In this study, Daniel et al. examined the mechanisms of macrophage polarization in general and alternative polarization in particular. Using unbiased epigenomic, molecular biology, and genetic GOF and LOF approaches, the authors show that the zinc finger transcription factor EGR2 acts as an essential and evolutionarily conserved broad-acting factor, linking transient polarization signals to stable epigenomic and transcriptional changes in macrophages.
Keywords: EGR2, IL-4, macrophage polarization, epigenomic regulation, transcription factor network
Abstract
Macrophages polarize into functionally distinct subtypes while responding to microenvironmental cues. The identity of proximal transcription factors (TFs) downstream from the polarization signals are known, but their activity is typically transient, failing to explain the long-term, stable epigenomic programs developed. Here, we mapped the early and late epigenomic changes of interleukin-4 (IL-4)-induced alternative macrophage polarization. We identified the TF, early growth response 2 (EGR2), bridging the early transient and late stable gene expression program of polarization. EGR2 is a direct target of IL-4-activated STAT6, having broad action indispensable for 77% of the induced gene signature of alternative polarization, including its autoregulation and a robust, downstream TF cascade involving PPARG. Mechanistically, EGR2 binding results in chromatin opening and the recruitment of chromatin remodelers and RNA polymerase II. Egr2 induction is evolutionarily conserved during alternative polarization of mouse and human macrophages. In the context of tissue resident macrophages, Egr2 expression is most prominent in the lung of a variety of species. Thus, EGR2 is an example of an essential and evolutionarily conserved broad acting factor, linking transient polarization signals to stable epigenomic and transcriptional changes in macrophages.
Macrophages (MFs) are integral parts of innate immunity and represent the first line of defense against invading pathogens. These activities require plasticity and rapid responses to environmental cues, leading to the deployment of epigenomic mechanisms resulting in specific and stable gene expression programs. MF gene expression is thought to be regulated at three levels. The first level is a permanent program established by the developmental regulation and is maintained by lineage-determining transcription factors (LDTFs) like PU.1, CEBP, and AP-1. The second level is an adaptation to a given microenvironment brought about by distinct tissue-specific polarization signals (Amit et al. 2016; Guilliams et al. 2020) modulating TFs’ activity (Bonnardel et al. 2019); i.e., singular polarizing cytokines such as IL-4 or IL-13 via signal transducer and activator of transcription 6 (STAT6). The third level of regulation takes place transiently upon pathogen encounter or inflammatory stimulation (Glass and Natoli 2016). Therefore, MF polarization is part of normal physiologic processes, actively contributing to controlling infection, cancer, and inflammatory disease progression (Lawrence and Natoli 2011). Consequently, investigation of the polarization process became an intensely researched topic in immunology with therapeutic ramifications.
The two end points of the contiguous spectrum of macrophage polarization are the classical polarization of macrophages (M1 MFs) via TLR or inflammatory cytokine signaling such as interferon γ or at the opposite end of the spectrum, alternative polarization (M2 MFs) induced by IL-4 or IL-13 (Murray et al. 2014). These two models are widely used to learn about the molecular events of polarization of MFs (Murray et al. 2014). Functionally, classically polarized MFs are characterized by high antimicrobial activities critical for host defense, while alternatively polarized MFs exhibit a characteristically more anti-inflammatory profile contributing to tissue-regeneration, tissue-remodeling, and the resolution of inflammation (Shapouri-Moghaddam et al. 2018). The in vivo roles of alternatively polarized MFs are highlighted in alveolar MFs in the steady-state and different pathological conditions including asthma and allergies, parasite infection, fibrosis, and cancer (Gordon and Martinez 2010; Cohen et al. 2018) . The cytokines IL-4 or IL-13 are the initiators of this stable gene expression program and cellular phenotype in its purest form and phosphorylated STAT6 acts as a downstream immediate transcriptional regulator. STAT6 is required for alternative polarization by IL-4 in bone marrow-derived macrophages (BMDM) and in vivo to mount effective antihelminth defense (Rückerl and Allen 2014). However, the requirements and contribution of myeloid or macrophage STAT6 are not as well understood due to the lack of genetic evidence establishing MF cell-autonomous function, as well as establishing the contribution of the various aspects of Th2 immune responses. Thus, BMDM represents the gold standard model to study the epigenomic and molecular details of polarization.
The TFs PU.1, AP-1, C/EBP, and IRF are all critical for MF development and function, establishing the MF-specific regulatory element landscape (lineage identity) (Heinz et al. 2015). Moreover, TF binding at low accessible genomic regions (labeled regulatory regions) also contributes to lineage identity in macrophages (Horvath et al. 2019), and form a molecular landing strip for signal-dependent transcription factors (SDTFs). These genomic loci are readily used during the initiation of polarization and confer plasticity. The early steps of alternative MF polarization have been studied, and the role of de novo/latent enhancers, mediating new cellular functions as well as providing cellular memory, has been established (Heinz et al. 2015). This is triggered by STAT6 homodimers that act in a rapid and transient fashion. DNA binding of STAT6 alters the expression of hundreds of genes within the first few hours of polarization, including several TFs. Due to its transient mechanism of action, the majority of STAT6 is released from the chromatin after 24 h of cytokine exposure (Czimmerer et al. 2018). Therefore, the polarization program enters into a self-sustained, largely STAT6-independent stable state. This observation leads to a so-far unresolved conundrum on how transiently acting STAT6 induces a stable and sustained polarization program.
Several STAT6-induced TFs have been described as essential regulators of the alternative MF phenotype, including the peroxisome proliferator-activated receptor γ (PPARG) (Odegaard et al. 2007), KLF4 (Liao et al. 2011), IRF4 (Satoh et al. 2010), MYC (Pello et al. 2012), and more recently BHLHE40 (Jarjour et al. 2019). The latter is reported to specifically control the proliferation of large peritoneal MFs when IL-4 is abundant (Jarjour et al. 2019). Our recent study suggests that the nuclear receptor PPARG is a late regulator of the alternative polarization program via ligand-regulated gene expression and transcriptional memory. Importantly, neither of these TFs possess widespread regulatory roles when MFs transit from the early to late polarization state (Daniel et al. 2018a).
Although the listed TFs are induced by STAT6 and their expression is sustained beyond STAT6's presence, their action is highly specific, controlling very confined parts of the broad polarization program. Therefore, there is a hiatus in our understanding, leading to the proposition that broader epigenomic mechanism(s) and/or factor(s) directly controlled by STAT6 must exist, acting as molecular linchpin(s) to link the early transient and late stable molecular events of polarization.
We took an unbiased systematic approach in the search for such epigenomic regulators, integrating both the mapping of genome activity patterns and de novo motif analysis. Using the combination of P300 (general coactivator with histone acetyltransferase activity) and H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq), we defined the early and late genome activity patterns of alternative polarization in BMDMs. Our analyses, which included genetic gain and loss-of-function studies, identified the TF EGR2 acting as a molecular linchpin between IL-4-activated STAT6 and a downstream stable transcriptional network of polarization.
Results
Mapping the temporal genome activity patterns of alternative MF polarization
In order to obtain robust and unbiased data sets to map the early transient and late stable events of alternative polarization we generated a time course of epigenomic, cistromic, and transcriptomic changes. We used murine bone marrow-derived macrophages (BMDMs referred to as MFs) and IL-4-mediated MF polarization as a model. Mature, unstimulated MFs received IL-4 for 1 h (short-term polarization) and 24 h (long-term polarization) to reveal early and late genome activities. We devised an approach that maps significant changes in P300 binding and its correlation with H3K27ac levels (histone modification correlating with P300 activity), using ChIP-seq to map active enhancers (Creyghton et al. 2010).
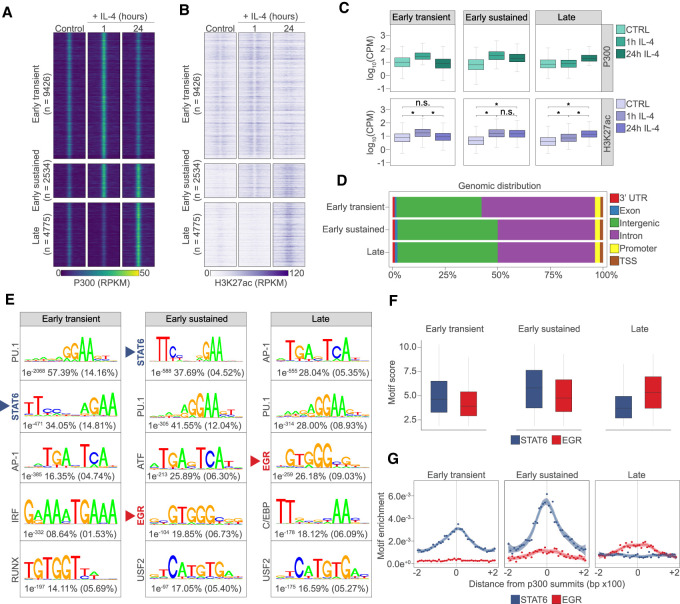
First, we identified genomic regions with differential P300 and H3K27ac levels followed by de novo motif discovery in the underlying genomic regions. This approach revealed the activated (n = 16,735) and repressed (n = 17,582) regulatory regions of polarization, while 14,353 sites showed no change (Fig. 1A; Supplemental Fig. S1A; Supplemental Table S1). Clustering the activity patterns of regulatory elements yielded three activated and three repressed groups. Activated regulatory elements exhibited “early transient” (activated at 1 h and lost activity by 24 h; n = 9426), “early sustained” (activated at 1 h and retained activity at 24 h; n = 2534), and “late” genomic activities (activated only by 24 h; n = 4775). Repressed regulatory elements followed very similar patterns by featuring “transiently” repressed (repressed at 1 h, but relieved of repression by 24 h; n = 12,496), “early” repressed (n = 1346) and “late” repressed genome activity sites (n = 3740) (Fig. 1A–C; Supplemental Fig. S1A,B). The majority of regulatory elements exhibiting increased genome activities were located on intergenic and intronic regions with marginal differences between the different categories (Fig. 1D), while a significant portion (∼40%) of transiently repressed genomic regions occurred at promoters (Supplemental Fig. S1C).These analyses uncovered the dynamics of the epigenomic changes, cataloged the immediate early and long-term, sustained genome activities of alternative MF polarization and prompted us to seek the drivers of the observed changes.
Figure 1.
Regulatory elements with distinct temporal genome activity patterns identify and link the EGR motifs to late genome activities. (A) Read distribution plot of P300 binding in the three groups showing distinct genome activity pattern in control and IL-4 polarized macrophage for the indicated periods of time. Results are represented in RPKM (reads per kilobase per million mapped reads) values. (B) Read distribution plot of H3K27ac signal in the three induced genome activity pattern groups in control and IL-4 polarized macrophage for the indicated periods of time. Signals are visualized around P300 summits. RPKM values are plotted. (C) Average binding signal for P300 and H3K27ac are represented as box plots on the genomic regions showing induced genome activity patterns. Average log10 CPM (counts per million mapped reads) values are plotted. Significant changes are determined with Wilcoxon test at P < 0.05. (D) Genomic distribution of the regulatory elements exhibiting distinct genome activity pattern. (E) Motif enrichment plots on the regulatory regions exhibiting different genome activity patterns. For each motif logo, P-value and percentage of genomic regions that contain the given motif in the target (P300-bound) and background genomic regions (in parenthesis) are shown. (F) Box plot representation of STAT6 and EGR motif scores on the induced genome activity pattern groups. Significant changes are determined with Wilcoxon test at P < 0.05. (G) Distribution of the enriched STAT6 and EGR motifs around the identified P300 summits are shown for the induced genome activity pattern groups in a ±200-bp window. Shaded areas represent the 95% confidence interval for LOESS smoothed means.
