Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) is a positive-sense single-stranded RNA (+ssRNA) that causes coronavirus disease 2019 (COVID-19). The viral genome encodes twelve genes for viral replication and infection. The third open reading frame is the spike (S) gene that encodes for the spike glycoprotein interacting with specific cell surface receptor – angiotensin converting enzyme 2 (ACE2) – on the host cell membrane. Most recent studies identified a single point mutation in S gene. A single point mutation in S gene leading to an amino acid substitution at codon 614 from an aspartic acid 614 into glycine (D614G) resulted in greater infectivity compared to the wild type SARS-CoV2. We were interested in investigating the mutation region of S gene of SARS-CoV2 from Korean COVID-19 patients. New mutation sites were found in the critical receptor binding domain (RBD) of S gene, which is adjacent to the aforementioned D614G mutation residue. This specific sequence data demonstrated the active progression of SARS-CoV2 by mutations in the RBD of S gene. The sequence information of new mutations is critical to the development of recombinant SARS-CoV2 spike antigens, which may be required to improve and advance the strategy against a wide range of possible SARS-CoV2 mutations.
Keywords: SARS coronavirus 2, Spike glycoprotein, COVID-19 receptor binding domain, COVID-19 mutation, COVID-19 antigens
INTRODUCTION
A positive-sense single-stranded RNA (+ssRNA) coronavirus causing severe acute respiratory syndrome (SARS-CoV) was first reported nearly two decades ago (1). Indeed, beta coronaviruses (CoV) have caused zoonotic epidemics or pandemics in man thrice in the past 20 years: SARS-CoV in 2002–2003, MERS-CoV in 2012, and SARS-CoV2 beginning in late 2019. However, the current outbreak SARS-CoV2 is highly contagious and has caused the worst pandemic on earth in the past 100 years, with the capacity to cause a severe respiratory illness with high mortality. In the absence of an effective treatment or vaccine, SARS-CoV2 has become a global pandemic with nearly 34.14 million cases and 1.018 million deaths as of early October 2020 (https://www.worldometers.info/coronavirus/).
The genome sequence of SARS-CoV2 shares approximately 80% identity with that of SARS-CoV. Furthermore, SARS-CoV2 uses the same cell entry receptor ACE2 as SARS-CoV (2). The result of SARS-CoV2 has emerged a pandemic although they share high similarity in genome sequence as well as the same receptor on host cell membrane. The S gene of SARS-CoV, which encodes for the spike glycoprotein on the viral envelope, recognizes a receptor on the membrane of specific host cells. The spike protein is cleaved into S1 and S2 subunits by the protease during SARS-CoV interaction with the cell surface ACE2 receptor (3,4,5,6).
Recent study showed that human ACE2 is the receptor for SARS-CoV2 by using S gene of pseudovirus system and spike pseudovirus entered 293T/hACE2 cells mainly via endocytosis [7]. The data also suggested that spike glycoprotein of SARS-CoV2 is less stable than that of SARS-CoV. Also, there is no antigen cross reactivity between SARS-CoV and SARS-CoV2 using polyclonal anti-SARS S1 subunit antibodies. The entry of SARS-CoV spike pseudovirions was sufficiently blocked, but the same polyclonal antibodies failed to block SARS-CoV2 infection. In addition, data showing that coronavirus disease 2019 (COVID-19) patients' sera with a limited neutralization effect implicate that recovery from a first infection may not be protective against subsequent infection (7).
The receptor binding domain (RBD 318-513 amino acids) within the S1 subunit of the SARS-CoV spike protein shares 73% identity with that the RBD (307-527 amino acids) of SARS-CoV2 (8), which is crucial for binding to ACE2 (9,10). While the RBD of S1 subunit binds to the peptidase domain (PD) of ACE2 on host cell membrane, the cleaved site of S2 subunit is exposed for further cleavage by host protease, which is essential for SARS-CoV2 infection (3,5,11). A recent study compared the cleavage site of SARS-CoV to that of SARS-CoV2 and found a difference in the cleavage of the S1 and S2 subunits of SARS-CoV and SARS-CoV2, which likely affects the viral infectivity (12). The extracellular domain of the SARS-CoV2 spike protein binds to the PD of ACE2 with a high affinity (13).
