Abstract
Anaerobic fungi (AF) play an essential role in feed conversion due to their potent fiber degrading enzymes and invasive growth. Much has been learned about this unusual fungal phylum since the paradigm shifting work of Colin Orpin in the 1970s, when he characterized the first AF. Molecular approaches targeting specific phylogenetic marker genes have facilitated taxonomic classification of AF, which had been previously been complicated by the complex life cycles and associated morphologies. Although we now have a much better understanding of their diversity, it is believed that there are still numerous genera of AF that remain to be described in gut ecosystems. Recent marker-gene based studies have shown that fungal diversity in the herbivore gut is much like the bacterial population, driven by host phylogeny, host genetics and diet. Since AF are major contributors to the degradation of plant material ingested by the host animal, it is understandable that there has been great interest in exploring the enzymatic repertoire of these microorganisms in order to establish a better understanding of how AF, and their enzymes, can be used to improve host health and performance, while simultaneously reducing the ecological footprint of the livestock industry. A detailed understanding of AF and their interaction with other gut microbes as well as the host animal is essential, especially when production of affordable high-quality protein and other animal-based products needs to meet the demands of an increasing human population. Such a mechanistic understanding, leading to more sustainable livestock practices, will be possible with recently developed -omics technologies that have already provided first insights into the different contributions of the fungal and bacterial population in the rumen during plant cell wall hydrolysis.
Keywords: anaerobic digestion, carbohydrate-active enzymes, food security, herbivores, methanogenesis, Neocallimastigomycota, rumen, sustainable agriculture
Introduction
The ability of herbivorous animals to ferment recalcitrant plant fiber into utilizable forms of energy, most cases in the form of volatile fatty acids (VFAs), has been attributed to the trillions of microbial cells that inhabit their gastrointestinal tract (Dearing and Kohl, 2017). Importantly, energy stored within complex plant carbohydrates is made accessible to the host animal only through the synergistic activity of its gut microbes (Morais and Mizrahi, 2019).
Independent of which type of mammalian herbivore digestive physiology is considered, all mammalian herbivores have evolved specialized gut compartments to give home to a complex microbial ecosystem of bacteria, anaerobic fungi (AF), protozoa, methanogenic archaea and bacteriophages (Morgavi et al., 2013). All three of the major types of herbivorous mammals depend on these microbiomes and their proper function to support their health and performance: ruminants (e.g., cattle, goats, and sheep), pseudoruminants (e.g., camelids and hippopotami) and hindgut herbivores (e.g., elephants, donkeys, horses and zebras). Furthermore, in foregut fermenting ruminants and pseudoruminants the forestomach microbes themselves also serve as a substantial source of protein and vitamins for the host unlike in hindgut herbivores (Mizrahi, 2013; Nagaraja, 2016). An excellent review of herbivore gastrointestinal physiology, including detailed drawings, and its role on microbial fermentation of plant biomass was summarized by Dehority (2002).
Foregut fermenters are capable of an enhanced degradation of plant biomass that is facilitated by a prolonged (60–90 h) retention time of the feed material in the rumen, the first and largest of three pre-gastric chambers of the foregut fermenter digestive tract. Gastric digestion in the ruminant digestive system occurs in the abomasum, which is the fourth chamber of the ruminant’s foregut. Plant material in the cecum and colon of a hindgut fermenter, on the other hand, is retained on average only half as long (i.e., 30–40 h in equines) as it is in the ruminant’s digestive tract and is consequently less completely digested (Trinci et al., 1994).
Despite the recent significant advances in our understanding of how bacteria and archaea influence the function, resilience, and the environmental footprint of herbivorous mammals such as ruminants (Kittelmann et al., 2014; Henderson et al., 2015; Liu et al., 2017; Huws et al., 2018; Wallace et al., 2019), our knowledge of AF and their influence on the host animal remains limited. This limited knowledge is perhaps not surprising considering that until the mid-20th century it was still believed that all fungi required oxygen (Trinci et al., 1994). It was only in 1975 that the ground-breaking work of Colin Orpin unequivocally confirmed the existence of AF, changing the accepted dogma of the time. Shortly after AF were first isolated and described in the rumen (Orpin, 1975, 1976, 1977a,b), they were also isolated from the horse cecum (Orpin, 1981). Since then, many more AF have been isolated from a wide range of domesticated and wild herbivores with eighteen different genera characterized to date (Hanafy et al., 2020).
