Abstract
Leukocyte immunoglobulin (Ig)-like receptors (LILRs), also known as CD85 and immunoglobulin-like transcripts (ILTs), play pivotal roles in regulating immune responses. These receptors define an immune checkpoint that immune therapy can target. Through cis or trans interactions with human leukocyte antigen (HLA)-G, the two most abundantly expressed inhibitory LILRs, LILRB1, and LILRB2 (LILRB1/2, also known as CD85j/d and ILT2/4), are involved in immunotolerance in pregnancy and transplantation, autoimmune diseases, and immune evasion by tumors. Although the discrete domains of LILRB1/2 are clear, the assembly mode of the four extracellular Ig-like domains (D1, D2, D3, and D4) remains unknown. Previous data indicate that D1D2 is responsible for binding to HLA class I (HLA-I), but the roles of D3D4 are still unclear. Here, we determined the crystal structure of the four Ig-like domain LILRB2 and four-domain LILRB1 in complex with HLA-G1. The angles between adjacent domains and the staggered assembly of the four domains suggest limited flexibility and limited plasticity of the receptors during ligand binding. The complex structure of four-domain LILRB1 and HLA-G1 supports the model that D1D2 is responsible for HLA-I binding, while D3D4 acts as a scaffold. Accordingly, cis and trans binding models for HLA-I binding to LILRB1/2 are proposed. The geometries of LILRB1/2 in complex with dimeric and monomeric HLA-G1 suggest the accessibility of the dimeric receptor, which in turn, transduces more inhibitory signals. The assembly of LILRB1/2 and its binding to HLA-G1 could aid in the design of immune regulators and benefit immune interference.
Keywords: LILRB1, LILRB2, HLA-G, checkpoint, structural studies
Subject terms: X-ray crystallography, Immunotherapy
Introduction
Leukocyte immunoglobulin-like receptors (LILRs/LIRs), also called immunoglobulin-like transcripts (ILTs) and CD85, are a family of receptors that regulate immune reactions and play pivotal roles in immunological homeostasis. Activating LILRs (LILRAs) contain a short cytoplasmic tail and are associated with the adaptor molecule FcεRγ, which has an immunoreceptor tyrosine-based activation motif (ITAM).1 By contrast, inhibitory LILRs (LILRBs) have an immunoreceptor tyrosine-based inhibitory motif in their cytoplasmic domain that interacts with tyrosine phosphatases and inhibits activating signals.1 Among the five LILRBs identified (LILRB1–5), LILRB1 (also called LIR1, ILT2, and CD85j), and LILRB2 (also called LIR2, ILT4, and CD85d) have been extensively studied.2,3
LILRB1 and LILRB2 (LILRB1/2) bind to multiple ligands and are involved in multiple physiological and pathological situations, which have been summarized in several excellent reviews.1–3 Among the ligands of LILRB1/2, human leukocyte antigen class I (HLA-I) is the most widely expressed. Upon binding to HLA-Is, LILRB1/2 generally inhibit the activities of immune cells, including antigen-presenting cells (APCs),4 CD8+ T cells5 and B cells.4,6 Moreover, LILRB1 inhibits the polarization of NK cell lytic granules and therefore the cytotoxicity of NK cells in response to target cells expressing HLA-I (in trans binding).4,7,8 LILRB1/2 also associate with HLA-Is expressed on the same cells to regulate mast cell activation and osteoclast development (in cis binding).9,10 Blocking the interaction between LILRB1 and HLA-I restores the cytotoxic activity of NK cells11 and potentiates macrophage phagocytosis of tumor cells.12 In addition to performing important functional regulation of other members in this family, LILRBs serve as crucial immune checkpoints that, like PD-1, PD-L1, and CTLA-4, could be targets for drug development to treat cancers.13
The LILR family contains 13 members, including two pseudogenes. Except for LILRA3 and LILRB4 which have two domains, LILRs contain four immunoglobulin (Ig)-like domains in their extracellular part, including one soluble member (LILRA3).2 Usually, the four domains are named domain D1, D2, D3, and D4 from distal-to-proximal relative to the membrane. Although LILRs are important regulatory receptors and 20 years have passed since their characterization, whole structures of LILRs with four domains have yet to be solved. Previous work has solved the structures of discrete domains of LILRB1/2 (D1D2 and D3D4) and delineated the interaction between D1D2 and HLA-I.14–18 However, the roles of D3D4 of LILRB1/2 in the interaction with HLA are still under debate. No substantial binding between LILRB1/2 D3D4 and HLA-I has been detected by surface plasmon resonance (SPR).19 Interestingly, variable binding affinities of LILRB2 to HLA-B*3501 and HLA-B*3503 (which differ in residue 114 or 116 being located in the α1α2 domain of HLA-I) have been reported.20–22 Mutated presented peptides in HLA-B27, HLA-A11, B8, and B7 were also likely to confer enhanced binding affinity to LILRB2 and might be related to HIV-1 escape.23,24 Hence, two models have been proposed:25,26 one model is that no interaction exists between D3D4 of LILRB2 and HLA-I and the other model is that D3D4 bend to interact with α1α2 and the peptides.25,26 Further studies are needed to determine which model best describes the interaction between LILRB2 and HLA-I.
Here, we determined the structure of four-Ig domain LILRB2 and a complex structure of HLA-G1 and LILRB1 containing four Ig-like domains. For the first time, the hinge region angles between the D2 and D3 domains in LILRB1 and LILRB2 were elucidated and were found to be ~60° and ~50°, respectively. The arrangements of the four domains in the long axis were determined, and a staggered assembly mode for LILRB1/2 was uncovered. Compared with LILRB2, LILRB1 D2D3 displayed more open angles, probably due to the steric hindrance of D3 by W284. In addition, the structure of LILRB1 in complex with HLA-G1 provided the first direct structural data supporting the model that D1D2 are responsible for HLA-I binding and D3D4 act as a scaffold. Accordingly, models for HLA-I binding to LILRB1/2 in cis and trans are proposed. The geometries of LILRB1/2 in binding to dimeric and monomeric HLA-G1 indicate more accessibility of LILRB1/2 to the dimeric form of HLA-G1, leading to the transduction of more inhibitory signals. The structural data reported here could help to better understand the structures and functions of the LILR family, which would aid in the design of immune regulators and support immune interference.
Results
Structures of HLA-G1-bound LILRB1 and ligand-free LILRB2
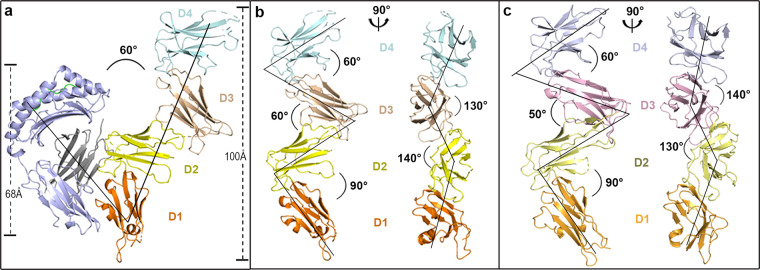
To determine the role of D3D4 in the HLA-I interaction, we solved the crystal structure of LILRB1 with four Ig domains in complex with HLA-G1 incorporating RIIPRHLQL (RL9 from histone H2A) at a resolution of 3.3 Å (Table 1). Two independent copies of the complex were found in the asymmetric unit, with a root mean square deviation (RMSD) of 0.479 Å (for 526 Cα atoms, without atoms from LILRB1 D3D4), and we chose the copy with the better electron density map for further analysis (Fig. S1). The overall structure of the complex demonstrates that the angle between HLA-G1 and LILRB1 is ~60°. In addition, the lengths of the four Ig-like domain LILRB1 and HLA-G1 are ~100 Å and ~68 Å, respectively (Fig. 1a).
