Abstract
The ongoing COVID-19 pandemic has piqued public interest in the properties, evolution, and emergence of viruses. Here, we discuss how these basic questions have surprisingly remained disputed despite being increasingly within the reach of scientific analysis. We review recent data-driven efforts that shed light into the origin and evolution of viruses and explain factors that resist the widespread acceptance of new views and insights. We propose a new definition of viruses that is not restricted to the presence or absence of any genetic or physical feature, detail a scenario for how viruses likely originated from ancient cells, and explain technical and conceptual biases that limit our understanding of virus evolution. We note that the philosophical aspects of virus evolution also impact the way we might prepare for future outbreaks.
Keywords: virus definition, virus origins, virus evolution, virion, COVID-19, virus-specific genes
Highlights
The distinctions between virions and viruses and modern and ancient cells are crucial to understand virus origins and evolution.
Viruses can be better defined by their generic features of genome propagation and dissemination rather than physical or biological properties of their virions or hosts.
Virus genomes are characterized by the abundance of virus-specific genes that lack detectable cellular homologs. Despite their abundance, virus-specific genes are rarely discussed in the models of virus origin and evolution.
The alignment-based methods are ill-suited for the origins of life research, especially when the objective is to place fast-evolving organisms or viruses in the tree of life.
Protein structures may provide a better alternative to resolve the very deep branches in the tree of life.
The Need to Redefine Viruses
The COVID-19 pandemic exemplifies the constant threat and pressure exerted by viruses on human health and the global economy. The pandemic has triggered an aggressive international response to contain virus spread, cure the disease, and prevent future infections. In parallel, it has rekindled public curiosity in virus definitions, origins, evolution, and their various modes of emergence. For example, Google search for ‘what is a virus’ reached peak popularity in March 2020 coinciding with the global rise in COVID-19 cases. Surprisingly, such fundamental questions have remained unsettled even among evolutionary virologists [1., 2., 3., 4., 5.] and cause confusion in the media portrayal and public perception. For instance, despite overwhelming scientific evidence supporting a natural zoonotic transmission of SARS-CoV-2 from animals to humans [6], many still suspect that the virus was purposefully engineered in laboratories. Similarly, viruses are generalized as noxious pathogens in common discussions and this focus greatly underestimates the many beneficial roles they play in the biosphere [7,8] and as mutualistic symbionts of many hosts (reviewed in [9,10]). In this article, we revisit fundamental questions about the nature, origins, and evolution of viruses during a time when public interest in virus biology is at its peak. We emphasize the need to rethink viruses in the light of new discoveries [2] and call for broader acceptance of new views that are resisted by (sometimes) century-old concepts established in early virology research (reviewed in [11]).
What Is a Virus?
Defining viruses is surprisingly controversial. This is largely because of the seemingly split nature of the virus reproduction cycle into two distinct stages: (i) an intracellular stage during which the virus reprograms the infected cell to produce viral particles or virions (see Glossary), and (ii) an extracellular stage during which virions escape the infected cells and persist in the external environment (similar to plant seeds [12]).1 Both stages, when considered separately, provide dramatically contrasting views about the nature and roles of viruses. For example, virions are metabolically inert infectious particles that do not meet any of the criteria we may use to define ‘life’ or living organisms [2]. However, since they can be purified, counted, and visualized under the microscope, their physical and biochemical properties (e.g., size, shape, metabolic capabilities, capsid) along with host/tissue specificity have become popular in the description, illustration, and naming of viruses (e.g., human immunodeficiency virus). These, in turn, have shaped our perceptions about viruses as nonliving inanimate biological objects that are, paradoxically, infectious.
Treating virions as viruses is a conceptual mistake [2,12., 13., 14., 15., 16.] that overlooks the dramatic changes viruses introduce inside infected cells. A virus-infected cell can effectively be transformed into a ‘hot spot’ for virion production [17] and can practically lose its identity (i.e., it now produces virions rather than two daughter cells) [18]. In some viral infections, large cell-like ‘virion factories’ are clearly visible [19]. This remarkable transformation is due to the virus-mediated manipulation and alteration of host metabolism and defenses [7]. The intracellular stage therefore involves substantial viral activity and is often the target of antiviral drugs to combat virus infection (e.g., antivirals that target HIV polymerase). Despite its immense role in establishing virus infection and existence inside the infected cells, it has unfortunately been referred to as the ‘eclipse’ or ‘vegetative’ phase [20,21] to indicate lack of hallmark signs of virus infection (e.g., virion production, plaques, and cell rupture) and ignored in the definitions and descriptions of viruses. As suggested by Jean-Michel Claverie, virion factory better represents the ‘virus self’ and virions are simply means to disseminate genetic information much like human gametes and plant seeds [12]. In other words, we should depart from the established usage of the word ‘virus’ as being synonymous to ‘virion’. The term ‘virus’ should refer to the process encompassing all phases of the virus infection cycle [3]. In this context, questioning the origin of ‘viruses’ takes a completely different and much broader meaning than simply questioning the origin of the virus particles [2,11,13,16].
