Rag2−/− mice (Supplemental Fig. S1G,H).
IL7-signaling drives malignant expansion of CD19+AA4.1hi cells in Ebf1+/−Pax5+/− mice
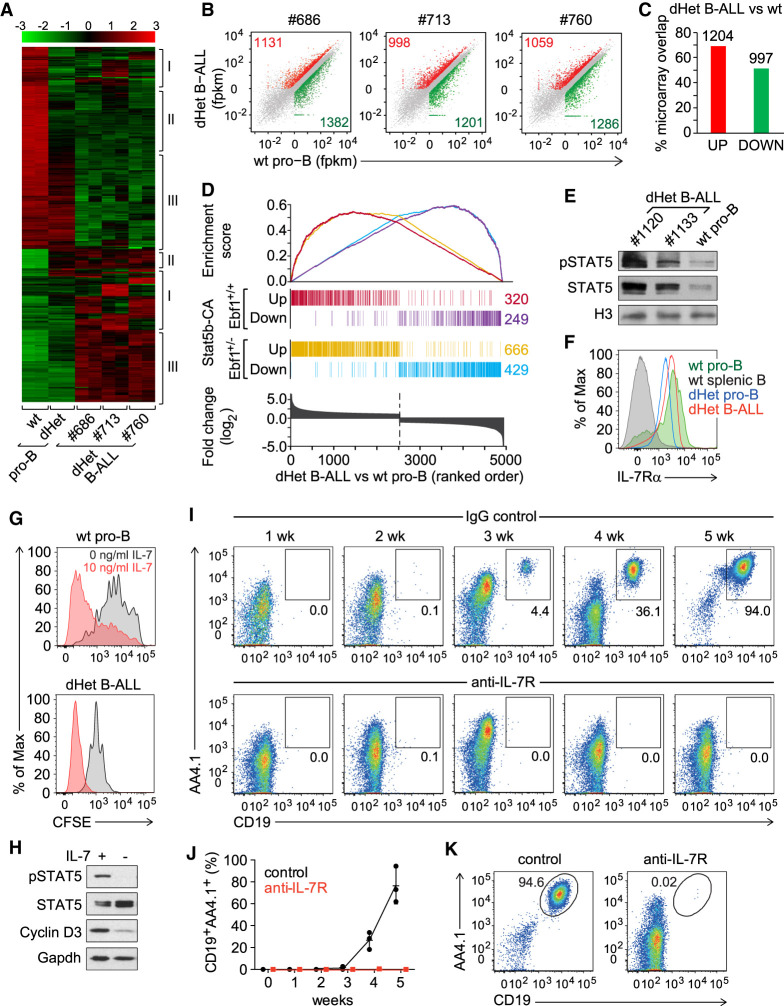
To understand the molecular network driving leukemia development in Ebf1+/−Pax5+/− mice, we performed a genome-wide microarray analysis of dHet leukemic cells from lymph nodes. To facilitate a comparison of the leukemic transcriptome with that of wild-type and nonleukemic dHet pro-B cells, we used sorted wild-type pro-B (Fr.B/C) and dHet pro-B cells at a similar developmental stage as found in dHet B-ALL cells. By using dHet pro-B cells as a nonleukemic control, we could identify genes dysregulated because of the combined Ebf1 and Pax5 heterozygosity and genes specific to the leukemic state of the cells. By using a stringent cutoff of fourfold differential expression, we identified 430 up-regulated and 554 down-regulated genes (Fig. 1A). We found clonal variations (cluster I), molecular signatures of Ebf1/Pax5 double heterozygosity (cluster II), and leukemia-specific signatures (cluster III).
Figure 1.
IL7-STAT5 signaling drives leukemia development. (A) Transcriptional profiling of genes differentially expressed (more than fourfold) between wild-type (wt) pro-B, Ebf1+/− Pax5+/− (dHet) pro-B and dHet B-ALL cells. B-ALL cells were derived from three independent mice (ID #686, #713, and #760). The expression values of the microarray analysis are scaled to z-score (top). The clusters of genes showing clonal variations (I), preleukemic-specific (II) and leukemia-specific (III) differences of expression are indicated. (B) RNA-seq scatter plots depicting the relative gene expression in dHet B-ALL cells (Y-axis) of three mice (ID #686, #713, and #760) relative to wt pro-B cells (X-axis). All significantly twofold up-regulated (red), twofold down-regulated (green), and unaltered genes (gray) are highlighted. (C) Percentage overlap of significantly up-regulated (red) or down-regulated (green) genes as determined by the RNA-seq analysis to the twofold up-regulated or down-regulated genes as determined by the microarray analysis, respectively. The numbers above the bars indicate the number of dysregulated genes in both analyses. (D) Overlap of up-regulated and down-regulated genes in Stat5b-CAtg and Stat5b-CAtg Ebf1+/− leukemic cells with genes that are differentially expressed in Ebf1+/−Pax5+/− dHet B-ALL cells relative to wt pro-B cells. (Bottom) The differentially expressed genes are ranked according to the fold change difference between dHet B-ALL and wt cells. (E) Immunoblot analysis of phospho-STAT5 and total STAT5 in dHet B-ALL cells (n = 2; mouse ID #1120 and #1133) and sorted wild-type fraction B and C cells. Histone3 (H3) was used as a loading control. (F) Flow cytometric analysis of IL-7Rα expression in dHet B-ALL cells (red), dHet pro-B (blue), wt pro-B (green), and wt splenic CD19+ B cells (gray). Wild-type pro-B and splenic B cells served as positive and negative controls, respectively. (G) Flow cytometric analysis of CFSE staining in wt pro-B cells (top) and dHet B-ALL cells (bottom). Cells were labeled with 5mM CFSE and cultured in the absence (gray) or presence (red) of IL-7 at 10 ng/mL for 4 d. (H) Immunoblot analysis of phospho-STAT5, total STAT5, and Cyclin D3 in dHet B-ALL cells cultured in the presence (+) or the absence (−) of IL-7 for 1 d. Gapdh was used as a loading control. (I,J) Longitudinal flow cytometric analysis of donor-derived CD19+AA4.1hi dHet B-ALL cells in peripheral blood of Rag2−/− recipient mice, injected with 1 mg of anti-IL-7 receptor antibody or control rat IgG antibody. The antibodies were injected 1 and 3 d before and every second day after the adoptive transfer of 1 × 105 CD19+AA4.1hi dHet B-ALL cells. The percentages of donor-derived dHet B-ALL cells in the peripheral blood were weekly determined up to 5 wk (n = 3). (K) Flow cytometry analysis of donor-derived CD19+AA4.1hi dHet B-ALL cells in the spleen of recipient mice, treated with control IgG or anti-IL7R antibody, at 5 wk after transplantation.
We also used RNA-seq for the analysis of the transcriptomes of these cells and observed an ∼60% overlap with the data sets of the microarray analysis (Fig. 1B,C). Among the genes with leukemia-specific signature, we detected several genes downstream from IL-7/STAT5 signaling, including Jak3, Mcl1, and Ccnd1 (Supplemental Fig. S2; Supplemental Table S1). Mice expressing a constitutively active form of STAT5 (STAT5-CA) have been reported to develop B-ALL with an enhanced onset in combination with an Ebf1 or Pax5 heterozygosity (Heltemes-Harris et al. 2011). Therefore, we compared the set of genes that is differentially expressed between dHet B-ALL cells and wt pro-B cells with the STAT5-CA leukemia mouse models. Of the 1055 dysregulated genes in leukemic cells of STAT-CA mice, 57% of up-regulated and 50% of down-regulated genes overlapped with genes that are twofold differentially expressed in dHet B-ALL cells versus wt pro-B cells (Fig. 1D). The combination of STAT5-CA with Ebf1+/− heterozygosity further increased the overlap of up-regulated and down-regulated genes with dHet B-ALL cells (Fig. 1D).
The constitutively active STAT5-CA mimics functionally the phosphorylated form of STAT5 (Ariyoshi et al. 2000). In dHet B-ALL cells, we detected enhanced phosphorylation and abundance of STAT5 relative to wt pro-B cells (Fig. 1E). Flow cytometric analysis revealed the expression of IL7R on the surface of dHet B-ALL at levels similar to wt pro-B cells (Fig. 1F). The removal of IL7 from the culture medium resulted in a fairly uniform proliferation stop of dHet B-ALL cells, as determined by CSFE dilution (Fig. 1G). In contrast, wt pro-B cells showed a more heterogeneous and less efficient stop of proliferation. Moreover, IL7 withdrawal from in vitro-cultured dHet B-ALL cells resulted in a loss of STAT5 phosphorylation and reduced cyclin D3 expression (Fig. 1H).
To determine the importance of IL7 signaling for the proliferative expansion of dHet B-ALL cells in vivo, we adoptively transferred 1 × 105 leukemic cells into Rag2-deficient recipient mice and injected the mice with IgG isotype control antibody or anti-IL-7R antibody. We determined the frequency of AA4.1+CD19+ leukemic cells by flow cytometry between 1 and 5 wk after transfer (Fig. 1I,J). In the IgG control-injected mice, AA4.1+CD19+ cells appeared in the blood after 3 wk and increased rapidly thereafter. At 5 wk, the frequency of these cells in the spleen increased to 95%, whereas virtually no AA4.1+CD19+ cells were detected in the spleen of mice that were injected with anti-IL7R antibody (Fig. 1K). Thus, IL7 signaling is necessary for the proliferative expansion of dHet B-ALL cells in vitro and in vivo.
In the transcriptome analysis, we also detected a reduced expression of genes encoding components of the pre-BCR signaling pathway, such as Cd79b, Blnk, Pik3ap1 (BCAP), Prkcb (p110β), and transcription-factor genes Foxo1 and its downstream target Bcl6 (Supplemental Fig. S2; Rickert 2013; Clark et al. 2014). The flow cytometric analysis indicated that the majority of dHet B-ALL cells represent the Fr. B/C pro-B-cell stage of differentiation, and we could not detect surface expression of the pre-BCR on dHet B-ALL cells (data not shown). Therefore, the reduced expression of components of the pre-BCR pathway could reflect the early differentiation state rather than an impaired signaling pathway.