Late and sustained genome activity patterns associate with the binding motif of the EGR transcription factor family
In order to identify candidate TFs mediating differential genome activity patterns, we turned our attention to the sequence determinants of the genomic regions exhibiting changes. Motif enrichment analysis identified the TF motifs associated with MF-specific regulatory regions across the three different genome activity groups (e.g. PU-1, AP-1, IRF, RUNX, and C/EBP) (Horvath et al. 2019). We could confirm that genomic regions of early activation (“early transient” and “early sustained”) were enriched for the STAT6 TF-binding motif, the initiator of the polarization process. Importantly, we observed the enrichment of novel TF motifs that have not been linked to genome activation in this model and were largely specific to genomic regions with sustained activity such as EGR and USF2 (Fig. 1E). Among these, EGR motifs were enriched specifically in the “early sustained” and “late” genome activity groups, indicating the potential importance of the respective TF family at these genomic regions (Fig. 1F,G). However, EGR motifs were not revealed using the enrichment analysis at repressed sites (Supplemental Fig. S1D), supervised motif score analysis at “transiently” repressed genomic regions uncovered similarly strong EGR motifs to the ones detected at “late” induced sites (Supplemental Fig. S1E). Interestingly, “transiently” repressed regions exhibited rapid loss of P300 and H3K27ac at 1 h but genome activity returned to the basal level by 24 h (Supplemental Fig. S1A). Altogether, this is indicative of EGR TFs being transcriptional activators of the late polarization program and may contribute to repression via indirect mechanisms. Prompted by these results, we decided to embark on evaluating the roles of EGRs in genome activation during alternative macrophage polarization.
EGR2 is a direct IL-4/STAT6 target with an expansive enhancer network
Our analyses thus identified an enrichment for the EGR motif at polarization-induced regulatory regions and predicted roles for this TF family in the late molecular program of polarization. The EGR TF family consists of four members with homologous DNA-binding domains (Poirier et al. 2007). Our most recent transcriptome-wide studies demonstrated the induced expression of Egr2 along with the marker genes (Retnla, Chil3, and Arg1) of alternative polarization (Czimmerer et al. 2018; Daniel et al. 2018b). Importantly, other EGR family members (Egr1, Egr3, and Egr4) were barely expressed and IL-4 had no impact on their expression (Supplemental Fig. S2A).
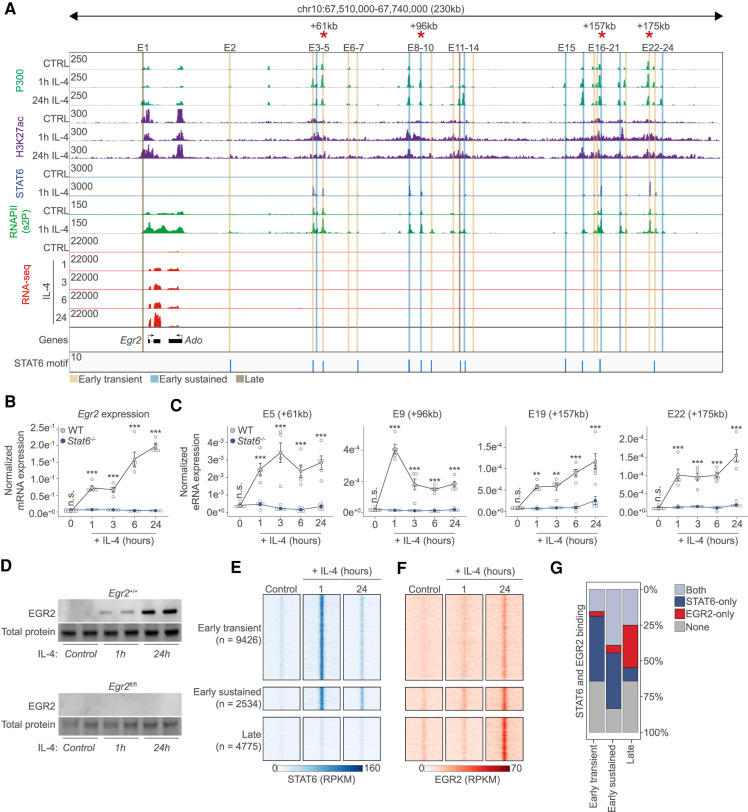
The Egr2 transcript was below our detection limit in unstimulated cells; however, it showed rapidly induced and sustained mRNA levels in the presence of IL-4. Thus, we took a closer look at the Egr2 locus utilizing ChIP-seq data for P300, H3K27ac, STAT6, and RNAPII-pS2 (elongating RNA polymerase II), which informed us about the existence of an enhancer cluster of ∼23 putative enhancer regions downstream from the gene and one enhancer located upstream (E1) (Fig. 2A; Supplemental Table S2). These putative enhancer regions contained STAT6 motifs (Fig. 2A) and exhibited slightly different genome activity patterns and STAT6 occupancies, with a few of them showing “late,” while most of them showed “early transient” and “early sustained” activities (presented in Figs. 1, 2A). Using Stat6−/− MFs we could demonstrate that Egr2 is an IL-4 and Stat6-dependent gene using a time-course experiment (1-, 3-, 6-, and 24-h-long exposure) (Fig. 2B). Also, we selected four distant enhancer regions (+61, +96, +157, and +175 kb) bound by STAT6 and having increased P300, H3K27ac, and RNAPII signals after 1-h IL-4 stimulation for validation purposes. Enhancer RNA synthesis, as a marker of enhancer activity, was rapidly induced at 1 h and maintained the induced levels over the time course in a strictly IL-4- and Stat6-dependent manner (Fig. 2C). Thus, Egr2 is an immediate early target of the IL-4/STAT6-activated transcriptional program and harbors an elaborate STAT6-bound enhancer network spanning almost 200 kb.
Figure 2.
The transcription factor EGR2 is a direct STAT6 target. (A) Genome browser view on the Egr2 locus. ChIP-seq results for P300, H3K27ac, STAT6, and RNAPII (s2P [2-phosphoserine form]) is shown in control (CTRL) and IL-4 polarized macrophages. RNA-seq results are also shown on the indicated IL-4 time course. Putative enhancers are highlighted (E1–E24) and their genome activity pattern is indicated as determined in Figure 1. Red asterisks indicate enhancers for which enhancer RNA levels are measured in C. (B) RT-qPCR measurements of the Egr2 mRNA levels on the indicated time course from wild-type (WT) and Stat6−/− macrophages. The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (C) RT-qPCR measurements of enhancer RNA levels on four distant enhancers over the indicated time course from WT and Stat6−/− macrophages. Experiments were repeated five times and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (D) Western blot of EGR2 expression in wild-type and Egr2fl/fl macrophages in the presence or absence of IL-4. Experiments were repeated four times. One representative blot is shown with duplicate samples. Total protein serves as a loading control. (E) Read distribution plot of STAT6 binding in the three induced genome activity pattern groups in control and IL-4 polarized macrophage for the indicated periods of time in a 2 kb window around the P300 summits. RPKM (reads per kilobase per million mapped reads) values are plotted. (F), Read distribution plot of EGR2 binding in the three induced genome activity pattern groups in control and IL-4 polarized macrophage for the indicated periods of time in a 2-kb window around the P300 summits. RPKM (reads per kilobase per million mapped reads) values are plotted. (G) Stacked bar plot representation of the cobinding properties of STAT6 and EGR2 in the three regulatory element groups showing induced genome activity patterns.
Polarization-induced EGR2 populates the macrophage genome largely avoiding the STAT6 cistrome due to its distinct temporal dynamics
The regulation and expression level of Egr2 are indicative of robust and long-term regulatory roles during the polarization process. In order to evaluate the protein levels and IL-4-mediated induction, we carried out Western blot analyses. EGR2 protein was not detectable in control macrophages, but IL-4 exposure led to robust induction (Fig. 2D; Supplemental Fig. S2B). Thus, we set out to generate a genome-wide map of EGR2-bound genomic regions after 1 and 24 h of IL-4 polarization using ChIP-seq analysis (referred to as the EGR2 cistrome). We performed a comparative analysis with the STAT6 cistrome in the same experimental system using our published data sets (Czimmerer et al. 2018). In unstimulated MFs, negligible binding was detected for both STAT6 and EGR2 (Supplemental Fig. S2C,D). As we and others reported previously, STAT6 rapidly populated the MF genome as early as 1 h after IL-4 exposure (∼21,000 binding sites) (Ostuni et al. 2013; Czimmerer et al. 2018). EGR2 binding was also measurable at this time point (∼10,000) binding sites), although very low binding signals could be detected. After 24 h, the STAT6 cistrome diminished substantially, while the EGR2 cistrome went through a remarkable expansion and binding signal increase representing close to ∼30,000 binding sites at this late time point (Supplemental Fig. S2B,C). These results show that there is temporal separation between the STAT6 and EGR2 cistromes. Side by side comparison of the STAT6 and EGR2 cistromes on the genomic regions having differential genome activities documented on Figure 1 also indicated strong spatial separation. Characteristically, STAT6 occupied sites with “early transient,” while EGR2 exhibited stronger binding signals at “early sustained” and “late” genome activity sites with very little overlap (Fig. 2E–G). The genomic regions bound by both factors belonged to “early sustained” genome activity regions, but even there, binding was temporally separated (Fig. 2E–G). These results clearly show that there is spatial and temporal separation between the TF cistromes, indicating the existence of an EGR2 dominated, late polarization program and making it unlikely that EGR2 acts as a collaborator factor of STAT6.
The IL-4/STAT6/EGR2 axis is required to regulate gene expression in alternatively polarized MFs
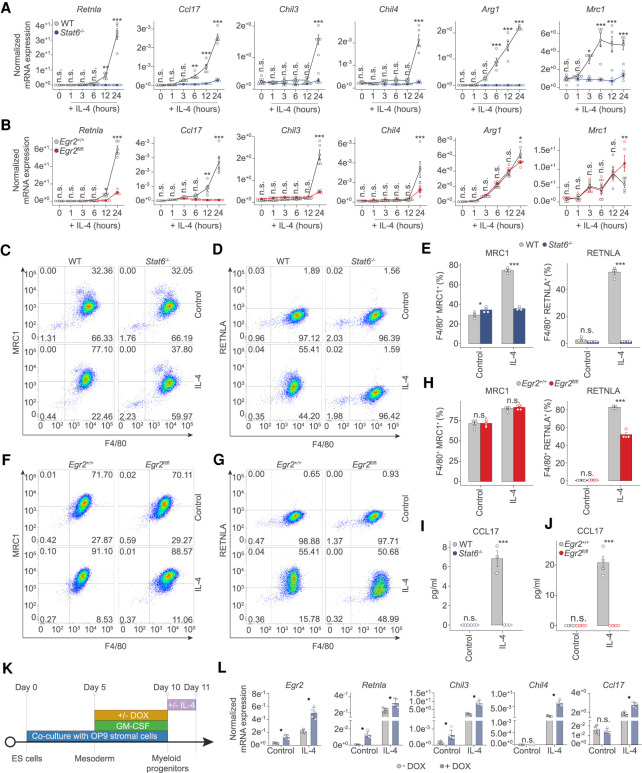
In order to probe whether the IL-4/STAT6/EGR2 axis is indeed functional in MFs, we performed a side-by-side comparison using MFs differentiated from the bone marrow of Stat6−/− (full-body knockout) and Egr2-deficient (LysM-Cre Egr2fl/fl [conditional knockout, referred to as Egr2fl/fl]) mice. We show that Egr2fl/fl MFs are devoid of Egr2 (Supplemental Fig. S3A). Furthermore, neither Stat6−/− nor Egr2fl/fl MFs were different from wild type regarding the expression of MF cell surface markers (ADGRE1: F4/80 and macrophage mannose receptor 1 [MRC1]) (Fig. 3C,F, control condition; Supplemental Fig. S3B–E) in the unstimulated state, thus representing fully mature cells. After a general characterization of the unstimulated MF state, we assessed the mRNA expression of a selected set of alternative polarization marker genes (Retnla, Ccl17, Chil3, Chil4, Arg1, and Mrc1) using samples collected at six different time points (1, 3, 6, 12, and 24 h) following IL-4 treatment. In the absence of Stat6, all six genes had abrogated response to IL-4 (Fig. 3A). Of these, Ccl17 showed total, Retnla, Chil3, and Chil4 close to absolute dependence on Egr2, while the induction of Arg1 and Mrc1 was independent of Egr2 (Fig. 3B). Next, we chose one representative gene from each of these classes and determined protein expression. Using flow cytometry, we studied the protein expression of the phagocytic receptor MRC1 and RETNLA (Resisitin-like molecule α) with roles in the resolution of inflammation as a secreted protein (Nair et al. 2009). Both MRC1 and RETNLA displayed IL-4/STAT6-dependent expression (Fig. 3C–E). Loss of Egr2 did not affect the expression of MRC1, but it was required for the full induction of RETNLA (Fig. 3F–H). Enzyme-linked immunosorbent assay (ELISA) for the CC chemokine CCL17 with roles in autoimmune diseases (Saeki and Tamaki 2006) and pulmonary fibrosis (Belperio et al. 2004) secreted by MFs exhibited an absolute dependence on both Stat6 and Egr2 (Fig. 3I,J). Thus, protein expressions followed changes in mRNA levels in all three cases indicating that MF function is likely to be substantially altered in the absence of Egr2. After establishing that Egr2 is required and necessary for many STAT6-regulated polarization-specific changes we sought to examine whether Egr2 was sufficient to bring about such changes.
