Abstract
RNA plays a well-established architectural role in the formation of membraneless interchromatin nuclear bodies. However, a less well-known role of RNA is in organizing chromatin, whereby specific RNAs have been found to recruit chromatin modifier proteins. Whether or not RNA can act as an architectural molecule for chromatin remains unclear, partly because dissecting the architectural role of RNA from its regulatory role remains challenging. Studies that have addressed RNA's architectural role in chromatin organization rely on in situ RNA depletion using Ribonuclease A (RNase A) and suggest that RNA plays a major direct architectural role in chromatin organization. In this review, we will discuss these findings, candidate chromatin architectural long non-coding RNAs and possible mechanisms by which RNA, along with RNA binding proteins might be mediating chromatin organization.
Keywords: architectural RNA, chromatin, compaction, heterochromatin, nuclear bodies, phase separation
Introduction
Of the three components of the central dogma (DNA, RNA and Protein), RNA is the most versatile molecule. RNA functions universally as the genetic messenger and as transfer RNAs that deliver amino acids to the site of protein synthesis on the RNA-scaffolded ribosome. In addition, ribozymes catalyze certain specific biochemical reactions, small RNAs (∼20–30 nucleotides) silence gene expression via transcriptional, post-transcriptional as well as chromatin-dependent gene silencing pathways and a functionally diverse class of >200 nucleotides long non-coding RNAs (lncRNAs) play roles in gene expression, genomic imprinting and genome organization [1–6]. Another distinct class of small RNAs (small nuclear RNAs and small nucleolar RNAs) of ∼ 150 nucleotides length, plays important roles in splicing of introns from pre-messenger RNA in the nucleus [7,8]. RNA also acts as the regulator of nuclear and chromatin organization [9]. Furthermore, RNA is the genetic material of retroviruses and other diverse viral clades [10,11].
RNA as an architectural molecule
RNA is an excellent architectural molecule due to its ability to form extensive secondary structures which provide surfaces for various RNA binding proteins (RBPs) thereby forming RNA–RBP scaffolds. These RNA–RBP architectural scaffolds are well known to form nuclear bodies, such as nucleoli, nuclear speckles and nuclear stress bodies, in the interchromatin space where RNA is highly abundant. Removal of RNA leads to collapse of these nuclear bodies providing clear evidence for the architectural role of RNA in formation of these structures [12–14]. A significant fraction of RNA has also been found to directly interact with chromatin and is proposed to have a structural role in chromatin organization [15–18]. However, unlike the well-established architectural role of RNA for nuclear bodies, its equivalent role in chromatin organization remains unclear. Direct chromatin structural modifiers identified to date are only proteins. Histone proteins package genomic DNA into nucleosomal arrays, which are further organized into more condensed heterochromatin and more open euchromatin by chromatin modifier proteins [19–24]. Heterochromatin protein 1 (HP1) and Polycomb Repressive Complex 1 (PRC1) condense nucleosomal arrays into compact heterochromatin, which limits access to DNA metabolism machineries. In contrast, euchromatic proteins maintain a more accessible chromatin environment that facilitates active transcription of underlying loci.
Here, we will discuss the potential role of RNA in maintaining the native structure of chromatin. RNA was shown to be a structural component of the nuclear matrix, a seemingly filamentous structure that spans the interchromatin space in electron microscopy images. However, the existence of the nuclear matrix as a discrete structure is now considered to have been a fixation artifact, as such structures were not visualized in living cells or using high-resolution cryo-electron tomography [25,26]. In contrast, membraneless nuclear bodies formed by architectural RNA are well characterized and can be cytologically visualized in the interchromatin space [12]. Most of these nuclear bodies are reservoirs of either small nucleolar RNA (snoRNA), transcribed mRNA and/or lncRNA, which maintain the integrity of these bodies by forming multivalent weak electrostatic or hydrophobic interactions with RBPs [13]. For example, associations between lncRNA NEAT1 and RNA-binding motifs containing proteins PSF/SFPQ, P54NRB/NONO, and PSPC1 create dynamic structural scaffolds that form paraspeckles [14,27]. The inducible transcription of NEAT1 followed by the direct visualization of the recruitment of paraspeckle proteins by live cell imaging has revealed that the act of NEAT1 transcription, and not lncRNAs alone, regulates paraspeckle maintenance [28]. Paraspeckles disappear after incubation with RNase A suggesting that the presence of RNA in paraspeckles is essential to maintain paraspeckle structure (DNase I does not affect their structural integrity) [27]. Paraspeckles also disassemble in the absence of active RNA Polymerase II transcription and subsequently reassemble on its restoration, suggesting that their integrity is not only dependent on the presence of RNA but also on RNA production [27]. Similarly, the nucleolus, which is formed by ribosomal RNA (rRNA) and RBPs (e.g. nucleophosmin, fibrillarin etc.), collapses into an irregular structure upon inhibition of transcription or by depletion of RNA [29]. Moreover, tethering RNA found in these nuclear bodies is sufficient to nucleate many of these bodies [12]. Given the structural role of RNA in the formation of nuclear bodies, the term architectural RNA has been attributed to RNAs associated with nuclear bodies [30,31]. Here, we will expand the use of the term architectural RNA to describe all RNA species that act as structural scaffolds for nuclear structures including chromatin.
