Abstract
ERK5 is a protein kinase that also contains a nuclear localisation signal and a transcriptional transactivation domain. Inhibition of ERK5 has therapeutic potential in cancer and inflammation and this has prompted the development of ERK5 kinase inhibitors (ERK5i). However, few ERK5i programmes have taken account of the ERK5 transactivation domain. We have recently shown that the binding of small molecule ERK5i to the ERK5 kinase domain stimulates nuclear localisation and paradoxical activation of its transactivation domain. Other kinase inhibitors paradoxically activate their intended kinase target, in some cases leading to severe physiological consequences highlighting the importance of mitigating these effects. Here, we review the assays used to monitor ERK5 activities (kinase and transcriptional) in cells, the challenges faced in development of small molecule inhibitors to the ERK5 pathway, and classify the molecular mechanisms of paradoxical activation of protein kinases by kinase inhibitors.
Keywords: extracellular signal-regulated kinases, kinases, paradoxical activation, small molecules
Introduction
Protein kinases play key roles in a variety of diseases including cancer and inflammation and have emerged as ‘druggable’ enzymes [1]. By 2019 some 48 small molecule protein kinase inhibitors had received USA FDA approval [2] with ∼175 more in clinical trials [3]. The principle of targeting protein kinases with small molecules is straightforward; block the active site so that it cannot bind ATP, prevent substrate binding, prevent binding of an up-stream activator or disrupt critical conformational changes. However, intensive research and clinical experience have identified two important limitations to the efficacy of kinase inhibitors. These are, innate or acquired resistance to the kinase inhibitor [4], and unintended activation of the target pathway, either by inhibition of negative feedback pathways [5] or through inhibitor binding to the kinase resulting in its paradoxical activation [6].
The ERK5 signalling pathway in health and disease
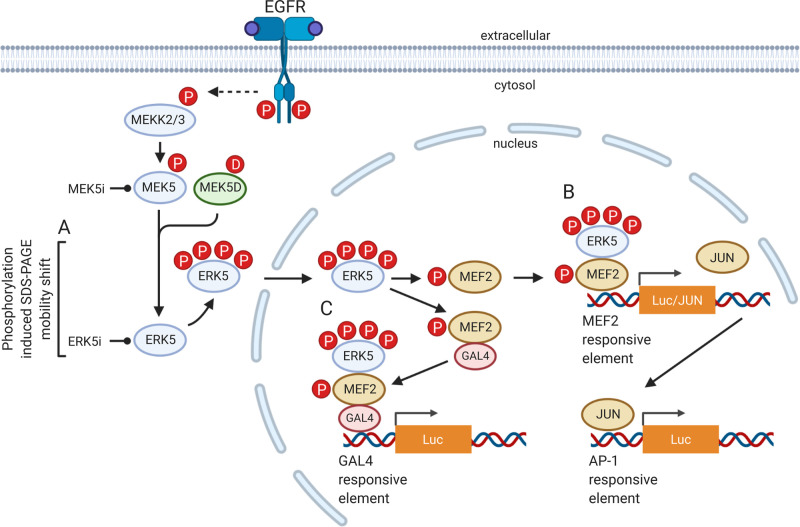
The mitogen-activated protein kinase (MAPK) family member, extracellular signal regulated kinase 5 (ERK5, also known as Big MAP Kinase 1 or BMK1) is encoded by the MAPK7 gene [7,8]. It is the effector kinase of a three-tiered MAPK pathway, comprising MEKK2 and MEKK3 (the MKKKs), MEK5 (MKK) and finally ERK5 (MAPK) (Figure 1). ERK5 contains an N-terminal kinase domain that shares 50% identity with ERK2 [7,8] and a large, unique C-terminal extension that includes a nuclear localisation signal (NLS) and a transcriptional activation domain (TAD) [9] (Figure 2). Upon cellular stimulation (by mitogens [10], Toll-like receptor ligands [11] or cellular stresses [12]) activated MEK5 phosphorylates the TEY motif in the ERK5 activation loop leading to activation of its kinase domain [13], much like activation of ERK1/2 by MEK1/2. ERK5 then auto-phosphorylates its C-terminus which promotes ERK5 translocation from the cytosol to the nucleus [14,15] where ERK5 binds, phosphorylates and activates MEF2 transcription factors such as MEF2D [10,16,17] (Figures 1 and 2). The ERK5 C-terminus is also phosphorylated by other protein kinases including ERK1/2 [18], CDK1 [19,20] and CDK5 [21]. Thus, the C-terminus both mediates a subset of the effects of the ERK5 kinase domain and integrates signals from other pathways. ERK5 nuclear translocation and transcriptional activity is also regulated by CDC37-induced dissociation of Hsp90 [22], and SUMOylation [23]. For an excellent review on the mechanisms that regulate ERK5 nuclear localisation see Tubita et al. [24].
Figure 1.
EGFR activation of the MEK5–ERK5 signalling pathway and the cell-based assays used to measure activation and inhibition by MEK5i and ERK5i (A) ERK5 autophosphorylation assay, (B) ERK5-driven MEF2 and AP-1 reporter assays, (C) ERK5-driven MEF2:GAL4 reporter assay. Figure created using bioRENDER.com.
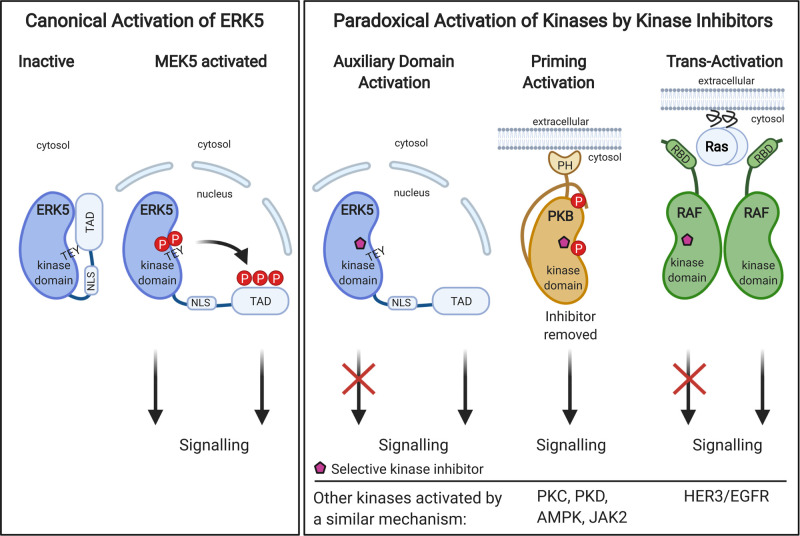
Figure 2. Canonical Activation of ERK5 and Paradoxical Activation of ERK5, PKB and RAF by kinase inhibitors.
TAD, transactivation domain; NLS, nuclear localisation signal; PH, pleckstrin homology domain; RBD, RAS binding domain. Figure created using bioRENDER.com.
Components of the ERK5 pathway are ubiquitously expressed in adult tissues [7,8,14]. In development, ERK5 regulates pluripotency in mouse embryonic stem cells [25], and MEK5 and ERK5 are required for blood vessel and cardiac development [26,27]. There is therapeutic potential in targeting the MEK5–ERK5 pathway in disease, especially cancer and inflammation. In this section, we have only considered data from knockdown and knockout experiments when assigning the therapeutic potential of inhibition of the ERK5 pathway. This is due to the pitfalls uncovered when generating ERK5i as therapeutics or tool compounds: off-target effects on kinases and bromo-domain containing proteins, and paradoxical activation (Table 1). We discuss these pitfalls in more detail later in the review. Knockdown of ERK5 or MEK5 by siRNA has shown anti-inflammatory effects in endothelial cells and monocytes [11,28]. Knockout of ERK5 in tumour-associated macrophages impedes the growth of melanoma and lung carcinoma in mouse models [29]. Furthermore, knockout of ERK5 in keratinocytes prevents inflammation-driven tumorigenesis [30]. Knockdown of ERK5 (or MEK5) by siRNA has also shown the therapeutic potential of the ERK5 pathway in mutant BRAF-driven melanoma [31,32,33], mutant KRAS-driven pancreatic ductal adenocarcinoma (PDAC) [34], as well as prostate [35], breast [36] and bladder cancers [37]. There are conflicting results obtained with ERK5 siRNA in hepatocellular carcinoma (HCC) [38,39]. In colorectal cancer, siRNA to ERK5 shows that ERK5 is required for the regulation of cancer stem-like cell properties and tumour-sphere growth [40], but not for inhibition of tumour cell proliferation [38].