Table 1.
Statistics for crystallographic data collection and structure refinement
| LILRB2 | LILRB1/HLA-G-RL9 | |
|---|---|---|
| Data collection | ||
| Space group | P61 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 172.20, 172.20, 65.60 | 69.68, 154.68, 98.20 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 102.24, 90.00 |
| Resolution (Å) | 50.00–3.80 (3.94–3.80)a | 50.00–3.30 (3.42–3.30) |
| Rp.i.m.b | 0.053 (0.519) | 0.105 (0.361) |
| CC1/2 | 0.980 (0.997) | 0.972 (0.995) |
| I/σI | 16.83 (1.85) | 7.89 (2.38) |
| Completeness (%) | 99.9 (100.0) | 99.9 (99.9) |
| Redundancy | 9.8 (10.0) | 4.2 (4.2) |
| Refinement | ||
| Resolution (Å) | 42.75–3.80 | 45.42–3.30 |
| No. reflections | 11,011 | 29,866 |
| Rwork/Rfreec | 0.264/0.287 | 0.220/0.267 |
| No. atoms | ||
| Protein | 2983 | 12,038 |
| Water | – | – |
| B-factors | ||
| Protein | 89.9 | 73.8 |
| Water | – | – |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.002 |
| Bond angles (°) | 0.702 | 0.631 |
| Ramachandran analysis | ||
| Favored (%) | 99.48 | 99.66 |
| Allowed (%) | 0.52 | 0.34 |
| Outliers (%) | 0.00 | 0.00 |
aValues in parentheses are for the highest-resolution shells
bRp.i.m. = ∑hkl[1/(N − 1)]1/2 ∑i|Ii − <I>|/∑hkl∑iIi, where Ii is the observed intensity and <I> is the average intensity from multiple measurements
cRwork = ∑||Fo| − |Fc| |/∑|Fo|, where Fo and Fc are the structure-factor amplitudes from the data and the model, respectively. Rfree is the R factor for a subset (5%) of reflections that were selected prior to the refinement calculations and were not included in the refinement
Fig. 1.
Overall structure of HLA-G1-bound LILRB1 and ligand-free LILRB2. a Geometry of the LILRB1 interaction with HLA-G1. The structure of LILRB1 D1 is shown in orange, D2 in yellow, D3 in wheat, and D4 in pale cyan. Light blue indicates the heavy chain of HLA-G1, light green indicates the presented peptide, and gray indicates β2m. b Cartoon backbone representation of LILRB1 displaying the angles between adjacent domains, which are marked the same as in a. c Cartoon backbone representation of LILRB2 displaying the angles between adjacent domains. The structure of LILRB2 D1 is shown in bright orange, D2 in pale yellow, D3 in light pink, and D4 in light blue
Compared to previous data, each of the four extracellular domains of HLA-G1-complexed LILRB1 maintain similar folds, with an RMSD of 0.28–0.59 (D1, compared with PDBs 1G0X, 1P7Q, 1VDG, 1UFU, 3D2U, 4NO0, and 5KNM), 0.34–0.73 (D2, compared with PDBs 1G0X, 1P7Q, 1VDG, 1UFU, 3D2U, 4NO0, and 5KNM), 0.40 (D3, compared with PDB 4LL9), and 0.41 (D4, compared with PDB 4LL9).14,16–18,27 Although they complex with HLA-G1, LILRB1 D1, and D2 have interdomain angles that resemble ligand-free D1D2 (PDB 1G0X), ~90°.27 The angle of the D3D4 hinge region is ~60°, similar to a previous report.25 For the first time, the angle between D2 and D3 is determined to be ~60° (the angle in the other copy is ~55°) (Fig. S1). The staggered arrangement of the four domains in the long axis of the molecule is also defined. Specifically, the angles between the axes formed by two adjacent domains are ~140° (the angle between the axis of D1D2 and D2D3) and ~130° (the angle between the axis of D2D3 and D3D4) (Fig. 1b).
The assembly of the four extracellular domains of LILRB2 was also illustrated in this study (Table 1). Each domain preserves the scaffold in the four Ig-like structure, as seen in the D1D2 or D3D4 structures, with RMSDs ranging from 0.51 to 1.03 for D1 (compared with PDBs 2GW5 and 2DYP), 0.43 to 0.55 for D2 (compared with PDBs 2GW5 and 2DYP), 0.43 for D3 (compared with PDB 4LLA), and 0.49 for D4 (compared with PDB 4LLA).15,25,28 The angles between LILRB2 D1D2 and D3D4 are ~90° and ~60°, respectively, similar to previous reports.15,25,28 The angle of the D2D3 hinge region is ~50°, which is slightly smaller than its counterpart in LILRB1. The four Ig-like domains in LILRB2 are also stacked, similar to LILRB1, at angles of ~130° (the angle between the axes of D1D2 and D2D3) and ~140° (the angle between the axes of D2D3 and D3D4) (Fig. 1c).
Structural comparisons of LILRB1/2 binding to different HLA alleles
Similar to previous reports,14,17 D1 interacts with the HLA-G1 α3 domain. Both D1 and D2, as well as the interdomain hinge region, form contacts with β2m, which noncovalently associates with the heavy chain of major histocompatibility complex class I molecules. (Fig. S2). Notably, LILRB1 includes Y38, which is conserved among group 1 LILRs (members that bind to HLA-Is) and makes contacts with HLA-G1 (Fig. S3). The phenyl rings of Y38 in D1 and F195 in HLA-G1 (HLA-G1 Y197 at the interface between LILRB2 and HLA-G1) probably form π–π stacking interactions and contribute to binding (Figs. S2A, S4, and S5A).
To date, six LILRB-HLA complex structures have been solved. These structures include LILRB1 D1D2 bound to two HLA-A2 (presenting two different peptides),14,18 HLA-F,17 and UL18 (an HCMV-encoded HLA-I homologue);16 LILRB2 D1D2 in complex with HLA-G1;15 and LILRB1 containing four Ig-like domains in complex with HLA-G1 in this report. Structural alignment along D1D2 shows that HLAs form different angles with D1D2, with HLA-A2 (PDB 1P7Q)14 and HLA-G1 (in this study) differing the most, by 15° (Fig. S6). HLA-A2 (PDB 4NO0),18 UL18,16 HLA-F,17 and HLA-G1 (in complex with LILRB2 D1D2)15 are interspersed, indicating the plasticity of LILRB1 in associating with HLA-I ligands (Fig. S6).