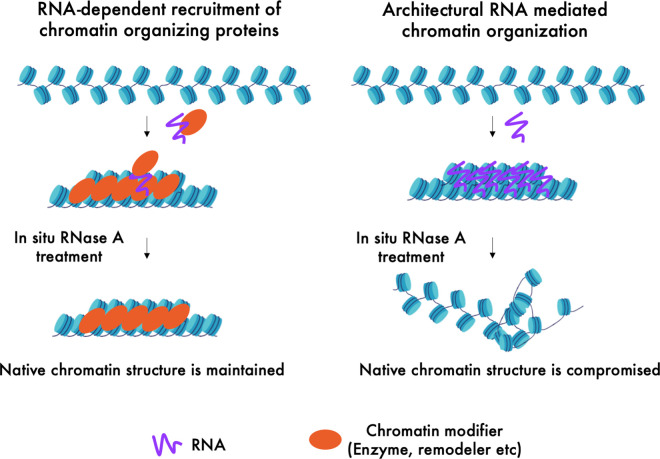
A significant fraction of RNA, mostly non-coding, has also been found to associate with chromatin [15–18,32,33]. RNA is the product of transcription, a process that in itself participates in chromatin organization [34]. Moreover, many non-coding RNAs contribute to chromatin organization by recruiting histone methyltransferases and other RBPs that are chromatin remodelers [35] (Figure 1). Dissecting these indirect regulatory roles from the direct architectural role remains a challenge, and as a result, architectural roles of RNA in chromatin organization are often unclear. Furthermore, unlike nuclear bodies, which are mostly filled with RNAs along with their protein binding partners, chromatin is associated with a low amount of RNA per given unit of chromatin (2%–5% of total nucleic acids in chromatin is RNA) [18]. Therefore, probing structural roles of RNA in chromatin organization requires more sensitive assays that can detect subtle architectural features of RNA in chromatin organization with high resolution in intact cells.
Figure 1. Schematic showing indirect regulatory and direct architectural role of RNA in chromatin organization.
RNA is known to recruit chromatin modifiers such as histone modifying enzymes, chromatin remodelers and chromatin compacting proteins (Left). RNA can also act as a tether to fold or compact chromatin by direct interactions (Right). In situ RNA depletion by RNase A treatment leads to loss of total nuclear RNA, while keeping the RNA-independent nuclear structures intact. The regions organized into specific chromatin configurations by architectural RNA will lose their structure upon RNA depletion while the regions maintained by chromatin modifiers proteins with not.
Evidence for structural roles of RNA in chromatin organization
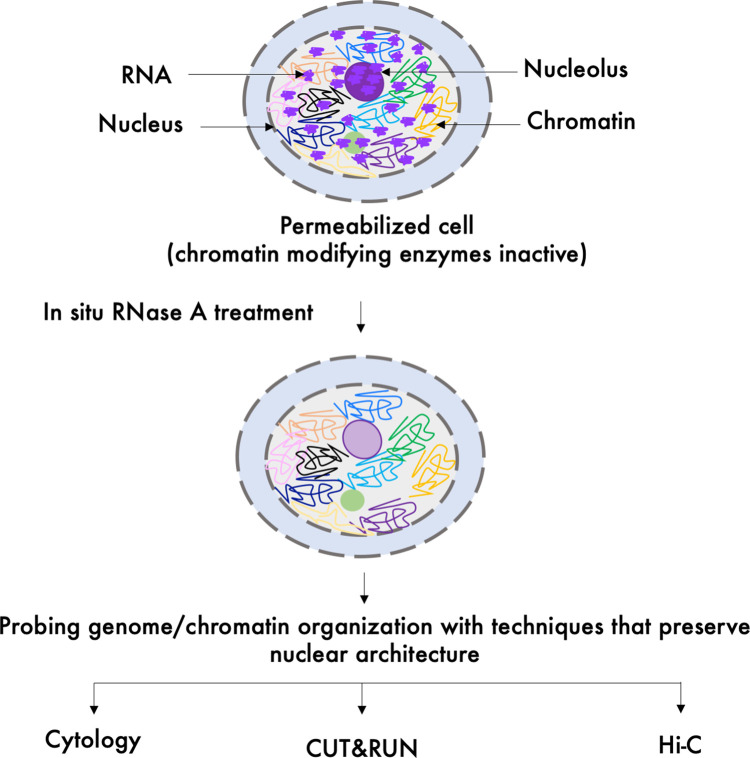
Each of the studies that have addressed the architectural contribution of RNA in chromatin integrity by either cytological visualization or other in situ assays rely on in situ RNA depletion using RNase A (Figure 2) [36–38]. RNase A digestion depletes total RNA, which limits further investigation of the role of specific RNAs in chromatin organization. However, in situ RNase A digestion provides a powerful system to identify an architectural RNA component in chromatin structure as it rules out indirect contributions of RNA by recruiting chromatin modifying proteins (e.g. histone methyltransferases, chromatin remodelers etc.), which become inactive after cells have been permeabilized with a detergent. In addition, both cytological visualization and in situ conformation capture assays (discussed below) preserve overall nuclear structures and can be analyzed in presence or absence of RNA.
Figure 2. In situ RNase A digestion approach to probe RNA's architectural role in chromatin/genome organization.
Cell permeabilization with detergents renders chromatin modifying enzymes inactivate. Upon in situ RNA depletion by RNase A treatment, histones remain stably bound to DNA in the nucleosomes suggesting that the primary chromatin structure remains unaffected by RNase A treatment [39]. However, the higher order chromatin structure may be affected by the loss of RNA. Whether or not architectural RNA maintains the higher order organization of nuclear structures including chromatin, can be investigated by in situ techniques such as cytological visualization, Cleavage Under Targets and Release Using Nuclease (CUT&RUN) [40] and Hi-C [41], in the control and RNase A treated cells.
One of the earliest reports of an architectural role of RNA came from observing RNase A treated Dinoflagellate cells/nuclei [40]. Dinoflagellate chromosomes are seen as condensed helical structures [40]. Cytological visualization of RNase A treated cells revealed decondensation, stretching and unwinding leading to irregular structures at all levels of chromosome organization [40]. Subsequently, visualization of electron micrographs of RNase A digested HeLa nuclei revealed that the global chromatin, seen as uniformly distributed granules in control cells, collapses into large clumps that fall either onto the nuclear lamina or the nucleolus [36]. This study led to the proposal that RNA is a structural component of the nuclear matrix and may contribute to higher order chromatin structure [36]. The first evidence for a structural role of RNA in maintaining a specific chromatin domain came from visualization of pericentric H3K9me-marked heterochromatin in RNase A treated mouse cells. Mouse pericentric heterochromatin domains are organized as large clusters called chromocenters around the nucleolus [42]. Cytological visualization of mouse nuclei stained using antibodies raised against a branched lysine-9 dimethylated H3 amino terminus peptide revealed a loss of the signals for branched chain epitopes upon in situ RNase A treatment as compared with the control untreated cells [37]. The four H3K9 dimethylated peptide ‘fingers’ mimicked the 3-D interaction points within a H3K9me2 heterochromatin domain. Addition of purified cellular RNA to RNase A treated cells restored H3K9me2 clusters, suggesting that the mode of action of architectural RNA is simple and direct [37]. Binding of H3K9me3 histone methyltransferase, SUV39H1, to its target site was lost upon RNase A treatment as well as in nucleic acid binding mutants of SUV39H1 [43]. SUV39H1 binds to nucleic acids with a higher binding affinity for RNA than DNA independent of its binding to H3K9me3 [44]. These findings suggest that direct binding to architectural RNA may help retain histone modifying enzymes onto the chromatin template.