Although ACE2 is the known receptor for SARS-CoV2, its primary role in host cells is in processing angiotensin, which controls vasoconstriction and blood pressure. ACE2 is a membrane protein expressed ubiquitously, but mainly lungs, heart, kidneys, and intestine (14,15). The reduction of ACE2 expression in cells may exacerbate cardiovascular diseases (1,16). The claw-like structure of PD in ACE2 and the complex structure of the RBD of SARS-CoV spike glycoprotein is supported by the molecular interactions in the binding sites between the RBD of spike protein and the PD of ACE2 (6,9,10,13,15).
The original report of the receptor for SARS-CoV is unclear because there were no solid biochemical results except immunoprecipitation and immunohistochemistry studies following cell infection (17) albeit there are different crystal structure studies (6,9,10,13,15). The present study with novel mutations in critical RBD of S gene may explain the high pathogenicity of SARS-CoV2 through precise biochemistry result with recombinant spike proteins as well as cell infectivity experiment.
MATERIALS AND METHODS
Ethical statement
This study was approved by the Institutional Review Board of Yeungnam University Medical Center, Korea (approval No. 2020-07-063).
Specimen collection
SARS-CoV2 infection in patients was confirmed by real-time RT-PCR according to the guidelines of Korea Centers for Disease Control & Prevention. All nasopharyngeal swab samples used for the clinical diagnostic tests were collected for further experimental analysis.
RT-PCR of SARS-CoV2 spike gene
Briefly, the synthesis of cDNA from RNA template of COVID-19 patients was performed using the MMLV Reverse Transcription kit (Millipore Sigma, St. Louis, MO, USA) according to the manufacturer's instructions. The forward and reverse primer sequences were used to amplify S gene of SARS-CoV2 are shown in Table 1. Normalization was based on the GAPDH using primers as follows; forward: 5′-ACCACAGTCCATGCCATCAC-3′ and reverse: 5′-TCCACCACCCTGTTGCTGTA-3′.
Table 1. Three pairs of forward and reverse primers used for SARS-CoV2 spike gene PCR amplification.
| Forward and reverse primers | ||
|---|---|---|
| 3,822 bp | ||
| CoV2F-1 | 5′ ATGTTTGTTTTTCTTGTTTTATTGC | |
| CoV2R-1 | 5′ TTATGTGTAATGTAATTTGACTCC | |
| 719 bp | ||
| CoV2F-2 | 5′ AACAATCTTGATTCTAAGGTTGGTGG | |
| CoV2R-2 | 5′ GTCTGAGTCTGATAACTAGCG | |
| 2,010 bp | ||
| CoV2F-3 | 5′ GCCACTAGTCTCTAGTCAGTG | |
| CoV2R-3 | 5′ CCGCCGAGGAGAATTAGTCT | |
The forward and reverse primer sequence were shown in three panels. The expected PCR product size of SARS-CoV2 spike gene was indicated on the top of each panel.
TA cloning and restriction enzyme map
The PCR product of the S gene of SARS-CoV2 was ligated into yT&A cloning vector according to the manufacturer’s instructions (DonginBiotech, Seoul, Korea). The positive plasmid vector (0.8 µg) was digested with Hind III restriction enzyme (7.5 units; Takara, Japan) with provided buffer to release the S gene of SARS-CoV2. The insert of S gene cDNA was detected by 1% agarose (Biobase, Minneapolis, MN, USA) photographed by Vilber lourmat Electronic Ballast transilluminators (Millipore Sigma).