Table 1. ERK5 inhibitors.
| Inhibitor /PubChem CID | Class of ERK5 inhibitor | IC50: assay | Other targets IC50: assay | Mechanism of action | Kinase inhibitor type [67] | Paradoxical activator (% of MEK5D effect) | Activity in tumour model(s) | Phase of development | References |
|---|---|---|---|---|---|---|---|---|---|
| BIX02188 /135398492 | MEK5 | 4.3 nM: in vitro kinase assay 0.82–1.15 µM: in cell ERK5:MEF2C |
In vitro kinase assays for: ERK5: 810 nM CSF1R:280 nM LCK: 390 nM KIT: 550 nM Kinases with >70% inhibition at 3 µM: Src Kinases with >70% inhibition at 10 µM: ABL CSF1R(FMS) KIT LCK using the University of Dundee and Invitrogen kinase selectivity testing |
ATP competitive | Type I‡ | NR | NT | Preclinical | [58] |
| BIX02189/135659062 | MEK5 | 1.5 nM: in vitro kinase assay 0.26–0.53 µM: in cell ERK5:MEF2C 49.5 nM: in cell ERK5:MEF2D |
In vitro kinase assays for: ERK5: 810 nM CSF1R:28 0nM LCK: 390 nM KIT: 550 nM Kinases with >70% inhibition at 3 µM: Src Kinases with >70% inhibition at 10 µM: ABL CSF1R(FMS) FGFR1 LCK RSK2 + 4 using the University of Dundee and Invitrogen kinase selectivity testing In cells: TGFβ-type 1R: 5 µM using a SMAD reporter |
ATP competitive | Type I‡ | NR | Yes | Preclinical | [55,58,85] |
| SC-1-181 | MEK5 | 10 µM decreases phospho-ERK5 by 59% | 10 µM had no effect on phospho-ERK1/2 No other data available |
Not ATP competitive |
Type III† | NR | NT | Preclinical | [61,62] |
| XMD8-92/46843772 | Dual ERK5/BRD4 | 190 nM: in cell lysate KiNativ 130 nM: in live cells KiNativ 364 nM: in vitro kinase assay 240 nM: In cell ERK5 band-shift |
LRRK2 (in vitro kinase assay): 59 nM BRD4 (BROMOscan assay): 170 nM Kinases with >90% displacement (KiNativ): DCAMKL1, TNK4 and PLK4 |
ATP competitive | Type I | 24% at 1µM | Yes | Preclinical | [63,64,83,86–89] |
| AX15836/122705989 | ERK5 | 8 nM: in cell lysate KiNativ 9 nM: in live cells KiNativ 31 nM: in cell ΔTAD-ERK5:MEF2D |
BRD4 BROMOscan assay: 3.6 µM No other kinases inhibited >70% at 1 µM by KiNativ |
ATP competitive | Type I* | 200% at 3 µM | NT | Preclinical | [55,64] |
| XMD17-109 (Cmpd26, ERK5-IN-1)/71604307 | Dual ERK5/BRD4 | 162 nM: in vitro kinase assay 90 nM: in cell EGF band-shift 4.2 µM: in cell AP-1 reporter 163 nM: in cell FL-ERK5:MEF2D 90 nM: in cell ΔTAD-ERK5:MEF2D |
In vitro LRRK2: 339 nM BRD4-1 Binding assay: 217 nM No other selectivity data published |
ATP competitive | Type I* | 40% at 1 µM | NT | Preclinical | [45,63] [55] |
| XMD17-26 (Cmpd 25) | Dual ERK5/BRD4* | 80 nM:in cell EGF band-shift 82 nM: in vitro kinase assay 152 nM: in cell FL-ERK5:MEF2D 28 nM: in cell ΔTAD-ERK5:MEF2D |
No selectivity data published | ATP competitive | Type I | 50% at 1 µM | NT | Preclinical | [44,55] |
| JWG-045 (XMD10-78) | ERK5/LRRK2 | 98 nM: in vitro kinase assay | BRD4 AlphaScreen™: 11 µM LRRK2 in vitro kinase assay: 289 nM No other selectivity data published |
ATP competitive | Type I* | NT | NT | Preclinical | [25,32,45] |
| JWG-071/131842089 | ERK5/LRRK2 | 88 nM: in vitro kinase assay | BRD4 AlphaScreen™: 5.42 µM LRRK2 in vitro kinase assay: 109 nM Kinases with >70% inhibition at 1 µM by KINOMEscan™ DCAMKL1/2 LRRK2 LRRK2 (G2019S) PLK4 |
ATP competitive | Type I* | NT | NT | Preclinical | [45] |
| Compound 46 | ERK5 | 820 nM: in vitro 3 µM: in cell FL-ERK5:MEF2D 1 µM: in cell ΔTAD-ERK5:MEF2D |
No binding to BRD4 at 20 µM. Kinases with ≥ 90% inhibition at 10 µM : DCAMKL3 JAK1 SLK MAP3K15 TYK2 JAK2 MST2 DCAMKL1 by KINOMEscan™ |
ATP competitive | Type I | 5% at 10 µM | Yes | Preclinical | [56] |
| BAY-885/134128280 | ERK5 | 35 nM: in vitro kinase assay 120 nM: in cell MEF2 reporter assay |
No other kinase inh >70% at 1 µM by Eurofins kinase panel No binding to BRD4 at >20 µM |
ATP competitive | Type I* | 20% at 1 µM | NT | Preclinical | [57] |
| ADTL-EI1712 | Dual ERK1/2 ERK5 |
1 µM KINOMEscan % inhibition ERK1 94% ERK2 91% ERK5 87% |
Kinases with >71% inhibition at 1 µM: KIT AXL KINOMEscan by Eurofins |
ATP competitive¶ | Type I¶ | NT | Yes | Preclinical | [66] |
| Compound 3 | ERK5 | 42 nM: in vitro | >20 other kinases inhibited at 10 µM using the ThermoFisher standard kinase panel Not tested against BRD4. |
ATP competitive | Type I | NT | NT | Preclinical | [68] |
| Compound 5 | ERK5 | 2.3 µM: in vitro | Kinases with >80% inh at 10 µM: CSF1R FLT4 GSK3α SYK TYK2 using the ThermoFisher standard kinase panel. Not tested against BRD4. |
Allosteric and ATP competitive |
Type IV | NT | NT | Preclinical | [68] |
| TG-02 (SB1317, Zotiraciclib)/16739650 | Multi-kinase inhibitor including ERK5 | 43 nM: in vitro |
In vitro kinase assays for: CDK9: 3 nM CDK5: 4 nM CDK2: 5 nM CDK3: 8 nM CDK1: 9 nM Lck: 11 nM TYK2: 14 nM Fyn: 15 nM JAK2: 19 nM FLT3: 19 nM FLT3 D835Y: 21 Fms: 27 nM TYRO3: 36 nM CDK7: 37 nM ERK5: 43 nM P38δ: 56 nM JAK1: 59 nM |
ATP competitive§ | Type I§ | NT | Yes | I/II | [90–92] |
NT, not tested; NR, not relevant; NP, not published.
No structural data available, based on binding mode of a close analogue;
No structural data available, based on binding mode derived from computational docking into AuroraA kinase [93];
No structural data available, based on binding mode derived from computational docking into ERK5 kinase domain [66];
No structural data available, based on binding mode derived from computational docking into a MEK5 homology model [61];
No structural data available, based on binding modes of close analogues to other kinases.
The challenge of monitoring ERK5 inhibition in cells
The use of protein kinase inhibitors in biological systems requires clear and specific biomarkers that confirm target inhibition and allow interpretation of the experiment. For example, inhibition of the RAS–RAF–MEK1/2–ERK1/2 pathway with a MEK1/2 or ERK1/2 inhibitor causes loss of phosphorylated ERK1/2 and/or RSK (direct substrates of MEK1/2 and ERK1/2, respectively). Monitoring the ERK1/2 pathway is made easier by the existence of more than 200 ERK1/2 substrates and interacting proteins [41].
Whilst inhibition of MEK5 is relatively easily assessed in cells by loss of ERK5 activation-loop TEY phosphorylation, the paucity of well-validated ERK5 substrates allowing reliable monitoring of ERK5 activity or inhibition has held back our understanding of ERK5 biology, and ERK5i development and disease positioning. Techniques that have been used to monitor ERK5i-dependent inhibition of ERK5 in cells include:
1. KiNativ™. This technique measures binding of small molecules to the kinase active site. It uses biotin-tagged acyl-phosphates of ATP and ADP as probes, which acylate the conserved active site lysines in protein kinases (and other ATP-dependent enzymes). Mass spectrometry is then used to identify streptavidin captured peptides following tryptic digestion. Pre-treatment of cells with and without inhibitors, followed by cell lysis and capture of non-inhibitor-bound ERK5 via the ATP-site probe allows the quantification of target engagement [42,43]. This technique does not measure kinase or transcriptional activity.
2. ERK5 autophosphorylation assay. MEK5-catalysed phosphorylation of the activation-loop TEY motif activates the ERK5 kinase domain, which subsequently drives autophosphorylation at multiple sites within the C-terminus (Figures 1 and 2). This multi-site phosphorylation causes a pronounced reduction in mobility of ERK5 on SDS–PAGE gels [44,45]. Since this ERK5 ‘band shift’ reflects kinase domain-catalysed autophosphorylation, its loss in cells treated with an ERK5i reports inhibition of ERK5 (Figure 1A). This assay is straightforward as it simply requires a reliable ERK5 antibody. However, at least some phosphorylation sites in the C-terminus are targeted in trans by other protein kinases. Furthermore, this assay does not take into account the function of the C-terminus and does not measure the activity of the ERK5 TAD.
