Abstract
Introduction
Venezuela and Colombia both adopted measures of containment early in response to the COVID-19 pandemic. However, Venezuela's ongoing humanitarian crisis has decimated its health care system, and forced millions of Venezuelans to flee through its porous border with Colombia. The extensive shared border, and illegal cross-border transit through improvised trails between the two countries are major challenges for public health authorities. We report the first SARS-CoV-2 genomes from Venezuela, and present a snapshot of the SARS-CoV-2 epidemiologic landscape in the Colombian-Venezuelan border region.
Methods
We sequenced and assembled viral genomes from total RNA extracted from nasopharyngeal (NP) clinical specimens using a custom reference-based analysis pipeline. Three assemblies obtained were subjected to typing using the Phylogenetic Assignment of Named Global Outbreak LINeages ‘Pangolin’ tool. A total of 376 publicly available SARS-CoV-2 genomes from South America were obtained from the GISAID database to perform comparative genomic analyses. Additionally, the Wuhan-1 strain was used as reference.
Results
We found that two of the SARS-CoV-2 genomes from Venezuela belonged to the B1 lineage, and the third to the B.1.13 lineage. We observed a point mutation in the Spike protein gene (D614G substitution), previously reported to be associated with increased infectivity, in all three Venezuelan genomes. Additionally, three mutations (R203K/G204R substitution) were present in the nucleocapsid (N) gene of one Venezuelan genome.
Conclusions
Genomic sequencing demonstrates similarity between SARS-CoV-2 lineages from Venezuela and viruses collected from patients in bordering areas in Colombia and from Brazil, consistent with cross-border transit despite administrative measures including lockdowns. The presence of mutations associated with increased infectivity in the 3 Venezuelan genomes we report and Colombian SARS-CoV-2 genomes from neighboring borders areas may pose additional challenges for control of SARS-CoV-2 spread in the complex epidemiological landscape in Latin American countries. Public health authorities should carefully follow the progress of the pandemic and its impact on displaced populations within the region.
Keywords: SARS-CoV-2, COVID-19, Colombia, Venezuela, Border, Coronavirus
Highlights
-
•
SARS-CoV-2 genomes obtained from Venezuela reveal the presence of B1 and B.1.13 lineages, suggesting two separate introductions.
-
•
All Venezuelan genomes carried a D614G substitution in the spike (S) protein, which has been linked to increased infectivity.
-
•
Three substitutions in the nucleocapsid (N) gene were additionally identified in one of the examined genomes.
1. Introduction
As Severe Acute Respiratory Syndrome Coronavirus −2 (SARS-CoV-2) spreads throughout the Western hemisphere, Latin America has become an epicenter for the Coronavirus Disease 2019 (COVID-19) pandemic. Since the report on March 13, 2020 of the first two cases diagnosed with COVID-19 in Venezuela, SARS-CoV-2 has spread rapidly across the country. The areas of highest transmission flank the Colombian-Venezuelan border, where the reported incidence reaches 47,9% with a 34,2% overall mortality (ESRI Venezuela, 2020).
Venezuela's ongoing humanitarian crisis has had a severe impact on its health care system: the country has experienced a mass exodus of medical personnel, shortages of treatment and supplies, as well as a systematic dismantling of public health infrastructure (Daniels, 2020). In addition, the increased poverty and violence, plus the abandonment of all epidemiological surveillance programs, has set the stage for the re-emergence of vaccine-preventable and vector-borne diseases (Grillet et al., 2019; Paniz-Mondolfi et al., 2019). The Venezuelan public health system cannot withstand current autochthonous threats, let alone emerging infectious agents such as SARS-CoV-2. Furthermore, the political and economic turmoil in Venezuela has precipitated one of the largest refugee crises witnessed in the hemisphere. This has resulted in the establishment of disease corridors to neighboring countries---particularly Colombia, which houses 1.4 million Venezuelan migrants (Daniels, 2020; Torres and Castro, 2019). Despite early implementation of lockdown and border restrictions starting in March 2020, control of cross-border migration has remained a challenge due to the length (approximately 2219 km) of the Venezuelan-Colombian border, and the unregulated transit of individuals through illegal trails known as “trochas”.
As of July 8th 2020, the majority of COVID-19 cases in Venezuela have been reported from three border states, Apure, Táchira and Zulia. (“Estadisticas Venezuela|COVID-19 en Venezuela, 2020”).