Possible architectural roles of specific transcripts have also been investigated for the binding of chromatin components at centromeres, DNA loci that mediate chromosome segregation by assembling a multproteinaceous structure called kinetochore, which then binds to spindles [45,46]. Centromeric chromatin comprises nucleosomes containing the centromere-specific histone variant CENP-A, which are tightly associated with other key centromeric DNA binding proteins CENP-B, CENP-C and CENP-T [47,48]. Centromeric DNA consists of megabase long arrays of tandem repeats called α-satellites, which transcribe into non-coding mature RNA that are physically associated with centromeric chromatin [49,50]. Targeted deletion of α-satellite transcripts results in a loss of CENP-A and CENP-C in cis [49]. Interestingly, in situ depletion of RNA by RNase A treatment reduces centromere binding of CENP-C significantly while leaving CENP-A levels unaffected [51]. These results suggest the presence of an architectural RNA component of centromeric chromatin. In addition, the finding that in vivo and not in situ RNA depletion alters CENP-A binding suggests that α-satellite transcripts might play both architectural and regulatory roles.
Recently, the Hi-C in situ chromosome conformation capture technique has been applied to test for a chromatin architectural role for RNA. Hi-C captures 3-D interactions by ligating proximal genomic regions and can be combined with in situ RNase digestion to probe the effect of total RNA depletion on genome organization [52]. Hi-C of control cells detected genomic compartments (compartment A corresponding to euchromatin and compartment B corresponding to heterochromatin) which are further subdivided into distinct long-range topologically associated domains (TADs) that define the set of interactions within a given region [41,53]. Hi-C analysis revealed that while TAD signals remained unaffected, compartment B signals were reduced upon RNA depletion [38]. These results suggest that 3-D genomic interactions between heterochromatic regions are maintained by architectural RNA.
Candidate architectural lncRNAs involved in chromatin organization
Although a small subset of total lncRNAs are known to play essential roles, many of them are involved in chromatin regulation by recruiting chromatin modifying proteins [54,55] (Table 1). LncRNAs exhibit properties that make them potential candidates for acting as architectural elements for chromatin organization [6]. RNA forms secondary structures that provide unique domains for interaction with specific proteins and other RNA molecules. A single lncRNA can act as an RNA scaffold either by interacting with multiple copies of the same protein or several different proteins at once [56–58].
Table 1. Long non-coding RNAs involved in chromatin organization.
| LncRNAs | Role in chromatin structure/function | References |
|---|---|---|
| XIST | - Localizes to the entire inactive X chromosomes and recruits machineries for imparting repressive chromatin marks. | [79] |
| KCNQ1OT1 | - Involved in bidirectional silencing of genes in the Kcnq1 domain. | [80] |
| - Interacts with the H3K9- and H3K27-specific histone methyltransferases G9a and the PRC2 complex, respectively in a lineage-specific manner in the placenta. | ||
| AIR | - Controls the Igf2r imprinting cluster in mouse by recruiting the G9a histone methyltransferase to silence target genes. | [81] |
| HOTAIR | - Transcribes from the HOXC locus on human Chr12 and represses transcription in trans across 40 kilobases of the HOXD locus on Chr2. | [66,82] |
| - Binds to the histone methyltransferase PRC2 complex and the histone demethylase LSD1/CoREST/REST complex at its 5′ domain 3′ ends, respectively. | ||
| MALAT1 | - Localizes to nuclear speckles and interacts with pre-mRNA splicing factors and active chromatin | [69] |
| NEAT1 | - Forms paraspeckles by interacting with RNA binding proteins. | [14,69] |
| SAT III | - Upon stress transcribes from pericentric repeats and forms nuclear stress bodies. | [83] |
| - Recruits RNA processing factors to nuclear stress bodies. | ||
| COLDAIR | - Represses a floral repressor FLOWERING LOCUS C (FLC) by interacting with PRC2 during vernalization in Arabidopsis thaliana. | [84] |
| roX1 and roX2 | - Recruit the dosage-compensation complex on the male X chromosome in a cell-type-specific fashion in Drosophila. | [85,86] |
The best-studied candidate chromatin architectural lncRNA is XIST, which coats the entire inactive X-chromosome (Xi) in therian female mammals [59]. XIST exploits 3-D proximal contacts to spread throughout the entire chromosome and recruits machinery for catalyzing repressive chromatin modifications [60,61]. XIST acts both in transcription regulation as well as in the direct architectural stabilization of the native Xi structure [62–64]. Another well-studied X-chromosomal lncRNA, FIRRE, establishes contacts with several autosomes, suggesting that it might be involved in bridging these contact sites to bring them together [65]. HOTAIR is a lncRNA that binds to two distinct histone modifying complexes, PRC2 and LSD1/CoREST/REST complex at its 5′ domain 3′ ends, respectively and therefore in principle can bring H3K27 methylated and H3K4 unmethylated sites in close proximity [66]. A large number of euchromatic proteins also bind RNA that originates from loci in trans, suggesting a role for RNA in holding together two or more distant sites [67–70]. For example, NEAT1 and MATAL1 lncRNAs, which are components of paraspeckles and nuclear speckles, respectively, interact with transcripts from several euchromatic loci [69]. Similarly, enhancer RNAs are involved in chromatin interactions such as looping, although it remains unclear whether chromatin interactions are due to these RNAs themselves or are merely a consequence of enhancer activation [70–72]. Besides ncRNAs, nascent pre-mRNAs along with regulatory proteins may also play an active role in chromatin regulation and have been reviewed elsewhere [73].