Avoid the Presence/Absence Criteria to Define Viruses
The virion- and host-centric virus definitions can cause ambiguities in distinguishing different viral lineages and even viruses from cellular organisms. For example, Forterre recently proposed to redefine viruses as ‘capsid-encoding organisms’ [22] and later as ‘virion-encoding organisms’ [2]. Both definitions recognize viruses as ‘organisms’ that produce capsids/virions and rightly put emphasis back on the intracellular stage of virus infection cycle. However, these views suffer from our ‘human’ habit of classifying biological entities based on the presence/absence or contrast of physical and genetic features. As we discuss later, such definitions rarely withstand the test of time and are vulnerable to change with new discoveries. For example, viruses were long considered tiny and submicroscopic biological entities (properties that describe virions not viruses!) before the discovery of ‘giant viruses’ with genomes and virions bigger than the genomes and sizes of many parasitic cells [23., 24., 25.]. In fact, holding onto the century-old size/shape virion-centric definitions delayed the discovery of giant viruses by more than a decade.2 Similarly, some scientists consider viruses ‘non-living’ because they do not encode metabolism-related genes [1]. However, this feature is neither unique nor common to all viruses. Many endosymbiotic cellular organisms are also characterized by extremely reduced metabolic and translational machineries [26., 27., 28.], and recent metagenomic surveys have verified the existence of several, and likely very ancient, metabolic genes in the genomes of giant viruses [7]. These genes likely help reconfigure the metabolism of infected cells during virus infection [7].
Using virion or capsid to distinguish viral lineages and viruses from cells can generate similar confusions. For example, it can complicate classifications for virus-like genetic elements and viruses that either lack virions (e.g., plasmids, viroids [29]) or encode only part of the virion (e.g., polydnaviruses). For example, the genome of polydnaviruses is dispersed within the genome of parasitoid wasps. The polydnavirus-associated wasps encode the virion packaging system and utilize virions as gene delivery vectors to infect caterpillars [30]. Since the virion is encoded by the wasp genome, polydnavirus-associated wasps may better resemble virion-encoding organisms under the virion-centric definition [31]. Similarly, virus-infected cells can excrete vesicles containing the virus nucleic acid, [32] and healthy cells routinely utilize extracellular vesicles for genetic communication [33]. These examples generalize the concept and morphology of a ‘virion’. Similarly, capsid-like compartments have been detected in cellular organisms where they perform functions such as storage of enzymes [34], and many viral capsid proteins either evolved directly from cellular proteins [35] or have distant homologs in cellular genomes [36]. These examples blur the separation of viruses from cells (and other parasitic genetic elements) based on the presence/absence of physical or genetic descriptors.
In sum, we discourage the use of any virus definition based on the presence or absence of any subset of genes or physical features (e.g., size, morphology, capsid proteins) because such definitions are often ambiguous, not broadly applicable, and more importantly prone to change with new discoveries. We assert that viruses can be better defined by their generic properties of genome dissemination and propagation [11]. Viruses replicate using the macromolecular machinery of other biological entities. This prong establishes absolute parasitism, which is a hallmark of viruses and virus-like genetic elements. Another feature of viruses is the ability to encapsulate and disseminate genomes in metabolically inert structures. This prong can also be generalized, and such structures could be any type of infectious particle without constraints of size, shape, or biochemical composition (e.g., vesicles) [37]. This definition encompasses both the encapsulated and non-encapsulated genomes (e.g., plasmids) and emphasizes the generic feature of how viruses propagate in cells rather than being dependent on the presence/absence of specific biomarkers [11].
Origins of Viruses: Which Hypothesis Is Biologically Plausible?
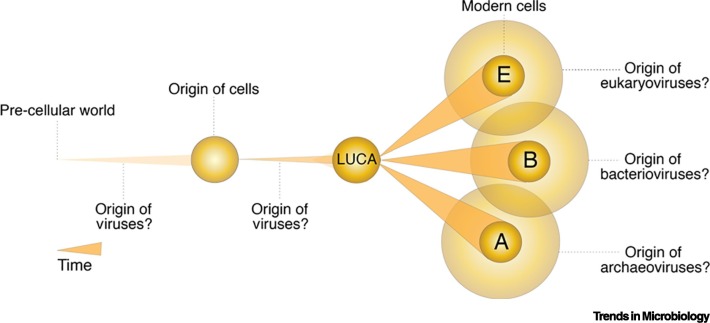
Under our generic definition, virus origin must mean the origin of parasitism and the subsequent ability of those parasitic entities to propagate via the production of metabolically inert structures. Since all modern-day viruses strictly parasitize cells (with the exception of virophages that parasitize the viral factory of other viruses) [38,39], we can assume that virus-mediated parasitism and propagation originated only after cells appeared in evolution as cells would provide both the resources to parasitize upon and the means for genome dissemination (e.g., capsids/vesicles). We therefore rule out virus existence in a ‘pre-cellular’ world as it would be incompatible with the proposed virus definition (Figure 1 for comparative scenarios).
Figure 1.
Different Scenarios for the Origin of Viruses.