To date, AF have been most extensively studied in ruminants, where they are recognized as an important microbe for good rumen function. This is primarily due to their role as highly efficient degraders of recalcitrant plant material (Li et al., 2017; Gilmore et al., 2019). Rumen AF are also syntrophic partners of the methanogenic archaea (Li et al., 2017; Gilmore et al., 2019). Additional insights into the currently understudied herbivore gut mycobiome has the potential to expand our scientific knowledge about life in the absence of oxygen, as well as open new avenues to improve the feed conversion efficiency of plant-based animal feeds. In this review, we discuss the current understanding of AF biology, ecology and their role in livestock production, along with future perspectives on how their true value can be realized.
Taxonomy
Anaerobic fungi were first documented over 100 years ago, when their flagellated zoospores were mistakenly identified as flagellate protozoa (Liebetanz, 1910; Braune, 1913). After first being incorrectly classified as Protozoa (Liebetanz, 1910; Braune, 1913), they were reclassified, due to the significant evidence collected over many years by Orpin, as belonging to the fungal phylum Chytridiomycetes (Barr, 1980, 1988). In 2007, they were acknowledged as being a distinct phylum, the Neocallimastigomycota (Hibbett et al., 2007). Recently, Tedersoo et al. (2018) proposed a new fungal subkingdom, Chytridiomycota, grouping the Neocallimastigomycota with two additional phyla, the aerobic Chytridiomycota and Monoblepharomycota, thereby acknowledging the monophyletic origin of the zoosporic chitinous fungi (Ebersberger et al., 2012).
The Neocallimastigomycota contains only one order (Neocallimastigales) and one family (Neocallimastigaceae) comprising eighteen genera; namely the monocentric rhizoidal Neocallimastix, Piromyces, Oontomyces, Buwchfawromyces, Pecoramyces, Liebetanzomyces, Feramyces, Agriosomyces, Aklioshbomyces, Capellomyces, Ghazallomyces, Joblinomyces, Khoyollomyces, and Tahromyces; the polycentric rhizoidal Anaeromyces and Orpinomyces; and the bulbous Caecomyces and Cyllamyces. Key morphological features of AF taxa, such as the number of flagella on zoospores, type of thallus and rhizoids, steps of zoosporangial development, and the shape of sporangia, are listed in Table 1.
TABLE 1.
Key morphological features of characterized genera of anaerobic fungi.
| Genus | Morphology [zoospore (z), thallus (t), rhizomycelium (r)] | Miscellaneous features | References |
| Agriosomyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Endogenous and exogenous zoosporangial development, rhizoids are swollen below the sporangial tightly constricted neck, swollen sporangiophores | Hanafy et al. (2020) |
| Aklioshbomyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Bi or triflagellate zoospores, endogenous and exogenous zoosporangial development, papillated sporangia, pseudo-intercalary endogenous sporangia occasionally, unbranched sporangiophores | Hanafy et al. (2020) |
| Anaeromyces | Uniflagellate (z) Polycentric (t) Filamentous (r) | Sporangia with acuminate (mucronate) apex, can be located on erect, solitary, unbranched sporangiophore, hyphae are highly branched, often with numerous constrictions (sausage-like appearance), sometimes with root-like appearance | Breton et al. (1990) |
| Buwchfawromyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Extensive rhizoidal system with twisted rhizoids, sporangia with no apical projections, septum can be visible, nuclei located in sporangia, but not observed in sporangiophores or rhizoids | Callaghan et al. (2015) |
| Caecomyces | Uniflagellate (z) Monocentric (t) Bulbous (r) | Bi or quadriflagellate zoospores, vegetative stage is absent of developed branching rhizoidal system, consists of spherical or ovoid bodies (holdfast or haustoria), tubular sporangiophores and bulbous rhizoids, nuclei usually present both in sporangia and vegetative cells | Gold et al. (1988) |
| Capellomyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Endogenous and exogenous zoosporangial development, unbranched sporangiophores can exhibit subsporangial swelling, zoospores released through apical pore | Hanafy et al. (2020) |
| Cyllamyces | Uniflagellate (z) Polycentric (t) Bulbous (r) | Bi or triflagellate zoospores, bulbous holdfast without rhizoids with multiple sporangia, which can be born on a single elongate or branched sporangiophore, nuclei present in bulbous holdfast and sporangiophores | Ozkose et al. (2001) |