Exact molecular mechanisms whereby lncRNAs might function as chromatin architectural elements remain speculative. Some lncRNAs such as HOTAIR that interact with two different chromatin modifying protein complexes, might act as bridges between two chromatin contact points [55]. RNA–RNA interactions are essential for maintaining ribonucleoprotein particles along with RNA–RBP interactions and can therefore play an important role in chromatin organization as well [74,75]. Such RNA–RNA interactions have been documented for XIST, which multimerizes via its A-repeat and circular RNAs that act as ‘sponges' for miRNAs [76–78].
Role of RNA in CTCF-mediated chromatin organization
LncRNAs also impact the 3-D organization of mammalian genomes by facilitating the function of architectural proteins involved in chromatin looping. The multiple zinc-finger architectural protein CCCTC-binding factor (CTCF) acts as an insulator between facultative heterochromatin and euchromatin and organizes them into spatially disjoint domains [87]. CTCF also promotes interactions between distant genomic elements by mediating chromatin loop formation [24,88]. A small fraction of CTCF sites also occur at TAD boundaries, which prevent chromatin loops formed by loop extrusion factors (cohesin or condensin) from growing further [53,89,90]. CTCF contains an RNA-binding domain, exhibits high-affinity for specific RNAs (Kd < 1 nM) and is known to interact with thousands of transcripts (including XIST, TSIX, and XITE) [91,92]. CTCF RNA binding mutants show compromised self-association, binding to chromatin and chromatin loop formation, suggesting that CTCF-RNA interactions regulate chromatin looping [92–94].
A possible role for architectural RNA in heterochromatin organization
Histone H3K9 methyl-marked constitutive heterochromatin is predominantly formed on repetitive DNA and is bound to the heterochromatin protein HP1 [95]. HP1 condenses in vitro reconstituted H3K9me-containing nucleosome arrays and bridges two H3K9me nucleosomes [96,97]. The ability to compact nucleosomal arrays of HP1 in vitro is only modest relative to the massive compaction at pericentric regions seen in vivo, suggesting that additional factors are required to condense H3K9me heterochromatin in the nucleus [96]. Interestingly, HP1 contains an RNA-binding domain and RNA binding is required for heterochromatin localization of mammalian HP1 to pericentric repetitive domains [98]. Drosophila HP1 is also known to interact with several RNAs originating mostly from repetitive regions [99]. In situ depletion of RNA leads to dispersion of H3K9me foci and compromises the ability of recombinant HP1 to bind to pericentric heterochromatin, raising the possibility that HP1 and RNA might act synergistically to compact chromatin [98]. Furthermore, specific depletion of pericentric major satellite (MajSat) transcripts with antisense oligonucleotides leads to a decrease in number of chromocenters (clusters of pericentric regions) per nucleus [100]. Chromocenters are also maintained by the RNA-binding SAF-B protein, which localizes to stress bodies and other locations as well [100]. These studies suggest that architectural RNAs together with RNA-binding proteins contribute to H3K9me heterochromatin compaction and possibly facilitate bridging between distant sites within megabase long heterochromatic domains. It is also possible that similar chromatin–RNA bridging interactions occur within domains of facultative heterochromatin compacted by the PRC1 complex, which is found to be associated with lncRNAs [101–103].
Components of membraneless RNA-filled nuclear bodies do not mix with the nucleoplasm because they undergo liquid-liquid phase separation (LLPS). Nuclear bodies, the best known examples of LLPS in the nucleus to date, phase-separate from the nucleoplasm due to multivalent weak interactions between RNA and RBPs [12,14,30,31,104,105]. The ability of RNA to form extensive secondary structures and establish multivalent interactions, and its net negative charge make RNA a potent modulator of LLPS. As a result, nuclear bodies such as the nucleolus and cytoplasmic P granules fulfil the most important criteria for classic LLPS behavior such as internal mixing, a critical concentration requirement, exchange of molecules with the nucleoplasm and ability to fuse with each other [29,105–108]. Other examples of RNA mediated LLPS are stress bodies that accumulate human pericentric repeat RNA (Sat III) [83,109]. RNA also buffers the phase separation of prion-like RBPs such that high RNA concentrations keep RBPs soluble and changes in RNA levels or RNA binding abilities of RBPs cause aberrant phase transitions [110].
Recent studies have attributed LLPS behavior to heterochromatin domains as well [111–114]. Components of both constitutive and facultative (HP1 and CBX2, respectively) form liquid droplets in vitro [111–115]. SAF-B also forms liquid droplets that are much more dynamic than those established by HP1 in vitro, and SAF-B knockdown results in a dispersed localization of pericentric heterochromatin suggesting that SAF-B-mediated phase separation may contribute to chromocenter condensates [100]. Unlike nuclear bodies, in vivo heterochromatin LLPS research is at an early stage and it remains to be determined how liquid droplets that form spontaneously from pure recombinant heterochromatin components in vitro, are regulated in vivo where one or more phase separating proteins (e.g. HP1 and SAF-B around pericentric regions) are present in a crowded environment of a given region. A recent investigation of biochemical and biophysical properties of in vivo HP1 foci showed a lack of LLPS behavior attributable to HP1 [116]. This suggests that the LLPS behavior of in vivo constitutive heterochromatin is much weaker when compared with nuclear bodies, and that the majority of the properties of heterochromatin can be explained by a classic condensed polymer model in which heterochromatin can be percolated with nucleoplasm.
To the extent that heterochromatin exhibits a degree of LLPS behavior, it is possible that architectural RNA is involved. HP1 and CBX2, respectively components of constitutive and facultative heterochromatin that compact nucleosome arrays, exhibit LLPS behavior in vitro and interact with several RNAs [96,111–113,117,118]. Recently, in vitro and in vivo LLPS behavior of HP1 and SAF-B was shown to be enhanced by pericentric MajSat RNA [100,119]. These initial reports hint at the possibility that heterochromatin LLPS involves multivalent weak RNA–RBP interactions (Figure 3).