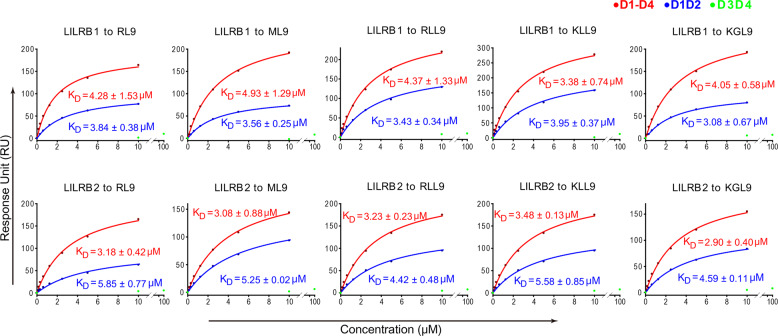
Among the reported complex structures, LILRB1 associates with UL18 with the highest binding affinity, with a nanomolar KD.16 Consistently, UL18 has the largest quantity of residues that interact with LILRB1. In addition, most of buried surface area, and most of H-bonds and van der Waals (vdw) contacts are between LILRB1 D1D2 and UL18 (Table S1). Notably, LILRB1 binds to other HLA-Is with similar binding affinities, which decrease by three orders of magnitude (KD = 2–7 μM) (Fig. 2 and Table S1). Compared with UL18, these complexes have reduced buried surface areas at the LILRB1 interface with HLA-Is, except LILRB2 in complex with HLA-G1. However, the binding of LILRB1 to other HLA-Is involves fewer contacts than the interactions between LILRB1 and UL18, including both vdw contacts and potential H-bonds, which account for the much lower binding affinities (Table S1).
Fig. 2.
Interaction between HLA-G1 and discrete domains of LILRB1/2. The binding affinities of LILRB1/2 with the monomeric HLA-G1 loading peptide were determined by SPR. The binding profiles are shown with the gradient concentrations of LILR proteins on the X axis and response units (RU) on the Y axis. The curves were obtained by fitting data to the 1:1 binding mode (BIAevaluation software). KD values are shown as the mean ± SEM of three independent experiments. The upper panel shows the four-Ig domain LILRB1 (red line), D1D2 (blue line), or D3D4 (green dots) interaction with HLA-G1 with the indicated peptides. The lower panel shows the four-Ig domain LILRB2 (red line), D1D2 (blue line), or D3D4 (green dots) interaction with HLA-G1 with the indicated peptides. Specifically, RL9 (RIIPRHLQL) originates from histone H2A, ML9 (MQPTHPIRL) originates from the HS1 protein and KGL9 (KGPPAALTL) originates from the cytokine receptor. However, the origins of RLL9 (RLPKDFRIL) and KLL9 (KLPAQFYIL) remain unclear
No interactions between LILRB1 D3D4 and HLA-G1
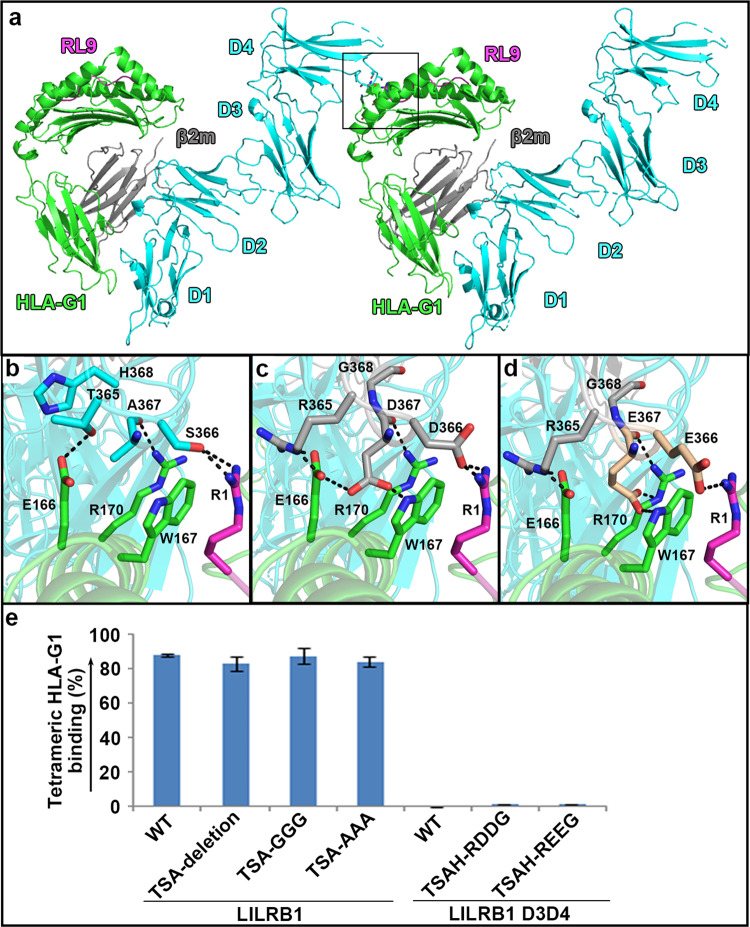
Further analysis of the complex structure between LILRB1 with four domains and HLA-G1 shows that there are no interactions between LILRB1 D3D4 and HLA-G1 in one asymmetry unit (Fig. 1). However, when symmetry mates were generated to analyze the crystal packing, LILRB1 D4 was observed to interact with another HLA-G1 peptide-presenting platform as well as the presenting peptide in the adjacent lattice (Fig. 3a). Residue T365 of LILRB1 D4 potentially forms a H-bond with E166 of HLA-G1, and A367 interacts with R170 of HLA-G1 via hydrogen-bonding interactions. Residue S366 binds to the RL9 peptide by forming two hydrogen bonds with the R1 residue of this peptide (Fig. 3b).
Fig. 3.
Assessment of the interaction between LILRB1 D3D4 and HLA-G1. a One symmetry mate of LILRB1 in complex with HLA-G1 was generated. LILRB1 D4 was observed to interact with HLA-G1 in the adjacent lattice. The region highlighted by the black square is enlarged in b. b The interaction network between D4 and the HLA-G1 peptide-presenting platform as well as the peptide. c, d To increase the interaction between D3D4 and HLA-G1, hydrophilic residues were introduced. 365TSAH368 were mutated to RDDG (c) or REEG (e). The probable conformations of the mutated residues are shown in c and d. f Evaluation of the interaction between tetrameric HLA-G1 incorporating RL9 and the indicated molecules was performed. 365TSA367 was deleted from LILRB1 either by deletion or mutation to AAA or GGG. RDDG or REEG were used to substitute 365TSAH368 in LILRB1 D3D4. The indicated LILRB1 proteins were transiently expressed on the membrane of HEK 293T cells and tagged with eGFP at its C-terminus. Then, the cells were sequentially collected and incubated with tetrameric HLA-G1 (RL9) and streptavidin-APC. eGFP-expressing cells were gated, and the proportion of APC+ cells was analyzed. The value of the column is the mean of triplicates (n = 3), and the bar represents the SEM value. The assays were independently performed twice. In panels a–e, green, gray, magenta, and cyan indicate the HLA-G1 heavy chain, β2m, the peptide, and LILRB1, respectively
To test whether this interaction is real or artificial due to crystal structure manipulation, we first evaluated the binding between LILRB1 D3D4 and HLA-G1. No interactions were detected between the two molecules in a SPR assay (Fig. 2), which is consistent with previous results.19 We also refolded HLA-G1 with five different peptides to assess the effect of the peptide on the association with LILRB1. As indicated in Fig. 2, LILRB1 had similar binding affinities to HLA-G1 incorporating various peptides, similar to LILRB1 D1D2, indicating that the peptides probably do not make additional contacts with LILRB1. We further deleted residues 365TSA367 in LILRB1, which were observed to interact with HLA-G1 in another lattice (Fig. 3b), by either deletion or mutation to AAA or GGG. Then, the mutated LILRB1 was expressed on the membrane of HEK 293T cells and tested for its interaction with the HLA-G1 tetramer with RL9 by flow cytometry. However, no significant differences were observed between wild-type and any mutated LILRB1 (Fig. 3e). Then, we changed the strategy from reducing the interaction mediated by LILRB1 to increasing the binding, if any, between D3D4 and HLA-G1. We introduced hydrophilic residues into D3D4 to form an electrostatic network with HLA-G1 and the peptides (Fig. 3c, d). Nevertheless, the mutants failed to bind to the HLA-G1 tetramer. Thus, we conclude that the observed interaction resulted from artificial results and that no interaction exists between LILRB1 D3D4 and HLA-G1.