Viruses originated either prior to or from cells. A pre-cellular scenario is incompatible with the proposed generic definition of virus propagation inside cells. In turn, the origin of archaeoviruses from Archaea, bacterioviruses from Bacteria, and eukaryoviruses from Eukarya also seems less likely as these viruses share several conserved protein folds involved in virion synthesis and other functions, indicating that they may have evolved prior to the diversification of LUCA into modern cells. These considerations support an intermediate timing for the origin of viruses, that is, from ancient cells that existed prior to LUCA. Modified from [82]. Abbreviation: LUCA, last universal common ancestor.
The next logical questions are the timings and mechanisms of when and how the first viruses appeared. The former question is relatively straightforward. In our view, viruses originated from ‘ancient’ cells that existed before the last universal common ancestor (LUCA) diversified into modern cells (i.e., the three superkingdoms, Archaea, Bacteria, and Eukarya) [40].3 There are multiple lines of evidence supporting this timing. For example, the genomes of archaeoviruses, bacterioviruses, and eukaryoviruses, are characterized by the abundance of virus-specific genes that lack detectable homologs in cellular genomes [41]. While these genes can be strain-specific with a recent de novo origin [42,43], their abundance and existence in diverse virus groups suggests their accumulation likely started very early in evolution. Similarly, viral lineages that infect distantly related hosts from all three superkingdoms share several conserved three-dimensional (3D) protein structural folds that also indicate that these lineages likely existed prior to LUCA diversification [44]. New viruses would then evolve from existing viruses via natural processes such as recombination and in response to new and emerging hosts.
The mechanisms of how ancient cells evolved into viruses are relatively less clear. Krupovic et al. recently proposed a hybrid model to answer this question [45]. According to their model, viral nucleic acids evolved in the pre-cellular world and virus propagation mechanisms evolved via the modification of cellular proteins to function as virus capsids once cells appeared in evolution. In their view, giant viruses such as pandoraviruses and Mimiviruses gradually became bigger due to frequent gene capture from host cells [46]. Their model thus explains the massive genetic diversity seen in virus replicons and proposes mechanisms for the origin of virus capsids and giant viruses. We disagree with the model on two major points.
First, it is unnecessary to invoke a pre-cellular world to explain the observed replicon and genetic diversity among modern-day viruses. This diversity can simply unfold in the pool of ancient cells that existed prior to LUCA. Second, the proposed incremental growth of viral genomes, especially large DNA viruses, via gene gain from hosts is incompatible with our knowledge of how endosymbiotic/parasitic cells evolve. Cells committed to obligate parasitism are characterized by extreme genomic and physical reduction as they increase dependency on their hosts [26,47]. It makes sense to think that viruses, which are the ultimate examples of parasitism, would also evolve similarly. This ‘reduction’ scenario is more parsimonious when one considers the gigantic genome sizes of pandoraviruses (~2500 genes). There is no clear incentive as to why a small-sized virus genome (~5 genes in papillomaviruses) would adopt a pathway towards gigantism, when it is already a well-established parasite. Moreover, regular and recent gene uptake from cells is expected to leave detectable similarity traces in the viral genomes. However, >90% of Pandoravirus genes show no similarity to cellular genes [24] (a feature conserved in many viruses [41], see also [48]) and viruses encode several protein fold structures that have never been detected in cells [41]. It is strange to think that regularly and recently captured viral genes are no longer recognizable whereas presumably ancient ‘core’ genes used to build virus phylogenies are readily recognizable.
Surprisingly, the existence and abundance of virus-specific genes (i.e., genes or protein folds detected only in viral genomes) are rarely discussed in the models of virus origin and evolution. Instead, homology of a subset of ‘core’ virus genes to their cellular counterparts is used to generalize the notion that viruses evolve by acquiring cellular genes [49]. This practice yields an incomplete view of the composition and evolution of virus genomes and ignores the significant de novo gene creation abilities in viruses, especially in pandoraviruses [42]. Moreover, ‘core’ genes describe the evolutionary histories of individual genes and not the whole organisms. They can be patchily distributed, similar to virus-specific genes, and the number of available core genes for phylogenetic studies is strongly dependent on the number of sampled genomes and their taxonomic range [50]. For example, no single gene or protein fold is conserved across all RNA and DNA viruses and very few are conserved across diverse virus families (e.g., DNA polymerase is conserved in many DNA viruses but not in papillomaviruses, and RNA polymerase is conserved in many RNA viruses but not in satellite viruses). The patchy distribution of both the core and virus-specific genes is expected from random reductive evolution where lost and conserved genes were randomly selected from ancient cells. We therefore propose that viruses, especially DNA viruses, evolved from one or multiple ancient cells via reduction [11,41,51,52]. This scenario better aligns with the evolutionary biology of endosymbiotic and parasitic cellular organisms and is more plausible considering the unique composition of virus genomes.
