Abstract
The pandemic of novel coronavirus disease (COVID-19) caused by the Severe Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) creates an immense menace to public health worldwide. Currently, the World Health Organization (WHO) has recognized the novel coronavirus as the main cause of global pandemic. Patients infected with this virus generally show fever, nausea, and respiratory illness, while some patients also manifest gastrointestinal symptoms such as abdominal pain, vomiting, and diarrhea. Traces of SARS-CoV-2 RNA have been found in gastrointestinal cells. Further angiotensin converting enzyme 2 (ACE2) the known receptor for the virus is extensively expressed in these cells. This implies that gastrointestinal tract can be infected and can also present them as a replication site for SARS-CoV-2, but since this infection may lead to multiple organ failure, therefore identification of another receptor is a plausible choice. This review aims to provide comprehensive information about probable receptors such as sialic acid and CD147 which may facilitate the virus entry. Several potential targets are mentioned which can be used as a therapeutic approach for COVID-19 and associated GI disorders. The gut microbiomes are responsible for high levels of interferon-gamma which causes hyper-inflammation and exacerbates the severity of the disease. Briefly, this article highlights the gut microbiome’s relation and provides potential diagnostic approaches like RDT and LC-MS for sensitive and specific identification of viral proteins. Altogether, this article reviews epidemiology, probable receptors and put forward the tentative ideas of the therapeutic targets and diagnostic methods for COVID-19 with gastrointestinal aspect of disease.
Keywords: COVID-19, SARS-CoV-2, Gastrointestinal disorders, Drugs, Diagnostic methods
Introduction
COVID-19 caused by novel SARS-CoV-2 is known to arrive from Wuhan wet market as an etiological agent leading to a global pandemic (2019–2020) [1]. According to the WHO as of September 17, 2020, this infection has more than 29 million active cases and about 0.58 million reported deaths in the form of viral pneumonia and affecting about 214 countries worldwide [2]. The pandemic of novel coronavirus is a great challenge because of exponential increase in patients, rate of infectivity, scarcity of resources, poor prognosis of disease, and ambiguity regarding disease pathogenesis [3].
Furthermore, SARS-CoV-2 is known to attack the lower respiratory system and cause viral pneumonia; however, it may also affect the gastrointestinal system [4]. Reports suggest that patients infected with nCoV showing digestive symptoms like diarrhea, vomiting may be among presenting features of disease [5]. However, the prognosis of COVID-19 patients with gastrointestinal symptoms is mostly unknown. In about 50% of COVID-19 cases, there is the presence of SARS-CoV-2 in fecal samples and its viral RNA has also been identified in intestinal mucosa suggesting that GI tract may be a probable route of infection [6]. Based on the studies in SARS-CoV-2, its receptor (ACE2) is known to be a critical component of gastric mucosa and gastrointestinal cells as well, due to which gastric mucosa or gastrointestinal tract may be considered as vulnerable site for SARS-CoV-2 infection [7, 8]. The lasting presence of virus in gastric mucosa may indicate the possibility of gastric glandular epithelial cells as an incubation site of virus. This can be further consolidated by the presence of viral nucleocapsid protein in the cytoplasm of gastrointestinal tract days after its clearance from respiratory sputum [9].
Since this virus arises from the family of Coronaviridae, it belongs to betacoronavirus genera. Moreover, the collectively known host receptor utilized by the SARS-CoV-2 is ACE2, sialic acids, and CD147 for the host cell infection [10–12]. Human leukocyte antigen (HLA) is an antigen-presenting factor for viral peptide and may be prioritized for the development of a vaccine against SARS-CoV-2 [13]. Both sialic acids and HLA are known to be important for Helicobacter pylori (H. pylori) mediated gastritis and gastric cancer (GC) respectively [14, 15]. Thus, they played an important role in causing gastrointestinal symptoms associated with COVID-19, with the emergence of the gastrointestinal tract as an important site for SARS-CoV-2 pathogenesis. A possible mechanism of viral interaction and pathology is not yet completely known. In this review, we summarize the association of gastrointestinal disorders with SARS-CoV-2, their association from the other coronaviruses along with the receptors which play a crucial role in facilitating the virus entry. Briefly, we also highlighted the gut microbe association in enhancing the infection, potential targets used as therapeutics and drugs which can be repurposed for the COVID-19 patients with gastric co-morbidities. This article highlights potential diagnostic approaches like RDT and LC-MS for sensitive and specific identification of viral proteins. Taken all together, this article reviews epidemiology, probable receptors and put forward the tentative ideas of the therapeutic targets, their drugs, and diagnostic tool for COVID-19 with gastrointestinal aspect of disease.
Coronavirus and Gastrointestinal Symptoms: a Long-Back Association
The coronaviruses comprise large, enveloped, positive-stranded RNA viruses. Coronavirus caused a broad spectrum of diseases in animals and humans [16]. Human coronaviruses (HCoV) can be classified into two serogroups with HCoV-229E and HCoV-NL63 included in serogroup one and HCoV-OC43 and HCoV-HKU1 falling in serogroup 2 [17]. The first two human coronaviruses, HCoV-229E and HCoV-OC43, are correlated with upper respiratory tract infections. Furthermore, HCoV-OC43 and HCoV-HKU1 cause gastrointestinal symptoms such as diarrhea, vomiting, nausea, and abdominal pain in up to 57% and 38% of infected people respectively [18, 19]. Therefore, gastrointestinal symptoms can be treated as evident as respiratory symptoms in coronavirus colds, often designated “gastric flu.”
Another coronavirus is the severe acute respiratory syndrome virus (SARS) which is also well known to affect the gut. Although infection results in diffuse alveolar damage, the changes in the gut are more precise which may include transmigration of intestinal bacteria and more lipopolysaccharide (LPS) permeability in the intestine. LPS causes an increase in the production of tumor necrosis factor (TNF), interleukin-1 (IL-1), and IL-6, thus resulting in aggravation of disease [20]. Furthermore, studies showed by Leung et al., states the SARS replication in the cells of small and large intestine of patients with accumulation of higher viral titer inside the endoplasmic reticulum (ER) and viral particles might leave from the apical membrane of the enterocytes. While the report suggests that there is only minimal disruption of intestinal cells caused by the virus despite the tropism, thus, diarrhea associated with SARS infection may be more related to proteins or toxins produced during viral replication than malabsorption or inflammation [21].
Middle East respiratory syndrome coronavirus (MERS-CoV), another virus of the coronaviridae family, known to cause human respiratory infections and gastrointestinal symptoms. Zhou et al. illustrated that intestinal organoids and human primary intestinal epithelial cells were immensely susceptible to MERS-CoV and can sustain vigorous viral replication [22]. Apart from the respiratory tract, the human alimentary tract may be analyzed as a potential site for viral entry. Enteric viruses (adenovirus, rotavirus) and some non-enteric viruses (adenovirus A12) can bypass the physical barriers and infect susceptible cells in the alimentary tract [23]. The gastrointestinal symptoms are generally seen in most human coronaviruses [24], so far, seven human coronaviruses have been known as the causative agents of mild or severe respiratory infections.
Epidemiology and Gastric Disorder as Clinical Predictor of SARS-CoV-2
SARS-CoV-2 shows the most common symptoms of the disease include pneumonia along with cough, sore throat, myalgia, diarrhea, nausea, vomiting, and fatigue [25, 26]. In adults, most common symptoms are anorexia (39.9–50.2%) and diarrhea (2–49.5%) while children suffered most from vomiting (6.5–66.7%). Furthermore, 34.3% of COVID-19 patients are having digestive symptoms that contribute to their delayed recovery, unlike the remaining patients who recover early [27], since the incubation time of a virus can range from 1 to 24 days, screening the patients becomes a tough task to handle [27, 28]. Moreover, several reports showed that viral RNA was detected in stool samples from 48.1% patients even in stool collected after the respiratory samples tested negative [27, 29]. This suggests that the virus utilized fecal-oral route for its transmission. Additional evidence of the association of SARS-CoV-2 and gut came when it was found in the endoscopy sample of a COVID-19 patient [9]. Wang et al. have cultured the SARS-CoV-2 from four different patients’ stool samples and found live virions in two samples through microscopy [30], although there are also reports that viruses are transmitted through fomites [31]. However, more research is required to confirm if this virus is viable in the stool and also to analyze the level of transmission through the fecal-oral route [32].
