Abstract
Depression is the single largest contributor to non-fatal health loss and affects 322 million people globally. The clinical heterogeneity of this disorder shows biological correlates and it makes the personalization of antidepressant prescription an important pillar of treatment. There is increasing evidence of genetic overlap between depression, other psychiatric and non-psychiatric disorders, which varies across depression subtypes. Therefore, the first step of clinical evaluation should include a careful assessment of psychopathology and physical health, not limited to previously diagnosed disorders. In part of the patients indeed the pathogenesis of depression may be strictly linked to inflammatory and metabolic abnormalities, and the treatment should target these as much as the depressive symptoms themselves. When the evaluation of the symptom and drug tolerability profile, the concomitant biochemical abnormalities and physical conditions is not enough and at least one pharmacotherapy failed, the genotyping of variants in CYP2D6/CYP2C19 (cytochromes responsible for antidepressant metabolism) should be considered. Individuals with altered metabolism through one of these enzymes may benefit from some antidepressants rather than others or need dose adjustments. Finally, if available, the polygenic predisposition towards cardio-metabolic disorders can be integrated with non-genetic risk factors to tune the identification of patients who should avoid medications associated with this type of side effects. A sufficient knowledge of the polygenic risk of complex medical and psychiatric conditions is becoming relevant as this information can be obtained through direct-to-consumer genetic tests and in the future it may provided by national health care systems.
Keywords: Major depressive disorder, Antidepressive agents, Genetics, Precision medicine.
INTRODUCTION
The total number of people living with depression in the world is 322 million, and it increased by 18.4% between 2005 and 2015, partly as a consequence of the increase in the age groups at which depression is more prevalent (55–74 years). Globally, depressive disorders are ranked as the single largest contributor to non-fatal health loss (7.5% of all years lived with disability [YLD] in 2015, for a total of over 50 million YLD) [1]. Depression, however, is also a major contributor to suicide, that is responsible for the death of a person every 40 seconds in the world [1].
Antidepressants are considered the first line treatment for moderate-severe major depressive disorder (MDD) [2]; however, about one third of patients shows symptom remission to the first antidepressant treatment and about one third do not reach remission after multiple antidepressant treatments [3]. There is still poor guidance in prescribing guidelines about the possible strategies to personalize antidepressant prescription, often resulting in the flattening of prescription towards a few options. Personalized prescription could improve remission rates and therefore recovery, that is the main goal of MDD treatment, as it is associated with better functioning and reduced risk of depressive relapse [4]. The availability of a large number of antidepressant medications (around 40 compounds) makes difficult to have a complete knowledge of their pharmacological properties and the scientific literature about them, therefore an easily accessible and practical guideline to evaluate at least the main criteria to personalize prescription would be very helpful for clinicians. Previous reviews provided suggestions about the personalization of antidepressant choice based on the individual symptom profile, in addition to drug tolerability profile, personal and family history, medical and psychiatric comorbidities, and concomitant medications [5]. However, a synthesis of clinical and genetic information to guide antidepressant choice at the individual level is lacking in the literature. The inter-individual variability in treatment response and side effects is indeed partly dependent from genetic variation, as it was demonstrated to be heritable [6]. Different symptom patterns were reported to have partially distinct genetic profiles and different overlap with psychiatric and immune-metabolic traits [7-9], suggesting the hypothesis of a link between the genetic factors influencing MDD pathogenesis and clinical manifestation and those modulating response to antidepressants. Therefore, clinical symptoms can be interpreted as signs of the individual genetic factors involved in disease pathogenesis and guide the choice of treatment beyond the approach of just prescribing an antidepressant that contrasts the individual clinical symptoms. This means that it is possible to extend the baseline evaluations applied to choose which antidepressant to prescribe without performing any genotyping. Despite there is no definitive evidence, currently genotyping of variants in cytochrome P450 (CYP450) genes is indeed suggested in patients who did not respond or tolerate at least one previous pharmacotherapy [10]. After the discussion of the interpretation of the individual symptom profile and the use of CYP450 genotyping to personalize antidepressant prescription, the present review will address the possible use of information derived from genome-wide data, a genotyping technology that has become relatively cheap and spread not only in research settings, but also as a direct-to-consumer product that provides information on ancestry, health conditions and genetic-based recommendations for lifestyles [11]. As of early 2019, more than 26 million people have taken at-home DNA tests using this type of product [11], suggesting that it will be more and more important for clinicians to provide adequate guidance to patients who want to know about the validity of the possible implications for their health. The use of clinical and genetic information to personalize treatment in MDD is presented in a stepped way that reflects their hierarchy and facilitate their application in clinical practice.
