Abstract
While horizontal gene transfer is prevalent across the biosphere, the regulatory features that enable expression and functionalization of foreign DNA remain poorly understood. Here, we combine high-throughput promoter activity measurements and large-scale genomic analysis of regulatory regions to investigate the cross-compatibility of regulatory elements (REs) in bacteria. Functional characterization of thousands of natural REs in three distinct bacterial species revealed distinct expression patterns according to RE and recipient phylogeny. Host capacity to activate foreign promoters was proportional to their genomic GC content, while many low GC regulatory elements were both broadly active and had more transcription start sites across hosts. The difference in expression capabilities could be explained by the influence of the host GC content on the stringency of the AT-rich canonical σ70 motif necessary for transcription initiation. We further confirm the generalizability of this model and find widespread GC content adaptation of the σ70 motif in a set of 1,545 genomes from all major bacterial phyla. Our analysis identifies a key mechanism by which the strength of the AT-rich σ70 motif relative to a host’s genomic GC content governs the capacity for expression of acquired DNA. These findings shed light on regulatory adaptation in the context of evolving genomic composition.
Subject terms: Bacterial genetics, Transcriptomics, Microbiome, Bacterial synthetic biology
Introduction
The impressive adaptability of microbes to novel and changing environments can be attributed to their genomic variability and their ability to acquire new capabilities via horizontal gene transfer (HGT) [1–3]. HGT is a prevalent microbial phenomenon whereby genetic materials are shuttled from one organism to another through mobile DNA such as transposons, plasmids, phage, and other vectors [4, 5]. Through HGT, organisms that diverged hundreds of millions of years ago can readily exchange genetic material and rapidly acquire new traits [6, 7]. HGT is also the major driver of the spread of genes associated with virulence and drug resistance, highlighting the importance of a deeper understanding of transfer processes [8–12].
In order to provide a fitness advantage to the host cell, horizontally acquired genes must be expressed at proper levels, since underexpression may be insufficient to improve fitness and overexpression may lead to cellular toxicity or burden, both of which could prevent long-term retention of horizontally acquired DNA [2, 13–16]. While transcription factors and their cis-regulatory elements (REs) tend to evolve faster than the target genes they regulate [17, 18], the core gene expression machineries (RNA polymerase, σ70) are highly conserved. However, there is some evidence that sequence specificities have diverged somewhat over evolutionary time and it is unclear how any potential differences may affect the functionalization of heterologous regulatory sequences [19–21]. Furthermore, bacterial species vary widely in their genomic repertoire of transcriptional regulators and GC contents, which dictate what types of binding sites can be targeted and the compositional context in which they exist. Variation in regulatory compatibility between species may impact the ability of organisms to express and functionalize exchanged genetic material, posing a fundamental question of how regulatory constraints may influence microbial evolution through HGT. However, there have been few studies that systematically examining the compatibility of regulatory sequences and how gene regulation may affect the successful utilization of transferred genes, especially between diverse organisms. We and others have observed that REs can behave differently in distinct host species [19, 22–24]. In our previous study [22], we characterized the activity levels of thousands of diverse natural REs in three species and found that the canonical σ70 was the primary driver of RE expression in each species tested. However, we also observed considerable differences between species in their ability to activate heterologous REs. Therefore, the factors that determine compatibility between heterologous REs and a host’s regulatory machinery remain unclear.
Here, we used a combination of experimental and computational approaches to investigate transcriptional regulatory compatibility between bacterial species. Using both existing and new high-throughput reporter data from thousands of natural REs from diverse prokaryotic species as well as mobile genetic elements [22], we examined the distinct expression capabilities of three bacterial species of distinct ecologies and GC contents. We identified a mechanism by which an organism’s σ70 stringency is adapted according to its genomic GC content. Importantly, this mechanism provides key insights into regulatory compatibility between diverse bacterial species. We found further support for this mechanism through analysis of 1545 diverse genomes from all major bacterial phyla. Together, these findings explain how bacteria encode regulatory information in varying compositional contexts and also how these mechanisms influence an organism’s capacity to activate horizontally acquired DNA.
