Versatile high-throughput method can isolate single cells based on spatiotemporal phenotypes.
Abstract
Unraveling the genetic and epigenetic determinants of phenotypes is critical for understanding and re-engineering biology and would benefit from improved methods to separate cells based on phenotypes. Here, we report SPOTlight, a versatile high-throughput technique to isolate individual yeast or human cells with unique spatiotemporal profiles from heterogeneous populations. SPOTlight relies on imaging visual phenotypes by microscopy, precise optical tagging of single target cells, and retrieval of tagged cells by fluorescence-activated cell sorting. To illustrate SPOTlight’s ability to screen cells based on temporal properties, we chose to develop a photostable yellow fluorescent protein for extended imaging experiments. We screened 3 million cells expressing mutagenesis libraries and identified a bright new variant, mGold, that is the most photostable yellow fluorescent protein reported to date. We anticipate that the versatility of SPOTlight will facilitate its deployment to decipher the rules of life, understand diseases, and engineer new molecules and cells.
INTRODUCTION
How genetic variation creates phenotypic diversity is a central question in the biomedical sciences. This relationship underlies efforts to understand the basic principles of cellular function and to decipher the causes and markers of diseases (1). The ability to map genotypes to phenotypes is also critical for biological engineering and synthetic biology applications such as library screening to develop new protein biosensors, transcriptional reporters, synthetic enzymes, RNA devices, genetic circuits, signaling pathways, and multicellular communities (2). In all cases, high-throughput screening approaches are desirable to sift through heterogeneous populations of cells and retrieve variants exhibiting the properties of interest. Fluorescence-Activated Cell Sorting (FACS) is widely used for this purpose due to its high throughput and its ability to assay multiple cell types such as bacteria, yeast, and mammalian cells. However, FACS cannot track temporal processes in single cells, and even advanced instruments are limited in their ability to screen based on subcellular features or morphology (3). Microscopy-based approaches are preferred when screening for temporal or spatial properties such as biosensor kinetics, enzymatic reaction rates, gene expression dynamics, and subcellular localization (4). However, unlike FACS that allows the high-throughput screening of pooled single-cell variants, conventional microscopy-based screening involves the slow sequential imaging of variants compartmentalized into separate wells of microplates (5).
The pressing need to genotype single cells based on complex and dynamic phenotypes has motivated new approaches. However, existing methods are limited in versatility, throughput, and/or ease of use. In situ genotyping after image-based analysis of pooled variants enables identifications of genes that affect spatiotemporal properties in vitro (6, 7). However, these approaches are slow; for example, a screen for expression of a fluorescent protein (FP) in bacteria transformed with a library of ~80,000 variants lasted ~40 hours and required 14 hybridization steps (6). Moreover, the need to transform and read DNA barcodes limits these methods to genetically tractable cells and complicates the extension of this technique to intact tissues and in vivo preparations. Other approaches rely on post-imaging retrieval of selected cells from pooled libraries using automated pipetting (8), laser-based extraction from microcapillaries (9), optical traps (10, 11), optical or optomagnetic tagging with laser-assisted binding of biotin conjugates (12, 13), or optically controlled adhesion using a synthetic chemical (14). While useful and innovative, these methods are limited to a relatively narrow range of experimental preparations. For example, they are specialized for assaying and retrieving only one or a few specific cell types, with none having demonstrated compatibility with both yeast and human cells. These methods are designed for screening cells isolated into specialized chambers, plated onto monolayers, and/or labeled with exogenous tags. Therefore, they cannot be easily extended to isolating cells from tissues such as organoids, explants, and in vivo preparations. Some techniques require fluorophores with excitation spectra that strongly overlap with green FPs (GFPs), complicating their application in experiments using green probes and indicators (12, 13). Furthermore, many of these newer approaches require custom hardware that can be time-consuming and difficult to implement in standard biomedical labs (8–10, 13).
To address the limitations of existing sorting techniques, we developed a general strategy for isolating single live cells with unique visual phenotypes. Several groups have reported the use of phototransformable FPs (15, 16) to label populations of cells with subsequent recovery by FACS (17–19). We sought to evaluate whether FP-based optical tagging and FACS-based retrieval could be extended to single cells, thus enabling the extraction of individual cells with rare visual phenotypes from large heterogeneous cultures. We call this general isolation method Single-cell Phenotypic Observation and Tagging with Light or SPOTlight (Fig. 1).
Fig. 1. Overall workflow and potential applications of the SPOTlight method.
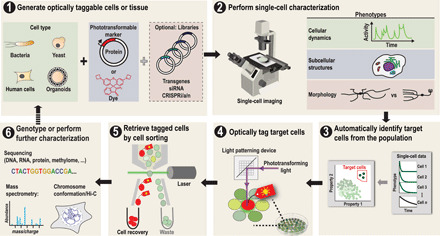
(Step 1) Cells of interest (e.g., bacteria, yeast, mammalian cells, or organoids) are made optically taggable by introducing a phototransformable FP or dye. These reagents can transform from a dim to bright state or from one color to another when excited with light of a specific wavelength. Some applications will screen naturally heterogeneous populations of cells, while others will screen libraries. siRNA, small interfering RNA. (Step 2) Cells are imaged for the phenotypes of interest. (Step 3) Phenotypes are quantified with single-cell resolution. Target cells are chosen based on the desired phenotypic profile. (Step 4) Target cells are optically tagged for retrieval by single-cell illumination. (Step 5) The tagged cells are detected and isolated using Fluorescence-activated cell sorting (FACS). (Step 6) The sorted cells are genotyped or further characterized as needed for the specific project. Additional rounds of screening can be performed if needed, for example, when conducting directed evolution experiments.
To illustrate the advantages of our approach, we first showed the generality of SPOTlight by demonstrating FP-based single-cell optical tagging in many experimental preparations including a variety of cell types and illumination methods. We also showed that single mammalian cells and whole organoids can be optically tagged by photoactivating an organic dye, thereby illustrating the applicability of our method to nongenetically tractable cells. To demonstrate the high-throughput nature of SPOTlight, we imaged more than 3 million cells expressing variations of a yellow FP (YFP) and simultaneously screened for photostability—a temporal property—and brightness. Using SPOTlight, we were able to identify a new YFP variant, mGold, that is up to ~5-fold more photostable and as bright as its parental protein, mVenus, making it the most photostable YFP to date. Because of the versatility and generality of SPOTlight, we anticipate its wide utilization for understanding and reengineering biological systems via high-throughput phenotypic screening of spatiotemporal properties.
RESULTS
Precise optical tagging of individual cells of various types within dense populations
While phototransformable FPs have been shown to enable optical highlighting of individual or population of cells (16–18, 20), we sought to determine whether these reagents could enable selective tagging of individual cells within crowded populations with high specificity and sufficient signal-to-background ratio to be detected by FACS. We focused on photoactivatable red FPs (PA-RFPs) given that they provide spectral compatibility with the large catalog of blue, cyan, green, and yellow fluorophores that are commonly used for spatiotemporal profiling of cellular activity (21). We benchmarked three PA-RFPs and selected PAmCherry1 (22) due to its faster photoactivation kinetics and larger fluorescence fold change upon activation compared with other PA-RFPs (fig. S1). We coexpressed PAmCherry1 and enhanced green FP (EGFP) in bacterial (Escherichia coli), yeast (Saccharomyces cerevisiae), and human cells [human embryonic kidney 293A (HEK293A)]. EGFP served as a marker to visualize cells under fluorescence microscopy. We arbitrarily selected cells in crowded fields of view and used a digital micromirror device (DMD) attached to a conventional widefield fluorescence microscope to restrict the photoactivating (violet) light to the preselected cells (fig. S2). For all the cell types tested, the tagged cells showed an increase in fluorescence of at least two orders of magnitude after 1.5 min of photoactivation (Fig. 2, A and B). Cells adjacent to the target cells showed minimal changes in fluorescence, confirming that we can achieve optical tagging with high spatial specificity in individual cells with a diameter ranging from ~1 μm (bacteria) to ~15 μm (human cells). We further showed that single cells expressing PAmCherry1 can be activated by two-photon laser scanning microscopy (fig. S3), a method of choice for deep-tissue imaging (23). However, initial illumination parameters produced lower contrast ratio than those obtained with one-photon illumination and will require further optimization.
Fig. 2. Spatially-precise single-cell optical tagging and retrieval can be achieved with multiple cell types.
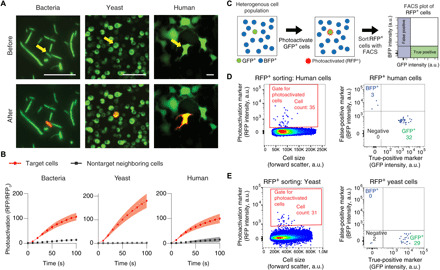
(A and B) Individual bacteria, yeast, and human cells can be selectively photoactivated in crowded cultures, with minimal spurious photoactivation of neighboring cells. (A) Representative examples. Yellow arrows point to targeted cells. Scale bars, 20 μm. (B) Mean photoactivation fold change of target cells compared with their closest neighboring cells. The shaded regions represent the standard error of the mean (SEM). n = 7 (bacteria), 18 (yeast), and 5 (human) target cells. Equal numbers of neighboring cells were quantified. RFPo is the RFP intensity at t = 0. (C to E) SPOTlight enables the identification of photoactivated cells with high precision. (C) Schematics of the experimental strategy. (D) Human cells expressing EGFP and PAmCherry (GFP+ cells) were diluted 20-fold with cells expressing TagBFP and PAmCherry1 (BFP+ cells). In this representative experiment, 56 GFP+ cells were photoactivated. Out of 35 cells detected in the RFP+ sorting gate (left), 32 (~91%) were true positives while 3 were false positives (right). (E) Yeast cells expressing EGFP and PAmCherry1 were diluted 500-fold with cells expressing TagBFP and PAmCherry1. In this representative experiment, 96 GFP+ cells were photoactivated. Of 31 cells detected in the RFP+ sorting gate (left), 29 (~94%) were true positives while 2 were false positives (right). The photoactivated cell gates for (D) and (E) were determined using controls shown in fig. S4C. a.u., arbitrary units.
We next sought to demonstrate that small populations of individually tagged cells can be isolated using FACS. We transiently transfected human cells with nucleus-localized fusions of PAmCherry1 with either GFP (GFP+ cells) or TagBFP [blue FP (BFP)+ cells]. GFP+ cells were diluted 20-fold with BFP+ cells. Automated image analysis was used to detect and photoactivate 50 to 100 GFP+ cells out of >100,000 cells under one-photon microscopy. We recovered 54 ± 10% (SD) of photoactivated cells using our RFP+ photoactivated cell gate with a precision [true positives/(true positives + false positives)] of 97 ± 5% (SD; Fig. 2, C and D, and fig. S4). To show that FACS-based detection of optically-tagged cells can be achieved with cells of different sizes and shapes, we also photoactivated 100 to 200 GFP+ yeast cells out of >600,000 cells containing a 500-fold excess of BFP+ cells. We recovered 26 ± 8% (SD) of photoactivated cells with a precision of 91 ± 2% (SD; Fig. 2, C and E, and fig. S4). Designing photoactivation gates to recover cells with weaker PA-RFP fluorescence increased recovery rates (sensitivity) but decreased precision (fig. S4E).
Optical tagging of human cells and intestinal organoids without genetic modification.