Importantly, multiple studies have found that the susceptible population for COVID-19 includes elderly mostly > 70 years of age, individuals with underlying disease, or weakened immune systems [33]. Furthermore, in a meta-analysis by Men and colleagues, 10% of patients were showing only gastrointestinal features upon infection. This may be due to delayed diagnosis which may lead to potential problems in the patient and the person who comes in contact with the patient. Another study suggests that about 3% of COVID-19 cases showed only digestive symptoms and no respiratory symptoms [8, 34]. H. pylori is a well-established gastrointestinal pathogen associated with multiple gastric disorders like chronic gastritis and gastric cancer. According to reports, SARS-CoV-2 and H. pylori infection are more likely to occur in patients with blood group A, thus increases the risk of gastrointestinal infection [35]. Furthermore, the patient having the history of H. pylori infection may become more susceptible to oral-fecal route transmission.
Nearly one-fifth of COVID-19 patients have reported with gastro-intestinal symptoms [36]. About 70% of patients with viral RNA shedding through gastrointestinal tract were reported which lasted for about 10 weeks after the symptom onset [31]. Pathophysiological reports have suggested that no considerable damage was observed in the mucosal epithelium of the esophagus, stomach, and duodenum tissues. However, major infiltration of lymphocytes is observed in squamous epithelium of lamina propria of the stomach, duodenum, and rectum which may cause abdominal pain [37]. Furthermore, abdominal pain is highly associated with COVID-19 severity along with nausea and vomiting which are comparatively less frequent. Considering the association of gut anomalies and its association with COVID-19 some of the symptoms like abdominal pain and diarrhea can also be considered COVID-19 symptoms or can be used as clinical predictors [36].
Receptor-Mediated Signaling with Gastrointestinal Disorders
It is well established that ACE2 is a receptor for SARS-CoV-2. Moreover, sialic acid, CD147 can also act as a receptor [10, 38]. However, the human coronavirus HKU1 and OC43 includes human leukocyte antigen (HLA) as its attachment factor and sialic acid as its receptor respectively [39, 40]. In SARS-CoV-2 too, HLA may present the viral peptides [41]. Moreover, another study by Ming et al. (2020) on COVID-19 patients from Wuhan has reported that there is an increased level of neutrophil, Interleukin-6 (IL-6), chemokine’s IP-10, MCP-1, MIP-1A, tumor necrosis factor-alpha (TNFα), and less expression of lymphocytes [42, 43]. Here we precisely explain the downstream signaling from the aforementioned factors concerning gastrointestinal disorders and COVID-19 taking SARS-CoV as a reference.
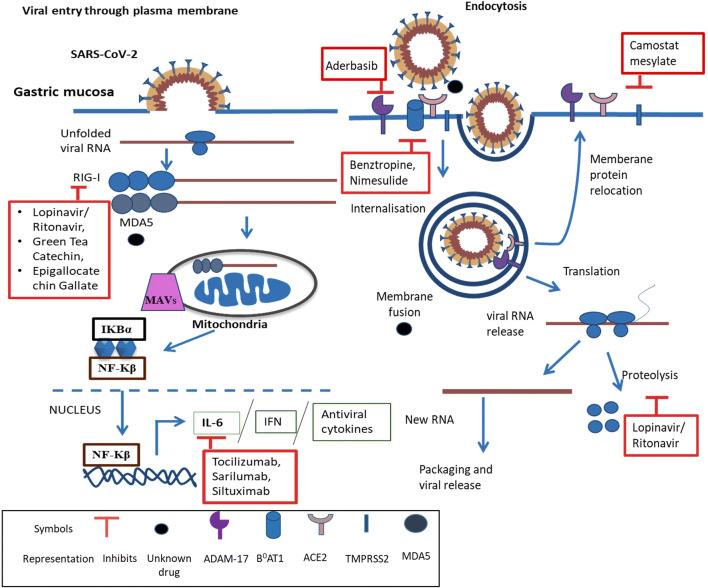
ACE2 is largely found in the gastrointestinal tract [44]. The messenger RNA of the ACE2 receptor is highly expressed and stabilized by neutral amino acid transporter B0AT1(SLC6A19) in the gastro-intestinal system [45]. SARS-CoV-2 utilizes spike protein for binding to its receptor ACE2, upon binding the plasma membrane fusion occurs and releases viral RNA. Further, viral RNA is detected as pathogen associated molecular pattern (PAMP’s) by pattern recognition receptor (mostly toll like receptor) [46]. The RNA released is recognized by the viral RNA receptor retinoic-acid inducible gene I (RIG-I), cytosolic receptor melanoma differentiation-associated gene5 (MDA5), nucleotidyltransferase cyclic GMP-AMP synthase (cGAS), and stimulator of interferon genes (STING) [40, 47]. Furthermore, this binding recruits TIR-domain-containing adaptor protein along with mitochondrial antiviral-signaling protein (MAVS) and induces downstream signaling which includes activation of nuclear factor-κB (NF-κB), interferon (IFN), and series of pro-inflammatory (IL-6) and antiviral cytokines (Fig. 1) [48, 49]. This viral entry can also be through endocytosis by clathrin dependent or independent pathway which may be used as a mechanism to avoid host detection (Fig. 1) [50, 51]. The upregulation of RIG-I and MDA-5 mediated through retinoic acid-inducible gene I-like receptors (RLRs) and toll-like receptors is also associated with gastric adenocarcinoma cells [52]. The role of ACE is controversial; it is known to have a significant role in gastric ulcer healing and can be related to virus mediated diarrhea [53]. Hence, studying ACE2 related signaling with gastric disorders and SARS-CoV-2 would provide better insight in determining therapeutic targets.
Fig. 1.
Potential colliding targets, their signaling, and inhibitors: SARS-CoV-2 RNA enters the cells through plasma membrane and endocytosis followed by recognition by RIG-I and MDA5 which binds to MAVs. Furthermore, this leads to the secretion of IL-6, IFN, and antiviral cytokines. Moreover, commercially available drugs and the targets with no known drugs have also been shown. Here ACE2, TMPRSS2, RIG-I, MDA5, IL-6, and ADAM-17 may serve as important targets concerning gastric disorder as well
Reports suggest that through in-silico analysis of viral peptides of major histocompatibility complex class I gene (MHC) ([HLA] A, B, C), HLA may present highly conserved SARS-CoV-2 peptides which suggest its ability to activate cross-protective T cell mediated immunity. These findings suggest that severity of SARS-CoV-2 may be affected by genetic variability of [HLA] A, B, and C [54]. Downregulation of HLA is a probable cause for poor prognosis in gastric and esophageal cancer. Though epigenetic and oncogenic studies of HLA are still ambiguous, reports suggest that MAPK and AKT (HER2) signaling regulates the expression of HLA in gastric and esophageal cancer [55]. Considering the importance of HLA and MAPK in the signaling pathway, they appear to be important targets for therapeutic use.
Sialic acid which is responsible for regulating various physiological and pathological processes is composed of a diversified family of acidic sugar [56]. This can be used for internalization of bacteria like H. pylori in gastric mucosa through sialic acid binding adhesin (SabA) for subsequent persistence of infection. H. pylori infection is considered a crucial risk factor in gastric carcinogenesis where only a subset of individuals develops tumor [57]. In healthy conditions, gastric mucosa mostly expresses neutral fucosylated glycans whose glycophenotype is modified by H.pylori infection which leads to overexpression of β3-N-acetylglucosaminyltransferase-5 (β3GnT5) which is followed by increased biosynthesis of sialyl-Lewis x [58]. This increased biosynthesis of sialyl-Lewis x further leads to successful strengthening of gastric epithelial attachment to H.pylori for efficient colonization, hence increasing the risk of gastric disorder. It has also been found that high levels of α 2,3 sialic acid residues were linked to GC cells invasion and metastasis [59]. Thus, the role of sialic acid in gastric comorbidities is evident; however, its role in SARS-CoV-2 is also emerging. The SARS-CoV-2 has very high infectivity due to its structure which contains various groups of terminal sialic acid [60, 61]. According to Menicagli et.al., one of the hypotheses states that sialic acid strengthens the capacity of diffusion which relies on the varied number of glycoproteins present on the COVID-19 capsule [62]. Additionally, there are certain sialic acids present on the host cell surface which act as additional receptors for binding sites of the S protein of SARS-CoV-2 [63], hence playing a role in the pathogenicity and epidemiology of the associated disease [63].