Here we report sequences for three of five SARS-CoV-2-positive samples, representing the first viral genomes from Venezuela, and providing a snapshot of the epidemiological landscape across the Colombian-Venezuelan border.
2. Methods
2.1. Ethical statement
The Colombian National Institute of Health (INS) is defined as the reference laboratory in Colombia. When a public health emergency occurs as is the COVID19, the national law 9–1979, decrees 786–1990 and 2323–2006, authorizes the INS to use the biological material and associated epidemiological information without informed consent, including the anonymous disclosure of results. This study was performed following the Declaration of Helsinki and its later amendments. The data of the patients was anonymized which do not represent any risk.
2.2. Patients, sampling and demographic data
Newly arrived migrants from Venezuela meeting case-definition criteria established by the Colombian Ministry of Health and Social Protection were screened for SARS-CoV-2 infection at different hospitals and healthcare centers in Norte de Santander and Bolivar Departments of Colombia between March 31st and May 1st, 2020. Molecular detection of SARS-CoV-2 in nasopharyngeal swab specimens in viral transport media (NP-VTM) was performed using the Berlin Charité protocol. SARS-CoV-2 -positive specimens from five individuals who were tested within 24 h of their arrival in Colombia were referred for further characterization. Four of the five individuals already were symptomatic at the time of arrival from Venezuela, and one was a close contact of a confirmed COVID-19 patient. Complete viral genomes were generated from specimens from three of these five cases. The patients, aged 30 (♂), 42 (♂) and 56 (♀) years, came from different regions, Cucuta, Norte de Santander, and Bolivar, (Fig. 1A). Sequencing was unsuccessful for the other 2 specimens, most likely due to insufficient target material.
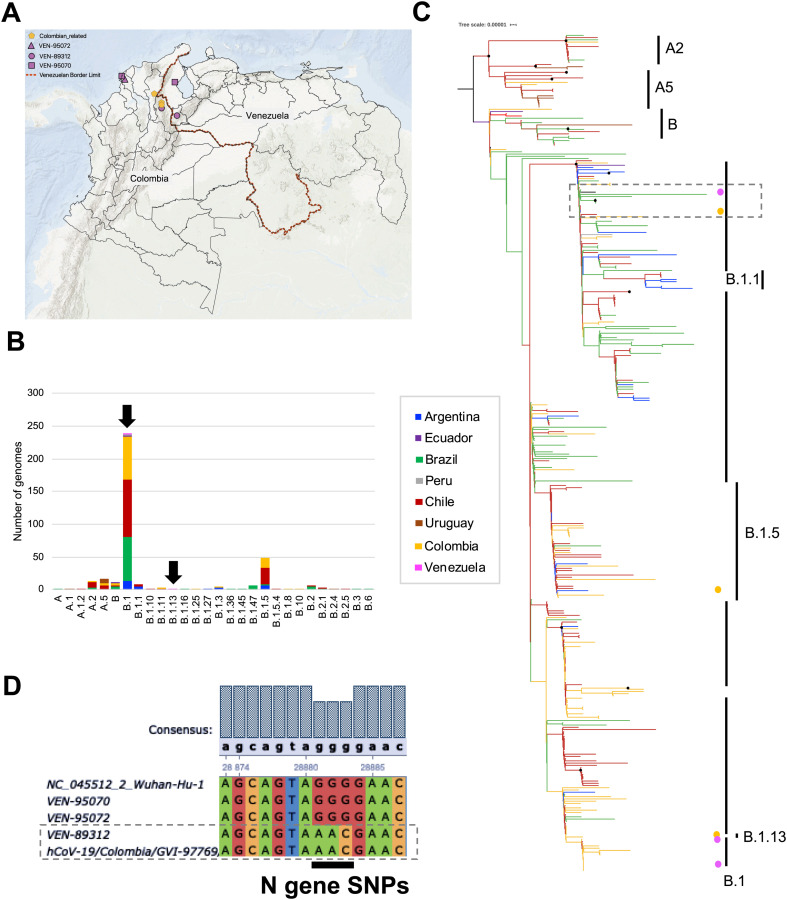
Fig. 1.