Having established single-cell optical tagging with PAmCherry1, we evaluated the applicability of our method for isolating nongenetically tractable cells using a photoactivatable dye [PA-JF549 (24)] rather than a photoactivatable FP. We stained human cells with PA-JF549 and targeted individual cells. After 1.5 min of photoactivation, the photoactivated human cells produced a ~130-fold increase in red fluorescence (Fig. 3, A and B), slightly larger than what we obtained with PAmCherry1 (Fig. 2B). Neighboring cells showed minimal changes in fluorescence, demonstrating the spatial precision of our technique. To verify that photoactivated cells can be detected using FACS, we tagged 112 of ~70,000 human cells and recovered 70 human cells, corresponding to a recovery rate of 62.5% (Fig. 3C). We confirmed by microscopy that the sorted HEK293A cells emitted bright red fluorescence expected of photoactivated cells (fig. S4F).
Fig. 3. A photoactivatable dye enables optical tagging and retrieval of human cells without genetically encoded transgenes.
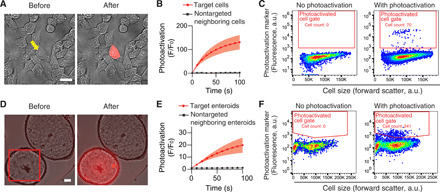
(A to C) Individual human cells stained with the photoactivatable dye PA-JF549 can be selectively tagged and retrieved. (A) Representative example of single-cell photoactivation. The yellow arrow points to the targeted cell. Merged images of the brightfield and red channels are shown. Scale bar, 20 μm. (B) Mean photoactivation fold change of target cells compared with their closest neighboring cells. F is the fluorescence; Fo is F at t = 0. The shaded regions represent the SEM. n = 12 cells per condition. (C) Photoactivated cells can be retrieved by FACS. In a representative example, 112 out of ~70,000 cells were individually photoactivated for 1 min. Seventy of the photoactivated cells (~63%) were recovered by FACS. (D to F) Whole enteroids stained with the photoactivatable dye PA-JF549 can be selectively tagged and their cells recovered by FACS. (D) Photoactivation of a representative enteroid. The red square shows the approximate area targeted for photoactivation. Merged images of the brightfield and red channels are shown. Scale bar, 50 μm. (E) Photoactivation fold change of target enteroids compared with their closest neighboring enteroids. The shaded regions represent the SEM. n = 11 enteroids per condition. (F) Cells from optically tagged whole enteroids can be recovered by FACS. Nine of 300 enteroids were each photoactivated for 1 min, pooled, and dissociated. A total of 241 individual cells were recovered by FACS.
We next sought to determine whether tissues could be stained with PA-JF549 and photoactivated, using human intestinal enteroids as a model. Photoactivation (1.5 min) of whole enteroids, which are typically composed of 500 to 1000 cells, produced a ~20-fold increase in red fluorescence with minimal activation of neighboring enteroids (Fig. 3, D and E). This fold increase in fluorescence was lower than observed with single human cells (Fig. 3B), possibly due to inefficient photoactivation of upper cell layers when using the bottom cell layer as the focal plane. We confirmed that photoactivated cells could be recovered by FACS by arbitrarily tagging 9 whole enteroids of a population of ~300, dissociating all enteroids into single cells, and sorting the resulting culture. A total of 241 individual photoactivated cells were recovered (Fig. 3F).
SPOTlight screening of ~3 million cells identified the most photostable YFP reported to date
While SPOTlight is a general strategy that can be applied to various visual phenotypes and cell types, we demonstrated its performance in a representative application that leverages its ability to screen for temporal properties. We focused on protein engineering given the large community seeking to develop improved antibodies, gene editing reagents, cell activity reporters, and protein-based optical and chemical switches. We decided to screen for improved FPs given the ubiquitous use of these reagents in biomedical research. A limitation of all FPs is that they suffer from photobleaching through repeated or prolonged illumination, thereby hindering their deployment in applications requiring long-term imaging of cellular activity (25) or high signal stability (26). We chose to improve mVenus (27, 28), a bright YFP widely used as an individual tag (29, 30) and within biosensors (31, 32).
Methods reported thus far for screening for photostability are limited in throughput. Photobleaching colonies on agar plates has low throughput because bacterial colonies must be sufficiently sparse to be isolated by picking and because plates are typically replaced manually. Moreover, the illumination must be spread over a wide area, resulting in low irradiances and, therefore, longer photobleaching times than achievable under microscopy (33). An alternative and innovative strategy is to photobleach and then sort single cells using a microfluidic device (34). However, as every cell is evaluated sequentially, high throughput can only be achieved using laser-based illumination at an irradiance that is orders of magnitude (~2 kW/cm2) above those commonly used for wide-field live cell imaging. Since photobleaching rates and mechanisms are dependent on illumination power and modality (35, 36), screening techniques with excitation power that more closely approximate common experimental methods would be preferred. An innovative image-based method based on single-cell barcoding was used to improve the photostability of the protein-fluorogen tag YFAST (6), but this method is laborious and time consuming as further discussed in the Introduction. Lastly, FACS is poorly adapted in screening for photostability given that it is a temporal property, requiring the comparison of fluorescence between the beginning and the end of illumination. Because SPOTlight is rapid and designed to screen for temporal properties, we proceeded to apply this method for improving the photostability of mVenus.
We sought to optimize YFPs for mammalian cell expression. Although Escherichia coli is the most common host for engineering FPs, we reasoned that yeast would be a more predictive host model given that it has similar protein synthesis machinery as mammalian cells (37). Moreover, beneficial mutations in bacteria may not extend to mammalian cells because mutations that increase maturation speed are predicted to have a larger increase in fluorescence when expressing FPs in faster-growing bacteria than in slower-growing mammalian cells (28). Screening in yeast, which has a growth rate between those of bacteria and mammalian cells, would mitigate this issue. Compared with screening in mammalian cells directly, yeast enables simpler production of large libraries. When commonly used FPs were expressed in yeast grown at 37°C and at pH 7, their relative brightness and photostability strongly predicted their relative performance in human cells, producing correlation coefficients (r) of 0.96 for brightness and 0.99 for photostability (fig. S5). These results validate our hypothesis that yeast cells grown under the right conditions are suitable to screen FPs for mammalian expression.
We sought to conduct a multiparameter screening of mVenus libraries given that simultaneous optimization of both brightness and photostability was previously shown to be critical to avoid selecting for photostable but dimmer variants (Fig. 4A) (36). We first developed an expression and screening vector coexpressing mVenus variants, PAmCherry1, and TagBFP from a strong constitutive promoter (Fig. 4B). TagBFP was used to normalize for cell-to-cell variations in plasmid copy number and expression capacity. We created libraries by multisite saturation mutagenesis of mVenus. Since different YFPs vary in their photostability, we targeted seven residues that showed high variation between these FPs (fig. S6A). In addition, as photobleaching can be affected by the conformational flexibility of the chromophore and by chemical reactions involving peri-chromophore residues (26), we also chose six residues that interact with the chromophore via hydrophobic interactions or hydrogen bonds (fig. S6B).
Fig. 4. Automated photostability and brightness screening of ~ 3 million cells encoding mVenus variants.
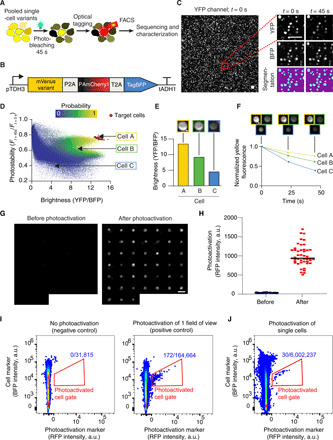
(A) Schematic of the experimental protocol. Photobleaching is performed with 508/25 nm light at 20 mW/mm2. (B) Expression cassette on the screening plasmid. (C to J) A representative round of screening. (C) Left: A representative field of view showing yeast cells expressing mVenus variants. Right: Magnified images and segmentation masks. Scale bars, 25 μm. (D) Photophysical properties of 287,186 individual cells from a representative library screening experiment. The color scheme represents the probability that each individual cell expresses a mutant with improved optical properties compared with its parental FP. The red circles indicate cells that were selected for optical tagging. (E and F) Representative example of three cells with different brightness (E) and photostability values (F). Cells are from the library depicted in (D). Scale bars, 1 μm. (G) A mosaic image of the 45 photoactivated cells before and after photoactivation. Scale bar, 10 μm. (H) RFP intensities of target cells before and after photoactivation. The black lines indicate the median values. (I) The photoactivation (RFP+) gate was designed using control samples with (left) and without (right) photoactivated cells. The ratio in blue indicates the number of cells inside the gate over the total number of cells analyzed. (J) Thirty of 45 optically tagged cells were detected and sorted by FACS.
We built eight mutagenesis libraries that targeted 21 residues: the 13 residues mentioned above and eight neighboring residues (fig. S6, C and D). We simultaneously targeted three residues per library, thus producing 8000 possible combinations. According to a probabilistic model (38), these libraries required screening of 12,000 clones to achieve a 95% probability of identifying one of the top three variants. We therefore sought to produce libraries with greater than 12,000 colonies. Using standard chemical transformation, we obtained up to 54,400 colonies (fig. S6E) that were pooled together for SPOTlight imaging. We imaged >10-fold more cells than colonies to enable excellent coverage of the colonies. The total number of individual cells assayed ranged from around 200,000 to more than 700,000 cells per library (fig. S6F).
Pooled single-cell variants were immobilized on an imaging plate to form a densely-packed monolayer of cells: ~5000 cells per field of view of ~0.43 mm2 (Fig. 4C). The cells were then photobleached with 508 nm light for 45 s at 20 mW/mm2, producing a ~40% average decrease in fluorescence. For each round of screening we photobleached and imaged 64 to 169 fields of view and conducted an automated image analysis of individual cells. As brightness and photostability can show variation between individual cells expressing the same FP, we quantified the probability that each cell was expressing a brighter and/or more photostable variant than the parental FP (Fig. 4, D to F). These statistics were used to establish selection thresholds for brightness and photostability. Up to 200 cells were automatically tagged using these pre-established thresholds for brightness and photostability (Fig. 4, G and H). We sorted photoactivated cells by FACS (Fig. 4, I and J), regrew each cell in a separate culture, and confirmed the improved brightness and/or photostability of the selected variants expressed by these homogenous populations.
We screened ~3 million single cells across eight libraries and identified several variants that showed improved photostability. Our best variant combined the mutations L46F and T63S (fig. S6G), resulting in up to ~5-fold longer photobleaching half-lives in yeast and human cells without any loss in brightness (Fig. 5, A and B, and fig. S7). To our knowledge, T63S has not previously been reported as a beneficial mutation. We named mVenus(L46F;T63S) “mGold” because it is a more (photo)durable protein. mGold has similar spectra, pKa (where Ka is the acid dissociation constant), and molecular brightness as mVenus (fig. S8, A to E, and table S1). mGold and mVenus are equally insensitive to chloride (fig. S8F and table S1) and have a similar profile of low cytotoxicity (fig. S8G). We demonstrated that mGold is monomeric (fig. S8H), has similar brightness as mVenus in bacteria (fig. S8, I and J), and can be used as a fusion partner in human cells (Fig. 5, C and D, and fig. S9). We also showed the suitability of mGold to visualize cellular dynamics by conducting 3- to 24-hour time-lapse imaging experiments with 30- to 180-s intervals. Using mGold fused with keratin, we visualized cell division (movie S1) and cell attachment (movie S2). When mVenus was imaged under the same conditions, it photobleached rapidly (Fig. 5C), highlighting the importance of photostability for time-lapse imaging.
Fig. 5. SPOTlight screening identified a YFP with up to fivefold improvement in photostability.
