Abstract
Condensin and cohesin, both members of the structural maintenance of chromosome (SMC) family, contribute to the regulation and structure of chromatin. Recent work has shown both condensin and cohesin extrude DNA loops and most likely work via a conserved mechanism. This review focuses on condensin complexes, highlighting recent in vitro work characterising DNA loop formation and protein structure. We discuss similarities between condensin and cohesin complexes to derive a possible mechanistic model, as well as discuss differences that exist between the different condensin isoforms found in higher eukaryotes.
Keywords: chromosomes, condensin, DNA binding, single-molecule, SMC, structural biology
Introduction
Organising DNA throughout the cell cycle to ensure correct gene expression, DNA replication, and chromosome division is a remarkable feat. Two structural maintenance of chromosomes complexes (SMCs), condensin and cohesin, contribute greatly to this process [1]. Accordingly, misregulation or mutation of either condensin or cohesin is associated with human disease, such as cancer and developmental disorders [2–5]. Condensin compacts DNA into chromosomes during mitosis and is also thought to play a role in genome architecture and transcriptional regulation [6–11]. Cohesin holds sister chromatids together and, along with CTCF, contributes to the formation of topologically associating DNA domains (TADs) [4,12,13]. Condensin and cohesin complexes share a similar molecular architecture (Figure 1A) [14,15] which, for simplicity, will be introduced using S. cerevisiae nomenclature (a list of homologues can be found in Table 1). Both complexes contain a pair of structural maintenance of chromosome (SMC) proteins, Smc2/4 and Smc1/3 for condensin and cohesin, respectively. SMC proteins are antiparallel coiled-coil proteins that hetero-dimerise via a hinge domain (Figure 1A). At the opposite end to the hinge is a split ATPase domain, harbouring two distinct ATP binding sites. While the first SMC head domain of the heterodimer binds ATP via a pocket containing the Walker A/B motifs, the second SMC head sandwiches the ATP molecule and provides the signature motif required for ATP hydrolysis. Mutation of key residues can prevent ATP binding (Q-loop), head engagement (signature motif) or slow ATP hydrolysis (EQ) (Figure 1B,C) [16–18]. A Kleisin protein, Brn1 in condensin and Scc1 in cohesin, binds to the SMC proteins to create a tripartite ring [19,20]. The Kleisin is bound by two heat repeat proteins [21], referred to as HAWKs [22] (HEAT proteins Associated With Kleisins). In condensin, the HAWKs are Ycg1 and Ycs4. In cohesin, however, the HAWKs are more diversified; Scc3 is bound to the C-terminal middle section of Scc1, while Scc2 and Pds5 compete to bind the N-terminal middle section of Scc1 [23]. Compared with yeast, metazoans have evolved additional condensin and cohesin isoforms. Humans have two isoforms of condensin; I and II, while cohesin has additional isoforms of the SMC1, STAG and PDS5 subunits (Table 1).
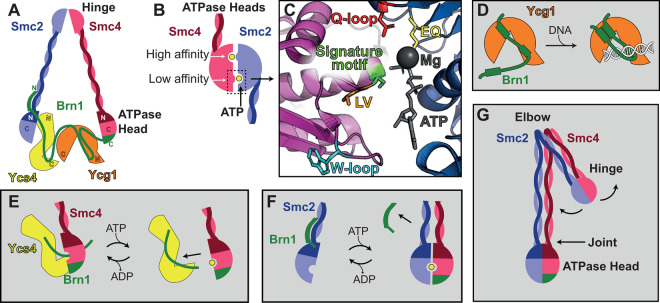
Figure 1. Condensin conformational changes.
(A) Schematic of the subunits that make up S. cerevisiae condensin, indicating the hinge dimerisation domain, and ATPase head domains. (B) Schematic of ATP bound ATPase head domain, indicating high and low affinity ATPase sites in Smc4 and Smc2, respectively. (C) An atomic model of engaged Smc2 and 4 ATPase heads, created by superimposition of C. thermophilum Smc2 and 4 crystal structures (6QJ0 and 6QJ2) [43] with C. thermophilum/S. cerevisiae chimeric engaged Smc1/3 heads (6QPW) [50]. Commonly used mutations to prevent ATP binding (Q-loop, red), prevent head dimerisation (signature motif, green) and slow ATP hydrolysis (EQ, yellow) are indicated, as well as W-loop (cyan) important for binding Ycs4 and the LV mutation (orange) which affects ATP hydrolysis if present in Smc2 and Z-loop formation if present in Smc4. (D) Brn1 creates a safety belt, locking DNA to the Ycg1 HAWK subunit. (E) Ycs4/Brn1 binds Smc4/Brn1, but ATP interferes with this interaction, creating a possible ATP dependent cycle. (F) Brn1 N-terminus binding to Smc2 is lost in the presence of ATP dependent head dimerisation, creating another possible ATP dependent cycle. (G) SMC dimers are able to bend at the elbow region such that the hinge moves towards the ATPase heads. SMC coiled-coils also have some flexibility at the joint region, near the ATPases heads.
Table 1. Equivalent subunits that make up non-meiotic condensin and cohesin complexes.
| Species | Name | SMC Subunits | Kleisin | HAWKS | ||
|---|---|---|---|---|---|---|
| S. cerevisiae | Condensin | Smc2 | Smc4 | Brn1 | Ycs4 | Ycg1 |
| S. pombe | Condensin | Cut14 | Cut3 | Cnd2 | Cnd1 | Cnd3 |
| H. sapiens | Condensin I | SMC2 | SMC4 | CAP-H | CAP-D2 | CAP-G |
| H. sapiens | Condensin II | SMC2 | SMC4 | CAP-H2 | CAP-D3 | CAP-G2 |
| S. cerevisiae | Cohesin | Smc3 | Smc1 | Scc1 | Scc2 or Pds5 | Scc3 |
| S. pombe | Cohesin | Psm3 | Psm1 | Rad21 | Mis4 or Pds5 | Psc3 |
| H. sapiens | Cohesin | SMC3 | SMC1a1 | RAD211 | NIBPL or PDS5A/B | STAG1/21 |
H. sapiens also have meiotic specific cohesin SMC, Kleisin and HAWK subunits, referred to as SMC1b, REC8 and STAG3, respectively.