CD147 also known as Basigin or EMMPRIN is a transmembrane glycoprotein known to bind to spike protein of SARS-CoV-2 and mediate in virus invasion and infection to other cells [11, 38]. Recently, a research team by Zhinan et al. after conducting surface Plasmon resonance analysis and competitive inhibition experiment found that CD147 anti-body competitively inhibited binding of CD147 and S protein [64]. Hence, it can be the potent target for therapeutics for COVID-19 patients. Moreover, CD147 is required for malaria parasite, Plasmodium falciparum invasion, which can explain the infection of SARS-CoV-2 in red blood cells [65]. However, it is also associated with gastric cancer invasion, metastasis and might be utilized for prognosis and indicator of tumor recurrence [66].
Increased level of IL-6 is often related to respiratory failure and acute respiratory distress syndrome (ARDS). It plays a crucial role in aggravating cytokine storms in COVID-19. It mainly follows two pathways cis and trans. In cis, IL-6 binds with membrane bound IL-6 receptor (mIL-6R) and gp130 which activates Janus kinases (JAKs) and signal transducer and activator of transcription 3 (STAT3) pathway. This JAK/STAT3 pathway then activates innate and acquired immunity causing cytokine release syndrome (CRS). In trans pathway, IL-6 binds to its soluble receptor (sIL-6R) which again activates JAK-STAT3 signaling. Further, it leads to secretion of vascular endothelial growth factor (VEGF), monocyte chemoattractant protein–1 (MCP-1), IL-8 and reduced E-cadherin expression on endothelial cells hence aggravating cytokine storm [67]. IL-6 R can also be used as a prognostic marker in gastric cancer. Hence, IL-6 can be a potent therapeutic target for COVID-19 patients suffering from gastric cancer [68].
Potential Colliding Targets and Associated Drugs
The potential targets like ACE-2, TMPRSS2, Cathepsin L/B, CD147, STING, RIG-I, MDA5, P38 MAPK, ADAM-17, sialic acid B0AT1, and IL-6 along with drugs can be used as therapeutics in COVID-19 and associated gastrointestinal disorders have been briefly explained.
SARS-CoV-2 spike protein binds to the ACE-2 receptor which may cause activation of p38 mitogen-activated protein kinase (MAPK), upregulate ADAM-17, and stimulate ROS formation [69, 70]. The Spike-ACE-2 complex is proteolytically processed by type 2 transmembrane protease (TMPRSS2) at its S1/S2 junction to release S2 subunit, which further facilitates viral and cell membrane fusion [71, 72]. Hence, to facilitate virus entry, TMPRSS2 and cathepsin L/B primes the S-protein of SARS-CoV-2 [71]. The commercially available drug for TMPRSS2 is camostat mesylate and nafamostat (Table 1), which is a clinically proven protease inhibitor and it can also improve reflux esophagitis, dyspepsia and inhibit SARS-CoV-2 infection [73–76]. Cathepsin L/B which also plays a crucial role in gastric cancer can be inhibited by cysteine protease inhibitor E64d (Table 1) [71, 99]. CD147 helps in P. falciparum invasion by binding to reticulocyte-binding protein 5 (Rh5) [95]. CD147 can also bind to the spike protein of SARS-CoV-2 for entry into the host cell [11, 38]. Targeting of CD147 through the meplazumab monoclonal antibody could be a possible potential therapy against COVID-19 disease (Table 1) [96].
Table 1.
Potential colliding targets and probable drugs with their phase I/II/III trials with their activity
| S. No | Target | Gastrointestinal disorder association | Drug/phaseI/II-(PI)Phase II/III-(PII) | Drug activity | References |
|---|---|---|---|---|---|
| 1 | ACE2 | Gastric ulcer healing, virus mediated diarrhea | PII-APN01 | ACE2 inhibitor | [7–10, 44, 53, 69, 70] |
| 2 | TMPRSS2 | Dyspepsia, reflux esophagitis | Camostat mesylate PII-NCT04352400 PII-nafamostat | Protease Inhibitor | [71–76] |
| 3 | (RIG-I) | H.pylori infection, gastric adenocarcinoma | Lopinavir/ritonavir, green tea catechin, epigallocatechin gallate PI-NCT03065023 | Protease Inhibitor | [47, 77–81] |
| 4 | MDA5 | Gastric adenocarcinoma | [47] | ||
| 5 | [HLA]A | Gastric cancer, esophageal cancer | No known commercial drug available | [13–15, 41, 54, 55] | |
| 6 | Sialic acid containing | H.pylori infection | Egg whites | [10–12, 14, 39, 56–63, 82, 83, 79] | |
| 7 | ADAM-17 | Gastric acid secretion | Aderbasib (INCB7839) | ADAM-17/TACE inhibitor | [69, 70, 84, 85] |
| 8 | P38 MAPK | Gastric cancer | PI-Ralimetinib (LY2228820) | P38 MAPK inhibitor | [69, 70, 85, 86] |
| 9 | B0AT1 (SLC6A19) | Benztropine, nimesulide | B0AT1 Inhibitor, Cyclooxygenase Inhibitor | [45, 87, 88] | |
| 10 | IL-6 | Chronic gastritis | Tocilizumab, sarilumab | Interleukin-6 receptor antagonist | [20, 39, 42, 43, 48, 49, 67, 68, 89–94] |
| 12 | CD147 | Gastric cancer | (PII) NCT04275245/meplazumab | anti-CD147 antibody | [11, 38, 64–66, 95, 96] |
| 13 | Stimulator of interferon genes (STING) | Gastric cancer | [47, 97, 98] | ||
| 14 | Cathepsin B | Digestive cancer | E64d | Cysteine protease inhibitor | [71, 99] |
STING (stimulator of interferon genes) is encoded by TMEM173, considered as a key adaptor molecule which links to the identification of cytosolic DNA leading to production of interferons (IFNs) and NFkB. STING can also identify infections by some RNA viruses. There are arguments that suggest the polymorphisms of the STING pathway could be involved in the pathogenesis of COVID-19 [97]. It is also reported that decreased STING is associated with poor prognosis of gastric cancer patients (Table 1) [98]. RIG-I precisely identifies the intracellular double-stranded viral RNA bearing 5′ triphosphate and invites molecules to activate antiviral signaling [77]. Hence, antiviral drugs would be useful to target its activity such as lopinavir with combination with ritonavir, along with them natural products like green tea catechin, epigallocatechin gallate are also been found to be effective [78], though the administration of lopinavir /ritonavir is often associated with drug-related diarrhea [79, 80]. Green tea catechin is considered a beneficial and effective way to prevent gastrointestinal disorders (Table. I) [81].
Sialic acid also plays a key role in eradicating H.pylori infection by inhibiting ROS production and NF-kB activation [82]. Hence, sialic acid mediated inhibitors may provide potent treatment to patients. Soluble macromolecules containing sialic acid can act as decoy receptors and competitively inhibit the receptor binding such as α2-macroglobulin, umifenovir, and other natural inhibitors include egg white which can be useful in COVID-19 patients having gastric disorders (Table 1) [82, 83]. B0AT1 is an amino acid transporter, abbreviation of major apical neutral amino acid transport system B0 and it belongs to the solute carrier family 6 (SLC6A19) [87]. It is the major Na+-dependent transporter for neutral amino acids in the small intestine and kidney [87]. The approved drug for B0AT1 inhibitor is Nimesulide (cyclooxygenase inhibitor) and Benztropine (Table 1) [88]. Increased secretion of pro-inflammatory cytokines like Interleukin-6 (IL-6) is a common factor in patients with gastritis and SARS-CoV-2 infection. IL-6 being a pro-inflammatory cytokine is linked to increased inflammation in chronic acute gastritis [89, 90]. IL-6 may act as a prognostic marker for gastric cancer and a potential biomarker for COVID-19 progression [68, 91]. Hence, tocilizumab and sarilumab are FDA-approved drugs which is an interleukin-6 receptor antagonist can be a potent therapeutic drug (Table 1) [92, 93]. Siltuximab is a monoclonal antibody against IL-6 (Table 1) [94].