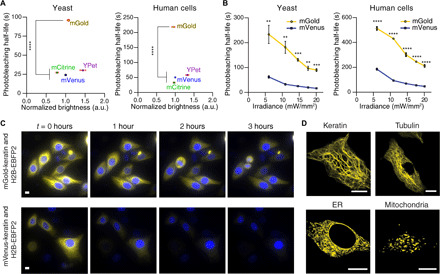
(A and B) In cellulo characterization of mGold, the best YFP variant identified from screening. (A) mGold was more photostable than commonly used YFPs and exhibited similar brightness. Cells were photobleached with 508/25-nm light at 20 mW/mm2. YFP brightness was normalized for cell-to-cell differences in protein expression (YFP/BFP). The square markers indicate the means from six yeast cultures or six independent transfections. For each replicate, the mean photobleaching half-life and brightness of several hundred yeast or human cells were determined. The error bars represent the SEM. P ≤ 0.0001 for Welch’s analysis of variance (ANOVA) for both yeast and human cell data. ****P ≤ 0.0001 for Dunnett’s T3 post hoc test. (B) mGold is more photostable than mVenus over a range of irradiance levels. The mean photobleaching half-lives were computed from three yeast cultures or three independent transfections per irradiance level. For each replicate, the mean photobleaching half-life of several thousand yeast cells or hundreds of human cells was determined. The error bars represent the SEM. P = 0.013 (yeast) and ≤ 0.0001 (human cells) for t tests comparing the areas under the curve. **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 for t tests corrected for multiple comparisons with the Holm-Šídák method. (C) Time-lapse imaging experiment of representative HeLa cells expressing a fusion of keratin with mGold (top) or mVenus (bottom). Nuclei were identified by coexpressing a fusion of H2B and EBFP2. Cells were imaged every 30 s for 3 hours. Scale bars, 20 μm. (D) Expressing fusions of mGold and subcellular localization tags produced the expected pattern of fluorescence in HeLa cells. ER, endoplasmic reticulum. Scale bars, 10 μm.
DISCUSSION
Mapping genotypes to phenotypes at the single cell level is critical to understanding and repurposing macromolecules, pathways, and organisms. Microscopy-based profiling of individual cells is now a routine, aided by automated hardware and image analysis tools for quantifying complex spatiotemporal phenotypes (21). The ability to retrieve specific cells with desired phenotypic profiles is a widespread need, bolstered by enthusiasm for single-cell multiomics technologies and cell atlas projects (39), but remains challenging. Advanced microscopy-based techniques have emerged for single-cell retrieval (8–14). However, existing approaches are best adapted to specific experimental contexts (e.g., cell type, imaging chamber, assay type and/or experimental preparation), limiting the breadth of their applications.
To address this problem, we developed SPOTlight, a general microscopy-based method for sorting individual cells based on visual phenotypes that has the potential to be applied to many different experimental contexts. SPOTlight combines microscopy-based imaging with single-cell optical tagging and FACS-based retrieval of tagged cells (Fig. 1). We illustrated the generality of our method by demonstrating that SPOTlight can be conducted with both yeast and human cells expressing PA-RFPs (Fig. 2) and nongenetically tractable human cells by replacing PA-RFPs with photoactivatable dyes (Fig. 3, A to D).
We expect that SPOTlight can be extended to bacterial cells given that they can also be photoactivated with high spatial precision (Fig. 2, A and B). However, attaching bacterial cells to glass using poly-l-lysine can interfere with cellular health (40) and in this work resulted in incomplete attachment of some cells on the plate. Experiments with bacteria would, therefore, benefit from microfluidics chambers or alternative strategies for planar imaging of cells. We also expect that SPOTlight can be adapted to retrieve single cells from tissue explants or in vivo, for example to isolate and determine the transcriptome of individual cells with interesting morphologies or activity patterns in a more physiological context. While we have demonstrated photoactivation and retrieval of populations of cells from human intestinal organoids (Fig. 3, E to G), the ability to extend these results to individual cells will require methods such as two-photon microscopy that can restrict photoactivation to single cells with high three-dimensional spatial precision. PA-RFPs can be photoactivated under two-photon illumination, as shown in populations of cells (41, 42) and in single cells (this work, fig. S3). However, conducting SPOTlight in tissues with two-photon excitation of individual cells will require improvements in the photoactivation contrast ratio and development of robust tissue dissociation protocols.
Retrieval of tagged cells after imaging allows the massively-parallel spatiotemporal screening of single-cell variants. As a demonstration that SPOTlight enables retrieval of individual cells based on temporal properties, we conducted a fully automated directed evolution of photostable and bright YFPs. We screened ~3 million single cells at a throughput of ~1500 to 2000 variants per min. This is the equivalent of ~16 to 21 96-well plates per min (assuming one variant per well), far exceeding the throughput possible with standard well-by-well approaches. The absolute throughput of SPOTlight will vary between assays depending on the properties being monitored, the number of cells per field of view (which, in turn, depends on the objective magnification, the detector size, and the cell density), and the speed at which a specific microscope and illumination method can scan fields of view. Compared with other screening approaches that compartmentalize single-cell variants into individual microfluidic chambers or microcapillaries (9–11), SPOTlight can work on densely packed populations of cells. As a result, a greater number of cells within a defined imaging area can be screened. For complex experiments, the FACS step of SPOTlight can be used as an opportunity to screen based on forward scatter, side scatter, or fluorescence in a different color channel. The latter would enable SPOTlight to be combined with assays such as antibody staining that are often more easily performed outside of the microscope stage.
While our manuscript was under revision, a study reported the application of a green-to-red photoconvertible protein for tagging and retrieving individual cells (43). Compared with our focus on a temporal property (photostability), the authors applied their method to spatial characteristics such as morphology and subcellular localization, demonstrating that our conceptual approach can be extended to a wide range of visual phenotypes. Our implementation with a single-color red photoactivable FP rather than a green-to-red photoconvertible FP enables spectral compatibility with cyan-to-yellow fluorophores. This is an important advantage, allowing screening with—or optimization of—many of the most commonly used fluorescent indicators of cellular activity (44, 45). SPOTlight can readily be extended to use photoactivatable FPs or dyes of other colors (20, 46, 47) when screening cells expressing or labeled with a red fluorophore.
Our screening experiments highlight the importance of multiparameter screening of both static and dynamic properties. One of the mutations of mGold, L46F, reverted a key mVenus mutation originally shown to increase fluorescence by more than 10-fold in E. coli at 37oC (48). Our results suggest that the use of F46L may come at the expense of photostability, a property that was not assayed in the original studies. In the context of mGold, L46F caused no significant decreases in brightness when expressed in bacteria, yeast, and human cells (Fig. 5A and fig. S8I). To the best of our knowledge, mGold is the most photostable YFP currently available. Therefore, beyond being a powerful illustration of the SPOTlight platform, we anticipate that mGold will facilitate experiments in which photostability is important, such as when conducting time-lapse imaging to follow cellular activity over prolonged durations or when high light power is necessary for detection of a small number of molecules.
Additional development of SPOTlight would also be useful. For example, a greater photoactivation contrast ratio would help improve sensitivity by increasing the detectability of optically tagged cells from the overall population (Fig. 2, D and E). This may be achieved by photoactivating cells with higher light power (43) or by developing new PA-RFPs with greater photoactivation efficiency and faster kinetics. Faster photoactivation would accelerate screening or enable tagging of more cells within the same duration. Optimizing the dissociation of cells from plates would be expected to increase the number of photoactivated cells that can be recovered.
Critically, SPOTlight can be rapidly disseminated to the broader community, as fluorescence microscopes can be easily retrofitted with photoactivation light sources and commercially available light patterning devices. Optical tagging can in principle be accomplished with a wide range of commercially-available devices that enable selective illumination of target cells such as digital micromirror devices (this work), galvano scanners, spatial light modulators, single-spot laser-based photoactivation units, or acousto-optic deflectors. Laser-scanning confocal microscopes, common in institutional core facilities, are often equipped with a 405 nm laser, which can photoactivate the PA-FPs and dyes described here. A key feature of many disruptive technologies is their utility across multiple fields and their compatibility with many experimental preparations (e.g., FPs and CRISPR-Cas9). Similarly, we believe that the versatility of SPOTlight for sophisticated phenotype-activated cell sorting will result in its broad adoption across the biomedical sciences, bioengineering, and synthetic biology. Overall, we anticipate that SPOTlight will become a method of choice for understanding and reengineering biological systems by mapping visual phenotypes to genotypes in individual cells.
METHODS
Post-publication updates and resources will be posted at www.spotlightscreening.tech.
Plasmid construction
Expression plasmids were cloned using standard cloning methods. Selected mGold expression plasmids and their sequences are available from Addgene (plasmid numbers 157995–158009). Additional plasmid sequences are available upon request.
For expression in bacteria, FPs were cloned in pNCS (49), a bacterial expression plasmid with a constitutive promoter (50), a T7 terminator, and an ampicillin resistance marker. For optical tagging experiments, EGFP and PAmCherry1 were coexpressed from the same promoter using separate ribosome binding sites (AAGAAGGAGATATACATATG).
To express constructs in yeast, we created the pJL1 plasmid by subcloning the strong constitutive pTDH3 promoter from the pYTK009 plasmid (51) in pDR196, a multicopy 2-micron plasmid with URA3 auxotrophic selection marker and the tADH1 terminator (52). Restriction sites for Nhe I and Bgl II were introduced in pDR196 to flank the pTDH3 promoter. Note that the Bgl II restriction site also functions as the Kozak sequence (51). Ribosome-skipping 2A sequences from porcine teschovirus-1 virus (P2A) and/or Thosea asigna virus (T2A) were used for multicistronic expression. To characterize PAmCherry1 expressed from the genome, genome integration was conducted as previously described (53).
To express constructs in human cells, we used a few different plasmid backbones. The experiments with Clover, EGFP, mNeonGreen, and Envy in fig. S5 were performed using the series of pcDNA3.1/puro-CAG-GFP-P2A-mCherry plasmids previously reported (54), as listed below (see the “Fluorescent proteins” section). The experiments with mClover3 and sfGFP were made with plasmids constructed by substituting Clover with these alternative GFPs. To determine the two-photon excitation spectra and cytotoxicity of FPs, we cloned FPs in the Nhe I and Hind III sites of pcDNA3.1/puro-CAG (55), a plasmid we have previously used for a similar purpose (56). For targeting mGold to subcellular regions, we replaced the native FP for mGold in Addgene plasmids #54134 (keratin), #56324 (endoplasmic reticulum), #56619 (mitochondria), #56618 (lysosome), #56332 (golgi), #55507(endosome), #55953 (nucleus), #56395 (actin), and #56399 (tubulin). For all other experiments, we used the pJL2 plasmid, which we created using the Nhe I and Kpn I restriction sites to replace the yeast expression cassette in pJL1 with an expression cassette for human cells. FPs were cloned between the strong constitutive cytomegalovirus promoter (subcloned from the pRS406 plasmid, Addgene #83410) and the bovine growth hormone polyadenylation signal (subcloned from the Duet011 plasmid, Addgene #17627).
Fluorescent proteins
The FPs used in this work are shown in Table 1.
Table 1. FPs used in this study.