The proposed reduction model is based on the generic definition of viruses and does not suggest that ancient cells reduced into virions. That would be mistaking viruses for their virions, a classical mistake that we have just criticized. Instead, we simply propose that ancient cells were the first to discover the benefits of parasitization and propagation via released particles (e.g., vesicles) [37]. Gradually, the ancient cells devolved as the released particles became fully capable of repeating the cycle of invasion and escape in coinhabiting cellular lineages. While the concept of a ‘virus-like cell’ or a ‘cellular ancestor of virus’ may be difficult to imagine, we already know several examples of viral genome endogenization into host DNA [53] and viral factories that alter the nature of the infected cells [12,18]. These modern-day events transiently restore the ancient ‘cellular self’ that may have been a more permanent feature in the past. In fact, cytoplasmic virion factories behave like a pseudo-nucleus where virus genome replication and translation are separated from host cytoplasm [54,55]. Some authors suggest that the eukaryotic nucleus likely evolved directly from an ancient viral factory [56,57].
Pathways to DNA Cells and Viruses
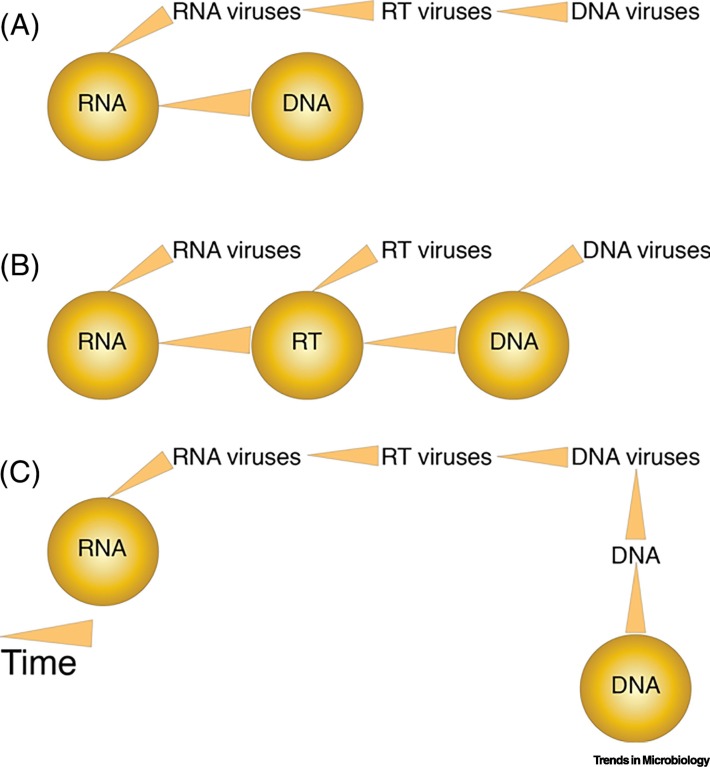
The ancient pre-LUCA cells likely harbored segmented RNA genomes [41,58,59]. It is therefore logical to think that RNA viruses evolved first from RNA cells, and that later, DNA viruses evolved directly from RNA viruses, and in parallel, DNA cells evolved from RNA cells (Figure 2A). This scenario implies that RNA viruses are the ancestors of DNA viruses and was supported in a recent phylogenomic analysis [41]. A second alternative could be that RNA viruses evolved directly from RNA cells, and DNA viruses evolved directly from DNA cells. Thus, both groups evolved independently from different cellular ancestors and possibly via different mechanisms (Figure 2B). Finally, a third alternative could be the evolution of RNA viruses from RNA cells. RNA viruses later invented DNA to escape the defenses of RNA cells. The invention of DNA was later picked up by RNA cells to become DNA cells [60,61] (Figure 2C). Testing these alternatives is challenging since molecular data are limited in their ability to resolve deep evolutionary events.
Figure 2.
Different Scenarios for the Evolution of Different Virus Replicon Groups.
(A) RNA viruses evolved from RNA cells and later evolved into retrotranscribing (RT) and DNA viruses. In parallel, RNA cells evolved into DNA cells. (B) The evolution of RNA, RT, and DNA viruses followed the emergence of RNA, RT, and DNA cells, respectively. (C) RNA viruses evolved from RNA cells and later evolved into RT and DNA viruses. RNA cells evolved into DNA cells once DNA was invented by viruses [60].