Mini-prep and DNA sequence analysis
The positive clone from colony PCR screening was cultured in 3 ml volume of LB broth for overnight at 37°C. The bacterial cells were used to isolate plasmid DNA according to manufacturer's instructions (Promega, Madison, WI, USA). The plasmid was digested with Hind III restriction enzyme to confirm the insert cDNA of SARS-CoV2 S gene. The positive SARS-CoV2 S gene plasmid was sent for DNA sequencing analysis (Cosmotech, Seoul, Korea).
RESULTS
RT-PCR of SARS-Cov2 S gene to obtain cDNA
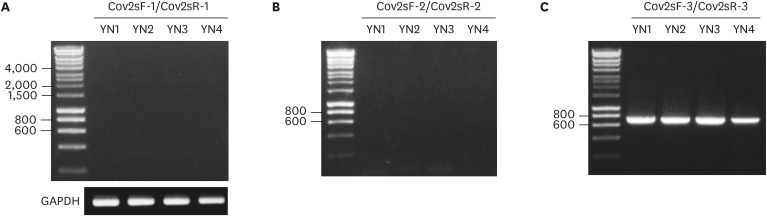
The nasopharyngeal swabs from patients were used to isolate the RNA samples to test SARS-CoV2 infection. The RNA samples from Yeungnam (YN) University Hospital were examined by RT-PCR according to the guidelines of Korea Centers for Disease Control and Prevention. SARS-CoV2 infection was confirmed by ~Ct value as shown in Table 2. We used four positive SARS-CoV2 RNA samples to isolate the S gene. The full length (FL) open reading frame (ORF) was comprised of 3,822 bp that encode 1,273 amino acid residues including the last stop “TAA” codon. The sense and reverse primers were designed (Table 1, upper panel) to amplify the FL S gene of SARS-CoV2. The pair of primer failed to amplify FL spike ORF of 3,822 bp (Fig. 1A) whereas a housekeeping gene GAPDH from the YN patients' RNA was amplified adequately in the bottom panel of Fig. 1A.
Table 2. The up to Ct value of four Korean COVID-19 YN patients.
| Patients | Up to Ct value |
|---|---|
| YN1 | 30.02 |
| YN2 | 21.86 |
| YN3 | 25.40 |
| YN4 | 27.34 |
Four Korean COVID-19 YN patients up to Ct value were shown on right column. YN2 patient was the lowest (21.86 cycles) whereas YN1 patient was the highest (30.02 cycles) by qPCR diagnosis according to the manufacture's instruction.
Figure 1. RT-PCR of SARS-CoV2 spike gene from Korean COVID-19 YN patients. The RNA samples of four Korean COVID-19 YN patients were converted into cDNA as described in Materials and Methods section. The stock of cDNA from four Korean COVID-19 YN patients was used as template to amplify SARS-CoV2 spike gene cDNA. (A) The pair of forward (CoV2sF-1) and reverse (CoV2sR-1) primer was used for PCR to synthesize 3,822 bp band. The PCR of housekeeping gene GAPDH was shown on the bottom. (B) The pair of forward (CoV2sF-2) and reverse (CoV2sR-2) primer was used for PCR to synthesize 719 bp band. (C) The pair of forward (CoV2sF-3) and reverse (CoV2sR-3) primer was used for PCR to generate 2,010 bp band. Representative data from 1 of 3 independent experiments performed are shown.
This finding suggested the presence of an unstable RNA transcript of SARS-CoV2 spike or positive-sense single-stranded RNA (+ssRNA SARS-CoV2) genome was not abundant enough to amplify the FL SARS-CoV2 S gene. Therefore, we focused on the specific mutation region of S gene in which aspartic acid 614 was mutated into glycine (D614G). Additional two pairs of forward and reverse primers were designed (Table 1, middle and bottom panel) to amplify the mutated region of the S gene. The forward CoV2sF-2 and reverse CoV2sR-2 primers successfully amplified a 719 bp PCR product from all four YN patients (Fig. 1B), but the forward CoV2sF-3 and CoV2R-3 primer failed to do it (Fig. 1C). The yield of PCR product in Fig. 1B was correlated to the GAPDH, but it was not corresponded to ~Ct value (Table 2).