Figure 3.
Both STAT6 and EGR2 are required to regulate gene expression during alternative macrophage polarization. (A) RT-qPCR measurements on the indicated marker genes of alternative macrophage polarization from WT and Stat6−/− macrophages over the indicated time course of IL-4 treatment. The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (B) RT-qPCR measurements on the indicated marker genes of alternative macrophage polarization from WT and Egr2fl/fl macrophages over the indicated time course of IL-4 treatment. The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (C) Representative FACS plot of MRC1 (CD206) and ADGRE1 (F4/80) expression in control and IL-4 polarized macrophages from WT and Stat6−/− animals. (D) Representative FACS plot of RETNLA and ADGRE1 (F4/80) expression in control and IL-4 polarized macrophages from WT and Stat6−/− macrophages. (E) Percentages of MRC1 (CD206), ADGRE1 (F4/80) and RETNLA, ADGRE1 (F4/80) double-positive macrophages in control and IL-4 treated conditions from WT and Stat6−/− macrophages. Experiments were repeated three times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (F) Representative FACS plot of MRC1 (CD206) and ADGRE1 (F4/80) expression in control and IL-4 polarized macrophages from Egr2+/+ and Egr2fl/fl animals. (G) Representative FACS plot of RETNLA and ADGRE1 (F4/80) expression in control and IL-4 polarized macrophages from Egr2+/+ and Egr2fl/fl animals. (H) Percentages of MRC1 (CD206), ADGRE1 (F4/80) and RETNLA, ADGRE1 (F4/80) double-positive macrophages in control and IL-4 treated conditions from Egr2+/+ and Egr2fl/fl animals. Experiments were repeated three times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (I,J) CCL17 levels determined by ELISA in control and IL-4-polarized WT, Stat6−/− and Egr2+/+, Egr2fl/fl macrophages. Experiments were performed three times for the Stat6−/−, while four times for the Egr2fl/fl conditions, and significant changes were identified by two-way analysis of variance (ANOVA). (K) Experimental scheme of the gain of function experiments using embryonic stem cells (ES) differentiated towards the myeloid lineage in coculture with OP9 stromal cells, in the presence of GM-CSF. Doxycycline (DOX) was used to induce Egr2 levels at the indicated time before IL-4 treatment (24-h exposure). Cells were harvested for gene expression analysis at day 11. (L) RT-qPCR measurements of the indicated genes from embryonic cell differentiated myeloid cells harboring a doxycycline (DOX)-inducible EGR2 expressing construct. Cells were polarized with IL-4 (24 h) or left untreated (control). The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times and significant changes were identified by unpaired t-test at P < 0.05.
Therefore, we performed gain-of-function experiments on myeloid cells differentiated from embryonic stem cells harboring a doxycycline (DOX)-inducible minigene encoding Egr2 (Fig. 3K; Bencsik et al. 2016). DOX efficiently induced the expression of EGR2, which was further augmented in the presence of IL-4 (Fig. 3L). Overexpression of Egr2 led to induced expression of Retnla and Chil3 even in the absence of IL-4. IL-4 exposure induced the expression of all the measured genes (Egr2, Retnla, Chil3, Chil4, and Ccl17), which was facilitated by the overexpression of Egr2 (Fig. 3L). Therefore, Egr2 is necessary and sufficient to induce the expression of at least a select set of polarization specific marker genes. However, this requirement has a range from none to substantial to absolute, depending on the gene. Thus, we set out to map the extent of this dependence in a genome-wide fashion.
Loss of EGR2 severely impacts the transcriptome of alternative polarization
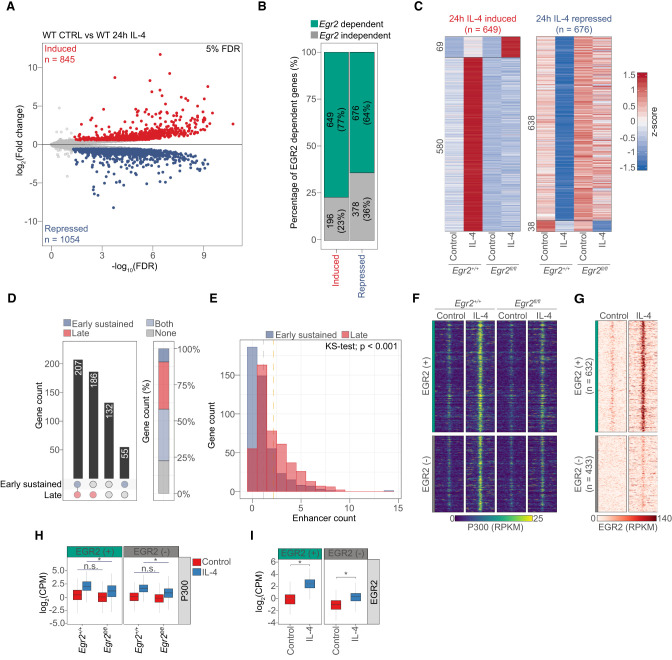
In order to shed light on the extent of the Egr2-dependent late polarization program, we performed RNA-seq experiments in wild-type and Egr2fl/fl macrophages following 24 h of IL-4 polarization. Principal component analysis and clustering of the samples based on Euclidean distance reported good reproducibility, no impact on the unstimulated MF state and the critical roles of IL-4 and EGR2 in bringing about the alternative polarization program (Supplemental Fig. S4A,B). Notably, members of the EGR family (Egr1, Egr3, and Egr4) do not compensate for the loss of Egr2 (Supplemental Fig. S4C). Global gene expression analysis identified 845 induced and 1054 repressed genes after 24 h of IL-4 treatment (Fig. 4A). Importantly, loss of Egr2 affected 77% (649/845) of the induced and 64% (676/1054) of the repressed gene program (n = 3; FDR 5%) (Fig. 4B; Supplemental Table S3). Of the 649 IL-4- and EGR2-induced genes, 580 showed lower or abolished sensitivity to IL-4, while 69 responded to a greater extent upon cytokine treatment in the absence of Egr2. The degree of repression also got diminished on 638 gene loci, while 38 genes showed facilitated repression in the Egr2−/− (Fig. 4C).
Figure 4.
EGR2 controls alternative macrophage polarization. (A) Volcano plot of IL-4-induced and -repressed genes in wild-type (WT) macrophages. (B) Stacked bar plot showing the percentages of the IL-4/Egr2-dependent and -independent induced and repressed genes. (C) Heat map representation of IL-4 altered, Egr2-dependent gene expression. IL-4-induced (left) and IL-4-repressed (right) gene expression is visualized in Egr2+/+ and Egr2fl/fl macrophages. (D) Classification of IL-4-induced and Egr2-dependent genes based on the annotated number of regulatory regions exhibiting early sustained or late genome activity patterns. Gene counts for each bar are represented and the circles below indicate the presence or absence of annotated regulatory elements from the genome activity groups. Percentage-wise distribution of the IL-4/Egr2-dependent genes with enhancer regions of the indicated genome activity patterns (stacked bar plot at the right). (E) Regulatory regions showing early sustained and late genome activity patterns are annotated to IL-4/Egr2-induced genes. The number of regulatory elements (enhancer count) annotated to the genes is shown. Differences between distributions were determined by Kolmogorov–Smirnoff test. (F) Read distribution plot of P300 ChIP-seq signal at EGR2 (+) and EGR2 (−) regions from Egr2+/+ and Egr2fl/fl macrophages in the absence (control) and presence of IL-4 (24 h). (G) Read distribution plot of EGR2 ChIP-seq signal at regulatory regions showing significant increment in P300 binding around IL-4/Egr2-induced genes. Regions of high EGR2-binding intensity [EGR2 (+)] and with negligible binding [EGR2 (−)] are annotated to the target genes based on genomic proximity. RPKM values are plotted. (H) Box plot representation of the average binding intensity of P300 ChIP-seq from the same conditions as presented in F. Log2 transformed CPM (counts per million) values are depicted. Significant changes are determined with Wilcoxon test at P < 0.05. (I) Box plot representation of the average binding intensity of EGR2 at EGR2 (+) and EGR2 (−) regions. Log2 transformed CPM (counts per million) values are depicted. Significant changes are determined with Wilcoxon test at P < 0.05.
Assessing the effects of the loss of Stat6 or Egr2 on the IL-4/EGR2-regulated gene programs revealed gene modules where the linkage of the two TFs can be appreciated using RNA-seq. We observed the following gene modules (all requiring STAT6) (Supplemental Fig. S4D): (1) IL-4/STAT6-induced genes, where EGR2 negatively impacts expression; (2) IL-4/STAT6/EGR2-induced genes; (3) IL-4/STAT6/EGR2-repressed genes; (4) IL-4/STAT6-repressed genes, where EGR2 has restraining activity on repression; (5) IL-4/STAT6-induced genes without EGR2-dependent regulation; and (6) IL-4/STAT6-repressed genes that lack EGR2 dependence. This analysis underlines the significant linkage between IL-4/STAT6 and EGR2, but also establishes that STAT6 is not entirely dependent on EGR2 to induce the late program.
Next, we turned our attention to the putative regulatory regions around the IL-4 and EGR2-induced genes displaying distinct genome activity patterns. Significantly, the majority of the EGR2-induced genes had regulatory elements (±100 kb around the transcription start site) exhibiting “early sustained,” “late,” or both of these in these genome activities (448/580) in agreement with the proposed late and broad stabilizing roles of EGR2. We could not assign such regulatory regions to 132 genes and 55 genes had regulatory elements bearing “early sustained” genome activity (Fig. 4D). The number of regulatory regions annotated to IL-4 and EGR2-induced genes showed that the largest gene set possessed only one regulatory element, and these had a bias towards having “early sustained” genome activity. Similar number of genes could be linked to two regulatory regions associating with slightly more genomic regions of “late” genome activity. This latter trend was, in general, true for all of the genes that had more than two annotated enhancers (Fig. 4E).
Thus, transcriptome analysis revealed the very large extent EGR2 is required for the IL-4 initiated late and stable alternative MF polarization program. Moreover, these genes are associating with genomic regions of “early sustained” and “late” genome activities strongly suggesting that the effect of EGR2 on polarization-specific gene regulation is direct. Therefore, we next wanted to identify the molecular mechanisms by which EGR2 impacts gene expression.