Results
Variation in capacity for heterologous regulatory DNA activation
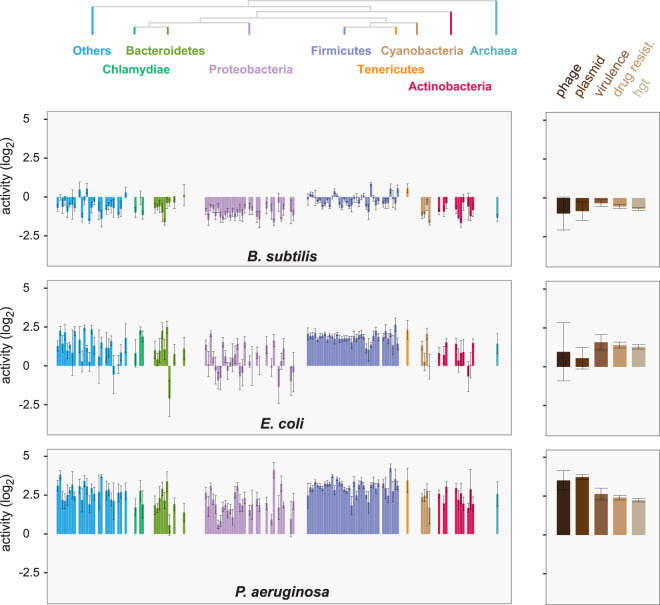
We previously developed a high-throughput sequencing-based approach to experimentally measure the transcriptional activities of thousands of natural REs from 184 prokaryotic genomes within 3 phylogenetically, ecologically, and compositionally distinct recipients: Bacillus subtilis (43% GC), Escherichia coli (51% GC), and Pseudomonas aeruginosa (67% GC) [22]. This library contained 29,249 barcoded REs attached to a GFP reporter, which enabled large-scale examination of heterologous promoter activity levels. Here, we have expanded this library by generating data for additional REs associated from bioinformatically identified HGT events (3818 sequences) [25], antibiotic resistance genes (1047 sequences) [26], virulence factors (336 sequences) [27], as well as plasmids and phage (2518 sequences) (Supplementary Table 1). Targeted RNA-seq of reporter mRNAs and amplicon sequencing of DNA were used to determine normalized transcriptional activities from library-harboring populations grown in rich media at mid-exponential phase (“Methods”). We observed that recipients have different capabilities in recognizing REs and distinct activity according to donor phylogeny (Fig. 1). Regulatory sequences from low GC species (e.g., Firmicutes) tended to be highly active across P. aeruginosa and E. coli and relatively higher in B. subtilis. When using the mean activity of endogenous REs from each recipient’s genus as a reference point (included in the library, “Methods”), most REs had lower activity than the average B. subtilis endogenous RE and the opposite was true for P. aeruginosa and its endogenous sequences. These same trends held true for REs from mobile elements, phage, and sequences from putative gene transfer events.
Fig. 1. Phylogenetic trends in RE library expression.
Mean transcription activities of regulatory elements from diverse genomes (left) and mobile DNA (right) are shown across the three distinct recipients as bar plots. Values represent the mean activity of REs from individual genomes and are normalized to endogenous reference sequences (“Methods”). Higher transcription is generally observed in P. aeruginosa compared with E. coli and B. subtilis. The donor phylogeny or library type is represented on the top, each bar represents one distinct donor. Error bars are two standard errors from the mean. Library represents 1859 (genomic) + 281 (mobile) REs that are informative in all recipients (RNA + DNA counts > 15) and at least 1 read of RNA and DNA. Only genomes with at least 3 informative RE measurements are displayed.
Positional patterns of transcription initiation
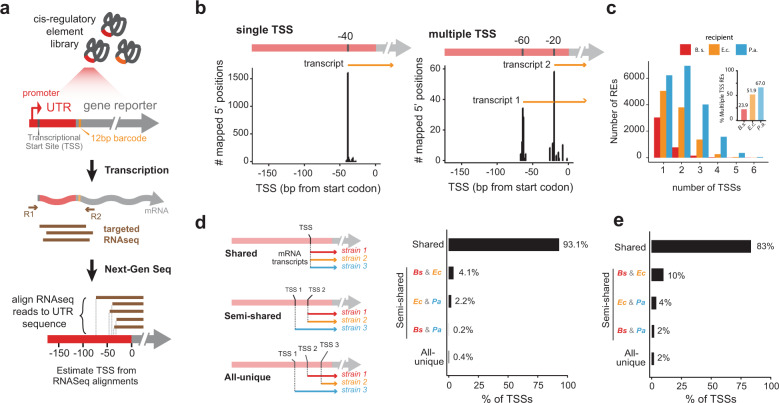
Gene expression in bacteria originates from transcription start sites (TSSs) that are found immediately downstream from the recognition sites of the acting sigma or transcription factors. To identify similarities and differences in how our three species utilize regulatory signals, we compared positional patterns of transcription initiation across our library. Our library sequencing strategy amplifies whole 5′ untranslated regions (UTRs) through adaptor ligation and targeted reverse transcription, enabling examination of upstream cis-regulatory region locations and compositions. By aligning RNA-seq reads associated with each barcoded mRNA to its reference sequences, we can systematically determine the transcription of each RE at TSS resolution (Fig. 2a, see “Methods”). Transcription initiation can occur from one or more distinct TSS locations within each RE (Fig. 2b). Indeed, 66.8% of the transcribed REs showed more than one measurable TSS in at least one recipient, and interestingly, the number of such multiple TSS REs varied depending on the recipient, with B. subtilis having the fewest followed by E. coli, with P. aeruginosa having the highest (Fig. 2c, p < 10−15, Fisher’s exact test), which follows a trend of increasing genomic GC content.
Fig. 2. Transcription start site utilization in RE library across hosts.
a TSS locations can be computed by aligning RNA-seq reads to their reference promoters. b TSS distributions show REs with single (left) as well as multiple (right) TSSs. c The distribution of number of TSSs per recipient. The fraction of promoters that contain multiple TSSs varies per recipient. Only REs with at least 10 RNA counts are considered, n = 3977, 10,515, and 19,154 B. subtilis, E. coli and P. aeruginosa, respectively. d Comparison of TSS location among different recipients for REs with single TSS (n = 367). Most REs (93.1%) are transcribed at the same TSS location, independent of the recipient species. e Same as (d), but including REs with multiple TSSs (n = 1404).