STEPS FOR PRECISION MEDICINE IN PSYCHIATRY
Step 1: Individual Symptom Profile from the Clinical and Genetic Perspective
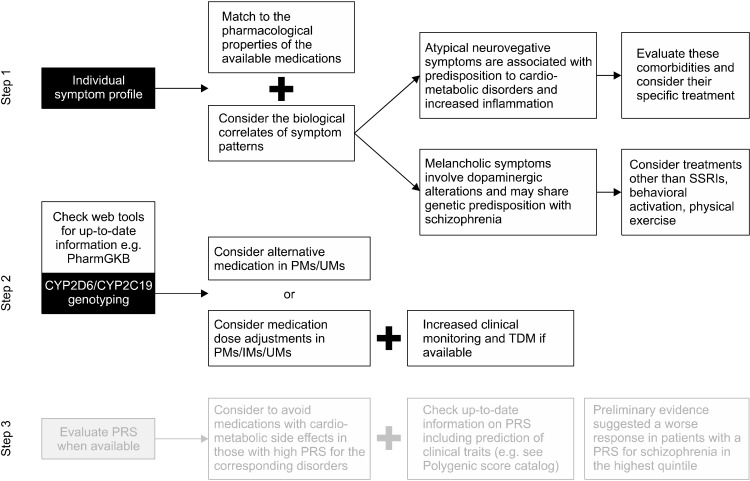
The individual profile of depressive symptoms has been suggested as an important criterion to guide antidepressant choice, as the available antidepressants show different pharmacological properties that make them different in terms of the most common side effects and targeted symptoms. This topic was deeply discussed in a previous review [5]; in this review, we discuss some examples that show the connection between the clinical-pharmacological and genetic perspective on depressive symptom profiles (Fig. 1) and represent cases when selective serotonin reuptake inhibitors (SSRIs), which are considered as the first choice in MDD [12], may have a lower chance of success.
Fig. 1.
Proposed steps for the personalization of treatment in depression. Step 3 is shadowed as its possible clinical utility has not been investigated yet in the treatment of patients with depression.
PRS, polygenic risk scores; TDM, therapeutic drug monitoring; SSRIs, selective serotonin reuptake inhibitors; PMs, poor metabolizers; IMs, intermediate metabolizers; UMs, ultrarapid metabolizers.
MDD with atypical neurovegetative symptoms: the importance of assessing and treating immune-metabolic abnormalities
The first example is represented by patients with MDD who report increased weight/appetite and hypersomnia, i.e., reversed neurovegetative symptoms. Based on the pharmacological properties of the available antidepressants, in order to reduce the risk of weight gain, the clinician should avoid medications with anti-histaminergic (e.g., mirtazapine), anti-5-HT2C (serotonin receptor 2C) (e.g., mirtazapine, paroxetine) and anti-alpha-adrenergic effects (e.g., tricyclic antidepressants or TCAs) [13]. Further, antidepressants with anti-cholinergic (e.g., TCAs) and anti-histaminergic properties should be avoided in order to not aggravate hypersomnia [5]. From a biological perspective, the results of genetic studies suggest that cases of MDD with reversed neurovegetative symptoms show higher genetic overlap with immune-metabolic traits compared with patients without these symptoms [7-9]. In detail, patients with these atypical depressive symptoms were demonstrated to have a higher genetic overlap with the genetic factors modulating C-reactive protein (CRP), body max index (BMI), and triglycerides [7-9]. These effects seem to be mostly dependent from the presence of the symptom weight gain during depression [7]. Over-weight and obesity have been causally associated with the risk of MDD and implicated in treatment-resistant depression (TRD) [14,15], possibly through the induction of pro-inflammatory and oxidative changes as well as dysregulations in dopaminergic neurotransmission [16]. There-fore, patients with reversed neurovegetative symptoms, particularly weight gain, should be evaluated for the presence of immune-metabolic alterations (e.g., increased plasma levels of CRP, cholesterol, triglycerides, glucose) and other risk factors for cardio-metabolic disorders (e.g., hypertension), as these are likely to contribute both to the pathogenesis of depressive symptoms and medical comorbidities in this group. The prevention and treatment of immune-metabolic abnormalities should be considered as a fundamental part of treatment, since it is likely required for a complete remission of depressive symptoms other than for promoting physical health. Interestingly, medications approved for the treatment of metabolic abnormalities such as proliferator-activated receptor (PPAR) agonists and statins were shown to have anti-inflammatory, neurotrophic and antidepressant effects. The PPAR agonists pioglitazone and rosiglitazone were demonstrated to have antidepressant effects in four open-label studies and in three out of four randomized controlled trials (RCTs) in patients with major depression [17]. Statins may have adjunctive antidepressant effects when used as add-on treatment to SSRIs according to preliminary RCTs [18]. BMI is positively correlated with CRP levels that were found to be higher in TRD compared to treatment-responsive depression [19]. Inflammation is associated with decreased dopaminergic neurotransmission and dopamine in turn regulates the immune system [20]; furthermore, in pro-inflammatory states, the metabolism of tryptophan is shifted from the production of serotonin to the neurotoxic metabolite kynurenine [19]. Consistently, treatment with bupropion (dopamine reuptake inhibitor)-SSRI combination was associated with lower depressive symptoms compared with SSRI monotherapy in patients with high CRP levels at baseline [20]. Similarly, pramipexole, a dopamine agonist with evidence of efficacy in TRD, has been shown to inhibit the production of interleukin 17 (IL-17) [21]. Therefore, antidepressants with dopaminergic activity should be preferred to serotonergic antidepressants in this group of patients. Another group of drugs under investigation is represented by compounds directly targeting inflammatory factors; in particular, a RCT found that the tumor necrosis factor (TNF) inhibitor infliximab was effective in reducing depressive symptoms in TRD when baseline CRP concentration was greater than 5 mg/L [22]. Finally, it is important to remember the anti-inflammatory effects of physical exercise, that was demonstrated to be especially beneficial in patients with higher levels of TNF-α at baseline [21].