Cloning the mutation region of SARS-CoV2 spike cDNA
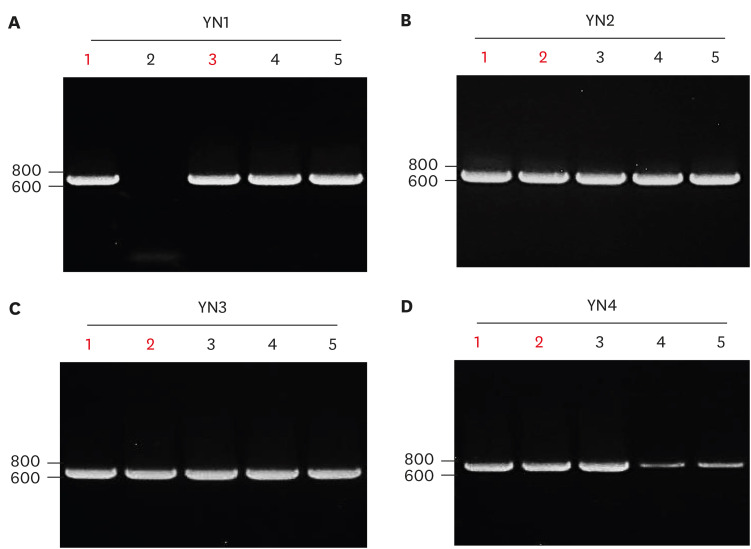
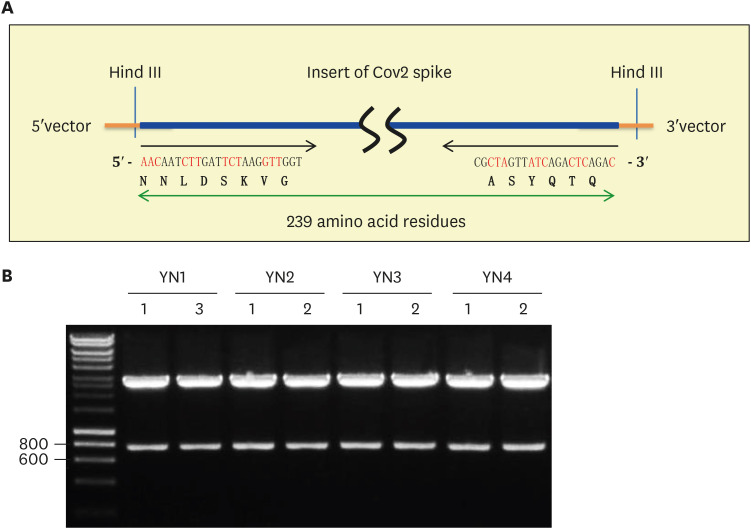
The PCR product of four patients was ligated into TA cloning vector and then the ligated samples were transformed into bacterial competent cells. Five white bacterial colonies from each patient were used to find a positive plasmid. The same primers were used for PCR screening to identify a positive plasmid clone containing SARS-CoV2 S gene cDNA. PCR screening identified different number of positive clones for example four clones of YN1 (Fig. 2A), five clones of YN2 (Fig. 2B), five clones of YN3 (Fig. 2C), and three clones of YN4 patient (Fig. 2D). Two positive plasmids indicated with red number from each patient were selected for the mini prep of plasmid purification (Fig. 2). As shown in Fig. 3A, Hind III restriction enzyme site is in both 15 bp of 5′ upstream and 44 bp of 3′ downstream in SARS-CoV2 spike mutation region. The isolated cDNA plasmid was digested with Hind III restriction enzyme to release the insert of S gene cDNA. The result showed a slightly higher band comparing to the 719 bp PCR product because additional 59 bp of vector sequence was released together with the S gene insert of SARS-CoV2 (Fig. 3B).