Gut Dysbiosis: Interaction of Microbiota with SARS-CoV-2
The human gut consists of 1014 microorganisms like bacteria, fungi, archaebacteria, and viruses [100]. These gut microbiomes have an important role in maintaining the health of the individual. The microbiomes and host have a symbiotic association in that the earlier gets food and shelter and in turn, helps the later in regulating physiological functioning like dietary digestion, and imparting protective immunity against pathogens [101]. Alterations of gut microbiota are known as “gut dysbiosis” which is associated with several diseases and disorders such as type 2 diabetes, IBD, cardiovascular disease, and depression [102–104]. Moreover, the COVID-19 treatment regime includes sets of drugs that have negative impacts on different organelles and may cause gut dysbiosis also [105].
Intriguingly, the pulmonary health is also affected through a vital cross-talk between the lungs and the gut microbiota known as the “gut-lung axis” [106]. This axis is bidirectional, i.e., microbial metabolites and the endotoxins can modulate the lung through blood, and during inflammation in the lung, it affects the gut microbiota [107]. This interdependency boosts a striking possibility that SARS-CoV-2 may affect the gut microbiota. Numerous reports have pointed out that alteration in the composition of the gut microbiota is correlated with respiratory infections [108]. Moreover, severe clinical outcomes in SARS-CoV-2 infected patients are associated with immune-compromisation and aging. Therefore, it is tempting to speculate the probable cross-talk between the gut microbiota and the lung in COVID-19 which may further influence the clinical manifestation.
One hypothesis regarding gut dysbiosis is microbiomes’ impacts on cytokines. Type II interferon (interferon-γ) is one such cytokine that plays important roles in antiviral responses [109]. Furthermore, microbial metabolic processes in the gut strongly impact the production of cytokines [101]. Generally, the microbiota can enhance chronic phase protein and interferon signaling in lung cells to protect it from viral infection [110]. However, in the case of SARS-CoV-2 infection, the body’s response to infection changes the scenario. Occasionally, COVID-19 patients’ immune response against the virus results in a cytokine storm ultimately leading to hyper-inflammation and multi-organ failure [111, 112], so far, a cytokine profile associated with COVID-19 severity has been characterized by increased interferon-γ inducible protein and many other cytokines. There is a lack of clinical evidence supporting the modulation of the gut microbiota that may have therapeutic value in COVID-19 patients, subjected to further research. From the current understanding, it can be speculated that the host cytokine molecular pathways, microbiota components, in association with cytokine responses, can be used as novel microbiome-based therapeutic approaches for SARS-CoV-2 infection [113].
Diagnostic Approaches
Due to the growing COVID-19 pandemic, there is a shortage of molecular testing capacity. Therefore, there is a need for new point of care immunodiagnostic tests for fast and accurate testing of the disease. However, they can only be used in research purposes and cannot be used for clinical decision making. There are 2 types of new point-of-care immunodiagnostic tests for antigen detection and host antibody detection.
Rapid Diagnostic Tests Based on Antigen Detection
COVID-19 virus proteins (antigens) present in a sample infected person are detected by rapid diagnostic test (RDT). If the concentration of target antigen is sufficient in the sample, the specific antibodies fixed to a paper strip enclosed in a plastic casing will bind to the target protein. This will generate a visually detectable signal, usually within 30 min. Actively replicating a virus expresses the antigen; hence, this test can be used for early detection of infection. However, the tests depend on various factors such as virus concentration, quality of sample, and precise formulation of reagents. Hence, its accuracy for SARS-CoV-2 virus can range from 34 to 80% [114].
Rapid Diagnostic Tests Based on Host Antibody Detection
This RDT detects the antibodies present in the blood of the patients believed to be infected COVID-19 with body produced in response to the infection [115]. The strength of antibodies produced depends on various factors such as severity of disease, age, nutritional status, and certain medication against infections like HIV [116]. Furthermore, most antibodies are produced in the second week of infection or may be generated in the recovery state of patients. However, one of the drawbacks of this test is that it may provide false-positive results by interacting with antibodies generated for other infections [114].
Proteomics of SARS-CoV-2 Infected Host Cells Reveal Their Potential Targeted Therapy
Reaction towards SARS-CoV-2 outbreak through expeditious, fast, and specific testing widely recognized as critical. Nowadays, most qRT-PCR–based methods are used for the testing of SARS-CoV-2, while the non-MS (mass spectrometry) methods such as enzyme-linked immunosorbent assays (ELISAs), western blots, and protein arrays depend on antibodies that were more successful during the outbreak of SARS-CoV in 2003 [117, 118]. Considering the immense variability in antibody production, the liquid chromatography coupled to mass spectrometry (LCMS) is an alternative attractive diagnostic approach for the identification of small molecules such as peptides and proteins in clinical settings with consistent results [119, 120].
These techniques measure the quantity of intact or proteolytically digested proteins with specificity, speed, sensitivity and in resolution up to the femto-gram [121]. The most of the LCMS techniques recruit tandem MS [122]. Furthermore, the measurement of fragment ions that is formed in tandem MS has their clinical significance due to its higher specificity and lesser chances of false-positive results [123]. Ihling et al. identified the SARS-CoV-2 virus nucleoproteins from diluted gargle solution of COVID-19 patients through the precipitation of protein followed by the proteolytic digestion through MS [124]. Study shows that the expression of ACE-2 receptor is high in heart tissue through the tandem-MS via tandem mass tag (TMT)-labeling and correlated with the higher heart failure. Bojkova et al. have isolated the SARS-CoV-2 and infected the human Caco-2 cell line which is a human colon epithelial cell and used proteome and translatome mass spectrometry to perform the cellular response [125]. Furthermore, they were identified as the key casualties of the host cell retaliation to infection. The above finding revealed the potential key molecules as a drug target for the SARS-CoV-2 infection [125].
One Dimensional and Two Dimensional Liquid Chromatography ESI/MS and Quantification of Virions
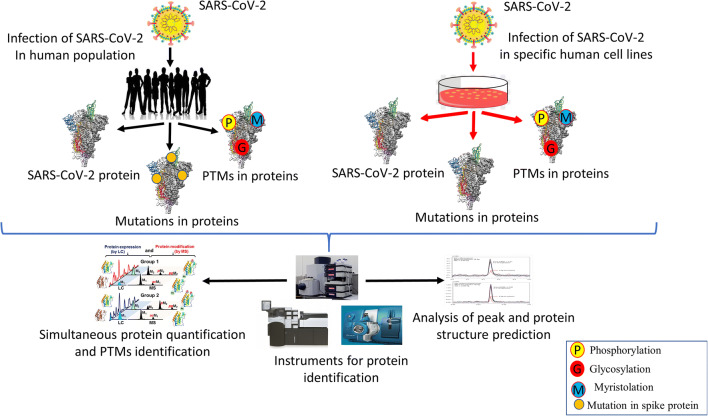
Although it is easy to identify the genome sequences of SARS-CoV, but the recognition of protein is difficult. Two different structural proteins such as spike and nucleocapsid that are encoded by SARS-CoV were identified by Krokhin et al. through the MS technique [126]. Intriguingly, Zeng et al. first time identified the four structural protein and cytosol and nucleus fractions of SARS-CoV infected vero E6 cells and also from the crude virion with shotgun strategy with 2D-LC-MS/MS followed by ESI-MS/MS or by one dimensional electrophoresis followed by ESI-MS/MS [127]. Post-translational modifications (PTMs) of viral proteins interfere with host cell signaling, cellular machinery hijacking and enhancing infectivity [128]. Thus, viruses like influenza, SARS-CoV, and SARS-CoV-2 utilize these PTM for enhancing the replication of their genome and for virion production. Moreover, the novel phosphorylation of structural proteins of SARS-CoV has been identified by this approach fig. 2 [129]. Heavy glycosylation of spikes may facilitate viral attachment, membrane fusion and critically stimulate the host immune response. There are about 22 potential N-glycosylation sites in S1 and S2 subunits of spike proteins. Shajahan et al. mapped the glycosylation sites of spike protein subunits S1 and S2 which are expressed on human cells through resolution MS [130]. Moreover, they have quantitatively characterized the N-glycosylation sites. Intriguingly, they have observed the unpredicted O-glycosylation modifications on the RBD domain of S1 subunit, spike protein. This is the first report where they have shown O-glycosylation on S1 subunit. Thus, this study might play their role in vaccine development through elucidation of the glycan attachment on spike protein of SARS-CoV-2 [130]. The limitations of LCMS in its medical setup, complex matrices, trace level analytes, and time-consuming sample preparations [131].
Fig. 2.