Note that Venus has incorrectly been used to refer to slightly different sequences in publications following the initial report of Venus (27, 28). The mVenus variant used in this work has been confirmed to be the monomeric (A206K) version of the original Venus by the first author of the publication reporting Venus (T. Nagai, pers. comm.) and has the same protein sequence (except A206K) as the Venus from the plasmid the Miyawaki group deposited on Addgene (#54859).
| Fluorescent protein | Type | Source |
| TagBFP | BFP | Gene synthesis using the sequence from Evrogen catalog no. FP171 as the template |
| EGFP | GFP | pcDNA3.1-Puro-CAG-EGFP-P2A-mCherry used in Ref. (55), a generous gift from Michael Lin (Stanford University) |
| Clover | GFP | pcDNA3.1-Puro-CAG-Clover-P2A-mCherry used in Ref. (54), a generous gift from Michael Lin (Stanford University) |
| mClover3 | GFP | Gene synthesis using the sequence in Ref. (54) as the template |
| sfGFP | GFP | Addgene #54579 |
| mNeonGreen | GFP | pcDNA3.1-Puro-CAG-mNeonGreen-P2A-mCherry used in Ref. (54), a generous gift from Michael Lin (Stanford University) |
| Envy | GFP | pcDNA3.1-Puro-CAG-Envy-P2A-mCherry used in Ref. (54), a generous gift from Michael Lin (Stanford University) |
| mVenus* | YFP | pNCS-mVenus, generous gift from Michael Lin (Stanford University) |
| mCitrine | YFP | Addgene #54723 |
| YPet | YFP | Gene synthesis using the sequence in Ref. (57) as the template |
| mCherry | RFP | pcDNA3.1-Puro-CAG-Clover-P2A-mCherry used in Ref. (54), a generous gift from Michael Lin (Stanford University) |
| tdTomato | RFP | Addgene #91767 |
| PAmCherry1 | PA-RFP | Addgene #44855 for yeast experiments, and Addgene #31928 for bacteria and mammalian cell experiments |
| PATagRFP | PA-RFP | Addgene #44854 |
| PAmKate | PA-RFP | Addgene #32692 |
Bacterial culture
E. coli XL10-Gold (catalog no. 200315, Agilent) was used for the optical tagging experiments and for general cloning purposes. Miller LB Broth (catalog no. BP1426, Fisher Scientific) and Miller LB Agar (catalog no. BP1425, Fisher Scientific) supplemented with appropriate antibiotics were used to grow bacteria and were prepared following the manufacturer’s instructions.
Yeast culture
All experiments with yeast cells were conducted using the S. cerevisiae BY4741 strain. All yeast expression plasmids used the URA3 gene as a selection marker. To avoid plasmid loss, cells were grown in yeast synthetic dropout medium prepared by mixing Minimal Synthetic Defined Base (catalog no. 630411, Takara Bio) and -Ura dropout Supplement (catalog no. 630416, Takara Bio) following the manufacturer’s instructions. For growth at pH 7.0, the synthetic dropout medium was supplemented with 10 mM HEPES (catalog no.H3375, Sigma-Aldrich) and adjusted pH to 7.0 using NaOH (catalog no. S5881, Sigma-Aldrich). To prepare yeast competent cells, yeast extract, peptone, and dextrose [YPD; catalog no. 242820, Becton Dickinson (BD)] was used. Agar plates were prepared by adding 25 g/liter of agar (catalog no. BP1423, Fisher Scientific) to liquid synthetic dropout and YPD media.
Human cell culture
HEK293A cells (RRID:CVCL_6910, catalog no. R70507, Thermo Fisher Scientific) and HeLa cells (from the Cell-Based Assay Screening Service Core, Baylor College of Medicine) were used for mammalian experiments. Human cells refer to HEK293A cells unless specified otherwise. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM)–high glucose (catalog no. D1145, Sigma-Aldrich) supplemented with 5% fetal bovine serum (FBS) (catalog no. F2442, Sigma-Aldrich), 2 mM l-glutamine solution (catalog no. G7513, Sigma-Aldrich), and 1% (% v/v) penicillin-streptomycin (catalog no. P4333, Sigma-Aldrich) at 37oC in air with 5% CO2. The cell lines were confirmed to be mycoplasma free using the MycoAlert Mycoplasma Detection Kit (catalog no. LT07-118, Lonza).
Enteroid culture
D103 duodenal enteroids were acquired from the Digestive Diseases Center Enteroid Core (Baylor College of Medicine). Enteroids were passaged in three dimensions suspended in phenol red–free, growth factor–reduced Matrigel (catalog no. 356231, Corning) with high Wnt complete media with growth factors (hW-CMGF+) medium (58) supplemented with 10 μmol of Y-27632 Rock inhibitor (catalog no. S1049, Selleck Chemicals) and normocin (100 μg/ml) (catalog no. ant-nr-1, InvivoGen) and split weekly.
Fluorescence imaging and optical tagging setup
General setup
Fluorescence imaging was performed using an inverted microscope (Eclipse Ti-E, Nikon Instruments) equipped with a motorized XY stage with linear encoders (H139E1N4, Prior Scientific), a hardware autofocus module (Perfect Focus System, Nikon Instruments), a multi-photon module (A1R-MP, Nikon Instruments), a 20× 0.75–numerical aperture (NA) objective (CFI Plan Apo Lambda, Nikon Instruments), and imaging software (NIS-Elements HC version 4.60, Nikon Instruments). For optically tagging whole enteroids, a 10× 0.45-NA objective (CFI Plan Apo Lambda, Nikon Instruments) was used. All power measurements were conducted at the sample plane using a power meter console (PM400, Thorlabs) connected to a sensor head (S170C, Thorlabs).
Widefield microscopy setup for SPOTlight experiments
Blue, green, yellow, and red FPs/dyes were excited with 395/50-nm (peak/bandwidth), 470/49-nm, 510/50-nm, and 550/40-nm light, respectively, from a solid-state multispectral light engine (Spectra X, Lumencor). Excitation and emission light were routed to and from the sample, respectively, using multi bandpass dichroic mirrors (for BFP and YFP: 69008bs, Chroma; for GFP and RFP: 89100bs, Chroma) located in a filter cube below the objective turret. Emission light was filtered with multiband filters (for BFP and YFP: 69008m, Chroma; for GFP and RFP: 89101m, Chroma). Although not necessary for this work, our microscope could record from two colors simultaneously using two scientific complementary metal-oxide-semiconductor (sCMOS) cameras (ORC Flash 4.0 V2, Hamamatsu) attached to a beamsplitter (TwinCam, Cairn Research). To separate blue and yellow fluorescence, we fitted the beamsplitter with a dichroic mirror (ZT488rdc, Chroma); BFP emission was further filtered by a 450/50-nm emission filter (ET450/50m, Chroma). To separate green and red fluorescence, the beamsplitter was fitted with a separate dichroic mirror (T565lpxr, Chroma), and RFP emission was additionally filtered by a 632/60-nm emission filter (ET632/60m, Chroma). To optically tag individual cells expressing photoactivatable FPs or stained with photoactivatable dyes, light from a high-power 405-nm light-emitting diode (UHP-F5-405, Prizmatix) was patterned using a digital micromirror device (600 × 800 micromirrors; 10.2 mm by 13.6 mm; TI-LA-DMD, Nikon Instruments).
Widefield microscopy setup for long-term time-lapse imaging experiments
For time-lapse imaging experiments with keratin-YFP fusions and a fusion of H2B and the enhanced blue FP 2 (H2B-EBFP2, used as nuclear marker), we used a different microscope setup for long-term imaging. We used an inverted microscope (Eclipse Ti-E, Nikon Instruments) equipped with a hardware autofocus module (Perfect Focus System, Nikon Instruments), a motorized XY stage with linear encoders (Ti-S-ER, Nikon Instruments), a 20× 0.75-NA objective (CFI Plan Apo VC DIC N2, Nikon Instruments), and imaging software (NIS-Elements HC version 5.20, Nikon Instruments). HeLa cells were maintained at 37oC in air with 5% CO2 using a stage top incubator (H301, Okolab). BFPs and YFPs were imaged with 405- and 520-nm laser lines, respectively, using a light engine that produces expanded beams for widefield microscopy (LDI-WF, 89 North). Excitation light was filtered (for BFP, ZET405/470/555/640x, Chroma; for YFP, ZET445/520x, Chroma). Excitation and emission lights were routed to and from the sample, respectively, using multiband dichroic mirrors (for BFP, ZT405/470/555/640rpc, Chroma; for YFP, ZT445/520rpc, Chroma). Emission light was filtered with multiband filters (for BFP, ZET405/470/555/640m, Chroma; for YFP, ZET445/520m, Chroma). Images were acquired using a sCMOS camera (pco.edge 4.2, PCO).
Two-photon microscopy setup
Two-photon imaging experiments were conducted on the same microscope described in the “Widefield microscopy setup for SPOTlight experiments” section. Galvanometric mirrors were used to steer a titanium:sapphire Chameleon Ultra II laser (Coherent). PAmCherry1 was excited with 800 nm light. A 560-nm long-pass dichroic mirror (Nikon Instruments) was used to separate green and red fluorescence. Emission light was further filtered by a 525/50-nm filter (GFP and YFP emission) or a 605/70-nm filter (PAmCherry1) and collected using gallium arsenide phosphide (GaAsP) detectors.
Characterization of optical tagging
Preparing cells for optical tagging
Yeast transformation was conducted using the LiAc/single-stranded (SS) carrier DNA/polyethylene glycol (PEG) method (59). One hundred microliters (3 × 108 cells) of frozen competent yeast cells were transformed with pJL1, a plasmid coexpressing EGFP and PAmcherry1. To maximize the transformation efficiency, we heat-shocked the cells for 1 hour and cotransformed cells with high purity single stranded DNA (ssDNA; catalog no. D9156, Sigma-Aldrich). Transformed yeast were plated on uracil dropout agar plates and incubated at 30°C. Seventy-two hours post-transformation, colonies were picked and grown overnight in uracil dropout medium at pH 4.3 and 30oC. Cells were diluted around ~20-fold and regrown to mid-log phase, corresponding to an OD600 (optical density at 600 nm) of 2.0, as measured using the SmartSpec3000 spectrophotometer (Bio-Rad). The cells were washed three times with water and plated on glass-bottom plates (catalog no. P24-1.5H-N, Cellvis) coated with a 0.1 mg/ml solution of poly-l-lysine. The stock solution of poly-l-lysine was prepared by dissolving poly-l-lysine (molecular weight, ≥300,000; catalog no. P1524, Sigma-Aldrich) to 1.0 mg/ml in a 0.1 mM boric acid–sodium tetraborate decahydrate solution (catalog nos. B0252 and B9876, Sigma-Aldrich) and adjusted pH to 8.5 using NaOH. The poly-l-lysine solution was diluted to its final concentration (0.1 mg/ml) using phosphate-buffered saline (PBS; catalog no. 806544, Sigma-Aldrich). The attached cells were washed twice with water. After the final wash, water was replaced with PBS.
To conduct experiments with human cells expressing PA-RFPs, HEK293A cells were cotransfected with pJL2-EGFP and pCMV-PAmCherry1 plasmids using the FuGENE HD Transfection Reagent (catalog no. E2311, Promega) following the manufacturer’s instructions, except that cells were transfected with 200 ng of DNA and 0.6 μl of FuGENE per well of a 24-well plate or 100 ng of DNA and 0.3 μl of FuGENE per well of a 96-well plate. Transfected cells were placed on a poly-l-lysine-coated 24-well or 96-well glass-bottom microplate (catalog no. P24-1.5H-N or P96-1.5H-N, Cellvis) and incubated at 37°C in air with 5% CO2 for 2 days. Only before imaging, cells were washed once with Dulbecco’s PBS (DPBS) (catalog no. 21-031-CV, Corning), and the growth medium was replaced with Hank’s balanced salt solution (HBSS) (catalog no. H8264, Sigma-Aldrich), supplemented with 10 mM HEPES.
For experiments with human cells stained with photoactivatable dyes, PA-JF549 dye (24) was diluted to 100 nM in supplemented DMEM. The medium used to culture HEK239A cells or enteroids was replaced by the diluted dye. The cells were stained for 15 min (HEK239A cells) or 30 min (enteroids) at 37oC in air with 5% CO2. Stained cells were washed once by replacing the diluted dye with fresh DMEM and by incubating the cells at 37oC in air with 5% CO2 for 15 min. After the wash, the culture medium was replaced with HBSS-HEPES solution.
Bacterial transformation was conducted using typical chemical transformation to introduce the pNCS plasmid coexpressing EGFP and PAmCherry1. Colonies were picked from the transformation plate and the cells were grown overnight at 37oC in LB medium supplemented with ampicillin (catalog no. BP176025, Fisher Scientific). Cells were diluted ~50-fold and regrown to mid-log phase (OD600, 0.5). Cells were immobilized on a glass-bottom plate as described above.