3. ERK5-driven AP-1 reporter assay. The AP-1 transcription factor consists of homo- and heterodimers of the bZIP transcription factors JUN (JUN, JUNB and JUND) and FOS (FOS, FOSB, FRA1 and FRA2) which bind to the core DNA sequence 5′-TGAG/CTCA-3′. A multimerised AP-1:Luc reporter can be stimulated by ERK5 when it is activated by co-transfected MEK5D (a constitutively active MEK5 mutant). This has been used to assess ERK5 activation and inhibition by ERK5i in cells [44]. However, the sheer complexity of AP-1 means it is very difficult to understand and interpret what is being measured in an ERK5-driven AP-1:Luc reporter assay. AP-1 activity is determined by dimer composition, the abundance of dimer partners (a function of de novo expression and proteasomal degradation) and their activity (controlled through phosphorylation, acetylation and protein:protein interactions). In terms of dimer composition, some FOS or JUN proteins can also heterodimerise with the ATF transcription factors (ATF2, ATF3 and B-ATF), MAFs (MAFB, MAFF, MAFG and MAFK) and the cAMP response element-binding proteins (CREBs). In terms of abundance and activity, the ERK1/2 [46,47,48] and JNK [49] pathways drive the expression and phosphorylation of multiple AP-1 proteins, increasing their abundance and activity. ERK1/2 signalling is a positive regulator of the FOS proteins, and ERK1/2 also phosphorylates JUNB and JUND; JNK phosphorylates and activates JUN and ATF2 [46,49].
The most likely mechanism by which ERK5 might regulate AP-1 is by promoting JUN expression. JUN expression is decreased in ERK5−/− MEFs and this is rescued by re-expression of ERK5 [50]. The JUN promoter is regulated by MEF2C, the best characterised substrate of ERK5; ERK5 phosphorylation of MEF2C enhances its transcriptional activity and promotes JUN expression [17,51]. ERK5 can also stimulate the phosphorylation and activation of CREB [52] and can phosphorylate ATF2 in vitro [20], however, ERK5 does not phosphorylate the Maf family member, MafA [53]. Taking these studies into account, ERK5-dependent regulation of AP-1:Luc is indirect, complex and mostly likely through an ERK5 → MEF2C → JUN pathway (Figure 1B), which is further complicated by other pathway inputs.
4. MEF2-dependent reporter assays. The best validated ERK5 interacting proteins and substrates are the MEF2A, C and D transcription factors [10,17,54]. Thus, the MEF2 proteins remain the most appropriate direct reporters of ERK5 activity and have been used in cellular reporter systems for monitoring MEK5 and/or ERK5 inhibition [55–57,58]. The simplest version consists of multimerised MEF2 binding sites and basic promoter elements to drive the expression of a reporter (typically firefly luciferase) in response to EGF (Figure 1B top) [57], making it a significant advance in ERK5 selectivity over AP-1. A potential disadvantage of this assay is that it may ‘report’ the activity of any one of the MEF2 proteins that can bind to the minimal binding site and it may also report on the activity of MEF2-interacting proteins that may be regulated by other pathways. This assay can be made more specific for ERK5 by driving ERK5 activation with co-transfected MEK5D.
A variation of this assay involves fusing the region of MEF2 housing ERK5 phosphorylation sites and the MEF2 TAD to the DNA-binding domain of the yeast transcription factor GAL4 (Figure 1C). This MEF2-GAL4 fusion can then be expressed together with a luciferase reporter controlled by multimerised GAL4 response elements (GREs), ERK5 and MEK5D to stimulate the pathway. This system has been shown to work for MEF2A, C and D [17] and although it has more components, it is a far more direct readout of ERK5 activation since ERK5 interacts directly with the MEF2-GAL4 reporter construct to drive luciferase expression. We have used this assay to measure the pharmacological inhibition of MEK5 and ERK5 [38,55,56].
Transcription-based readouts of ERK5 activity do not readily differentiate between the contribution of the kinase and TAD activities when a full length construct of (or endogenous) ERK5 is assessed. For example, we have found that a truncation mutant of ERK5 that lacked the TAD could drive MEF2D:GAL4 activity when co-transfected with MEK5D, but also that a kinase dead full-length form of ERK5 (that contains the TAD) could drive MEF2D:GAL4 luciferase activity following ERK5i treatment (see below) [55]. However, by using these different constructs, the contribution of the kinase domain and TAD can be delineated.
Development of MEK5 and ERK5 kinase inhibitors
By analogy with the RAF–MEK1/2–ERK1/2 pathway, one could conceive of targeting the ERK5 pathway at any step from MEKK2/3 to MEK5 or ERK5 itself (Figure 1). However, unlike RAF, which appears to be very selective for activation of MEK1/2, MEKK2/3 are able to activate other MKKs in addition to MEK5 [59] such that inhibition of MEKK2/3 may impact on JNK, p38 or ERK1/2 activity as well as ERK5. This may explain why efforts to selectively target ERK5 signalling have focused on MEK5 or ERK5. A summary of current MEK5 and ERK5 inhibitors is presented in Table 1 and Figure 3.
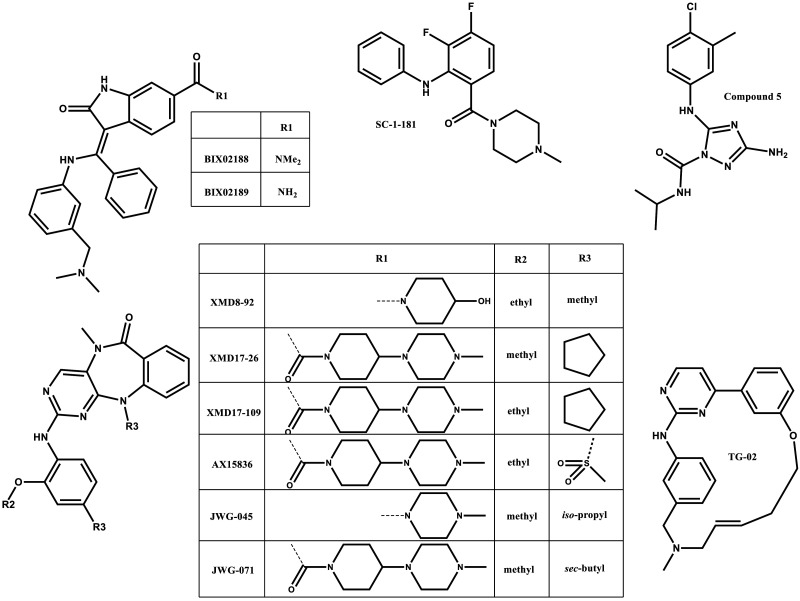
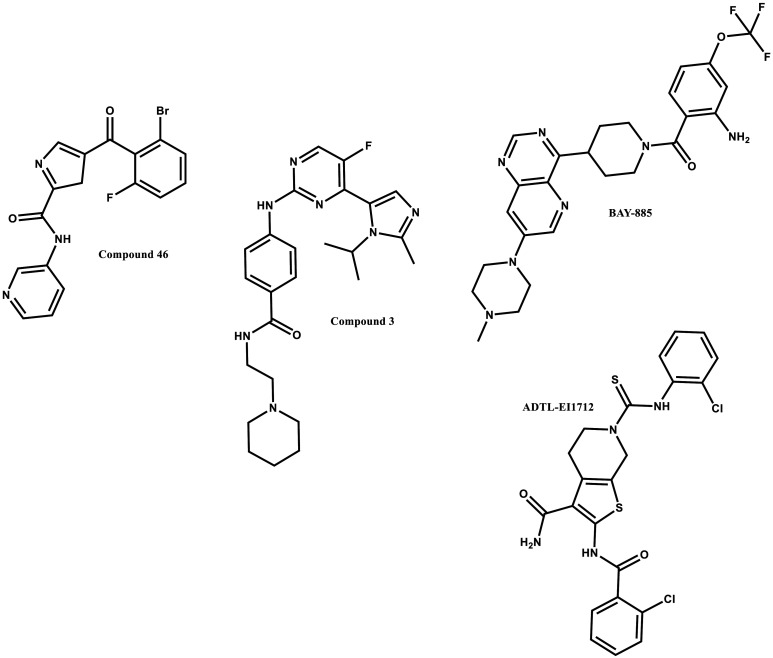
Figure 3. ERK5 inhibitors.
The first pathway inhibitors to be reported were the MEK5 inhibitors, BIX02188 and BIX02189. These inhibitors are relatively selective for MEK5, with the strongest off-target effect on Src [58]. They have been useful for dissecting the role of the MEK5–ERK5 pathway in cells [28,60]. More recently, SC-1-181 has been described; however, the selectivity data have yet to be reported [61,62].