Loss and gain of EBF1 and Pax5 binding in dHet B-ALL cells
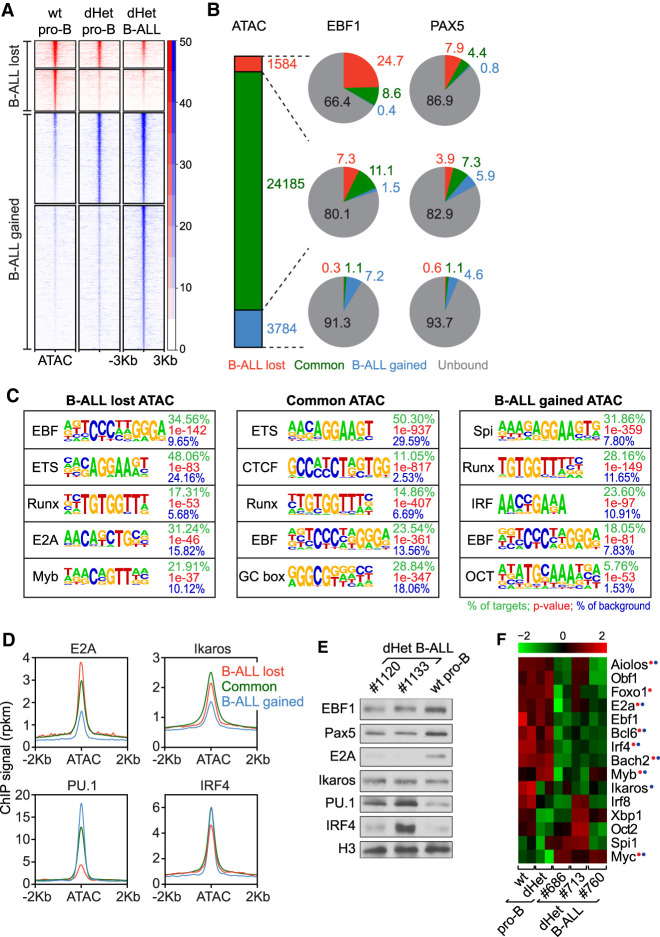
To identify EBF1- and/or Pax5-mediated regulators of leukemia development, we performed a genome-wide comparative EBF1 and Pax5 ChIP-seq analysis in wild-type pro-B, dHet pro-B, and dHet B-ALL cells (Supplemental Fig. S3A,B). For these experiments, we used primary B-ALL cells and in vitro cultured wt and dHet pro-B cells that were derived from Rag2−/− mice to arrest the cells at a similar developmental stage as found in dHet B-ALL mice. We identified 7503, 7245, and 5890 EBF1-occupied sites, shared in two biological replicates of wt pro-B, dHet pro-B, and dHet B-ALL cells, respectively (Supplemental Fig. S3A). Approximately 47% of the EBF1-bound peaks were commonly detected in wt pro-B, dHet pro-B, and dHet B-ALL cells. A similar fraction of EBF1-bound sites was lost in dHet cells relative to wt pro-B cells, whereby 1658 peaks were lost specifically in dHet B-ALL cells. Notably, we also observed a gain of EBF1 occupancy in dHet cells, whereby 1223 peaks were specific for dHet B-ALL cells.
To gain some insight into the mechanism underlying the unexpected gain of EBF1 binding in dHet B-ALL cells, we examined whether EBF1-bound sites in these cells overlap with sites that are transiently occupied by EBF1 during induction of EBF1 in Ebf1 knockout pre-pro-B cells (Li et al. 2018). Notably, we observed an almost 30% overlap of B-ALL gained EBF1 occupancy with EBF1 peaks that are transiently detected during the pre-pro-B to pro-B-cell transition (Supplemental Fig. 3C). In contrast, the majority (∼80%) of EBF1-occupied sites that are common to wt pro-B, dHet pro-B, and dHet B-ALL cells, coincide with persistent EBF1 peaks that are detected upon EBF1 induction and are maintained throughout the pro-B-cell stage (Supplemental Fig. 3C).
In the genome-wide Pax5 ChIP-seq analysis, we identified 1088 and 1220 Pax5-occupied sites that were lost and gained specifically in dHet B-ALL cells (Supplemental Fig. 3B). We also interrogated both ChIP-seq data sets to identify sites that are cobound by EBF1 and Pax5 (Supplemental Fig. S3D). In wt pro-B and dHet B-ALL cells, we detected only a small proportion of co-occupied sites (12% and 8.7%, respectively). Thus, the combined heterozygosity of Ebf1 and Pax5 affects predominantly distinct sets of individually occupied sites rather than the co-occupancy.
To gain further insight into the role of other transcription factors regulating the leukemia development and to understand the gain and loss of chromatin accessibility on EBF1- and Pax5-bound sites in dHet B-ALL cells, we performed genome-wide ATAC (assay for transposase-accessible chromatin) sequencing (Fig. 2A). In a comparison between the leukemic and wild-type peak sets, we identified 3784 peaks specific to dHet B-ALL cells and 1584 peaks specific to wt pro-B cells (Fig. 2A; Supplemental Fig. 3E). Approximately 25% of the wild-type-specific ATAC peaks (lost in both dHet pro-B and dHet B-ALL cells) overlapped with wt-specific EBF1 occupancy, whereas 8% of these ATAC peaks overlapped with Pax5-bound sites (Fig. 2B). Of the B-ALL-gained ATAC peaks, ∼7% and ∼5% overlapped with the B-ALL-specific EBF1-bound and Pax5-bound sites, respectively. Thus, the genome-wide ATAC-seq profiling correlates, at least in part, with the gain and loss of EBF1 and Pax5 binding in leukemic cells.
Figure 2.
Changes in the transcriptional network of dHet B-ALL. (A) Heat map of ATAC signals ∼3 kb from the center of ATAC peaks that are lost (red) or gained (blue) in dHet B-ALL cells relative to wild-type pro-B cells. The clusters are further grouped based on the overlap with ATAC peaks in dHet pro-B cells. The data represent two biological replicates. The RPKM count value is scaled to the heat map signal intensity. (B) The percentage overlap of EBF1 (middle) and Pax5 (right) peaks to the ATAC peaks (left) is shown in the pie charts. The gained, common, and lost peaks in dHet B-ALL cells relative to wild type are highlighted in red, green, and blue, respectively. EBF1- or Pax5-unbound ATAC peaks are shown in gray. (C) Analysis of transcription factor-binding motifs at ATAC peaks. The top five motifs enriched in ATAC peaks that are lost (left), retained (middle), and gained (right) in B-ALL cells relative to wild-type pro-B cells are shown. The percentage of peaks having the motif, the P-value, and percentage detected in the background are indicated for each motif. (D) Distribution of previously published E2A, Ikaros, IRF4 and PU.1 ChIP signals in pro-B cells, ∼2 kb of the ATAC peak centers, in B-ALL lost (red), retained (green) and gained (blue) ATAC clusters. (E) Immunoblot analysis of EBF1, Pax5, E2A, Ikaros, PU.1, and IRF4 in dHet B-ALL cells (n = 2; mouse ID #1120 and #1133) and sorted wild-type pro-B cells. Histone 3 (H3) served as the loading control. (F) RNA-seq expression profiles of B lineage transcription factors in wt pro-B, dHet pro-B, and dHet B-ALL cells derived from three mice (mouse ID #686, #713, and #760). (Top) The FPKM expression values are scaled to the z-score. Genes having differential (B-ALL-gained or B-ALL-lost) EBF1 binding (red) and/or Pax5 binding (blue) are highlighted.
De novo motif prediction analysis indicated that ∼35% of the wild-type-specific (B-ALL lost) ATAC peaks and 18% of the leukemia-specific (B-ALL gained) ATAC peaks contain the EBF1-binding motif (Fig. 2C). In addition, we identified binding sites for ETS, Runx, E2A, and Myb among the top DNA motifs in B-ALL lost ATAC peaks, whereas we detected an enrichment of the Spi/PU.1 and IRF motifs in the B-ALL gained ATAC peaks (Fig. 2C). We did not detect the Pax5 motif due to limitations of tools in identifying complex motifs. We extended this analysis by comparing the ATAC-seq clusters with previously published binding profiles of B-cell transcription factors in pro-B cells (Lin et al. 2010; Revilla et al. 2012; Schwickert et al. 2014). Consistent with the motif enrichment analysis, we detected a reduced distribution of E2A and Ikaros ChIP signals in B-ALL gained ATAC peaks relative to the B-ALL lost ATAC peak cluster, whereas we observed an inverse relationship for PU.1 and IRF4 ChIP signals (Fig. 2D). Specifically, the wt-specific ATAC peaks showed a 32% overlap with E2A occupancy and 14% overlap with Ikaros occupancy, whereas the B-ALL-specific ATAC peaks had only a 13% overlap with E2A binding and 6% overlap with Ikaros binding (Supplemental Fig. S3F). Conversely, PU.1 occupancy overlapped with 26% of B-ALL-lost and 45% of B-ALL-gained ATAC peaks, suggesting a premarking of these sites by PU.1 and IRF4 (Supplemental Fig. S3F,G).
The changes in the occupancy of these transcription factors could be accounted for by an altered transcript and protein abundance (Fig. 2E,F). Tcf3 (E2A) transcript and protein levels were lower in dHet B-ALL cells relative to wt pro-B and dHet pro-B cells. Although Ikaros transcript abundance was reduced in dHet B-ALL cells, we did not detect an obvious change in Ikaros protein level. Conversely, we detected increased PU.1 transcript and protein levels in dHet B-ALL cells. We also observed enhanced IRF4 protein levels in some B-ALL clones despite reduced Irf4 transcript levels (Fig. 2E,F). Together, these data indicate that the loss of ATAC peaks in leukemic cells is not only related to the diminished binding of EBF1 and Pax5 but also to a reduced E2A and Ikaros occupancy. In contrast, the gain of EBF1 occupancy and ATAC peaks in B-ALL cells is associated with enhanced PU.1 binding and may reflect a transient developmental stage that is stabilized by the leukemic transformation.