Analysis of SARS-CoV-2 proteins, mutations, and post-translational modifications through LCMS followed by ESI/MS for effective therapeutics
Conclusion
We conclude that SARS-CoV-2 is a causative agent of COVID-19 and its association with the GI tract is well known from earlier coronaviruses leaving a long-lasting impact on patients. In fact, the severity will increase in patients having GI disorders. The attribute of gastrointestinal symptoms existing in COVID-19 is more subtle than the respiratory symptoms; hence, they are easily ignored. However, during the entire course of COVID-19, patients might have only gastrointestinal symptoms and may shed the virus in feces, even though their respiratory samples tests negative. Thus, it is pivotal to observe these gastrointestinal symptoms with caution in the early stage of COVID-19. Furthermore, dynamic monitoring of the digestive system and cytokines is also required during clinical practice to decrease the chances of the complications and mortality of COVID-19 patients. Moreover, the detection of SARS-CoV-2 in fecal samples is essential for clinical practice along with routine testing, particularly for patients with atypical symptoms before leaving the hospital to confirm viral clearance. The ACE2 receptor is ubiquitously found on the surface of the GI tract; thus, it is a potential replication site for SARS-CoV-2. Another receptor sialic acid is used by SARS-CoV-2 for its entry and HLA as its attachment factor. We have mentioned several potential targets that could be used as possible therapeutics. COVID-19 patients have suffered from hyper-inflammation due to which gut microbes will further exacerbate the infection. In addition to inflammation, the current treatment regimens can also negatively affect gut microbiota and cause digestive complications. We have also aimed to provide insight techniques like RDT, LC-MS which can be used for diagnosis and target the viral proteins with high sensitivity. Hence, this review intends to provide comprehensive information on SARS-CoV-2 concerning GI disorders.
Acknowledgments
We are thankful to CSIR for a scholarship to Charu Sonkar, Dharmendra Kashyap, University Grants Commission, and DST-inspire for fellowship to Budhadev Baral and Nidhi Varshney respectively in the form of research stipend. We appreciate our lab colleagues for insightful discussions and advice. We gratefully acknowledge the Indian Institute of Technology Indore for providing facilities and support.
Abbreviations
- (COVID-19)
Coronavirus disease 2019
- (SARS-CoV-2)
Severe acute respiratory syndrome coronavirus 2
- (SARS)
Severe acute respiratory syndrome virus
- (GC)
Gastric cancer
- (H. pylori)
Helicobacter pylori
- (IFN) (STING)
Stimulator of interferon
- (HLA)
Human leukocyte antigen
- (TGEV)
Transmissible gastroenteritis virus
- (LPS)
lipopolysaccharide
- (TNF)
Tumor necrosis factor
- (IL-1)
Interleukin-1
- (IL-6)
Interleukin-6
- (MERS-CoV)
Middle East respiratory syndrome coronavirus
- (PAMP’s)
Type II interferon (interferon-γ), pathogen associated molecular pattern
- (RIG-I)
Retinoic-acid inducible gene I
- (MDA5)
Melanoma differentiation-associated gene5
- (RLRs)
Retinoic acid-inducible gene I-like receptors
- (cGAS)
cyclic GMP-AMP synthase
- (TMPRSS2)
Type 2 transmembrane protease
- (MHC)
Major histocompatibility complex class I gene
- (β3GnT5)
β3-N-acetylglucosaminyltransferase-5
- (9-OAc-Sia)
9-O-acetylated sialic acid
- (SLC6A19)
Solute carrier family 6
- (JAKs)
Janus kinases
- (STAT3)
Signal transducer and activator of transcription 3
- (RDT)
Rapid diagnostic test
- (ELISAs)
Enzyme-linked immunosorbent assays
- (LCMS)
Liquid chromatography coupled to mass spectrometry
- (MS)
Mass spectrometry
- (TMT)
Tandem mass tag
- (PTMs)
Post-translational modifications
- (AT1)
Angiotensin II receptor type 1
Authors’ Contributions
HCJ: supervision and reviewing and editing; CS: conceptualization, methodology, data curation, writing—original draft preparation, and reviewing and editing; DK: writing—original draft preparation, reviewing and editing, and data curation; NV: methodology, original draft preparation, reviewing and editing, and data curation; BB: visualization and reviewing and editing.
Funding
This study is funded by the Council of Scientific and Industrial Research grant no 37(1693)/17/EMR-II, Department of Science and Technology as Ramanujan fellowship grant no. SB/S2/RJN-132/20/5. The funding organization has not played any role in the study design or the preparation of the manuscript.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
This article is part of the Topical Collection on Covid-19
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dharmendra Kashyap and Nidhi Varshney contributed equally to this work.
References
- 1.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 2.WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int. Accessed 17 Sep 2020.
- 3.Endeman H, van der Zee P, van Genderen ME, van den Akker JPC, Gommers D. Progressive respiratory failure in COVID-19: a hypothesis. Lancet Infect Dis. 2020; S1473309920303662. [DOI] [PMC free article] [PubMed]
- 4.Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020; 92:833–840. 10.1002/jmv.25825. [DOI] [PubMed]
- 5.Pan L, Mu M, Yang P, Sun Y, Wang R, Yan J, et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: a descriptive, cross-sectional, multicentre study. Am J Gastroenterol. 2020;115: 766–773. [DOI] [PMC free article] [PubMed]
- 6.Ng SC, Tilg H. COVID-19 and the gastrointestinal tract: more than meets the eye. Gut. 2020;69:973–974. doi: 10.1136/gutjnl-2020-321195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Fan J, Wu F, Huang Q, Guo M, Lv Z, et al. The ACE2/angiotensin-(1–7)/mas receptor axis: pleiotropic roles in cancer. Front Physiol. 2017;8. 10.3389/fphys.2017.00276. [DOI] [PMC free article] [PubMed]
- 8.Mao R, Qiu Y, He JS, Tan JY, Li XH, Liang J, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID- 19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020; 5:667–678 [DOI] [PMC free article] [PubMed]
- 9.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morniroli D, Giannì ML, Consales A, Pietrasanta C, Mosca F. Human Sialome and coronavirus disease-2019 (COVID-19) pandemic: an understated correlation? Front Immunol. 2020;11:1480. doi: 10.3389/fimmu.2020.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulrich H, Pillat MM. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev Rep. 2020;16:434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, Bucci E, Piacentini M, Ippolito G, Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan CM, Lau SKP, Woo PCY, Tse H, Zheng B-J, Chen L, Huang JD, Yuen KY. Identification of major histocompatibility complex class I C molecule as an attachment factor that facilitates coronavirus HKU1 spike-mediated infection. JVI. 2009;83:1026–1035. doi: 10.1128/JVI.01387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unemo M, Aspholm-Hurtig M, Ilver D, Bergström J, Borén T, Danielsson D, Teneberg S. The sialic acid binding SabA adhesin of Helicobacter pylori is essential for nonopsonic activation of human neutrophils. J Biol Chem. 2005;280:15390–15397. doi: 10.1074/jbc.M412725200. [DOI] [PubMed] [Google Scholar]
- 15.Xu D-P, Shi W-W, Zhang T-T, Lv H-Y, Li J-B, Lin A, Yan WH. Elevation of HLA-G-expressing DC-10 cells in patients with gastric cancer. Hum Immunol. 2016;77:800–804. doi: 10.1016/j.humimm.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Li C, Zhao G, Chu H, Wang D, Yan HH-N, et al. Human intestinal tract serves as an alternative infection route for Middle East respiratory syndrome coronavirus. Sci Adv. 2017;3:eaao4966. doi: 10.1126/sciadv.aao4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esper F, Ou Z, Huang YT. Human coronaviruses are uncommon in patients with gastrointestinal illness. J Clin Virol. 2010;48:131–133. doi: 10.1016/j.jcv.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanwar A, Selvaraju S, Esper F. Human coronavirus-HKU1 infection among adults in Cleveland. Ohio Open Forum Infect Dis. 2017;4:ofx052. doi: 10.1093/ofid/ofx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumthip K, Khamrin P, Ushijima H, Maneekarn N. Enteric and non-enteric adenoviruses associated with acute gastroenteritis in pediatric patients in Thailand, 2011 to 2017. PLoS One. 2019;14:e0220263. doi: 10.1371/journal.pone.0220263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To K, Tong JH, Chan PK, Au FW, Chim SS, Allen Chan K, et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: anin-situ hybridization study of fatal cases. J Pathol. 2004;202:157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung WK, To K. Chan PKS, Chan HLY, Wu AKL, Lee N, et al. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection1 1The authors thank Man-yee Yung, Sara Fung, Dr. Bonnie Kwan, and Dr. Thomas Li for their help in retrieving patient information. Gastroenterology. 2003;125:1011–1017. doi: 10.1016/j.gastro.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Liu SM, Yu XH, Tang SL, Tang CK. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int J Antimicrobial Agents. 2020; 55:105951 [DOI] [PMC free article] [PubMed]