We selected REs expressed in the three recipients and classified the ones with the same TSS in all recipients as having universally shared TSS, those with shared TSS in two recipients and distinct TSS in one recipient as semi-shared TSS, and those with distinct TSS in all three recipients as unique TSS (see “Methods”). We classify REs to contain a single TSS when at least 90% of reads align within 5 bps of median TSS value. For REs that have a single TSS in each recipient, 93% were universally shared across all three recipients, while the rest showed varying degrees of similarity or recipient specificity (4.1% P. aeruginosa-specific, 0.2% E. coli-specific, and 2.2% B. subtilis-specific, 0.4% unique in all three species) (Fig. 2d). REs with multiple TSSs also showed similarly high levels of positional similarity across all unique TSS positions in each RE across recipients (Fig. 2e), suggesting that the majority of universally-active sequences share common regulatory signals, with additional TSSs being utilized depending on the species. De novo motif finding on transcriptionally active REs from this expanded library identified the canonical σ70 motif (TTGACA and TATAAT for −35 and −10 sequences, respectively) as the only identifiable signature involved in expression of acquired REs, in line with our previous findings [22]. Furthermore, we find here that the majority of experimentally identified TSSs are located at a distance of ~34 bp downstream from a sigma70 motif predicted in silico, corresponding to the expected distance of the 5′ boundary of the −35 hexamer from the first transcribed nucleotide (Supplementary Fig. S1).
Genomic GC content and σ70 stringency
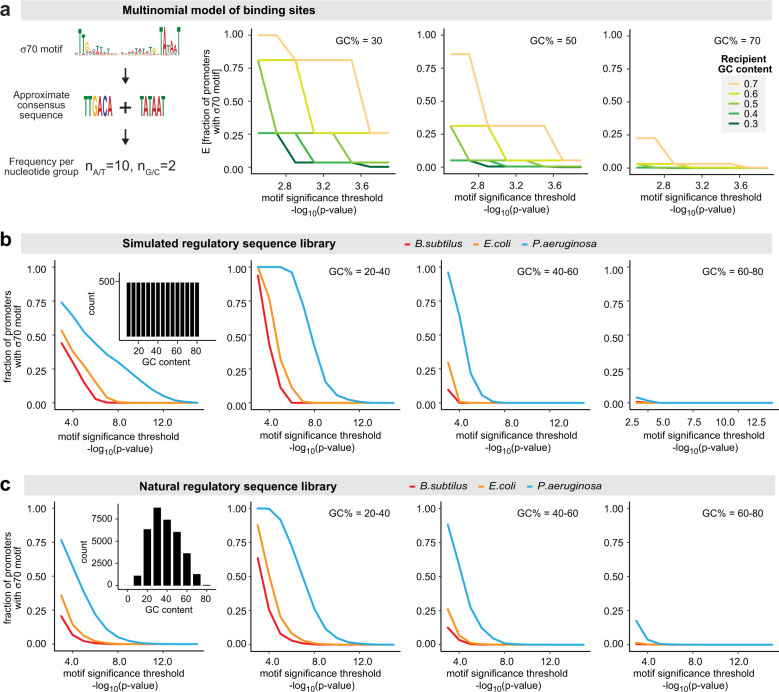
Donor-recipient regulatory compatibility may be evaluated in terms of (1) the likelihood that a donor RE is active in a recipient (i.e., promoter stringency) and (2) the ability of the recipient’s transcriptional machinery to transcribe a variety of foreign REs (i.e., recipient promiscuity). It may be beneficial for an organism to possess flexible regulation such that it can activate acquired genes from diverse sources. Bacterial intergenic regions tend to be lower in GC content than coding sequences (Supplementary Fig. S2). Due to its AT-rich nature, σ70-like sequences are more likely to occur by chance throughout the genomes of low GC REs. This fact, combined with the capacity of σ70 to bind to degenerate binding sites may lead to a high potential for off target initiation in low GC species [28]. To avoid this, we reasoned that individual species may require different levels of stringency in recognizing σ70 sites in order to properly distinguish promoters from their background genomic context. Under this paradigm, low GC species may require higher consensus binding to avoid spurious intragenic expression, while GC-rich species may have more lax requirements when recognizing binding sites since AT-rich sequences occur less frequently in their genomes (Fig. 3a and Supplementary Fig. S3).
Fig. 3. Recipient and donor RE GC content govern σ70 motif encoding within regulatory sequences.
a A simplified model that approximates the σ70 motif to an AT-rich multinomial sequence. The expected fraction of promoters with a motif match is plotted as a function of motif significance threshold for this simplified model. Each line represents a different recipient GC content, panels represent RE GC contents of 0.3, 0.5, and 0.7. Low GC promoters are more likely to contain σ70 motifs and GC-rich recipients are more likely to identify significant σ70 motifs. b The fraction of promoters with σ70 motif for a simulated regulatory library of random GC content follows similar trend observed in the simplified model represented in (a). Random promoters were generated with GC content in bins of 0.05 ranging from 0.1 to 0.8, 500 promoters for each bin, totaling 7500 promoters. c The fraction of promoters with σ70 motif in the library of regulatory sequences is consistent with the predictions from the simplified model (a) and random library (b).
To determine if altered σ70 stringency can explain the differences in capacity for activating heterologous promoters among our three species, we quantitatively evaluated σ70 stringency by controlling for each host’s genomic GC content background when scanning for binding sites in library REs (see “Methods”). For comparison, we also generated simulated libraries of sequences of varying composition in silico. Indeed, increasing host GC content was associated with a higher abundance of σ70 hits across a wide range of significance thresholds in both simulated sequences and the natural REs in our library (Fig. 3b,c). This trend held up across a range of RE compositions, although significant σ70 hits were nearly exclusive to P. aeruginosa at high RE GC contents. These results suggest that the stringency required for an organism to recognize and also encode σ70 binding sites within regulatory regions is balanced to its genomic GC content.