Altered transcriptional networks in dHet leukemic cells
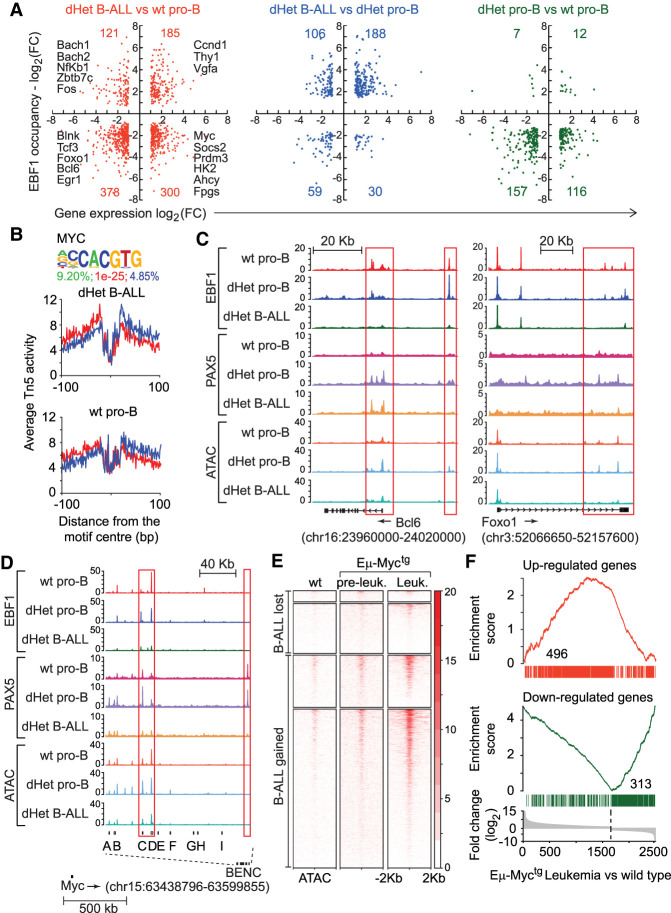
By interrogating the genome-wide ChIP-seq and microarray transcriptome analysis, we identified a leukemia-specific signature of EBF1 and Pax5 occupancy and the associated gene expression program. We normalized EBF1 binding in all three cell types and calculated the fold-change in EBF1 binding among the cell types. The overlap of altered gene expression and differential EBF1 occupancy indicated that the gain or loss of EBF1 binding is associated with both the up-regulated and down-regulation of gene expression (Fig. 3A; Supplemental Table S2). A total of 378 genes that were down-regulated in leukemic cells relative to wt pro-B cells also showed reduced EBF1 occupancy (Fig. 3A, left panel). This set includes genes encoding the pre-BCR signaling components Blnk, Pik3r1, Cd79b, Nfatc1, and Prkcb, as well as the B lineage transcription factors Tcf3 (E2a), Foxo1, Bcl6, Ikzf3, Myb, Fli1, Egr1, and Ets2 (Supplemental Table S1). In addition, 121 down-regulated genes that gained EBF1 occupancy in leukemic cells included Bach1, Bach2, Hlx, Nfkb1, Fos, Dtx1, and Zbtb7c. The up-regulated genes included 185 genes that gained and 300 genes that lost EBF1 binding in leukemic cells. Among the 300 up-regulated genes with reduced EBF1 occupancy, we identified Myc, components of glycolysis (Hk2 and Pgm2), one-carbon metabolism (Ahcy, Fpgs, and Mthfd1l) and cell proliferation (Socs2, Prmt3, Cux1, and Fyn). The fourth gene set with a gain in expression and EBF1 occupancy included Ccnd1, Thy1, Ass1, and Vgfa. The comparison of dHet B-ALL cells with dHet pro-B and wild-type pro-B cells indicated that the majority of dysregulated genes are associated with enhanced and reduced EBF1 occupancy in leukemic cells and with reduced EBF1 occupancy in dHet pro-B cells (Fig. 3A, middle and right panels). Thus, the dysregulation of genes with the gain of EBF1 occupancy is confined to the leukemic state of cells.
Figure 3.
The activation of Myc and its targets in dHet B-ALL. (A) Overlap of genes differentially expressed (X-axis) and differentially bound by EBF1 (Y-axis) in dHet B-ALL cells relative to wild-type pro-B cells (left), dHet B-ALL cells relative to dHet pro-B cells (middle), and dHet pro-B cells relative to wild-type pro-B cells (right). The key differentially regulated genes between dHet B-ALL and wt pro-B cells are highlighted. (B) The abundance of the Myc-binding motif in the B-ALL gained ATAC peaks (top) and digital genomic footprinting analysis showing normalized Tn5 insertion profiles, in dHet B-ALL and wt pro-B cells, around footprinted Myc motifs identified in dHet B-ALL cells (bottom). Insertions on the forward and reverse strands are indicated in red and blue, respectively. The percentage of peaks having the motif, the P-value, and percentage detected in the background are indicated. (C,D) Screenshots showing the distribution of EBF1 and Pax5 occupancy and chromatin accessibility, as determined by ChIP-seq and ATAC-seq analysis, on the Bcl6 gene (C, left), Foxo1 gene (C, right), and BENC regulatory elements of the Myc locus (D) in wt pro-B, dHet pro-B, and dHet B-ALL cells. The ChIP and ATAC signals are normalized to 10 million reads (Y-axis). (E) Heat map of Myc ChIP signals, from the wt, preleukemic (Eµ-Myctg), and leukemic cells (Eµ-Myctg), 2 kb around the center of dHet B-ALL-lost and B-ALL-gained ATAC peaks. (F) Overlap of genes up-regulated (top) or down-regulated (bottom) in dHet B-ALL cells to the genes differentially expressed in the Eµ-Myctg leukemic cells. The differentially expressed genes are ranked according to the fold change difference between Eµ-Myctg leukemic cells and wt cells.
In the cluster of ATAC peaks that were gained specifically in dHet B-ALL cells, we also detected an enrichment of binding sites of Stat5 and Myc, as well as an enhanced occupancy of these binding sites as determined by digital footprinting (Fig. 3B; Supplemental Fig. S3H). This enhanced Myc binding is consistent with the up-regulation of Myc expression in dHet B-ALL cells (Fig. 2F; Supplemental Fig. S2). In pre-B cells, the expression of Myc is repressed by Bcl6 and in a subset of human B-ALL, a positive feedback loop between pre-BCR and Bcl6 has been detected (Nahar et al. 2011; Geng et al. 2015). Bcl6 and its transcriptional activator Foxo1 were both found to be down-regulated in dHet B-ALL cells (Fig. 2F; Supplemental Fig. S2). Therefore, we examined whether the transcriptional network of Foxo1, Bcl6, and Myc may be affected by altered binding of EBF1 and/or Pax5. In dHet B-ALL cells, we observed a reduced binding of EBF1 and Pax5 and reduced ATAC sites in the Foxo1 and Bcl6 genes (Fig. 3C). In addition, we observed a diminished binding of EBF1 and Pax5 at regulatory elements C and D of the blood enhancer cluster (BENC) downstream to the Myc locus (Bahr et al. 2018), suggesting a direct role of EBF1 and Pax5 in regulating the Foxo1, Bcl6, and Myc network in B-ALL cells (Fig. 3D).
Similar to the Ebf1/Pax5 dHet mouse model, mice constitutively expressing Myc in the B-cell lineage, driven by immunoglobulin heavy chain enhancer, develop leukemia (Harris et al. 1988). Previous genome-wide Myc binding profile in the wt pro-B, preleukemic, and leukemic cells indicated a gain of Myc binding in leukemic cells (Sabò et al. 2014; Tesi et al. 2019). By profiling the distribution of Myc binding reported in Eµ-Myctg mice around the ATAC-seq peaks that are gained in Ebf1/Pax5 dHet mice, we found a significant enrichment of Myc binding on the ATAC peaks in the B-ALL gained cluster, relative to B-ALL lost cluster (Fig. 3E). The majority of sites were specifically gained in Eµ-Myctg leukemic cells but not in preleukemic cells, suggesting that Myc may act as a key downstream driver of leukemia in Ebf1/Pax5 dHet mice. We also compared the gene expression profiles of both the mouse model and found a 30% (496/1673) overlap of up-regulated and 37% (313/852) overlap of down-regulated genes (Fig. 3F). Taken together, these data suggest a role of Myc in the disease development of Ebf1+/− Pax5+/− mouse model.
Altered central carbon metabolism in dHet B-ALL cells
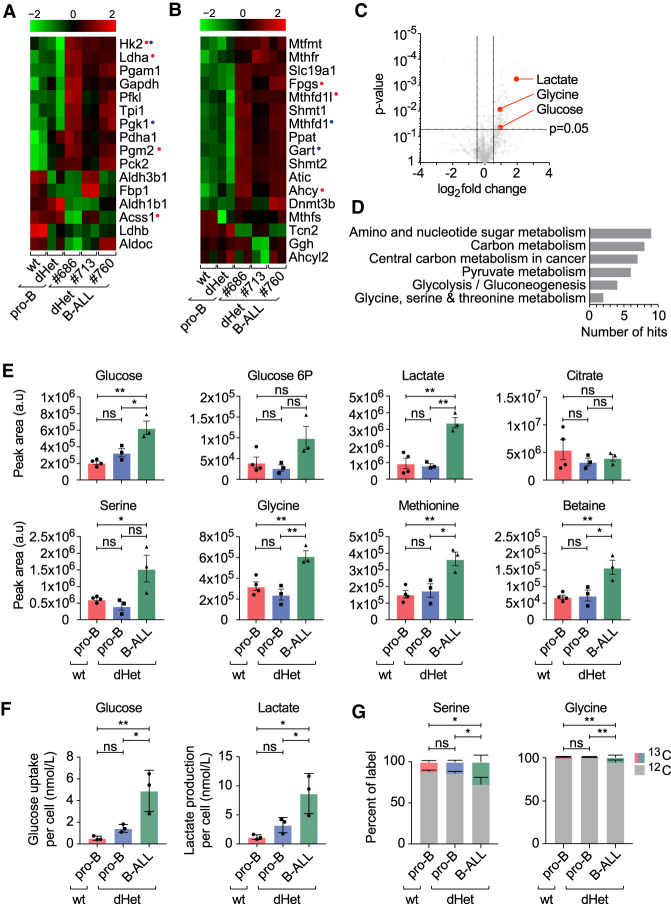
To identify pathways promoting leukemia development, we subjected the genes that are up-regulated and down-regulated in dHet B-ALL to a gene set enrichment analysis. Interestingly, we found glycolysis, folate biosynthesis, and one-carbon metabolism among the pathways highly enriched in up-regulated gene sets (Supplemental Fig. S4A). Comparative mRNA analysis of wt pro-B cells, preleukemic dHet pro-B cells, and leukemic dHet B-ALL cells showed a marked alteration in the expression of genes involved in central carbon metabolism (Fig. 4A). Whereas some of the metabolic genes were abundantly expressed in wild-type pro-B cells (Aldh3b1, Aldh1b1, Acss1, and Ldhb), their expression is down-regulated in leukemic B-ALL. However, genes encoding key enzymes in glycolysis (Hk2, Pfkl, Tpi1, Gapdh, Pgam1, Ldha, and Pdha1) were markedly up-regulated in leukemic B-ALL cells, in line with the engagement of Warburg metabolism in rapidly proliferating malignant cells (Vander Heiden et al. 2009). Only a small proportion of these genes were similarly up-regulated in the dHet premalignant cells, whereas the expression of a large proportion of key glycolytic modulators was not yet prevalent. This suggested that the loss of one copy of Ebf1 and Pax5 can alter the expression of genes encoding key metabolic enzymes, but that the engagement of Warburg metabolism might be selected for during progressive transformation in vivo.