The majority of effort has focused on the development of small molecule ERK5i, probably because ERK5 is the ‘effector kinase’ in the pathway. However, ERK5 is proving to be a challenging target. The first ERK5i to be described were XMD8-92 [63], cpd 25 (XMD17-26) and cpd 26 (also known as XMD17-109 and ERK5-IN-1) [44]. Although selective for ERK5 over other kinases, they had off-target effects on the bromo-domain containing protein, BRD4 [64] — an epigenetic reader involved in transcriptional regulation [65]. AX15836, an analogue of XMD8-92, was engineered to lack bromo-domain activity and is a potent and selective inhibitor of ERK5 kinase activity. However, cell-based experiments showed that AX15836 did not phenocopy genetic ERK5 knockdown. This brings into question results obtained using XMD8-92. Even though XMD8-92 phenocopies the biological effects of siRNA knockdown of ERK5, it is likely this arises through different mechanisms; XMD8-92, through binding to bromo-domain containing proteins, and siRNA to ERK5 by ablating the kinase domain and the C-terminus (including the TAD). This study also suggested that the ERK5 C-terminal domain is important for the biological function of ERK5 [64]. Heedful researchers have since used BRD4 inhibitors, such as JQ1, to delineate the role of BRD4 in their systems [25,45,55,56] and testing of ERK5 inhibitors against BRD4 is now an essential step in ERK5 drug discovery. Adding to the complexity, we have found that cpd 26 (XMD17-109) and AX15836, cause a conformational change in the kinase domain which leads to exposure of the C-terminal NLS and paradoxical activation of the ERK5 TAD [55] (see below and Figure 2).
Additional ERK5i in the public domain include JWG-045 and JWG-071, a further development of the XMD8-92 series. Both have selectivity over BRD4 but inhibit LRRK2 and are classed as dual ERK5/LRRK2 inhibitors. The kinase selectivity data for JWG-071 have been published, showing that it only inhibits three other kinases (LRRK2, PLK2, DCAMKL1 and 2) at low micro molar concentrations. In cell-based assays, an inhibitor that blocks LRRK2 activity (such as JWG-048 [45]), and not ERK5 activity, could be used to confirm ERK5 kinase involvement. A multi-site collaboration reported cpd 46; this inhibitor is distinct from the XMD8-92 series, has no activity towards BRD4 or LRRK2, is highly selective for ERK5 and is also suitable for use in animal studies [56]. Bayer have reported BAY-885, which like AX15836 is very selective for ERK5 and did not significantly inhibit other kinases or BRD4 [57]. However, like AX15386 it also paradoxically activates ERK5 transcriptional activity (Figure 4A and Table 1).
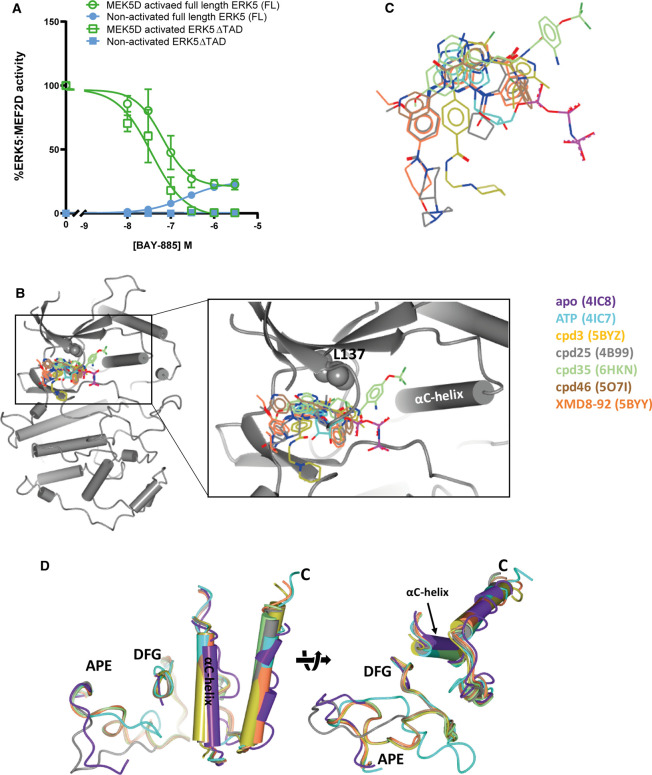
Figure 4. ERK5 small molecule inhibitor binding to the ERK5 kinase domain.
(A) BAY-885 induces transcriptional activity in the ERK5:MEF2D reporter system. HEK293 cells were transfected with GAL4-MEF2D, GAL4:LUC and CMV:Renilla, together with either wild-type HA-ERK5 (full length) or HA-ERK5ΔTAD and EGFP-MEK5D or EGFP (control) as indicated. Four hours post-transfection, cells were treated with either DMSO (control) or BAY-885. Twenty-four hours post transfection, cells were lysed and firefly luciferase activity was measured and normalised to Renilla. The results are presented as the mean of three independent experiments ± SEM. For full method see [55]. (B,C) Comparison of the binding mode of selected ERK5 inhibitors with ATP highlights the divergence in solvent channel groups and the extension of the BAY series (exemplified by cpd35) toward the back pocket. (B) Inhibitors shown within the context of ERK5 (cartoon representation); the gatekeeper residue (L137) is shown as spheres and the αC-helix is labelled. (C) ATP-binding site viewed from the N-terminal lobe of ERK5; protein atoms omitted for clarity. (D) Comparison of the conformations of the ERK5 activation loop (N-terminal DFG motif to C-terminal APE motif), αC- and C-terminal helices in the presence and absence of ATP and selected inhibitors demonstrates the similarity in positioning of the αC- and C-terminal helices in the inhibitor- and ATP-bound structures and their divergence from the apo structure, whilst the majority of inhibitor-bound activation loops adopt a similar conformation, they all differ from both the apo and ATP-bound states. In panels (B–D), inhibitor carbon atoms (B,C) or protein cartoon (D) are coloured as indicated in the key. Crystal structures of ERK5 kinase domain alone and in complex with ATP or selected ATP-competitive inhibitors were overlaid using the ssm algorithm in CCP4MG [94]. Co-ordinates were extracted from the PDBe [https://www.ebi.ac.uk/pdbe/] using the entry codes indicated in the key.
Recently a triple ERK1/2/5 inhibitor, ADTL-EI1712, has been reported [66]. This was developed to simultaneously block the ERK1/2 and ERK5 pathways in cancers where ERK5 activity can compensate for inhibition of the RAS–RAF–MEK1/2–ERK1/2 pathway [31,34]. Two other kinases, Kit and Axl are also inhibited to a similar extent as ERK1, ERK2 and ERK5, but BRD4 engagement was not assessed [66].
All the above inhibitors are type I inhibitors, and have either been shown experimentally or predicted to bind to the active form of ERK5 where the phenylalanine from the activation-loop DFG motif is ‘in’, the catalytic αC-helix is ‘in’, the inhibitors occupy part of the adenine binding pocket and form hydrogen bonds with the kinase hinge region (which connects the N and C-terminal lobes of the enzyme) (Figure 4B). For a review on the classification of kinase inhibitors see [67]. A type IV ERK5 inhibitor, cpd 5, has been identified by Chen et al. [68]. Type IV inhibitors do not bind the ATP or peptide substrate binding sites and are considered to act as allosteric inhibitors. Although cpd 5 does not bind directly to the ATP-binding site, it displaces the kinase P-loop into the ATP-binding site and thus is ATP competitive. This inhibitor has off-target effects against five other kinases at concentrations required to inhibit ERK5 in cells [68] but it has not been tested against BRD4.
Some ERK5i have anti-tumour activity (Table 1). As XMD8-92, ADTL-EI1712 and TG-02 all have off-target effects (either on BRD4 or other kinases), it is impossible to conclude whether or not this is a direct consequence of ERK5 kinase inhibition (even when it phenocopies knockout techniques, for the reasons discussed above). Cpd 46 is a selective ERK5i, with mild paradoxical activation effects, that prevents tumour growth by inhibiting angiogenesis [56]. These findings show that it is important to understand how targeting ERK5 works — inhibition of tumour cell proliferation or survival, inhibition of tumour angiogenesis, or in appropriate models, reducing the tumour promoting role of the immune system.
Paradoxical signalling induced by ERK5 kinase inhibitors differs from other kinases — classification of kinase inhibitor-induced paradoxical activation
The unintended activation of kinase signalling through inhibitor binding to the kinase, termed paradoxical activation, has emerged as a significant challenge to kinase inhibitor development. The first kinase that was reported to be paradoxically activated by selective small molecule inhibitors was PKB. Inhibitor binding promotes PKB membrane localisation, regulatory site phosphorylation (T308 and S473) by PDK1 and mTORC2, and acquisition of a phosphatase-resistant conformation; consequently, when the inhibitor is removed the ‘primed’ kinase is fully active [69,6]. Other kinases that undergo ‘priming paradoxical activation’ (Figure 2) include PKC [70], PKD [71], AMPK [72] and JAK2 [73,74,75]. This mechanism can have severe physiological consequences; for example, a life-threatening cytokine-rebound syndrome occurs when the JAK2 inhibitor, ruxolitinib, is withdrawn too quickly and this is due to priming paradoxical activation of JAK2 [73,74,75].