MDD with melancholic symptoms: deficits in the reward system and genetic overlap with schizophrenia
Symptoms of melancholic depression, namely anhedonia, weight/appetite loss, insomnia, and psychomotor disturbances, are also relevant for guiding treatment choice in MDD. Anhedonia is characterized by non-reactive and pervasive impairment of the capacity to experience or anticipate pleasure; it was associated with poor antidepressant response, particularly to SSRIs, and it often persists as residual symptom. In depressed patients with prominent anhedonia, clinicians should consider treatment strategies such as behavioral activation, physical exercise, and/or antidepressant combination (consider antidepressants with dopaminergic or noradrenergic activity) or augmentation strategies [23]. Consistently with the hypothesis of a central role of dopamine in reward processing, variants in the gene coding for the dopamine D2 receptor (DRD2) and that coding for the metabotropic glutamate receptor GRM5, implicated in striatal dopamine depletion, were associated with anhedonia [24]. Anhedonia and the other cited symptoms of melancholic depression were demonstrated to share genetic factors with schizophrenia [9,24,25] and alcohol dependence [7,25]. In another study, high genetic loading for variants associated with schizophrenia was associated with both poor antidepressant response and a trend of more frequent melancholic MDD [26], suggesting that genetic loci associated with schizophrenia may mediate both the risk of melancholic symptoms and poor response to treatment, though the presence and direction of causal relationships cannot be assumed based on these findings. However, these results support the appropriateness of considering alternative treatment strategies in patients with melancholic symptoms, as suggested above. Comorbidity of MDD and alcohol use disorders is also associated with poorer clinical outcomes [27] and previous studies showed that symptoms of melancholic depression, particularly psychomotor disturbances, are associated with substance use disorders compared to other depressive subtypes [28,29], though other factors such as the inclusion of bipolar patients may affect the direction of this association [30]. Therefore, this represents a relevant variable to assess in this group, as it influences treatment and prognosis. Another interesting finding is that the genetic factors modulating predisposition to alcohol dependence seem to be largely distinct from those implicated in the frequency of alcohol consumption. As a matter of fact, alcohol use disorders have a positive genetic correlation with MDD, particularly with MDD showing typical neurovegetative symptoms, while the genetics of alcohol consumption is negatively correlated with atypical neurovegetative symptoms and cardio-metabolic disorders [7,31], suggesting the possibility of broad metabolic abnormalities in patients with atypical depressive symptoms.
Step 2: Cytochrome P450 Genotyping
If the initial treatment choice, based on the individual symptom profile, drug tolerability and other clinical factors, is not effective or tolerated, clinicians should consider the genotyping of variants in CYP2D6 and CYP2C19 genes (Fig. 1). These two CYP450 isoenzymes are largely responsible for the metabolism of most antidepressants and the corresponding genes are highly polymorphic, i.e., they have a number of known genetic variants, many altering the transcription/function of the coded enzyme [32]. The type and number of variants in genes coding for CYP450 enzymes identify four main metabolizing groups, namely: 1) extensive metabolizers (EMs, no genetic variants altering enzyme function), 2) poor metabolizers (PMs, carrying two inactive alleles), 3) intermediate metabolizers (IMs, with one inactive or two partially active alleles), and 4) ultrarapid metabolizers (UMs, usually carrying gene duplications) [33]. Based on studies demonstrating a relationship between genotype-based metabolizing group and drug/metabolite plasma levels, dose adjustments were calculated in PMs and UMs, and over 10 antidepressants have prescribing recommendations in terms of drug choice and dose, with different level of recommendation [32]. Overall, guidelines suggest to consider an antidepressant drug not metabolized by a defective CYP450 enzyme (PMs or IMs) or CYP450 enzyme with increased activity (UMs), in order to avoid severe side effects or treatment failure, respectively [32]. Alternatively, the drug dose should be adjusted and increased monitoring applied, using also therapeutic drug monitoring (TDM) if available. For some TCAs, namely amitriptyline and nortriptyline, and for the SSRI paroxetine, these recommendations have a strong level of evidence according to the guideline curated by the Clinical Pharmacogenetics Implementation Consortium [32]. In other cases, such as for citalopram and escitalopram (mostly metabolized by CYP2C19), the level of recommendation is moderate, in line with the evidence that the risk of side effects is moderately increased in CYP2C19 PMs treated with standard doses (particularly, sexual side effects), but also symptom improvement and symptom remission are better compared to EMs [34]. In the case of antidepressants associated with the risk of QTc prolongation such as citalopram and escitalopram [35], the adjustment of the dose seems a good and prudent practice in PMs, as well as a baseline ECG and assessment of cardiac comorbidities. To summarize, if there is no strong evidence against the selection of a certain antidepressant based on the individual metabolizing status for CYP2D6/CYP2C19 and the clinical history/symptom profile supports the choice of that drug, it seems a reasonable option to go ahead and adjust the drug dose according to guidelines, inform the patient about potential risks, increase clinical monitoring and use TDM if available.