DNA compaction and loop-extrusion
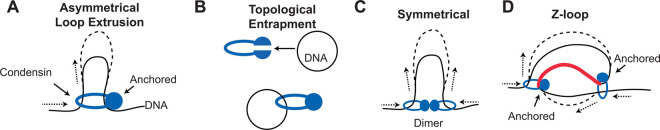
DNA loop extrusion is a process whereby a loop of DNA is extruded through the SMC ring (Figure 2). Several recent studies using in vitro single-molecule approaches illustrate S. cerevisiae, X. laevis and H. sapiens condensin and cohesin complexes compact DNA and extrude DNA loops in an ATP dependent manner [17,24–31]. Cohesin activity requires the presence of the loader complex Scc2/NIPBL [29,30], while loop extrusion cannot be observed in the presence of PDS5A/B [28]. Both H. sapiens condensin and cohesin can compact DNA in the presence of nucleosomes, suggesting DNA bound proteins may not be obstacles to activity [27,29]. Loop-extrusion assays using cell lysates demonstrate that condensin and cohesin activity is cell cycle regulated; cohesin is responsible for the formation of the majority of loops in interphase, while condensin is responsible for loop establishment in mitotic extracts [31]. Additionally, single-molecule experiments using S. cerevisiae cohesin observe that two separate pieces of DNA can be tethered together by cohesin, which is thought to occur via topological entrapment of DNA, where the SMC ring opens such that DNA enters (Figure 2B). However, while DNA tethering is not observed in similar assays performed with condensin, condensin is able to translocate along one piece of DNA while transporting a separate piece [24,27,30]. There is also evidence that multiple pentamers can work together, with oligomerisation of S. cerevisiae condensin increasing DNA compaction activity in magnetic tweezer experiments [32] and single-molecule fluorescence studies suggest dimers of H. sapiens condensin and cohesin extrude symmetrical DNA loops (Figure 2C) [27,29]. In contrast, single-molecule fluorescence assays have shown S. cerevisiae condensin complexes predominantly extrude loops asymmetrically (Figure 2A), and when multiple condensin complexes collide on DNA they create higher-order Z loops, in which three double-stranded DNA helices align in parallel with one condensin at each edge (Figure 2D) [33]. Although single-molecule studies have shown loop-extrusion in real-time, the mechanistic role of each subunit and ATP remains largely unknown, however, recent structural studies have begun to address this.
Figure 2. SMC loop extrusion.
(A) Asymmetric loop extrusion, where a loop of DNA passes through a condensin ring anchored to DNA on one side. (B) Topological loading of SMC complexes, such as cohesin, is achieved by opening of the SMC-Kleisin ring to allow DNA to enter. (C) Symmetrical loop extrusion by a dimer of condensin complexes. For this to be achieved, one condensin complex extrudes DNA on each side, with neither condensin complex anchored to the DNA. (D) A condensin Z-loop, formed when two condensin complexes pass each other on DNA. The region of DNA in red has been compacted by both condensin complexes, hence the total amount of DNA compacted by a Z-loop is less than if two condensin complexes formed two separate loops.
Brn1 buckles DNA to Ycg1
The crystal structure of the S. cerevisiae condensin HAWK, Ycg1 and Kleisin, Brn1 in complex with DNA suggest that Brn1 folds over and interact with DNA via a patch of conserved positively charged residues [34]. This structure appears to be conserved from yeast to human condensin I, with the H. sapiens Ycg1 homologue, CAP-G, having the same fold [35]. Mutation of positive-patch residues in the Kleisins Brn1 or CAP-H in yeast and human cells, respectively, results in loss of condensin chromosomal localisation [34]. Condensin complexes that lack Ycg1 or harbour mutations in the Brn1 positive patch are unable to compact DNA in magnetic tweezer experiments [17], resulting in a model where Brn1 acts as a ‘safety belt', tethering DNA to Ycg1 (Figure 1D). This is thought to enforce asymmetric loop extrusion (Figure 2A), however, translocation has also been observed for S. cerevisiae condensin [24] and symmetrical loop extrusion (Figure 2C) has been observed in H. sapiens condensin complexes [27]. Possible mechanisms we can propose to explain this are that DNA can slide beneath the latched safety-belt or that the safety belt opens and closes during loop extrusion. Chromosome assembly assays performed by Kinoshita et al. show that condensin I complexes lacking the CAP-G subunit can localise to DNA and form chromosomes, suggesting human tetrameric condensin complexes may maintain some functionality. However, these chromosomes are both longer and thinner than those formed by pentameric complexes, suggesting CAP-G plays a key role in shaping chromosomes [36]. Additionally, Brn1 may help regulate Ycg1 function by increasing Ycg1 rigidity and preventing Ycg1 oligomerisation and aggregation [37].
The Ycg1/Brn1/DNA structure bears striking similarity with the S. cerevisiae cohesin subunits Scc3 and Scc1 bound to DNA, in which the Kleisin Scc1 also contributes to the DNA binding affinity [38]. However, contrary to what is observed in condensin complexes, there is little evidence that the cohesin Kleisin subunit folds over DNA. In single-molecule experiments, while condensin remains bound to DNA after ATP is washed out [17], loss of ATP or NIPBL/MAU2 results in human cohesin loop release [28], suggesting that cohesin is less stably tethered to DNA. This could be explained by the discovery of the ‘gripping state' where Scc2/NIPBL within a cohesin complex firmly grips DNA in the presence of ATP [39–41]. In the case of human cohesin, lower DNA association stability might be compensated for by additional DNA binding factors, such as CTCF, which binds directly at the Scc3/Scc1 interface [42].
Ycs4 binding to Smc4 is an ATP sensitive switch
The recent crystal structure of the other condensin HAWK, Ycs4, from C. thermophilum suggests it plays an essential role in regulating the Smc4 ATPase domain [43]. The co-crystal structure of Ycs4/Brn1 bound to the Smc4 ATPase, suggests that Ycs4/Brn1 binds to Smc4 via conserved sites on Ycs4 and Smc4, referred to as the KG-loop and W-loop, respectively. Comparison of this structure with the S. cerevisiae engaged Smc1 ATPase domain homodimer structure (PDB: 1W1W [44]) suggests that Smc4 can not bind to Ycs4 at the same time as Smc2. Furthermore, the addition of ATP inhibits Ycs4/Smc4 complex formation and mutation of either the KG-loop or the W-loop reduces the ATPase rate, suggesting that Ycs4/Brn1 can regulate the ATP binding cycle (Figure 1E). Both the KG and W-loops are conserved in humans and mutation of homologous residues in human CAP-D2 impairs loading onto chromosomes, suggesting this layer of regulation is likely to be conserved in humans [43]. Sequence analysis by Hassler et al. suggests that the W-loop could also be present in the cohesin subunit Smc1, and if we examine crosslinking data from Bürmann et al. [45] we find the W-loop of Smc1 crosslinks to Scc2 (Smc1:1124-Scc2:1193), suggesting that in the absence of ATP, these might interact similarly to Smc4 and Ycs4. Furthermore, Scc2 plays a role in regulating the ATP hydrolysis rate and is required for maximal ATPase activity of cohesin [46]. However, the cryo-EM structures of H. sapiens and S. pombe cohesin in an ATP-bound state show that Scc2 homologous subunit contacts SMC1 even in the presence of engaged heads [39,40]. This could reflect differences between the cohesin and condensin complexes, or simply a difference in how subunits behave in the presence or absence of DNA.