| Feramyces | Polyflagellate (z) Monocentric (t) Filamentous (r) | Extensive highly branched rhizoidal system with wide and narrow hyphae, wide hyphae with constrictions at irregular intervals, single terminal sporangium per thallus with the occasional formation of pseudo-intercalary sporangia, sporangiophores frequently coiled or wide and flattened, often forming an apophysis-like or eggcup-like swelling below the sporangium, both endogenous and exogenous zoosporangial development, zoospores are released through apical pore with the sporangial wall staying intact, or through detachment of the whole sporangium | Hanafy et al. (2018) |
| Ghazallomyces | Polyflagellate (z) Monocentric (t) Filamentous (r) | Endogenous and exogenous zoosporangial development, highly branched rhizoids, unbranched sporangiophores, pleomorphic sporangia with septum, sporangial necks constricted with narrow port, zoospores released through apical pore | Hanafy et al. (2020) |
| Joblinomyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Biflagellate zoospores, both endogenous and exogenous zoosporangial development, sporangiophores vary in length, zoospores released through wide apical pore resulting in empty cup-shaped sporangium | Hanafy et al. (2020) |
| Khoyollomyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Endogenous and exogenous zoosporangial development, highly branched rhizoids, intercalary swellings in broad hyphae, multisporangiate thallus, branched sporangiophores with two to four sporangia, zoospores released through wide apical pore | Hanafy et al. (2020) |
| Liebetanzomyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Endogenous and exogenous zoosporangial development, extensive anucleate rhizoidal system without constrictions, single terminal sporangium per thallus, sporangium with septum on sporangiophore of variable length, sometimes forming eggcup-like structure below the sporangium or showing cyst-like structure. Pleomorphism in sporangial and rhizoidal structures on different substrates is typical | Joshi et al. (2018) |
| Neocallimastix | Polyflagellate (z) Monocentric (t) Filamentous (r) | Rhizoid tubular or inflated below the neck of sporangia, sporangia located on unbranched or branched sporangiophores | Heath et al. (1983) |
| Oontomyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Intercalary rhizoidal swellings, sporangia never mucronated, formed terminally, long sporangiophores can be separated from the rhizomycelium by distinct constriction | Dagar et al. (2015) |
| Orpinomyces | Polyflagellate (z) Polycentric (t) Filamentous (r) | Polynucleate rhizomycelium of extensively branched hyphae, wider hyphae can have tightly constricted points at close intervals (bead-like or sausage-like appearance) | Barr et al. (1989) |
| Pecoramyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Biflagellate zoospores, both endogenous and exogenous zoosporangial development, single terminal sporangium formed per thallus, sporangiophores unbranched, often forming apophysis-like or eggcup-like swelling below sporangium. Extensive anucleate rhizoidal system lacks rhizoidal swellings or constrictions | Hanafy et al. (2017) |
| Piromyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Bi or quadriflagellate zoospores, both endogenous and exogenous zoosporangial development, rhizoids with or without subsporangial swelling, septum often in mature zoosporangia | Barr et al. (1989) |
| Tahromyces | Uniflagellate (z) Monocentric (t) Filamentous (r) | Bi or triflagellate zoospores, both endogenous and exogenous zoosporangial development, branched rhizoids, short swollen sporangiophores, sporangia with septum, sporangial necks constricted | Hanafy et al. (2020) |
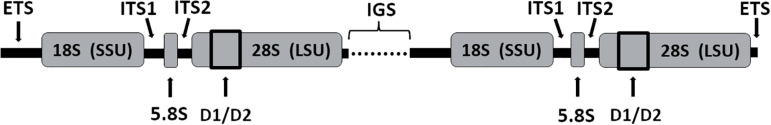
Although morphological features have been crucial for the classification of AF in the past, this approach is encumbered with difficulties due to the extensive morphological variations, pleomorphism in sporangial and rhizoidal structures, similarities in morphological features of monocentric/uniflagellate genera, failure to produce sporangia, and the absence of zoosporogenesis in some polycentric species. Hence, ribosomal RNA (rRNA) operon-based analysis is also needed to verify and support classification of AF. The topology of the ribosomal RNA (rrn) operon, indicating regions that have been used for taxonomic classification, is shown in Figure 1. This culture-independent approach, which in many cases is based on the nucleotide sequence of the internal transcribed spacer 1 (ITS1) region, suggests that the digestive tract of wild and domestic herbivores harbors several clades and genera-equivalent groups within the Neocallimastigaceae that have not yet been cultured (Koetschan et al., 2014; Paul et al., 2018).