D2D3 interdomain interface in LILRB1 and LILRB2
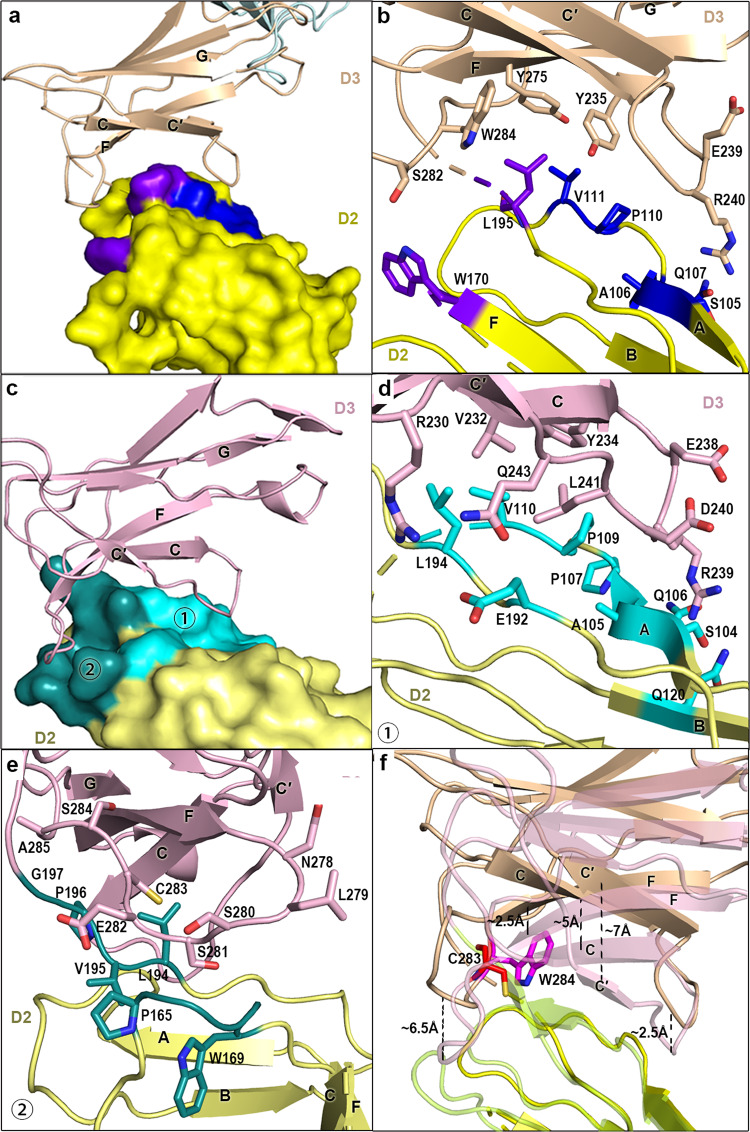
The domain interface between LILRB1 D2D3 is formed by interactions between the C–C′ loop, F strand and F–G loop of D3 with the A strand, A–B loop of D2 and connecting region between D2 and D3 (G–A loop) (Fig. 4a, b), respectively. Compared to the accessible buried area in the interdomain interface between D1D2 (1233.5 Å2) and D3D4 (1120.6 Å2), the value of D2D3 was calculated to be 696.5 Å2. A hydrophobic network is formed by D3 W284, Y235, and Y275 with D2 P110, V111, and L195 at the center of the contact region (Figs. 4b and S5B). In addition, S282 in the D3 F–G loop interacts with W170 of the F strand of D2. R240, located in the D3 C–C′ loop, interacts with multiple residues in D2 (S105, A106, and Q107), forming a small hydrophilic patch. (Fig. 4b). Due to the low electron densities of three amino acids in the loop linking D2 and D3, the second structure of this region is invisible, indicating the flexibility of this region.
Fig. 4.
D2D3 interface in LILRB1 and LILRB2. a An overview of the binding interface between LILRB1 D2 and D3. D2 is shown in surface representation. D3 is shown in cartoon backbone representation. The sites of contact are further delineated in b with the details of the amino acid interactions. b Contact networks between LILRB1 D2 and D3. The residues involved in the interaction are shown in stick representation. c An overview of the binding interface between LILRB2 D2, shown in surface representation, and D3, shown in cartoon representation. The contacting residues (indicated by numbers 1 and 2) are further delineated in d and e. d, e Sites of interaction between D2 and D3, indicated with numbers 1 and 2 in c. f The variation of the angles between D2 and D3 in LILRB1 and LILRB2. The two structures are aligned with respect to D2. Stick representations of C283 in LILRB2 and W284 in LILRB1 are shown. The structures of LILRB2 are displayed at 50% transparency
Compared with LILRB1, more residues and interactions are involved in the LILRB2 D2 and D3 interface. LILRB2 238ERDL241 and Q243, which are in the C–C′ loop, interact with 104SAQP107 and 109PV110, which are located on the A strand and A–B loop (Figs. 4c, d and S5C), respectively. This region confers 94 vdw contacts. On the other patch, the D3 F–G loop, constituted by 278NLSSECSA285, makes 75 and 97 vdw contacts with the D2 E–F loop (165PNRRW169) and the interdomain loop linking D2 and D3 (194LVPG197), respectively (Fig. 4c, e). D3 Q243 interacts with E192. L241 contacts D2 P109 and V110. In addition, residues R230, V232, and Y234 on the D3 C strand contribute to the D2 and D3 interaction through binding to P109, E192, and L194 (Fig. 4d). Accordingly, the stronger interaction between LILRB2 D2 and D3 leads to the more compact assembly of D2D3, resulting in a buried area of 1475.4 Å2 in the D2D3 interface.