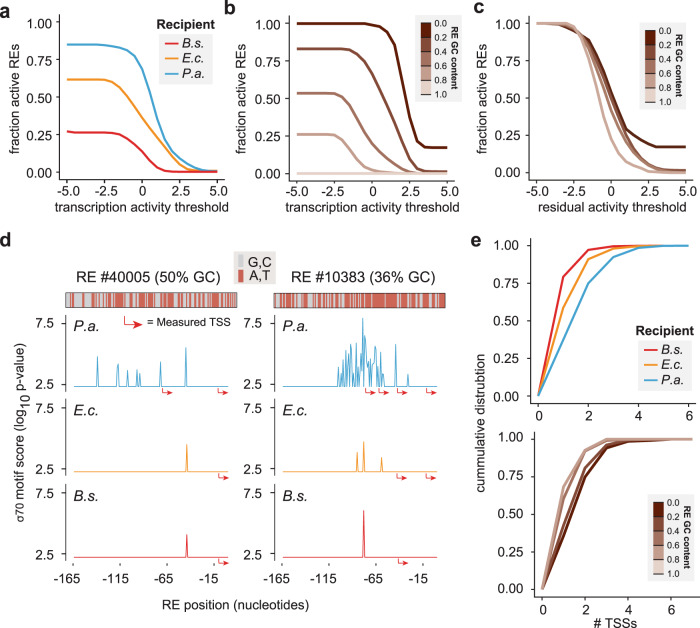
Several trends observed in our experimental measurements of heterologous RE activities can be explained through our model of GC content balanced σ70 stringency. First, the stark differences in capacity to activate diverse REs (Fig. 4a) align with our prediction that endogenous genomic GC content dictates the minimum activation requirements for transcriptional initiation (Fig. 3b, c). Similarly, the model explains why the GC content of regulatory sequences is inversely proportional to their activity levels (Fig. 4b and Supplementary Fig. 4a,b) and also the observations that REs from low GC bacterial groups like the Firmicutes have a higher likelihood and magnitude of expression across recipients, while the activation of higher GC REs from Actinobacteria and other high-GC species are rare in B. subtilis and more common in the other two hosts. We computed the residual activity (observed minus expected) by controlling for factors previously characterized to be predictive of RE expression [22], namely the σ70 best motif score and the stability of the 5′ mRNA secondary structure and observed that the RE GC content has no association with the residual expression magnitude in B. subtilis and P. aeruginosa and only weak association with E. coli (Fig. 4c and Supplementary Fig. 4c,d). These factors largely explain the observed differences in donor-recipient compatibility (Supplementary Figs. 5 and 6). In short, these analyses show that the AT-rich nature of the σ70 motif is the key factor underlying the association of RE GC content with expression.
Fig. 4. Promiscuity and stringency in transcriptional activation depends on recipient and RE GC compositions.
a The fraction of active promoters for different species show recipient promiscuity (P. aeruginosa > E. coli > B. subtilis) in proportion to genomic GC content. Transcription activity is defined in log scale units normalized by the activity of endogenous promoters, such as that a value of 0 indicates the average endogenous activity (Eq. (4)), n = 20,327, 28,608, and 24,473 for B. subtilis, E. coli, and P. aeruginosa, respectively. b The fraction of active promoters across a range of activity thresholds is shown for different promoter GC content. Low GC promoters show more promiscuous activity than high-GC promoters in our regulatory library. c Transcriptional profile as a function of residual activity (i.e., measured minus expected activity based on a regression model with the variables σ70 motif and mRNA 5’end stability) barely shows GC association, in contrast to (b). d Two examples of regulatory sequences showing expected σ70 motifs (spike plot) and experimentally identified TSS locations (red arrows). Low GC content promoters (right) are more likely than higher GC promoters (left) to contain a σ70-like motif. GC-rich recipients (e.g., P. aeruginosa) are more likely to contain σ70-like signals. e Density of number of TSSs per regulatory sequences is higher in GC-rich recipients. Low GC promoters are more likely to contain multiple TSS signals. b, c and low panel in e show results for E. coli. Data for B. subtilis and P. aeruginosa are shown in Supplementary Fig. 8.
Altered σ70 stringency also explains the differences in utilization of multiple TSSs observed in these species. We reasoned that the lower stringency requirements for σ70-driven transcription initiation in high-GC organisms may lead to the utilization of additional binding sites at distinct positions that are unrecognized by low GC species. Indeed, the higher frequency of multiple TSSs in P. aeruginosa, and to a lesser extent E. coli (Fig. 2c), is largely the result of less stringent recognition of additional σ70 binding sites (Fig. 4d). This effect is particularly evident in low GC REs, which have a large difference in GC content with the P. aeruginosa genome (Fig. 4e).
To further demonstrate promiscuity, we also cloned five degenerate libraries of GC contents ranging from 33 to 66% with downstream ribosome binding sites into our three hosts using the same reporter scheme. The activity distributions of each library were characterized through flow cytometric analysis of sfGFP expression. No significant expression was observed by flow cytometry in any library in B. subtilis, while most of the random constructs in the lowest GC content library were active in P. aeruginosa, indicating a higher capacity for transcriptional activation in the latter (Supplementary Fig. 7). Together these results show that while the canonical σ70 motif is highly conserved, variation in initiation stringency arising from adaptation of the transcriptional machinery to host genomic GC content can lead to different positional patterns and levels of gene expression.