FIGURE 1.
Topology of the ribosomal RNA (rrn) operon. Genes of the small (SSU; 18S), large (LSU; 28S) and 5.8S subunit, internal transcribed spacer 1 (ITS1) and 2 (ITS2), flanked by the external transcribed spacer (ETS) regions and linked by the intergenic spacer (IGS).
Although the ITS1 region is currently the molecular marker of choice to assign taxonomy to AF, there are some limitations to this marker, including a high (up to 13%) variation of the ITS1 sequence of clones from a single culture (Callaghan et al., 2015). This variability makes classification of new isolates specifically challenging. Furthermore, the ITS1 region itself ranges in size (Edwards et al., 2008) and recent work of Edwards et al. (2019) indicated preferential amplification of smaller sized ITS1 regions. PCR primer choice is also crucial, which was highlighted by the finding that the sequence amplified by a widely used primer (i.e., MN100F) is not conserved in all AF (Kittelmann et al., 2013; Callaghan et al., 2015).
These problematic aspects of using the ITS1 as phylogenetic marker gene, and the resulting instability of the ITS1 phylogeny, has led to recent efforts to explore the potential of using the sequence of the large 28S rRNA subunit (LSU) as a phylogenetic marker instead (as the 18S rRNA gene is too conserved in AF). It appears that the D1/D2 region of the LSU has a taxonomic resolution similar to the ITS1 region (Wang et al., 2017). However, the more conserved size of the D1/D2 marker and lower heterogeneity within individual cultures enables a more stable phylogenetic backbone for AF classification. Since it provides molecular support for all currently accepted genera of AF, and provides a higher resolution compared to the ITS1 region, utilization of the D1/D2 region of the LSU as a phylogenetic marker seems favorable (Dagar et al., 2011; Wang et al., 2017; Hanafy et al., 2020).
A number of AF specific primers targeting the LSU region have been published (Dollhofer et al., 2016; Nagler et al., 2018), and employed to study AF community composition of environmental samples (Dollhofer et al., 2017; Nagler et al., 2019). However, a sufficiently large reference database for these taxonomic marker genes from previously characterized taxa and their type specimens is still lacking (Wang et al., 2017). There is also a challenge of how to relate LSU sequences to the clades that until now only contain “unculturable” representatives and that were identified by environmentally derived ITS1 sequences (Edwards et al., 2019). Despite the advances of the LSU D1/D2 region as a phylogenetic marker, ITS1 still remains the most accepted phylogenetic marker for AF. As such, it is likely that ITS1 will continue to be used in the near future as the primary barcode to assess AF diversity and community structure in environmental samples. However, it is undisputable that to advance a more accurate and higher resolution classification of members belonging to the AF, a continuous expansion and curation of an AF LSU reference database is need. An alternative option could also be a more complex database that integrates the use of different reference taxonomic marker genes (Wurzbacher et al., 2019). A comprehensive ITS1 sequence database and associated taxonomy files (Koetschan et al., 2014), including the most recently described genera, are available from the AF Network website1.
Latest DNA sequence technologies (i.e., PacBio and Nanopore sequencing) generating long reads appear to offer solutions to overcome some of these challenges, specifically the incompleteness, fragmentation and non-overlapping of extant ribosomal data. These new sequencing techniques enable the generation of full-length reference sequences (up to 10 kb in length) that span several regions suitable for taxonomic classification and intraspecies assignment, including the above mentioned ITS1 and LSU gene. Primers targeting the whole fungal ribosomal tandem repeat region, consisting of ETS, SSU, ITS1, 5.8S, ITS2, LSU, and IGS (Figure 1), have already been successfully applied to specimens of three fungal phyla, including early diverging fungi (Wurzbacher et al., 2019). A stable and reliable AF classification system would be enormously facilitated by the ability to widely utilize the complete ribosomal operon sequence as phylogenetic marker, assuming a corresponding well-curated reference database will be available in the future.