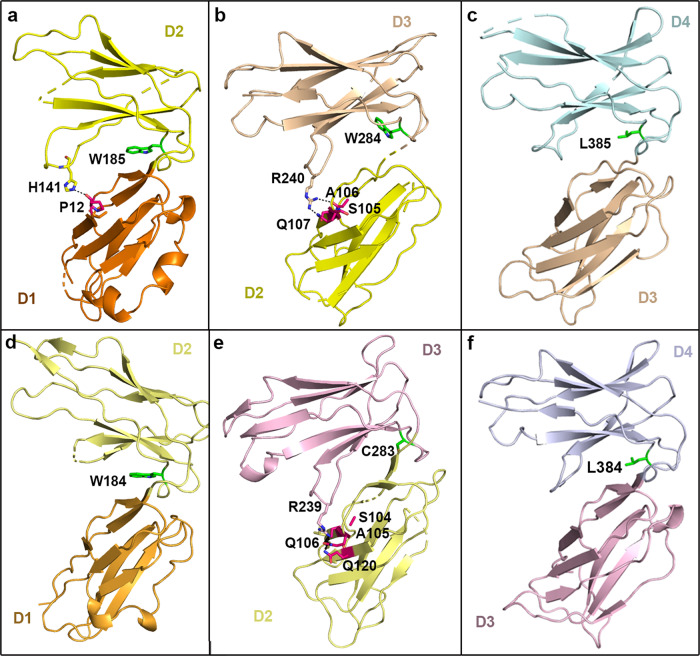
Notably, in the center of the D2D3 contact region, the aromatic W284 in LILRB1 is substituted by the less bulky C283 in LILRB2 (Fig. 4f). Compared with C283 in LILRB2, W284 contributes more to the hydrophobic interaction. However, it may also confer more steric hindrance to the strands of F and C in D3. As shown in Fig. 4f, strand F, together with the loop extending from LILRB1 strand F (F–G loop), shifts away from D2 by ~2.5 Å and ~6.5 Å compared with LILRB2. Concomitantly, LILRB1 strand C moves away from D2 by ~5 Å. Because of the interaction between antiparallel strands of C and C′, strand C′ and the loop linking the two strands shift away from D2 by ~7 Å and ~2.5 Å in LILRB1 (Fig. 4f), respectively. Taken together, the residues in the F–G loop, especially those in the center of the interface (W284/C283 in LILRB1/2), affect the conformation of adjacent secondary structural elements, including the C–C′ loop, causing their shift. These shifts in LILRB1 may be synergized, leading to the reduced interaction between D2 and D3 in LILRB1 and, consequently, the more open angle than in LILRB2.
Structural predictions for other LILR family members
Due to sequence similarities, domains in other LILR members will fold into tertiary structures similar to those of the Ig domains, such as the domains in LILRB1 and LILRB2. Thus, the structures of extracellular regions of other LILR proteins, especially those that have four Ig-like domains, will depend on the nature of the interfaces between adjacent domains.
Similar to the previous results of the discrete domains, most residues responsible for D1D2 interactions are conserved among LILRs27 (Fig. S3). Notably, bulky W185/W184 are located in the center of the D1D2 interface of LILRB1/2 (Fig. 5a, d). In addition, H141 in the D2 C–C′ loop interacts with the main chain of P12 (7 vdw contacts with 4.5 Å cutoff) and likely helps to stabilize the interaction, thereby stabilizing the angle between D1 and D2 (Fig. 5a). The same pair of residues (H140 and P12) is conserved in LILRB2, but the D2 C–C′ loop in this receptor was invisible due to the low electron density (Fig. 5d). Interestingly, W185 and P12 are conserved among the four Ig-domains containing LILRs, while H141 is present in group 1 LILRs (Fig. S3). However, the equivalent position is substituted by L141 in group 2 LILRs, which is unable to associate with HLA-Is (Fig. S3). L141 might hydrophobically interact with the pyrrolidine ring of P12. These data indicate that the angle of D1D2 in four-Ig domain LILRs might be conserved to be ~90°.
Fig. 5.
Binding modes and the interaction details between adjacent domains in LILRs. All of the structures presented are shown with alignment to superimposed LILRB1 D1. a–c The upper panel indicates the interdomain angles of D1D2, D2D3, and D3D4 in LILRB1. d–f The lower panel indicates the interdomain angles of D1D2, D2D3, and D3D4 in LILRB2. A green stick representation indicates that the residue in the F–G loop is located in the center of the contact region. The residues in the A strand or A–B loop that are responsible for the interaction are shown in hot pink stick representation
As indicated in Fig. S3, the majority of residues in the interface between D2 and D3 are conserved among LILRs. Specifically, all four Ig-domains containing LILRs include W at the same position as W284 in LILRB1, except LILRB2 (C283) (Figs. 5b, e and S3). In LILRB1, R240 in the C–C′ loop potentially forms 21 vdw contacts as well as two potential H-bonds with the side chain of Q107 and the main chain of A106, respectively (Fig. 5b). Similar pairs of H-bonds between R239 and Q106 and A105 are present in LILRB2 (Fig. 5e). R239 interacts with most of the residues in strand A (51 vdw contacts) and contributes more interactions than R240 in LILRB1 (Fig. 5d). These strong interactions between the D3 C–C′ loop and the D2 A strand might partially explain why D2D3 has a more closed conformation than D1D2, although both interfaces possess an aromatic W in the center of the contact region. Although R240 in LILRB1 is conserved in most LILRs (except LILRB5 with H235 and LILRA4 with D235), LILRB1 Q107 is substituted by L in four other Ig-domain LILRs (Fig. S3). The equivalent R240 might still interact with the main chain atoms of the residues in the A strand, such as the equivalent A106 in LILRB1, but the binding strength decreases. This interaction more closely resembles the counterpart of the D1D2 interface (H141 with the main chain of P12). Thus, the interdomain angle of D2D3 in LILRA1, A2, A3, A6, and B3 seem to be similar to that of the LILRB1/2 D1D2 interface (~90°). In LILRB5 and A4, due to the substitution of residues with short side chains, the interaction between D2 and D3 might further decrease and exhibit the largest angles.
In the interface between D3 and D4, less bulky L385/384 is present in the center of the contact region in LILRB1/2 (Fig. 5c, f). Similarly, all of the LILRs display conservation at the interacting positions (Fig. S3), indicating that they form similar hydrophobic associations in the center of the contact region. In addition, L also confers less steric hindrance than W. Thus, other LILRs might adopt similar angles in D3D4 as LILRB1/2 (~60°).
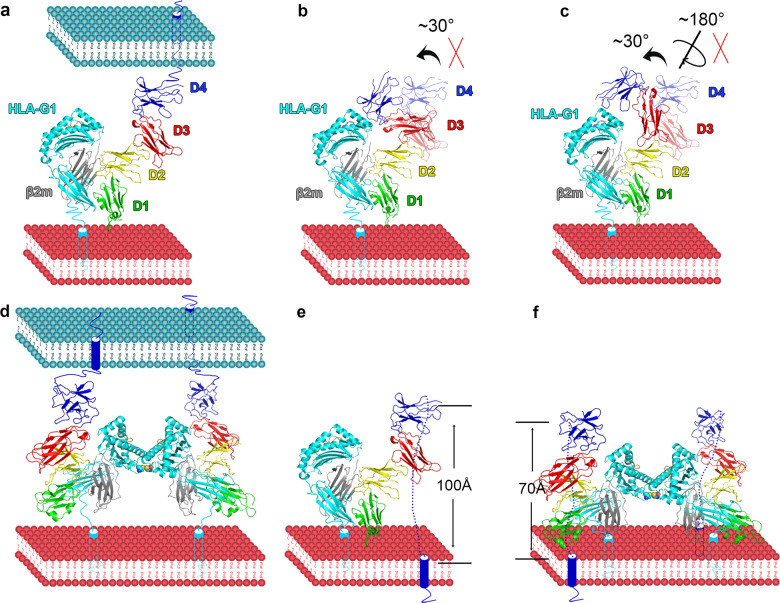
Notably, based on the above analysis, strong interactions are present in each adjacent domain. Thus, as observed in the structure of the complex (Figs. 1a and 6a), HLA-G1 exclusively binds to D1D2 of LILRB1 and D3D4 of this receptor is unlikely to bend or rotate to interact with the peptide or peptide-presenting platform (Fig. 6b, c). Similarly, LILRB1/2 likely adopts the same binding mode to interact with other HLA-Is (Fig. S7), and D3D4 does not contact HLA-Is.