Existing Methods Are Ill-Suited to Study Virus Origins
In standard phylogenetic analyses, gene and protein sequences are aligned to elucidate the phylogenetic history of a group of organisms. This alignment is used to infer a phylogenetic tree using various methods [62]. While the alignment-dependent methods work very well in resolving the evolutionary relationships among closely-related (micro)organisms and have significant other applications, they are probably not suited for the origins or ‘tree of life’ research [63]. This is especially true when the objective is to place fast-evolving organisms and viruses in the tree of life [64]. First, the subset of virus genes for which reliable homologs can be found is extremely small [48]. This fact greatly limits the choice and the number of available orthologous genes to be used in phylogeny reconstruction. This sometimes leads authors to resort to subjective, nonstatistically supported approaches to suggest distant homology relationships [65]. Another problem is the recovery of a reliable alignment of homologous genes from a diverse set of genomes. In general, statistically detectable sequence similarity fades over evolutionary time, sometimes leading to complete loss of evolutionary signal due to mutation saturation [66,67]. In addition, protein domain (i.e., structural and functional units within proteins) gains, losses, rearrangements, duplications, and transfers are frequent events in the evolution of genes and genomes [68,69]. These events can happen at different rates in different lineages and thus add many unaligned or poorly aligned regions in sequence alignments [70]. Recovery of a reliable alignment therefore often requires significant manual curation (e.g., removal of a large proportion of poorly aligned sites), which impacts reproducibility by introducing subjectivity [71], and may even be impossible for diverse RNA virus groups [64]. Indeed, the genomes of RNA and retrotranscribing viruses exhibit very high mutation rates [72]. HIV lineages evolving within the same host can differ by 5–10% whereas intrahuman genetic variation could be <0.1% even after ~2.5 million years [73]. These facts greatly limit our ability to reconstruct past evolutionary events using molecular sequence information alone and prompt us to evaluate the potential of alternative, more conserved, molecular characters such as protein structures [74] (Box 1 ).
Box 1. Protein Structures Can Improve Deep Evolutionary Inferences.
Advancements in structural biology allow us to explore and utilize new sets of molecular characters to study deep evolution. Protein function is usually determined directly by the 3D shape of the protein. This fact constrains the preservation of protein structure over longer periods of time, as tampering with the structure could lead to loss of function and could be quite damaging [63,75,76]. As of 26 May 2020, there are ~160 000 protein structural entries in the RCSB Protein Data Bank [77]. These structures correspond to ~1400 protein folds [78], indicating that protein structure space is relatively well sampled and possibly finite [79]. Illergård et al. showed that protein structures evolve at least three to ten times slower than protein sequences [74]. Protein folds are thus (apparently) advantageous as they are remarkably conserved across all species, (and even) viruses, as revealed by their use in recent studies [80,81]. It is possible that a large number of protein folds we see today are very ancient, and even predated LUCA [37,41]. Their use could thus be extremely powerful if subjected to careful phylogenomic and comparative analyses. However, much like the resistance to accepting emerging viewpoints on viruses, protein structures have been rarely utilized in evolutionary studies. This remains another major roadblock in our understanding of virus origins and evolution.
Alt-text: Box 1
Concluding Remarks
Viruses can be better defined based on the generic features of genome dissemination rather than specific virion-associated or physical (size) properties. The defining feature of virus genomes is the existence and abundance of virus-specific genes and protein folds that have no homologs in the cellular world. These genes are rarely discussed in the models of virus origin and evolution, and instead most evolutionary studies rely on a very small subset of viral genes for which we can find reliable cellular homologs. Often such homologies are interpreted as gene uptake from cells by viruses, which is an oversimplified notion for the evolution of virus genomes. Moreover, the fast mutation rates of RNA and retrotranscribing viruses almost make it impossible to recover a reliable alignment for deep virus evolutionary studies. In this regard, focusing on alternative molecular characters that are better conserved in evolution (e.g., protein structures) can possibly provide better solutions. Viruses are likely very old and originated from ancient RNA cells that predated LUCA. They continue to play important roles in the evolution of cells and exert enormous pressure on human health and the global economy. Updating our views on the origins and definitions of viruses (e.g., the distinction between virion and virus) may also help to clarify our thinking about the risk of emergence and spread of new viral diseases (see Outstanding Questions).
Outstanding Questions.
How to visualize and illustrate viruses (virus factories) if virions cannot and should not be used to describe viruses?
How are RNA and DNA viruses evolutionarily related?
Do well defined boundaries exist between cellular and viral lineages?
How do we explain the origin and abundance of virus-specific genes in viral genomes?
Can protein structure-based methods improve the evolutionary studies of viruses?
Alt-text: Outstanding Questions
Acknowledgments
The authors would like to thank Chantal Abergel for continuous discussions and advice. A.N. is supported by the US Department of Energy Laboratory Directed Research and Development (LDRD) program at the Los Alamos National Laboratory (20180751PRD3). E.R.-S. is supported by the LDRD program under project number 20180612ECR and by NIH/NIAID grant R01AI087520 to Thomas Leitner.
Glossary
- Endosymbiosis
the intimate existence of organisms inside the cells or body of other organisms. Notable examples include endosymbiosis of the ancestors of mitochondria and chloroplasts by proto-eukaryotes or the ancestors of eukaryotes.
- Last universal common ancestor (LUCA)
the common ancestor of modern cells, Archaea, Bacteria, and Eukarya. LUCA was not the first cell. It was the last population of cells that diversified into modern cells.
- Orthologous
refers to genes that diverged from the common ancestor as a result of speciation.
- Tree of life
a diagram that describes the evolutionary history among modern species using the metaphors of branching patterns, roots, and leaves to represent evolutionary relationships, ancestors, and modern species, respectively. The topology of the tree of life and the place of viruses in the tree are hotly debated topics.
- Virion
virus particle that can be purified and visualized. The core of a virion comprises the virus nucleic acid (DNA or RNA) enclosed inside a protein shell called a capsid.