The role of ATP
Recent work by Hassler et al. has provided mechanistic insights into condensin architecture and regulation by solving the crystal structures of the ATPase domains of Smc2 and Smc4 from C. thermophilum. Comparison of the Smc2 structure to ATPγS bound dimers of cohesin Smc1 heads suggests key differences in the ATP binding site of Smc2, resulting in Smc2 having a low binding affinity for ATP. In contrast, Smc4 displays high affinity for ATP, and ATP binding to this site alone is sufficient to promote ATPase head dimerisation, which in turn stimulates binding of ATP to Smc2 (Figure 1B) [43]. Consistent with this finding, extensive screening of Smc4 ATPase site mutants identified specific mutations that reduced condensin activity, while leaving the Smc2 ATPase domain unperturbed [47]. Interestingly, Elbatsh et al. found that mutation of a conserved leucine residue to valine near the signature loop (referred to in the text as LV, Figure 1C) of SMC2 or SMC4 results in markedly different phenotypes in human cells. Smc2-LV reduces the ATP hydrolysis rate of S. cerevisiae condensin, and SMC2-LV results in fuzzy, poorly condensed chromosomes, while mutation of SMC4-LV did not significantly affect ATPase rate and results in highly condensed chromosomes. This is consistent with Smc4 and Smc2 having high and low-affinity ATP binding sites, respectively. Both mutations in S. cerevisiae condensin were able to compact DNA and extrude DNA loops, with Smc2-LV mutant having slower DNA compaction and loop extrusion rates, in line with its reduction in ATPase rate. Smc4-LV however, was able to compact DNA faster, while still displaying the same loop extrusion rate. Further investigation found that Smc4-LV could not form Z-loops, and where two independent DNA loops result in more DNA compaction than two condensin complexes creating a Z-loop, this could account for faster DNA compaction while maintaining the same loop extrusion rate (Figure 2D). Similar results were observed in human cells, as Hi-C data suggests that SMC4-LV creates larger DNA loops than wild-type [48]. The mechanism behind how two condensin complexes interact to form Z-loops is not known, but if we examine the location of the Smc4-LV mutation, we see it is proximal to the W-loop of Smc4 found to bind Ycs4 (Figure 1C). Hence, we speculate that Smc4-LV might alter the Smc4/Ycs4 interaction in a way that prevents two condensin complexes passing each other on DNA, whether this is within one condensin pentamer or possible interactions between pentamers.
Hassler et al. also presents an NMR structure of a fusion made from the N-terminus of Brn1 with two helices of the Smc2 coiled-coil proximal to the ATPase domain. Overlay of the Brn1/Smc2 fusion structure with the crystal structure of C. thermophilum Smc2, suggests that Brn1 binding results in a conformational change in the Smc2 coiled-coil helices. Using a pentameric complex with a TEV protease cleavage site on Brn1 between the Smc2 and Ycs4 binding sites, the authors proceed to show that binding of ATP results in loss of the N-terminal fragment of Brn1, while mutations of Smc2/4 preventing ATP binding or reducing head engagement retain Brn1. This suggests a mechanism whereby ATP binding and head engagement opens the condensin ring by releasing the N-terminus of Brn1 (Figure 1F) [43].
Similarly in cohesin, the N-terminal region of the Kleisin Scc1 releases upon addition of ATP or ATPyS [49,50]. This is associated with cohesin unloading in the presence of Pds5 and Wapl [49,51–53] where a fusion of the Smc3 C-terminus to the Scc1 N-terminus reduces unloading [54]. However, this has recently been implicated in S. pombe cohesin loading, where FRET experiments detect N-terminal Kleisin release upon ATP binding and an increase in N-terminal Kleisin occupancy in the presence of Mis4, DNA and non-hydrolysable ATP [40]. Release of the Kleisin is not thought to be required for loop extrusion activity, as a trimeric fusion of SMC3–RAD21–SMC1a with crosslinked hinge domains are still able to extrude DNA loops [28]. However, cryo-EM structures of H. sapiens and S. pombe cohesin with engaged ATPase heads clearly show the N-terminus of the Kleisin, Rad21 binding to SMC3. This suggests either a conformational change of coiled-coils results in the temporary release of Rad21 or that the presence of DNA and/or the loader NIBPL/Mis4 prevents release or contribute to rebinding of Rad21 to SMC3/Psc3 [39,40]. Based on similarity, the release of N-terminal Brn1 could contribute to loss of condensin DNA loops, and if ATP binding to Smc2 promotes release, it might explain why condensin has evolved such that Smc2 has low ATP binding affinity. However, unlike cohesin, there is little evidence that condensin has release factors and the proposed mechanisms for DNA decompactions include post-translational modifications of condensin subunits, alteration at the protein level of condensin subunits and degradation [55–58].
SMC coiled-coil conformation
A striking feature of recent H. sapiens and S. cerevisiae condensin structural work is a bend in the coiled coils ∼15 nm from the hinge [27,59]. This bend, referred to as the ‘elbow', has also been observed in EM analysis and crosslinking data of S. cerevisiae and S. pombe cohesin and E. coli MukBEF SMC-Kleisin, and results in the hinge bending to contact the SMC arms near the ATPase domains [40,60]. In cohesin, crosslinking and FRET based studies support the hypothesis that the hinge may fold to contact S. pombe Pds5, Psc3 or Mis4 [49,61,62] and cryo-EM structures of H. sapiens, S. cerevisiae and S. pombe suggest that the hinge folds to contact the HAWKs [39–41]. Folding of the SMC arms is observed in recent AFM data of S. cerevisiae condensin, showing that the hinge can fold towards the ATPase domains, however, does so with the SMC arms open, transitioning from an open O to a B shaped conformation [63]. Hence, folding of the SMC hinge towards the globular region (Figure 1G) is a conserved feature in SMC complexes. As the SMC hinge domains have been found to bind DNA [64–68] and S. cerevisiae condensin and cohesin have been observed to interact with DNA via both their hinge and globular domains [41,63], folding is likely to have a key role in the loop extrusion mechanism.