Widespread GC-adaptation in σ70 encoding within bacterial genomes
In order to assess conservation of the canonical σ70 motif across bacterial phylogenetic groups, we analyzed putative σ70 binding sites in regulatory regions for a set of 1545 genomes representing diversity from all major bacterial phyla. We first computationally extracted all non-convergent intergenic regions from each genome and used FIMO to identify putative σ70 binding sites (see “Methods”). The maximum σ70 motif –log10(p value) was taken as the σ70 score for each intergenic region. Usage of σ70 was examined using the entire set of intergenic regions as well as the subset associated with housekeeping genes of COG categories J (translation) and K (transcription). We tested the hypothesis of whether the average σ70 score from natural sequences were significantly different from the one observed in shuffled sequences and assumed evidence of σ70 motif usage at an FDR threshold of 0.01 (“Methods”). We found evidence of significant σ70 motif usage in a substantial fraction of bacterial species (1303 out of 1545 organisms). The majority of cases (1171) have intergenic regions significantly enriched in σ70 matches and fewer organisms (132) showed depleted signal for σ70 matches (Supplementary Fig. 8).
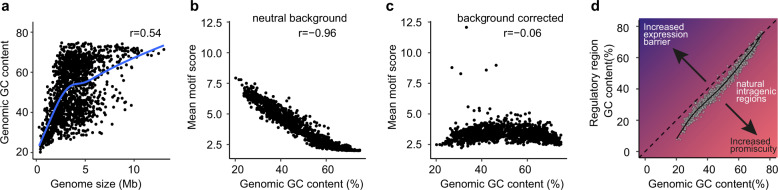
Next, we investigated the relationship between σ70 motif composition and genomic GC content in our set of 1545 genome sequences. Consistent with previous studies, genome size had a strikingly strong correlation with genome GC content (Pearson R = 0.54) [29], indicating an underlying link between GC content and genome expansion (Fig. 5a). Further we compared the motif score of regulatory regions obtained by FIMO assuming the near-neutral GC content background of E. coli (51%) or controlled by the genomic GC content of each species (see “Methods”). The mean intergenic region σ70 score of each genome was strongly and negatively correlated with genomic GC content when calculated using the nearly uniform background nucleotide probabilities of E. coli MG1655 (Pearson correlation r = −0.96, Fig. 5b). However, the average σ70 scores were relatively uniform when the motif scanning corrects for each organism’s GC content (Pearson correlation = −0.06, Fig. 5c), highlighting the importance of considering background genomic GC content composition when assessing putative σ70 binding sites. These findings suggest that our proposed mechanism of GC-adapted cis-regulatory encoding of σ70 binding sites is generalizable throughout the bacterial kingdom (Fig. 5d).
Fig. 5. The signal for σ70 motif is nearly uniform among 1,545 representative genomes after background GC content correction.
a GC content is highly correlated with genome size in bacteria (blue curve shows LOESS smooth). b, c Mean σ70 motif scores across non-convergent intergenic regions > 100 bp in length from 1545 genomes. A strong bias in σ70 motif score and GC content occurs when motif is scanned assuming a neutral background (b), but this bias disappears when motif scoring is corrected by genome GC content (c). d Schematic representation of expression barrier and promiscuity in terms of REs and recipient GC content. Gray points represent the GC contents of individual genomes and their intergenic regions. Pearson correlation coefficient values (r) are displayed in (a)–(c).
Discussion
Here, we have identified a mechanism to explain regulatory compatibility between bacteria of diverse genomic compositions. Through large-scale characterization of natural promoters from diverse bacterial species and mobile elements, we identified a relationship between host genomic GC content and regulatory sequence composition that determines the likelihood and magnitude of transcriptional strength. We suggest a model whereby the stringency required for σ70-mediated transcription initiation may vary across different bacterial species. The need for bacteria to distinguish true binding sites from spurious σ70-like motifs throughout their genomes requires tuning promoter stringency to each species’ genomic GC content. This model explains why GC-rich species have a high capacity for activating diverse promoters and also why low GC content sequences are easier to be expressed by diverse recipients. The model also suggests that while most promoters are transcribed at shared TSSs across different recipients, a GC-rich host like P. aeruginosa can recognize additional σ70-signals, leading to more observed TSSs. By analyzing 1545 diverse genomes, we found widespread evidence for GC-adaptation of σ70 stringency throughout the bacterial kingdom.
Future studies could examine the mechanistic basis of altered σ70 stringency. Interestingly, while alternative sigma factors tend to require stringent binding site matches to initiate transcription from their target promoters, certain mutations to their protein sequences enable recognition of binding sites with more degeneracy [30]. These findings suggest that similar evolutionary tuning may enable GC-adaptation of σ70 stringency in concert with evolving GC content. In line with our proposed mechanism, promoters from GC-rich Rhizobium and Streptomyces species (61.5 and 72.1 %GC, respectively) [31, 32] have higher levels of degeneracy in their σ70 motifs than low GC organisms like Clostridium fermentans (GC% = 35) [33], consistent with less stringent requirements for initiation of transcription. A study of synthetic promoter sequences in industrial Clostridium species with similarly low GC contents identified a strong preference for AT-rich promoters [23], providing further support for GC-adaptation of promoter stringency. Future studies combining the high-throughput approaches as described here and rationally designed libraries may help reveal the biophysical basis of promoter promiscuity and its evolution [34, 35]. Furthermore, deeper understanding of promoter stringency and host promiscuity may aid efforts to tailor transcriptional regulation in synthetic gene circuits for more reliable engineering of diverse organisms of ranging GC compositions.