Regional comparative genomic analysis of SARS-CoV-2. A. Geographical distribution of the sequences from Venezuelan genomes and the available Colombian genomes analyzed in this study. The pink symbols indicate the Venezuelan patients identified in Colombia with their respective Venezuelan state origin. B. Stacked bar plot of the number of genomes per lineage determined using Phylogenetic Assignment of Named Global Outbreak LINeages ‘Pangolin’ tool. Three SARS-CoV-2 genomes from Venezuela were compared with 376 assemblies from other 7 South American countries (Argentina, Brazil, Chile, Colombia, Ecuador, Peru and Uruguay) using the publicly available GISAID EpiCoV™ database (https://www.gisaid.org/). Frequencies are discriminated by country of origin. The Venezuelan lineages are identified by black arrows. C. Maximum likelihood tree built in IQtree shows the phylogenetic relationships between genomes from Venezuela (pink dots) and the closest Colombian regions (yellow dots) with other South American genomes. The branches were colored according to the country of origin, using the color code of A panel. The clustering of the most frequent pangolin lineages (n > 10) is represented on the right side. The black dots represent highly supported nodes. D. Multiple alignment of the Nucleocapsid gene showing the substitutions found using the Wuhan-1 sequence as reference.
2.3. Phylogenetic analysis
We sequenced and assembled viral genomes from total RNA extracted from NP viral transport medium (VTM) clinical specimens. Sample preparation for sequencing was done using whole-genome amplification with custom designed tiling primers and the Artic Consortium protocol (https://artic.network/ncov-2019) with a read length of 150, and the modifications previously described (Gonzalez-Reiche et al., 2020). Amplicon libraries were prepared using the Nextera XT DNA Sample Preparation kit (Illumina, cat. FC-131-1096), as recommended by the manufacturer that then were sequenced by Illumina sequencing.
The initial bioinformatic analysis was previously described (Gonzalez-Reiche et al., 2020). Briefly, the primer sequences were trimmed using cutadapt version 2.8, and were aligned to SARS-CoV-2 genome MN908947.3 using minimap2 version 2.17-r941, to call a consensus sequence using Pilon v1.23, allowing for all variant types (single nucleotide variants, small insertions/deletions, large insertions/deletions or block substitution events, and local mis-assemblies). Regions with less than 10 fold-coverage were masked and unamplified regions at the end of the viral genome were also removed. Finally, consensus sequences were annotated using Prokka v1.14.6 and a custom SARS-CoV-2 reference annotation file. The complete genomes were typed using the Phylogenetic Assignment of Named Global Outbreak LINeages ‘Pangolin’ tool (Rambaut et al., 2020).
A total of 376 publicly available SARS-CoV-2 genomes encompassing the lineage diversity from South America were downloaded from the GISAID EpiCoV™ database (https://www.gisaid.org/) for comparative genomic analyses (Table S1). These sequences were aligned in MAFFT (Katoh et al., 2018), using the Wuhan-1 strain (NC_045512.2) as reference. The complete data set was subjected to the same typing scheme using Pangolin tool. Thus far, 28 pangolin lineages have been reported for SARS-CoV2 strains circulating in South American countries (Fig. 1B), with B1 as the predominant lineage representing 62.9% of the total reported genomes. Four other lineages include B.1.5 (12.5%), A.5 (4.5%), A.2 and B (3.4% each) account for an additional 23.8%. Each of the remaining 15 lineages accounted for fewer than 10 genomes, with several having only a single representative. A time-scaled maximum likelihood (ML) phylogeny based on TreeTime built in IQTREE (Rambaut et al., 2016; Sagulenko et al., 2018) revealed that there was no clustering by originating country in the reconstruction, although a general clustering by pangolin lineages was observed (Fig. 1C). These analyses are in agreement with the simultaneous circulation of SARS-CoV-2 lineages from different geographical origins.
A detailed screening of single-nucleotide polymorphisms (SNPs) in important open reading frames (ORFs) of SARS-CoV-2 was then conducted, and substitutions in Spike and Nucleocapsid sequences were evaluated. The alignment was inspected using Ugene (http://ugene.net/). The regions of interest were exported considering the ORFs described for the reference strain Wuhan-1 (NC_045512.2), as previously described in NCBI: https://www.ncbi.nlm.nih.gov/nuccore/?term = Severe + acute + respiratory + syndrome + coronavirus +2 + isolate + Wuhan-Hu-1.