EGR2 induces epigenomic remodeling by direct and indirect means
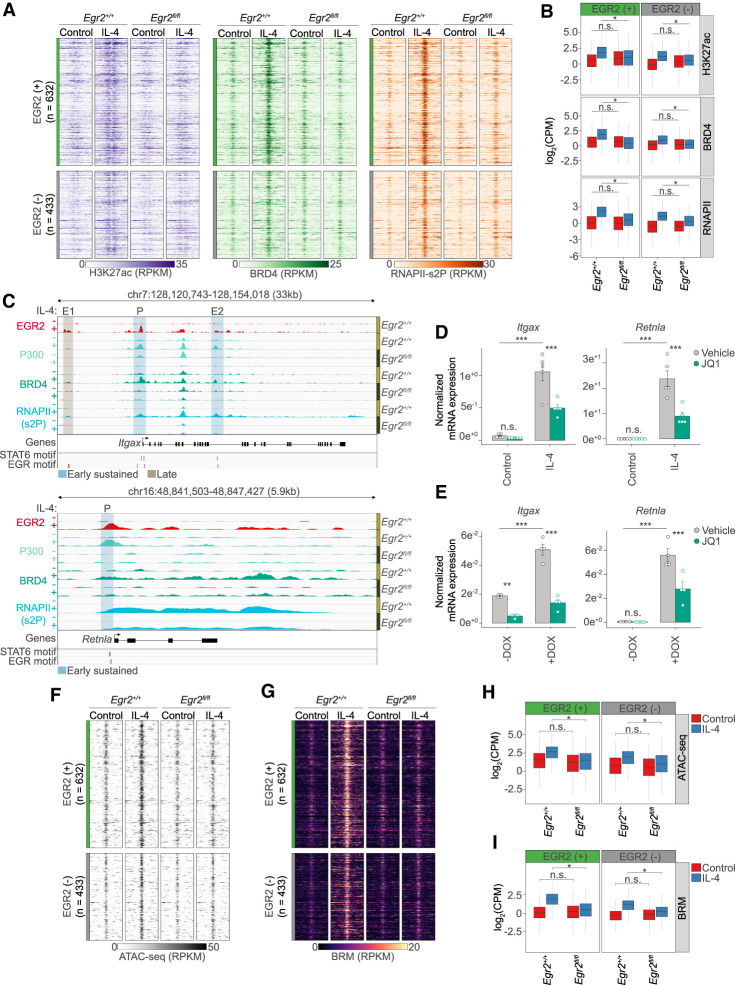
Identification of the genes with IL-4/STAT6 and EGR2-dependent induction allowed us to take a closer look at the epigenetic state of the surrounding regulatory regions. For these analyses, we used 580 genes showing reduced IL-4-induced expression in Egr2−/− MFs. We annotated 1065 regulatory regions falling into a ±100-kb genomic window around the transcription start site of these genes and exhibiting IL-4- and EGR2-dependent P300 recruitment based on ChIP-seq data (Fig. 4F,H). Of these regions, 59% (632/1065) bound EGR2 (+), while 41% (433/1065) showed negligible binding signal; thus, we designated them EGR2 negative (−) (Fig. 4G,I). Importantly, EGR2 (+) sites were annotated to 356 EGR2-dependent genes indicating direct regulatory roles, accounting for 54% of the total EGR2-induced gene program. Regarding genomic localization, 95% of these genomic regions were distal sites, while 5% were promoter proximal sites (Supplemental Fig. S4E). EGR2 (+) genomic regions showed strong enrichment for the EGR TF binding motif. STAT6 and AP-1 motifs were present to similar degrees under both EGR2 (+) and (−) sites. In addition, EGR2 (−) sites showed enrichment for the TF motif, MITF, which has been linked to MF functions by physically interacting with the myeloid LDTF, PU.1 (Supplemental Fig. S4F; Luchin et al. 2001). This classification provided the basis of our analysis to examine the roles of EGR2 at these putative regulatory elements around EGR2-dependent genes.
Assessing the active enhancer mark H3K27ac reported that both EGR2 (+) and (−) regions had elevated acetylation levels in an IL-4 and EGR2-dependent manner (Fig. 5A,B). These data implicate the binding of acetylated histone reader proteins to facilitate transcriptional initiation and elongation. Thus, we examined whether EGR2 was required for the recruitment of the acetylated histone reader, Bromodomain-containing protein 4 (BRD4) and whether it correlated with the presence of the elongating RNA polymerase II (RNAPII). BRD4 is required for the phosphorylation of the C-terminal domain of RNAPII on Ser-2 by recruiting the P-TEFb complex to favor transcriptional elongation (RNAPII-pS2) (Jang et al. 2005). ChIP-seq analyses for both BRD4 and RNAPII-pS2 showed IL-4-induced enrichment at EGR2 (+) and (−) sites, where IL-4-mediated recruitment was EGR2-dependent. (Fig. 5A,B). To asses whether inhibition of BRD4 affects the IL-4 and EGR2-dependent program, we applied the JQ-1 inhibitor, which ablates the binding ability of BRD4 to acetylated histones in the paradigm of alternative polarization. The inhibitor was added in the last 12 h of the 24-h long IL-4 treatment paradigm in order to avoid the inhibition of the early transcriptional events (Supplemental Fig. S5A). We examined two representative genes (Itgax and Retnla) where EGR2 is both required for IL-4-induced transcription and also for the recruitment of P300, BRD4, and RNAPII-pS2 (Fig. 5C; Supplemental Table S2). The genes showed very low expression levels in the unstimulated state, but IL-4 robustly induced transcription, which was dampened by BRD4 inhibition (Fig. 5D). Similarly, we confirmed these results in our gain of function model system (Supplemental Fig. S5B) by showing that the exclusively EGR2-mediated induction of Itgax and Retnla was impaired upon BRD4 inhibition (Fig. 5E), while it does not have a negative effect on DOX-induced EGR2 expression (Supplemental Fig. S5C). Our results establish the requirement for EGR2 in both the transcriptional and epigenetic programming of IL-4 polarized MFs. However, it remained unclear whether EGR2 also contributed to chromatin remodeling.
Figure 5.
EGR2 establishes an enhancer network in the proximity of its target genes and remodels chromatin. (A) Read distribution plot of H3K27ac, BRD4, and RNAPII-pS2 ChIP-seq signal at EGR2 (+) and EGR2 (−) regions from Egr2+/+ and Egr2fl/fl macrophages in the absence (control) and presence of IL-4 (24 h). (B) Box plot representation of the average binding intensities of H3K27ac, BRD4, and RNAPII-pS2 ChIP-seq from the same conditions as presented in A. Log2 transformed CPM (counts per million) values are depicted. Significant changes are determined by Wilcoxon test at P < 0.05. (C) Genome browser view on the Itgax and Retnla loci. ChIP-seq results for EGR2 in wild-type macrophages (Egr2+/+) and P300, BRD4, and RNAPII-pS2 in both Egr2+/+ and Egr2fl/fl macrophages are shown. Polarized (24 h of IL-4 treatment) and control conditions are depicted for each protein. (D) RT-qPCR measurements of Itgax and Retnla mRNA levels in JQ1 (BRD4 inhibitor) treated control and IL-4 polarized macrophages. Vehicle indicates solvent control for the JQ1 treatment. The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by, two-way analysis of variance (ANOVA). (E) RT-qPCR measurements of Itgax and Retnla mRNA levels in JQ1 (BRD4 inhibitor) treated myeloid cells possessing a doxycycline sensitive, EGR2 expressing construct. Experiments were performed in the absence (−) and presence (+) of DOX. Vehicle indicates solvent control for the JQ1 treatment. The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (F) Read distribution plot of ATAC-seq signal at EGR2 (+) and EGR2 (−) regions from Egr2+/+ and Egr2fl/fl macrophages in the absence (control) and presence of IL-4 (24 h). RPKM values are plotted. (G) Read distribution plot of BRM ChIP-seq signal at EGR2 (+) and EGR2 (−) regions from Egr2+/+ and Egr2fl/fl macrophages in the absence (control) and presence of IL-4 (24 h). (H) Box plot representation of the average signal intensity of ATAC-seq at EGR2 (+) and EGR2 (−) regions. Log2 transformed CPM (Counts Per Million) values are depicted. Significant changes are determined with Wilcoxon test at P < 0.05. (I) Box plot representation of the average signal intensity of BRM ChIP-seq at EGR2 (+) and EGR2 (−) regions. Log2 transformed CPM (counts per million) values are depicted. Significant changes are determined with Wilcoxon test at P < 0.05.
EGR2 is required for alternative polarization-induced chromatin remodeling
To examine EGR2's role in chromatin remodeling during polarization, we assessed chromatin accessibility using assay for transposase-accessible chromatin sequencing (ATAC-seq) (Buenrostro et al. 2013). Both EGR2 (+) and EGR2 (−) genomic regions exhibited weak ATAC-seq signals in the unstimulated state, but IL-4 very substantially induced accessibility. Polarization-induced chromatin changes were abrogated in the absence of Egr2 (Fig. 5F,G). In search of a mechanism, we were wondering whether members of the SWI/SNF, ATP-dependent chromatin remodeling complex are also recruited to these regions and exhibit similar dependencies. Thus, we assessed the genome-wide localization of Brahma (BRM or SMARCA2), having key ATPase activities in the complex, changing chromatin structure by altering the contacts of nucleosomes with DNA (Gatchalian et al. 2020). ChIP-seq unequivocally supported our hypothesis and showed IL-4 and EGR2-dependent chromatin binding of BRM at both EGR2 (+) and EGR2 (−) sites (Fig. 5G,I), perfectly aligning with our ATAC-seq results. Genes (Retnla and Itgax) where EGR2 alone could induce gene expression in the gain-of-function paradigm appeared to be very sensitive to the loss of Egr2 and were unable to recruit BRM to their promoters correlating with complete loss of promoter accessibility (Supplemental Fig. S5D).
These findings, along with the previously observed effects on genome activity patterns highlight the requirement for EGR2 during alternative polarization as a mediator of transcriptional activation and chromatin remodeling, acting immediately downstream from STAT6. The fact that there are similar changes at genomic regions lacking EGR2 binding raised the possibility that these indirect effects are carried out by EGR2-induced TFs further downstream, leading to the establishment of an at least three-tiered transcriptional cascade.