Figure 2. T&A cloning of the amplified SARS-CoV2 spike gene PCR product from Korean COVID-19 YN patients. The PCR product of SARS-CoV2 spike gene from (A) YN1, (B) YN2, (C) YN3, and (D) YN4 was ligated into T&A cloning plasmid vector. The same forward (CoV2sF-2) and reverse (CoV2sR-2) primer amplified 719 bp of SARS-CoV2 spike gene PCR product from four YN patients. Representative data from 1 of 2 independent experiments performed are shown.
Figure 3. Restriction map identifying the 719 bp of SARS-CoV2 spike gene insert from Korean COVID-19 YN patients. (A) The diagram depicted restriction enzyme Hind III cleavage site at 5′ and 3′ of SARS-CoV2 spike gene insert containing 239 amino acid residues. The beginning and end of DNA sequence as well as amino acid sequence was shown. (B) As described (A), Hind III restriction enzyme released the 778 bp of SARS-CoV2 spike gene insert, which is slightly larger than the 719 bp PCR product due to the 59 bp of 5′ and 3′ TA vector sequence. All positive clones release 778 bp of SARS-CoV2 spike gene insert. Representative data from 1 of 2 independent experiments performed are shown.
Sequencing data of CoV2 spike cDNA in mutation region
DNA sequencing of the S gene cDNA revealed three mutations in addition to the known D614G mutation. The N-terminal region of the SARS-CoV2 S gene was translated into amino acid sequence and deposited into the NCBI database (GenBank accession number: MW052550). The alignment of four SARS-CoV2 spike amino acid sequences compared to the wild type sequence revealed that there are two additional mutations in the critical RBD and another mutation in subdomain (SD) 2, which is very close to the known mutation residue D614G (Figs. 4). The known mutation (D614G) residue highlighted by light blue with red amino acid residue. New three mutations residues were highlighted by green with red amino acid residue. Eight cysteine residues were marked by yellow as well as the potential glycosylation was indicated by gray (Fig. 4). The abundant cysteine residues are probably involved in an inter- or intra-disulfide bond for the ternary structure of spike glycoprotein. Also, the three potential glycosylation sites may influence structure and stability of spike glycoprotein on lipid envelope of SARS-CoV2. Interestingly, the known mutation site D614G residue including three novel mutation sites are conserved between severe acute respiratory syndrome (SARS) and SARS-CoV2 in Fig. 4B.
Figure 4. The partial 239 Amino acid sequence alignment of SARS-CoV2 spike gene from Korean COVID-19 YN patients comparing to that of wild type. (A) The align of Korean COVID-19 and wild ype SARS-CoV2 spike gene amino acid sequence revealed three new mutations (G504D/V524D/P579L) highlighted by green with red letter. The known mutation (D614G) marked by blue with red letter. Three potential glycosylation sites highlighted by gray and Cysteine was indicated with yellow highlight. (B) The align of SARS-CoV, SARS-CoV2, and Korean COVID-19 YN patients revealed the four mutation sites are conserved between SARS-CoV and SARS-CoV2. The four mutation residues are indicated by yellow with red residue.
Chromatographic DNA sequencing result of CoV2 spike mutation sites
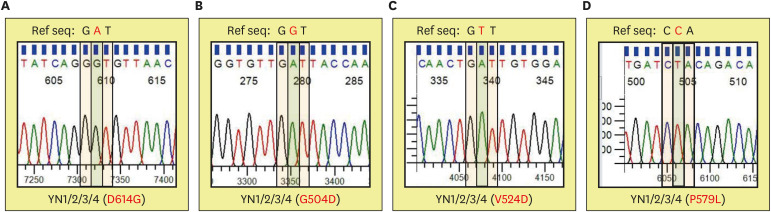
The known single point mutation at D614G in SD2 of spike has been reported by different groups. aspartic acid 614 codon “GAT” (reference sequence) was substituted by glycine 614 codon “GGT” (D614G) by chromatographic DNA sequencing result (Fig. 5A). This known D614G mutation was present in all four patients (Fig. 4). Additional three amino acid residue mutations were analyzed. Interestingly, these mutations were present as common mutation in all four patients (Fig. 4). The chromatographic DNA sequencing result of the four patients showed glycine 504 codon “GGT” (reference sequence) was substituted by aspartic acid 504 codon “GAT” (G504D) (Fig. 5B). Chromatographic DNA sequencing data of the four patients exhibited valine 524 codon “GTT” (reference sequence) that was substituted by aspartic acid 524 codon “GAT” (V524D) (Fig. 5C). The last mutation site was due to proline 579 codon “CCA” (reference sequence) being replaced by leucine 579 codon “CTA” (P579L) in all four patients (Fig. 5A and D).