3. Results
An average of 113,821.3 reads were obtained per sample reaching 960.52× of average depth of coverage in the genomes analyzed. We found that two of the SARS-CoV-2 genomes obtained (VEN-89312 and VEN-95072) were identical and belonged to the B1 lineage, while the third (VEN-95070) belonged to the B.1.13 lineage, suggesting two separate introductions (da Candido et al., 2020; Ramirez et al., 2020). Phylogenetic analysis revealed that two of the Venezuelan SARS-CoV-2 genomes (VEN-95070 and VEN-95072) closely resembled genomes from neighboring Colombia (Cesar and Norte de Santander Departments) (Ramirez et al., 2020) while the third (VEN-89312) was related to genomes from Brazil (Fig. 1C) (da Candido et al., 2020).
The three Venezuelan genomes carried a G-to-A point mutation at position 23,403 resulting in a D614G substitution in the spike (S) protein. This mutation characterizes the B.1 lineages and has been associated with enhanced viral entry into host cells, and potentially with increasing infectivity and transmissibility (Bhattacharyya et al., 2020; Korber et al., 2020). Additionally, we identified three substitutions in the nucleocapsid (N) gene of VEN-89312 changing GGG-to-AAC at positions 28,616–28,618 resulting R203K/G204R substitutions according to the whole-genome position after removing the 5'UTR. These R203K/G204R substitutions, which have been reported previously in other South American genomes, were absent from the two other Venezuelan genomes we sequenced. The alignment is shown highlighting the SNPs found in the N gene (Fig. 1D) (Crooks et al., 2004).
4. Discussion
Herein, we report for the first time three genome sequences of SARS-CoV-2 from patients in the Colombian-Venezuelan border. Despite the small number of samples, this data provides valuable initial information about potential transmission routes and vulnerabilities, particularly in the context of government limitations on access to data related to SARS-CoV-2 in Venezuela. Access to additional genomic information will be needed to enable robust conclusions regarding the spread of SARS-CoV-2 across the Colombian-Venezuelan border.
The presence of mutations previously reported to be associated with increased infectivity and transmissibility in the genomic sequences we determined for SARS-CoV-2 from both Venezuela and neighboring border regions of Colombia may represent an additional challenge for control of the COVID-19 pandemic in Latin America. The specter of a SARS-CoV-2 variant with increased infectivity further complicates the already-complex epidemiological landscape with its chronic limitations of biomedical understaffing, poor healthcare infrastructure, limited or inadequate diagnostic capacities, and poor compliance of the general population with disease containment measures (Miller et al., 2020).
Following the first report of SARS-CoV-2 in Brazil in late February 2020 (Andrus et al., 2020; Rodriguez-Morales et al., 2020), SARS-CoV-2 has spread rapidly across the region, resulting in more than 7 million cases as of September 21st, 2020, with Brazil and Colombia reporting the highest burden of disease (“Epidemic Diseases - Cumulative suspected and confirmed COVID-19 cases reported by countries and territories in the Americas, 2020”).
Although Colombia and Venezuela both implemented early lockdown and containment strategies since March 10th and March 17th, respectively, (“CORONAVIRUS (COVID-19), 2020”, “Presidente Maduro anuncia que este martes todo el país entra en cuarentena social • Ministerio del Poder Popular para Relaciones Exteriores, 2020”), according to current official records there have been over 770,500 COVID-19 cases in Colombia and over 66,600 cases in Venezuela (“CORONAVIRUS (COVID-19), 2020”, “Estadisticas Venezuela|COVID-19 en Venezuela, 2020”). However, these numbers have to be contextualized because: (i) Colombia has exclusively performed state-of-the-art RT-PCR-based SARS-CoV-2 diagnosis whereas Venezuelan authorities have relied mostly on serological testing that are considerably less specific (Chia et al., 2020) and less useful than molecular methods for acute case detection; (ii) Venezuela has precarious epidemiological surveillance and contact tracing systems; and (iii) Venezuela has reduced international traffic through airports secondary to extensive departure of multiple airlines.