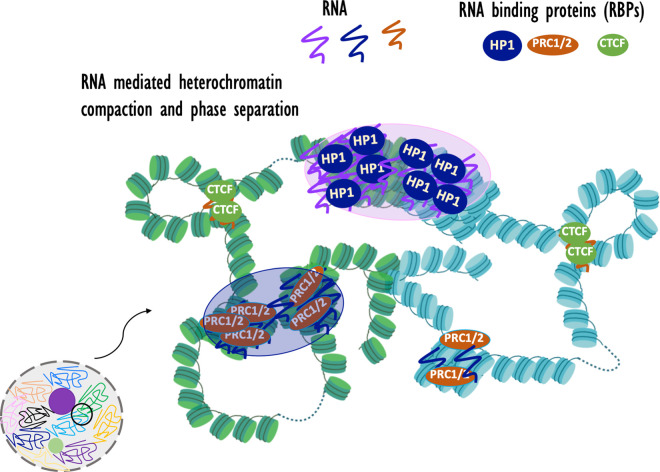
Figure 3. Architectural RNA and chromatin organization.
Architectural RNAs may contribute to heterochromatin organization by facilitating compaction and possibly LLPS and to chromatin loop formation by tethering CTCF to distant loci. Nucleosomal arrays are shown from two different chromosomes that are marked in two different colors. Large transparent purple bubbles represent LLPS of compact heterochromatin.
Perspectives
The ability of RNA to participate in multivalent interactions via its sequence and secondary structures makes it an excellent architectural molecule, as is evident from its scaffolding role for nuclear bodies [31]. Architectural RNA may help to maintain heterochromatin compaction and centromeric chromatin structures. Architectural RNA–RBP interactions may also contribute to chromatin looping and intrachromosomal interactions in euchromatic regions.
A next step in understanding the roles of architectural RNAs in maintaining chromatin structure is to apply genome-scale approaches that deplete specific RNAs from the intact nucleus in situ and that map RNA-chromatin interactions, such as GRID-seq, PIRCh-seq and CUT&RUN [120–122]. Characterization of specific chromatin architectural RNAs will set the stage for asking: 1) Which architectural RNA act in cis and which act in trans? 2) How exactly does a given architectural RNA make contact with the chromatin? Which one among RNA–RBP, RNA–RNA, or RNA–DNA (e.g. R-loops and G-quadruplexes) interactions are used by a given architectural lncRNA?
Understanding the role of RNA mediated compaction in the formation of heterochromatic foci such as chromocenters in the mouse nucleus should shed light on the possible role of LLPS in heterochromatin condensation.
Finally, RNA components of heterochromatin assemblies might provide sequence specificity to RBPs in heterochromatin, including CENPs, HP1, SAF-B and PRC1, and understanding the mechanistic basis for these interactions is an important goal of chromatin research.
Acknowledgements
We acknowledge Henikoff lab members for insightful discussions and funding from the Howard Hughes Medical Institute and a grant from the National Institutes of Health (R01 HG010492 to S.H.).
Abbreviations
- CTCF
CCCTC-binding factor
- HP1
heterochromatin protein 1
- LLPS
liquid-liquid phase separation
- PRC1
Polycomb Repressive Complex 1
- RBP
RNA binding protein
- TAD
topologically associated domain
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Authors Contributions
J.T and S.H. wrote the manuscript.
References
- 1.Engreitz J.M., Ollikainen N. and Guttman M. (2016) Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- 2.Bergmann J.H., Li J., Eckersley-Maslin M.A., Rigo F., Freier S.M. and Spector D.L. (2015) Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 25, 1336–1346 10.1101/gr.189027.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schalch T. and Steiner F.A. (2017) Structure of centromere chromatin: from nucleosome to chromosomal architecture. Chromosoma 126, 443–455 10.1007/s00412-016-0620-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matranga C. and Zamore P.D. (2007) Small silencing RNAs. Curr. Biol. 17, R789–R793 10.1016/j.cub.2007.07.014 [DOI] [PubMed] [Google Scholar]
- 5.Bühler M. (2009) RNA turnover and chromatin-dependent gene silencing. Chromosoma 118, 141–151 10.1007/s00412-008-0195-z [DOI] [PubMed] [Google Scholar]
- 6.Ransohoff J.D., Wei Y. and Khavari P.A. (2018) The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157 10.1038/nrm.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valadkhan S. and Gunawardane L.S. (2013) Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem. 54, 79–90 10.1042/bse0540079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J.A. and Manley J.L. (1991) Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature 352, 818–821 10.1038/352818a0 [DOI] [PubMed] [Google Scholar]
- 9.Sawyer I.A. and Dundr M. (2017) Chromatin loops and causality loops: the influence of RNA upon spatial nuclear architecture. Chromosoma 126, 541–557 10.1007/s00412-017-0632-y [DOI] [PubMed] [Google Scholar]
- 10.Vargiu L., Rodriguez-Tomé P., Sperber G.O., Cadeddu M., Grandi N., Blikstad V. et al. (2016) Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13, 7 10.1186/s12977-015-0232-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood L., Booth D.G., Vargiu G., Ohta S., deLima Alves F., Samejima K. et al. (2016) Auxin/AID versus conventional knockouts: distinguishing the roles of CENP-T/W in mitotic kinetochore assembly and stability. Open Biol. 6, 150230 10.1098/rsob.150230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shevtsov S.P. and Dundr M. (2011) Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- 13.Mao Y.S., Zhang B. and Spector D.L. (2011) Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond C.S. and Fox A.H. (2009) Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644 10.1083/jcb.200906113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang R.C. and Bonner J. (1965) Histone-bound RNA, a component of native nucleohistone. Proc. Natl Acad. Sci. U.S.A. 54, 960–967 10.1073/pnas.54.3.960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonner J. and Widholm J. (1967) Molecular complementarity between nuclear DNA and organ-specific chromosomal RNA. Proc. Natl Acad. Sci. U.S.A. 57, 1379–1385 10.1073/pnas.57.5.1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes D.S., Mayfield J.E., Sander G. and Bonner J. (1972) Chromosomal RNA: its properties. Science 177, 72–74 10.1126/science.177.4043.72 [DOI] [PubMed] [Google Scholar]
- 18.Rodríguez-Campos A. and Azorín F. (2007) RNA is an integral component of chromatin that contributes to its structural organization. PLoS ONE 2, e1182 10.1371/journal.pone.0001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Folco H.D., Pidoux A.L., Urano T. and Allshire R.C. (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319, 94–97 10.1126/science.1150944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lachner M., O'Carroll D., Rea S., Mechtler K. and Jenuwein T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 10.1038/35065132 [DOI] [PubMed] [Google Scholar]
- 21.Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C. et al. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 10.1038/35065138 [DOI] [PubMed] [Google Scholar]