Fig. 6.
Models of full-length LILR binding to HLA-I in cis and trans. The green, yellow, red, and blue colors indicate the four domains in LILRB1/2. The cyan, gray, and magenta represent the HLA heavy chain, β2m, and peptide, respectively. a Trans interaction between LILRB1/2 and HLA-I. b The proposed interaction mode in which D3 and D4 bend to interact with HLA-I and the peptide. c The proposed interaction mode in which the acute interchain angle between D3 and D4 faces the HLA-I and peptide, allowing the interaction between D3D4 and the peptide as well as HLA-I. In b and c, D3D4 in the complex structure between LILRB1 and HLA-G1 are displayed at 50% transparency, while those with no transparency indicate D3D4 in the two proposed models.25,26 d Trans interaction between LILRB1/2 and an HLA-G1 dimer. e Cis interaction between LILRB1/2 and HLA-I. f Cis interaction between LILRB1/2 and an HLA-G1 dimer
Cis and trans interactions between LILRB1/2 and HLA-G1
The interactions between LILRB1/2 and HLA-Is are reported to be either cis or trans interactions. In addition, HLA-G1 could form homodimers on cells (PDB 2D31).29 Comparison between monomeric and dimeric HLA-G1 indicates that there are no significant conformational changes at LILRB1/2 binding sites (Fig. S8). Thus, dimeric HLA-G1 can interact with either one or two molecules of LILRB1/2 expressed on other cells through a trans interaction (Fig. 6a, d). As assessed by the SPR assay, dimeric HLA-G1 presenting variable peptides displayed similar binding strengths for LILRB1 and LILRB2 (with four Ig-like domains or D1D2), indicating that the peptides seem to have no effect on the interactions. Furthermore, no detectable interactions were observed between D3D4 and dimeric HLA-G1 (Fig. S9).
Of note, for a cis interaction, the C-terminus of D4 of LILRB1/2, which binds to monomeric HLA-G1, needs to be elongated by ~100 Å (Fig. 6e). By contrast, due to dimer formation, the binding sites on dimeric HLA-G1 are tilted and the distance of the C-terminus of interacting LILRB1/2 D4 is ~70 Å, which is shorter than that of monomeric HLA-G1 (Figs. 6f and S10).
Discussion
The LILR family is a group of immune receptors that regulate immune reactions and maintain immune hemostasis.1–3 Structural studies help to understand the functions of these immune receptors.
Previous studies indicate that D3D4 may bend or turn to interact with the peptide and α1α2 of HLA-Is.25,26 However, the structure of the HLA-G1 complex with four-Ig domain LILRB1 reported here indicated that there were no interactions between D3D4 and HLA-G1. Consistently, we did not observe an association between D3D4 (either in LILRB1 or LILRB2) and the respective HLA-G1 incorporating five different peptides. The binding affinities of the four-Ig domain LILRB1/2 to HLA-G1 with different peptides did not display obvious variation. Importantly, the structural studies reported here showed that residues in the interface of D2D3 formed both hydrophobic and hydrophilic interactions and thereby stabilized the interdomain angle of D2D3. This stabilization most likely prevents D3 from rotating anticlockwise with respect to D2 to allow either D3 or D4 to make contacts with HLA-Is. In addition, the results suggest that D3 is prevented from rotating along the D1D2 axis to force the acute angle of D3D4 to face HLA-Is and interact with them. According to sequence alignment and interdomain interface analysis, all of the LILRs, especially those in group I, probably adopt similar angles in D1D2 and D3D4 as LILRB1/2. Although D2D3 is proposed to be relatively flexible in LILRA1, A2 and A3, they still might have a similar interdomain angle as LILRB1/2 D1D2 (~90°). Thus, they are unlikely to utilize D3D4 to interact with the peptide or HLA-I α1α2 domains. The underlying mechanisms by which the mutated peptides that presented by HLA-Is or polymorphisms in the HLA-Is α1 or α2 domains modify the immune reactions through LILRB2 require further study.
LILRBs are believed to function by binding to HLA-Is in both cis and trans.5,6,9,10,30 The question is the interaction modes by which LILRBs bind to HLA-I in cis and trans, especially for HLA-G1, which also forms dimers on cells. Structural data in this study indicate that through trans interaction, one HLA-G1 monomer binds to one LILRB1/2 molecule. One HLA-G1 dimer simultaneously interacts with two LILRB1/2 molecules and amplify LILRB-related inhibition signals.
Notably, the stalk regions (segments from the C-terminus of D4 to the transmembrane domain) of LILRB1 and LILRB2 are relatively longer (consisting of 44 and 43 residues, respectively) than other type I transmembrane proteins, such as HLA-I molecules. These stalk regions in the two receptors are full of G, S, P, and T residues, indicating their flexibility. Taking the 48KVEHSDL54 loop in β2m as an example, the main chain of the seven residues is ~21 Å. Theoretically, these 43–44 residues could constitute a loop limited to 120 Å, which might support the return and insertion of the C-terminus of LILRB1 and LILRB2 into the membrane.
Due to the different configurations of HLA-G1 between monomers and dimers, interacting LILRB1/2 in cis exhibit different angles relative to the membrane. Consequently, the distance of LILRBs’ C-terminus to the membrane is different in the two binding modes (~100 Å and 70 Å, respectively), indicating that HLA-G1 dimers are more accessible for LILRB1/2 binding than HLA-G1 monomers. This difference might also explain why dimers are the major functional form of HLA-G1 and lead to much stronger inhibition of LILRB than monomers.
LILRB1 functions as a checkpoint through its interaction with HLA-Is, relying on the invariant β2m subunit. LILRB1 cannot interact with β2m-free heavy chains, in contrast to LILRB2. Further structural studies have revealed that D1D2 of LILRB1 employs interdomain loops to interact with β2m allocated to different HLA-Is. The LILRB1 with four Ig-like domains reported in this study was shown to use a similar mode to interact with HLA-G1. These structural data suggest that the interdomain region could be an important target to block the interaction with β2m, thereby inhibiting the binding of LILRB1 by HLA-Is.
In conclusion, we reported the structures of LILRB1 and LILRB2 with four Ig-like domains and observed how the four domains are arranged. The complex structure reported here provides the first direct evidence that D1D2 is responsible for all of the interactions with HLA-Is, while D3D4 is not the reason to explain why HLA-Is that carry a single residue substitution in their α1α2 domains or in the presented peptides display variable binding affinities to LILRB2. The assembly of four other Ig-domain LILRs was speculated based on sequence and structural comparisons. This structural information will help to better understand the structures of the LILR family and therefore their functions.
Materials and methods
Gene cloning
The coding regions for each protein, including LILRB1 D1D2 (1-198), LILRB1 D3D4 (199-394), LILRB2 D1D2 (1-196), and LILRB2 D3D4 (197-393), were cloned into pET21a. Four extracellular domains of both LILRB1 (1-394) and LILRB2 (1-393) were cloned into the pFastBac1 plasmid and subjected to insect cell expression. An N-terminal GP67 signal peptide and a C-terminal 6× His were added to facilitate protein secretion and purification.