- Virion factories
intracellular compartments, formed inside virus-infected cells, that increase virus replication.
Footnotes
In addition, a third stage may exist if the virus genome either integrates into the host DNA or becomes part of the host cytoplasm. Such examples may not lead to virion production or diseases. Because classical signs of virus infection (e.g., virion production, cell rupture) may not be obvious, it is possible that we have massively underestimated nonharmful virus–cell interactions involving virus genome endogenization and domestication by cells [83].
The first giant virus, Acanthamoeba polyphaga mimivirusi, was initially mistaken for a Gram-positive bacterium. It was first discovered in 1992 during a pneumonia outbreak in Bradford, UK and the large size of its virion misled scientists to believe that it must be a bacterium (called 'Bradfordcoccus'). Its virus nature was finally revealed in 2003 [84] and the virus was aptly named ‘mimivirus’ for 'bacteria-mimicking virus'. This is a famous example where adhering to century-old virion and size-based virus definitions delayed a significant discovery.
In the 1970s, Carl Woese pioneered the method of using molecular sequences to study evolution. His work led to the recognition of Archaea [85], then called the ‘third domain’ of life [86]. Archaea have recently taken center stage in evolutionary debates regarding the origin of eukaryotes. There is great controversy on whether Archaea were the first group of diversified organisms on Earth [87], are a sister group to eukaryotes [88,89], or are our ancestors [90,91].
References
- 1.Moreira D., Lopez-Garcia P. Ten reasons to exclude viruses from the tree of life. Nat. Rev. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- 2.Forterre P. To be or not to be alive: How recent discoveries challenge the traditional definitions of viruses and life. Stud. Hist. Phil. Biol. Biomed. Sci. 2016;59:100–108. doi: 10.1016/j.shpsc.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Dupré J., Guttinger S. Viruses as living processes. Stud. Hist. Phil. Biol. Biomed. Sci. 2016;59:109–116. doi: 10.1016/j.shpsc.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Claverie J.M., Abergel C. Open questions about giant viruses. Adv. Virus Res. 2013;85:25–56. doi: 10.1016/B978-0-12-408116-1.00002-1. [DOI] [PubMed] [Google Scholar]
- 5.Van Regenmortel M.H. The metaphor that viruses are living is alive and well, but it is no more than a metaphor. Stud. Hist. Phil. Biol. Biomed. Sci. 2016;59:117–124. doi: 10.1016/j.shpsc.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Andersen K.G. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moniruzzaman M. Dynamic genome evolution and complex virocell metabolism of globally-distributed giant viruses. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-15507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz F. Giant virus diversity and host interactions through global metagenomics. Nature. 2020;578:432–436. doi: 10.1038/s41586-020-1957-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roossinck M.J. The good viruses: viral mutualistic symbioses. Nat. Rev. Microbiol. 2011;9:99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 10.Roossinck M.J. Move over, bacteria! viruses make their mark as mutualistic microbial symbionts. J. Virol. 2015;89:6532–6535. doi: 10.1128/JVI.02974-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claverie J.M., Abergel C. Giant viruses: The difficult breaking of multiple epistemological barriers. Stud. Hist. Phil. Biol. Biomed. Sci. 2016;59:89–99. doi: 10.1016/j.shpsc.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Claverie J.M. Viruses take center stage in cellular evolution. Genome Biol. 2006;7:110. doi: 10.1186/gb-2006-7-6-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandea C.I. A new theory on the origin and the nature of viruses. J. Theor. Biol. 1983;105:591–602. doi: 10.1016/0022-5193(83)90221-7. [DOI] [PubMed] [Google Scholar]
- 14.Van Regenmortel M.H. Logical puzzles and scientific controversies: The nature of species, viruses and living organisms. Syst. Appl. Microbiol. 2009;33:1–6. doi: 10.1016/j.syapm.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Boycott A.E. The transition from live to dead: the nature of filtrable viruses. Proc. R. Soc. Med. 1928;22:55–69. doi: 10.1177/003591572802200121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandea C.I. The origin and evolution of viruses as molecular organisms. Nat. Prec. 2009 doi: 10.1038/npre.2009.3886.1. Published October 23, 2009. [DOI] [Google Scholar]
- 17.Lwoff A. Principles of classification and nomenclature of viruses. Nature. 1967;215:13–14. doi: 10.1038/215013a0. [DOI] [PubMed] [Google Scholar]
- 18.Forterre P. Manipulation of cellular syntheses and the nature of viruses: The virocell concept. C. R. Chim. 2011;14:392–399. [Google Scholar]
- 19.Suzan-Monti M. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lwoff A. The concept of virus. J. Gen. Microbiol. 1957;17:239–253. doi: 10.1099/00221287-17-2-239. [DOI] [PubMed] [Google Scholar]
- 21.Jacob F., Wollman E. Viruses and genes. Sci. Am. 1961;204:93–107. [PubMed] [Google Scholar]
- 22.Raoult D., Forterre P. Redefining viruses: lessons from Mimivirus. Nat. Rev. 2008;6:315–319. doi: 10.1038/nrmicro1858. [DOI] [PubMed] [Google Scholar]