- 22.Hahn M., Dambacher S., Dulev S., Kuznetsova A.Y., Eck S., Wörz S. et al. (2013) Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev. 27, 859–872 10.1101/gad.210377.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuettengruber B., Bourbon H.M., Di Croce L. and Cavalli G. (2017) Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171, 34–57 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 24.Phillips J.E. and Corces V.G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 10.1016/j.cell.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pederson T. (2000) Half a century of ‘the nuclear matrix’. Mol. Biol. Cell 11, 799–805 10.1091/mbc.11.3.799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H. and O'Shea C.C. (2017) ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, eaag0025 10.1126/science.357.6346.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox A.H., Bond C.S. and Lamond A.I. (2005) P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell 16, 5304–5315 10.1091/mbc.e05-06-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao Y.S., Sunwoo H., Zhang B. and Spector D.L. (2011) Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13, 95–101 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boisvert F.M., van Koningsbruggen S., Navascués J. and Lamond A.I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- 30.Chujo T., Yamazaki T. and Hirose T. (2016) Architectural RNAs (arcRNAs): a class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim. Biophys. Acta 1859, 139–146 10.1016/j.bbagrm.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 31.Chujo T. and Hirose T. (2017) Nuclear bodies built on architectural long noncoding RNAs: unifying principles of their construction and function. Mol. Cells 40, 889–896 10.14348/molcells.2017.0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bynum J.W. and Volkin E. (1980) Chromatin-associated RNA: differential extraction and characterization. Biochim. Biophys. Acta 607, 304–318 10.1016/0005-2787(80)90083-0 [DOI] [PubMed] [Google Scholar]
- 33.Holoubek V., Deacon N.J., Buckle D.W. and Naora H. (1983) A small chromatin-associated RNA homologous to repetitive DNA sequences. Eur. J. Biochem. 137, 249–256 10.1111/j.1432-1033.1983.tb07822.x [DOI] [PubMed] [Google Scholar]
- 34.van Steensel B. and Furlong E.E.M. (2019) The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 20, 327–337 10.1038/s41580-019-0114-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grewal S.I. (2010) RNAi-dependent formation of heterochromatin and its diverse functions. Curr. Opin. Genet. Dev. 20, 134–141 10.1016/j.gde.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nickerson J.A., Krochmalnic G., Wan K.M. and Penman S. (1989) Chromatin architecture and nuclear RNA. Proc. Natl Acad. Sci. U.S.A. 86, 177–181 10.1073/pnas.86.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maison C., Bailly D., Peters A.H., Quivy J.P., Roche D., Taddei A. et al. (2002) Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334 10.1038/ng843 [DOI] [PubMed] [Google Scholar]
- 38.Barutcu A.R., Blencowe B.J. and Rinn J.L. (2019) Differential contribution of steady-state RNA and active transcription in chromatin organization. EMBO Rep. 20, e48068 10.15252/embr.201948068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jitendra Thakur, H.F., Llagas T., Disteche C.M. and Henikoff S. (2019) Architectural RNA is required for heterochromatin organization. bioRxiv 10.1101/784835 [DOI] [Google Scholar]
- 40.Marie odile Soyer-Gobillard M.H. (1985) The native structure of dinoflagellate chromosomes. Involvement of structural RNA. Eur. J. Cell Biol. 36, 334–342 https://pascal-francis.inist.fr/vibad/index.php?action=getRecordDetail&idt=9229335 [Google Scholar]
- 41.Dekker J., Marti-Renom M.A. and Mirny L.A. (2013) Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 10.1038/nrg3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guenatri M., Bailly D., Maison C. and Almouzni G. (2004) Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166, 493–505 10.1083/jcb.200403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson W.L., Yewdell W.T., Bell J.C., McNulty S.M., Duda Z., O'Neill R.J. et al. (2017) RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife 6, e25299 10.7554/eLife.25299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirai A., Kawaguchi T., Shimojo H., Muramatsu D., Ishida-Yonetani M., Nishimura Y. et al. (2017) Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. eLife 6, e25317 10.7554/eLife.25317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malik H.S. and Henikoff S. (2009) Major evolutionary transitions in centromere complexity. Cell 138, 1067–1082 10.1016/j.cell.2009.08.036 [DOI] [PubMed] [Google Scholar]
- 46.Westhorpe F.G. and Straight A.F. (2014) The centromere: epigenetic control of chromosome segregation during mitosis. Cold Spring Harb. Perspect. Biol. 7, a015818 10.1101/cshperspect.a015818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakur J. and Henikoff S. (2016) CENPT bridges adjacent CENPA nucleosomes on young human α-satellite dimers. Genome Res. 26, 1178–1187 10.1101/gr.204784.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thakur J. and Henikoff S. (2018) Unexpected conformational variations of the human centromeric chromatin complex. Genes Dev. 32, 20–25 10.1101/gad.307736.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McNulty S.M., Sullivan L.L. and Sullivan B.A. (2017) Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell 42, 226–40.e6 10.1016/j.devcel.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willard H.F. (1985) Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 37, 524–532 PMID: [PMC free article] [PubMed] [Google Scholar]
- 51.Wong L.H., Brettingham-Moore K.H., Chan L., Quach J.M., Anderson M.A., Northrop E.L. et al. (2007) Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17, 1146–1160 10.1101/gr.6022807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong S. and Zhang Y. (2019) Deciphering Hi-C: from 3D genome to function. Cell Biol. Toxicol. 35, 15–32 10.1007/s10565-018-09456-2 [DOI] [PubMed] [Google Scholar]
- 53.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y. et al. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D. et al. (2017) CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, aah7111 10.1126/science.aah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra K. and Kanduri C. (2019) Understanding long noncoding RNA and chromatin interactions: what We know So Far. Noncoding RNA 5, 54 10.3390/ncrna5040054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allen T.A., Von Kaenel S., Goodrich J.A. and Kugel J.F. (2004) The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 11, 816–821 10.1038/nsmb813 [DOI] [PubMed] [Google Scholar]