The extracellular domain of HLA-G1 was cloned into pET21a. The biotinylation tag (GLNDIFEAQKIEWHE) was linked to the 3′ end of HLA-G1. In addition, C42S was also prepared based on the pET21a expression vector.
To evaluate the interaction between four-Ig domain LILRB1 or LILRB1 D3D4 and HLA-G1, the coding regions for each protein, including LILRB1 (1-627) and LILRB1 D3D4 (199-627) with its original N-terminal signal peptide (MTPILTVLICLGLSLGPRTHVQA), were cloned into the pEGFP-N1 vector to produce proteins fused with eGFP expressed on the cell membrane.
Protein expression and purification
Recombinant proteins were refolded and purified as previously reported.25,31,32 Briefly, inclusion bodies including LILRB1 D1D2, D3D4, LILRB2 D1D2, D3D4, wild-type HLA-G1 heavy chains with a biotinylation tag, C42S mutants with or without the biotinylation tag and β2m were isolated from cell pellets by sonication and washed with washing buffer (0.5% Triton X-100, 50 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10 mM EDTA, 10 mM β-mercaptoethanol (β-ME), and 0.1% NaN3) and resuspension buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10 mM EDTA, 10 mM β-ME, and 0.1% NaN3) and then dissolved overnight in a denaturing buffer (8 M Urea, 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 10 mM EDTA, 10% (v/v) glycerol, and 10 mM DTT).
Then, the proteins were refolded by dilution against a refolding buffer (100 mM Tris, pH 8.0; 400 mM l-arginine; 5 mM EDTA-Na; 5 mM glutathione; 0.5 mM glutathione disulfide). For HLA-G1 refolding, inclusion bodies containing β2m, peptides, and inclusion bodies containing the heavy chain were sequentially added. The peptides used for HLA-G1 refolding include RL9 (RIIPRHLQL from histone H2A), ML9 (MQPTHPIRL from HS1 protein), RLL9 (RLPKDFRIL, unknown), KLL9 (KLPAQFYIL, unknown), and KGL9 (KGPPAALTL from cytokine receptor). After 12 h of slow stirring at 4 °C, the refolded protein was concentrated and changed to 20 mM Tris-HCl (pH 8.0) and 150 mM NaCl buffer and further analyzed by gel filtration (Superdex® 200 column, GE Healthcare).
Both LILRB1 (1–394) and LILRB2 (1–393) were expressed and purified as previously reported.33 Briefly, recombinant bacmids were prepared and then transfected into sf9 cells to obtain a baculovirus stock, which was then used to infect High5 cells for protein expression. Target proteins in the supernatant were sequentially collected, affinity purified by a HisTrap HP column (GE Healthcare), and further purified via gel filtration (Superdex® 200 column, GE Healthcare).
HLA-G1 with the biotinylation tag was refolded as described above. After purification, HLA-G1 was biotinylated using the BirA enzyme (Avidity). APC-tagged streptavidin (BioSource International) was added at a molar ratio of 4:1 to refolded HLA-G1, with the biotin-streptavidin interaction causing four HLA-G1 monomers to bind to streptavidin and create a tetramer. The HLA-G1 tetramers were then used to stain cells.
Binding analysis using SPR
Soluble LILRB1 and LILRB2 expressed in insect cells and refolded D1D2 and D3D4 of both LILRB1 and LILRB2 were exchanged into HBS-EP buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 0.005% Tween 20). Monomeric HLA-G1 (C42S) or dimeric HLA-G1 (wild type), which contained the biotin tag at the C-terminus and incorporated the indicated peptide (RL9, ML9, RLL9, KLL9, and KGL9), was immobilized on an SA chip to ~400 response units. Full-length or discrete LILRB proteins were serially diluted and injected. Specifically, both LILRB1 and LILRB2 were loaded at concentrations of 0.156, 0.3125, 0.625, 1.25, 2.5, 5, and 10 μM. Both LILRB1 D1D2 and LILRB2 D1D2 were injected at concentrations of 0.156, 0.3125, 0.625, 1.25, 2.5, 5, 10, and 20 μM. Both LILRB1 D3D4 and LILRB2 D3D4 were loaded at concentrations of 0, 10, and 100 μM. The binding responses were recorded. SPR experiments were performed using a BIAcore® 3000 system (BIAcore). KD values were calculated using the model of steady-state affinity. Data were analyzed by BIAevaluation (BIAcore) and SigmaPlot 10.
Flow cytometry
HEK 293T cells transiently transfected with the pEGFP-N1-LILRB1 and pEGFP-N1-D3D4 plasmids and indicated mutants (TSA deletion, TSA-AAA and TSA-GGG in LILRB1; TSAH-REEG and TSAH-RDDG in LILRB1 D3D4) were used for the binding test. At 48 h post transfection, cells (2 × 105) were collected and stained with tetrameric HLA-G1 incorporating RL9 (RIIPRHLQL) at a final concentration of 1 μg/mL on ice for 30 min. After washing, streptavidin-conjugated APC (1 μg/mL) was added. Then, the cells were subjected to analysis using a BD FACSCalibur. Only eGFP+ cells were gated, and the fluorescence shift for APC was analyzed. FlowJo 7.6 was used for data analysis.
Crystallization, data collection, and structure determination
Crystallization trials were set up with commercial crystallization kits (Hampton Research and Molecular Dimensions) using the sitting-drop vapor-diffusion method. Normally, 1 μL protein at the corresponding concentrations was mixed with 1 μL reservoir solution. Purified HLA-G1 (RL9) was mixed with LILRB1 at a molar ratio of 1:1. Crystals with reflection at high resolutions were obtained under the condition of 0.2 M imidazole malate, 15% w/v PEG 4000, and pH 6.0 at a concentration of 5 mg/mL at 4 °C. Diffractable crystals of LILRB2 were finally obtained under the condition consisting of 0.1 M Tris, pH 8.5, and 25% v/v tert-butanol at a concentration of 10 mg/mL at 4 °C. Crystals were cryoprotected in reservoir solution containing 20% (v/v) glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected at Shanghai Synchrotron Radiation Facility (SSRF) BL17U and processed with HKL2000.34 The structures were determined by molecular replacement using Amore in the CCP4 suite with the coordinates of LILRB2 D1D2 and HLA-G1 (PDB 2DYP), LILRB1 D3D4 (PDB 4LL9), and LILRB2 D3D4 (PDB: 4LLA) as search probes. The atomic models were completed with COOT35 and refined with phenix.refine in Phenix,36 and the stereochemical qualities of the final models were assessed with MolProbity.37 Data collection, processing, and the refinement statistics are summarized in Table 1.
Supplementary information
Acknowledgements
We thank the staff of BL17U beamline at SSRF. We are grateful to Zheng Fan from the Institute of Microbiology Chinese Academy of Sciences (CAS) for technical assistance with the SPR experiments. This work was supported by the Strategic Priority Research Program of CAS (grant no. XDA12020358), the National Basic Research Program (973) of China (grant no. 2015CB910503), and the National Natural Science Foundation of China (NSFC, grant nos 31502078 and 31390432). Q.W. is supported by the Youth Innovation Promotion Association CAS (grant no. 2018119). H.S. is supported by the Young Elite Scientist Sponsorship Program by CAST (2016QNRC001) and the Youth Innovation Promotion Association CAS (2017117). G.F.G. is a leading principal investigator of the NSFC Innovative Research Group (Grant no. 81621091).