Life Cycle
One of the additional and major challenges that contribute to the current lack of a consistent and standardized taxonomic classification system for AF is the morphological transformations AF undergo throughout their corresponding life cycles (Table 1). AF reproduce asexually and they alternate between a motile zoospore and a non-motile vegetative stage. Flagellate zoospores, which are released from mature sporangia, actively move toward freshly ingested plant tissues in the rumen; a chemotactic response triggered by soluble sugars (Orpin and Bountiff, 1978) and phenolic acids (Wubah and Kim, 1996). Zoospore liberation is influenced by diet, and in ruminants is induced by water-soluble haems and other porphyrins (Orpin and Greenwood, 1986; Orpin, 1994). Although zoospores are motile for several hours (Lowe et al., 1987a), they tend to attach to plant fragments within 30 min after being released from a sporangium (Heath et al., 1986; Edwards et al., 2008). After attachment, the zoospores shed their flagella and form a cyst. The encysted fungus then germinates to produce a fungal thallus that is composed of the sporangium and a filamentous rhizomycelium or a bulbous holdfast.
In most of the monocentric AF, thallus development is of the endogenous type, where on germination, the zoospore cyst develops a germ tube that branches and grows into a rhizoid system. The nucleus remains in the zoospore cyst, which develops into a new sporangium and the anucleate rhizoids. Since the zoospore cyst retains the nucleus and develops into a sporangium, the sporangial development type is referred to as endogenous, and since this type of development results in a single sporangium per thallus, it is said to be monocentric. In some of the monocentric AF, such as the Piromyces spp., thallus development is of the exogenous type, which involves a two-sided germination of the zoospore cyst. During this process, a germ tube develops into a rhizoid system during the endogenous sporangial development, but once a substantial rhizoid has developed, a tubular outgrowth (the sporangiophore or sporangial stalk) emerges on the side opposite of the main rhizoid and the sporangium develops at the apex of the outgrowth. As the original nucleus escapes the zoospore cyst and develops elsewhere, the sporangial development is said to be exogenous (Barr et al., 1989). Both types are strictly determinate. In other members of the monocentric AF, such as the Capellomyces, both exogenous and endogenous sporangial development takes place (Hanafy et al., 2017, 2018, 2020; Joshi et al., 2018).
Polycentric AF display an exogenous thallus development, during which a one-sided germination of the zoospore cyst occurs. During the germination process, the content of the zoospore cyst migrates into germ tube which then develops into a nucleated branched rhizomycelium capable of developing multiple sporangia. Currently it appears that the remaining cyst, which has been emptied, has no further function. Development of thalli of polycentric fungi is said to be non-determinate.
In the case of the bulbous genera, nuclei are observed in the vegetative parts of the thallus (holdfast/branched sporangiophores), consistent with exogenous development. Thallus development in the bulbous genera is of a limited polycentric type, where the encysted zoospore forms a bulbous holdfast without rhizoids. Bulbous holdfasts give rise to single or multiple sporangia including branched sporangiophores. Growth in these fungi is not non-determinate like the thalli of polycentric fungi, but not as strictly determinate as in the case of the monocentric filamentous AF (Ozkose et al., 2001).
Our current understanding of the AF life cycle is based on what has been learned from rumen AF, but it is likely that AF associated with pseudoruminants pass through similar, if not even identical, life cycles stages. However, there are numerous aspects of AF biology that remain to be reassessed for hindgut herbivores. The cause of zoospore release in the hindgut is unclear, as it is not known if the known triggering compounds in the rumen (i.e., water-soluble haems and other porphyrins) can survive passage through the gastric stomach and small intestine. Whereas zoospores that are released within the rumen locate freshly ingested plant material chemotactically using soluble sugars (Orpin and Bountiff, 1978) and phenolic acids (Wubah and Kim, 1996), it is unclear to what extent these chemotactic signals are available to (and used by) AF in the cecum and colon of hindgut herbivores.
Regardless of herbivore type, the life cycle of AF has been proposed to contain a resting phase. While resting structures are still not fully understood, they provide a compelling explanation for why some of the currently known AF can be cultured from fecal material after prolonged periods of desiccation and oxygen exposure (Milne et al., 1989; Davies et al., 1993; McGranaghan et al., 1999; Griffith et al., 2009). To date resting cysts (Orpin, 1981), melanized resistant sporangia (Wubah et al., 1991) and multi-chambered spore-like structures (Brookman et al., 2000) have been described in different AF taxa. Although it may well be the case that there is no resting structure common to all AF, with taxon-specific structures instead, resting structures are thought to play an important role in the inter-animal transfer of AF. For example, it has been suggested that the survival of AF in saliva is likely to be an important transfer mechanism in ruminants and pseudoruminants (Lowe et al., 1987b). In hindgut fermenters, feces may play a more important role as a transfer mechanism between animals than saliva, particularly as certain hindgut fermenters, like foals, exhibit coprophagic behavior (Marinier and Alexander, 1995).