Though the available data suggests that patients who did not respond or tolerate at least one previous pharmacotherapy are the group mostly likely to benefit from CYP2D6/CYP2C19 genotyping [10,36], there is still no definitive evidence, and different recommendations can be found. As a matter of fact, according to the French National Network of Pharmacogenetics (Réseau national de pharmacogénétique [RNPGx]), CYP2D6/CYP2C19 geno-typing is advisable before initiating an antidepressant treat-ment, especially in patients with a high risk of toxicity [37].
CYP2D6/CYP2C19 genotyping is not available in most clinical services and it is often not reimbursed by national health care systems, therefore if a patient is willing to fund their own genetic testing, commercial pharmacogenetic tests are an option. These tests have a median cost of CAD 499 (range from CAD 199 to CAD 2,310) according to a recent review referred to the tests available in Canada (however, prices may not be the same in other countries) [38]. When considering this possibility, clinicians should keep in mind that usually commercial pharmacogenetic tests include also genetic variants not endorsed by guidelines [39], and two main points should be considered to guide the choice: 1) the test has to include all the genetic variants endorsed by guidelines (see [32]), pay particular attention if the patient is of non-European ancestry, as the frequency of variants in CYP2D6/CYP2C19 genes varies across ethnicities, sometimes of a great extent [40]; 2) the report has to be sufficiently detailed and show the results per each gene and variant tested, therefore the level of evidence of each of them according to guidelines can be considered when choosing the antidepressant.
Step 3: Possible Utility of Polygenic Risk Scores
As genome-wide genotyping is becoming cheaper and cheaper, direct-to-consumer companies offering genetic testing services is exploding. The price range is GBP 50–129 (in the United Kingdom, prices may vary in other countries) and they provide different type of information, often including health conditions. As of early 2019, more than 26 million people have taken one of such tests, with numbers that are increasing exponentially [11]. Genome-wide arrays provide genotypes of hundreds of thousands or millions of genetic variants throughout the genome, and these data can be used to calculate polygenic risk scores (PRS) for psychiatric and non-psychiatric traits, a measure reflecting the genetic loading conferred by the combined set of risk variants associated with a trait (e.g., antidepressant non-response, risk of a certain disease) (Fig. 1). While PRS did not show strong prediction of psychiatric diagnoses or response to antidepressants [26,41,42], similarly to the PRS of non-psychiatric traits, they may helpful when used in combination with non-genetic risk factors, though evidence in this regard is still lacking. However, a proper knowledge about the interpretation and possible clinical implications of PRS is going to be important in clinical settings, since the exponential diffusion of direct-to-consumer products. Currently, PRS were shown to add information to non-genetic risk factors for coronary artery disease (CAD), type 2 diabetes (T2D), breast and prostate cancers, and Alzheimer’s disease [43], the first two being especially relevant in MDD, since the high comorbidity rates [44,45]. The PRS of CAD was shown to add prediction power to the traditional risk factors such as blood pressure, cholesterol levels and smoking habits, and to the identification of patients more likely to benefit from statins (those with the highest CAD PRS); interestingly, disclosing CAD genetic risk to patients when deciding whether to initiate statin therapy resulted in improved benefits [43]. Therefore, the availability of CAD PRS in a patient with MDD could represent an additional information to guide treatment choice, as the prescription of medications associated with higher risk of cardio-metabolic side effects should be considered carefully in case of high CAD PRS, and include of course the evaluation of non-genetic risk factors for CAD. The hazard for incident T2D was found to be 3.45 times higher in the highest T2D PRS quintile compared with the lowest quintile, after adjusting for body mass index and other known predictors. Adding PRS to other variables in a prediction model for 5-year T2D risk resulted in continuous net reclassification improvement of 0.32 (95% confidence interval: 0.21–0.44) [46]. Therefore, the PRS of T2D has a potential utility in guiding antidepressant prescription and clinical monitoring, similarly to what we discussed for CAD PRS, though there is still no evidence about what PRS threshold should warn against the prescription of medications associated with the risk of cardio-metabolic side effects.