- 57.Ilik I. and Akhtar A. (2009) Rox RNAs: non-coding regulators of the male X chromosome in flies. RNA Biol. 6, 113–121 10.4161/rna.6.2.8060 [DOI] [PubMed] [Google Scholar]
- 58.Samata M. and Akhtar A. (2018) Dosage compensation of the X chromosome: a complex epigenetic assignment involving chromatin regulators and long noncoding RNAs. Annu. Rev. Biochem. 87, 323–350 10.1146/annurev-biochem-062917-011816 [DOI] [PubMed] [Google Scholar]
- 59.Pinheiro I. and Heard E. (2017) X chromosome inactivation: new players in the initiation of gene silencing. F1000Res. 6, F1000 Faculty Rev-344 10.12688/f1000research.10707.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon M.D., Pinter S.F., Fang R., Sarma K., Rutenberg-Schoenberg M., Bowman S.K. et al. (2013) High-resolution xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504, 465–469 10.1038/nature12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C. et al. (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clemson C.M., McNeil J.A., Willard H.F. and Lawrence J.B. (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275 10.1083/jcb.132.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tattermusch A. and Brockdorff N. (2011) A scaffold for X chromosome inactivation. Hum. Genet. 130, 247–253 10.1007/s00439-011-1027-4 [DOI] [PubMed] [Google Scholar]
- 64.Creamer K.M. and Lawrence J.B. (2017) RNA: a window into the broader role of RNA in nuclear chromosome architecture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160360 10.1098/rstb.2016.0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L. et al. (2014) Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F. et al. (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A. et al. (2013) Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 10.1038/nature11884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y. et al. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I. et al. (2014) The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 55, 791–802 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai Z., Cao C., Ji L., Ye R., Wang D., Xia C. et al. (2020) RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 582, 432–437 10.1038/s41586-020-2249-1 [DOI] [PubMed] [Google Scholar]
- 71.Hou Y., Zhang R. and Sun X. (2019) Enhancer LncRNAs influence chromatin interactions in different ways. Front. Genet. 10, 936 10.3389/fgene.2019.00936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y. et al. (2013) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 10.1038/nature12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Skalska L., Beltran-Nebot M., Ule J. and Jenner R.G. (2017) Regulatory feedback from nascent RNA to chromatin and transcription. Nat. Rev. Mol. Cell Biol. 18, 331–337 10.1038/nrm.2017.12 [DOI] [PubMed] [Google Scholar]
- 74.Ferrandon D., Koch I., Westhof E. and Nüsslein-Volhard C. (1997) RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16, 1751–1758 10.1093/emboj/16.7.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Treeck B., Protter D.S.W., Matheny T., Khong A., Link C.D. and Parker R. (2018) RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl Acad. Sci. U.S.A. 115, 2734–2739 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duszczyk M.M., Wutz A., Rybin V. and Sattler M. (2011) The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17, 1973–1982 10.1261/rna.2747411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K. et al. (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 78.Panda A.C. (2018) Circular RNAs act as miRNA sponges. Adv. Exp. Med. Biol. 1087, 67–79 10.1007/978-981-13-1426-1_6 [DOI] [PubMed] [Google Scholar]
- 79.Loda A. and Heard E. (2019) Xist RNA in action: past, present, and future. PLoS Genet. 15, e1008333 10.1371/journal.pgen.1008333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pandey R.R., Mondal T., Mohammad F., Enroth S., Redrup L., Komorowski J. et al. (2008) Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246 10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- 81.Nagano T., Mitchell J.A., Sanz L.A., Pauler F.M., Ferguson-Smith A.C., Feil R. et al. (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 10.1126/science.1163802 [DOI] [PubMed] [Google Scholar]
- 82.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A. et al. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biamonti G. (2004) Nuclear stress bodies: a heterochromatin affair? Nat. Rev. Mol. Cell Biol. 5, 493–498 10.1038/nrm1405 [DOI] [PubMed] [Google Scholar]
- 84.Kim D.H., Xi Y. and Sung S. (2017) Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 13, e1006939 10.1371/journal.pgen.1006939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park Y., Kelley R.L., Oh H., Kuroda M.I. and Meller V.H. (2002) Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298, 1620–1623 10.1126/science.1076686 [DOI] [PubMed] [Google Scholar]
- 86.Cheetham S.W. and Brand A.H. (2018) RNA-DamID reveals cell-type-specific binding of roX RNAs at chromatin-entry sites. Nat. Struct. Mol. Biol. 25, 109–114 10.1038/s41594-017-0006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O. et al. (2015) CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 10.1126/science.1262088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ong C.T. and Corces V.G. (2014) CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 10.1038/nrg3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nora E.P., Goloborodko A., Valton A.L., Gibcus J.H., Uebersohn A., Abdennur N. et al. (2017) Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169, 930–44.e22 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Phillips-Cremins J.E., Sauria M.E., Sanyal A., Gerasimova T.I., Lajoie B.R., Bell J.S. et al. (2013) Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 10.1016/j.cell.2013.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kung J.T., Kesner B., An J.Y., Ahn J.Y., Cifuentes-Rojas C., Colognori D. et al. (2015) Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol. Cell 57, 361–375 10.1016/j.molcel.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saldaña-Meyer R., González-Buendía E., Guerrero G., Narendra V., Bonasio R., Recillas-Targa F. et al. (2014) CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 28, 723–734 10.1101/gad.236869.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saldaña-Meyer R., Rodriguez-Hernaez J., Escobar T., Nishana M., Jácome-López K., Nora E.P. et al. (2019) RNA interactions Are essential for CTCF-mediated genome organization. Mol. Cell 76, 412–22.e5 10.1016/j.molcel.2019.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hansen A.S., Hsieh T.S., Cattoglio C., Pustova I., Saldaña-Meyer R., Reinberg D. et al. (2019) Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol. Cell 76, 395–411.e13 10.1016/j.molcel.2019.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Almouzni G. and Probst A.V. (2011) Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus 2, 332–338 10.4161/nucl.2.5.17707 [DOI] [PubMed] [Google Scholar]