Figure 5. Chromatographic data of DNA sequencing form Korean COVID-19 YN patients. The representative chromatography of D614G (A), G504D (B), V524D (C), and P579L (D) mutation was shown from the DNA sequencing results of four Korean COVID-19 patients. The codon of reference sequence is on the top replaced by a red letter, which changed amino acid residue indicated at the bottom. Representative data from 1 of 3 independent experiments performed are shown.
DISCUSSION
The COVID-19 pandemic began near the end of 2019 and has caused extraordinary medical, social, and economic destruction in many parts of the world (12). So far there is no specific treatment and vaccine for COVID-19 although there are promising leads on several lines of investigations. Elucidating more precisely of how SARS-CoV2 enters host cells is a priority as disruption of this entry can mitigate the replication and spread of SARS-CoV2. It is fairly well-established that following the cleavage of the SARS-CoV2 envelope spike protein by the host cell protease (3,5,11), and the interaction of RBD to the host cell receptor ACE2, the virus gains entry into cells (4,6,7,9,10,12,13).
A recent study reported a single point mutation in which amino acid residue aspartic acid 614 replaced by glycine (D614G) in S gene, resulting in enhanced infectivity of SARS-CoV2 (18). However, the significance of this mutation is uncertain since the D614G mutation is located in the SD2 – which has no known specific function – rather than the RBD of S gene (Fig. 6). We investigated the RBD of SARS-CoV2 S gene from four Korean COVID-19 patients. The analysis of DNA sequencing revealed the D614G mutation in all four Korean COVID-19 patients (Fig. 4) suggesting that Korean COVID-19 SARS-CoV2 probably came from the same origin.
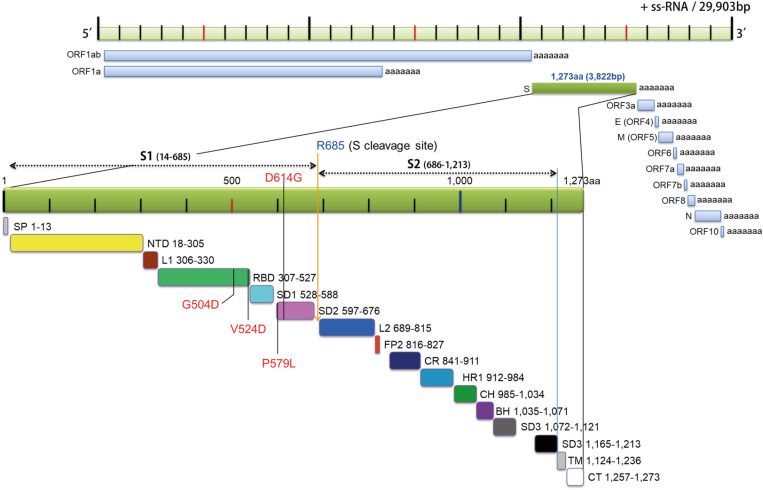
Figure 6. Schematic draw of SARS-CoV2 genome and spike gene. NCBI (accession number: NC_045512) of 29,903 bp SARS-CoV2 genome was depicted physical map in genetics according to twelve ORF gene with poly+A sequence. The third ORF spike gene containing 3,822 bp codes 1,273 amino acid residues including “TAA” stop codon. The spike gene was shown by green bar expanded for sixteens subdomains with or without function. S1 and S2 subdomain cleavage site was indicated by blue letter (R685). The specific mutation sites were indicated by black line with red letter.