The second and best-known example of paradoxical kinase activation is seen with the first generation BRAFV600E/K inhibitors vemurafenib and dabrafenib. These inhibitors were developed to selectively inhibit BRAFV600E/K and although they are effective in treating BRAFV600E/K-driven melanoma they cause adventitious tumour progression in non-melanoma tissue. This is because inhibitor binding to wild-type RAF isoforms induces RAS-GTP-dependent CRAF homodimers or BRAF-CRAF heterodimers in which the drug-bound protomer trans-activates the drug-free promoter leading to MEK1/2–ERK1/2 activation [76,77] (Figure 2). Another example of this ‘paradoxical activation by trans-activation’ is bosutinib binding to the pseudokinase HER3 which induces an EGFR-dependent proliferative signal [78,79] (Figure 2). These examples highlight the importance of anticipating whether a kinase is susceptible to inhibitor-induced paradoxical activation.
We have recently shown that the binding of cpd 25 (XMD17-26), 26 (XMD17-109) or AX15836 to the ERK5 kinase domain promotes NLS and TAD exposure which in turn promotes nuclear localisation and paradoxical activation of ERK5 transcriptional activity [55]. The ERK5 kinase domain and the NLS and TAD normally inhibit each other through an intramolecular interaction that is relieved by ERK5i binding (Figure 2). Generation of inhibitor-resistant ERK5 mutants confirmed that this was an ‘on target’ effect of ERK5i binding to the kinase domain. This represents a new mechanism redolent of steroid hormone receptor activation, where ligand binding induces nuclear translocation and transcriptional activity. Thus, we have termed this ‘auxiliary domain paradoxical activation’ (Figure 2).
Do all ERK5i induce paradoxical activation? We have subsequently tested XMD8-92, cpd 46 and BAY-885 in the MEF2D:GAL4 assay (Figure 1). All three paradoxically activate ERK5 in the assay, albeit to differing degrees (Figure 4A, Table 1). We have compared the binding of XMD8-92, cpd 25 (XMD17-26), cpd 35 (an analogue of BAY-885) and cpd 46 to ERK5 to ATP (Figure 4B,C). These ERK5i all occupy a similar space within the adenosine-binding region of the ATP-binding site (Figure 4B,C) and engage in hydrogen bonds with the hinge region, whilst they differ somewhat in the positioning of their solvent-exposed moieties. In addition, cpd 35 extends to occupy an area termed the ‘back pocket’. This region, which lies between the ‘gatekeeper’ residue (L137 in ERK5) and the αC helix, is not occupied by ATP. Comparison of the conformation of the ERK5 αC helix and activation loop in the presence and absence of inhibitors or ATP (Figure 4D) reveals that the inhibitors bind to a more ‘active-like’ conformation of ERK5. Furthermore, our structural simulations with AX15836 showed a reduction in flexibility of the activation loop [55]. The exact mechanism that causes NLS exposure and TAD paradoxical activation when ERK5i bind to the ERK5 kinase domain remains elusive. It could be conformational changes elicited by inhibitor binding, solvent-exposed moieties preventing auto-inhibition, effects on Hsp90 binding, post-translational modifications or a combination of events. We ruled out changes in phosphorylation of known key sites on ERK5 [55], but ERK5 SUMOylation remains to be tested.
Promoting nuclear localisation of ERK5 is an undesirable consequence of ERK5i. Nuclear ERK5 is linked to a high Gleason grade, bone metastasis and ultimately a poor prognosis in prostate cancer [35,80–82]. In HCC there is an increase in ERK5 nuclear localisation but no increase in ERK5 kinase activity [83], and overexpression of the oncogene, Cdc37, promotes kinase-inactive nuclear localisation of ERK5 while increasing cell proliferation [22]. Thus, ERK5i that promote nuclear localisation in tumours with predominately cytosolic ERK5, may exacerbate tumour progression. If tumours have kinase active nuclear ERK5, ERK5i may have some effect if the kinase activity is driving tumorigenesis, but not if ERK5 transcriptional activity is required. Some tumours already have nuclear kinase-inactive ERK5 [83], here ERK5i may increase its transcriptional activity. This highlights the importance of understanding the mechanism of action of ERK5i and determining whether ERK5 kinase and/or transcriptional activity is required for the cellular role of ERK5.
Conclusions
These findings show that it is essential that all ERK5i are tested not just for off-target activity against kinases and bromo-domain containing proteins, but also for promotion of nuclear localisation and paradoxical transcriptional activation using appropriately sensitive assays. For ERK5 this can be assessed using the ERK5:MEF2D reporter assays, such as the GAL4-MEF2D driven GRE reporter (Figures 1B,C). It should be noted that when using exogenously expressed ERK5, a large tag on the N-terminus, such as GST or GFP will hide this effect and should be avoided (PAL, unpublished results). It also remains to be seen whether other kinases with auxiliary functional domains are activated by small molecules in a similar way to ERK5. To delineate the role of the ERK5 kinase domain in cells, AX15836, cpd 46 and BAY-885 are ERK5 kinase selective inhibitors, but all cause TAD paradoxical activation (to varying degrees) so care must be taken when interpreting results. This highlights the need to find a selective paradox-breaking ERK5i or to use an alternative approach such as a PROTAC [84] to completely remove ERK5 from cells. Critically, these results also raise the question of the relative importance of the ERK5 kinase domain and NLS/TAD functions in normal ERK5 biology and in diseases where ERK5 is implicated.
Dedication
We dedicate this article to the memory of Michael Wakelam a dear friend, mentor, colleague, a world leader in lipid signalling and lipidomics, and Director of the Babraham Institute until his death in the 2020 Covid-19 pandemic.
Perspectives
Importance of the field: Protein kinase inhibitors can induce unintended paradoxical signalling. Paradoxical kinase activation can exacerbate disease, such as stimulating tumour growth. ERK5 nuclear localisation and transcriptional activity is paradoxically stimulated by ERK5i.
Current thinking: Characterising the mode of action of kinase inhibitors on their targets using appropriately sensitive assays is critical for all drug discovery programmes. Determining whether ERK5i promote transcriptional activation of ERK5 is essential for any new potential ERK5 therapeutic.
Future directions: Activation of ERK5 transcriptional activity by ERK5i raises significant questions about the role of ERK5 kinase and transcriptional activities in ERK5 biology, including disease biology and therapeutics.
Acknowledgements
We would like to thank the members of the Cook Laboratory for their support and encouragement.
Abbreviations
- ABL
Abelson murine leukaemia homolog
- AMPK
5′ AMP-activated protein kinase
- AP-1
activator protein-1
- ATF
activating transcription family
- AXL
AXL, coming from the Greek word ‘anexelekto’, means uncontrolled
- BRD4
bromo-domain-containing protein 4, a transcriptional and epigenetic regulator
- bZIP
basic leucine zipper domain
- CDC37
cell division cycle 37
- CDK1/2/3/4/5/7/9
cyclin dependent kinase1/2/3/4/5/7/9
- CREB
cAMP response element-binding protein
- CSF1R(FMS)
colony-stimulating factor 1 receptor (Feline McDonough Sarcoma)
- DCAMKL1/2
doublecortin and CaMK (Ca2+/calmodulin-dependent protein kinase)-like 1/2/3
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ERK1/2
extracellular signal-regulated protein kinase 1/2
- ERK5
extracellular signal-regulated protein kinase 5
- ERK5i
small molecule ERK5 inhibitor
- FGFR1
fibroblast growth factor receptor 1
- FLT3/4
FMS (Feline McDonough Sarcoma)-like tyrosine kinase 3/4
- FOS
cellular homolog of the viral oncoprotein v-fos
- FRA
Fos-related antigen
- Fyn
src family kinase
- GAL4
yeast transcription factor
- GSK3α
glycogen synthase kinase 3α
- HER3
human epidermal growth factor receptor 3
- Hsp90
heat-shock protein 90
- JAK1/2
Janus kinase 1/2
- JNK
c-Jun N-terminal kinases
- JUN
cellular homolog of the viral oncoprotein v-jun
- LCK
lymphocyte-specific protein tyrosine kinase
- LRRK2
leucine-rich repeat serine/threonine-protein kinase 2
- MAF
cellular homolog of the viral oncoprotein v-maf (musculoaponeurotic fibrosarcoma)
- MAPK
mitogen-activated protein kinase
- MAP3K15
mitogen-activated protein kinase kinase kinase 15
- MEF2 A, C and D
myocyte enhancer factor-2 A, C and D
- MEKK2/3
MAPK/extracellular signal-regulated kinase kinase 2/3
- MEK5
MAPK kinase 5
- MEK5D
constitutively active form of MEK5 where the regulatory activation-loop phosphorylation sites are mutated to aspartic acid to mimic phosphorylation
- MKK
MAP kinase kinase
- MKKK
MAP kinase kinase kinase
- MST2
mammalian serine/threonine 2 kinase
- mTORC2
mammalian target of rapamycin complex 2
- NLS
nuclear localisation signal
- P38δ
class of mitogen-activated protein kinases that are responsive to stress stimuli
- PDK1
phosphoinositide-dependent kinase-1
- PH
pleckstrin homology
- PKC
protein kinase C
- PKD
protein kinase D
- PROTAC
proteolysis targeting chimeras
- RAF
rapidly accelerated in fibrosarcoma kinase
- RAS
Rat sarcoma small GTPase
- RBD
RAS binding domain
- RSK2/4
ribosomal protein S6 kinase 2/4
- SLK
Ste20-like serine/threonine protein kinase
- Src
short for sarcoma kinase
- STK
spleen tyrosine kinase
- TAD
trans-activation domain
- TYK2
non-receptor tyrosine-protein kinase 2
- TYRO3
tyrosine-protein kinase receptor
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Open Access
Open access for this article was enabled by the participation of University of York in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
Funding
This work was supported by a BBSRC project grant BB/N015886/1 (P.A.L. and S.J.C), an MRC-DPFS award MR/K007580/1 (S.J.C., J.A.T and P.A.L.) and Institute Strategic Programme Grants BB/J004456/1 and BB/P013384/1 from BBSRC (S.J.C.).