Our proposed mechanism may help explain patterns observed in bioinformatic analyses of HGT in bacterial genomes. For example, it has been observed that horizontally acquired regions have often lower GC content than the host background genome, even after long-term retention [36]. This pattern may result from low GC DNA having an inherent tendency toward constitutive expression when transferred to new hosts, thus facilitating functionalization of encoded genes. Relatedly, mobile DNA encoded on phage and plasmids have also been shown to be more AT-biased than their hosts [37]. We speculate these sequences may be under selection for lower GC content to ensure transcriptional activity in various recipient strains. Furthermore, our findings help explain the prevalence of DNA silencing mechanisms in the bacterial kingdom. DNA silencing proteins are known to suppress the fitness burden caused by uncontrolled expression of horizontally acquired DNA. In particular, H-NS-like proteins transcriptionally repress long stretches of horizontally acquired DNA that are more AT-rich than the endogenous genome [29, 38–40] and there are no known mechanisms for silencing GC-rich DNA [29, 41]. The long-term retention of GC content bias in HGT regions may be contingent to the process in which they are wired in the host genome. DNA silencing proteins may initially buffer fitness cost at the time of acquisition [42] and later become the regulator in regions of horizontally acquired DNA [43].
GC-adaptation of σ70 encoding to host GC content may provide a driving force for shaping genomic GC evolution in bacteria [44–46]. Given the GC-to-AT mutational bias that often occurs in bacteria [47, 48], there must be a counteracting selective force that balances the GC compositions to what is observed within genomes. As such, transcriptional promiscuity could be a major contributor to this selective force. In this context, an increase in AT content could increase the rate of nonspecific transcription, resulting in a fitness burden that may be especially higher for GC-rich species. This idea is supported by studies showing that synonymously recoding genes to a higher AT content generally reduces the fitness of the host organism in synthetic studies [16, 38, 47]. It has also been proposed that E. coli has evolved to avoid intragenic σ70 sites through its codon choices [49]. We expect that low GC organisms have developed a highly stringent threshold for activation of σ70 motifs to avoid spurious transcription initiation from the frequent promoter-like sequences found in their genomes. Furthermore, while natural sequences may have selected against spurious internal promoters, synthetic biologists may wish to take promoter promiscuity and regulatory stringency requirements into account in order to avoid inadvertent regulatory signals in codon-optimized versions of gene.
Promoter promiscuity may also be the reason for the striking positive correlation between genome size and GC content [29]. Previous theoretical work suggested that a high-GC genome may experience a stronger barrier to generate new regulatory binding sites since regulatory regions tend to be AT-rich [50]. Our data however show that, in the case of σ70, binding site signal must be significantly distinct from organism’s background genome in order to be recognized and expressed. According to our model, low GC content bacteria have evolved stronger transcription stringency to compensate for the background σ70 motif signal encoded in their AT-rich genome. This mechanism would cause a difficulty to low GC organisms in functionalizing horizontally acquired DNA from higher GC organism and ultimately limit genome expansion. In contrast, high-GC genomes contain fewer AT-rich sequences and have a more relaxed activation requirement that may facilitate successful functionalization and incorporation of acquired genes from diverse organisms, thus enabling genome expansion.
Finally, it is possible that regulatory compatibility may constrain the rates and directionality of HGT between species of different GC compositions. One study observed that 86% of transfers occurred between species within 5% GC content difference [51]. Promoter promiscuity and regulatory stringency may impact the rates and directionality of transfers between compositionally distinct species. Bioinformatically identified HGT events are biased toward compatible exchanges and may not be representative of all attempted transfers that occur in nature. In order to investigate the mechanisms associated with HGT, new approaches that measure real-time HGT in natural or synthetic microbial communities will enable deeper characterization of transfer processes, from acquisition to functionalization [52].
Methods
We previously generated and characterized the gene expression properties of 29,249 natural REs from 184 diverse prokaryotic genomes [22]. In this study, we describe the further characterization of five additional RE libraries from bioinformatically identified gene transfer events (n = 3818) [25], antibiotic resistance genes (n = 1047, from ARDB) [26], virulence factors (n = 336, from VFDB) [27], as well as from plasmids and phage genomes (n = 2518, from IMG) [53]. Transformation and activity measurements were performed as described in previous studies [22, 54]. Briefly, synthesized REs were cloned as a pooled library upstream of a GFP reporter, transformed into target bacteria. Library cultures were grown to mid-exponential phase, divided, pelleted, and subjected to DNA and RNA purification. Targeted RNA-seq of sfGFP mRNA and amplicon DNA-seq libraries were prepared from total RNA and purified library DNA respectively and were sequenced on Illumina HiSeq platforms. Reads were processed bioinformatically and filtered to account for synthesis errors. Transcription activity levels were computed according to the log transformed relative abundances of RNA and DNA reads. Transcription values were also normalized according to those of endogenous control sequences from each target organism’s genus that were included in the library. For TSS analysis, we only considered promoters with at least 10 RNA reads. For Transcription activity, we only considered promoters with at least 15 total count of RNA + DNA reads (Supplementary Fig. 9). The number of TSSs per promoter per recipient was computed using k-means clustering. The MEME package [55] was used to identify regulatory motifs in our dataset. The FOLD function from the RNAstructure algorithm [56] was used to compute mRNA stability. Null model of best kmer motif degeneracy was obtained according to an extreme value distribution [57]. The random regulatory libraries were designed with defined proportions of GC content and evaluated according to its GFP fluorescence using a BD Fortessa flow cytometer. For our large-scale in silico analysis of regulatory regions, we downloaded the entire set of 1545 representative complete bacterial genomes from NCBI and reannotated them using Prokka [58]. We annotated functional categories using Diamond [59] to map genes into COG database [60] and extracted non-convergent intergenic regions for motif analysis. Data used in this paper are available in supporting data.