SP, signal peptide; NTD, N-terminal domain; L1, loop 1; RBD, receptor binding domain; L2, loop 2; FP, fusion peptide; CR, connected region; HR, heptad repeat; CH, central helix; BH, b-hairpin; TM, transmembrane domain; CT, cytosolic domain.
The D614G mutation has been reported from different regions of the world including continental Europe, Asia, North America, and other continents with fewer number of COVID-19 (18). The D614G mutation in the S gene of SARS-CoV2 dramatically increased after March 21, 2020 compared with before March 1, 2020 [18]. There were many SARS-CoV2 isolates with the D614G mutation in Europe prior to March 1, 2020, but there are not many in China and North America. However, the D614G mutation is dominant all over the world between March 21–30, 2020. What is occurring now? The present study with respiratory samples from Korean COVID-19 patients suggest a rapid progression of SARS-CoV2.
It is interesting that the D614G mutation exists in all four Korean COVID-19 patients. In addition, we found two additional mutations G504D/V524D within the critical RBD domain of S gene (4,6,7,9,10,12,13). A third P579L mutation is located near the known D614G mutation in SD2 but without known specific function. Therefore, it is necessary to investigate the function of different domains in spike gene to explain the significance of the mutations in SARS-CoV2 pathogenicity.
Fig. 6 illustrates the genetic map of SARS-CoV2, which is a positive-sense single-stranded RNA (+ssRNA) virus with 29,903 bp. The genome of SARS-CoV2 consists of twelve ORF encoding an independent transcript that was depicted according to its location in the single-stranded genomic RNA. The S gene is located in the 3rd ORF that contains 3,822 bp encoding for 1,273 amino acid residues (Fig. 6, green bar). The large glycoprotein spike protein is divided by 16 subdomains including a hydrophobic signal peptide and transmembrane domain as described by its physical map in genetics. An exact subdomain was marked by the number of amino acid residue using recent study (8,19).
The progression of SARS-CoV2 in Korean COVID-19 patients was demonstrated by novel mutations in the critical RBD of the S gene of SARS-CoV2. The D614G mutation was indicated on the top of spike gene with red letter. The D614G and P579L mutations present in the SD2 (Fig. 6, pink bar) region of the C-terminus end of the RBD. However, two novel mutations in S gene of Korean COVID-19 in the Korean patients were present in RBD (Fig. 6, green bar), a site that is important for binding to ACE2 receptor on the host cell membrane. There were no mutations in the SD1 region (Fig. 6, light blue) between RBD and SD2. Unfortunately, there is not enough functional study of the SD1 and SD2 regions of S gene. Previous report analyzing the architecture of SARS-CoV2 transcriptome using advance technology sequenced the whole genome of SARS-CoV2 from Korean COVID-19 patients, but this study did not mention the four mutations found in S gene from Korean COVID-19 patients (20).
Studies are underway to identify the precise functional significance of novel mutations in S gene by expressing these mutants in mammalian as well as in bacterial cells. The furin or other host enzyme cleavage site was indicated as S cleavage site on N-terminal of Arg (R) 685. Unlike SARS-CoV2, SARS-CoV does not need proteolytic cleavage to enter host cells suggesting a low infectivity compared to SARS-CoV2 (3,5). Interestingly, the novel P579L mutation is close to S cleavage site that may influence the infectivity of SARS-CoV2 as reported in previous study (12).
Since viruses can proliferate only in live cells, they generally do not want to eliminate the host prior to replication and infection of another host. While viral mutation is generally associated with increasing pathogenicity – through immune evasion or drug resistance – it is important to be cognizant of the teleological concept that viruses also need the host to survive. Therefore, it is necessary to thoroughly investigate whether SARS-CoV2 mutations enhance or decrease viral infectivity and pathogenicity. Future study with S gene mutation eventually defines how virus and human adapt each other as well as helps understand viral mutation is beneficial or deleterious to the host.