Author Contributions
P.A.L. conducted the BAY-885 ERK5 paradoxical activation assay. J.A.T. performed the structural analysis. S.J.C., J.A.T. and P.A.L. wrote and revised the manuscript for critically important intellectual content.
References
- 1.Ferguson F.M. and Gray N.S. (2018) Kinase inhibitors: the road ahead. Nat. Rev. Drug Discov. 17, 353–377 10.1038/nrd.2018.21 [DOI] [PubMed] [Google Scholar]
- 2.Roskoski R J. (2019) Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 144, 19–50 10.1016/j.phrs.2019.03.006 [DOI] [PubMed] [Google Scholar]
- 3.Carles F., Bourg S., Meyer C. and Bonnet P. (2018) PKIDB: a curated, annotated and updated database of protein kinase inhibitors in clinical trials. Molecules 23, 908 10.3390/molecules23040908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Little A.S., Balmanno K., Sale M.J., Newman S., Dry J.R., Hampson M. et al. (2011) Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci. Signal. 4, ra17 10.1126/scisignal.2001752 [DOI] [PubMed] [Google Scholar]
- 5.O'Reilly K.E., Rojo F., She Q.B., Solit D., Mills G.B., Smith D. et al. (2006) mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 66, 1500–1508 10.1158/0008-5472.CAN-05-2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okuzumi T., Fiedler D., Zhang C., Gray D.C., Aizenstein B., Hoffman R. et al. (2009) Inhibitor hijacking of Akt activation. Nat. Chem. Biol. 5, 484–493 10.1038/nchembio.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.D., Ulevitch R.J. and Han J. (1995) Primary structure of BMK1: a new mammalian map kinase. Biochem. Biophys. Res. Commun. 213, 715–724 10.1006/bbrc.1995.2189 [DOI] [PubMed] [Google Scholar]
- 8.Zhou G., Bao Z.Q. and Dixon J.E. (1995) Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270, 12665–9 10.1074/jbc.270.21.12665 [DOI] [PubMed] [Google Scholar]
- 9.Kasler H.G., Victoria J., Duramad O. and Winoto A. (2000) ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell Biol. 20, 8382–8389 10.1128/MCB.20.22.8382-8389.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato Y., Kravchenko V.V., Tapping R.I., Han J., Ulevitch R.J. and Lee J.D. (1997) BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 16, 7054–7066 10.1093/emboj/16.23.7054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilhelmsen K., Mesa K.R., Lucero J., Xu F. and Hellman J. (2012) ERK5 protein promotes, whereas MEK1 protein differentially regulates, theToll-like receptor 2 protein-dependent activation of human endothelial cells and monocytes. J. Biol. Chem. 287, 26478–26494 10.1074/jbc.M112.359489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe J., Kusuhara M., Ulevitch R.J., Berk B.C. and Lee J.D. (1996) Big mitogen-activated protein kinase 1 (BMK1) is a redox-sensitive kinase. J. Biol. Chem. 271, 16586–16590 10.1074/jbc.271.28.16586 [DOI] [PubMed] [Google Scholar]
- 13.Mody N., Campbell D.G., Morrice N., Peggie M. and Cohen P. (2003) An analysis of the phosphorylation and activation of extracellular-signal-regulated protein kinase 5 (ERK5) by mitogen-activated protein kinase kinase 5 (MKK5) in vitro. Biochem. J. 372(Pt 2), 567–575 10.1042/bj20030193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buschbeck M. and Ullrich A. (2005) The unique C-terminal tail of the mitogen-activated protein kinase ERK5 regulates its activation and nuclear shuttling. J. Biol. Chem. 280, 2659–2667 10.1074/jbc.M412599200 [DOI] [PubMed] [Google Scholar]
- 15.Morimoto H., Kondoh K., Nishimoto S., Terasawa K. and Nishida E. (2007) Activation of a C-terminal transcriptional activation domain of ERK5 by autophosphorylation. J. Biol. Chem. 282, 35449–35456 10.1074/jbc.M704079200 [DOI] [PubMed] [Google Scholar]
- 16.Kato Y., Chao T.H., Hayashi M., Tapping R.I. and Lee J.D. (2000) Role of BMK1 in regulation of growth factor-induced cellular responses. Immunol. Res. 21, 233–237 10.1385/IR:21:2-3:233 [DOI] [PubMed] [Google Scholar]
- 17.Kato Y., Zhao M., Morikawa A., Sugiyama T., Chakravortty D., Koide N. et al. (2000) Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275, 18534–18540 10.1074/jbc.M001573200 [DOI] [PubMed] [Google Scholar]
- 18.Honda T., Obara Y., Yamauchi A., Couvillon A.D., Mason J.J., Ishii K. et al. (2015) Phosphorylation of ERK5 on Thr732 is associated with ERK5 nuclear localization and ERK5-dependent transcription. PLoS One 10, e0117914 10.1371/journal.pone.0117914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz-Rodriguez E. and Pandiella A. (2010) Multisite phosphorylation of Erk5 in mitosis. J. Cell Sci. 123(Pt, 3146–3156 10.1242/jcs.070516 [DOI] [PubMed] [Google Scholar]
- 20.Inesta-Vaquera F.A., Campbell D.G., Tournier C., Gomez N., Lizcano J.M. and Cuenda A. (2010) Alternative ERK5 regulation by phosphorylation during the cell cycle. Cell Signal. 22, 1829–1837 10.1016/j.cellsig.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 21.Zhuang K., Zhang J., Xiong M., Wang X., Luo X., Han L. et al. (2016) CDK5 functions as a tumor promoter in human colorectal cancer via modulating the ERK5-AP-1 axis. Cell Death Dis. 7, e2415 10.1038/cddis.2016.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erazo T., Moreno A., Ruiz-Babot G., Rodriguez-Asiain A., Morrice N.A., Espadamala J. et al. (2013) Canonical and kinase activity-independent mechanisms for extracellular signal-regulated kinase 5 (ERK5) nuclear translocation require dissociation of Hsp90 from the ERK5-Cdc37 complex. Mol. Cell. Biol. 33, 1671–1686 10.1128/MCB.01246-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erazo T., Espinosa-Gil S., Dieguez-Martinez N., Gomez N. and Lizcano J.M. (2020) SUMOylation is required for ERK5 nuclear translocation and ERK5-mediated cancer cell proliferation. Int. J. Mol. Sci. 21, 2203 10.3390/ijms21062203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tubita A., Lombardi Z., Tusa I., Dello Sbarba P. and Rovida E. (2020) Beyond kinase activity: ERK5 nucleo-cytoplasmic shuttling as a novel target for anticancer therapy. Int. J. Mol. Sci. 21, 938 10.3390/ijms21030938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams C.A., Fernandez-Alonso R., Wang J., Toth R., Gray N.S. and Findlay G.M. (2016) Erk5 is a key regulator of naive-primed transition and embryonic stem cell identity. Cell Rep. 16, 1820–1828 10.1016/j.celrep.2016.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regan C.P., Li W., Boucher D.M., Spatz S., Su M.S. and Kuida K. (2002) Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc. Natl. Acad. Sci. U.S.A. 99, 9248–9253 10.1073/pnas.142293999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Merritt A.J., Seyfried J., Guo C., Papadakis E.S., Finegan K.G. et al. (2005) Targeted deletion of mek5 causes early embryonic death and defects in the extracellular signal-regulated kinase 5/myocyte enhancer factor 2 cell survival pathway. Mol. Cell. Biol. 25, 336–345 10.1128/MCB.25.1.336-345.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilhelmsen K., Xu F., Farrar K., Tran A., Khakpour S., Sundar S. et al. (2015) Extracellular signal-regulated kinase 5 promotes acute cellular and systemic inflammation. Sci. Signal. 8, ra86 10.1126/scisignal.aaa3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giurisato E., Xu Q., Lonardi S., Telfer B., Russo I., Pearson A. et al. (2018) Myeloid ERK5 deficiency suppresses tumor growth by blocking protumor macrophage polarization via STAT3 inhibition. Proc. Natl. Acad. Sci. U.S.A. 115, E2801–E2E10 10.1073/pnas.1707929115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finegan K.G., Perez-Madrigal D., Hitchin J.R., Davies C.C., Jordan A.M. and Tournier C. (2015) ERK5 is a critical mediator of inflammation-driven cancer. Cancer Res. 75, 742–753 10.1158/0008-5472.CAN-13-3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song C., Wang L., Xu Q., Wang K., Xie D., Yu Z. et al. (2017) Targeting BMK1 impairs the drug resistance to combined inhibition of BRAF and MEK1/2 in melanoma. Sci. Rep. 7, 46244 10.1038/srep46244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusa I., Cheloni G., Poteti M., Gozzini A., DeSouza N.H., Shan Y. et al. (2018) Targeting the extracellular signal-regulated kinase 5 pathway to suppress human chronic myeloid leukemia stem cells. Stem Cell Rep. 11, 929–943 10.1016/j.stemcr.