Optical tagging under widefield illumination
The digital micromirror device was used to produce illumination areas at the sample plane of 4, 16, and 64 μm2 for photoactivation of bacteria, yeast, and human cells, respectively, unless noted otherwise. The power levels at the sample planes were 1.2, 1.4, and 2.1 μW, respectively. Individual cells at the center of the fields of view (i.e., target cells) were photoactivated for 240 s and RFP images (67 mW/mm2; 50-ms exposure time) were taken every 10 s. GFP images were taken before photobleaching to identify the cells closest to the target cells for evaluation of nonspecific photoactivation. For optical tagging of cells stained with dyes, brightfield images were taken to identify the cells closest to the target cells. To optically tag enteroids, illumination regions were manually defined to redirect the photoactivating light to different parts of the enteroids or whole enteroids.
Optical tagging under two-photon illumination
To optically tag individual cells, we used the same 20× 0.75-NA objective (CFI Plan Apo Lambda, Nikon) as elsewhere. A zoom size of ~30 was used to scan single cells with 800 nm light at 2% power with a pixel dwell time of 0.125 μs and a scanning rate of 8 Hz. The total duration of photoactivation was 12 s. To monitor red fluorescence generated by photoactivation, images were taken every 3 s using 1050-nm light at 30% power and a pixel dwell time of 16 μs.
Flow cytometry
To recover yeast cells, a fluorescence-activated cell sorter (SH800S, Sony Biotechnology) equipped with 405-nm, 488-nm, and 561-nm laser was used to detect blue, yellow and red fluorescence, respectively. 450/50-nm, 525/50-nm, and 600/60-nm emission filters were used to filter blue, yellow, and red fluorescence, respectively. To recover human cells, a different cell sorter (FACSAriaII, BD Biosciences) was used because of internal regulations of our cytometry core that prohibited the use of human cells with the SH800S sorter. The FACSAriaII was equipped with a laser with the same excitation wavelengths as with the SH800S sorter. 450/50-nm, 530/30-nm, 610/20-nm emission filters were used to filter blue, yellow, and red fluorescence, respectively. For the FP cytotoxicity assay, a flow cytometer (Attune NxT, Thermo Fisher Scientific) was used. This flow cytometer was equipped with 405-nm and 488-nm lasers. The 405-nm laser with a 512/25-nm emission filter was used to detect green fluorescence while the 488-nm laser with a 530/30-nm emission filter was used to detect yellow fluorescence.
Yeast and human cells were prepared for flow cytometry first by detaching the cells on imaging plates or culture dishes with trypsin-EDTA solution (catalog no. T3924, Sigma-Aldrich) for 5 min at 37oC, deactivating trypsin by adding culture medium, washing the cells once with PBS, and finally resuspending the cells in PBS. To prepare 3D enteroids for flow cytometry, we dissociated the 3D enteroids into single cells first by lifting the enteroids from Matrigel using ice-cold 0.5-mM EDTA in PBS. Lifted enteroids were then dissociated with trypsin-EDTA for 4 min at 37oC. Trypsin was deactivated by adding enteroid culture medium. Cell solution was filtered using a 40-μm cell strainer (catalog no. 352340, Corning). Cells were washed once with PBS and resuspended in PBS.
Simulating the recovery of photoactivated cells
The best-fit lines of sensitivity versus precision plot were determined by simulating the recovery of photoactivated cells using flow cytometry data. The cell count, means, and SDs of logged red fluorescence intensities of nonphotoactivated cells were used to simulate a Gaussian distribution of the nonphotoactivated cells. The number of the photoactivated cells, the fraction of imaged cells that were also analyzed by flow cytometry, and the means and SDs of logged red fluorescence intensities of the photoactivated cells were used to simulate a Gaussian distribution of the photoactivated cells. The ratio of GFP+ cells in the nonphotoactivated population was estimated from the actual flow cytometry results, and the ratio of the GFP+ cells in the photoactivated population was estimated as the largest precision from each trial. Various red fluorescence thresholds were applied to define photoactivated and nonphotoactivated cells to calculate the true-positive (GFP+ and RFP+) cells and false-positive (GFP− and RFP+) cells in the actual flow cytometry results and the simulated populations. These cell counts were used to calculate the sensitivity and precision at each red fluorescence threshold. To fit a recovery simulation model with the actual data, the SD of the logged red fluorescence intensities of the photoactivated population and the retrieval rate were optimized to minimize the sum of the squared offsets between the fitting curve and the actual data points.
Brightness and photostability screening using SPOTlight
Constructing, transforming, and plating yeast libraries
We developed mutagenesis libraries by simultaneously randomizing three predefined residues per library. Degenerate codons (NNS) were used to randomize each position with all 20 amino acids during a polymerase chain reaction. The overlap regions between inserts and linearized vector were designed as 18 base pairs in length. In-Fusion HD Cloning Kit was used for seamless DNA cloning (catalog no. 639650, Takara Bio). The In-Fusion reaction comprised the following components: 250 ng of linear vector (digested with Bgl II and Pst I), gel extracted and purified DNA insert fragments (2:1 to 6:1 molar ratio of insert to vector), 1.2 μl 5× In-Fusion HD enzyme premix, and sterilized distilled water to a total volume of 6 μl. The In-Fusion reaction mixture was incubated at 50°C for 15 min according to the user manual. The yeast transformation was conducted by adding 24 μl In-Fusion reaction mixture (~1 to1.5 μg of vector and fragments) into 100 μl of frozen yeast competent cells (3 × 108 cells) using the LiAc/SS carrier DNA/PEG method (59). To maximize the transformation efficiency, we heat-shocked the cells for 1 hour and cotransformed cells with the high-purity ssDNA. Transformed yeast were plated on uracil dropout agar plates and incubated at 37°C instead of the typical 30oC yeast culturing condition to better mimic mammalian culture conditions. For yeast library transformation, we did not mimic the typical pH of mammalian cell culture because plating transformed yeast cells on agar plates with pH 7.0 produced few colonies.
Seventy-two hours after transformation, the colonies on the transformation plates (typically several thousand per 10-cm agar plate) were collected using a cell scraper (catalog no. 08-771-1A, Fisher Scientific) into 1 ml of sterile water. The cells were washed three times with water and plated on glass-bottom plates (catalog no. P24-1.5H-N or P06-1.5H-N, Cellvis) coated with a 0.1 mg/ml solution of poly-l-lysine prepared as described above (see the “Preparing cells for optical tagging” section). The attached cells were washed twice with water. After the final wash, water was replaced with PBS.
Imaging and photobleaching yeast libraries
Immobilized single cell libraries were imaged by widefield fluorescence using the microscope described above (see the “Fluorescence imaging and optical tagging setup” section). Automated imaging was performed by sequentially scanning 64 to 169 nonoverlapping fields of view. Because the illuminated area (~1.2 mm2) was larger than the field of view captured by the camera (~0.43 mm2), we spaced the fields of view to avoid imaging previously illuminated areas. Yellow and blue fluorescence images were acquired for each field of view at t = 0, 22.5, and 45 s; we minimized the number of data points to reduce unnecessary data storage. Yellow fluorescence was imaged using 508/25-nm excitation light at 20 mW/mm2 and a 50-ms exposure time. To image blue fluorescence, we used 395/25-nm light with lower irradiance (3.2 mW/mm2) to minimize the nonselective photoactivation of PAmCherry1 during imaging. To ensure sufficient signal, a longer exposure time (400 ms) was used.
Photobleaching was performed by illuminating 508/25-nm excitation light at 20 mW/mm2 for 45 s. Excitation light irradiance was calculated by dividing the measured power with the illumination area. For each objective used, the illumination area was determined by photoactivating a field of view of a dense culture of PAmCherry1-expressing yeast cells; only the photoactivated cells emit red fluorescence, and therefore, the region that shows red fluorescence corresponds to the illuminated area.
Analysis of yeast images
Images were segmented to identify individual cells. Supervised pixel classification was performed on a single representative image of the blue channel (reference TagBFP image) at t = 0 s using the machine learning–based segmentation software ilastik (version 1.3.2) (60), which generated a binary segmentation mask. To accelerate the generation of segmentation masks for all additional fields of view, we developed a custom program in MATLAB (version r2019b, MathWorks) that would extract segmentation parameters from the initial segmentation mask and conduct segmentation of several images in parallel. Depending on the total number of fields of view and the degree of parallelization, we could achieve a segmentation speed of ~3 s per field of view or ~5 min for a typical library. Because cells were immobilized and exhibited minimal movement during imaging, the segmentation mask from the initial time point could be used for later time points. However, since a two-camera setup was used to enable dual-color imaging, channel registration between the blue and yellow channels was performed before applying the segmentation mask to the yellow channel.
To analyze the individual cells identified above, the mean yellow and blue channel pixel intensities [F(Y) and F(B), respectively] of each cell was computed for each time point (t). The brightness (B) of each cell were defined as the ratio of its yellow and blue mean pixel intensities at t0, the initial time point, i.e., B = F(Y, t0)/F(B, t0). Normalization by blue fluorescence was performed to reduce the cell-to-cell variation caused by the variation of plasmid copy number. For SPOTlight screening experiments, the photostability (P) was defined as the fluorescence remaining after a period of continuous illumination, expressed as a fraction of the initial fluorescence, i.e., P = F(Y, tf)/F(Y, t0), where tf is the final time point.
To increase the chance that the selected variant cells would have greater brightness or photostability than the parental cells, a probabilistic model was generated. After brightness and photostability values were quantified for all cells, the joint cumulative distribution function (JCDF) of these two scores of the parental cells was computed using kernel density estimation. Using the brightness and photostability values of each cell of a given library, we used the JCDF to compute the probability of the library cells, displaying better characteristics than the parental cells. Depending on the number of promising candidates, 60 to 200 cells with high probability of being better variants (>90 to 95%) were selected for optical tagging and recovery. Cell selection was automated but could be refined by manual inspection if desired.
Optical tagging and recovery of target yeast cells
The locations of the target cells were determined from the images’ metadata and used to automatically position the microscope stage so that target cells were located in the center of the field of view. Using the DMD, photoactivation was performed on a 4 μm by 4 μm region centered on the target cells. The photoactivation light irradiance measured at the sample plane was 88 mW/mm2. Forty-five to 60 s of photoactivation was used as a compromise between photoactivation fold change and viability. The microscope automatically and sequentially photoactivated target cells. For automated single-cell photoactivation, any step that introduces small offsets (several micrometers) between the proposed cell locations (calculated from the image metadata) and actual cell locations can reduce the optical tagging efficiency. These offsets can accumulate through frequent movements of the microscope stage. To accurately locate the target cells for photoactivation, we introduced an additional layer of analysis. Before photoactivating each cell, we used the General Analysis function in NIS-Elements (Nikon Instruments) to locate the nearest cell to the center of the field of view and moved the stage to the centroid position of the cell. This ensured that each target cell was located exactly at the center of the field of view before photoactivation. In addition, to minimize the false-positive rate, if the analysis code detected more than one cell equally close to the center of the field of view, then the code skipped photoactivation of that target cell and moved to the next target cell. For each target cell, red fluorescence images were taken using 550/30-nm light at 67 mW/mm2 before and after photoactivation and the emitted light was filtered by a 632/60-nm filter (ET632/60m, Chroma).
After photoactivation, cells were detached from the glass-bottom plates by removing the imaging solution and incubating the cells in the trypsin-EDTA solution for 10 min at 37°C. Cells were pipetted up and down vigorously to promote detachment and resuspension into single cells. Detached cells were washed once with PBS and resuspended in PBS. The SH800S cell sorter was used to detect and retrieve cells that were optically tagged. FlowJo (version 10.6.1, FlowJo LLC) was used to analyze flow cytometry data.