- 23.Verity R, Okell LC, Dorigatti I, Winskill P, Whittaker C, Imai N, Cuomo-Dannenburg G, Thompson H, Walker PGT, Fu H, Dighe A, Griffin JT, Baguelin M, Bhatia S, Boonyasiri A, Cori A, Cucunubá Z, FitzJohn R, Gaythorpe K, Green W, Hamlet A, Hinsley W, Laydon D, Nedjati-Gilani G, Riley S, van Elsland S, Volz E, Wang H, Wang Y, Xi X, Donnelly CA, Ghani AC, Ferguson NM. Estimates of the severity of coronavirus disease 2019: a model-based analysis. Lancet Infect Dis. 2020;20:669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portes SAR, Volotão E d M, Rocha MS, Rebelo MC, Xavier M d PTP, de Assis RM, et al. A non-enteric adenovirus A12 gastroenteritis outbreak in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz. 2016;111:403–406. doi: 10.1590/0074-02760160030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian Y, Rong L, Nian W, He Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment Pharmacol Ther. 2020;51:843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Zai J, Wang X, Li Y. Potential of large “first generation” human-to-human transmission of 2019-nCoV. J Med Virol. 2020;92:448–454. doi: 10.1002/jmv.25693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020; 159:81–95 [DOI] [PMC free article] [PubMed]
- 28.Lee I-C, Huo T-I, Huang Y-H. Gastrointestinal and liver manifestations in patients with COVID-19. J Chin Med Assoc. 2020;83:521–523. doi: 10.1097/JCMA.0000000000000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, Guo Q, Sun X, Zhao D, Shen J, Zhang H, Liu H, Xia H, Tang J, Zhang K, Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020; 323:1843–1844 [DOI] [PMC free article] [PubMed]
- 31.Xing Y-H, Ni W, Wu Q, Li W-J, Li G-J, Wang W-D, Tong JN, Song XF, Wing-Kin Wong G, Xing QS. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect. 2020;53:473–480. doi: 10.1016/j.jmii.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta S, Parker J, Smits S, Underwood J, Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces – a rapid review. Color Dis. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pk C, Da W, Lk T, Sm I, Ac L, Cs L, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363:1699–1700. doi: 10.1016/S0140-6736(04)16255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma C, Cong Y, Zhang H. COVID-19 and the digestive system. Am J Gastroenterol. 2020; 115:1003–1006 [DOI] [PMC free article] [PubMed]
- 35.Uno Y. Why does SARS-CoV-2 invade the gastrointestinal epithelium? Gastroenterology. 2020;159:1622–1623. doi: 10.1053/j.gastro.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry BM, de Oliveira MHS, Benoit J, Lippi G. Gastrointestinal symptoms associated with severity of coronavirus disease 2019 (COVID-19): a pooled analysis. Intern Emerg Med. 2020;15:857–859. doi: 10.1007/s11739-020-02329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camargo SMR, Singer D, Makrides V, Huggel K, Pos KM, Wagner CA, et al. Tissue-specific amino acid transporter partners ACE2 and collectrin differentially interact with hartnup mutations. Gastroenterology. 2009;136:872–882.e3. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. preprint. Microbiology. 2020. 10.1101/2020.03.14.988345.
- 39.Pöhlmann S, Gramberg T, Wegele A, Pyrc K, van der Hoek L, Berkhout B, et al. Interaction between the spike protein of human coronavirus NL63 and its cellular receptor ACE2. In: Perlman S, Holmes KV, et al., editors. The Nidoviruses. Boston: Springer; 2006. pp. 281–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zahn LM. HLA genetics and COVID-19. Science. 2020;368:841.2–84841. [Google Scholar]
- 42.Ni M, Tian F, Xiang D, Yu B. Characteristics of inflammatory factors and lymphocyte subsets in patients with severe COVID-19. J Med Virol. 2020. 10.1002/jmv.26070. [DOI] [PMC free article] [PubMed]
- 43.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine Growth Factor Rev. 2020; 54: 62–75 [DOI] [PMC free article] [PubMed]
- 44.Sungnak W, Huang N, Bécavin C, Berg M, Queen R, Litvinukova M, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang Y, Rose AJ, Sijmonsma TP, Bröer A, Pfenninger A, Herzig S, Schmoll D, Bröer S. Mice lacking neutral amino acid transporter B0AT1 (Slc6a19) have elevated levels of FGF21 and GLP-1 and improved glycaemic control. Molec Metab. 2015;4:406–417. doi: 10.1016/j.molmet.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 47.Essig M, Matt M, Massy Z. The COVID-19 outbreak and the angiotensin-converting enzyme 2: too little or too much? Nephrol Dial Transplant. 2020;35:1073–1075. doi: 10.1093/ndt/gfaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawlik MW, Kwiecien S, Ptak-Belowska A, Pajdo R, Olszanecki R, Suski M, et al. The renin-angiotensin system and its vasoactive metabolite angiotensin-(1-7) in the mechanism of the healing of preexisting gastric ulcers. The involvement of mas receptors, nitric oxide, prostaglandins and proinflammatory cytokines. J Physiol Pharmacol. 2016;67:75–91. [PubMed] [Google Scholar]
- 49.Fujita M, Hayashi I, Yamashina S, Fukamizu A, Itoman M, Majima M. Angiotensin type 1a receptor signaling-dependent induction of vascular endothelial growth factor in stroma is relevant to tumor-associated angiogenesis and tumor growth. Carcinogenesis. 2005;26:271–279. doi: 10.1093/carcin/bgh324. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuba K, Imai Y, Ohto-Nakanishi T, Penninger JM. Trilogy of ACE2: a peptidase in the renin-angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol Ther. 2010;128:119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qu J, Hou Z, Han Q, Zhang C, Tian Z, Zhang J. Poly(I:C) exhibits an anti-cancer effect in human gastric adenocarcinoma cells which is dependent on RLRs. Int Immunopharmacol. 2013;17:814–820. doi: 10.1016/j.intimp.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 53.D’Amico F, Baumgart DC, Danese S, Peyrin-Biroulet L. Diarrhea during COVID-19 infection: pathogenesis, epidemiology, prevention, and management. Clin Gastroenterol Hepatol. 2020;18:1663–1672. doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, Thompson RF. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J Virol 2020; 94: e00510–20 [DOI] [PMC free article] [PubMed]
- 55.Qu J-L, Qu X-J, Zhao M-F, Teng Y-E, Zhang Y, Hou K-Z, Jiang YH, Yang XH, Liu YP. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41:875–880. doi: 10.1016/j.dld.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 56.Jun L, Yuanshu W, Yanying X, Zhongfa X, Jian Y, Fengling W, Xianjun Q, Kokudo N, Wei T, Weixia Z, Shuxiang C. Altered mRNA expressions of sialyltransferases in human gastric cancer tissues. Med Oncol. 2012;29:84–90. doi: 10.1007/s12032-010-9771-1. [DOI] [PubMed] [Google Scholar]
- 57.Pinho SS, Carvalho S, Marcos-Pinto R, Magalhães A, Oliveira C, Gu J, Dinis-Ribeiro M, Carneiro F, Seruca R, Reis CA. Gastric cancer: adding glycosylation to the equation. Trends Mol Med. 2013;19:664–676. doi: 10.1016/j.molmed.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Marcos NT, Magalhães A, Ferreira B, Oliveira MJ, Carvalho AS, Mendes N, et al. Helicobacter pylori induces β3GnT5 in human gastric cell lines, modulating expression of the SabA ligand sialyl–Lewis x. J Clin Invest. 2008;118:2325–2336. doi: 10.1172/JCI34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang F-L, Cui S-X, Sun L-P, Qu X-J, Xie Y-Y, Zhou L, Mu YL, Tang W, Wang YS. High expression of α 2, 3-linked sialic acid residues is associated with the metastatic potential of human gastric cancer. Cancer Detect Prev. 2009;32:437–443. doi: 10.1016/j.cdp.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Robson B. Bioinformatics studies on a function of the SARS-CoV-2 spike glycoprotein as the binding of host sialic acid glycans. Comput Biol Med. 2020;122:103849. doi: 10.1016/j.compbiomed.2020.103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fantini J, Di Scala C, Chahinian H, Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55:105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menicagli R, Limodio M. COVID-19 solution. Int J Prev Med. 2020;11(6):11–73. doi: 10.4103/ijpvm.IJPVM_227_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Devaux CA, Rolain J-M, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020;55:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.The second key protein of SARS-CoV-2 infection, CD147 - CLOUD-CLONE CORP.(CCC). http://www.cloud-clone.com/topic/The-second-key-protein-of-SARS-CoV-2-infection%2D%2DCD147.html. Accessed 16 Jul 2020.