EGR2 binds and activates enhancers on the Egr2 locus indicative of autoregulation
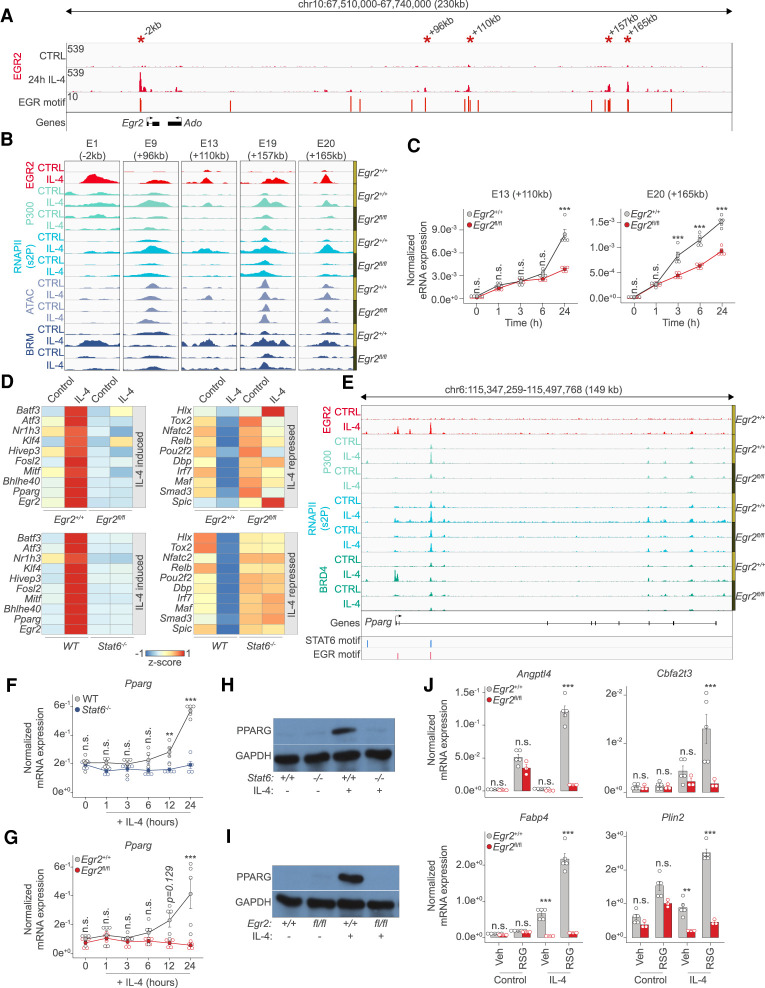
We found so far that EGR2 is induced by the transiently acting STAT6, but its cistrome remains stable, raising the question of what maintains its expression at later time points once STAT6 is gone? We took a closer look at the putative enhancer regions of the Egr2 gene locus and observed that EGR2 binds to at least five of these regulatory regions (E9: +96 kb, E13: +110 kb, E19: +159 kb, E20: +165 kb), including one near its promoter (E1) (Fig. 6A). Moreover, all of these regulatory elements possess strong EGR motifs. Looking at our ChIP-seq and ATAC-seq results informed us about the IL-4 and EGR2-dependent recruitment of P300, RNAPII, and BRM, while de novo chromatin opening of the E13 enhancer got abrogated in the absence of EGR2 (Fig. 6B). As a proof of principle, we selected two of these enhancers (E13 and E20) and determined their transcriptional activities during the polarization time course. Enhancer RNA measurements confirmed the presumed role of EGR2 in the induction of both E13 and E20. Specifically, E13 exhibited late EGR2-dependence (24 h), while E20 appeared to be EGR2 regulated at a very early (3-h) time point as well (Fig. 6C). Importantly, the earliest enhancer activities (1 h) were not affected by the loss of Egr2, in agreement with the previous findings that STAT6 initiates the expression of Egr2 (Fig. 6C). Thus, long-term, high expression level of Egr2 is likely to be at least in part, maintained by an autoregulatory feedback loop, where EGR2 occupies some of its own enhancers, sustaining its own expression and converting a transient signal to a stable one.
Figure 6.
The IL-4/STAT6/EGR2 axis controls the transcription factor cascade of alternative macrophage polarization. (A) Genome browser view on the Egr2 locus. Shown are EGR2 ChIP-seq results in unstimulated (CTRL) and IL-4 polarized macrophages. Motif scores of the EGR motifs overlapping with putative enhancer elements are indicated. Asterisks indicate EGR2 bound regions magnified in B. (B) Genome browser images on the enhancer regions highlighted in A. Shown are ChIP-seq results for EGR2, P300, RNAPII(s2P), and BRM in Egr2+/+ and Egr2fl/fl macrophages in the absence (control) and presence of IL-4 (24 h). ATAC-seq results in the same conditions are also shown. (C) Enhancer RNA expression detected by RT-qPCR on the indicated loci. The level of eRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (D) Heat map representation of IL-4 altered (induced and repressed), Egr2-dependent (top) and Stat6-dependent (bottom) transcription factor gene expression. IL-4-induced (left) and IL-4-repressed (right) gene expression is visualized in the two genotypes. (E) Genome browser view on the Pparg gene locus. ChIP-seq data for EGR2 in wild type macrophages (Egr2+/+) and P300, RNAPII(s2P) and BRD4 in both Egr2+/+ and Egr2fl/fl macrophages are shown. Polarized (24 h of IL-4 treatment) and control conditions are depicted for each track. Regulatory regions with STAT6 and EGR motifs are highlighted. (F) RT-qPCR measurements of Pparg mRNA levels in wild-type (WT) and Stat6−/− control and IL-4 polarized macrophages over the indicated time course. The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (G) RT-qPCR measurements of Pparg mRNA levels in Egr2+/+ and Egr2fl/f control and IL-4 polarized macrophages over the indicated time course. The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (H) Western blot of PPARG expression in wild-type (Stat6+/+) and Stat6−/− macrophages in the presence or absence of IL-4. Experiments were repeated four times. One representative blot is shown. GAPDH serves as a loading control. (I) Western blot of PPARG expression in Egr2+/+ and Egr2fl/f macrophages in the presence or absence of IL-4. Experiments were repeated four times. One representative blot is shown. GAPDH serves as a loading control. (J) RT-qPCR measurements of Angptl4, Cbfa2t3, Fabp4, and Plin2 mRNA levels in Egr2+/+ and Egr2fl/fl control and IL-4 polarized macrophages exposed to either solvent control (vehicle) or rosiglitazone (RSG). The level of mRNA is normalized to the expression of Ppia. Experiments were repeated five times, and significant changes between groups were calculated by, two-way analysis of variance (ANOVA).
EGR2 regulates the transcription factor cascade of polarization
Next, we assessed the role of EGR2 in the regulation of the downstream TF cascade of alternative polarization. We used the TF database called TFcheckpoint consisting of 1020 TFs (Chawla et al. 2013). Overlaying this list with all the expressed genes (n = 10,943) in macrophages yielded 339 TFs. Of these, 173 showed IL-4 dependent regulation and 76% (n = 132) were Egr2-dependent. Loss of Egr2 had minimal enhancing effects on IL-4-mediated TF gene activation or repression (facilitated repression n = 4, facilitated activation n = 6), while it was required for IL-4-mediated repression of 72 and activation of 50 TF genes, respectively (Supplemental Fig. S6A,B; Supplemental Table S4). Of the IL-4-induced TFs, more have been reported as important regulators of alternative polarization for instance Pparg, Klf4, Myc, and Bhlhe40 but our analyses point to other potential candidates, all downstream from STAT6 and EGR2 (Fig. 6D).
Next, as a proof of principle and to establish the hierarchical relationship between STAT6, EGR2, and a specialized downstream TF, we focused on PPARG due to its roles in the maintenance of alternative macrophage polarization and lipid ligand-induced gene expression and the fact that it was believed to be regulated by STAT6 (Lavin et al. 2014; Dai et al. 2017; Murray 2017). On the Pparg gene locus, EGR2 binds to several putative enhancers, recruiting P300, BRD4, and RNAPII (Fig. 6E). Gene expression measurements validated the absolute IL-4/STAT6 and EGR2-dependent regulation of Pparg (Fig. 6F,G). In addition, protein expression of PPARG was also entirely Stat6- and Egr2-dependent in the presence of IL-4 (Fig. 6H,I; Supplemental Fig. S6C,D). Moreover, IL-4 facilitated ligand-induced transcription on some of the canonical PPARG target genes (Angptl4, Cbfa2t3, Fabp4, and Plin2) (Daniel et al. 2018b) in an EGR2-dependent manner (Fig. 6J). These results lend strong support to the notion that EGR2 acts immediately downstream from STAT6 and acts upstream of a large number of TFs establishing at least three layers of regulation or an even more complex TF cascade of alternative macrophage polarization.
The IL-4/STAT6-induced Egr2 axis is evolutionarily conserved in mouse and human macrophages
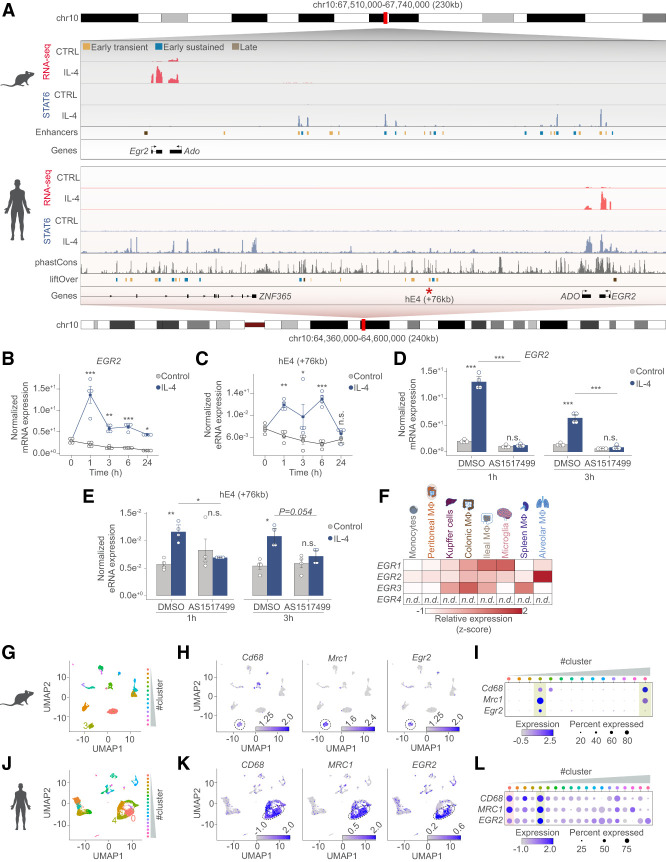
EGR2 appears to be a critical regulator of IL-4-induced polarization of murine macrophages. However, alternative polarization is quite divergent between mice and humans (Noël et al. 2004). Thus, we decided to assess whether IL-4 is able to induce EGR2 in human CD14+ monocyte-derived differentiating macrophages (Supplemental Fig. S7A). First, we examined the mouse and human Egr2 gene loci that exhibited considerable conservation as revealed by phastCons tracks between the two species (Fig. 7A). Second, we carried out extensive gene expression measurements in the two species. For these experiments, we also included the other major Th2 cytokine IL-13, which robustly induces Egr2 expression in BMDMs (Supplemental Fig. S7B). Human macrophages showed similarly robust and sustained EGR2 expression upon either IL-4 or IL-13 exposure (Fig. 7B; Supplemental Fig. S7C). Moreover, we observed conservation between some of the distant regulatory regions of the mouse and human Egr2 loci. One of these enhancers that is located +76 kb from the human EGR2 gene exhibited strong IL-4-induced enhancer RNA expression (Fig. 7C). Next, we used a specific STAT6 inhibitor (AS1517499) as a pharmacological loss-of-function tool to assess the roles of STAT6 in IL-4-induced EGR2 expression in human MFs. We found that inhibition of STAT6 completely abolished the transcription of both the EGR2 gene and the +76-kb enhancer, thus the regulation of EGR2 by IL-4-activated STAT6 appears to be conserved between mouse and human MFs (Fig. 7D,E). These results strongly positioned EGR2 as an evolutionarily conserved mediator of STAT6-dependent polarization in primary murine and human cells ex vivo but left us with a question regarding its role in vivo in tissue MFs.
Figure 7.