Holocomplex structure
While individual crystal structures have provided much needed molecular detail of subunit interfaces, full understanding of the mechanism requires structural information on the intact complex in different functional states. Recent work by Lee and Merkel et al. [59] have begun to address this gap in knowledge by determining cryo-EM structures of S. cerevisiae condensin in the presence and absence of ATP. Despite using pentameric complexes for much of their analysis, only one HAWK domain is visible in each structure, suggesting the HAWKs are highly dynamic. Two conformations were determined in the absence of ATP (Apo). In both conformations Ycs4 binds the Smc4 ATPase head, consistent with the Ycs4/Smc4 crystal structure presented by Hassler et al. [43], however the conformation of the ATPase heads with respect to each other differs. In one conformation the heads are close together, but not near enough to sandwich ATP, while in the other the ATPase heads are separated by ∼10 nm, with Ycs4 bridging the gap by binding Smc2. In the ATP bound structure, the ATPase heads are engaged and no density is visible for the N-terminal region of Brn1, again consistent with Hassler et al. [43]. This structure reveals that Ycg1 interacts with the Smc2 ATPase head. Mutation of this interaction region in yeast results in reduced cell viability, suggesting it could be physiologically relevant. Further experiments determined that binding of Ycs4 to Smc4 in the absence of ATP is mutually exclusive to Ycg1 binding to Smc2 in the presence of ATP. This suggests a domain ‘flip-flop' mechanism, where HAWKs flip in and out to allow loop extrusion. However, none of the recent condensin EM structures were determined in the presence of DNA, hence we can only speculate where DNA resides within the structure during ATP hydrolysis and loop extrusion.
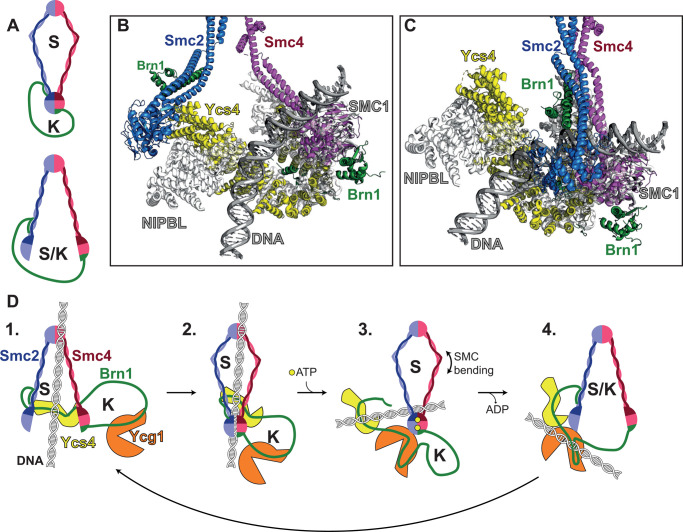
Current models of S. cerevisiae cohesin and B. subtilis Smc-ScpA [69,70] suggest SMC complexes have of two distinct entrapment compartments, referred to as the S compartment (as it is formed by the SMCs), and the K compartment (formed by Kleisin), which can fuse to create a larger S/K ring (Figure 3A). The investigation into K and S compartments in S. cerevisiae cohesin suggest Scc2 promotes ATP independent DNA entry into both the S and K compartments, but not the S/K ring, while Scc3 and ATP hydrolysis is required for DNA entry into the K-compartment [41]. Cryo-EM work in this study, and by others on S. pombe and H. sapiens cohesin, show that DNA is gripped by Scc2 on one side and trapped by engaged ATPase heads on the other [39–41], hence, DNA could gain entry to the S and K compartments by DNA binding to Scc2, before the ATPase heads close around it. Overlay of the S. cerevisiae Apo condensin structures with NIPBL/SMC1/DNA from H. sapiens cohesin structure [39], suggest condensin could bind DNA in a similar manner (Figure 3B,C). While no structural data exist for Ycs4 directly binding to DNA, several evidences suggest that Ycs4 play a central role in condensin binding to DNA: a positively charged patch defines the Ycs4 putative DNA binding surface [59], Ycs4 is able to efficiently bind DNA in gel shift assays [34], and the H. sapiens condensin I Ycs4 equivalent, CAP-D2 is essential for chromosome formation in chromosome assembly assays [36], suggesting that this is essential in condensin binding to DNA.
Figure 3. A potential mechanism for condensin DNA binding.
(A) SMC complexes can create two distinct DNA entrapment compartments, the S-compartment (S), made by the SMCs and the K-compartment (K), made by the Kleisin. Opening of the SMC ATPase heads could result in fusion of these compartments (S/K). (B) Overlay of the S. cerevisiae bridged Apo condensin structure (coloured by domain, PDB: 6YVV) with NIPBL, SMC1 ATPase domain and DNA from the H. sapiens cohesin structure (shown in white/grey, PDB: 6WG3). This places DNA in contact with the top of the Smc4 ATPase domain and a region of Ycs4 with positive surface charge, discussed by Lee and Merkel et al. [59]. (C) Overlay of the S. cerevisiae Apo condensin structure (PDB: 6YVU) with NIPBL, SMC1 ATPase domain and DNA from the H. sapiens cohesin structure (PDB: 6WG3), suggesting the ATPases heads clamp around DNA after it is bound by Ycs4. (D) The proposed mechanism for condensin DNA binding during ATP turn over.
Based on condensin existing structural work, and taking into consideration a general functional homology with cohesin, we propose the following steps in the mechanism of condensin loop extrusion (Figure 3D):
DNA binds to Ycs4 in the condensin bridged state. The hinge could additionally contribute to DNA binding.
DNA binding promotes closure of the ATPase heads, resulting in DNA trapped within both the K and the S compartment.
Proximity of ATPase heads promote ATP binding, release of Ycs4 and binding of Ycg1 to the Smc2 ATPase heads. SMC coiled coils bending could contribute to the relocation of DNA within the S compartment.
ATPase heads open after ATP hydrolysis, creating a temporary S/K ring, enabling DNA to pass into the K compartment. Once DNA is in the K-compartment it can bind Ycg1 and the Brn1 safety belt can close. The cycle can then repeat when Ycs4 binds the Smc4 ATPase head, displacing Ycg1.