Detailed description of methods for library design, data acquisition and analysis are provided as supporting material. Raw sequencing reads can be found at PRJNA431139.
Supplementary information
Acknowledgements
We thank members of the Wang lab for helpful scientific discussions and feedback. HHW acknowledges funding support from the NIH (1DP5OD009172-02, 1U01GM110714-01A1, 1R01AI132403-01), NSF (MCB‐1453219), Sloan Foundation (FR-2015-65795), DARPA (W911NF-15-2-0065), and ONR (N00014-15-1-2704). NIJ was supported by a NSF Graduate Research Fellowship (DGE-16-44869).
Author contributions
ALCG, NIJ, and HHW designed the study with help from CSS, MBS, and EJA; ALCG, NIJ, and AY performed the experiments and analyzed the data. AG, NIJ, and HHW wrote the paper with input from all authors.
Compliance with ethical standards
Conflict of interest
HHW is a member of the Scientific Advisory Board of SNIPR Biome. The authors declare no other competing financial interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Antonio L. C. Gomes, Nathan I. Johns
Supplementary information
The online version of this article (10.1038/s41396-020-0696-1) contains supplementary material, which is available to authorized users.
References
- 1.Polz MF, Alm EJ, Hanage WP. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 2013;29:170–5. doi: 10.1016/j.tig.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park C, Zhang J. High expression hampers horizontal gene transfer. Genome Biol Evolut. 2012;4:523–32. doi: 10.1093/gbe/evs030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–87. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 4.Rosen MJ, Davison M, Bhaya D, Fisher DS, et al. Fine-scale diversity and extensive recombination in a quasisexual bacterial population occupying a broad niche. Science (New York, N.Y.) 2015;348:1019–23. doi: 10.1126/science.aaa4456. [DOI] [PubMed] [Google Scholar]
- 5.McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH, et al. High frequency of horizontal gene transfer in the oceans. Science. 2010;330:50. doi: 10.1126/science.1192243. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–8. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 7.Hehemann J-H, Kelly AG, Pudlo NA, Martens EC, Boraston AB, Davies G. Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc Natl Acad Sci USA. 2012;109:19786–91. doi: 10.1073/pnas.1211002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes ALC, Galagan JE, Segrè D. Resource competition may lead to effective treatment of antibiotic resistant infections. PLoS ONE. 2013;8:e80775. doi: 10.1371/journal.pone.0080775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–20. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 10.Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol. 2011;14:236–43. doi: 10.1016/j.mib.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–11. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baltrus DA. Exploring the costs of horizontal gene transfer. Trends Ecol Evolut. 2013;28:489–95. doi: 10.1016/j.tree.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 14.San Millan A, Toll-Riera M, Qi Q, MacLean RC. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat Commun. 2015;6:6845. doi: 10.1038/ncomms7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 16.Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. Genome-wide experimental determination of barriers to horizontal gene transfer. Science. 2007;318:1449–52. doi: 10.1126/science.1147112. [DOI] [PubMed] [Google Scholar]
- 17.Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol. 2006;358:614–33. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol. 2004;14:283–91. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. RNA polymerases from Bacillus subtilis and Escherichia coli differ in recognition of regulatory signals in vitro. J Bacteriol. 2000;182:6027–35. doi: 10.1128/JB.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yus E, Yang J-S, Sogues A, Serrano L. A reporter system coupled with high-throughput sequencing unveils key bacterial transcription and translation determinants. Nat Commun. 2017;8:368. doi: 10.1038/s41467-017-00239-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vingadassalom D, Kolb A, Mayer C, Rybkine T, Collatz E, Podglajen I. An unusual primary sigma factor in the Bacteroidetes phylum. Mol Microbiol. 2005;56:888–902. doi: 10.1111/j.1365-2958.2005.04590.x. [DOI] [PubMed] [Google Scholar]
- 22.Johns NI, Gomes ALC, Yim SS, Yang A, Blazejewski T, Smillie CS, et al. Metagenomic mining of regulatory elements enables programmable species-selective gene expression. Nat Methods. 2018;15:323–9. doi: 10.1038/nmeth.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mordaka PM, Heap JT. Stringency of synthetic promoter sequences in clostridium revealed and circumvented by tuning promoter library mutation rates. ACS Synth Biol. 2018;7:672–81. doi: 10.1021/acssynbio.7b00398. [DOI] [PubMed] [Google Scholar]
- 24.Jensen PR, Hammer K. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol. 1998;64:82–7. doi: 10.1128/AEM.64.1.82-87.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480:241–4. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 26.Liu B, Pop M. ARDB-antibiotic resistance genes database. Nucleic Acids Res. 2009;37:D443–7. doi: 10.1093/nar/gkn656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Xiong Z, Sun L, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012;40:D641–5. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hook-Barnard IG, Hinton DM. Transcription initiation by mix and match elements: flexibility for polymerase binding to bacterial promoters. Gene Regul Syst Biol. 2007;1:275–93. [PMC free article] [PubMed] [Google Scholar]