The IL-4/STAT6 signaling axis regulates EGR2 expression in human monocytes and EGR2 expression marks lung-resident macrophage subsets. (A) Genome browser views on the Egr2 loci in mouse and human macrophages. Shown are RNA-seq and STAT6 ChIP-seq results from the two species. Annotated enhancers are highlighted and color-coded according to their activity patterns. Conservation scores of the Egr2 locus is presented in the phastCons track followed by the liftOver track that shows the annotation of the conserved mouse enhancer regions to the human genome. The enhancer located +76 kb from the human EGR2 gene is marked with an asterisk because it is conserved between the two species and binds STAT6 as well. (B) RT-qPCR measurements of EGR2 mRNA levels in control and IL-4 polarized human differentiating macrophages over the indicated time course. The level of mRNA is normalized to the expression of PPIA. Experiments were repeated four times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (C) RT-qPCR measurements of the synthesized enhancer RNA levels from the conserved enhancer located +76 kb from the human EGR2 gene. The level of eRNA from control and IL-4 polarized human differentiating macrophages over the indicated time course is shown, normalized to the expression of PPIA. Experiments were repeated four times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (D) RT-qPCR measurements of EGR2 mRNA levels in control and IL-4 polarized human differentiating macrophages in the presence or absence (DMSO [dimethyl sulfoxide as vehicle control]) of the STAT6 inhibitor (AS1517499) at the two indicated time points. The level of mRNA is normalized to the expression of PPIA. Experiments were repeated four times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (E) RT-qPCR measurements of enhancer RNA levels, synthesized from the +76-kb enhancer in control and IL-4 polarized human differentiating macrophages. Experiments were performed in the presence or absence (DMSO [dimethyl sulfoxide as vehicle control]) of the STAT6 inhibitor (AS1517499) at the two indicated time points. The level of eRNA is normalized to the expression of PPIA. Experiments were repeated four times, and significant changes between groups were calculated by two-way analysis of variance (ANOVA). (F) Heat map representation of the mRNA expression levels (RNA-seq GSE63340) of the EGR TF family members in the indicated cell types. Egr4 transcript was not detected (n.d.) in any of the cell types. (G) UMAP depicting the different cell clusters of the mouse lung defined by single cell RNA-seq (GSE133747). Cluster 3 is highlighted, which corresponds to the macrophage cluster having high Cd68 and Mrc1 expression. (H) Feature plots depicting the expression of Cd68, Mrc1, and Egr2 expression in the mouse data set. Cluster 3 is encircled to highlight the macrophage population. Log normalized expression values are plotted. (I) Dot plot representation of the average expression and percentage wise expression of Cd68, Mrc1, and Egr2 in the different cell clusters of the mouse data set. (J) UMAP depicting the different cell clusters of the human lung defined by single cell RNA-seq (GSE128033). Clusters 0 and 4 are highlighted, which corresponds to the macrophage clusters having high CD68 and MRC1 expression. (K) Feature plots depicting the expression of CD68, MRC1, and EGR2 expression in the human data set. Clusters 0 and 4 are encircled to highlight the macrophage populations. Log normalized expression values are plotted. (L) Dot plot representation of the average expression and percentage wise expression of CD68, MRC1, and EGR2 in the different cell clusters of the human data set.
Egr2 expression marks lung-resident MF subsets in different species
We sought to answer this question by analyzing published RNA-seq data sets (GSE63340) from mouse monocytes and various tissue resident MF subsets to obtain information on the tissue-specific expression of Egr TF genes (Lavin et al. 2014). It appears that most Egr TF genes were widely expressed in tissue-specific MF subsets, but Egr2 had a narrower expression pattern that was largely restricted to alveolar MFs. Moreover, other EGR family members were barely or not expressed in this MF subset, suggesting a more specific role for EGR2 in these cells (Fig. 7F). This observation raised another question concerning the evolutionarily conserved expression of Egr2 in the lung-resident MF subsets of other species.
Single-cell RNA-seq data sets from mouse, human, pig, and rat healthy lungs were used from recently published work to address this question (Morse et al. 2019; Raredon et al. 2019). We analyzed these data and performed uniform manifold approximation and projection for dimensionality reduction (UMAP) (Becht et al. 2018) using all of the cells from the different species (Fig. 7G,J; Supplemental Fig. S7D,G). Regarding the human lung analyses, we integrated data sets from five healthy lung samples from the following study (GSE128033) (Supplemental Fig. S7J; Morse et al. 2019), while lung data sets of the other species are derived from GSE133747 (Raredon et al. 2019). After dimensionality reduction, we called clusters and identified MFs based on Cd68, Mrc1 (mouse: cluster3, human: clusters 0 and 4, and rat: cluster 2), or CD163, MRC1 (pig: cluster 7) expression (Fig. 7H,I,K,L). After the identification of the MF clusters, we plotted the expression of EGR2, which was specific to at least one macrophage population in the different species (Fig. 7H,I,K,L; Supplemental Fig. S7E,F,H,I). Importantly, high EGR2 expression appeared to be relatively specific to the MF compartment in all the studied species. Thus, besides being required for IL-4/STAT6-induced polarization in murine MFs, EGR2's induction by STAT6 appears to be evolutionarily conserved between mouse and human MFs. In addition, bulk and single-cell RNA-seq data point to EGR2 as a regulator of lung-resident MF function in the steady state, which should be investigated in future studies.
Discussion
Understanding the hierarchical transcriptional program of MF polarization in health and disease will ultimately lead to a better knowledge of cell type specification and targeted therapies, aiming to reprogram MFs to exploit beneficial phenotypes and to avoid harmful ones. Here, we identified the zinc finger TF, EGR2 as an essential component of the IL-4-mediated and STAT6-dependent alternative polarization program linking the early transient events to late, stable ones establishing key hierarchical relationships between layers of TFs.
Here we show that (1) EGR2 is an evolutionarily conserved direct target of STAT6 in murine and human MFs; (2) EGR2 and STAT6 establish temporally and spatially distinct cistromes during polarization; (3) around three-quarters (77%) of STAT6-regulated genes depend on the presence of EGR2; (4) EGR2 is sufficient to directly regulate target genes as well as recruit chromatin remodeling and histone-modifying complexes, including an autoregulatory loop inducing its own expression; (5) EGR2 controls a downstream TF cascade of process-specific TFs, including PPARG, and thus works as a molecular linchpin to connect the transient STAT6 signal to long-lasting epigenomic changes to support stable gene expression; and (6) EGR2 marks lung-resident macrophage subsets in multiple species. The importance of STAT6 downstream regulators, acting when STAT6 is excluded from the nucleus, has been raised by prior research in the establishment and maintenance of the late polarization program (Murray 2017). However, so far, no broad acting transcriptional and/or epigenomic mechanism was identified to explain this conundrum and establish the sequence of events. We took advantage of mapping the genome activity (using the active enhancer markers P300 and H3K27ac) patterns of macrophage polarization over a time course, where both the immediate early and long-term (likely indirect) effects of IL-4-activated STAT6 can be studied. This strategy proved to be fruitful and led to the identification of a singular very potent transcriptional regulator, EGR2 with thus far unknown function in myeloid cells. Although transcriptome-wide analyses reported Egr2 as an IL-4-induced gene in macrophages, it remained hidden in the vast data sets without any detail about the nature and extent of its roles (Jablonski et al. 2015; Roy et al. 2015). More recently, EGR2 has been linked to the induction of a few marker genes of alternative polarization but neither its genome-wide contribution to the epigenomic and transcriptomic program, nor its mechanism of action was revealed in great detail (Veremeyko et al. 2018).
The EGR TF family members possess a highly homologous DNA-binding domain, thus recognize the same DNA sequence, implying the potentially overlapping or redundant roles of the family members (Poirier et al. 2007). In our model, Egr genes are not or barely expressed in unstimulated macrophages. IL-4 selectively induce Egr2, and no other members of the family are responsive to IL-4. Moreover, the genetic deletion of Egr2 does not affect the expression of the other Egr genes; thus, there is no redundancy and other EGRs do not compensate for the loss of Egr2. This is a key feature of our study because it allowed the thorough characterization of EGR2's role without other family members’ interference. EGR2 was found to be dispensable for macrophage differentiation (Carter and Tourtellotte 2007), and our results confirm these findings by showing that the lack of Egr2 does not impact the unstimulated MF phenotype, and therefore EGR2 is not needed for MF differentiation per se. However, EGR family members are primarily linked to memory formation and learning in the central nervous system (CNS) (Poirier et al. 2007), several studies reveal their critical roles in the immune system (Schneider et al. 2014), including T cells, suggesting that they have roles in establishing transcriptional programs beyond the CNS. Our study is supporting this notion and establishes EGR2 as an essential regulator of MF polarization with the role to imprint the long-term epigenetic and transcriptomic program of IL-4.
In terms of its precise participation within the sequence of events in alternative polarization program, Egr2 is turned on by IL-4-activated STAT6 during the first hour of cytokine exposure. This immediate early response and induction allows EGR2 to take the baton from STAT6, which is being released from the chromatin by the time EGR2 populates the MF genome. The cistromic behavior of STAT6 and EGR2 are rather different and almost mutually exclusive, displaying remarkable spatial and temporal separation. Therefore, EGR2 connects the early STAT6-driven gene expression program to the late mostly EGR2-driven program. Importantly, these data imply that mechanistically EGR2 is neither a cofactor, nor a collaborator factor of STAT6. Regarding the impact of EGR2 on polarization we could show that besides the canonical alternative polarization marker genes (Retnla, Chil3, Chil4, and Mmp12), EGR2 controls 77% of the late gene expression program of polarization, including TFs (Klf4, Myc, Pparg, and Bhlhe40) with already described downstream roles indicating its broad impact. As a proof of principle, we show that the nuclear receptor PPARG is one of these TFs, which has been linked to alternative polarization and is vital for alveolar MF development (Odegaard et al. 2007; Schneider et al. 2014). We reported previously that IL-4-activated STAT6 facilitates ligand responses of PPARG by increasing the level and chromatin binding of the protein (Szanto et al. 2010; Daniel et al. 2018a). Here we extend this model with a critical new piece by showing that in the absence of EGR2, IL-4-activated STAT6 is no longer able to induce the expression of PPARG. Thus, EGR2 is immediately downstream from STAT6 and indispensable for the augmented ligand responsiveness of PPARG by regulating the receptor's expression. Moreover, EGR2 binds and regulates its own enhancers’ activities indicative of autoregulatory role, which might be important in carrying out its broad, long-term effects and converting a transient STAT6 signal to a permanent, sustained epigenomic imprint (Fig. 8). Therefore, EGR2 is not part of the transcriptional machinery regulating STAT6-dependent target genes. Instead, it is a strictly downstream epigenomic effector. This reveals an at least three-tiered transcriptional cascade with IL-4-regulated STAT6 on top turning on hundreds of genes, a fraction of them independent of EGR2. Then, EGR2 as a “transcriptional second messenger” is robustly induced, regulates its own expression, and can provide stable epigenomic changes along with the induction of TFs with much narrower specificity such as PPARG, KLF4, or BHLHE40. It is possible that additional levels of regulation exist and that the process gets “lateral input” from additional signaling pathways. Transcriptional cascades have been identified in classical polarization using LPS signaling as a model (Tong et al. 2016). However, in that case, no broad-acting secondary TF has been identified, but rather several members of the NFkB family have unique and also overlapping roles. EGR2 acquires this central and broad role by a multitude of regulatory activities: (1) chromatin remodeling and recruitment of the ATP-dependent chromatin remodeling complex (SWI/SNF) member BRM (SMARCA2), (2) de novo enhancer formation and recruitment of the acetyltransferase P300 to deposit H3K27ac on enhancers, and (3) probably as a consequence of the previous two, EGR2 attracts BRD4 and facilitates transcriptional elongation (Fig. 8). The exact order and molecular details, including the contribution of the specific domains of EGR2, need to be uncovered in future studies.
Figure 8.