Several control samples were prepared to calibrate flow cytometry experiments and analyze the resulting data. Yeast cells expressing single FPs—parental YFP only, TagBFP only, and photoactivated PAmCherry1 only—or empty vectors were prepared to calculate and compensate for bleed-through between the channels. To help determine the appropriate range of fluorescence that best distinguishes photoactivated cells from nonphotoactivated cells (i.e., to find appropriate FACS gates for photoactivated cells), three photoactivation standard samples were prepared: one sample with no photoactivated cells, one sample with ~100 photoactivated cells, and one sample with ~200 photoactivated cells. We determined the range of fluorescence that resulted in the appropriate ratio of cells (0:1:2) among the three samples. Photoactivated cells were sorted in a 10 ml tube containing the synthetic dropout medium and plated on a synthetic dropout agar plate. We sorted all the photoactivated cells into a single tube, plated this mixed preparation onto an agar plate, and incubated this plate at 37oC for 72 hours to enable colony formation. We selected to sort cells into a single tube over sorting each cell into individual wells (index sorting) due to improved yield. The total number of cells sorted by FACS typically exceeded the number of cells imaged by microscopy because imaging was only performed on a subset of plated cells. Colonies grown from the sorted sample were grown in synthetic dropout medium for population-level analysis. Plasmid DNA was prepared from promising variants using a yeast plasmid miniprep kit (catalog no. D2001, Zymo Research) and sequenced to identify novel mutations.
In cellulo FP characterization
Preparation of yeast cells
Yeast cells expressing FPs were streaked on synthetic dropout agar plates from glycerol stocks. Individual colonies were picked and grown overnight at 37°C in a synthetic dropout medium buffered at pH 7.0 with 10 mM HEPES. The growth temperature and pH were not standard for yeast (which are typically grown at 30°C and at acidic pH between 4 and 6) but were chosen so that our results would be most relevant to expression in mammalian systems that are normally cultured at 37oC and pH 7.3. We used pH 7.0 instead of pH 7.3 because we observed that pH higher than 7.0 severely stunt the growth of yeast cells. For each variant, three colonies were selected and grown in separate culture tubes overnight until saturation. Cells were diluted ~20-fold and regrown to mid-log phase, which corresponded to an OD600 of 0.5 when measured with the SmartSpec 3000 spectrophotometer. Note that the OD600 at mid-log phase of yeast culture grown in pH 7.0 was smaller than that of yeast culture grown in pH 4.3. Cells were washed three times with sterile water and immobilized on the poly-l-lysine–coated 96-well glass-bottom microplate for 15 min. Water was replaced with PBS for imaging.
Preparation of human cells
FP expression plasmids were transfected in HEK293A cells using the FuGENE HD Transfection Reagent following the manufacturer’s instructions, except that cells were transfected with 200 ng of DNA and 0.6 μl of FuGENE per well of a 24-well plate, or 100 ng of DNA and 0.3 μl of FuGENE per well of a 96-well plate. Transfected cells were placed on a poly-l-lysine-coated 96-well glass-bottom microplate and incubated at 37°C in air with 5% CO2 for 2 days. Only before imaging, cells were washed once with DPBS, and the growth medium was replaced with HBSS supplemented with 10 mM HEPES.
Photobleaching under widefield illumination
The hardware setup was identical as described for screening experiments. Slight changes were made to the photobleaching and imaging conditions used for the SPOTlight screening. Each well of microplate contained a population of cells expressing the same variant. For each well, two to three fields of view were photobleached with 508/25-nm light for 420 s at 20 mW/mm2. During photobleaching, yellow fluorescence images were taken every 10 s at 2.9 mW/mm2 with a 50-ms exposure time. An image of the reference FP (TagBFP) was acquired once for each field of view before photobleaching.
Characterization of GFPs in yeast and human cells was conducted similarly to YFP characterization. Cells were photobleached for 480 s with 470/50 nm light at 35 mW/mm2. During photobleaching, green fluorescence images were taken every 10 s at 13 mW/mm2 with a 50 ms exposure time. To minimize spectral overlap with GFPs, mCherry was used as a reference FP instead of a BFP. RFP images were acquired once before photobleaching.
Photobleaching under laser-scanning illumination
mGold photostability was also characterized under laser scanning microscopy. HEK293A cells transiently expressing mVenus or mGold were prepared as described above. Photobleaching was conducted using a high-speed confocal microscope (LSM880 with Airyscan, Zeiss) using a 40× 1.4-NA oil immersion objective (Plan-Apochromat DIC M27, Zeiss). Photobleaching was conducted for 250 s by unidirectional scanning a 53.14 μm by 53.14 μm area at 3.18 Hz with a 514 nm argon laser (LGK7812, Lasos) at 5% power (32 μW) and a pixel dwell time of 0.67 μs. Pixels (512 × 512) with an image depth of 12 bits were acquired continuously during photobleaching using a pinhole size of 600.6 μm. Images were analyzed using ImageJ (61) by manually drawing masks around the cells and tracking the fluorescence of individual cells over time.
Image analysis of brightness and photostability
For each variant, population-level analysis was conducted. The fields of view were segmented (see the “Analysis of yeast images” section) to compute single-cell brightness and photostability scores. All the segmented cells in the two to three fields of view were combined, leading to a total of ~500 to 8000 yeast cells and ~100 to 200 human cells. The brightness and photostability of individual cells were computed after background subtraction and outliers were removed using the robust regression followed by outlier identification (ROUT) method (Q = 1%) method (62). Mean brightness and photostability values were determined to obtain a population-level performance of the variants. The brightness of individual cells was calculated as described in yeast screening. We also computed the photobleaching half-life, that is, the time required for the fluorescence to decrease by half, as a standard metric for photostability. For each variant, yeast cultures were grown from three independent colonies, and three independent transfections were conducted in human cells. The mean and SD of the brightness and photostability from these triplicates were calculated and compared for statistical analysis.
Obtaining excitation spectra under two-photon illumination
HEK293A cells were transiently transfected with pcDNA3.1/puro-CAG plasmids expressing mGold or mVenus. Two days after transfection, the cells were washed with and imaged in extracellular solution. This solution contained 110 mM NaCl (catalog no. S3014, Sigma-Aldrich), 26 mM sucrose (catalog no. S0389, Sigma-Aldrich), 23 mM glucose (catalog no. G8270, Sigma-Aldrich), 5 mM KCl (catalog no. P9541, Sigma-Aldrich), 2.5 mM CaCl2 dihydrate (catalog no. 223506, Sigma-Aldrich), and 1.3 mM MgSO4 (catalog no. M2643, Sigma-Aldrich), and was adjusted to pH 7.4. Imaging was performed using the microscope described above (see the “Two-photon microscopy setup” section). Images were taken using various wavelengths from 700 to 1080 nm with 10 nm increments. The laser power was 10 mW for all wavelengths. Images were background corrected and manually segmented. The mean fluorescence of cells at each wavelength was calculated and plotted. We imaged at 900 nm with regular intervals during the spectral scanning and confirmed there was minimal photobleaching during the scanning. Small deviations in the actual power from the target power of 10 mW were corrected by assuming a quadratic dependence of fluorescence on illumination power.
Evaluating FP cytotoxicity
To evaluate FP cytotoxicity, we adapted the assay conducted by Shemiakina et al. (63). FP expression plasmids expressing EGFP, mGold, or mVenus were transfected separately in HeLa cells. For each FP plasmids, cells were transfected following the manufacturer’s instructions and used 2 μg of DNA and 6 μl FuGENE. Transfected cells were placed in a well of a 6-well microplate (catalog no. 3516, Corning) and incubated at 37°C in air with 5% CO2. Two days after transfection, cells were detached by incubating the cells in a trypsin-EDTA solution for ~5 min. EGFP cells were mixed with mGold or mVenus cells to produce two mixed populations: (i) EGFP+ and mGold+ mixed cells and (ii) EGFP+ and mVenus+ mixed cells. The proportion of GFP+ and YFP+ cells in each mixed cell population was analyzed using the Attune NxT flow cytometer. Mixed population of cells were diluted 10-fold and plated into three wells of a six-well plate. Five days after transfection, mixed cells were detached and the proportion of GFP+ and YFP+ cells was analyzed using flow cytometry. For both flow cytometry analyses, single-FP controls (cells expressing only EGFP, mVenus, or mGold) and negative control (cells that were not transfected) were prepared to compensate for the spectral overlap between EGFP and YFPs and to design analysis gates for GFP+ and YFP+ cells. Only live cells were analyzed by identifying and removing dead cells from analysis. NucBlue DAPI (4′,6-diamidino-2-phenylindole) stain (catalog no. R37606, Thermo Fisher Scientific) was used to stain dead cells. Cytotoxicity was calculated using the flow cytometry data from both day 2 and day 5 using the following formula: ((% of YFP+ cells at day 2) / (% of YFP+ cells at day 5))/((% of EGFP+ cells at day 2) / (% of EGFP+ cells at day 5)).
Imaging mGold fusion constructs
Confocal imaging was conducted to evaluate whether mGold fused to different organelle/cytoskeleton-localization tags/proteins would produce fluorescence with the expected pattern of subcellular localization. HeLa cells were transfected with FuGENE as described above (see the “preparation of human cells” section). Cells were placed on the glass-bottom 24-well plates without poly-l-lysine coating because we observed that poly-l-lysine unexpectedly seems to promote detachment with HeLa cells, producing rounder cells after the PBS wash step. Transfected HeLa cells were washed with PBS and resuspended in HBSS supplemented with 10 mM HEPES. Laser-scanning confocal images were obtained using a high-speed confocal microscope (LSM880 with Airyscan, Zeiss) driven by the Zen software (version 2.3 SP1, Zeiss). Images were acquired with a 40× 1.1-NA water immersion objective (LD C-Apochromat Korr M27, Zeiss), a 488 nm argon laser (LGK7812, Lasos) at 3% power, and a per-pixel dwell time of 2 μs. Emission light was filtered using a multipass beamsplitter (MBS 488/561/633, Zeiss) and acquired with a 32 channel GaAsP detector (Airyscan, Zeiss) with a detector gain of 740 and 1–Airy unit pinhole size. To increase the signal-to-noise ratio, two scans were performed and averaged for each image.
In vitro FP characterization
In vitro characterization of FPs was conducted by adapting published methods (64).
Expressing and purifying FPs
Bacterial cells from the E.coli strain BL21 were grown to an OD600 of 0.1 and induced using 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, catalog no. 11411446001, Roche) for 8 hours. FPs were purified using the Ni-NTA Fast Start Kit (catalog no. 30600, Qiagen) and dialyzed in 5 or 50 mM tris(hydroxymethyl)aminomethane (tris) buffer at pH 7.5. Tris buffer was prepared by mixing Trizma base (catalog no. T6066, Sigma-Aldrich) with Trizma hydrochloride (catalog no. T3253, Sigma-Aldrich). Protein concentrations were determined using the Pierce BCA Protein Assay Kit (catalog no. 23225, Thermo Fisher Scientific) according to the manufacturer’s instructions.
Obtaining excitation and emission spectra under one-photon illumination
FPs were diluted in 50 mM tris buffer at varying concentrations. A plate reader (Cytation 5, BioTek) was used to produce excitation and emission spectra. Excitation scan data used excitation wavelengths from 400 to 555 nm and collected emission intensity at 580/10 nm. Emission scan data used an excitation wavelength of 475/10 nm and used emission wavelengths from 500 to 700 nm. Measurements were also collected for a control well with only 50 mM tris buffer and no FPs. We then created excitation and emission curves by subtracting the blanks from each sample, normalizing each individual well to have a maximum fluorescent intensity value of 1, and then averaging the curves (7 to 9 technical replicates) for each FP. The excitation and emission peaks were determined from these averaged curves.