The evidence supporting a possible utility of PRS in predicting antidepressant response is poor [26,47]. Preliminary results support the hypothesis that the odds ratio (OR) of non-response is 2.23 (95% confidence interval: 1.21–4.10) in the highest quintile of schizophrenia PRS compared to the lowest quintile [26]. A study in a larger sample reported consistent results [6]. Higher genetic loading for schizophrenia was shown to predict poorer response to lithium in bipolar disorder [48], extending the previous findings that were referred to MDD only. Another preliminary finding of interest suggests that patients with MDD in the lowest quintile of schizophrenia PRS may respond better to antidepressant monotherapy compared to antidepressant augmentation with atypical antipsychotics, while patients in the highest quintile of schizophrenia PRS responded poorly to both therapeutic strategies [26].
DISCUSSION
The treatment of MDD is still largely based on a trial and error approach or local/personal practices, with the risk of suboptimal response and common lack of symptom remission, that results in residual functional impairment and risk of relapse/recurrence. This review proposes a possible step by step approach to guide clinicians on the criteria and actions to take into consideration when prescribing treatments to patients with MDD, with particular attention to cases when SSRI may have a lower chance of efficacy and general comorbidities are contributing to disease pathogenesis (Fig. 1). The proposed steps are organized in a hierarchical order: the first one is the base of the process, representing the essential evaluation that should be performed in all patients, and it consists in the clinical assessment of the individual symptom profile, that can be matched with the pharmacological properties of the available medications [5], but also interpreted in the light of the genetic heterogeneity of depressive symptoms. This last point is particularly relevant because different depressive symptom profiles were linked to only partially overlapping genetic factors, for example the genetic correlation between depression with atypical and typical neurovegetative symptoms is “only” 0.54 (standard error = 0.14, where a genetic correlation of one indicates complete genetic overlap) [7], and these MDD subtypes show genetic overlap with distinct traits, with potential implica-tions on treatment. Genetic studies have indeed demonstrated that the genetic variants modulating the risk of MDD are shared to various extent with other psychiatric and non-psychiatric traits [49], and in this review we discussed that MDD includes clinically and genetically heterogeneous subtypes. When the first step of the proposed process is not successful, and despite at least one adequate treatment in terms of duration and dose the patient continues to suffer from depression, or the patient presents non-tolerated side effects, clinicians should consider the genotyping of CYP2C19/CYP2D6 variants. This can provide complementary information to guide the following prescription decisions, as individuals with altered drug metabolism may respond or tolerate better some antidepressants rather than others. If even the addition of prescribing recommendations based on CYP2C19/CYP2D6 genotyping does not help the identification of an effective and tolerated treatment, there are no other criteria with evidence of benefits in guiding the choice of specific medications in MDD, but a range of possibilities with variable evidence of efficacy in TRD [12]. However, as step 3, we suggested the evaluation of the possible clinical implications of PRS, in case they are available (i.e., if the patient decided to take one of the direct-to-consumer genetic tests and is willing to share the results with the treating physician). PRS have currently limited clinical applications and in fields of medicine other than psychiatry, though they may be helpful to identify patients at risk of cardio-metabolic disorders and avoid medications with this type of side effects in this group. Genomic medicine is in a phase of rapid evolution, therefore the predictive accuracy of PRS and the corresponding clinical applications may change rapidly in the next future. There are useful web tools to stay updated with the latest information in this regard; for example the Polygenic Score (PGS) Catalog includes information on over 200 PRS for 100 traits, including MDD and schizophrenia, and it provides information on PRS metrics, such as the AUC (area under the receiver operating characteristic curve [ROC]), an estimation of the percentage of individuals that are correctly classified as cases by the PRS [50]. Despite there is still no PRS available for antidepressant response, since the insufficient power of the previous samples [47], recent results on a larger sample size suggest that PRS significantly predicts symptom improvement and remission to antidepressant treatment, thought the variance explained is still very low [6]. Another interesting web tool to guide individuals in the interpretation of their genome-wide genotyping data is impute.me, a resource that provides up-to-date information on the effect of single genetic variants and PRS for a range of traits, currently including psychiatric disorders and response to clozapine in schizophrenia [51]. It is also possible for the user to upload their genome-wide data and know their PRS for each of the available traits and where their PRS value lies compared to a reference population. At least a basic knowledge of these resources and the way to use them will be more and more relevant in the clinical practice, as the number of individuals taking at-home-DNA tests is increasing exponentially and guidance by their treating physicians on the interpretation of results is advisable in order to avoid unneeded worrying, unnecessary clinical testing or treat-ments. The rapid drop of genotyping and DNA sequencing costs is also going to facilitate their inclusion among services provided by national health care systems, starting from individuals with rarer and severe disorders, then possibly extending it to larger fractions of the population including healthy individuals, as effective preventive strategies for most diseases may become available.