Human condensin and chromosome structure
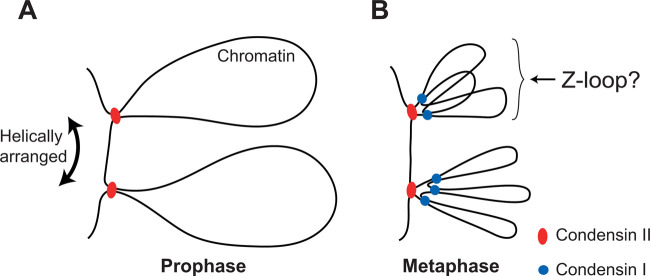
H. sapiens condensin I and II are spatially separated during interphase; condensin II localises within the nucleus throughout the cell cycle, while condensin I is cytoplasmic during interphase and only gains access to the DNA after nuclear envelope break down, allowing condensin II to start loading before condensin I [71]. Depletion of either condensin I or II results in different chromosome morphology and genomic instability phenotypes. Condensin I depletion results in wider chromosomes and the formation of ultrafine bridges at anaphase, while condensin II depletion results in elongated, curly chromosomes and larger chromatin bridges [7,48,72–74]. Condensin I and II are also present on chromosomes in different ratios throughout mitosis, starting at roughly 2 times more condensin I than condensin II in prophase, going up to ∼6 times more in anaphase [75]. This ratio shapes chromosomes, where altering the ratio to 1 : 1 causes chromosomes to become shorter and thicker [76]. Despite condensin I being more abundant, condensin II seems to contribute more to the rigidity of chromosomes. Whole chromosome stretching assays indicate that condensin II-depleted chromosomes have a much larger decrease in elastic modulus than condensin I-depleted chromosomes [77]. Super-resolution microscopy has shown that both condensin I and II localise to an axis formed at the centre of chromosomes, with condensin II localising at the core of the axis, surrounded by condensin I. This work also suggested that in prometaphase condensin II forms loops ∼450 kb in size, while condensin I forms loops ∼90 kb in size [75]. In agreement with this work, in silico chromatin modelling and analysis of Hi-C data from DT-40 chicken cells where condensin I or II was rapidly depleted by auxin-inducible degradation of CAP-H or CAP-H2, respectively, suggest that in prometaphase condensin II promoted the formation of ∼400 kbp loops, while condensin I forms ∼80 kbp loops. Furthermore, this work suggests that condensin I and II loops form a helical axis at the core of chromosomes with ∼12 Mbp per helical turn, which results from the combined activity of condensin I and II. Condensin I and II alone form only narrow or wide helical axes, respectively, creating distinct diagonal stripes in the Hi-C plot [7]. Collectively, these studies suggest a model for mitotic chromosome formation, where first condensin II makes large DNA loops (Figure 4A), followed by condensin I making multiple smaller loops nested within the larger condensin II loops (Figure 4B). As Elbatsh et al. [48] demonstrated, mutations that prevent Z-loop formation cause a hyper condensed phenotype in human HAP1 cells deficient in condensin II, it is conceivable that some condensin I loops could take the form of Z-loops, and we speculate how Z-loops might contribute to chromosome structure (Figure 4B).
Figure 4. Chromosome formation by condensin loop extrusion.
(A) At the start of mitosis in prophase, before nuclear envelope breakdown, condensin II is able to start loading on chromatin, resulting in the formation of large loops. (B) After nuclear envelope break down in metaphase, condensin I gains access to DNA, and forms multiple loops nested within the larger condensin II loops. Some condensin I loops could take the form of Z-loops.
Altogether, these studies suggest the activities of condensin I and II differ in vivo. In agreement with this notion, purified recombinant condensin I and II also display different activity. Condensin II binds to DNA with higher affinity and compacts DNA with slower velocity than condensin I, despite a similar bulk ATPase activity [27]. Whether the intrinsic different biophysical properties of condensin I and II are sufficient to explain the differences observed in vivo remain to be addressed. In fact, the actions of condensin I and II in vivo could also be further modulated and regulated by co-factors present in the cellular environment. There is an ever-increasing list of condensin I or II specific co-factors including but not limited to, the chromo kinesin KIF4A, telomere-associated TRF1 and TANK1, cell cycle factors RB1, pRB and Plk1, chromatin modeller component, Arid1a, the transcription factor, TFIIIC and DNA damage response factor, MCPH1 [78–87]. The role of condensin I and II binding partners in the structural organisation of the genome is largely unknown.
Future directions
There is still much work to be done in order to fully understand how condensin complexes work in the cellular environment. However, recent work has shown that throughout evolution, condensin complexes, as well as cohesin, can extrude DNA loops, a fundamental function which has remained conserved throughout evolution. Individual conformational changes that subunits of condensin undergo in the presence of ATP and DNA have started to be identified, but the full range of steps elucidating the locations of DNA binding surfaces throughout the ATPase cycle still need to be determined. While both condensin and cohesin can compact nucleosome-bound DNA, further work is required to better understand how these complexes works on chromatin. Recent work also poses a number of new questions. Does condensin (and cohesin) work symmetrically as dimers, asymmetrically as monomers or both? What is the mechanism underlying the formation of Z-loops? Do human condensin complexes and cohesin form Z-loops?
Although many questions remain, the recent advances described here illustrate the strength of integrating complementary approaches, such as in vitro biochemistry, Cryo-EM, single-molecule experiments and Hi-C methods. These results form a strong foundation to help build a detailed understanding of how condensin complexes function to organise chromatin in cells.
Perspectives
Importance of the field: Condensin complexes are essential for organising DNA into chromosomes during mitosis, and alterations in condensin function are associated with genome instability and cancer. Hence, structural and mechanistic understanding of condensin function is crucial for understanding its role in human disease.
Current thinking: Condensin complexes extrude DNA loops using a mechanism that involves multiple inter-subunit interactions that undergo conformational changes in a DNA or/and ATP dependent manner.
Future directions: High resolution Cryo-EM structures of condensin complexes bound to DNA throughout the ATPase cycle are needed to provide insight into the holoenzyme mechanism. Building on previous results by adding co-factors and chromatin will further enhance our understanding of how condensin works in cells and contributes to human disease.
Acknowledgements
We would like to thank Prof. Luis Aragon, Dr. Martin Houlard and Dr. Helen King for the helpful comments during the preparation of this manuscript.
Abbreviations
- AFM
atomic force microscopy
- EM
electron microscopy
- HAWK
HEAT proteins associated with Kleisins
- SMC
structural maintenance of chromosome
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work is supported by a Cancer Research UK Programme Foundation (CR-UK C47547/A21536) and a Wellcome Trust Investigator Award to A.V. (200818/Z/16/Z).
Author Contributions
E.E.C. conceived the review, prepared the figures and wrote the manuscript. A.V. contributed to manuscript writing.