The concurrent humanitarian crisis has forced millions of Venezuelans to flee to neighboring countries ---mainly Colombia--- seeking economic and social stability (Torres and Castro, 2019; Tuite et al., 2018). Venezuelans with ongoing health issues also travel to Colombia to obtain high-quality healthcare such as surgical procedures and hemodialysis (Daniels, 2020). However, massive Venezuelan migration has resulted in an unprecedented infectious disease exodus, representing one of the most concerning public health threats in the region (Grillet et al., 2019; Torres and Castro, 2019). COVID-19 has deepened the situation and has prompted xenophobia and further marginalization of Venezuelan migrants and refugees at the Venezuelan-Colombian border (Daniels, 2020).
The shared presence of B lineages in SARS-CoV-2 from Venezuela and Colombia reinforces the close interactions of persons living in border regions and the difficulty of containment across a porous border. Similarly, the detection of the B.1.13 lineage, only previously described in cases in Spain, England and Australia (Batty et al., 2020) further underlines the rapid global spread of SARS-CoV-2 through interconnected populations. Additionally, the presence of substitution D614G in the spike protein of the three viruses from patients residing in the current hotspots of COVID-19 in Venezuela may correlate with the reported increased infectivity (Korber et al., 2020) observed in SARS-CoV-2-infected patients in the state of Zulia (“Zulia suma 15 muertes confirmadas por coronavirus y seis no reportadas - Efecto Cocuyo, 2020”).
An important limitation to our study is that the small number of genomes currently available from Venezuela could potentially result in sampling bias. Given the difficulty in obtaining samples, the extent to which our findings truly reflect Venezuela's overall phylogenetic landscape remains to be determined. Future studies are needed to expand the SARS-CoV-2 genome repertoire in Venezuela and related areas, and to enable better understanding of the interplay between genotype and phenotype, and their relevance for disease surveillance and containment. Additionally, collection and sequencing of SARS-CoV-2 from all geographical regions (Colombia and Venezuela) will be needed to understand the true spread between these two neighbors.
5. Conclusion
The Venezuelan humanitarian and refugee crisis, coupled with uncontrolled migration across the Colombian-Venezuelan border, is a devastating reminder of the potential effects of infectious disease spillover on the already vulnerable public health systems of neighboring countries. As SARS-CoV-2 continues to spread across Latin America, public health authorities and the international community should carefully follow the impact of the pandemic on displaced populations. Intensive efforts are urgently needed to help minimize the impact of Venezuela's crisis on the COVID-19 pandemic crisis. Future efforts to obtain additional viral genomes will be essential to fully uncover the genomic epidemiology of SARS-CoV-2 in the Colombian-Venezuelan border.
Funding
This work was supported by the University of Glasgow, Scottish Funding Council and the Global Challenges Research Fund (GCRF) and GCRF Research Network EP/T003782/1.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104616.
Appendix A. Supplementary data
Supplementary Table S1. GISAID accession numbers of the 376 SARS-CoV-2 genomes used in this study.
References
- Andrus J.K., Evans-Gilbert T., Santos J.I., Guzman M.G., Rosenthal P.J., Toscano C., Valenzuela M.T., Siqueira M., Etienne C., Breman J.G. Perspectives on battling COVID-19 in countries of Latin America and the Caribbean. Am. J. Trop. Med. Hyg. 2020;1–4 doi: 10.4269/ajtmh.20-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty E.M., Kochakarn T., Panthan B., Kumpornsin K., Jiaranai P., Wangwiwatsin A., Kotanan N., Jaruampornpan P., Watthanachockchai T., Rakmanee K., Sensorn I., Sungkanuparph S., Pasomsub E., Chookajorn T., Chantratita W. Genomic surveillance of SARS-CoV-2 in Thailand reveals mixed imported populations, a local lineage expansion and a virus with truncated ORF7a. medRxiv. 2020 doi: 10.1101/2020.05.22.20108498. 2020.05.22.20108498. [DOI] [Google Scholar]