As zoospores and fungal thalli represent different parts of the same AF life cycle, consistent and accurate enumeration of AF is challenging. Approaches used to count exclusively free zoospores (France et al., 1990), fungal colonies on agar strips (Ushida et al., 1989), or both morphologies in culture supernatants (Theodorou et al., 1990; Obispo and Dehority, 1992; Griffith et al., 2009), and chitin measurements (Gay, 1991; Rezaeian et al., 2004b) have, in recent years, been widely replaced by molecular quantification via real-time PCR. This method overcomes the contrast within the life cycle between low zoospore numbers yet high AF vegetative biomass, as well as the paucity or absence of zoosporogenesis observed in some polycentric axenic cultures (Ho and Bauchop, 1991). On the other hand, the real-time PCR approach to quantify fungal biomass possesses its own challenges, such as the varying amount of fungal biomass produced by monocentric, polycentric and bulbous genera relative to DNA content. This makes translating quantitative estimates derived from real-time PCR into fungal biomass very challenging (Denman and McSweeney, 2006; Edwards et al., 2008). Estimating AF abundance by quantifying the number of ITS1 spacer regions [(Mura et al., 2019) or rRNA genes, i.e., 5.8S rRNA gene (Edwards et al., 2008) or LSU (Nagler et al., 2018)] seems to be taxon independent. However, it still remains to be determined if all AF have the same copy number of the rrn operon.
Influence of Host on Anaerobic Fungi Community
Although diet has a significant effect on the structure of the AF community, host animal phylogeny has been shown to be a more important factor (Liggenstoffer et al., 2010; Kittelmann et al., 2012). Kittelmann et al. (2012) provided strong evidence that both diet and ruminant species, as well as interactions between those two parameters, affect the diversity of the AF community by subjecting cattle, sheep and deer to three different diets (summer pasture, winter pasture, and silage). Recent studies revealed that the genetic background of the host animal can influence the activity of the entire rumen microbiota, including the community of rumen AF (Roehe et al., 2016). In addition to the genetic background, several breed-associated phenotypes, such as eating frequency, dry matter intake, and rumen size potentially contribute to the variations in rumen microbiota that is observed among various breeds (Wallace et al., 2019; Zhang et al., 2020).
Anaerobic fungi were reported to be present in domesticated and non-domesticated equine species, with the AF community composition in horses and ponies being more similar to zebras than donkeys (Edwards et al., 2020b). In a separate study, AF diversity in donkeys was shown to be higher when compared to that of ponies and pony × donkey hybrids (Edwards et al., 2020a). Several studies have revealed that the genus Khoyollomyces (formerly known as AL1) is almost exclusively found in equines (Liggenstoffer et al., 2010; Mura et al., 2019; Edwards et al., 2020b; Hanafy et al., 2020). This may be due to the growth or metabolic characteristics of Khoyollomyces, making it more adapted to growth in the equine hindgut. Metabolic differences have previously been reported for equine and rumen strains of Piromyces, with equine strains possessing faster growth and higher fiber degradation capacity compared to rumen isolates (Julliand et al., 1998). This is perhaps not surprising considering the fundamental differences between ruminants (where freshly ingested feed directly enters the rumen), and hindgut herbivores (where feed first passes through the stomach and small intestine before reaching the hindgut) in terms of the main gut site where fiber degradation primarily occurs. There is also evidence indicating that the genus Oontomyces is specific to camelids (Dagar et al., 2015). Further research is needed to understand key factors that limit, or conversely broaden, the host distribution of certain AF taxa.
Role of Anaerobic Fungi in Methanogenesis
Ruminants are a significant source of anthropogenic methane (CH4) producing ∼90 Tg of CH4 annually (Reay et al., 2018), with exact values differing based on the methodology employed to quantify emissions (Capper and Bauman, 2013; Lyu et al., 2018). Although methanogenesis occurs in the rumen, there appears to be no direct benefit of the microbial generated CH4 (which is released into the environment) to the animal itself. The ruminant provides the anaerobic environment that is necessary for the archaea, first described as archaebacteria in the late 1970s by Woese and Fox (1977), that are capable of CH4 production. Due to the complex nature of the rumen microbiome and the interactions between the individual community members and their biochemistry, the role of AF in methanogenesis can only be understood in light of knowledge about archaea and processes facilitated by them. There are three different biochemical routes by which archaea can produce CH4: (1) acetoclastic methanogenesis, (2) hydrogenotrophic methanogenesis, and (3) methylotrophic methanogenesis (Borrel et al., 2013; Offre et al., 2013).