Figure 4.
Changes in the metabolic pathways of dHet B-ALL. (A,B) RNA-seq expression profile of genes involved in glycolysis (A) and one-carbon metabolism (B) in wild-type (wt) pro-B, dHet pro-B, and dHet B-ALL cells. Data were derived from three mice (mouse ID #686, #713, and #760). (Top) The FPKM expression values are scaled to the z-score. Genes having differential (B-ALL gained or B-ALLlost) EBF1 binding (red) and Pax5 binding (blue) are highlighted. (C) Untargeted metabolic analysis (using XCMS) of polar metabolites extracted from 5 × 106 wild-type pro-B, dHet pro-B, and dHet B-ALL cells that were cultured in the presence of IL-7. (D) Pathway analysis of significantly different metabolites using the KEGG pathway mapper tool. The total number of significantly different metabolites found in each pathway is indicated. (E) Targeted analysis of metabolic pathways of interest identified in D. Histograms compare metabolites in glycolysis (glucose, glucose-6P, and lactate) and one-carbon metabolism (serine, glycine, methionine, and betaine) extracted from 5 × 106 wt pro-B (red), dHet pro-B (blue), and dHet B-ALL (green) cells. (F) Glucose uptake (left) and lactate release (right) of 0.4 × 106 wt pro-B, dHet pro-B and dHet B-ALL cells cultured for 24 h. (G) 13C-glucose tracing analysis depicting the relative levels of 13C and 12C serine (left) and glycine (right) in 2 × 106 wt pro-B, dHet pro-B and dHet B-ALL cells cultured with 13C-glucose for 10 min.
We also observed the dysregulation of genes associated with one-carbon metabolism (Fig. 4B). To further validate the transcriptional data showing enhanced one-carbon metabolism and glycolysis in dHet B-ALL cells, we carried out an untargeted metabolomics analysis. We identified lactate, glycine, and glucose with significantly different abundance in wt pro-B and dHet B-ALL cells (Fig. 4C). KEGG pathway analysis of significantly altered metabolite abundance showed that carbon metabolism, one-carbon metabolism (glycine and serine), and glycolysis pathways were overrepresented in dHet B-ALL cells, in keeping with our transcriptional data (Fig. 4D).
Untargeted analysis of glycolysis indicated that intracellular glucose and glucose-6-phosphate were increased in dHet B-ALL cells, relative to wt pro-B cells and nonleukemic dHet pro-B cells (Fig. 4E). We also observed increased levels of glucose uptake and lactate release in the dHet B-ALL cells relative to wt pro-B and dHet pro-B cells (Fig. 4F). The key enzyme promoting the lactate production under high glycolytic flux Ldha is up-regulated in dHet B-ALL cells. The significant increase in the lactate level in dHet B-ALL cells suggests that both glucose uptake and glucose metabolism via glycolysis are augmented in dHet B-ALL cells. However, citrate levels remained similar between leukemic and nonleukemic cells, perhaps reflective of selected glucose carbon allocation to lactate in dHet B-ALL cells. Upstream of lactate, glycolytic carbons can be diverted into anabolic pathways, including the pentose phosphate pathway and one-carbon metabolism. The glycolytic intermediate 3-phosphoglycerate can be converted to serine and further metabolized to glycine, thereby donating one carbon to 5,10-methylene tetrahydrofolate to synthesize tetrahydrofolate as part of the folate cycle. Both serine and glycine levels were increased in dHet B-ALL cells, indicating enhanced engagement of the folate cycle. We also detected increased levels of methionine and betaine in dHet B-ALL cells, indicating an enhanced donation of carbon from the folate cycle to the methionine cycle. Moreover, we traced the fate of glucose in wt pro-B, dHet pro-B, and dHet B-ALL cells by using 13C-labeled glucose. In line with our metabolomics results, we found an increased utilization of glucose for serine and glycine synthesis in dHet B-ALL cells relative to wt pro-B and dHet pro-B cells (Fig. 4G). Overall, this analysis shows a metabolic remodeling in dHet B-ALL cells to increase carbon allocation to one-carbon metabolism, likely to support the gain of leukemia-specific metabolic properties of these cells.
Enhanced one-carbon metabolism promotes leukemic cell survival
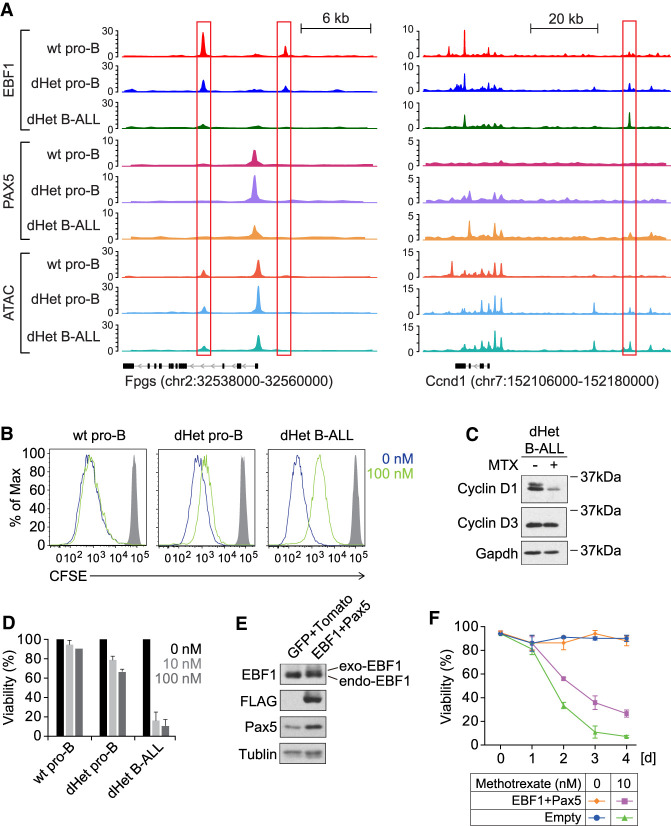
Bioinformatic analysis of publicly available RNA expression data of pretreatment diagnostic samples from a childhood high-risk B-ALL cohort, COG-P9906, revealed an elevated expression of one-carbon pathway genes in one-third (68/207) of patients (Supplemental Fig. S4B). Thus, the up-regulation of one-carbon metabolism may be a key feature of many B-ALL. The one-carbon metabolism is involved in DNA synthesis and can be altered in leukemic cells by treating cells with methotrexate (MTX), a structural analog of folic acid (Kager et al. 2005). In cells, MTX is polyglutamated and competes with cellular folate cofactors, thereby inhibiting enzymes in thymidine and de novo purine biosynthesis (Kager et al. 2005). We found several genes in this pathway that are up-regulated in B-ALL cells and differentially occupied by EBF1 or Pax5 (Fig. 3A; Supplemental Table S1). For example, FPGS encoding folylpolyglutamate synthetase, which is essential for folate homeostasis and polyglutamation of MTX, is up-regulated and loses EBF1 occupancy in B-ALL cells (Fig. 5A, left panel). To assess the efficiency of MTX treatment on the proliferation of wt pro-B, dHet pro-B, and dHet B-ALL cells, we labeled MTX-treated or untreated cells with CFSE and monitored CFSE dilution by flow cytometric analysis (Fig. 5B). We detected no difference in the proliferation of MTX-treated wt pro-B cells relative to untreated cells and only modestly reduced proliferation of dHet pro-B cells after MTX treatment. However, MTX-treated dHet B-ALL cells proliferated much slower compared with their untreated counterparts. Notably, MTX treatment of dHet B-ALL cells markedly reduced the level of cyclin D1, but not that of cyclin D3 (Fig. 5C). The abundant cyclin D1 expression in untreated dHet B-ALL cells relative to wt pro-B cells, which do not express cyclin D1 (Cooper et al. 2006), correlates with a gain of EBF1 and Pax5 occupancy at the Ccnd1 gene locus specifically in dHet B-ALL cells (Fig. 5A, right panel).
Figure 5.
Enhanced folate pathway and methotrexate sensitivity in dHet B-ALL cells. (A) Distribution of EBF1 occupancy, Pax5 occupancy, and chromatin accessibility in Fpgs (left) and Ccnd1 (right) loci as determined by ChIP-seq and ATAC-seq analysis of wt pro-B, dHet pro-B, and dHet B-ALL cells. The ChIP and ATAC signals are normalized to 10 million reads (Y-axis). (B) Flow cytometry analyzing cell proliferation by CFSE dilution in wt pro-B, dHet pro-B, and dHet B-ALL cells, cultured in the absence (blue) or presence of 100 nM methotrexate (MTX, green) for 3 d. Gray histogram shows undivided negative control. (C) Immunoblot analysis of Cyclin D1 and Cyclin D3 levels in untreated and MTX-treated (100 nM) dHet B-ALL cells. Gapdh served as a loading control. (D) Analysis of the viability of wt pro-B, dHet pro-B, and dHet B-ALL cells upon treatment with methotrexate (0, 10, and 100 nM) for 3 d. Relative cell viability was measured by flow cytometry as the frequency of PI-negative cells. (E) Immunoblot analysis to detect EBF1 and Pax5 in dHet B-ALL cells transduced with bicistronic retroviruses expressing FLAG-EBF1 along with GFP and Pax5 along with Tomato. Tubulin served as a control. (F) Time-course analysis of the viability of MTX-treated (10 nM) and untreated dHet B-ALL cells cotransduced with EBF1- and Pax5-expressing retroviruses or with empty control virus. Cell viabilities were calculated by flow cytometry as frequencies of PI-negative cells.