- 23.Raoult D. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 24.Philippe N. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 25.Legendre M. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Madrigal S. Complete genome sequence of 'Candidatus Tremblaya princeps' strain PCVAL, an intriguing translational machine below the living-cell status. J. Bacteriol. 2011;193:5587–5588. doi: 10.1128/JB.05749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luef B. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015;6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 28.Omsland A. Chlamydial metabolism revisited: Interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol. Rev. 2014;38:779–801. doi: 10.1111/1574-6976.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato Y. Hadaka virus 1: a capsidless eleven-segmented positive-sense single-stranded RNA virus from a phytopathogenic fungus, Fusarium oxysporum. mBio. 2020;11 doi: 10.1128/mBio.00450-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke G.R., Strand M.R. Polydnaviruses of parasitic wasps: domestication of viruses to act as gene delivery vectors. Insects. 2012;3:91–119. doi: 10.3390/insects3010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Desjardins C.A. Unusual viral genomes: Mimivirus and the polydnaviruses. In: Beckage N.E., Drezen J.M., editors. Parasitoid Viruses: Symbionts and Pathogens. Elsevier; 2012. pp. 115–125. [Google Scholar]
- 32.Gill S. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol. Rev. 2019;43:273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soler N., Forterre P. Vesiduction: the fourth way of HGT. Environ. Microbiol. 2020;22:2457–2460. doi: 10.1111/1462-2920.15056. [DOI] [PubMed] [Google Scholar]
- 34.Sutter M. Structural basis of enzyme encapsulation into a bacterial nanocompartment. Nat. Struct. Mol. Biol. 2008;15:939–947. doi: 10.1038/nsmb.1473. [DOI] [PubMed] [Google Scholar]
- 35.Krupovic M., Koonin E.V. Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2401–E2410. doi: 10.1073/pnas.1621061114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasir A., Caetano-Anollés G. Identification of capsid/coat related protein folds and their utility for virus classification. Front. Microbiol. 2017;8:380. doi: 10.3389/fmicb.2017.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nasir A. Untangling the origin of viruses and their impact on cellular evolution. Ann. N. Y. Acad. Sci. 2015;1341:61–74. doi: 10.1111/nyas.12735. [DOI] [PubMed] [Google Scholar]
- 38.La Scola B. The virophage as a unique parasite of the giant mimivirus. Nature. 2008;455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 39.Claverie J.M., Abergel C. Mimivirus and its virophage. Annu. Rev. Genet. 2009;43:49–66. doi: 10.1146/annurev-genet-102108-134255. [DOI] [PubMed] [Google Scholar]
- 40.Forterre P. The two ages of the RNA world, and the transition to the DNA world: a story of viruses and cells. Biochimie. 2005;87:793–803. doi: 10.1016/j.biochi.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 41.Nasir A., Caetano-Anollés G. A phylogenomic data-driven exploration of viral origins and evolution. Sci. Adv. 2015;1 doi: 10.1126/sciadv.1500527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Legendre M. Vol. 10. 2019. Pandoravirus celtis illustrates the microevolution processes at work in the giant Pandoraviridae genomes; pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legendre M. Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 2018;9:2285. doi: 10.1038/s41467-018-04698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abrescia N.G.A. Structure unifies the viral universe. Annu. Rev. Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 45.Krupovic M. Origin of viruses: primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 2019;17:449–458. doi: 10.1038/s41579-019-0205-6. [DOI] [PubMed] [Google Scholar]
- 46.Koonin E.V., Yutin N. Evolution of the large nucleocytoplasmic DNA viruses of eukaryotes and convergent origins of viral gigantism. Adv. Virus Res. 2019;103:167–202. doi: 10.1016/bs.aivir.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 47.McCutcheon J.P., Moran N.A. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 48.Boratto P.V.M. Yaravirus: A novel 80-nm virus infecting Acanthamoeba castellanii. Proc. Natl. Acad. Sci. U. S. A. 2020;117:16579–16586. doi: 10.1073/pnas.2001637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreira D., López-García P. Evolution of viruses and cells: do we need a fourth domain of life to explain the origin of eukaryotes? Philos. Trans. R. Soc. B Biol. Sci. 2015;370:20140327. doi: 10.1098/rstb.2014.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dagan T., Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nasir A. Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya. BMC Evol. Biol. 2012;12:156. doi: 10.1186/1471-2148-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claverie J.M. Mimivirus and the emerging concept of 'giant' virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 53.Feschotte C., Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 54.Forterre P. Defining life: The virus viewpoint. Orig. Life Evol. Biosph. 2010;40:151–160. doi: 10.1007/s11084-010-9194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaikeeratisak V. Assembly of a nucleus-like structure during viral replication in bacteria. Science. 2017;355:194–197. doi: 10.1126/science.aal2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forterre P., Gaïa M. Giant viruses and the origin of modern eukaryotes. Curr. Opin. Microbiol. 2016;31:44–49. doi: 10.1016/j.mib.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Bell P.J.L. Viral eukaryogenesis: was the ancestor of the nucleus a complex DNA virus? J. Mol. Evol. 2001;53:251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- 58.Woese C.R. The primary lines of descent and the universal ancestor. In: Bendall D.S., editor. Evolution from Molecules to Men. Cambridge University Press; 1983. pp. 209–233. [Google Scholar]