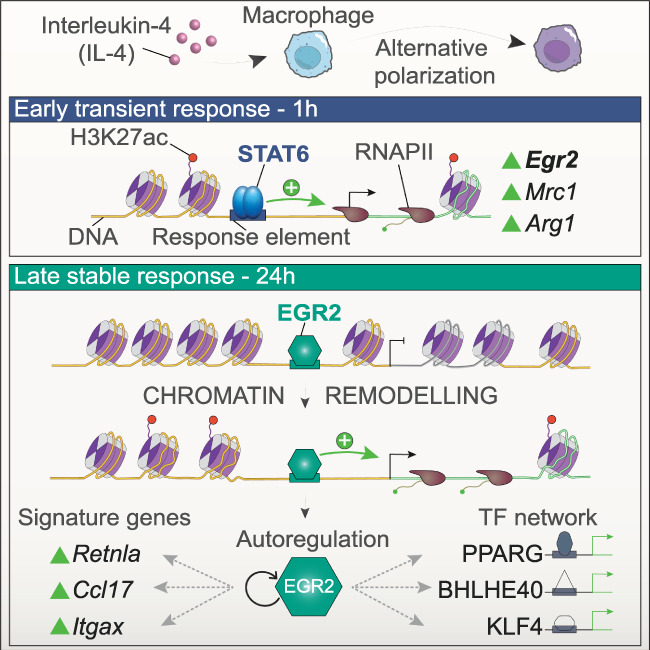
The role of Egr2 in the transcriptional cascade mediating IL4/STAT6-induced alternative polarization. The Th2-type cytokine IL-4 induces the phenotypic switch of macrophages to become alternatively polarized (top). The immediate early transcriptional regulator of alternative polarization STAT6 orchestrates a rapidly developing but transient gene expression program including the marker genes of polarization (Mrc1 and Arg1). The transcription factor EGR2 is also subject to IL-4/STAT6-mediated early induction via enhancer activation (H3K27ac) and RNAPII recruitment (middle panel). Once the polarization program proceeds to the late stages, EGR2 becomes a central component by guiding the transition between early and late polarization, acting as a “molecular linchpin” via forming an extensive cistrome and chromatin remodeling activity. EGR2 regulates the vast majority of the late polarization program including the signature genes of this stage (Retnla, Ccl17, and Itgax). Moreover, EGR2 controls the expression of an entire transcription factor cascade including PPARG, BHLHE40, and KLF4 with important roles in alternative macrophage polarization. This program is sustained partly by EGR2's autoregulatory activity.
The IL-4/STAT6/EGR2 axis is conserved between mice and humans and the expression of EGR2 can be induced by both Th2-type cytokines (IL-4 and IL-13) in both species. Thus, EGR2 belongs to the otherwise remarkably low number of alternative polarization-specific marker genes shared by these two species such as Mrc1, Arg1, and fibronectin (Fn1) (Noël et al. 2004), adding to its presumed significance. In terms of tissue-specific activity and relevance, alternative MF polarization has been observed in a variety of disease conditions like allergic airway disease (Gour and Wills-Karp 2015), helminth infection, fibrosis, and cancer (Van Dyken and Locksley 2013), therefore these are obvious places and processes EGR2's role could and should be studied in. Our efforts are hindered though by the facts that alternative polarization is altered drastically by exposure to other cytokines and signals, the lack of clear evidence to establish the role of MF-specific STAT6 in these processes due to the unavailability of tissue-specific knockouts. In addition, redundancy, that is, the induction and compensatory activity of other EGR family members cannot be easily excluded. Without a better understanding of the role of STAT6 and the integration of other signals with it working out EGR2's role appears to be rather futile. Nonetheless, the reanalysis of mouse tissue-resident macrophage RNA-seq data revealed a lung-restricted expression pattern for Egr2 in the steady state. This along with the finding of basophil-derived IL-13 playing a role in alveolar macrophage function (Cohen et al. 2018) lend further support to EGR2's in vivo role. Further analysis of published single-cell RNA-seq data from mouse, rat, pig, and human healthy lung samples not only confirmed the expression of Egr2, but also revealed the evolutionarily conserved presence of Egr2 in lung-resident macrophage populations. Therefore, work can be initiated to define in vivo roles of Egr2 in the lung-resident macrophage subsets. In summary, we discovered EGR2, a molecular linchpin connecting the early and late alternative polarization program, with broad specificity and molecular functions inducing chromatin opening and histone acetylation, representing an evolutionarily conserved component of mouse and human MF responses to Th2-type cytokine rich milieu.
Materials and methods
Mouse strains
Male, wild-type 3-mo-old mice were used and bred under specific-pathogen-free (SPF) conditions. All the strains were kept on the C57BL/6 genetic background. The Egr2fl/fl animals were a generous gift from Patrick Charnays laboratory. We crossed these animals with lysozyme-Cre (LysCre)+ animals to establish the conditional knockout strain (Egr2fl/fl LysCre). These mice were backcrossed to the C57BL/6J strain for eight generations. As controls we used Egr2+/+ LysCre littermates. Full-body Stat6−/− (Jackson Laboratory) animals were maintained by breeding knockout male and female mice. Wild-type C57BL/6 animals were used as controls. Animals were handled according to the regulatory standards of the animal facilities of the University of Debrecen and Johns Hopkins All Children's Hospital.
Bone marrow-derived macrophages
Isolation and differentiation were completed as described earlier (Daniel et al. 2014b). Isolated bone marrow-derived cells were differentiated for 6 d in the presence of L929 supernatant. On the third day of differentiation, supernatant was collected centrifuged and replated into the same plates in fresh media and differentiation continued for three more days.
Treatment conditions
For description regarding treatment conditions, see the Supplemental Material.
ChIP (chromatin immunoprecipitation)
ChIP was performed with minor modifications of the previously described protocol (Daniel et al. 2014a). We lowered the sonication strength to low and shearing was performed in two consecutive rounds of 5 min (total 10 min). Libraries were prepared with Ovation Ultralow Library Systems V2 (Nugen) according to the manufacturer's instructions. The following antibodies were used: EGR2 (sc-20690), P300 (sc-585), BRM (ab15597), BRD4 (A301-985A100), H3K27ac (ab4729), RNAPII-pS2 (ab5095).
RNA-seq
Wild-type and Egr2fl/fl macrophages were differentiated in the presence of L929 cell supernatants for 6 d on 15-cm dishes. On the sixth day cells were replated onto six-well plates at a 2 × 106 cells/mL density and treated with IL-4 (20 ng/mL) for 24 h or left untreated. After 24 h, RNA was collected and isolated with Trizol. For additional details regarding RNA-seq, see the Supplemental Material.
Western blot
Whole-cell lysates were resolved by electrophoresis in 10% polyacrylamide gel and then transferred to immobilon-P transfer membrane. Membranes were probed with anti-PPARγ (81B8) anti-Egr2 (ab108399) and anti-GAPDH (AM4300) antibodies as indicated according to manufacturer's recommendations.
Monocyte isolation and differentiation
Human monocytes were isolated from peripheral blood mononuclear cells (PBMC) of healthy volunteers. Buffy coats were obtained from the Regional Blood Bank. For additional details regarding monocyte isolation and differentiation, see the Supplemental Material.
Real-time quantitative PCR for enhancer RNA and mRNA detection (qPCR)
RNA was isolated with Trizol reagent (Ambion). RNA was reverse-transcribed with high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's protocol. Transcript quantification was performed by qPCR reactions using SYBR Green master mix (Roche). Transcript levels were normalized to Ppia and PPIA. Primers are available upon request.
Generation of ES-derived myeloid progenitors
To generate chemically inducible ESC clones, a p2lox targeting vector was engineered that carry the murine Egr2 coding sequence. The targeting vector was delivered into the ZX1 ESCs for generation of doxycycline-inducible ESC clones as described. For additional details regarding generation of ES-derived myeloid progenitors, see the Supplemental Material.
ELISA
Culture supernatants from cells were collected at the indicated times. Samples were centrifuged at 1000g for 10 min at 4°C, and the supernatants were separated and stored at −20°C until analysis. These supernatants were then probed for the presence of the following cytokine using ELISA kits according to the manufacturer's instructions: thymus and activation-regulated chemokine (CCL17/TARC; DuoSet ELISA DY529; R&D Systems). Plates were read using the Thermo Multiskan Ascent microplate reader and analyte concentrations were calculated with ProcartaPlex Analyst 1.0 software from Affymetrix.
Flow cytometry
For flow cytometric measurements, cells were labeled with Zombie Green fixable viability dye (Biolegend) for 15 min at room temperature and then blocked with anti CD16/32 (2.4G2) in staining buffer (1× PBS + 2%FBS) for 15 min at 4°C. Cell surface staining was performed in staining buffer to F4/80 (Biolegend BM8), CD206 (AbD Serotec MR5D3) for 30 min at 4°C. For intracellular detection of RELMα, Zenon Alexa fluor 350 rabbit IgG labeling kit (Thermo Fischer Scientific) was used to add fluorescent label to RELMα (Peprotech) antibody, BD Cytofix Cytoperm fixation/permeabilization solution kit (BD Biosciences) was used for fixation/permeabilization and staining according to the manufacturer's recommendations. Samples were acquired on a BD FACS Aria III (BD Biosciences) cell sorter using BD FACSDiva 6.0 software (BD Biosciences). Data analysis was performed using FlowJo v10 software (Becton Dickinson and Company).
ATAC-seq
ATAC-seq was carried out as described earlier with minor modification. For additional details regarding generation of ATAC-seq, see the Supplemental Material.
RNA-seq mapping and gene expression quantification
Sequencing quality was evaluated by FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were aligned to the mouse reference genome (mm10) using the default parameters of HISAT2 (Kim et al. 2019). For additional details regarding RNA-seq mapping and gene expression quantification, see the Supplemental Material. Differentially expressed genes are reported in Supplemental Tables S3 and S4.
ATAC-seq and ChIP-seq analyses
Sequencing quality was evaluated by FastQC software. The primary analysis of ATAC-seq and ChIP-seq reads was carried out using our ChIP-seq command line pipeline (Barta 2011). For additional details regarding ATAC-seq and ChIP-seq analyses, see the Supplemental Material.
P300 differential binding analysis
To determine significant genome activity patterns, we first used DiffBind (Ross-Innes et al. 2012) to generate a P300 consensus peak sets with minOverlap = 2 to generate the raw count matrix for these regions. Peaks with at least 10 CPM (counts per million) in two samples were kept for the downstream analyses. For additional details regarding P300 differential binding analysis, see the Supplemental Material.
De novo motif analysis and motif enrichment
We used HOMER's findMotifsGenome.pl script to search for de novo motif enrichments with mm10 -len 10,12,14 -size 200 -dumpFasta -bits -homer2 parameters. The resulting STAT6 and EGR position weight matrices (PWMs) were used as an input for annotatePeaks.pl (mm10 -size 500 -hist 10) to calculate motif enrichment scores and histograms. Data were visualized in R with ggplot2. Genomic positions of the motives were calculated using scanMotifGenomeWide.pl with default parameters (mm10).
Analyses of publicly available single-cell RNA-seq data
We used the following data sets: GSE133747 and GSE128033. Analysis was performed using Seurat (version 3.1.2) (Stuart et al. 2019). For additional details regarding P300 differential binding analysis, see the Supplemental Material.
Statistical analysis
Statistical analyses were performed in R 3.6.2. qPCR measurements were presented as means ± SD. We generated at least three biological replicates. The exact replicate numbers are indicated in the figure legends. Differences between groups were calculated by two-way analysis of variance [ANOVA; stats::aov() function] with Tukey's post hoc test [multcomp::glht() function] or Kolmogorov-Smirnov test. Statistical significance is depicted as follows: P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***). ATAC-seq and ChIP-seq densities presented on box plots were analyzed with Wilcoxon test in R, using random resampling approach. Statistical parameters are reported in the figure legends and also in the methods section under each specific method description.
Data and software availability
Publicly available, published data sets can be accessed on the following GEO accession numbers: single-cell RNA-seq data sets GSE133747 and GSE128033 and published bulk RNA-seq of tissue-resident macrophage populations GSE63340. Sequencing data sets performed in this study are available at the NCBI GEO under accession number GSE151015.
Supplementary Material
Acknowledgments
We thank members of the Nagy laboratory for discussions and comments on the manuscript. We also thank Dr. Katalin Sandor (Stanford University) for assistance with analyses of the single-cell RNA-seq data sets, and Dr. Michael Kyba (University of Minnesota) for providing the OP9 cells, the ZX1 mouse ES cell line, and the p2Lox targeting plasmid. B.D. was supported by a postdoctoral fellowship from the American Heart Association (17POST33660450), and L.N. was supported by the National Institutes of Health (R01DK115924). Support for this work in the Nuclear Receptor Research Laboratory was provided by the National Research, Development, and Innovation Office (KKP129909, K124298, and PD124843 to G.N., FK132185 to Z.C., and K124890 to I.S. and P.B.) and the European Union and the European Regional Development Fund (GINOP-2.3 2-15-2016-0006). Part of the sequencing was performed at the Centre National de Genotypage (CNG) Evry by Steven McGinn, Anne Boland, Doris Lechner, and Marie Therese Bihoreau, and was supported by the European Sequencing and Genotyping Infrastructure (funded by the European Commission, FP7/2007-2013) under grant agreement 26205 (ESGI), as part of the ADIPOMACTX transnational access program. Z.C. was supported by the Premium Postdoctoral Fellowship Program of the Hungarian Academy of Sciences, and G.N. was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences and the ÚNKP-19-4-DE-173 New National Excellence Program of the Ministry of Human Capacities.