- Bhattacharyya C., Das C., Ghosh A., Singh A.K., Mukherjee S., Majumder P.P., Basu A., Biswas N.K. Global spread of SARS-CoV-2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. bioRxiv. 2020 doi: 10.1101/2020.05.04.075911. 2020.05.04.075911. [DOI] [Google Scholar]
- Chia W.N., Tan C.W., Foo R., Kang A.E.Z., Peng Y., Sivalingam V., Tiu C., Ong X.M., Zhu F., Young B.E., Chen M.I.C., Tan Y.-J., Lye D.C., Anderson D.E., Wang L.-F. Serological differentiation between COVID-19 and SARS infections. Emerg. Microb. Infect. 2020:1–23. doi: 10.1080/22221751.2020.1780951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CORONAVIRUS (COVID-19) WWW Document. 2020. https://www.minsalud.gov.co/salud/publica/PET/Paginas/Covid-19_copia.aspx (accessed 7.8.20)
- Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Candido D.S., Claro I.M., de Jesus J.G., de Souza W.M., Moreira F.R.R., Dellicour S., Mellan T.A., du Plessis L., Pereira R.H.M., da Sales F.C.S., Manuli E.R., Theze J., de Almeida L., Menezes M.T., Voloch C.M., Fumagalli M.J., de Coletti T.M., Silva C.A.M., Ramundo M.S., Amorim M.R., Hoeltgebaum H., Mishra S., Gill M., Carvalho L.M., Buss L.F., Prete C.A., Ashworth J., Nakaya H., da Peixoto P.S., Brady O.J., Nicholls S.M., Tanuri A., Rossi A.D., Braga C.K.V., Gerber A.L., Guimaraes A.P., Gaburo N., Alencar C.S., de Ferreira A.C.S., Lima C.X., Levi J.E., Granato C., Ferreira G.M., da Francisco R.S., Granja F., Garcia M.T., Moretti M.L., Perroud M.W., Castineiras T.M.P.P., Lazari C.D.S., Hill S.C., de Santos A.A.S., Simeoni C.L., Forato J., Sposito A.C., Schreiber A.Z., Santos M.N.N., Sa C.Z., Souza R.P., Resende Moreira L.C., Teixeira M.M., Hubner J., Leme P.A.F., Moreira R.G., Nogueira M.L., Ferguson N., Costa S.F., Proenca-Modena J.L., Vasconcelos A.T., Bhatt S., Lemey P., Wu C.-H., Rambaut A., Loman N.J., Aguiar R.S., Pybus O.G., Sabino E.C., Faria N.R. Evolution and epidemic spread of SARS-CoV-2 in Brazil. Science. 2020;2020 doi: 10.1126/science.abd2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J.P. Venezuelan migrants “struggling to survive” amid COVID-19. Lancet (Lond. Engl.) 2020;395:1023. doi: 10.1016/S0140-6736(20)30718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epidemic Diseases - Cumulative suspected and confirmed COVID-19 cases reported by countries and territories in the Americas WWW Document. 2020. https://ais.paho.org/phip/viz/COVID19Table.asp (accessed 6.26.20)
- Estadisticas Venezuela|COVID-19 en Venezuela. 2020. https://covid19.patria.org.ve/estadisticas-venezuela/ [WWW Document] (accessed 7.8.20)
- Gonzalez-Reiche A.S., Hernandez M.M., Sullivan M.J., Ciferri B., Alshammary H., Obla A., Fabre S., Kleiner G., Polanco J., Khan Z., Alburquerque B., van de Guchte A., Dutta J., Francoeur N., Melo B.S., Oussenko I., Deikus G., Soto J., Sridhar S.H., Wang Y.-C., Twyman K., Kasarskis A., Altman D.R., Smith M., Sebra R., Aberg J., Krammer F., García-Sastre A., Luksza M., Patel G., Paniz-Mondolfi A., Gitman M., Sordillo E.M., Simon V., van Bakel H. Introductions and early spread of SARS-CoV-2 in the New York City area. Science. 2020 doi: 10.1126/science.abc1917. eabc1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet M.E., Hernández-Villena J.V., Llewellyn M.S., Paniz-Mondolfi A.E., Tami A., Vincenti-Gonzalez M.F., Marquez M., Mogollon-Mendoza A.C., Hernandez-Pereira C.E., Plaza-Morr J.D., Blohm G., Grijalva M.J., Costales J.A., Ferguson H.M., Schwabl P., Hernandez-Castro L.E., Lamberton P.H.L., Streicker D.G., Haydon D.T., Miles M.A., Acosta-Serrano A., Acquattela H., Basañez M.G., Benaim G., Colmenares L.A., Conn J.E., Espinoza R., Freilij H., Graterol-Gil M.C., Hotez P.J., Kato H., Lednicky J.A., Martinez C.E., Mas-Coma S., Morris J.G., Navarro J.C., Ramirez J.L., Rodriguez M., Urbina J.A., Villegas L., Segovia M.J., Carrasco H.J., Crainey J.L., Luz S.L.B., Moreno J.D., Noya Gonzalez O.O., Ramírez J.D., Alarcón-de Noya B. Venezuela’s humanitarian crisis, resurgence of vector-borne diseases, and implications for spillover in the region. Lancet Infect. Dis. 2019;19:e149–e161. doi: 10.1016/S1473-3099(18)30757-6. [DOI] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2018;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C., Group, S.C.