- 65.Muramatsu T. Basigin: a multifunctional membrane protein with an emerging role in infections by malaria parasites. Expert Opin Ther Targets. 2012;16:999–1011. doi: 10.1517/14728222.2012.711818. [DOI] [PubMed] [Google Scholar]
- 66.Chu D, Zhu S, Li J, Ji G, Wang W, Wu G, et al. CD147 Expression in human gastric cancer is associated with tumor recurrence and prognosis. PLoS ONE. 2014;9:e101027. doi: 10.1371/journal.pone.0101027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.COVID-19 and the cytokine storm the crucial role of IL-6 - Enzo Life Sciences. https://www.enzolifesciences.com/science-center/technotes/2020/april/covid-19-and-the-cytokine-storm-the-crucial-role-of-il-6/. Accessed 16 Jul 2020.
- 68.Simondurairaj C, Krishnakumar R, Sundaram S, Venkatraman G. Interleukin-6 receptor (IL-6R) expression in human gastric carcinoma and its clinical significance. Cancer Investig. 2019;37:293–298. doi: 10.1080/07357907.2019.1638395. [DOI] [PubMed] [Google Scholar]
- 69.Grimes JM, Grimes KV. p38 MAPK inhibition: a promising therapeutic approach for COVID-19. J Mol Cell Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gwathmey TM, Pendergrass KD, Reid SD, Rose JC, Diz DI, Chappell MC. Angiotensin-(1-7)-angiotensin-converting enzyme 2 attenuates reactive oxygen species formation to angiotensin II within the cell nucleus. Hypertension. 2010;55:166–171. doi: 10.1161/HYPERTENSIONAHA.109.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong J-C, Turner AJ, Raizada MK, Grant MB, Oudit GY. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rabi FA, Al Zoubi MS, Kasasbeh GA, Salameh DM, Al-Nasser AD. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9:231. doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kono K, Takahashi A, Sugai H, Umekawa T, Yano T, Kamiyasu K, Teramatsu M, Fujii H. Oral trypsin inhibitor can improve reflux esophagitis after distal gastrectomy concomitant with decreased trypsin activity. Am J Surg. 2005;190:412–417. doi: 10.1016/j.amjsurg.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 75.Sai JK, Suyama M, Kubokawa Y, Matsumura Y, Inami K, Watanabe S. Efficacy of camostat mesilate against dyspepsia associated with non-alcoholic mild pancreatic disease. J Gastroenterol. 2010;45:335–341. doi: 10.1007/s00535-009-0148-1. [DOI] [PubMed] [Google Scholar]
- 76.Yamamoto M, Matsuyama S, Li X, Takeda M, Kawaguchi Y, Inoue J, Matsuda Z. Identification of nafamostat as a potent inhibitor of Middle East respiratory syndrome coronavirus S protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ranjith-Kumar CT, Murali A, Dong W, Srisathiyanarayanan D, Vaughan R, Ortiz-Alacantara J, Bhardwaj K, Li X, Li P, Kao CC. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009;284:1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ranjith-Kumar CT, Lai Y, Sarisky RT, Cheng KC. Green tea catechin, epigallocatechin gallate, suppresses signaling by the dsRNA innate immune receptor RIG-I. PLoS One. 2010;5:e12878. doi: 10.1371/journal.pone.0012878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cvetkovic RS, Goa KL. Lopinavir/ritonavir. Drugs. 2003;63:769–802. doi: 10.2165/00003495-200363080-00004. [DOI] [PubMed] [Google Scholar]
- 80.Chandwani A, Shuter J. Lopinavir/ritonavir in the treatment of HIV-1 infection: a review. Ther Clin Risk Manag. 2008;4:1023–1033. doi: 10.2147/tcrm.s3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koo MWL, Cho CH. Pharmacological effects of green tea on the gastrointestinal system. Eur J Pharmacol. 2004;500:177–185. doi: 10.1016/j.ejphar.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 82.Matrosovich M, Klenk H-D. Natural and synthetic sialic acid-containing inhibitors of influenza virus receptor binding. Rev Med Virol. 2003;13:85–97. doi: 10.1002/rmv.372. [DOI] [PubMed] [Google Scholar]
- 83.Leneva IA, Burtseva EI, Yatsyshina SB, Fedyakina IT, Kirillova ES, Selkova EP, Osipova E, Maleev VV. Virus susceptibility and clinical effectiveness of anti-influenza drugs during the 2010–2011 influenza season in Russia. Int J Infect Dis. 2016;43:77–84. doi: 10.1016/j.ijid.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 84.Murumkar PR, Ghuge RB, Chauhan M, Barot RR, Sorathiya S, Choudhary KM, et al. Recent developments and strategies for the discovery of TACE inhibitors. Expert Opin Drug Discov. 2020;15:779–801 [DOI] [PubMed]
- 85.Smolka AJ, Backert S. How Helicobacter pylori infection controls gastric acid secretion. J Gastroenterol. 2012;47:609–618. doi: 10.1007/s00535-012-0592-1. [DOI] [PubMed] [Google Scholar]
- 86.Vergote I, Heitz F, Buderath P, Powell M, Sehouli J, Lee CM, Hamilton A, Fiorica J, Moore KN, Teneriello M, Golden L, Zhang W, Pitou C, Bell R, Campbell R, Farrington DL, Bell-McGuinn K, Wenham RM. A randomized, double-blind, placebo-controlled phase 1b/2 study of ralimetinib, a p38 MAPK inhibitor, plus gemcitabine and carboplatin versus gemcitabine and carboplatin for women with recurrent platinum-sensitive ovarian cancer. Gynecol Oncol. 2020;156:23–31. doi: 10.1016/j.ygyno.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 87.Bröer S, Gether U. The solute carrier 6 family of transporters. Br J Pharmacol. 2012;167:256–278. doi: 10.1111/j.1476-5381.2012.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng Q, Shah N, Bröer A, Fairweather S, Jiang Y, Schmoll D, Corry B, Bröer S. Identification of novel inhibitors of the amino acid transporter B0AT1 (SLC6A19), a potential target to induce protein restriction and to treat type 2 diabetes. Br J Pharmacol. 2017;174:468–482. doi: 10.1111/bph.13711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bauditz J, Ortner M, Bierbaum M, Niedobitek G, Lochs H, Schreiber S. Production of IL-12 in gastritis relates to infection with Helicobacter pylori. Clin Exp Immunol. 1999;117:316–323. doi: 10.1046/j.1365-2249.1999.00968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang M, Dong D, Qi D, Ling C, Zhang D. Hp infection and expression of TGF-βR II,IL-6 and TNF-α in patients with chronic atrophic gastritis. Chin J Immunol. 2018;34:751–756. [Google Scholar]
- 91.Ulhaq ZS, Soraya GV. Interleukin-6 as a potential biomarker of COVID-19 progression. Med Mal Infect. 2020;50:382–383. doi: 10.1016/j.medmal.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tu Y-F, Chien C-S, Yarmishyn AA, Lin Y-Y, Luo Y-H, Lin Y-T, Lai WY, Yang DM, Chou SJ, Yang YP, Wang ML, Chiou SH. A review of SARS-CoV-2 and the ongoing clinical trials. Int J Mol Sci. 2020;21:2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fu B, Xu X, Wei H. Why tocilizumab could be an effective treatment for severe COVID-19? J Transl Med. 2020;18:164. doi: 10.1186/s12967-020-02339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.van Rhee F, Fayad L, Voorhees P, Furman R, Lonial S, Borghaei H, Sokol L, Crawford J, Cornfeld M, Qi M, Qin X, Herring J, Casper C, Kurzrock R. Siltuximab, a novel anti-interleukin-6 monoclonal antibody, for Castleman’s disease. J Clin Oncol. 2010;28:3701–3708. doi: 10.1200/JCO.2009.27.2377. [DOI] [PubMed] [Google Scholar]
- 95.van Ooij C. Basigin opens the door to malaria. Nat Rev Microbiol. 2012;10:3–3. doi: 10.1038/nrmicro2715. [DOI] [PubMed] [Google Scholar]