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tusa I., Gagliardi S., Tubita A., Pandolfi S., Urso C., Borgognoni L. et al. (2018) ERK5 is activated by oncogenic BRAF and promotes melanoma growth. Oncogene 37, 2601–2614 10.1038/s41388-018-0164-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaseva A.V., Blake D.R., Gilbert T.S.K., Ng S., Hostetter G., Azam S.H. et al. (2018) KRAS suppression-induced degradation of MYC is antagonized by a MEK5-ERK5 compensatory mechanism. Cancer Cell 34, 807–822.e7 10.1016/j.ccell.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clape C., Fritz V., Henriquet C., Apparailly F., Fernandez P.L., Iborra F. et al. (2009) miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One 4, e7542 10.1371/journal.pone.0007542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortiz-Ruiz M.J., Alvarez-Fernandez S., Parrott T., Zaknoen S., Burrows F.J., Ocana A. et al. (2014) Therapeutic potential of ERK5 targeting in triple negative breast cancer. Oncotarget 5, 11308–11318 10.18632/oncotarget.2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noguchi S., Mori T., Hoshino Y., Maruo K., Yamada N., Kitade Y. et al. (2011) MicroRNA-143 functions as a tumor suppressor in human bladder cancer T24 cells. Cancer Lett. 307, 211–220 10.1016/j.canlet.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 38.Lochhead P.A., Clark J., Wang L.Z., Gilmour L., Squires M., Gilley R. et al. (2016) Tumor cells with KRAS or BRAF mutations or ERK5/MAPK7 amplification are not addicted to ERK5 activity for cell proliferation. Cell Cycle 15, 506–518 10.1080/15384101.2015.1120915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zen K., Yasui K., Nakajima T., Zen Y., Gen Y., Mitsuyoshi H. et al. (2009) ERK5 is a target for gene amplification at 17p11 and promotes cell growth in hepatocellular carcinoma by regulating mitotic entry. Genes Chromosomes Cancer 48, 109–120 10.1002/gcc.20624 [DOI] [PubMed] [Google Scholar]
- 40.Pereira D.M., Gomes S.E., Borralho P.M. and Rodrigues C.M.P. (2019) MEK5/ERK5 activation regulates colon cancer stem-like cell properties. Cell Death Discov. 5, 68 10.1038/s41420-019-0150-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Kriegsheim A., Baiocchi D., Birtwistle M., Sumpton D., Bienvenut W., Morrice N. et al. (2009) Cell fate decisions are specified by the dynamic ERK interactome. Nat. Cell Biol. 11, 1458–1464 10.1038/ncb1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patricelli M.P., Szardenings A.K., Liyanage M., Nomanbhoy T.K., Wu M., Weissig H. et al. (2007) Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 46, 350–358 10.1021/bi062142x [DOI] [PubMed] [Google Scholar]
- 43.Patricelli M.P., Nomanbhoy T.K., Wu J., Brown H., Zhou D., Zhang J. et al. (2011) In situ kinase profiling reveals functionally relevant properties of native kinases. Chem. Biol. 18, 699–710 10.1016/j.chembiol.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elkins J.M., Wang J., Deng X., Pattison M.J., Arthur J.S., Erazo T. et al. (2013) X-ray crystal structure of ERK5 (MAPK7) in complex with a specific inhibitor. J. Med. Chem. 56, 4413–4421 10.1021/jm4000837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J., Erazo T., Ferguson F.M., Buckley D.L., Gomez N., Munoz-Guardiola P. et al. (2018) Structural and atropisomeric factors governing the selectivity of pyrimido-benzodiazipinones as inhibitors of kinases and bromodomains. ACS Chem. Biol. 13, 2438–2448 10.1021/acschembio.7b00638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karin M., Liu Z. and Zandi E. (1997) AP-1 function and regulation. Curr. Opin. Cell Biol. 9, 240–246 10.1016/S0955-0674(97)80068-3 [DOI] [PubMed] [Google Scholar]
- 47.Madrigal P. and Alasoo K. (2018) AP-1 takes centre stage in enhancer chromatin dynamics. Trends Cell Biol. 28, 509–511 10.1016/j.tcb.2018.04.009 [DOI] [PubMed] [Google Scholar]
- 48.Eferl R. and Wagner E.F. (2003) AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3, 859–868 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- 49.Karin M. (1996) The regulation of AP-1 activity by mitogen-activated protein kinases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 351, 127–134 10.1098/rstb.1996.0008 [DOI] [PubMed] [Google Scholar]
- 50.Gilley R., March H.N. and Cook S.J. (2009) ERK1/2, but not ERK5, is necessary and sufficient for phosphorylation and activation of c-Fos. Cell Signal. 21, 969–977 10.1016/j.cellsig.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 51.Kayahara M., Wang X. and Tournier C. (2005) Selective regulation of c-jun gene expression by mitogen-activated protein kinases via the 12-o-tetradecanoylphorbol-13-acetate- responsive element and myocyte enhancer factor 2 binding sites. Mol. Cell. Biol. 25, 3784–3792 10.1128/MCB.25.9.3784-3792.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finegan K.G., Wang X., Lee E.J., Robinson A.C. and Tournier C. (2009) Regulation of neuronal survival by the extracellular signal-regulated protein kinase 5. Cell Death Differ. 16, 674–683 10.1038/cdd.2008.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benkhelifa S., Provot S., Nabais E., Eychene A., Calothy G. and Felder-Schmittbuhl M.P. (2001) Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol. Cell. Biol. 21, 4441–4452 10.1128/MCB.21.14.4441-4452.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang C.C., Ornatsky O.I., McDermott J.C., Cruz T.F. and Prody C.A. (1998) Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res. 26, 4771–4777 10.1093/nar/26.20.4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lochhead P.A., Tucker J.A., Tatum N.J., Wang J., Oxley D., Kidger A.M. et al. (2020) Paradoxical activation of the protein kinase-transcription factor ERK5 by ERK5 kinase inhibitors. Nat. Commun. 11, 1383 10.1038/s41467-020-15031-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Myers S.M., Miller D.C., Molyneux L., Arasta M., Bawn R.H., Blackburn T.J. et al. (2019) Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38alpha and BRD4. Eur. J. Med. Chem. 178, 530–543 10.1016/j.ejmech.2019.05.057 [DOI] [PubMed] [Google Scholar]
- 57.Nguyen D., Lemos C., Wortmann L., Eis K., Holton S.J., Boemer U. et al. (2019) Discovery and characterization of the potent and highly selective (Piperidin-4-yl)pyrido[3,2- d]pyrimidine based in vitro probe BAY-885 for the kinase ERK5. J. Med. Chem. 62, 928–940 10.1021/acs.jmedchem.8b01606 [DOI] [PubMed] [Google Scholar]
- 58.Tatake R.J., O'Neill M.M., Kennedy C.A., Wayne A.L., Jakes S., Wu D. et al. (2008) Identification of pharmacological inhibitors of the MEK5/ERK5 pathway. Biochem. Biophys. Res. Commun. 377, 120–125 10.1016/j.bbrc.2008.09.087 [DOI] [PubMed] [Google Scholar]
- 59.Todd D.E., Densham R.M., Molton S.A., Balmanno K., Newson C., Weston C.R. et al. (2004) ERK1/2 and p38 cooperate to induce a p21CIP1-dependent G1 cell cycle arrest. Oncogene 23, 3284–3295 10.1038/sj.onc.1207467 [DOI] [PubMed] [Google Scholar]
- 60.Amano S., Chang Y.T. and Fukui Y. (2015) ERK5 activation is essential for osteoclast differentiation. PLoS One 10, e0125054 10.1371/journal.pone.0125054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chakrabarty S., Monlish D.A., Gupta M., Wright T.D., Hoang V.T., Fedak M. et al. (2018) Structure activity relationships of anthranilic acid-based compounds on cellular and in vivo mitogen activated protein kinase-5 signaling pathways. Bioorg. Med. Chem. Lett. 28, 2294–2301 10.1016/j.bmcl.2018.05.029 [DOI] [PubMed] [Google Scholar]
- 62.Wright T.D., Raybuck C., Bhatt A., Monlish D., Chakrabarty S., Wendekier K. et al. (2020) Pharmacological inhibition of the MEK5/ERK5 and PI3K/Akt signaling pathways synergistically reduces viability in triple-negative breast cancer. J. Cell Biochem. 121, 1156–1168 10.1002/jcb.29350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng X., Elkins J.M., Zhang J., Yang Q., Erazo T., Gomez N. et al. (2013) Structural determinants for ERK5 (MAPK7) and leucine rich repeat kinase 2 activities of benzo[e]pyrimido-[5,4-b]diazepine-6(11H)-ones. Eur. J. Med. Chem. 70, 758–767 10.1016/j.ejmech.2013.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin E.C., Amantea C.M., Nomanbhoy T.K., Weissig H., Ishiyama J., Hu Y. et al. (2016) ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proc. Natl. Acad. Sci. U.S.A. 113, 11865–11870 10.1073/pnas.1609019113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Donati B., Lorenzini E. and Ciarrocchi A. (2018) BRD4 and cancer: going beyond transcriptional regulation. Mol. Cancer 17, 164 10.1186/s12943-018-0915-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang G., Zhao Y., Liu Y., Sun D., Zhen Y., Liu J. et al. (2020) Discovery of a novel dual-target inhibitor of ERK1 and ERK5 that induces regulated cell death to overcome compensatory mechanism in specific tumor types. J. Med. Chem. 63, 3976–3995 10.1021/acs.jmedchem.9b01896 [DOI] [PubMed] [Google Scholar]
