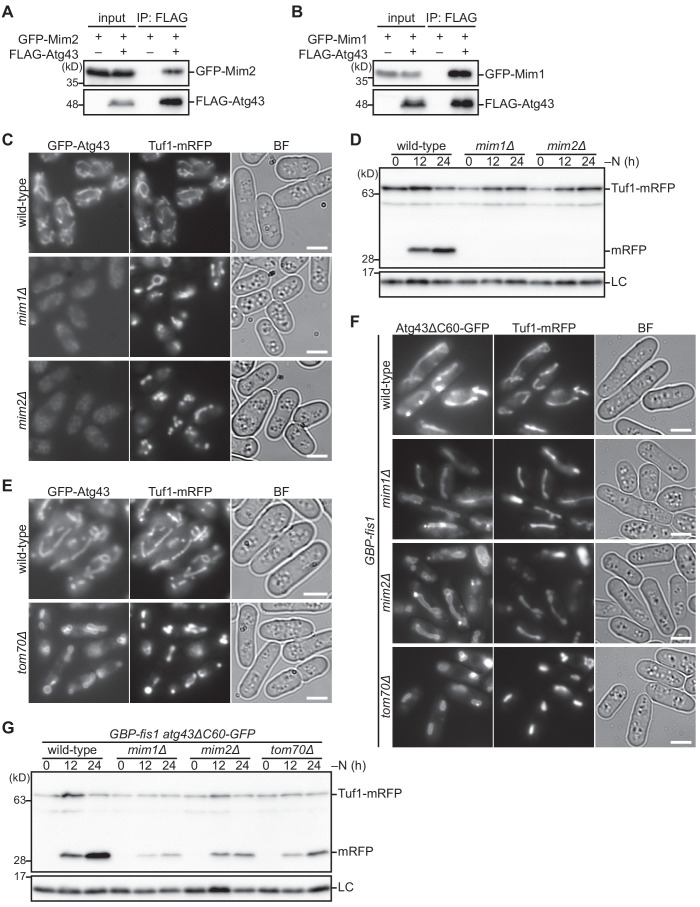
Figure 5. The MIM complex facilitates mitophagy through the loading of Atg43 to the MOM.
(A, B) Crude cell lysates (input) were prepared from cells co-expressing GFP-fused Mim2 or Mim1 with (+) or without (–) FLAG-tagged Atg43. Anti-FLAG immunoprecipitants (IP: FLAG) were analyzed by immunoblotting. (C, E) The indicated strains co-expressing GFP-Atg43 and Tuf1-mRFP were grown in EMM for microscopy. (D) The indicated strains expressing Tuf1-mRFP were collected at the indicated time points after shifting to nitrogen starvation medium and were used for immunoblotting. (F) The indicated strains co-expressing a GFP-fused C-terminal truncated form of Atg43 (Atg43ΔC60-GFP) and Tuf1-mRFP were grown in EMM for microscopy. (G) Wild-type, mim1Δ, mim2Δ, and tom70Δ cells co-expressing GBP-fused Fis1 and GFP-fused Atg43 lacking the 60 C-terminal aa (Atg43ΔC60-GFP) were collected at the indicated time points after shifting to nitrogen starvation medium. Cells were then used for immunoblotting. Histone H3 was used as a loading control (LC) for immunoblotting. Scale bars represent 5 µm. BF, bright-field image.