We also monitored the viability of wt pro-B, dHet pro-B, and dHet B-ALL cells that were treated with 10 nM or 100 nM MTX (Fig. 5D). The dHet B-ALL cells were very sensitive to MTX and ∼80% of the cells died within 4 d, whereas wt and dHet pro-B cells did not significantly respond to the drug. To further address the direct importance of EBF1 and Pax5 for the folate pathway-dependent survival of B-ALL cells, we transduced dHet B-ALL cells with bicistronic retroviral constructs expressing EBF1 or Pax5 along with GFP or Tomato, respectively. As a control, we transduced an empty vector expressing GFP and Tomato (Fig. 5E). Upon MTX treatment, we observed a significant difference in the viability of EBF1 and Pax5 transduced leukemic cells relative to empty vector-transduced cells but no obvious change in proliferation (Fig 5F; data not shown). Four days after the treatment, we detected ∼30% viability in EBF1 and Pax5 cotransduced cells, whereas empty vector-transduced cells showed an ∼4% survival. This result suggests that the restoration of EBF1 and Pax5 expression in dHet B-ALL attenuates the effects of MTX on cell viability. Thus, the folate pathway is important for the survival of dHet B-ALL cells.
Single-cell sequencing reveals heterogeneity and progression of leukemic state
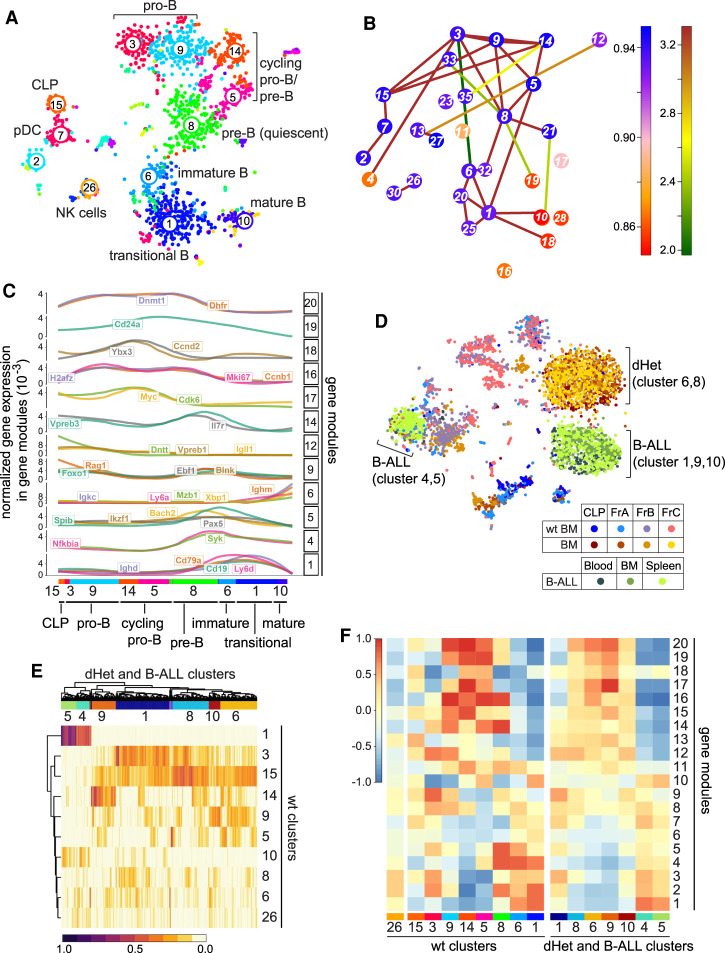
To obtain further insight into the composition and developmental stages of leukemic cells, we performed single-cell RNA sequencing of sorted cells from wild-type, preleukemic, and leukemic mice. First, we analyzed single-cell RNA sequencing data of FACS-sorted common lymphoid progenitors (CLPs; Lin− Scaint Kitint Flt3+), fraction A pre-pro-B cells (B220+CD43+NK1.1−BST2−HSA−BP1−), and fraction B/C pro-B cells (B220+CD43+NK1.1− BST2−HSA+BP1+) from wild-type mice and used RaceID3 (Herman et al. 2018) to generate clusters representing the major developmental stages (Fig. 6A). We performed one-versus-one differential gene expression analysis of the wild-type clusters to identify marker genes indicative of B-cell differentiation stages (Supplemental Fig. S5A). We were able to distinguish CLP (cluster 15) expressing Flt3 and Il7r, pro-B cells marked by Rag1, Il7r, Dntt (clusters 3 and 9), and cycling pro-B and pre-B cells expressing Cdk6, Myc, and pre-BCR components (clusters 14 and 5). We also identified pre-B cells initiating recombination of the light chain loci marked by Rag1 and Il2ra expression (cluster 8). On the basis of the expression levels of Ly6d, Ighd, and Igkc, we identified immature B cells (cluster 6), transitional B cells (cluster 1), and mature B cells (cluster 10), respectively. In this analysis, we also recovered a few NK-cells (cluster 26) and plasmacytoid dendritic cells (clusters 2 and 7).
Figure 6.
Leukemic cells of Ebf1+/−Pax5+/− mice have mixed signatures of early pro-B cells. (A) t-SNE plot representation of the single-cell RNA-seq analysis of wild-type bone marrow B cells. Each dot represents a cell. The numbers and different colors represent clusters of similar cells defined by the RaceID algorithm. The clusters are assigned to different developmental stages or cell types based on the gene expression profile. (B) Graphic representation of the lineage inference using StemID. The colors of the connections depict the −log10 P-value and the colors of the nodes indicate Δ-entropy of the cluster with the corresponding number. (C) Pseudotemporal ordering of gene expression changes along a putative differentiation trajectory of clusters 15, 3, 9, 14, 5, 8, 6, 1, and 10 as inferred in B. Genes are grouped into modules (left Y-axis) with similar expression profiles using self-organizing maps. Selected gene expression profiles along the differentiation trajectory are shown. Gene expression for every gene is normalized to total gene expression. (D) t-SNE map representing the clustering of cells in fractions A, B, and C and CLP sorting gates from wild-type and dHet bone marrow cells as well as from dHet B-ALL cells that were derived from blood, bone marrow, and spleen. The cells were clustered using RaceID. The cells from different sorting gates and their phenotypes are color-coded and indicated in the key. (E) Heat map representing the clustering of leukemic cells (columns) to wt cluster medoids (rows) using quadratic programming. The color-coded scale represents the weights from 0 to 1, which indicate the similarities of leukemic cells toward the wt cluster medoids. (Top) The hierarchical clustering of the weights yielded seven major leukemic clusters. (F) Heat map representing the aggregated expression values of genes defining 20 modules (rows) in each wt cluster (left X-axis) and dHet cluster (right X-axis). The aggregated expression value of each module is scaled as a z-score between 1 and −1.
In addition, we applied the StemID algorithm (Grün et al. 2016) to derive a putative differentiation trajectory of wild-type B lineage cells (Fig. 6B; Supplemental Fig. S5B). This analysis uncovered a pseudotemporal differentiation trajectory along the clusters 15, 3, 9, 14, 5, 8, 6, 1, and 10 and was in agreement with the different B-cell differentiation stages for the inferred clusters. The links between earlier and later stages (e.g., clusters 3 and 8) are most likely due to the recurrent expression of recombination-associated genes and other similarities in these clusters that confounded the computational analysis. This pseudotemporal order was used to derive gene expression modules with similar gene expression patterns along the pseudotemporal axis, which included known B-cell differentiation marker genes (Fig. 6C; Supplemental Table S3). The differentiation trajectory starts at the CLP stage (cluster 15) and continues through the stages of pro-B cells (clusters 3 and 9), pre-B cells (clusters 14, 5, and 8), immature B cells (cluster 6), transitional B cells (cluster 1), and mature B cells (cluster 10). The expression dynamics of selected marker genes are in agreement with the B-cell differentiation stages (Fig. 6C).
To better understand the leukemic transformation, we performed single-cell RNA sequencing of CLPs, pre-pro-B cells, and pro-B cells (fractions B and C) isolated from a 22-wk-old preleukemic dHet mouse, prior to the detection of AA4.1+CD19+ B-ALL cells in the blood and with a modest accumulation of cells in the bone marrow (data not shown). In addition, we used dHet B-ALL cells from the blood, bone marrow, and spleen of a 26-wk-old leukemic dHet mouse with the accumulation of AA4.1+CD19+ cells in secondary lymphoid organs. Upon clustering, preleukemic and leukemic cells grouped separately from the wild-type cells and from each other (Fig. 6D). Although the overall transcriptome of the preleukemic and leukemic cells seemed to be changed based on the clustering result, we reasoned that we could use the wild-type clustering information to better understand the cell differentiation stage of the leukemic cells. To this end, we described every leukemic cell as a linear combination of the wild-type cluster medoids; i.e., the most representative cell for each of the wild-type clusters 15, 3, 9, 14, 5, 8, 6, 1, and 10. After optimizing the weight contributions for each wild-type cluster medoid by quadratic programming (see the Materials and Methods), we visualized these weights across all leukemic cells (Fig. 6E; Supplemental Fig. S5D,E). Moreover, we interrogated the gene modules from the wild-type pseudotemporal differentiation trajectory with leukemic clusters. The expression pattern of the leukemic gene modules recapitulated the pattern of wild-type cells but also showed the coexpression of gene modules from later differentiation stages (Fig. 6F). The derivation of marker genes by pairwise comparisons of wild-type versus dHet B-ALL clusters confirmed the assignment of leukemic clusters to specific stages of B-cell differentiation (Fig. 6D; Supplemental Fig. S5E). In particular, this analysis indicated that dHet pro-B-cell clusters 6 and 8 differ from two major B-ALL populations consisting of clusters 1, 9, and 10 and 4 and 5, which represent an early stage and a transitional B stage, respectively. Taken together, these data suggest that the altered gene expression pattern in the majority of leukemic cells carries hallmarks of early stages of B-cell differentiation that differ from the gene expression signature detected in accumulated dHet cells in the bone marrow. In addition, a smaller population of leukemic cells in the periphery shows a gene expression signature of later stages of differentiation.