Acknowledgments
Chiara Fabbri is supported by Fondazione Umberto Veronesi.
Footnotes
Conflicts of Interest
Alessandro Serretti is or has been consultant/speaker for: Abbott, Abbvie, Angelini, Astra Zeneca, Clinical Data, Boheringer, Bristol Myers Squibb, Eli Lilly, GlaxoSmithKline, Innovapharma, Italfarmaco, Janssen, Lundbeck, Naurex, Pfizer, Polifarma, Sanofi, Servier. Chiara Fabbri has no potential conflicts of interest.
Author Contributions
Conceptualization: Alessandro Serretti. Writing−original draft: Chiara Fabbri. Writing−review & editing: Alessandro Serretti.
References
- 1.World Health Organization, author. Depression and other common mental disorders: global health estimates [Internet] World Health Organization; Geneva: 2017. [cited at 2020 Jul 20]. https://apps.who.int/iris/bitstream/handle/10665/254610/WHO-MSD-MER-2017.2-eng.pdf;jsessionid=2A52412EC4FD7E2707B9228C4ED61604?sequence=1 . [Google Scholar]
- 2.Kennedy SH, Lam RW, McIntyre RS, Tourjman SV, Bhat V, Blier P, et al. CANMAT Depression Work Group, author. Canadian Network for Mood and Anxiety Treatments (CANMAT) 2016 clinical guidelines for the management of adults with major depressive disorder: section 3. pharmacological treatments. Can J Psychiatry. 2016;61:540–560. doi: 10.1177/0706743716659417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huynh NN, McIntyre RS. What are the implications of the STAR*D trial for primary care? A review and synthesis. Prim Care Companion J Clin Psychiatry. 2008;10:91–96. doi: 10.4088/PCC.v10n0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaynes BN, Warden D, Trivedi MH, Wisniewski SR, Fava M, Rush AJ. What did STAR*D teach us? Results from a large-scale, practical, clinical trial for patients with depression. Psychiatr Serv. 2009;60:1439–1445. doi: 10.1176/ps.2009.60.11.1439. [DOI] [PubMed] [Google Scholar]
- 5.Serretti A. The present and future of precision medicine in psychiatry: focus on clinical psychopharmacology of antide-pressants. Clin Psychopharmacol Neurosci. 2018;16:1–6. doi: 10.9758/cpn.2018.16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pain O, Hodgson K, Trubetskoy V, Baune B, Biernacka J, Fabbri C, et al. Investigating the common genetic basis of antidepressant response. Abstract NR 57; 27th World Congress of Psychiatric Genetics (WCPG); Oct 26-31, 2019; Los Angeles, USA. pp. S91–S92. [DOI] [Google Scholar]
- 7.Badini I, Coleman JRI, Hagenaars SP, Hotopf M, Breen G, Lewis CM, et al. Depression with atypical neurovegetative symptoms shares genetic predisposition with immuno-metabolic traits and alcohol consumption. Psychol Med. 2020 doi: 10.1017/S0033291720002342. doi: 10.1017/S0033291720002342. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milaneschi Y, Lamers F, Peyrot WJ, Baune BT, Breen G, Dehghan A, et al. CHARGE Inflammation Working Group and the Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, author. Genetic association of major depression with atypical features and obesity-related immunometabolic dysregulations. JAMA Psychiatry. 2017;74:1214–1225. doi: 10.1001/jamapsychiatry.2017.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milaneschi Y, Lamers F, Peyrot WJ, Abdellaoui A, Willemsen G, Hottenga JJ, et al. Polygenic dissection of major depression clinical heterogeneity. Mol Psychiatry. 2016;21:516–522. doi: 10.1038/mp.2015.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.International Society of Psychiatric Genetics, author. Genetic Testing Statement [Internet] International Society of Psychiatric Genetics; Brentwood (TN): 2019. [cited at 2019 Mar 14]. https://ispg.net/genetic-testing-statement/ [Google Scholar]
- 11.Chatzou M. The ultimate guide on how to offer genetic testing services or boost your current offerings in no time [Internet] Lifebit Blog; London: 2019. [cited 2020 Jul 21]. https://blog.lifebit.ai/2019/09/04/ultimate-guide-how-to-offer-genetic-testing-services/ [Google Scholar]
- 12.Taylor DM, Barnes TRE, Young AH. The Maudsley prescribing guidelines in psychiatry. 13th ed. Wiley-Blackwell; Hoboken: 2018. [Google Scholar]