References
- 1.Dekker J. and Mirny L. (2016) The 3D genome as moderator of chromosomal communication. Cell 164, 1110–1121 10.1016/j.cell.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin C.-A., Murray J.E., Carroll P., Leitch A., Mackenzie K.J., Halachev M. et al. (2016) Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 30, 2158–2172 10.1101/gad.286351.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leiserson M.D.M., Vandin F., Wu H.-T., Dobson J.R., Eldridge J V., Thomas J.L. et al. (2015) Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet. 47, 106–114 10.1038/ng.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Losada A. (2014) Cohesin in cancer: chromosome segregation and beyond. Nat. Rev. Cancer 14, 389–393 10.1038/nrc3743 [DOI] [PubMed] [Google Scholar]
- 5.Horsfield J.A., Print C.G. and Mönnich M. (2012) Diverse developmental disorders from the one ring: distinct molecular pathways underlie the cohesinopathies. Front. Genet. 3, 171 10.3389/fgene.2012.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swedlow J.R. and Hirano T. (2003) The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell 11, 557–569 10.1016/S1097-2765(03)00103-5 [DOI] [PubMed] [Google Scholar]
- 7.Gibcus J.H., Samejima K., Goloborodko A., Samejima I., Naumova N., Nuebler J. et al. (2018) A pathway for mitotic chromosome formation. Science 359, eaao6135 10.1126/science.aao6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwasaki O., Corcoran C.J. and Noma K.I. (2016) Involvement of condensin-directed gene associations in the organization and regulation of chromosome territories during the cell cycle. Nucleic Acids Res. 44, 3618–3628 10.1093/nar/gkv1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Bortle K., Nichols M.H., Li L., Ong C., Takenaka N., Qin Z.S. et al. (2014) Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 15, R82 10.1186/gb-2014-15-5-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., Hu Y., Oh S., Ma Q., Merkurjev D., Song X. et al. (2015) Condensin I and II complexes license full estrogen receptor α-dependent enhancer activation. Mol. Cell 59, 188–202 10.1016/j.molcel.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki O., Tanizawa H., Kim K.D., Kossenkov A., Nacarelli T., Tashiro S. et al. (2019) Involvement of condensin in cellular senescence through gene regulation and compartmental reorganization. Nat. Commun. 10, 5688 10.1038/s41467-019-13604-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nasmyth K. and Haering C.H. (2009) Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- 13.Merkenschlager M. and Nora E.P. (2016) CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17, 17–43 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- 14.Nasmyth K. and Haering C.H. (2005) The structure and function of Smc and Kleisin complexes. Annu. Rev. Biochem. 74, 595–648 10.1146/annurev.biochem.74.082803.133219 [DOI] [PubMed] [Google Scholar]
- 15.Schleiffer A., Kaitna S., Maurer-Stroh S., Glotzer M., Nasmyth K. and Eisenhaber F. (2003) Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell 11, 571–575 10.1016/S1097-2765(03)00108-4 [DOI] [PubMed] [Google Scholar]
- 16.Lammens A., Schele A. and Hopfner K.-P. (2004) Structural biochemistry of ATP-Driven dimerization and DNA-stimulated activation of SMC ATPases. Curr. Biol. 14, 1778–1782 10.1016/j.cub.2004.09.044 [DOI] [PubMed] [Google Scholar]
- 17.Eeftens J.M., Bisht S., Kerssemakers J., Kschonsak M., Haering C.H. and Dekker C. (2017) Real-time detection of condensin-driven DNA compaction reveals a multistep binding mechanism. EMBO J. 36, 3448–3457 10.15252/embj.201797596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano M. and Hirano T. (2004) Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 23, 2664–2673 10.1038/sj.emboj.7600264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson D.E., Losada A., Erickson H.P. and Hirano T. (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156, 419–424 10.1083/jcb.200111002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haering C.H., Löwe J., Hochwagen A. and Nasmyth K. (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788 10.1016/S1097-2765(02)00515-4 [DOI] [PubMed] [Google Scholar]
- 21.Neuwald A.F. and Hirano T. (2000) HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 10, 1445–1452 10.1101/gr.147400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wells J.N., Gligoris T.G., Nasmyth K.A. and Marsh J.A. (2017) Evolution of condensin and cohesin complexes driven by replacement of kite by Hawk proteins. Curr. Biol. 27, R17–R18 10.1016/j.cub.2016.11.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi S., Borek D.M., Otwinowski Z., Tomchick D.R. and Yu H. (2016) Crystal structure of the cohesin loader Scc2 and insight into cohesinopathy. Proc. Natl Acad. Sci. U.S.A. 113, 12444–9 10.1073/pnas.1611333113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terakawa T., Bisht S., Eeftens J.M., Dekker C., Haering C.H. and Greene E.C. (2017) The condensin complex is a mechanochemical motor that translocates along DNA. Science 358, 672–676 10.1126/science.aan6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganji M., Shaltiel I.A., Bisht S., Kim E., Kalichava A., Haering C.H. et al. (2018) Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105 10.1126/science.aar7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strick T.R., Kawaguchi T. and Hirano T. (2004) Real-time detection of single-molecule DNA compaction by condensin I. Curr. Biol. 14, 874–880 10.1016/j.cub.2004.04.038 [DOI] [PubMed] [Google Scholar]
- 27.Kong M., Cutts E.E., Pan D., Beuron F., Kaliyappan T., Xue C. et al. (2020) Human condensin I and II drive extensive ATP-dependent compaction of nucleosome-bound DNA. Mol. Cell 79, 99–114.e9 10.1016/j.molcel.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davidson I.F., Bauer B., Goetz D., Tang W., Wutz G. and Peters J.M. (2019) DNA loop extrusion by human cohesin. Science 366, 1338–1345 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- 29.Kim Y., Shi Z., Zhang H., Finkelstein I.J. and Yu H. (2019) Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutierrez-Escribano P., Newton M.D., Llauró A., Huber J., Tanasie L., Davy J. et al. (2019) A conserved ATP- and Scc2/4-dependent activity for cohesin in tethering DNA molecules. Sci. Adv. 5, eaay6804 10.1126/sciadv.aay6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golfier S., Quail T., Kimura H. and Brugués J. (2020) Cohesin and condensin extrude DNA loops in a cell-cycle dependent manner. Elife. 9, 821306 10.7554/eLife.53885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keenholtz R.A., Dhanaraman T., Palou R., Yu J., D'amours D. and Marko J.F. (2017) Oligomerization and ATP stimulate condensin-mediated DNA compaction. Sci. Rep. 7, 14279 10.1038/s41598-017-14701-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim E., Kerssemakers J., Shaltiel I.A., Haering C.H. and Dekker C. (2020) DNA-loop extruding condensin complexes can traverse one another. Nature 579, 438–442 10.1038/s41586-020-2067-5 [DOI] [PubMed] [Google Scholar]