A small subset of bone marrow pro-B cells shares several hallmarks with leukemic cells
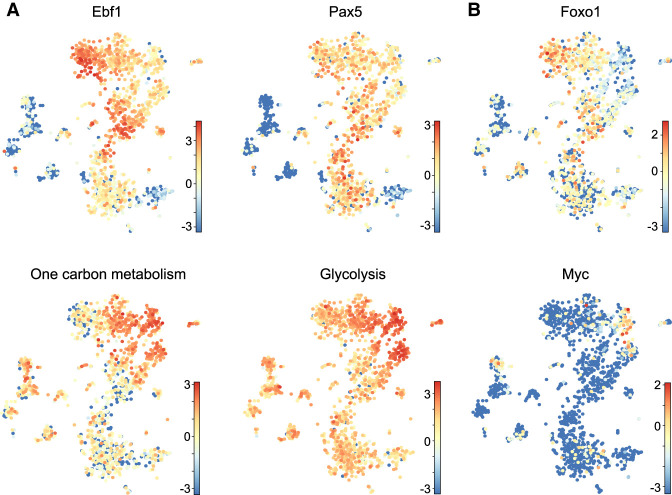
The single-cell RNA-seq profiling of wt, dHet bone marrow cells and B-ALL cells indicated a mixed expression status of the B-ALL clusters relative to the wt clusters. To identify potential precursors to leukemic cells in the wt bone marrow, we interrogated the single-cell RNA-seq analysis of wt pro-B cells with the expression pattern of genes differentially expressed in the gene expression analysis of wt pro-B and dHet B-ALL cells. Interestingly, we found a reduced expression of Ebf1 and Pax5 in the wt clusters 5 and 14, relative to the neighboring clusters 3, 8, and 9 (Fig. 7A, top panels). We also observed abundant expression of genes associated with one-carbon metabolism and glycolysis in clusters 5 and 14 (Fig. 7A, bottom panels, Supplemental Fig. S6A). This observation suggests that the cells in clusters 5 and 14 may have a high metabolic activity similar to that of dHet B-ALL cells. To extend this analysis, we pooled the clusters 5 and 14 and searched for differentially expressed genes relative to the neighboring clusters 8 and 9. By this comparison, we identified 774 differentially expressed genes (Supplemental Table S4). Of these genes, 39% (113/287) of up-regulated and 58% (281/487) of down-regulated genes overlapped with genes that are differentially expressed in dHet B-ALL cells relative to wt pro-B cells (Supplemental Fig. S6B). In addition to key metabolic genes, clusters 5 and 14 include genes with an expression pattern similar to that of leukemic cells. In particular, we found low levels of Foxo1 and high levels of Myc in clusters 5 and 14 (Fig. 7B). Thus, the dynamic down-regulation of Ebf1 in cycling pro-B cells may lead to the transient up-regulation of Myc and metabolic changes that are also observed in dHet B-ALL cells.
Figure 7.
dHet B-ALL cells originate from a subpopulation of pro-B cells. (A,B) t-SNE plots representing the expression of genes encoding EBF1, Pax5, components of glycolysis and one-carbon metabolism (A) and Foxo1 and Myc (B) in wild-type bone marrow B lineage cells. Each dot represents a cell and the differential expression values are color-coded with the z-score values. The cumulative expression values of genes involved in one-carbon metabolism and glycolysis are indicated.
Discussion
B lymphopoiesis requires a complex regulatory network of transcription factors that ensures the coordination of cell proliferation, differentiation, and changes in gene expression, chromatin structure, and metabolism. Disruptions of the gene regulatory circuitry by mutations or altered gene dosage have been associated with impaired differentiation and/or cellular transformation. In particular, mutations of the transcription factor IKZF1, PAX5, and EBF1 have been associated with B progenitor acute lymphoblastic leukemia (for review, see Somasundaram et al. 2015; Chan and Müschen 2017; Roberts and Mullighan 2019). This malignancy has been shown to involve multiple changes in cell proliferation, transcriptional circuits, signaling, and the metabolic state.
Here, we use the Ebf1+/−Pax5+/− mouse model for B-ALL to explore further the mechanism of cellular transformation by the altered gene dosage of Ebf1 and Pax5. Previous analysis of these mice indicated that developmentally arrested leukemic cells show an increase in DNA damage and lineage infidelity (Prasad et al. 2015; Ungerbäck et al. 2015; Somasundaram et al. 2016). The current analysis indicated that the proliferative expansion of Ebf1+/−Pax5+/− leukemic cells is dependent on IL7R signaling in vitro and in vivo. In particular, injection of anti-IL7R antibody into mice, adoptively transferred with dHet B-ALL cells, prevented the expansion of the leukemic cells. Moreover, the activation of the IL7-responsive transcription factor STAT5 acts in concert with a haploinsufficiency for Pax5 and Ebf1 to initiate leukemia (Heltemes-Harris et al. 2011). Activated STAT5 also synergizes with defects in components of pre-BCR signaling to initiate B-ALL (Katerndahl et al. 2017). The IL7R signaling pathway rivals with signaling by the pre-B-cell receptor to ensure the proper balance of proliferation and differentiation (for review, see Clark et al. 2014). The balance of IL7Rα signaling and pre-BCR signaling depends on a dynamic and complex gene regulatory circuitry in which EBF1 and Pax5 activate components of the pre-BCR and the signaling by the pre-BCR enhances the expression of Pax5, Irf4, Irf8, and Foxo1 (Ochiai et al. 2012). In Ebf1+/−Pax5+/− leukemic cells, we observed a reduced expression of genes encoding these transcription factors and signaling components, which frequently corresponded to a diminished binding of EBF1 and/or Pax5 at regulatory regions associated with these genes. Thus, the leukemic state of dHet B-ALL cells involves a major change in the gene regulatory circuitry in which IL7R signaling is enhanced at the expense of pre-BCR-driven cell differentiation.
Our analyses of chromatin accessibility, EBF1, and Pax5 occupancy in wt pro-B cells, preleukemic dHet pro-B cells, and dHet B-ALL cells identified additional down-regulated transcription factor genes in B-ALL cells that show an altered occupancy by EBF1 and/or Pax5. In particular, Ikaros, which binds and functionally defines pre-B cell superenhancers (Hu et al. 2016), is down-regulated in B-ALL cells and shows a reduced Pax5 occupancy. The role of Ikaros in early B-cell differentiation and B-ALL involves the regulation of cell adhesion and changes in the metabolic and cell cycle states (Joshi et al. 2014; Hu et al. 2016; Schjerven et al. 2017; Witkowski et al. 2017; Ferreirós-Vidal et al. 2019). Ikaros was also shown to activate the expression of Foxo1 and repress the Myc gene. The analysis of Ikaros targets that either respond or do not respond to Myc indicated that metabolic housekeeping genes are enriched among Myc-sensitive Ikaros targets (Ferreirós-Vidal et al. 2019).
Myc is up-regulated in dHet B-ALL cells and both EBF1 and Pax5 bind to the blood enhancer cluster (BENC) in wt pro-B cells, but not in dHet B-ALL cells. The BENC superenhancer, located 1.7 Mb downstream from the Myc gene, is essential for the maintenance of MLL-AF9-driven leukemia in mice and confers hematopoietic expression (Bahr et al. 2018). Specifically, the C and D modules of the BENC, which are bound by EBF1, Pax5, Myb, Erg, and Gfi1b, are required for normal B-cell differentiation and Myc expression. We found that EBF1 binds both the C and the D modules in wt pro-B and dHet pro-B cells but not in dHet B-ALL cells. Moreover, the occupancy of Pax5 at the C module is markedly reduced in dHet B-ALL cells, suggesting that the binding of EBF1 and Pax5 safeguard wt pro-B cells from high Myc expression. Notably, our single-cell analysis of wt pro-B cells identified a small subset with a gene signature resembling that of leukemic B-ALL cells, including a low level of Ebf1 expression and high expression of Myc and genes involved in glycolysis and one-carbon metabolism.
Increased glycolysis, glucose uptake and ATP levels have been identified as metabolic hallmarks of the majority of B-ALL, whereby Pax5 and Ikaros act as metabolic gatekeepers (Chan and Müschen 2017; Xiao et al. 2018). Moreover, abundant Myc expression has been linked to the activation of metabolic networks during cell cycle entry in fibroblasts (Morrish et al. 2009). In particular, Myc increases the oxidative metabolism of glucose and regulates the partitioning of glucose carbons into the folate and pentose pathways. Moreover, Myc activates many genes involved in one-carbon metabolism, including Shtm2, and the overexpression of Shtm2 is sufficient to overcome the growth defects of Myc-deficient cells (Nikiforov et al. 2002; Morrish et al. 2008). Finally, the relationship between Myc and the folate pathways is extended by the observation that Myc+/+ cells are more susceptible to MTX treatment than Myc−/− cells (Morrish et al. 2009). In addition to the BENC regulatory element of Myc, many of the folate pathway genes are differentially bound by EBF1 and/or Pax5 in dHet B-ALL cells versus dHet pro-B cells, suggesting that an altered regulatory network of EBF1, Pax5, and Myc underlies the up-regulation of the folate pathway in leukemic cells.
The scRNA-seq analysis also provided some initial insight into the progression of leukemia in this mouse model as we detected different signatures of early stage B cells in accumulated preleukemic dHet cells in the bone marrow and the majority of leukemic B-ALL cells in the spleen and blood. In cluster 1 of the major leukemic cell population, we detected cells with a signature of NK and CD8+ T cells (Klrb1c and Klrd1 expression), consistent with the previous observation that Ebf1/Pax5 dHet pro-B cells can be converted into T cells by providing Notch signals in vitro. These in vitro converted T cells maintain a leukemic state upon transplantation into recipient mice (Ungerbäck et al. 2015; Somasundaram et al. 2016). Thus, the Ebf1/Pax5 heterozygosity may allow for changes in cell identity during disease progression in vivo. However, the presence of late and early stage B-ALL cells in leukemic dHet mice is likely due to different developmental arrests.
In conclusion, the combined heterozygosity of Ebf1 and Pax5 results in altered STAT5- and Myc-centered regulatory networks that may facilitate leukemic transformation by up-regulating the glycolytic and folate metabolism and by keeping B lymphoid cells in an IL7-driven progenitor stage with some plasticity of cell identity.