Although most gut methanogens are thought to be hydrogenotrophic (Ferry, 2010; Borrel et al., 2013; Lyu et al., 2018), the majority of the global microbial CH4 is produced via acetoclastic methanogenesis (Ferry, 2010; Lyu et al., 2018). In the rumen ecosystem, both acetate and hydrogen are produced (Baldwin and Allison, 1983) and are available for methanogenesis. However, since their turnover rates in the rumen are high, the contribution of CH4 produced via the acetoclastic pathway accounts only for a small fraction of the overall CH4 produced in the rumen (Baldwin and Allison, 1983; Janssen, 2010). During hydrogenotrophic CH4 synthesis H2 and CO2 are combined to yield CH4 (Baldwin and Allison, 1983), with contributions from the newly described methylotrophic methanogens (Poulsen et al., 2013).
Under normal circumstances, H2 seldom reaches high concentrations in the rumen and the dissolved H2 concentration is usually about 0.1–50 μM, which is 0.014–6.8% of its maximum solubility at 39°C and 1 atm (Hegarty and Gerdes, 1999; Janssen, 2010). The scarcity and poor water solubility of H2 limits the access of methanogens to molecular hydrogen, necessitating the development of close physical contact and intimate syntrophic partnership between H2 producers and H2 metabolizers. Extreme forms of these interspecies H2-transfer are the methanogenic endo- and ecto-symbionts of rumen protozoa (Embley and Finlay, 1994). The close association seen between rumen ciliates and methanogens (Vogels et al., 1980) seems a more general feature of anaerobic ciliates, aimed at boosting their metabolic rate (Rotterová et al., 2020). Although AF are not known to have methanogenic endo- or ecto-symbionts, they do contain modified mitochondria known as hydrogenosomes (van der Giezen, 2009), and there is in vitro based evidence of cross-feeding (syntrophy) between hydrogenic AF and methanogenic archaea in the herbivore gut ecosystem (Yarlett et al., 1986).
Differential centrifugation of cellular fractions revealed that fungal hydrogenosomes convert malate or pyruvate under anaerobic conditions into H2, CO2, and acetate with the concomitant production of ATP (Yarlett et al., 1986; Marvin-Sikkema et al., 1993). This is similar to the process found in trichomonads, anaerobic urogenital parasites, where hydrogenosomes were first discovered (Müller, 1993). The H2 produced is the result of the action of the oxygen-sensitive enzyme hydrogenase (Davidson et al., 2002), which is the terminal electron acceptor for the metabolism coming from pyruvate. The excess H2 produced by AF can be used by methanogens to regenerate oxidized nucleotides (e.g., NAD, NADP) (Yarlett et al., 1986). Indeed, methanogenic archaea have even been found attached to the surface of fungal rhizoids and sporangia (Bauchop and Mountfort, 1981; Jin et al., 2011), which is likely to improve interspecies hydrogen transfer. Within the rumen microbiome, symbiotic ecto- and endosymbiotic partnership have been reported for rumen protozoa and methanogens (Finlay et al., 1994; Irbis and Ushida, 2004; Valle et al., 2015). Since other fungi have been shown to accommodate prokaryotic endosymbionts (Partida-Martinez et al., 2007), there is the possibility that methanogenic archaea can also exist intracellularly within AF, although such an intimate symbiotic relationship has not been reported until today.
There have been numerous co-culture studies, particularly with hydrogenotrophic Methanobrevibacter isolates and representatives of the genera Piromyces, Neocallimastix, Orpinomyces, Caecomyces, and Anaeromyces (Edwards et al., 2017). Methane, CO2, formate and acetate are the main products when AF are grown in the presence of methanogens, whereas H2, lactate, succinate and ethanol production is drastically decreased compared to the corresponding pure anaerobic fungal cultures (Bauchop and Mountfort, 1981; Marvin-Sikkema et al., 1990). This difference between co- and pure cultures is due to the inter-species hydrogen transfer in the methanogenic co-cultures influencing the efficiency of anaerobic fungal fermentation. This shifts AF product formation away from more oxidized end-products (i.e., lactate and ethanol) and toward the production of more reduced products (i.e., formate and acetate). Recently the life cycle stage has also been shown to shift metabolite production of AF grown in methanogenic co-culture (Li et al., 2019).