- 13.Nihalani N, Schwartz TL, Siddiqui UA, Megna JL. Weight gain, obesity, and psychotropic prescribing. J Obes. 2011;2011:893629. doi: 10.1155/2011/893629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyrrell J, Mulugeta A, Wood AR, Zhou A, Beaumont RN, Tuke MA, et al. Using genetics to understand the causal influence of higher BMI on depression. Int J Epidemiol. 2019;48:834–848. doi: 10.1093/ije/dyy223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvi SJ, Grima E, Tan M, Rotzinger S, Lin P, Mcintyre RS, et al. Treatment-resistant depression in primary care across Canada. Can J Psychiatry. 2014;59:349–357. doi: 10.1177/070674371405900702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemp DE, Schinagle M, Gao K, Conroy C, Ganocy SJ, Ismail-Beigi F, et al. PPAR-γ agonism as a modulator of mood: proof-of-concept for pioglitazone in bipolar depression. CNS Drugs. 2014;28:571–581. doi: 10.1007/s40263-014-0158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colle R, de Larminat D, Rotenberg S, Hozer F, Hardy P, Verstuyft C, et al. PPAR-γ agonists for the treatment of major depression: a review. Pharmacopsychiatry. 2017;50:49–55. doi: 10.1055/s-0042-120120. [DOI] [PubMed] [Google Scholar]
- 18.Köhler-Forsberg O, Gasse C, Berk M, Østergaard SD. Do statins have antidepressant effects? CNS Drugs. 2017;31:335–343. doi: 10.1007/s40263-017-0422-3. [DOI] [PubMed] [Google Scholar]
- 19.Chamberlain SR, Cavanagh J, de Boer P, Mondelli V, Jones DNC, Drevets WC, et al. Treatment-resistant depression and peripheral C-reactive protein. Br J Psychiatry. 2019;214:11–19. doi: 10.1192/bjp.2018.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jha MK, Minhajuddin A, Gadad BS, Greer T, Grannemann B, Soyombo A, et al. Can C-reactive protein inform antidepressant medication selection in depressed outpatients? Findings from the CO-MED trial. Psychoneuroendocrinology. 2017;78:105–113. doi: 10.1016/j.psyneuen.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jha MK, Trivedi MH. Personalized antidepressant selection and pathway to novel treatments: clinical utility of targeting inflammation. Int J Mol Sci. 2018;19:233. doi: 10.3390/ijms19010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raison CL, Rutherford RE, Woolwine BJ, Shuo C, Schettler P, Drake DF, et al. A randomized controlled trial of the tumor necrosis factor antagonist infliximab for treatment-resistant depression: the role of baseline inflammatory biomarkers. JAMA Psychiatry. 2013;70:31–41. doi: 10.1001/2013.jamapsychiatry.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perna G, Alciati A, Daccò S, Grassi M, Caldirola D. Personalized psychiatry and depression: the role of sociodemographic and clinical variables. Psychiatry Investig. 2020;17:193–206. doi: 10.30773/pi.2019.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward J, Lyall LM, Bethlehem RAI, Ferguson A, Strawbridge RJ, Lyall DM, et al. Novel genome-wide associations for anhedonia, genetic correlation with psychiatric disorders, and polygenic association with brain structure. Transl Psychiatry. 2019;9:327. doi: 10.1038/s41398-019-0635-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorp JG, Marees AT, Ong JS, An J, MacGregor S, Derks EM. Genetic heterogeneity in self-reported depressive symptoms identified through genetic analyses of the PHQ-9. Psychol Med. 2019 doi: 10.1017/S0033291719002526. doi: 10.1017/S0033291719002526. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Fanelli G, Benedetti F, Kasper S, Kautzky A, Zohar J, Souery D, et al. Higher polygenic risk scores for schizophrenia may be suggestive of treatment non-response in major depressive disorder. [cited 2020 Jul 21];medRxiv.2020.01.15.20017699 [Preprint] 2020 doi: 10.1101/2020.01.15.20017699. http://medrxiv.org/lookup/doi/10.1101/2020.01.15.20017699 . [DOI] [PubMed]
- 27.DeVido JJ, Weiss RD. Treatment of the depressed alcoholic patient. Curr Psychiatry Rep. 2012;14:610–618. doi: 10.1007/s11920-012-0314-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marmorstein NR. Associations between subtypes of major depressive episodes and substance use disorders. Psychiatry Res. 2011;186:248–253. doi: 10.1016/j.psychres.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leventhal AM, Francione Witt C, Zimmerman M. Associa-tions between depression subtypes and substance use dis-orders. Psychiatry Res. 2008;161:43–50. doi: 10.1016/j.psychres.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brailean A, Curtis J, Davis K, Dregan A, Hotopf M. Characteristics, comorbidities, and correlates of atypical depression: evidence from the UK Biobank Mental Health Survey. Psychol Med. 2020;50:1129–1138. doi: 10.1017/S0033291719001004. [DOI] [PubMed] [Google Scholar]
- 31.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10:1499. doi: 10.1038/s41467-019-09480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.PharmGKB, author. Clinical guideline annotations [Internet] PharmGKB; Stanford (CA): 2020. [cited 2020 Jul 21]. https://www.pharmgkb.org/guidelineAnnotations . [Google Scholar]
- 33.Porcelli S, Fabbri C, Spina E, Serretti A, De Ronchi D. Genetic polymorphisms of cytochrome P450 enzymes and antidepressant metabolism. Expert Opin Drug Metab Toxicol. 2011;7:1101–1115. doi: 10.1517/17425255.2011.597740. [DOI] [PubMed] [Google Scholar]