- 96.Kumar R, Gupta N, Kodan P, Mittal A, Soneja M, Wig N. Battling COVID-19: using old weapons for a new enemy. Trop Dis Travel Med Vaccines. 2020;6:6. doi: 10.1186/s40794-020-00107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berthelot J-M, Lioté F. COVID-19 as a STING disorder with delayed over-secretion of interferon-beta. EBioMedicine. 2020;56:102801. doi: 10.1016/j.ebiom.2020.102801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Song S, Peng P, Tang Z, Zhao J, Wu W, Li H, Shao M, Li L, Yang C, Duan F, Zhang M, Zhang J, Wu H, Li C, Wang X, Wang H, Ruan Y, Gu J. Decreased expression of STING predicts poor prognosis in patients with gastric cancer. Sci Rep. 2017;7:39858. doi: 10.1038/srep39858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen S, Dong H, Yang S, Guo H. Cathepsins in digestive cancers. Oncotarget. 2017;8:41690–41700. doi: 10.18632/oncotarget.16677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dhar D, Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285:198018. doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, et al. Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD Treatment Targeting the Gut Microbiome. Pathogens. 2019;8:126. doi: 10.3390/pathogens8030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, Shulzhenko N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tang WHW, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120:1183–1196. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jakhmola S, Indari O, Baral B, Kashyap D, Varshney N, Das A, Chatterjee S, Jha HC. Comorbidity assessment is essential during COVID-19 treatment. Front Physiol. 2020;11:984. doi: 10.3389/fphys.2020.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Keely S, Talley NJ, Hansbro PM. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. 2012;5:7–18. doi: 10.1038/mi.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dumas A, Bernard L, Poquet Y, Lugo-Villarino G, Neyrolles O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. 2018;20:e12966. doi: 10.1111/cmi.12966. [DOI] [PubMed] [Google Scholar]
- 108.Groves HT, Higham SL, Moffatt MF, Cox MJ, Tregoning JS. Respiratory viral infection alters the gut microbiota by inducing inappetence. mBio. 2020;11:e03236–e03219. doi: 10.1128/mBio.03236-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, et al. Linking the human gut microbiome to inflammatory cytokine production capacity. Cell. 2016;167:1125–1136.e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Trottein F, Sokol H. Potential causes and consequences of gastrointestinal disorders during a SARS-CoV-2 infection. Cell Reports. 2020;32:107915 [DOI] [PMC free article] [PubMed]
- 111.Kalantar-Zadeh K, Ward SA, Kalantar-Zadeh K, El-Omar EM. Considering the effects of microbiome and diet on SARS-CoV-2 infection: nanotechnology roles. ACS Nano. 2020;14:5179–5182. doi: 10.1021/acsnano.0c03402. [DOI] [PubMed] [Google Scholar]
- 112.Jakhmola S, Indari O, Chatterjee S, Jha HC. SARS-CoV-2, an underestimated pathogen of the nervous system. SN Compr Clin Med. 2020. 10.1007/s42399-020-00522-7. [DOI] [PMC free article] [PubMed]
- 113.Mendes V, Galvão I, Vieira AT. Mechanisms by which the gut microbiota influences cytokine production and modulates host inflammatory responses. J Interf Cytokine Res. 2019;39:393–409. doi: 10.1089/jir.2019.0011. [DOI] [PubMed] [Google Scholar]
- 114.Advice on the use of point-of-care immunodiagnostic tests for COVID-19. https://www.who.int/news-room/commentaries/detail/advice-on-the-use-of-point-of-care-immunodiagnostic-tests-for-covid-19. Accessed 16 Jul 2020.
- 115.Ravi N, Cortade DL, Ng E and Wang SX. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens Bioelectron. 2020;165:112454 [DOI] [PMC free article] [PubMed]
- 116.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases, 2020;ciaa344 [DOI] [PMC free article] [PubMed]
- 117.Yip TTC, Cho WCS, Cheng WW, Chan JWM, Ma VWS, Yip T-T, Lau Yip CNB, Ngan RKC, Law SCK. Application of ProteinChip array profiling in serum biomarker discovery for patients suffering from severe acute respiratory syndrome. In: Rampal JB, editor. Microarrays: volume 2: applications and data analysis. Totowa: Humana Press; 2007. pp. 313–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yip TT, Chan JW, Cho WC, Yip T-T, Wang Z, Kwan T-L, Law SC, Tsang DN, Chan JK, Lee KC, Cheng WW, Ma VW, Yip C, Lim CK, Ngan RK, Au JS, Chan A, Lim WW, Ciphergen SARS Proteomics Study Group Protein Chip Array profiling analysis in patients with severe acute respiratory syndrome identified serum amyloid a protein as a biomarker potentially useful in monitoring the extent of pneumonia. Clin Chem. 2005;51:47–55. doi: 10.1373/clinchem.2004.031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kammila S, Das D, Bhatnagar PK, Sunwoo HH, Zayas-Zamora G, King M, Suresh MR. A rapid point of care immunoswab assay for SARS-CoV detection. J Virol Methods. 2008;152:77–84. doi: 10.1016/j.jviromet.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cho S-J, Woo H-M, Kim K-S, Oh J-W, Jeong Y-J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J Biosci Bioeng. 2011;112:535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pitt JJ. Principles and applications of liquid chromatography-mass spectrometry in clinical biochemistry. Clin Biochem Rev. 2009;30:19–34. [PMC free article] [PubMed] [Google Scholar]
- 122.Grebe SKG, Singh RJ. Clinical peptide and protein quantification by mass spectrometry (MS) TrAC Trends Anal Chem. 2016;84:131–143. [Google Scholar]
- 123.Liebler DC, Zimmerman LJ. Targeted quantitation of proteins by mass spectrometry. Biochemistry. 2013;52:3797–3806. doi: 10.1021/bi400110b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ihling C, Tänzler D, Hagemann S, Kehlen A, Hüttelmaier S, Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. Journal of Proteome Research. 2020. 10.1021/acs.jproteome.0c00280 [DOI] [PubMed]
- 125.Bojkova D, Klann K, Koch B, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472 [DOI] [PMC free article] [PubMed]
- 126.Krokhin O, Li Y, Andonov A, Feldmann H, Flick R, Jones S, Stroeher U, Bastien N, Dasuri KVN, Cheng K, Simonsen JN, Perreault H, Wilkins J, Ens W, Plummer F, Standing KG. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol Cell Proteomics. 2003;2:346–356. doi: 10.1074/mcp.M300048-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zeng R, Ruan H-Q, Jiang X-S, Zhou H, Shi L, Zhang L, Sheng QH, Tu Q, Xia QC, Wu JR. Proteomic analysis of SARS associated coronavirus using two-dimensional liquid chromatography mass spectrometry and one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by mass spectrometric analysis. J Proteome Res. 2004;3:549–555. doi: 10.1021/pr034111j. [DOI] [PubMed] [Google Scholar]
- 128.Fung TS, Liu DX. Post-translational modifications of coronavirus proteins: roles and function. Futur Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fukushi M, Yoshinaka Y, Matsuoka Y, Hatakeyama S, Ishizaka Y, Kirikae T, Sasazuki T, Miyoshi-Akiyama T. Monitoring of S protein maturation in the endoplasmic reticulum by Calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus. J Virol. 2012;86:11745–11753. doi: 10.1128/JVI.01250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shajahan A, Supekar NT, Gleinich AS, Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;cwaa042 [DOI] [PMC free article] [PubMed]
- 131.Peng J, Tang F, Zhou R, Xie X, Li S, Xie F, Yu P, Mu L. New techniques of on-line biological sample processing and their application in the field of biopharmaceutical analysis. Acta Pharm Sin B. 2016;6:540–551. doi: 10.1016/j.apsb.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]