- 59.Poole A.M. The path from the RNA world. J. Mol. Evol. 1998;46:1–17. doi: 10.1007/pl00006275. [DOI] [PubMed] [Google Scholar]
- 60.Forterre P. Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: A hypothesis for the origin of cellular domain. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3669–3674. doi: 10.1073/pnas.0510333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claverie J.M., Abergel C. Mimivirus: The emerging paradox of quasi-autonomous viruses. Trends Genet. 2010;26:431–437. doi: 10.1016/j.tig.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 62.Kapli P. Phylogenetic tree building in the genomic age. Nat. Rev. Genet. 2020;21:428–444. doi: 10.1038/s41576-020-0233-0. [DOI] [PubMed] [Google Scholar]
- 63.Caetano-Anollés G., Nasir A. Benefits of using molecular structure and abundance in phylogenomic analysis. Front. Genet. 2012;3:172. doi: 10.3389/fgene.2012.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holmes E.C., Duchêne S. Can sequence phylogenies safely infer the origin of the global virome? mBio. 2019;10 doi: 10.1128/mBio.00289-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krupovic M. Evolution of a major virion protein of the giant pandoraviruses from an inactivated bacterial glycoside hydrolase. Virus Evol. 2020 doi: 10.1093/ve/veaa059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sober E., Steel M. Testing the hypothesis of common ancestry. J. Theor. Biol. 2002;218:395–408. [PubMed] [Google Scholar]
- 67.Nasir A. Arguments reinforcing the three-domain view of diversified cellular life. Archaea. 2016;2016:1851865. doi: 10.1155/2016/1851865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nasir A. Global patterns of protein domain gain and loss in superkingdoms. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore A.D., Bornberg-Bauer E. The dynamics and evolutionary potential of domain loss and emergence. Mol. Biol. Evol. 2012;29:787–796. doi: 10.1093/molbev/msr250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Caetano-Anollés G. Rooting phylogenies and the tree of life while minimizing ad hoc and auxiliary assumptions. Evol. Bioinform. 2018;14 doi: 10.1177/1176934318805101. 1176934318805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Philippe H. Pitfalls in supermatrix phylogenomics. Eur. J. Taxon. 2017;283:1–25. [Google Scholar]
- 72.Sanjuán R. Viral mutation rates. J. Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leitner T. The puzzle of hiv neutral and selective evolution. Mol. Biol. Evol. 2018;35:1355–1358. doi: 10.1093/molbev/msy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Illergård K. Structure is three to ten times more conserved than sequence--a study of structural response in protein cores. Proteins. 2009;77:499–508. doi: 10.1002/prot.22458. [DOI] [PubMed] [Google Scholar]
- 75.Chothia C. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 76.Gerstein M., Hegyi H. Comparing genomes in terms of protein structure: Surveys of a finite parts list. FEMS Microbiol. Rev. 1998;22:277–304. doi: 10.1111/j.1574-6976.1998.tb00371.x. [DOI] [PubMed] [Google Scholar]
- 77.Burley S.K. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andreeva A. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020;48:D376–D382. doi: 10.1093/nar/gkz1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caetano-Anollés G. The origin, evolution and structure of the protein world. Biochem. J. 2009;417:621–637. doi: 10.1042/BJ20082063. [DOI] [PubMed] [Google Scholar]
- 80.Mughal F. The origin and evolution of viruses inferred from fold family structure. Arch. Virol. 2020:1–15. doi: 10.1007/s00705-020-04724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bokhari R.H. Bacterial origin and reductive evolution of the CPR group. Genome Biol. Evol. 2020;12:103–121. doi: 10.1093/gbe/evaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nasir A. Viral evolution Primordial cellular origins and late adaptation to parasitism. Mob. Genet. Elements. 2012;2:247–252. doi: 10.4161/mge.22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nasir A. Long-term evolution of viruses: A Janus-faced balance. BioEssays. 2017;39 doi: 10.1002/bies.201700026. [DOI] [PubMed] [Google Scholar]
- 84.La Scola B. A giant virus in amoebae. Science. 2003;299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 85.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Woese C.R. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Staley J.T., Caetano-Anollés G. Archaea-First and the co-evolutionary diversification of domains of life. BioEssays. 2018;40:1800036. doi: 10.1002/bies.201800036. [DOI] [PubMed] [Google Scholar]
- 88.Da Cunha V. Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Da Cunha V. Asgard archaea do not close the debate about the universal tree of life topology. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zaremba-Niedzwiedzka K. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 91.Williams T.A. Phylogenomics provides robust support for a two-domains tree of life. Nat. Ecol. Evol. 2020;4:138–147. doi: 10.1038/s41559-019-1040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]