Materials and methods
Mice
Ebf1+/−Pax5+/− double-heterozygous mice were obtained by crossing Ebf1+/− and Pax5+/− mice (Urbanek et al. 1994; Lin and Grosschedl 1995). The mice were maintained under the specific pathogen-free conditions in the animal facility of Max Planck Institute of Immunology and Epigenetics. All mouse experiments were carried out in accordance with the guidelines of the Federation of European Laboratory Animal Science Association (FELASA) and following legal approval of the Animal Committee in Freiburg, Germany.
Flow cytometry and adoptive transfers
Single-cell suspensions were subjected to the lysis of red blood cells and the cells were stained with fluorochrome-conjugated antibodies: CD19, AA4.1 (CD93), B220, HSA (CD24), BP-1, and IL-7R (CD127). The stained cells were analyzed by BD LSRFortessa or sorted using BD FACSAria. To detect the apoptotic cells, single-cell suspensions were subjected to the incubation with Annexin V and 7-ADD according to the manufacturer's instruction (BD Bioscience). Data were analyzed by FlowJo software. For adoptive transfers, CD19+ cells were sorted from the spleen of wild-type C57BL/6 mice, while CD19+AA4.1hi cells were sorted from the spleen of Ebf1+/−Pax5+/− mice. Purified cells (1 × 105) were injected intravenously into 4.5-Gy irradiated Rag2−/− mice.
Single-cell RNA-seq
Cells were sorted on a BD Influx cell sorter into 384-well plates. The single-cell RNA-seq was performed as using the CEL-Seq2 protocol as described previously (Hashimshony et al. 2016) but adapted for the use with a nanoliter pipetting robot (mosquito HTS, TTP Labtech). In short, the libraries were sequenced on an Illumina HiSeq 2500 system in high-output run mode at a depth of ∼200,000 reads per cell and analyzed as described previously (Derecka et al. 2020). The analysis by RaceID3 and StemID for for pseudotemporal ordering and inference of gene expression modules is described in the Supplemental Material.
Bioinformatics analysis and data integration
The microarray expression data of the Stat5-CA mouse models were used for overlapping with our dHet B-ALL mouse model (Katerndahl et al. 2017). The raw Affymetrix array data were obtained from NCBI-GEO (GSE25643) and the expression analysis was performed using affylmGUI application. The arrays were background-corrected and RMA-normalized, and differential expression was calculated for each contrast with the linear model fit functions available in the package. The differentially expressed genes of each comparison were filtered with the 0.01 P-value and twofold change cutoff. The up-regulated and down-regulated genes were compared with the ranked list of differentially expressed genes in dHet B-ALL cells using the GSEA tool (Subramanian et al. 2005). The Myc-regulated differentially expressed genes in Eμ-Myc transgenic leukemic mice were obtained from the previously published study, GSE51011 (Sabò et al. 2014). The ranked list of Myc-bound and differentially expressed genes in Eμ-Myc leukemic cells was overlapped with up-regulated and down-regulated genes in dHet B-ALL cells using the GSEA tool. The differential occupancy of EBF1 was calculated using diffBind tool. The peaks identified by the MACS2 tool were merged and used for calculating the differential occupancy of EBF1. The read counts of the peaks for every sample was calculated and normalized using dba.count function. The differential binding analysis was performed for the contrast matrix using dba.analyze function, which internally runs the DESeq2 to identify differentially occupied peaks, with the FDR cut-off 0.05. The overlap of differentially expressed and differentially EBF1-regulated genes was visualized using R visualization tools. A similar approach was followed to identify differentially induced ATAC peaks.
The differentially induced ATAC peaks identified by MACS2 and diffBind were merged and subjected to the de novo motif prediction using Homer (v4.7) tools (Heinz et al. 2010). The de novo motifs were ranked based on the P-value and assigned to the best matching known motif in B cells. The EBF1/PAX5/ATAC peaks were assigned to the nearest TSS of genes within 100-kb distance. The distribution of RPKM-normalized ChIP/ATAC signals, with the bin size 10, around the peaks was visualized using the plotheatmap function available in the deepTools package (Ramírez et al. 2016). The pathway enrichment analysis was performed for the up-regulated and down-regulated genes in the KEGG pathways database of MSigDB. The top 10 up-regulated or down-regulated pathways with the high NES cutoff were ranked based on the enrichment score. The transiently and consistently occupied EBF1 sites were obtained from a previously published study (Li et al. 2018). All of the data sets were normalized to 10 million reads using Homer tools while visualizing each individual locus using R packages Gviz and GenomicRanges (Lawrence et al. 2013). The pediatric childhood B-ALL expression data were obtained from the TARGET initiative (phs000463). The Myc ChIP data of the Eμ-Myctg mice were obtained from GSE51011 and visualized around B-ALL-gained or B-ALL-lost ATAC peaks (Sabò et al. 2014).
Metabolite extraction and metabolite measurement by LC-MS
For metabolite analysis, cells were washed in ice-cold PBS and metabolites were extracted in 100 µL of extraction buffer (50:30:20, methanol:acetonitrile:water) per million cells. Extraction buffer was cooled for 30 min on dry ice beforehand. Samples were centrifuged at maximum speed for 10 min to remove protein debris and stored at −80°C until acquisition.
LC-MS was carried out using an Agilent 1290 Infinity II UHPLC inline with a Bruker Impact II QTOF operating in negative ion mode. Scan range was from 30 to 1050 Da. Mass calibration was performed at the beginning of each run. LC separation was on a Phenomenex Luna propylamine column (50 × 2 mm, 3 µm particles) using a solvent gradient of 100% buffer B (5 mM ammonium carbonate in 90% acetonitrile) to 90% buffer A (10 mM NH4 in water). Flow rate was from 1000 to 750 L/min. Autosampler temperature was 5°C and injection volume was 2 µL.
Data availability
Data sets generated in this study are available as a superseries in the GEO database under accession number GSE158673.
Supplementary Material
Acknowledgments
We thank I. Falk, H.J. Schwarz, D. Komandin, F. Ludin, and J. Curtis for the excellent technical assistance. We thank members of R.G.’s laboratory for discussions, and M. Rott for help with manuscript preparation. We are grateful to J. Büscher for analyzing metabolites. We thank the Bioinformatics, Deep Sequencing, Metabolomics, Imaging, FACS, and Animal Facilities of the Max Planck Institute of Immunobiology and Epigenetics. This work was supported by funds from the Max Planck Society and German Research Foundation. K.K. was supported by the Uehara Memorial Foundation, Japan. D.G. was supported by the German Research Foundation (DFG; GR4980/3-1).
Author contributions: S.R. designed and performed the bioinformatics, data analysis, and integration. K.K. designed and performed cellular and mouse experiments. J.S.H. performed single-cell RNA-seq analysis. M.B. performed cellular and immunoblot experiments. S.B. and R.L. conducted ChIP-seq and ATAC-seq. H.R., J.E.-H., and R.K.-G. performed the metabolomics. E.L.P. supervised J.E.-H. and R.K.-G. D.G. supervised J.S.H. R.G. conceived and supervised the study. R.G. and S.R. wrote the manuscript with input from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Artilce and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.340216.120.
Freely available online through the Genes & Development Open Access option.
References
- Åhsberg J, Ungerbäck J, Strid T, Welinder E, Stjernberg J, Larsson M, Qian H, Sigvardsson M. 2013. Early B-cell factor 1 regulates the expansion of B-cell progenitors in a dose-dependent manner. J Biol Chem 288: 33449–33461. 10.1074/jbc.M113.506261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenzana TL, Schjerven H, Smale ST. 2015. Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev 29: 1801–1816. 10.1101/gad.266999.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi K, Nosaka T, Yamada K, Onishi M, Oka Y, Miyajima A, Kitamura T. 2000. Constitutive activation of STAT5 by a point mutation in the SH2 domain. J Biol Chem 275: 24407–24413. 10.1074/jbc.M909771199 [DOI] [PubMed] [Google Scholar]
- Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, et al. 2018. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553: 515–520. 10.1038/nature25193 [DOI] [PubMed] [Google Scholar]
- Boller S, Grosschedl R. 2014. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol Rev 261: 102–115. 10.1111/imr.12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller S, Ramamoorthy S, Akbas D, Nechanitzky R, Burger L, Murr R, Schübeler D, Grosschedl R. 2016. Pioneering activity of the C-terminal domain of EBF1 shapes the chromatin landscape for B cell programming. Immunity 44: 527–541. 10.1016/j.immuni.2016.02.021 [DOI] [PubMed] [Google Scholar]
- Chan LN, Müschen M. 2017. B-cell identity as a metabolic barrier against malignant transformation. Exp Hematol 53: 1–6. 10.1016/j.exphem.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MR, Mandal M, Ochiai K, Singh H. 2014. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat Rev Immunol 14: 69–80. 10.1038/nri3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M. 2007. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature 449: 473–477. 10.1038/nature06159 [DOI] [PubMed] [Google Scholar]
- Cooper AB, Sawai CM, Sicinska E, Powers SE, Sicinski P, Clark MR, Aifantis I. 2006. A unique function for cyclin D3 in early B cell development. Nat Immunol 7: 489–497. 10.1038/ni1324 [DOI] [PubMed] [Google Scholar]
- Derecka M, Herman JS, Cauchy P, Ramamoorthy S, Lupar E, Grün D, Grosschedl R. 2020. EBF1-deficient bone marrow stroma elicits persistent changes in HSC potential. Nat Immunol 21: 261–273. 10.1038/s41590-020-0595-7 [DOI] [PubMed] [Google Scholar]
- Ferreirós-Vidal I, Carroll T, Zhang T, Lagani V, Ramirez RN, Ing-Simmons E, Gómez-Valadés AG, Cooper L, Liang Z, Papoutsoglou G, et al. 2019. Feedforward regulation of Myc coordinates lineage-specific with housekeeping gene expression during B cell progenitor cell differentiation. PLoS Biol 17: e2006506 10.1371/journal.pbio.2006506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S, Goloviznina NA, Chen WY, Huan J, LaTocha D, et al. 2015. Self-enforcing feedback activation between BCL6 and pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell 27: 409–425. 10.1016/j.ccell.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Muraro MJ, Boisset JC, Wiebrands K, Lyubimova A, Dharmadhikari G, van den Born M, van Es J, Jansen E, Clevers H, et al. 2016. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19: 266–277. 10.1016/j.stem.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RR, Kincade PW, Dorshkind K. 2007. The protean nature of cells in the B lymphocyte lineage. Immunity 26: 703–714. 10.1016/j.immuni.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. 1988. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med 167: 353–371. 10.1084/jem.167.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, Gennert D, Li S, Livak KJ, Rozenblatt-Rosen O, et al. 2016. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-seq. Genome Biol 17: 77 10.1186/s13059-016-0938-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Kunisada T, Ogawa M, Sudo T, Kodama H, Suda T, Nishikawa S, Nishikawa S. 1990. Stepwise progression of B lineage differentiation supported by interleukin 7 and other stromal cell molecules. J Exp Med 171: 1683–1695. 10.1084/jem.171.5.1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N, Thomas DA, Koeuth T, Baechler EC, Kornblau SM, et al. 2011. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med 208: 1135–1149. 10.1084/jem.20101947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JS, Sagar, Grün D. 2018. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat Methods 15: 379–386. 10.1038/nmeth.4662 [DOI] [PubMed] [Google Scholar]