-19 G Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020 doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.J., Loaiza J.R., Takyar A., Gilman R.H. Covid-19 in latin america: novel transmission dynamics for a global pandemic? PLoS Negl. Trop. Dis. 2020;14:3–7. doi: 10.1371/JOURNAL.PNTD.0008265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniz-Mondolfi A.E., Tami A., Grillet M.E., Márquez M., Hernández-Villena J., Escalona-Rodríguez M.A., Blohm G.M., Mejías I., Urbina-Medina H., Rísquez A., Castro J., Carvajal A., Walter C., López M.G., Schwabl P., Hernández-Castro L., Miles M.A., Hotez P.J., Lednicky J., Morris J.G., Crainey J., Luz S., Ramírez J.D., Sordillo E., Llewellyn M., Canache M., Araque M., Oletta J. Resurgence of vaccine-preventable diseases in Venezuela as a regional public health threat in the Americas. Emerg. Infect. Dis. 2019;25:625–632. doi: 10.3201/eid2504.181305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presidente Maduro anuncia que este martes todo el país entra en cuarentena social • Ministerio del Poder Popular para Relaciones Exteriores, 2020.Presidente Maduro anuncia que este martes todo el país entra en cuarentena social • Ministerio del Poder Popular para Relaciones Exteriores WWW Document. 2020. http://mppre.gob.ve/2020/03/16/Venezuela-cuarentena-social/ (accessed 6.26.20)
- Rambaut A., Lam T.T., Carvalho L.M., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly path-O-gen) Virus Evol. 2016;2:1–7. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., Hill V., O’Toole Á., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv. 2020 doi: 10.1101/2020.04.17.046086. 2020.04.17.046086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J.D., Florez C., Munoz M., Hernandez C., Castillo A., Gomez S., Rico A., Pardo L., Barros E.C., Castaneda S., Ballesteros N., Martinez D., Vega L., Jaimes J., Cruz-Saavedra L., Patino L.H., Teheran A., Gonzalez-Reiche A., Hernandez M., Sordillo E., Simon V., van Bakel H., Paniz-Mondolfi A. The arrival and spread of SARS-CoV2 in Colombia. J. Med. Virol. 2020;2020 doi: 10.1002/jmv.26393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Morales A.J., Gallego V., Escalera-Antezana J.P., Méndez C.A., Zambrano L.I., Franco-Paredes C., Suárez J.A., Rodriguez-Enciso H.D., Balbin-Ramon G.J., Savio-Larriera E., Risquez A., Cimerman S. COVID-19 in Latin America: the implications of the first confirmed case in Brazil. Travel Med. Infect. Dis. 2020;35:101613. doi: 10.1016/j.tmaid.2020.101613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko P., Puller V., Neher R.A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:1–9. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J.R., Castro J.S. Venezuela’s migration crisis: a growing health threat to the region requiring immediate attention. J. Travel Med. 2019;26 doi: 10.1093/jtm/tay141. [DOI] [PubMed] [Google Scholar]
- Tuite A.R., Thomas-Bachli A., Acosta H., Bhatia D., Huber C., Petrasek K., Watts A., Yong J.H.E., Bogoch I.I., Khan K. Infectious disease implications of large-scale migration of Venezuelan nationals. J. Travel Med. 2018;25:1–8. doi: 10.1093/jtm/tay077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulia suma 15 muertes confirmadas por coronavirus y seis no reportadas - Efecto Cocuyo WWW Document. 2020. https://efectococuyo.com/coronavirus/zulia-suma-15-muertes-confirmadas-por-coronavirus-y-seis-no-reportadas/ (accessed 6.27.20)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. GISAID accession numbers of the 376 SARS-CoV-2 genomes used in this study.