- 34.Fabbri C, Tansey KE, Perlis RH, Hauser J, Henigsberg N, Maier W, et al. Effect of cytochrome CYP2C19 metabolizing activity on antidepressant response and side effects: meta-analysis of data from genome-wide association studies. Eur Neuropsycho-pharmacol. 2018;28:945–954. doi: 10.1016/j.euroneuro.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Howland RH. A critical evaluation of the cardiac toxicity of citalopram: part 1. J Psychosoc Nurs Ment Health Serv. 2011;49:13–16. doi: 10.3928/02793695-20111011-01. [DOI] [PubMed] [Google Scholar]
- 36.Greden JF, Parikh SV, Rothschild AJ, Thase ME, Dunlop BW, DeBattista C, et al. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: a large, patient- and rater-blinded, randomized, controlled study. J Psychiatr Res. 2019;111:59–67. doi: 10.1016/j.jpsychires.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Picard N, Boyer JC, Etienne-Grimaldi MC, Barin-Le Guellec C, Thomas F, Loriot MA French National Network of Pharmaco-genetics (RNPGx), author Pharmacogenetics-based personalized therapy: levels of evidence and recommendations from the French Network of Pharmacogenetics (RNPGx) Therapie. 2017;72:185–192. doi: 10.1016/j.therap.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Maruf AA, Fan M, Arnold PD, Müller DJ, Aitchison KJ, Bousman CA. Pharmacogenetic testing options relevant to psychiatry in Canada: options de tests pharmacogénétiques pertinents en psychiatrie au Canada. Can J Psychiatry. 2020;65:521–530. doi: 10.1177/0706743720904820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabbri C, Zohar J, Serretti A. Pharmacogenetic tests to guide drug treatment in depression: comparison of the available testing kits and clinical trials. Prog Neuropsychopharmacol Biol Psychiatry. 2018;86:36–44. doi: 10.1016/j.pnpbp.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Bousman C, Maruf AA, Müller DJ. Towards the integration of pharmacogenetics in psychiatry: a minimum, evidence-based genetic testing panel. Curr Opin Psychiatry. 2019;32:7–15. doi: 10.1097/YCO.0000000000000465. [DOI] [PubMed] [Google Scholar]
- 41.So HC, Sham PC. Exploring the predictive power of polygenic scores derived from genome-wide association studies: a study of 10 complex traits. Bioinformatics. 2017;33:886–892. doi: 10.1093/bioinformatics/btw745. [DOI] [PubMed] [Google Scholar]
- 42.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambert SA, Abraham G, Inouye M. Towards clinical utility of polygenic risk scores. Hum Mol Genet. 2019;28:R133–R142. doi: 10.1093/hmg/ddz187. [DOI] [PubMed] [Google Scholar]
- 44.Wu Q, Kling JM. Depression and the risk of myocardial infarction and coronary death: a meta-analysis of prospective cohort studies. Medicine (Baltimore) 2016;95:e2815. doi: 10.1097/MD.0000000000002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holt RI, de Groot M, Golden SH. Diabetes and depression. Curr Diab Rep. 2014;14:491. doi: 10.1007/s11892-014-0491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Läll K, Mägi R, Morris A, Metspalu A, Fischer K. Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet Med. 2017;19:322–329. doi: 10.1038/gim.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García-González J, Tansey KE, Hauser J, Henigsberg N, Maier W, Mors O, et al. Pharmacogenetics of antidepressant response: a polygenic approach. Prog Neuropsychopharmacol Biol Psychiatry. 2017;75:128–134. doi: 10.1016/j.pnpbp.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 48.Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, et al. International Consortium on Lithium Genetics (ConLi+Gen), author Association of polygenic score for schizophrenia and HLA antigen and inflammation genes with response to lithium in bipolar affective disorder: a genome-wide association study. JAMA Psychiatry. 2018;75:65–74. doi: 10.1001/jamapsychiatry.2017.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, author. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert SA, Gil L, Jupp S, Ritchie SC, Xu Y, Buniello A, et al. The polygenic score catalog: an open database for reproducibility and systematic evaluation. [cited 2020 Jul 21];medRxiv. 2020.05.20.2010821 [Preprint] 2020 doi: 10.1038/s41588-021-00783-5. http://medrxiv.org/lookup/doi/10.1101/2020.05.20.20108217 . [DOI] [PMC free article] [PubMed]
- 51.Folkersen L, Pain O, Ingason A, Werge T, Lewis CM, Austin J. Impute.me: an open-source, non-profit tool for using data from direct-to-consumer genetic testing to calculate and interpret polygenic risk scores. Front Genet. 2020;11:578. doi: 10.3389/fgene.2020.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]