- 29.Navarre WW, Mcclelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS—facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA; 2007. 1456–71. 10.1101/gad.1543107.evolution. [DOI] [PubMed]
- 30.Koo B-M, Rhodius VA, Nonaka G, deHaseth PL, Gross CA. Reduced capacity of alternative sigmas to melt promoters ensures stringent promoter recognition. Genes Dev. 2009;23:2426–36. doi: 10.1101/gad.1843709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez-Romero MA. The Rhizobium etli sigma70 (SigA) factor recognizes a lax consensus promoter. Nucleic Acids Res. 2006;34:1470–80. doi: 10.1093/nar/gkl023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong Y, Kim J-N, Kim MW, Bucca G, Cho S, Yoon YJ, et al. The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2) Nat Commun. 2016;7:11605. doi: 10.1038/ncomms11605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boutard M, Ettwiller L, Cerisy T, Alberti A, Labadie K, Salanoubat M, et al. Global repositioning of transcription start sites in a plant-fermenting bacterium. Nat Commun. 2016;7:13783. doi: 10.1038/ncomms13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urtecho G, Tripp AD, Insigne KD, Kim H, Kosuri S. Systematic dissection of sequence elements controlling σ70 promoters using a genomically encoded multiplexed reporter assay in Escherichia coli. Biochemistry. 2019;58:1539–51. doi: 10.1021/acs.biochem.7b01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Einav T, Phillips R. How the avidity of polymerase binding to the −35/−10 promoter sites affects gene expression. Proc Natl Acad Sci USA. 2019;116:13340–5. doi: 10.1073/pnas.1905615116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daubin V, Lerat E, Perrière G. The source of laterally transferred genes in bacterial genomes. Genome Biol. 2003;4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rocha EPC, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–4. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 38.Lamberte LE, Baniulyte G, Singh SS, Stringer AM, Bonocora RP, Stracy M, et al. Horizontally acquired AT-rich genes in Escherichia coli cause toxicity by sequestering RNA polymerase. Nat Microbiol. 2017;2:16249. doi: 10.1038/nmicrobiol.2016.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Rueda E, Ibarra JA. Distribution of putative xenogeneic silencers in prokaryote genomes. Comput Biol Chem. 2015;58:167–72. doi: 10.1016/J.COMPBIOLCHEM.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Flores-Ríos R, Quatrini R, Loyola A. Endogenous and foreign nucleoid-associated proteins of bacteria: occurrence, interactions and effects on mobile genetic elements and Host’s biology. Comput Struct Biotechnol J. 2019;17:746–56. doi: 10.1016/J.CSBJ.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeifer E, Hünnefeld M, Popa O, Frunzke J. Impact of xenogeneic silencing on phage–host interactions. J Mol Biol. 2019. 10.1016/J.JMB.2019.02.011. [DOI] [PMC free article] [PubMed]
- 42.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. 2007:315:251–2. 10.1126/science.1137550. [DOI] [PubMed]
- 43.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JCD. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lassalle F, Périan S, Bataillon T, Nesme X, Duret L, Daubin V. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLoS Genet. 2015;11:1–20. doi: 10.1371/journal.pgen.1004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hildebrand F, Meyer A, Eyre-walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed]
- 46.Gophna U. The unbearable ease of expression—how avoidance of spurious transcription can shape G+C content in bacterial genomes. FEMS Microbiol Lett. 2018;365. 10.1093/femsle/fny267. [DOI] [PubMed]
- 47.Raghavan R, Kelkar YD, Ochman H. A selective force favoring increased G+C content in bacterial genes. Proc Natl Acad Sci USA. 2012;109:14504–7. doi: 10.1073/pnas.1205683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long H, Sung W, Kucukyildirim S, Williams E, Miller SF, Guo W, et al. Evolutionary determinants of genome-wide nucleotide composition. Nat Ecol Evolut. 2018;2:237–40. doi: 10.1038/s41559-017-0425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yona AH, Alm EJ, Gore J. Random sequences rapidly evolve into de novo promoters. Nat Commun. 2018;9:1530. doi: 10.1038/s41467-018-04026-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cordero OX, Hogeweg P. The consequences of base pair composition biases for regulatory network organization in prokaryotes. Mol Biol Evolut. 2009;26:2171–3. doi: 10.1093/molbev/msp132. [DOI] [PubMed] [Google Scholar]
- 51.Popa O, Hazkani-covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21:599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munck C, Sheth RU, Freedberg DE, Wang HH. Recording mobile DNA in the gut microbiota using an Escherichia coli CRISPR-Cas spacer acquisition platform. Nat Commun. 2020;11:95. doi: 10.1038/s41467-019-14012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–22. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci USA. 2013;110:14024–9. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathews DH. RNA secondary structure analysis using RNA structure. Curr Protoc Bioinforma. 2014;46:12.16.11–12.16.25. doi: 10.1002/0471250953.bi1206s46. [DOI] [PubMed] [Google Scholar]
- 57.Gomes AL, Abeel T, Peterson M, Azizi E, Lyubetskaya A, Carvalho L, et al. Decoding ChIP-Seq with a double-binding signal refines binding peaks to single-nucleotides and predicts cooperative interaction. Genome Res. 2014:1686–97. 10.1101/gr.161711.113. [DOI] [PMC free article] [PubMed]
- 58.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 59.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 60.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–9. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.