Quantifying the extinction coefficient and quantum yield
We first used the SmartSpec 3000 spectrophotometer to measure the absorbance at 475 nm of purified FPs and of a reference dye, rhodamine 123 (catalog no. R302, Thermo Fisher Scientific). For each FP and the dye, we then conducted a serial dilution to achieve 5 to 6 diluted samples with various absorbances. The absorbance measured using the spectrometer was used to ensure all the dilutions would have absorbances less than 0.05 to minimize the inner filter effect. The Cytation5 plate reader was then used to scan the emission spectrum (500 nm to 700 nm with 1 nm steps; excitation wavelength of 475/10 nm). The area under the curve was calculated for each emission spectrum and plotted against absorbance. The slope of each sample’s emission versus absorbance curves were determined using linear regression (intercept = 0). Quantum yields (QY) were calculated using the formula QYFP = QYSt × (SSt/SFP) × (RFP/RSt), where S is the slope of the sample’s curve and R is the refractive index of the solvent used. The FP subscript refers to the FP sample, while St refers to the rhodamine 123 standard. Extinction coefficients were determined with the SmartSpec 3000 spectrophotometer by finding the peak absorption, the wavelength of which was determined earlier using an absorbance sweep reading with the plate reader. Three dilutions of each FP variant in 50 mM tris were measured with the spectrophotometer in 1-cm quartz cuvettes (catalog no. 9109252, Bio-Rad). The extinction coefficient (ε) was determined using the Beer-Lambert law [A = ε × l × C or ε = A/(l × C); where A is absorbance, l is path length, and C is molar concentration] after correcting for the dilution of the sample. ε was calculated by averaging the values obtained at different dilutions.
Quantifying the pKa
To determine the pKa, a series of buffers were prepared with pH in the range of 3 to10 with an increment of 0.5. Buffers with pH 3 to pH 5.5 were made with 100 mM citric acid (catalog no. C0759, Sigma-Aldrich) and 100 mM sodium citrate (catalog no. S1804, Sigma-Aldrich), buffers with pH 6 to pH 8 were made with 100 mM KH2PO4 (catalog no. P5379, Sigma-Aldrich) and 100 mM Na2HPO4 (catalog no. S7907, Sigma-Aldrich), and buffers with pH 8.5 to pH 12 were made with 100 mM glycine (catalog no. 16372-A4, Fisher Scientific) and 100 mM NaOH. All buffers were then adjusted to the desired pH using HCl (catalog no. 320331, Sigma-Aldrich) or NaOH. One hundred microliters of each pH buffer were loaded into a 96-well glass-bottom imaging plate (catalog no. P96-1.5H-N, Cellvis) with 10 μl of 1 μM protein samples dialyzed in 5 mM tris in four replicates. The Cytation 5 plate reader was then used to determine emission intensity of each FP at 530 nm using 500 nm excitation light. Emission intensity versus pH was then normalized to the intensity value at pH 10 and plotted. Linear interpolation was used to determine the pKa, defined as the pH at which fluorescence is half of its maximum.
Quantifying photostability
We constructed a photobleaching chamber using a glass microscope slide (catalog no. 7101, Henso) as the base and then mounting a coverslip (catalog no. 48366-227, VWR) on top of two additional spacer coverslips attached to the base with tape. We then mixed a 1 μM dilution of each FP with 20% (w/v) acrylamide/bis-acrylamide, 30% (w/v) solution (catalog no. A3699, Sigma-Aldrich), 3 μl of 10% ammonium persulfate (catalog no. A3678, Sigma-Aldrich), and 0.5 μl of N,N,N′,N′-tetramethylethylenediamine (TEMED) (catalog no. T9281, Sigma-Aldrich) in a 1.5-ml tube. Right after adding TEMED, we transferred ~80 μl of the solution to the space between the top coverslip and glass slide and waited ~5 min for the gel to polymerize. We then sealed the edges with Cytoseal 60 (catalog no. 8310-4, Thermo Fisher Scientific). After 12 hours, the slides were continuously photobleached for 7 min with 5-s-interval image captures using widefield 510/25-nm light with 62.5 mW/mm2 (using a 60× 1.4-NA oil objective; CFI Plan Apo VC, Nikon Instruments). For each FP, nine fields of view were tested. For each field of view, the photobleaching half-life was calculated by determining the time at which the fluorescence reached half of the initial fluorescence. The mean photobleaching half-life values were calculated and plotted.
Determining oligomeric state
Size exclusion chromatography was conducted to determine the oligomeric state of mGold. mGold (10 μM) and size standards—mCherry (monomeric, 10 μM), mVenus (monomeric, 10 μM), and tdTomato (dimeric, 6.5 μM)—were analyzed using a fast protein liquid chromatography instrument (NGC Chromatography, Bio-Rad) with a 30 cm × 10 cm (length × diameter) gel filtration column (Superdex 200 10/30 GL, GE Healthcare). One hundred microliters of samples were injected and ran separately at a flow rate of 0.5 ml/min. The flow buffer was 50 mM tris buffer at pH 7.5 supplemented with 100 mM NaCl. The FPs were detected by measuring the absorbance at 515 nm (mGold and mVenus), 587 nm (mCherry), and 555 nm (tdTomato).
Determining chloride sensitivity
Chloride titration assays were conducted to determine the chloride sensitivity of mGold. Chloride solutions with various concentrations (0, 0.25, 0.5, and 1.0 M) were prepared by mixing appropriate amounts of KCl with MOPS buffer. MOPS buffer was prepared by adding MOPS (20 mM, catalog no. M1254, Sigma-Aldrich), sodium acetate (5 mM, catalog no. S8750, Sigma-Aldrich), and Na2 EDTA (1 mM catalog no. E5134, Sigma-Aldrich) in water; pH was adjusted to 7.0 using NaOH. To keep the ionic strength among the solutions constant, appropriate amounts of potassium gluconate (catalog no. P1847, Sigma-Aldrich) was added to the 0, 0.25, and 0.5 M KCl solutions so that the total amounts of ions in these solutions were the same as that in 1.0 M KCl solution. A total of 1.5 μg of purified mGold (or mVenus) was added to 100 μl of KCl solutions and placed in wells of the 96-well glass-bottom plate. The Cytation 5 plate reader was used to measure the fluorescence of mGold or mVenus. The excitation wavelength of 508/10 nm and emission wavelength of 530/10 nm were used. MOPS buffer without FPs was used as a blank. The fluorescence value of the blank was subtracted from that of each sample and the fluorescence values were normalized to that of 0 M KCl sample.
Statistics
Details of the statistical analyses performed for each figure are provided separately (see Supplementary Statistics). Prism (version 8.2.1, GraphPad) was used to conduct statistical analysis. Briefly, for statistical analysis with a sample size of >30, parametric analysis was used without checking for normality because violation of normality has a small effect on parametric analysis when the sample size is large (65). For a sample size of ≤30, the normality was checked using the Shapiro-Wilk normality test, visually checking the distribution using the Tukey boxplot, and plotting the QQ plot. We used two-sided analysis of variance (ANOVA) or t test for parametric analysis and the Mann Whitney test for nonparametric analyses. For datasets that showed lognormal distribution, we applied a log transformation before conducting a statistical analysis. To decide whether to apply Welch’s correction to account for difference in variances, we used the Brown-Forsythe test (for ANOVAs) or the F test (for t tests). If the variances of samples were significantly different, then the Welch’s correction was applied. Statistical analyses on trends (e.g., photoactivation ratio versus time) were conducted using the areas under the curve (AUCs).
Supplementary Material
Acknowledgments
We thank S. Di Rienzi and R. Britton (Baylor College of Medicine, BCM) for providing human intestinal enteroids and guidance in conducting experiments using the enteroids. We thank K. Rai (Rice University) for assistance with the quantification of purified mGold. We thank A. Papusha, Y. Yu, and G. Ira (BCM) for assistance with yeast culture. We thank members of the St-Pierre lab for feedback on experiments and the manuscript, with special thanks to M. Land for assistance with the figures. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the CPRIT Core Facility Support Award (CPRIT-RP18-672), the NIH (P30 CA125123 and S10 RR024574) and assistance of J. M. Sederstrom for flow cytometry experiments. We thank the Optical Imaging & Vital Microscopy Core and J. Kirk at Baylor College of Medicine for guidance with confocal imaging experiments. We also thank the Integrated Microscopy Core and F. Stossi at Baylor College of Medicine for assistance with using Cytation 5 plate reader. Funding: F.S.-P. was supported by the McNair Medical Foundation, start-up funds from Baylor College of Medicine, the Klingenstein-Simons Fellowship Award in Neuroscience, a Welch Foundation grant (Q-2016-20190330), NIH grants (R01EB027145, U01NS113294, and U01NS118288), and NSF grants (NeuroNex 1707359, and IdeasLab 1935265). Author contributions: F.S.-P. conceived the overall project with substantial intellectual input and leadership from J.L. J.L., P.H.S., S.G., X.L., and F.S.-P. developed and validated the overall SPOTlight pipeline. Z.L. and J.L. developed the automated analysis methods. J.L. and F.S.-P. designed the experiments. J.L., J.F.A., and S.L. conducted the FP screening experiments. J.L., J.F.A., and Z.L. analyzed the screening experiment data. J.L. and X.L. characterized the FPs in cellulo. J.F.A. and J.L. characterized the FPs in vitro. J.L. and F.S.-P. wrote the manuscript. All authors contributed to editing the manuscript. Competing interest: F.S.-P., J.L., Z.L., and P.H.S. have filed a provisional patent based on this work. The authors declare no other competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The analysis code used in this study can be provided by the corresponding author upon reasonable request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/43/eabb7438/DC1
REFERENCES AND NOTES
- 1.Shendure J., Findlay G. M., Snyder M. W., Genomic medicine–progress, pitfalls, and promise. Cell 177, 45–57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tizei P. A. G., Csibra E., Torres L., Pinheiro V. B., Selection platforms for directed evolution in synthetic biology. Biochem. Soc. Trans. 44, 1165–1175 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nitta N., Sugimura T., Isozaki A., Mikami H., Hiraki K., Sakuma S., Iino T., Arai F., Endo T., Fujiwaki Y., Fukuzawa H., Hase M., Hayakawa T., Hiramatsu K., Hoshino Y., Inaba M., Ito T., Karakawa H., Kasai Y., Koizumi K., Lee S. W., Lei C., Li M., Maeno T., Matsusaka S., Murakami D., Nakagawa A., Oguchi Y., Oikawa M., Ota T., Shiba K., Shintaku H., Shirasaki Y., Suga K., Suzuki Y., Suzuki N., Tanaka Y., Tezuka H., Toyokawa C., Yalikun Y., Yamada M., Yamagishi M., Yamano T., Yasumoto A., Yatomi Y., Yazawa M., Carlo D. D., Hosokawa Y., Uemura S., Ozeki Y., Goda K., Intelligent Image-Activated Cell Sorting. Cell 175, 266–276.e13 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Rimon N., Schuldiner M., Getting the whole picture: Combining throughput with content in microscopy. J. Cell Sci. 124, 3743–3751 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Boutros M., Heigwer F., Laufer C., Microscopy-based high-content screening. Cell 163, 1314–1325 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Emanuel G., Moffitt J. R., Zhuang X., High-throughput, image-based screening of pooled genetic-variant libraries. Nat. Methods 14, 1159–1162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman D., Singh A., Schmid-Burgk J. L., Carlson R. J., Mezger A., Garrity A. J., Zhang F., Blainey P. C., Optical pooled screens in human cells. Cell 179, 787–799.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piatkevich K. D., Jung E. E., Straub C., Linghu C., Park D., Suk H.-J., Hochbaum D. R., Goodwin D., Pnevmatikakis E., Pak N., Kawashima T., Yang C.-T., Rhoades J. L., Shemesh O., Asano S., Yoon Y.-G., Freifeld L., Saulnier J. L., Riegler C., Engert F., Hughes T., Drobizhev M., Szabo B., Ahrens M. B., Flavell S. W., Sabatini B. L., Boyden E. S., A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 14, 352–360 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen B., Lim S., Kannan A., Alford S. C., Sunden F., Herschlag D., Dimov I. K., Baer T. M., Cochran J. R., High-throughput analysis and protein engineering using microcapillary arrays. Nat. Chem. Biol. 12, 76–81 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luro S., Potvin-Trottier L., Okumus B., Paulsson J., Isolating live cells after high-throughput, long-term, time-lapse microscopy. Nat. Methods 17, 93–100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovac J. R., Voldman J., Intuitive, image-based cell sorting using optofluidic cell sorting. Anal. Chem. 79, 9321–9330 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binan L., Mazzaferri J., Choquet K., Lorenzo L.