Author contributions: B.D., Z.C., and L.N. directed the study and wrote the manuscript. B.D., Z.C., P.B., Z.K., S.P., P.T., G.H., T.C., A.N., I.S., and A.B. performed the experiments. L.H. designed the bioinformatic approaches and carried out the analyses. G.N. and A.H. analyzed data. S.S. and J.F.-D. directed the sequencing analyses. L.N. obtained funding and provided supervision.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.343038.120.
References
- Amit I, Winter DR, Jung S. 2016. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol 17: 18–25. 10.1038/ni.3325 [DOI] [PubMed] [Google Scholar]
- Barta E. 2011. Command line analysis of ChIP-seq results. EMBnet J 17: 13–17. 10.14806/ej.17.1.209 [DOI] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2018. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37: 38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Belperio JA, Dy M, Murray L, Burdick MD, Xue YY, Strieter RM, Keane MP. 2004. The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. J Immunol 173: 4692–4698. 10.4049/jimmunol.173.7.4692 [DOI] [PubMed] [Google Scholar]
- Bencsik R, Boto P, Szabó RN, Toth BM, Simo E, Bálint BL, Szatmari I. 2016. Improved transgene expression in doxycycline-inducible embryonic stem cells by repeated chemical selection or cell sorting. Stem Cell Res 17: 228–234. 10.1016/j.scr.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Bonnardel J, T'Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, Vanneste B, De Prijck S, Nedospasov SA, Kremer A, et al. 2019. Stellate cells, hepatocytes, and endothelial cells imprint the kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity 51: 638–654 e9. 10.1016/j.immuni.2019.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JH, Tourtellotte WG. 2007. Early growth response transcriptional regulators are dispensable for macrophage differentiation. J Immunol 178: 3038–3047. 10.4049/jimmunol.178.5.3038 [DOI] [PubMed] [Google Scholar]
- Chawla K, Tripathi S, Thommesen L, Lægreid A, Kuiper M. 2013. TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics 29: 2519–2520. 10.1093/bioinformatics/btt432 [DOI] [PubMed] [Google Scholar]
- Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, Miklosi A, Salame TM, Halpern KB, David E, et al. 2018. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell 175: 1031–1044 e18. 10.1016/j.cell.2018.09.009 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czimmerer Z, Daniel B, Horvath A, Rückerl D, Nagy G, Kiss M, Peloquin M, Budai MM, Cuaranta-Monroy I, Simandi Z, et al. 2018. The transcription factor STAT6 mediates direct repression of inflammatory enhancers and limits activation of alternatively polarized macrophages. Immunity 48: 75–90 e6. 10.1016/j.immuni.2017.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Bhargava P, Stanya KJ, Alexander RK, Liou YH, Jacobi D, Knudsen NH, Hyde A, Gangl MR, Liu S, et al. 2017. Macrophage alternative activation confers protection against lipotoxicity-induced cell death. Mol Metab 6: 1186–1197. 10.1016/j.molmet.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel B, Balint BL, Nagy ZS, Nagy L. 2014a. Mapping the genomic binding sites of the activated retinoid X receptor in murine bone marrow-derived macrophages using chromatin immunoprecipitation sequencing. Methods Mol Biol 1204: 15–24. 10.1007/978-1-4939-1346-6_2 [DOI] [PubMed] [Google Scholar]
- Daniel B, Nagy G, Hah N, Horvath A, Czimmerer Z, Poliska S, Gyuris T, Keirsse J, Gysemans C, Van Ginderachter JA, et al. 2014b. The active enhancer network operated by liganded RXR supports angiogenic activity in macrophages. Genes Dev 28: 1562–1577. 10.1101/gad.242685.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel B, Nagy G, Czimmerer Z, Horvath A, Hammers DW, Cuaranta-Monroy I, Poliska S, Tzerpos P, Kolostyak Z, Hays TT, et al. 2018a. The nuclear receptor PPARγ controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity 49: 615–626 e6. 10.1016/j.immuni.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel B, Nagy G, Horvath A, Czimmerer Z, Cuaranta-Monroy I, Poliska S, Hays TT, Sauer S, Francois-Deleuze J, Nagy L. 2018b. The IL-4/STAT6/PPARγ signaling axis is driving the expansion of the RXR heterodimer cistrome, providing complex ligand responsiveness in macrophages. Nucleic Acids Res 46: 4425–4439. 10.1093/nar/gky157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J, Liao J, Maxwell MB, Hargreaves DC. 2020. Control of stimulus-dependent responses in macrophages by SWI/SNF chromatin remodeling complexes. Trends Immunol 41: 126–140. 10.1016/j.it.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Natoli G. 2016. Molecular control of activation and priming in macrophages. Nat Immunol 17: 26–33. 10.1038/ni.3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. 2010. Alternative activation of macrophages: mechanism and functions. Immunity 32: 593–604. 10.1016/j.immuni.2010.05.007 [DOI] [PubMed] [Google Scholar]
- Gour N, Wills-Karp M. 2015. IL-4 and IL-13 signaling in allergic airway disease. Cytokine 75: 68–78. 10.1016/j.cyto.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, Thierry GR, Bonnardel J, Bajenoff M. 2020. Establishment and maintenance of the macrophage niche. Immunity 52: 434–451. 10.1016/j.immuni.2020.02.015 [DOI] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16: 144–154. 10.1038/nrm3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Daniel B, Szeles L, Cuaranta-Monroy I, Czimmerer Z, Ozgyin L, Steiner L, Kiss M, Simandi Z, Poliska S, et al. 2019. Labelled regulatory elements are pervasive features of the macrophage genome and are dynamically utilized by classical and alternative polarization signals. Nucleic Acids Res 47: 2778–2792. 10.1093/nar/gkz118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado Jde D, Popovich PG, Partida-Sanchez S, Guerau-de-Arellano M. 2015. Novel markers to delineate murine M1 and M2 macrophages. PLoS One 10: e0145342 10.1371/journal.pone.0145342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19: 523–534. 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Jarjour NN, Schwarzkopf EA, Bradstreet TR, Shchukina I, Lin CC, Huang SC, Lai CW, Cook ME, Taneja R, Stappenbeck TS, et al. 2019. Bhlhe40 mediates tissue-specific control of macrophage proliferation in homeostasis and type 2 immunity. Nat Immunol 20: 687–700. 10.1038/s41590-019-0382-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. 2014. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159: 1312–1326. 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, Natoli G. 2011. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 11: 750–761. 10.1038/nri3088 [DOI] [PubMed] [Google Scholar]
- Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et al. 2011. Kruppel-like factor 4 regulates macrophage polarization. J Clin Invest 121: 2736–2749. 10.1172/JCI45444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchin A, Suchting S, Merson T, Rosol TJ, Hume DA, Cassady AI, Ostrowski MC. 2001. Genetic and physical interactions between Microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. J Biol Chem 276: 36703–36710. 10.1074/jbc.M106418200 [DOI] [PubMed] [Google Scholar]
- Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, Jiang Y, Kass DJ, Gibson K, Chen W, et al. 2019. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J 54: 1802441 10.1183/13993003.02441-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ. 2017. Macrophage polarization. Annu Rev Physiol 79: 541–566. 10.1146/annurev-physiol-022516-034339 [DOI] [PubMed] [Google Scholar]
- Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, et al. 2014. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41: 14–20. 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair MG, Du Y, Perrigoue JG, Zaph C, Taylor JJ, Goldschmidt M, Swain GP, Yancopoulos GD, Valenzuela DM, Murphy A, et al. 2009. Alternatively activated macrophage-derived RELM-α is a negative regulator of type 2 inflammation in the lung. J Exp Med 206: 937–952. 10.1084/jem.20082048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël W, Raes G, Hassanzadeh Ghassabeh G, De Baetselier P, Beschin A. 2004. Alternatively activated macrophages during parasite infections. Trends Parasitol 20: 126–133. 10.1016/j.pt.2004.01.004 [DOI] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. 2007. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 447: 1116–1120. 10.1038/nature05894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. 2013. Latent enhancers activated by stimulation in differentiated cells. Cell 152: 157–171. 10.1016/j.cell.2012.12.018 [DOI] [PubMed] [Google Scholar]
- Pello OM, De Pizzol M, Mirolo M, Soucek L, Zammataro L, Amabile A, Doni A, Nebuloni M, Swigart LB, Evan GI, et al. 2012. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 119: 411–421. 10.1182/blood-2011-02-339911 [DOI] [PubMed] [Google Scholar]
- Poirier R, Cheval H, Mailhes C, Charnay P, Davis S, Laroche S. 2007. Paradoxical role of an Egr transcription factor family member, Egr2/Krox20, in learning and memory. Front Behav Neurosci 1: 6 10.3389/neuro.08.006.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raredon MSB, Adams TS, Suhail Y, Schupp JC, Poli S, Neumark N, Leiby KL, Greaney AM, Yuan Y, Horien C, et al. 2019. Single-cell connectomic analysis of adult mammalian lungs. Sci Adv 5: eaaw3851 10.1126/sciadv.aaw3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Schmeier S, Arner E, Alam T, Parihar SP, Ozturk M, Tamgue O, Kawaji H, de Hoon MJ, Itoh M, et al. 2015. Redefining the transcriptional regulatory dynamics of classically and alternatively activated macrophages by deepCAGE transcriptomics. Nucleic Acids Res 43: 6969–6982. 10.1093/nar/gkv646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückerl D, Allen JE. 2014. Macrophage proliferation, provenance, and plasticity in macroparasite infection. Immunol Rev 262: 113–133. 10.1111/imr.12221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Tamaki K. 2006. Thymus and activation regulated chemokine (TARC)/CCL17 and skin diseases. J Dermatol Sci 43: 75–84. 10.1016/j.jdermsci.2006.06.002 [DOI] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, et al. 2010. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol 11: 936–944. 10.1038/ni.1920 [DOI] [PubMed] [Google Scholar]
- Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. 2014. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol 15: 1026–1037. 10.1038/ni.3005 [DOI] [PubMed] [Google Scholar]
- Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A. 2018. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol 233: 6425–6440. 10.1002/jcp.26429 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902 e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, et al. 2010. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity 33: 699–712. 10.1016/j.immuni.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AJ, Liu X, Thomas BJ, Lissner MM, Baker MR, Senagolage MD, Allred AL, Barish GD, Smale ST. 2016. A stringent systems approach uncovers gene-specific mechanisms regulating inflammation. Cell 165: 165–179. 10.1016/j.cell.2016.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken SJ, Locksley RM. 2013. Interleukin-4- and interleukin-13-mediated alternatively activated macrophages: roles in homeostasis and disease. Annu Rev Immunol 31: 317–343. 10.1146/annurev-immunol-032712-095906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veremeyko T, Yung AWY, Anthony DC, Strekalova T, Ponomarev ED. 2018. Early growth response gene-2 is essential for M1 and M2 macrophage activation and plasticity by modulation of the transcription factor CEBPβ. Front Immunol 9: 2515 10.3389/fimmu.2018.02515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available, published data sets can be accessed on the following GEO accession numbers: single-cell RNA-seq data sets GSE133747 and GSE128033 and published bulk RNA-seq of tissue-resident macrophage populations GSE63340. Sequencing data sets performed in this study are available at the NCBI GEO under accession number GSE151015.