Author contributions
Y.S. and G.F.G. initiated and coordinated the project. Q.W., H.S., Y.S., J.Y. and G.F.G. designed the experiments. Q.W. and H.S. conducted the experiments with the assistance of G.N. Q.W. obtained diffractable complex crystals of LILRB1 and HLA-G1, and H.S. obtained diffractable crystals of free LILRB2. J.Q. solved the crystal structures. Q.W., H.S., H.C., Y.S., J.Y. and G.F.G. analyzed the data. Q.W. wrote the manuscript. H.S., S.T., J.W., M.F., Z.T., X.C., Z.A., J.Y. and G.F.G revised the manuscript.
Data availability
The accession numbers for the atomic coordinates and diffraction data reported in this paper are PDB 6AED (crystal structure of LILRB2) and 6AEE (crystal structure of LILRB1/HLA-G1 complex), respectively.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Qihui Wang, Hao Song
Contributor Information
Qihui Wang, Email: wangqihui@im.ac.cn.
George F. Gao, Email: gaof@im.ac.cn
Supplementary information
The online version of this article (10.1038/s41423-019-0258-5) contains supplementary material.
References
- 1.Brown D, Trowsdale J, Allen R. The LILR family: modulators of innate and adaptive immune pathways in health and disease. Tissue Antigens. 2004;64:215–225. doi: 10.1111/j.0001-2815.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Nakamura A. Regulation of immune and neural function via leukocyte Ig-like receptors. J. Biochem. 2017;162:73–80. doi: 10.1093/jb/mvx036. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, et al. Leukocyte immunoglobulin-like receptors in human diseases: an overview of their distribution, function, and potential application for immunotherapies. J. Leukoc. Biol. 2017;102:351–360. doi: 10.1189/jlb.5MR1216-534R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonna M, et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiroishi M, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl Acad. Sci. USA. 2003;100:8856–8861. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merlo A, et al. Inhibitory receptors CD85j, LAIR-1, and CD152 down-regulate immunoglobulin and cytokine production by human B lymphocytes. Clin. Diagn. Lab. Immunol. 2005;12:705–712. doi: 10.1128/CDLI.12.6.705-712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Favier B, LeMaoult J, Lesport E, Carosella ED. ILT2/HLA-G interaction impairs NK-cell functions through the inhibition of the late but not the early events of the NK-cell activating synapse. FASEB J. 2010;24:689–699. doi: 10.1096/fj.09-135194. [DOI] [PubMed] [Google Scholar]
- 8.Ponte M, et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc. Natl Acad. Sci. USA. 1999;96:5674–5679. doi: 10.1073/pnas.96.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuda A, Nakamura A, Maeda T, Sakamoto Y, Takai T. Cis binding between inhibitory receptors and MHC class I can regulate mast cell activation. J. Exp. Med. 2007;204:907–920. doi: 10.1084/jem.20060631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori Y, et al. Inhibitory immunoglobulin-like receptors LILRB and PIR-B negatively regulate osteoclast development. J. Immunol. 2008;181:4742–4751. doi: 10.4049/jimmunol.181.7.4742. [DOI] [PubMed] [Google Scholar]
- 11.Roberti MP, et al. Overexpression of CD85j in TNBC patients inhibits Cetuximab-mediated NK-cell ADCC but can be restored with CD85j functional blockade. Eur. J. Immunol. 2015;45:1560–1569. doi: 10.1002/eji.201445353. [DOI] [PubMed] [Google Scholar]
- 12.Barkal, A. A. et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. 19, 76–84 Nat. Immunol. (2017). [DOI] [PMC free article] [PubMed]
- 13.Kang X, et al. Inhibitory leukocyte immunoglobulin-like receptors: Immune checkpoint proteins and tumor sustaining factors. Cell Cycle. 2016;15:25–40. doi: 10.1080/15384101.2015.1121324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat. Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 15.Shiroishi M, et al. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc. Natl Acad. Sci. USA. 2006;103:16412–16417. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Bjorkman PJ. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc. Natl Acad. Sci. USA. 2008;105:10095–10100. doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dulberger CL, et al. Human leukocyte antigen F presents peptides and regulates immunity through interactions with NK cell receptors. Immunity. 2017;46:1018–1029 e1017. doi: 10.1016/j.immuni.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohammed F, et al. The antigenic identity of human class I MHC phosphopeptides is critically dependent upon phosphorylation status. Oncotarget. 2017;8:54160–54172. doi: 10.18632/oncotarget.16952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–613. doi: 10.1016/S1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, et al. HLA-B*35-Px-mediated acceleration of HIV-1 infection by increased inhibitory immunoregulatory impulses. J. Exp. Med. 2009;206:2959–2966. doi: 10.1084/jem.20091386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 22.Steinle A, et al. Motif of HLA-B*3503 peptide ligands. Immunogenetics. 1996;43:105–107. doi: 10.1007/BF00186615. [DOI] [PubMed] [Google Scholar]
- 23.Lichterfeld M, et al. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 2007;204:2813–2824. doi: 10.1084/jem.20061865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Huang J, Toth I, Lichterfeld M, Yu XG. Mutational escape in HIV-1 CTL epitopes leads to increased binding to inhibitory myelomonocytic MHC class I receptors. PLoS One. 2010;5:e15084. doi: 10.1371/journal.pone.0015084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam G, et al. Crystal structures of the two membrane-proximal Ig-like domains (D3D4) of LILRB1/B2: alternative models for their involvement in peptide-HLA binding. Protein Cell. 2013;4:761–770. doi: 10.1007/s13238-013-3908-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichterfeld M, Yu XG. The emerging role of leukocyte immunoglobulin-like receptors (LILRs) in HIV-1 infection. J. Leukoc. Biol. 2012;91:27–33. doi: 10.1189/jlb.0811442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman, T. L., Heikema, A. P., West, A. P. & Bjorkman, P. J. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2). Immunity. 13, 727–736 (2000). [DOI] [PubMed]
- 28.Willcox BE, et al. Crystal structure of LIR-2 (ILT4) at 1.8 Å: differences from LIR-1 (ILT2) in regions implicated in the binding of the human cytomegalovirus class I MHC homolog UL18. BMC Struct. Biol. 2002;2:6. doi: 10.1186/1472-6807-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiroishi M, et al. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J. Biol. Chem. 2006;281:10439–10447. doi: 10.1074/jbc.M512305200. [DOI] [PubMed] [Google Scholar]
- 30.Colonna M, et al. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J. Exp. Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, et al. Crystal structure of myeloid cell activating receptor leukocyte Ig-like receptor A2 (LILRA2/ILT1/LIR-7) domain swapped dimer: molecular basis for its non-binding to MHC complexes. J. Mol. Biol. 2009;386:841–853. doi: 10.1016/j.jmb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 32.Cheng H, et al. Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance. J. Biol. Chem. 2011;286:18013–18025. doi: 10.1074/jbc.M111.221028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang W, et al. Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus. Protein Cell. 2010;1:459–467. doi: 10.1007/s13238-010-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams CJ, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the atomic coordinates and diffraction data reported in this paper are PDB 6AED (crystal structure of LILRB2) and 6AEE (crystal structure of LILRB1/HLA-G1 complex), respectively.