-E., Wang Y. C., Affar E. B., De Koninck Y., Ragoussis J., Kleinman C. L., Costantino S., Live single-cell laser tag. Nat. Commun. 7, 11636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binan L., Bélanger F., Uriarte M., Lemay J. F., Pelletier de Koninck J. C., Roy J., Affar E. B., Drobetsky E., Wurtele H., Costantino S., Opto-magnetic capture of individual cells based on visual phenotypes. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien M.-P., Werley C. A., Farhi S. L., Cohen A. E., Photostick: A method for selective isolation of target cells from culture. Chem. Sci. 6, 1701–1705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X. X., Lin M. Z., Photoswitchable fluorescent proteins: Ten years of colorful chemistry and exciting applications. Curr. Opin. Chem. Biol. 17, 682–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukyanov K. A., Chudakov D. M., Lukyanov S., Verkhusha V. V., Innovation: Photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 6, 885–891 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Victora G. D., Schwickert T. A., Fooksman D. R., Kamphorst A. O., Meyer-Hermann M., Dustin M. L., Nussenzweig M. C., Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medaglia C., Giladi A., Stoler-Barak L., De Giovanni M., Salame T. M., Biram A., David E., Li H., Iannacone M., Shulman Z., Amit I., Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 358, 1622–1626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomura M., Yoshida N., Tanaka J., Karasawa S., Miwa Y., Miyawaki A., Kanagawa O., Monitoring cellular movement in vivo with photoconvertible fluorescence protein “Kaede” transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 105, 10871–10876 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson G. H., Lippincott-Schwartz J., A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Specht E. A., Braselmann E., Palmer A. E., A critical and comparative review of fluorescent tools for live-cell imaging. Annu. Rev. Physiol. 79, 93–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subach F. V., Patterson G. H., Manley S., Gillette J. M., Lippincott-Schwartz J., Verkhusha V. V., Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmchen F., Denk W., Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Grimm J. B., English B. P., Choi H., Muthusamy A. K., Mehl B. P., Dong P., Brown T. A., Lippincott-Schwartz J., Liu Z., Lionnet T., Lavis L. D., Bright photoactivatable fluorophores for single-molecule imaging. Nat. Methods 13, 985–988 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Yang H. H., St-Pierre F., Genetically encoded voltage indicators: Opportunities and challenges. J. Neurosci. 36, 9977–9989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamontova A. V., Grigoryev A. P., Tsarkova A. S., Lukyanov K. A., Bogdanov A. M., Struggle for photostability: Bleaching mechanisms of fluorescent proteins. Russ. J. Bioorg. Chem. 43, 625–633 (2017). [Google Scholar]
- 27.Kremers G.-J., Goedhart J., van Munster E. B., Gadella T. W. J., Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45, 6570–6580 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Balleza E., Kim J. M., Cluzel P., Systematic characterization of maturation time of fluorescent proteins in living cells. Nat. Methods 15, 47–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padamsey Z., Jeans A., Imaging synaptic vesicles using VGLUT1-venus knock-in mice: Insights into the dynamic nature of intersynaptic vesicle exchange. J. Neurosci. 32, 3284–3286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata S., Yasuda A., Renault-Mihara F., Suyama S., Katoh H., Inoue T., Inoue Y. U., Nagoshi N., Sato M., Nakamura M., Akazawa C., Okano H., Sox10-Venus mice: A new tool for real-time labeling of neural crest lineage cells and oligodendrocytes. Mol. Brain 3, 31 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larrieu A., Champion A., Legrand J., Lavenus J., Mast D., Brunoud G., Oh J., Guyomarc’h S., Pizot M., Farmer E. E., Turnbull C., Vernoux T., Bennett M. J., Laplaze L., A fluorescent hormone biosensor reveals the dynamics of jasmonate signalling in plants. Nat. Commun. 6, 6043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marvin J. S., Scholl B., Wilson D. E., Podgorski K., Kazemipour A., Müller J. A., Schoch S., Quiroz F. J. U., Rebola N., Bao H., Little J. P., Tkachuk A. N., Cai E., Hantman A. W., Wang S. S.-H., DePiero V. J., Borghuis B. G., Chapman E. R., Dietrich D., DiGregorio D. A., Fitzpatrick D., Looger L. L., Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 15, 936–939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaner N. C., Lin M. Z., McKeown M. R., Steinbach P. A., Hazelwood K. L., Davidson M. W., Tsien R. Y., Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dean K. M., Lubbeck J. L., Davis L. M., Regmi C. K., Chapagain P. P., Gerstman B. S., Jimenez R., Palmer A. E., Microfluidics-based selection of red-fluorescent proteins with decreased rates of photobleaching. Integr. Biol. 7, 263–273 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cranfill P. J., Sell B. R., Baird M. A., Allen J. R., Lavagnino Z., de Gruiter H. M., Kremers G.-J., Davidson M. W., Ustione A., Piston D. W., Quantitative assessment of fluorescent proteins. Nat. Methods 13, 557–562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean K. M., Lubbeck J. L., Binder J. K., Schwall L. R., Jimenez R., Palmer A. E., Analysis of red-fluorescent proteins provides insight into dark-state conversion and photodegradation. Biophys. J. 101, 961–969 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokmakov A. A., Kurotani A., Takagi T., Toyama M., Shirouzu M., Fukami Y., Yokoyama S., Multiple post-translational modifications affect heterologous protein synthesis. J. Biol. Chem. 287, 27106–27116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nov Y., When second best is good enough: Another probabilistic look at saturation mutagenesis. Appl. Environ. Microbiol. 78, 258–262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chappell L., Russell A. J. C., Voet T., Single-Cell (Multi)omics Technologies. Annu. Rev. Genomics Hum. Genet. 19, 15–41 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Dubois A. V., Midoux P., Gras D., Si-Tahar M., Bréa D., Attucci S., Khelloufi M.-K., Ramphal R., Diot P., Gauthier F., Hervé V., Poly-L-Lysine compacts DNA, kills bacteria, and improves protease inhibition in cystic fibrosis sputum. Am. J. Respir. Crit. Care Med. 188, 703–709 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Hartwich T. M. P., Subach F. V., Cooley L., Verkhusha V. V., Bewersdorf J., Determination of two-photon photoactivation rates of fluorescent proteins. Phys. Chem. Chem. Phys. 15, 14868–14872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rompolas P., Mesa K. R., Kawaguchi K., Park S., Gonzalez D., Brown S., Boucher J., Klein A. M., Greco V., Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasle N., Cooke A., Srivatsan S., Huang H., Stephany J. J., Krieger Z., Jackson D., Tang W., Pendyala S., Monnat R. J. Jr., Trapnell C., Hatch E. M., Fowler D. M., High-throughput, microscope-based sorting to dissect cellular heterogeneity. Mol. Syst. Biol. 16, e9442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enterina J. R., Wu L., Campbell R. E., Emerging fluorescent protein technologies. Curr. Opin. Chem. Biol. 27, 10–17 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Germond A., Fujita H., Ichimura T., Watanabe T. M., Design and development of genetically encoded fluorescent sensors to monitor intracellular chemical and physical parameters. Biophys. Rev. 8, 121–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sansalone L., Tang S., Garcia-Amorós J., Zhang Y., Nonell S., Baker J. D., Captain B., Raymo F. M., A photoactivatable far-red/near-infrared BODIPY to monitor cellular dynamics in vivo. ACS Sens. 3, 1347–1353 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Piatkevich K. D., Subach F. V., Verkhusha V. V., Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat. Commun. 4, 2153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A., A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Bajar B. T., Wang E. S., Zhang S., Lin M. Z., Chu J., A guide to fluorescent protein FRET pairs. Sensors 16, 1488 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam A. J., St-Pierre F., Gong Y., Marshall J. D., Cranfill P. J., Baird M. A., McKeown M. R., Wiedenmann J., Davidson M. W., Schnitzer M. J., Tsien R. Y., Lin M. Z., Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005–1012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee M. E., DeLoache W. C., Cervantes B., Dueber J. E., A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 4, 975–986 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Meyer A., Eskandari S., Grallath S., Rentsch D., AtGAT1, a high affinity transporter for γ-aminobutyric acid in Arabidopsis thaliana. J. Biol. Chem. 281, 7197–7204 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flagfeldt D. B., Siewers V., Huang L., Nielsen J., Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 26, 545–551 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Bajar B. T., Wang E. S., Lam A. J., Kim B. B., Jacobs C. L., Howe E. S., Davidson M. W., Lin M. Z., Chu J., Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St-Pierre F., Marshall J. D., Yang Y., Gong Y., Schnitzer M. J., Lin M. Z., High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 17, 884–889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chamberland S., Yang H. H., Pan M. M., Evans S. W., Guan S., Chavarha M., Yang Y., Salesse C., Wu H., Wu J. C., Clandinin T. R., Toth K., Lin M. Z., St-Pierre F., Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. eLife 6, e25690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nguyen A. W., Daugherty P. S., Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 23, 355–360 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Chang-Graham A. L., Danhof H. A., Engevik M. A., Tomaro-Duchesneau C., Karandikar U. C., Estes M. K., Versalovic J., Britton R. A., Hyser J. M., Human intestinal enteroids with inducible neurogenin-3 expression as a novel model of gut hormone secretion. Cell. Mol. Gastroenterol. Hepatol. 8, 209–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gietz R. D., Schiestl R. H., Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 1–4 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Berg S., Kutra D., Kroeger T., Straehle C. N., Kausler B. X., Haubold C., Schiegg M., Ales J., Beier T., Rudy M., Eren K., Cervantes J. I., Xu B., Beuttenmueller F., Wolny A., Zhang C., Koethe U., Hamprecht F. A., Kreshuk A., ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., Eliceiri K. W., ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Motulsky H. J., Brown R. E., Detecting outliers when fitting data with nonlinear regression – a new method based on robust nonlinear regression and the false discovery rate. BMC Bioinformatics 7, 123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shemiakina I. I., Ermakova G. V., Cranfill P. J., Baird M. A., Evans R. A., Souslova E. A., Staroverov D. B., Gorokhovatsky A. Y., Putintseva E. V., Gorodnicheva T. V., Chepurnykh T. V., Strukova L., Lukyanov S., Zaraisky A. G., Davidson M. W., Chudakov D. M., Shcherbo D., A monomeric red fluorescent protein with low cytotoxicity. Nat. Commun. 3, 1204 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Ai H.-W., Baird M. A., Shen Y., Davidson M. W., Campbell R. E., Engineering and characterizing monomeric fluorescent proteins for live-cell imaging applications. Nat. Protoc. 9, 910–928 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Ghasemi A., Zahediasl S., Normality tests for statistical analysis: A guide for non-statisticians. Int. J. Endocrinol. Metab. 10, 486–489 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henikoff S., Henikoff J. G., Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 89, 10915–10919 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/43/eabb7438/DC1