- 34.Kschonsak M., Merkel F., Bisht S., Metz J., Rybin V., Hassler M. et al. (2017) Structural basis for a safety-belt mechanism that anchors condensin to chromosomes. Cell 171, 588–600.e24 10.1016/j.cell.2017.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hara K., Kinoshita K., Migita T., Murakami K., Shimizu K., Takeuchi K. et al. (2019) Structural basis of HEAT-kleisin interactions in the human condensin I subcomplex. EMBO Rep. 20, e47183 10.15252/embr.201847183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinoshita K., Kobayashi T.J. and Hirano T. (2015) Balancing acts of two HEAT subunits of condensin I support dynamic assembly of chromosome axes. Dev. Cell 33, 94–107 10.1016/j.devcel.2015.01.034 [DOI] [PubMed] [Google Scholar]
- 37.Manalastas-Cantos K., Kschonsak M., Haering C.H. and Svergun D.I. (2019) Solution structure and flexibility of the condensin HEAT-repeat subunit Ycg1. J. Biol. Chem. 294, 13822–9 10.1074/jbc.RA119.008661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Muir K., Bowler M.W., Metz J., Haering C.H. and Panne D. (2018) Structural basis for Scc3-dependent cohesin recruitment to chromatin. eLife 7, e38356 10.7554/eLife.38356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Z., Gao H., Bai X. and Yu H. (2020) Cryo-EM structure of the human cohesin-NIPBL-DNA complex. Science 368, 1454–1459 10.1126/science.abb0981 [DOI] [PubMed] [Google Scholar]
- 40.Higashi T.L., Eickhoff P., Sousa J.S., Locke J., Nans A., Flynn H.R. et al. (2020) A structure-based mechanism for DNA entry into the cohesin ring. Mol. Cell 79, 917–933.e9 10.1016/j.molcel.2020.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collier J.E., Lee B.-G., Roig M.B., Yatskevich S., Petela N.J., Metson J. et al. (2020) Transport of DNA within cohesin involves clamping on top of engaged heads by Scc2 and entrapment within the ring by Scc3. Elife. 9, 264–277 10.7554/eLife.59560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Haarhuis J.H.I., Sedeño Cacciatore Á., Oldenkamp R., van Ruiten M.S., Willems L. et al. (2020) The structural basis for cohesin–CTCF-anchored loops. Nature 578, 472–476 10.1038/s41586-019-1910-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassler M., Shaltiel I.A., Kschonsak M., Simon B., Merkel F., Thärichen L. et al. (2019) Structural basis of an asymmetric condensin ATPase cycle. Mol. Cell 74, 1175–1188.e9 10.1016/j.molcel.2019.03.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haering C.H., Schoffnegger D., Nishino T., Helmhart W., Nasmyth K. and Löwe J. (2004) Structure and stability of cohesin's Smc1-kleisin interaction. Mol. Cell 15, 951–964 10.1016/j.molcel.2004.08.030 [DOI] [PubMed] [Google Scholar]
- 45.Bürmann F., Lee B.-G., Than T., Sinn L., O'Reilly F.J., Yatskevich S. et al. (2019) A folded conformation of MukBEF and cohesin. Nat. Struct. Mol. Biol. 26, 227–236 10.1038/s41594-019-0196-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petela N.J., Gligoris T.G., Metson J., Lee B.G., Voulgaris M., Hu B. et al. (2018) Scc2 Is a potent activator of cohesin's ATPase that promotes loading by binding Scc1 without Pds5. Mol. Cell 70, 1134–1148.e7 10.1016/j.molcel.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palou R., Dhanaraman T., Marrakchi R., Pascariu M., Tyers M. and D'Amours D. (2018) Condensin ATPase motifs contribute differentially to the maintenance of chromosome morphology and genome stability. PLoS Biol. 16, e2003980 10.1371/journal.pbio.2003980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elbatsh A.M.O., Kim E., Eeftens J.M., Raaijmakers J.A., van der Weide R.H., García-Nieto A. et al. (2019) Distinct roles for condensin's two ATPase sites in chromosome condensation. Mol. Cell 76, 724–737.e5 10.1016/j.molcel.2019.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murayama Y. and Uhlmann F. (2015) DNA entry into and exit out of the cohesin ring by an interlocking gate mechanism. Cell 163, 1628–1640 10.1016/j.cell.2015.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muir K.W., Li Y., Weis F. and Panne D. (2020) The structure of the cohesin ATPase elucidates the mechanism of SMC–kleisin ring opening. Nat. Struct. Mol. Biol. 27, 233–239 10.1038/s41594-020-0379-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huis in ‘t Veld P.J., Herzog F., Ladurner R., Davidson I.F., Piric S., Kreidl E. et al. (2014) Characterization of a DNA exit gate in the human cohesin ring. Science 346, 968–972 10.1126/science.1256904 [DOI] [PubMed] [Google Scholar]
- 52.Murayama Y., Samora C.P., Kurokawa Y., Iwasaki H. and Uhlmann F. (2018) Establishment of DNA–DNA interactions by the cohesin ring. Cell 172, 465–477.e15 10.1016/j.cell.2017.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ciosk R., Shirayama M., Shevchenko A., Tanaka T., Toth A., Shevchenko A. et al. (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5, 243–254 10.1016/S1097-2765(00)80420-7 [DOI] [PubMed] [Google Scholar]
- 54.Chan K.L., Roig M.B., Hu B., Beckouët F., Metson J. and Nasmyth K. (2012) Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell 150, 961–974 10.1016/j.cell.2012.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buster D.W., Daniel S.G., Nguyen H.Q., Windler S.L., Skwarek L.C., Peterson M. et al. (2013) SCFSlimb ubiquitin ligase suppresses condensin II-mediated nuclear reorganization by degrading Cap-H2. J. Cell Biol. 201, 49–63 10.1083/jcb.201207183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wei-Shan H., Amit V.C. and Clarke D.J. (2019) Cell cycle regulation of condensin Smc4. Oncotarget 10, 263–276 10.18632/oncotarget.26467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doughty T.W., Arsenault H.E. and Benanti J.A. (2016) Levels of Ycg1 limit condensin function during the cell cycle. PLoS Genet. 12, 1–23 10.1371/journal.pgen.1006216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thattikota Y., Tollis S., Palou R., Vinet J., Tyers M. and D'Amours D. (2018) Cdc48/VCP promotes chromosome morphogenesis by releasing condensin from self-entrapment in chromatin. Mol. Cell 69, 664–676.e5 10.1016/j.molcel.2018.01.030 [DOI] [PubMed] [Google Scholar]
- 59.Lee B., Merkel F., Allegretti M., Hassler M., Cawood C., Lecomte L. et al. (2020) Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism. Nat. Struct. Mol. Biol. 27, 743–751 10.1038/s41594-020-0457-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bürmann F., Shin H.-C., Basquin J., Soh Y.-M., Giménez-Oya V., Kim Y.-G. et al. (2013) An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat. Struct. Mol. Biol. 20, 371–379 10.1038/nsmb.2488 [DOI] [PubMed] [Google Scholar]