ACKNOWLEDGEMENTS
This work was supported by BK21 plus project fund and the National Research Foundation of Korea (NRF-2015R1A2A2A01003472, NRF-2014M3A6A4075058, NRF-2015R1A2A1A15051472). This paper was supported by Konkuk University in 2019. Siyoung Lee was supported by NRF-2019R1I1A1A01057699. Lin-Woo Kang was supported by Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ013284012020) from Rural Development Administration, Republic of Korea.
Abbreviations
- ACE2
angiotensin converting enzyme 2
- Ang
angiotensin
- COVID-19
coronavirus disease 2019
- FL
full length
- ORF
open reading frame
- PD
peptidase domain
- RBD
receptor binding domain
- S
spike
- SARS-CoV2
severe acute respiratory syndrome coronavirus 2
- SD
subdomain
- ssRNA
positive-sense single-stranded RNA
- YN
Yeung Nam
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Lee JH, Bae S, Yum H, Lee Y, Chan E, Yu L, Azam T, Kim YD, Yeom SC, Kang LW, Shin KC, Shim S, Kim S2.
- Data curation: Kim S1, Lee JH, Lee S, Nguyen T, Hwang J, Kim H, Jhun H, Lee Y, Kang LW, Shin KC, Shim S, Kim S2.
- Formal analysis: Kim S1, Lee JH, Lee S, Hwang J, Kim H, Choi YO, Hong J, Bae S, Jhun H, Yum H, Lee Y, Chan E, Yu L, Kim YD, Yeom SC, Kang LW, Shin KC, Kim S2.
- Funding acquisition: Lee S, Shim S, Kim S2.
- Investigation: Kim S2.
- Methodology: Kim S1, Lee JH, Nguyen T, Hwang J, Kim H, Jhun H, Lee Y, Kang LW, Kim S2.
- Project administration: Kim S2.
- Resources: Lee JH, Lee Y, Shin KC.
- Software: Kim S2.
- Supervision: Kim S2.
- Validation: Lee JH, Lee S, Hwang J, Bae S, Lee Y, Kang LW, Shin KC, Kim S2.
- Visualization: Kim S1, Lee JH, Lee Y, Kang LW, Shin KC, Kim S2.
- Writing - original draft: Kim S2.
- Writing - review & editing: Chan E, Azam T, Kim S2.
Kim S1, Sinae Kim; Kim S2, Soohyun Kim.
References
- 1.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, Oliveira-dos-Santos AJ, da Costa J, Zhang L, Pei Y, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 2.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallagher TM, Buchmeier MJ. Coronavirus spike proteins in viral entry and pathogenesis. Virology. 2001;279:371–374. doi: 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simmons G, Zmora P, Gierer S, Heurich A, Pöhlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song W, Gui M, Wang X, Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lokman SM, Rasheduzzaman M, Salauddin A, Barua R, Tanzina AY, Rumi MH, Hossain MI, Siddiki AM, Mannan A, Hasan MM. Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: a computational biology approach. Infect Genet Evol. 2020;84:104389. doi: 10.1016/j.meegid.2020.104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 10.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donoghue M, Wakimoto H, Maguire CT, Acton S, Hales P, Stagliano N, Fairchild-Huntress V, Xu J, Lorenz JN, Kadambi V, et al. Heart block, ventricular tachycardia, and sudden death in ACE2 transgenic mice with downregulated connexins. J Mol Cell Cardiol. 2003;35:1043–1053. doi: 10.1016/s0022-2828(03)00177-9. [DOI] [PubMed] [Google Scholar]
- 15.Li G, He X, Zhang L, Ran Q, Wang J, Xiong A, Wu D, Chen F, Sun J, Chang C. Assessing ACE2 expression patterns in lung tissues in the pathogenesis of COVID-19. J Autoimmun. 2020;112:102463. doi: 10.1016/j.jaut.2020.102463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oudit GY, Crackower MA, Sarao R, Yagil Y, Chappell M, Backx PH, Penninger JM. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Circulation. 2002;106:315–315. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]