- Herzog S, Reth M, Jumaa H. 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol 9: 195–205. 10.1038/nri2491 [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhang Z, Kashiwagi M, Yoshida T, Joshi I, Jena N, Somasundaram R, Emmanuel AO, Sigvardsson M, Fitamant J, et al. 2016. Superenhancer reprogramming drives a B-cell–epithelial transition and high-risk leukemia. Genes Dev 30: 1971–1990. 10.1101/gad.283762.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yoshida T, Georgopoulos K. 2017. Transcriptional circuits in B cell transformation. Curr Opin Hematol 24: 345–352. 10.1097/MOH.0000000000000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi I, Yoshida T, Jena N, Qi X, Zhang J, Van Etten RA, Georgopoulos K. 2014. Loss of ikaros DNA-binding function confers integrin-dependent survival on pre-B cells and progression to acute lymphoblastic leukemia. Nat Immunol 15: 294–304. 10.1038/ni.2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kager L, Cheok M, Yang W, Zaza G, Cheng Q, Panetta JC, Pui CH, Downing JR, Relling MV, Evans WE. 2005. Folate pathway gene expression differs in subtypes of acute lymphoblastic leukemia and influences methotrexate pharmacodynamics. J Clin Invest 115: 110–117. 10.1172/JCI22477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katerndahl CDS, Heltemes-Harris LM, Willette MJL, Henzler CM, Frietze S, Yang R, Schjerven H, Silverstein KAT, Ramsey LB, Hubbard G, et al. 2017. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat Immunol 18: 694–704. 10.1038/ni.3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Cauchy P, Ramamoorthy S, Boller S, Chavez L, Grosschedl R. 2018. Dynamic EBF1 occupancy directs sequential epigenetic and transcriptional events in B-cell programming. Genes Dev 32: 96–111. 10.1101/gad.309583.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Grosschedl R. 1995. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376: 263–267. 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. 2010. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643. 10.1038/ni.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukin K, Fields S, Lopez D, Cherrier M, Ternyak K, Ramírez J, Feeney AJ, Hagman J. 2010. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc Natl Acad Sci 107: 7869–7874. 10.1073/pnas.1003525107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, et al. 2004. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol 5: 1069–1077. 10.1038/ni1119 [DOI] [PubMed] [Google Scholar]
- Mansson R, Welinder E, Ahsberg J, Lin YC, Benner C, Glass CK, Lucas JS, Sigvardsson M, Murre C. 2012. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci 109: 21028–21033. 10.1073/pnas.1211427109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Neretti N, Sedivy JM, Hockenbery DM. 2008. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle 7: 1054–1066. 10.4161/cc.7.8.5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. 2009. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene 28: 2485–2491. 10.1038/onc.2009.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764. 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of ikaros. Nature 453: 110–114. 10.1038/nature06866 [DOI] [PubMed] [Google Scholar]
- Nahar R, Ramezani-Rad P, Mossner M, Duy C, Cerchietti L, Geng H, Dovat S, Jumaa H, Ye BH, Melnick A, et al. 2011. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood 118: 4174–4178. 10.1182/blood-2011-01-331181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechanitzky R, Akbas D, Scherer S, Györy I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R. 2013. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol 14: 867–875. 10.1038/ni.2641 [DOI] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Zhang J, Georgopoulos K. 2009. Genome-wide lineage-specific transcriptional networks underscore ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity 30: 493–507. 10.1016/j.immuni.2009.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O'Connell B, Petrenko O, Kotenko I, Beavis A, Sedivy JM, Cole MD. 2002. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol 22: 5793–5800. 10.1128/MCB.22.16.5793-5800.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Kee BL. 2007. The transcriptional regulation of B cell lineage commitment. Immunity 26: 715–725. 10.1016/j.immuni.2007.05.010 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401: 556–562. 10.1038/44076 [DOI] [PubMed] [Google Scholar]
- Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. 2012. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol 13: 300–307. 10.1038/ni.2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okosun J, Bödör C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 46: 176–181. 10.1038/ng.2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Riordan M, Grosschedl R. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11: 21–31. 10.1016/S1074-7613(00)80078-3 [DOI] [PubMed] [Google Scholar]
- Pang SH, Minnich M, Gangatirkar P, Zheng Z, Ebert A, Song G, Dickins RA, Corcoran LM, Mullighan CG, Busslinger M, et al. 2016. PU.1 cooperates with IRF4 and IRF8 to suppress pre-B-cell leukemia. Leukemia 30: 1375–1387. 10.1038/leu.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol 9: 203–215. 10.1038/ni1555 [DOI] [PubMed] [Google Scholar]
- Prasad MA, Ungerbäck J, Åhsberg J, Somasundaram R, Strid T, Larsson M, Månsson R, De Paepe A, Lilljebjörn H, Fioretos T, et al. 2015. Ebf1 heterozygosity results in increased DNA damage in pro-B cells and their synergistic transformation by Pax5 haploinsufficiency. Blood 125: 4052–4059. 10.1182/blood-2014-12-617282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. Deeptools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla IDR, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, Smeenk L, Trupke J, Sommer A, Jaritz M, et al. 2012. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J 31: 3130–3146. 10.1038/emboj.2012.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC. 2013. New insights into pre-BCR and BCR signalling with relevance to B cell malignancies. Nat Rev Immunol 13: 578–591. 10.1038/nri3487 [DOI] [PubMed] [Google Scholar]
- Roberts KG, Mullighan CG. 2019. The biology of B-progenitor acute lymphoblastic leukemia. Cold Spring Harb Perspect Med 10: a034835 10.1101/cshperspect.a034835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Györy I, Imhof S, Spivakov M, Williams RR, Busslinger M, Fisher AG, Grosschedl R. 2007. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol 27: 579–594. 10.1128/MCB.01192-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabò A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, Morelli MJ, Bora P, Doni M, Verrecchia A, et al. 2014. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511: 488–492. 10.1038/nature13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjerven H, Ayongaba EF, Aghajanirefah A, McLaughlin J, Cheng D, Geng H, Boyd JR, Eggesbø LM, Lindeman I, Heath JL, et al. 2017. Genetic analysis of Ikaros target genes and tumor suppressor function in BCR-ABL1+ pre-B ALL. J Exp Med 214: 793–814. 10.1084/jem.20160049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Tagoh H, Gültekin S, Dakic A, Axelsson E, Minnich M, Ebert A, Werner B, Roth M, Cimmino L, et al. 2014. Stage-specific control of early B cell development by the transcription factor Ikaros. Nat Immunol 15: 283–293. 10.1038/ni.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C, Wechsler J, Yang J, Hayes J, Klein RJ, et al. 2013. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 45: 1226–1231. 10.1038/ng.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M. 2018. Molecular regulation of differentiation in early B-lymphocyte development. Int J Mol Sci 19: 1928 10.3390/ijms19071928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Pongubala JM, Medina KL. 2007. Gene regulatory networks that orchestrate the development of B lymphocyte precursors. Adv Exp Med Biol 596: 57–62. 10.1007/0-387-46530-8_5 [DOI] [PubMed] [Google Scholar]
- Somasundaram R, Prasad MA, Ungerbäck J, Sigvardsson M. 2015. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood 126: 144–152. 10.1182/blood-2014-12-575688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaram R, Åhsberg J, Okuyama K, Ungerbäck J, Lilljebjörn H, Fioretos T, Strid T, Sigvardsson M. 2016. Clonal conversion of B lymphoid leukemia reveals cross-lineage transfer of malignant states. Genes Dev 30: 2486–2499. 10.1101/gad.285536.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesi A, de Pretis S, Furlan M, Filipuzzi M, Morelli MJ, Andronache A, Doni M, Verrecchia A, Pelizzola M, Amati B, et al. 2019. An early Myc-dependent transcriptional program orchestrates cell growth during B-cell activation. EMBO Rep 20: e47987 10.15252/embr.201947987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, et al. 2007. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity 26: 335–344. 10.1016/j.immuni.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET, Grosschedl R. 2010. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity 32: 714–725. 10.1016/j.immuni.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Ungerbäck J, Åhsberg J, Strid T, Somasundaram R, Sigvardsson M. 2015. Combined heterozygous loss of Ebf1 and Pax5 allows for T-lineage conversion of B cell progenitors. J Exp Med 212: 1109–1123. 10.1084/jem.20132100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79: 901–912. 10.1016/0092-8674(94)90079-5 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilagos B, Hoffmann M, Souabni A, Sun Q, Werner B, Medvedovic J, Bilic I, Minnich M, Axelsson E, Jaritz M, et al. 2012. Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med 209: 775–792. 10.1084/jem.20112422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski MT, Hu Y, Roberts KG, Boer JM, McKenzie MD, Liu GJ, Le Grice OD, Tremblay CS, Ghisi M, Willson TA, et al. 2017. Conserved IKAROS-regulated genes associated with B-progenitor acute lymphoblastic leukemia outcome. J Exp Med 214: 773–791. 10.1084/jem.20160048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G, Chan LN, Klemm L, Braas D, Chen Z, Geng H, Zhang QC, Aghajanirefah A, Cosgun KN, Sadras T, et al. 2018. B-Cell-Specific diversion of glucose carbon utilization reveals a unique vulnerability in B cell malignancies. Cell 173: 470–484.e18. 10.1016/j.cell.2018.02.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. 2008. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol 181: 3364–3372. 10.4049/jimmunol.181.5.3364 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sets generated in this study are available as a superseries in the GEO database under accession number GSE158673.