- 67.Roskoski R J. (2016) Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol. Res. 103, 26–48 10.1016/j.phrs.2015.10.021 [DOI] [PubMed] [Google Scholar]
- 68.Chen H., Tucker J., Wang X., Gavine P.R., Phillips C., Augustin M.A. et al. (2016) Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site. Acta Crystallogr. D Struct. Biol. 72(Pt, 682–693 10.1107/S2059798316004502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan T.O., Zhang J., Rodeck U., Pascal J.M., Armen R.S., Spring M. et al. (2011) Resistance of Akt kinases to dephosphorylation through ATP-dependent conformational plasticity. Proc. Natl. Acad. Sci. U.S.A. 108, E1120–7 10.1073/pnas.1109879108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cameron A.J., Escribano C., Saurin A.T., Kostelecky B. and Parker P.J. (2009) PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat. Struct. Mol. Biol. 16, 624–630 10.1038/nsmb.1606 [DOI] [PubMed] [Google Scholar]
- 71.Kunkel M.T. and Newton A.C. (2015) Protein kinase d inhibitors uncouple phosphorylation from activity by promoting agonist-dependent activation loop phosphorylation. Chem. Biol. 22, 98–106 10.1016/j.chembiol.2014.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ross F.A., Hawley S.A., Auciello F.R., Gowans G.J., Atrih A., Lamont D.J. et al. (2017) Mechanisms of paradoxical activation of AMPK by the kinase inhibitors SU6656 and Sorafenib. Cell Chem. Biol. 24, 813–24.e4 10.1016/j.chembiol.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koppikar P., Bhagwat N., Kilpivaara O., Manshouri T., Adli M., Hricik T. et al. (2012) Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 489, 155–159 10.1038/nature11303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tvorogov D., Thomas D., Liau N.P.D., Dottore M., Barry E.F., Lathi M. et al. (2018) Accumulation of JAK activation loop phosphorylation is linked to type I JAK inhibitor withdrawal syndrome in myelofibrosis. Sci. Adv. 4, eaat3834 10.1126/sciadv.aat3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andraos R., Qian Z., Bonenfant D., Rubert J., Vangrevelinghe E., Scheufler C. et al. (2012) Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent. Cancer Discov. 2, 512–523 10.1158/2159-8290.CD-11-0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hatzivassiliou G., Song K., Yen I., Brandhuber B.J., Anderson D.J., Alvarado R. et al. (2010) RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 464, 431–435 10.1038/nature08833 [DOI] [PubMed] [Google Scholar]
- 77.Poulikakos P.I., Zhang C., Bollag G., Shokat K.M. and Rosen N. (2010) RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 464, 427–430 10.1038/nature08902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Claus J., Patel G., Autore F., Colomba A., Weitsman G., Soliman T.N. et al. (2018) Inhibitor-induced HER2-HER3 heterodimerisation promotes proliferation through a novel dimer interface. eLife 7, e32271 10.7554/eLife.32271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levinson N.M. and Boxer S.G. (2014) A conserved water-mediated hydrogen bond network defines bosutinib's kinase selectivity. Nat. Chem. Biol. 10, 127–132 10.1038/nchembio.1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahmad I., Singh L.B., Yang Z.H., Kalna G., Fleming J., Fisher G. et al. (2013) Mir143 expression inversely correlates with nuclear ERK5 immunoreactivity in clinical prostate cancer. Br. J. Cancer 108, 149–154 10.1038/bjc.2012.510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCracken S.R., Ramsay A., Heer R., Mathers M.E., Jenkins B.L., Edwards J. et al. (2008) Aberrant expression of extracellular signal-regulated kinase 5 in human prostate cancer. Oncogene 27, 2978–2988 10.1038/sj.onc.1210963 [DOI] [PubMed] [Google Scholar]
- 82.Ramsay A.K., McCracken S.R., Soofi M., Fleming J., Yu A.X., Ahmad I. et al. (2011) ERK5 signalling in prostate cancer promotes an invasive phenotype. Br. J. Cancer 104, 664–672 10.1038/sj.bjc.6606062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rovida E., Di Maira G., Tusa I., Cannito S., Paternostro C., Navari N. et al. (2015) The mitogen-activated protein kinase ERK5 regulates the development and growth of hepatocellular carcinoma. Gut 64, 1454–1465 10.1136/gutjnl-2014-306761 [DOI] [PubMed] [Google Scholar]
- 84.Paiva S.L. and Crews C.M. (2019) Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol. 50, 111–119 10.1016/j.cbpa.2019.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park S.J., Choi Y.S., Lee S., Lee Y.J., Hong S., Han S. et al. (2016) BIX02189 inhibits TGF-beta1-induced lung cancer cell metastasis by directly targeting TGF-beta type I receptor. Cancer Lett. 381, 314–322 10.1016/j.canlet.2016.08.010 [DOI] [PubMed] [Google Scholar]
- 86.Yang Q., Deng X., Lu B., Cameron M., Fearns C., Patricelli M.P. et al. (2010) Pharmacological inhibition of BMK1 suppresses tumor growth through promyelocytic leukemia protein. Cancer Cell 18, 258–267 10.1016/j.ccr.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Al-Ejeh F., Miranda M., Shi W., Simpson P.T., Song S., Vargas A.C. et al. (2014) Kinome profiling reveals breast cancer heterogeneity and identifies targeted therapeutic opportunities for triple negative breast cancer. Oncotarget 5, 3145–3158 10.18632/oncotarget.1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sureban S.M., May R., Weygant N., Qu D., Chandrakesan P., Bannerman-Menson E. et al. (2014) XMD8-92 inhibits pancreatic tumor xenograft growth via a DCLK1-dependent mechanism. Cancer Lett. 351, 151–161 10.1016/j.canlet.2014.05.011 [DOI] [PubMed] [Google Scholar]
- 89.Umapathy G., El Wakil A., Witek B., Chesler L., Danielson L., Deng X. et al. (2014) The kinase ALK stimulates the kinase ERK5 to promote the expression of the oncogene MYCN in neuroblastoma. Sci. Signal. 7, ra102 10.1126/scisignal.2005470 [DOI] [PubMed] [Google Scholar]
- 90.Alvarez-Fernandez S., Ortiz-Ruiz M.J., Parrott T., Zaknoen S., Ocio E.M., San Miguel J. et al. (2013) Potent antimyeloma activity of a novel ERK5/CDK inhibitor. Clin. Cancer Res. 19, 2677–2687 10.1158/1078-0432.CCR-12-2118 [DOI] [PubMed] [Google Scholar]
- 91.Goh K.C., Novotny-Diermayr V., Hart S., Ong L.C., Loh Y.K., Cheong A. et al. (2012) TG02, a novel oral multi-kinase inhibitor of CDKs, JAK2 and FLT3 with potent anti-leukemic properties. Leukemia 26, 236–243 10.1038/leu.2011.218 [DOI] [PubMed] [Google Scholar]
- 92.William A.D., Lee A.C., Goh K.C., Blanchard S., Poulsen A., Teo E.L. et al. (2012) Discovery of kinase spectrum selective macrocycle (16E)-14-methyl-20-oxa-5,7,14,26-tetraazatetracyclo[19.3.1.1(2,6).1(8,12)]heptaco sa-1(25),2(26),3,5,8(27),9,11,16,21,23-decaene (SB1317/TG02), a potent inhibitor of cyclin dependent kinases (CDKs), Janus kinase 2 (JAK2), and fms-like tyrosine kinase-3 (FLT3) for the treatment of cancer. J. Med. Chem. 55, 169–196 10.1021/jm201112g [DOI] [PubMed] [Google Scholar]
- 93.Poulsen A., William A., Blanchard S., Nagaraj H., Williams M., Wang H. et al. (2013) Structure-based design of nitrogen-linked macrocyclic kinase inhibitors leading to the clinical candidate SB1317/TG02, a potent inhibitor of cyclin dependant kinases (CDKs),Janus kinase 2 (JAK2), and Fms-like tyrosine kinase-3 (FLT3). J. Mol. Model. 19, 119–130 10.1007/s00894-012-1528-7 [DOI] [PubMed] [Google Scholar]
- 94.McNicholas T.P., Yum K., Ahn J.H., Mu B., Plettenburg O., Gooderman A. et al. (2012) Structure and function of glucose binding protein-single walled carbon nanotube complexes. Small 8, 3510–3516 10.1002/smll.201200649 [DOI] [PubMed] [Google Scholar]