- 61.Intyre J M., Muller E.G.D., Weitzer S., Snydsman B.E., Davis T.N. and Uhlmann F. (2007) In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 26, 3783–3793 10.1038/sj.emboj.7601793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murayama Y. and Uhlmann F. (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505, 367–371 10.1038/nature12867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryu J.-K., Katan A.J., van der Sluis E.O., Wisse T., de Groot R., Haering C. et al. (2019) AFM images of open and collapsed states of yeast condensin suggest a scrunching model for DNA loop extrusion. bioRxiv 10.1101/2019.12.13.867358 [DOI] [Google Scholar]
- 64.Griese J.J., Witte G. and Hopfner K.-P.K.P. (2010) Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 38, 3454–3465 10.1093/nar/gkq038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soh Y.M., Bürmann F., Shin H.C., Oda T., Jin K.S., Toseland C.P. et al. (2015) Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol. Cell 57, 290–303 10.1016/j.molcel.2014.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uchiyama S., Kawahara K., Hosokawa Y., Fukakusa S., Oki H., Nakamura S. et al. (2015) Structural basis for dimer formation of human condensin SMC and its implications for single strand DNA recognition. J. Biol. Chem. 290, 29461–29477 10.1074/jbc.M115.670794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Srinivasan M., Scheinost J.C., Petela N.J., Gligoris T.G., Wissler M., Ogushi S. et al. (2018) The cohesin ring uses Its hinge to organize DNA using non-topological as well as topological mechanisms. Cell 173, 1508–1519.e18 10.1016/j.cell.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiu A., Revenkova E. and Jessberger R. (2004) DNA interaction and dimerization of eukaryotic SMC hinge domains. J. Biol. Chem. 279, 26233–26242 10.1074/jbc.M402439200 [DOI] [PubMed] [Google Scholar]
- 69.Vazquez Nunez R., Ruiz Avila L.B. and Gruber S. (2019) Transient DNA occupancy of the SMC interarm space in prokaryotic condensin. Mol. Cell 75, 209–223.e6 10.1016/j.molcel.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chapard C., Jones R., van Oepen T., Scheinost J.C. and Nasmyth K. (2019) Sister DNA entrapment between juxtaposed Smc heads and Kleisin of the cohesin complex. Mol. Cell 75, 224–237.e5 10.1016/j.molcel.2019.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ono T., Fang Y., Spector D.L. and Hirano T. (2004) Spatial and temporal regulation of condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell 15, 3296–3308 10.1091/mbc.e04-03-0242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Green L.C., Kalitsis P., Chang T.M., Cipetic M., Kim J.H., Marshall O. et al. (2012) Contrasting roles of condensin I and condensin II in mitotic chromosome formation. J. Cell Sci. 125, 1591–1604 10.1242/jcs.097790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F. and Hirano T. (2003) Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115, 109–121 10.1016/S0092-8674(03)00724-4 [DOI] [PubMed] [Google Scholar]
- 74.Ono T., Sakamoto C., Nakao M., Saitoh N. and Hirano T. (2017) Condensin II plays an essential role in reversible assembly of mitotic chromosomes in situ. Mol. Biol. Cell 28, 2875–2886 10.1091/mbc.E17-04-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Walther N., Hossain M.J., Politi A.Z., Koch B., Kueblbeck M., Ødegård-Fougner Ø. et al. (2018) A quantitative map of human condensins provides new insights into mitotic chromosome architecture. J. Cell Biol. 217, 2309–2328 10.1083/jcb.201801048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shintomi K. and Hirano T. (2011) The relative ratio of condensing I to II determines chromosome shapes. Genes Dev. 25, 1464–1469 10.1101/gad.2060311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun M., Biggs R., Hornick J. and Marko J.F. (2018) Condensin controls mitotic chromosome stiffness and stability without forming a structurally contiguous scaffold. Chromosome Res. 26, 277–295 10.1007/s10577-018-9584-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lipp J.J., Hirota T., Poser I. and Peters J.-M. (2007) Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. J. Cell Sci. 120, 1245–1255 10.1242/jcs.03425 [DOI] [PubMed] [Google Scholar]
- 79.Wallace H.A., Klebba J.E., Kusch T., Rogers G.C. and Bosco G. (2015) Condensin II regulates interphase chromatin organization through the Mrg-binding motif of Cap-H2. G3 5, 803–817 10.1534/g3.115.016634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wallace H.A., Rana V., Nguyen H.Q. and Bosco G. (2019) Condensin II subunit NCAPH2 associates with shelterin protein TRF1 and is required for telomere stability. J. Cell. Physiol. 234, 20755–20768 10.1002/jcp.28681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marshall A.E., Ishak C.A. and Dick F.A. (2019) An RB-Condensin II complex mediates long-Range chromosome interactions and influences expression at divergently paired genes. Mol. Cell. Biol. 40, e00452-19 10.1128/mcb.00452-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu S., Fatkhutdinov N., Rosin L., Luppino J.M., Iwasaki O., Tanizawa H. et al. (2019) ARID1A spatially partitions interphase chromosomes. Sci. Adv. 5, eaaw5294 10.1126/sciadv.aaw5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Daniloski Z., Bisht K.K., McStay B. and Smith S. (2019) Resolution of human ribosomal dna occurs in anaphase, dependent on tankyrase 1, condensin II, and topoisomerase IIα. Genes Dev. 33, 276–281 10.1101/gad.321836.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yuen K.C., Slaughter B.D. and Gerton J.L. (2017) Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci. Adv. 3, e1700191 10.1126/sciadv.1700191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longworth M.S., Herr A., Ji J.-Y. and Dyson N.J. (2008) RBF1 promotes chromatin condensation through a conserved interaction with the condensin II protein dCAP-D3. Genes Dev. 22, 1011–1024 10.1101/gad.1631508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abe S., Nagasaka K., Hirayama Y., Kozuka-Hata H., Oyama M., Aoyagi Y. et al. (2011) The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 25, 863–874 10.1101/gad.2016411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wood J.L., Liang Y., Li K. and Chen J. (2008) Microcephalin/MCPH1 associates with the condensin II complex to function in homologous recombination repair. J. Biol. Chem. 283, 29586–29592 10.1074/jbc.M804080200 [DOI] [PMC free article] [PubMed] [Google Scholar]