- 96.Azzaz A.M., Vitalini M.W., Thomas A.S., Price J.P., Blacketer M.J., Cryderman D.E. et al. (2014) Human heterochromatin protein 1α promotes nucleosome associations that drive chromatin condensation. J. Biol. Chem. 289, 6850–6861 10.1074/jbc.M113.512137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Machida S., Takizawa Y., Ishimaru M., Sugita Y., Sekine S., Nakayama J.I. et al. (2018) Structural basis of heterochromatin formation by human HP1. Mol. Cell 69, 385–97.e8 10.1016/j.molcel.2017.12.011 [DOI] [PubMed] [Google Scholar]
- 98.Muchardt C., Guilleme M., Seeler J.S., Trouche D., Dejean A. and Yaniv M. (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3, 975–981 10.1093/embo-reports/kvf194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alekseyenko A.A., Gorchakov A.A., Zee B.M., Fuchs S.M., Kharchenko P.V. and Kuroda M.I. (2014) Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev. 28, 1445–1460 10.1101/gad.241950.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huo X., Ji L., Zhang Y., Lv P., Cao X., Wang Q. et al. (2020) The nuclear matrix protein SAFB cooperates with major satellite RNAs to stabilize heterochromatin architecture partially through phase separation. Mol. Cell 77, 368–83.e7 10.1016/j.molcel.2019.10.001 [DOI] [PubMed] [Google Scholar]
- 101.Francis N.J., Kingston R.E. and Woodcock C.L. (2004) Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 10.1126/science.1100576 [DOI] [PubMed] [Google Scholar]
- 102.Boettiger A.N., Bintu B., Moffitt J.R., Wang S., Beliveau B.J., Fudenberg G. et al. (2016) Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 10.1038/nature16496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ray M.K., Wiskow O., King M.J., Ismail N., Ergun A., Wang Y. et al. (2016) CAT7 and cat7l long non-coding RNAs tune polycomb repressive complex 1 function during human and zebrafish development. J. Biol. Chem. 291, 19558–19572 10.1074/jbc.M116.730853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sawyer I.A., Bartek J. and Dundr M. (2019) Phase separated microenvironments inside the cell nucleus are linked to disease and regulate epigenetic state, transcription and RNA processing. Semin. Cell Dev. Biol. 90, 94–103 10.1016/j.semcdb.2018.07.001 [DOI] [PubMed] [Google Scholar]
- 105.Weber S.C. and Brangwynne C.P. (2012) Getting RNA and protein in phase. Cell 149, 1188–1191 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- 106.Mitrea D.M., Cika J.A., Guy C.S., Ban D., Banerjee P.R., Stanley C.B. et al. (2016) Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5, e13571 10.7554/eLife.13571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y.J., Lee C. et al. (2010) A three-dimensional model of the yeast genome. Nature 465, 363–367 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peng A. and Weber S.C. (2019) Evidence for and against liquid-Liquid phase separation in the nucleus. Noncoding RNA. 5, 50 10.3390/ncrna5040050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guillén-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlüßler R. et al. (2020) RNA-Induced Conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181, 346–61.e17 10.1016/j.cell.2020.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I. et al. (2018) RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921 10.1126/science.aar7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L. et al. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Plys A.J., Davis C.P., Kim J., Rizki G., Keenen M.M., Marr S.K. et al. (2019) Phase separation of polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 33, 799–813 10.1101/gad.326488.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X. and Karpen G.H. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tatavosian R., Kent S., Brown K., Yao T., Duc H.N., Huynh T.N. et al. (2019) Nuclear condensates of the polycomb protein chromobox 2 (CBX2) assemble through phase separation. J. Biol. Chem. 294, 1451–1463 10.1074/jbc.RA118.006620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sanulli S., Trnka M.J., Dharmarajan V., Tibble R.W., Pascal B.D., Burlingame A.L. et al. (2019) HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 10.1038/s41586-019-1669-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Erdel F., Rademacher A., Vlijm R., Tünnermann J., Frank L., Weinmann R. et al. (2020) Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-Liquid phase separation. Mol. Cell 78, 236–49.e7 10.1016/j.molcel.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grau D.J., Chapman B.A., Garlick J.D., Borowsky M., Francis N.J. and Kingston R.E. (2011) Compaction of chromatin by diverse polycomb group proteins requires localized regions of high charge. Genes Dev. 25, 2210–2221 10.1101/gad.17288211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Canzio D., Chang E.Y., Shankar S., Kuchenbecker K.M., Simon M.D., Madhani H.D. et al. (2011) Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41, 67–81 10.1016/j.molcel.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clara Lopes Novo E.W., Hockings C., Poudel C., Sheekey E., Walker S., Kaminski Schierle G.S. et al. (2020) Satellite repeat transcripts modulate heterochromatin condensates and safeguard chromosome stability in mouse embryonic stem cells. bioRxiv 10.1101/2020.06.08.139642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou B., Li X., Luo D., Lim D.H., Zhou Y. and Fu X.D. (2019) GRID-seq for comprehensive analysis of global RNA-chromatin interactions. Nat. Protoc. 14, 2036–2068 10.1038/s41596-019-0172-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fang J., Ma Q., Chu C., Huang B., Li L., Cai P. et al. (2019) PIRCh-seq: functional classification of non-coding RNAs associated with distinct histone modifications. Genome Biol. 20, 292 10.1186/s13059-019-1880-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Skene P.J. and Henikoff S. (2017) An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6, e21856 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]