In exchange for the excess H2 produced by AF, methanogens have a beneficial effect on AF growth and activity. This is evidenced in terms of increased cellulolytic enzyme activity and dry matter disappearance in methanogenic co-cultures compared to anaerobic fungal cultures alone (Bauchop and Mountfort, 1981; Jin et al., 2011). The ability of AF and methanogen co-cultures to rapidly convert lignocellulose containing plant material into CH4 also has good potential for biotechnological applications (Cheng et al., 2009; Jin et al., 2011; Peng et al., 2016), particularly in terms of biogas production from lignocellulosic waste streams. Conversely, in ruminants CH4 production is viewed as an unfavorable outcome of the fermentation. In ruminants, eructation of H2 in the form of large amounts of CH4 represents a loss of energy for the animal in addition to being a significant source of anthropogenic CH4.
Several rodent hindgut fermenters and non-ruminant foregut fermenters also emit CH4 of a magnitude as high as ruminants, but in contrast equids, macropods and rabbits produce much less (Clauss et al., 2020). There has been a move in recent years to understand why some domesticated ruminants produce lower CH4 compared to others (Shi et al., 2014), and a subsequent push to utilize feed-based approaches to decrease ruminal methanogenesis (Jeyanathan et al., 2014). However, it remains to be determined to what extent the growth and activity of rumen AF is affected by reducing the number and/or activity of methanogenic archaea using these approaches. Being able to understand the interdependencies of these populations will be important for informing a holistic understanding of the microbiome as it pertains to ruminant function. Current knowledge regarding interactions of AF with bacteria and protozoa is more limited, and has been reviewed elsewhere (Gordon and Phillips, 1993; Edwards et al., 2017).
Role of Anaerobic Fungi in Plant Cell-Wall Degradation
In vitro studies suggest that the contribution of AF to the ruminal degradation of plant material could be more significant than the contribution of cellulolytic rumen bacteria (Joblin et al., 1989; Lee et al., 2000b). This is likely due to the broad range of enzymes that are produced by the AF combined with their physical ability to break open fibrous materials through their penetrating hyphal tips. These tips have high concentrations of fibrolytic enzymes, whose enzymatic activity subsequently also increases nutrient access for other cellulolytic microbes (Ho et al., 1988; Ljungdahl, 2008; Haitjema et al., 2014; Dagar et al., 2015; Solomon et al., 2016). Whilst rumen zoospore numbers are low compared to counts of bacteria and archaea, AF have been shown to represent up to 20% of the rumen microbial biomass (Rezaeian et al., 2004a) and 10–16% of rRNA transcript abundance (Elekwachi et al., 2017). The observation that some AF species are capable of releasing up to 95% of the fermentable sugars from untreated plant leaves during a 4-day incubation period (Sijtsma and Tan, 1996) further highlights their critical role during the rumen digestion of fibrous plant biomass. These findings have led to the general belief that AF have been essential in the successful evolution of mammalian herbivores (Wang et al., 2019).
Understanding how AF and their carbohydrate-active enzymes (CAZymes) contribute to the degradation of indigested biomass in the herbivore gut is rather limited compared to the knowledge of the role of bacteria. This is partially due to the more extensive genomic resources that exist for rumen bacteria compared to AF (Hess et al., 2011; Seshadri et al., 2018; Stewart et al., 2019). AF genome information that is currently publicly available is summarized in Table 2, and includes the AF strain host isolation source and number of CAZymes identified.
TABLE 2.
Publicly available genomes of anaerobic fungi.
| Organism | Strain | Host | Genome sizea [base pairs] | Gene counta | CAZyme counta | References |
| Anaeromyces robustus | S4 | Sheep | 71,685,009 | 12,832 | 1,766 | Haitjema et al. (2017) |
| Caecomyces churrovis | – | Sheep | 165,495,782 | 15,009 | NDb | Henske et al. (2017) |
| Neocallimastix californiae | G1 | Goat | 193,495,782 | 20,219 | 2,743 | Haitjema et al. (2017) |
| Pecoramyces ruminantium | C1A | Cow | 100,954,185 | 18,936 | 2,029 | Youssef et al. (2013) |
| (formerly Orpinomyces sp.) | ||||||
| Piromyces finnis | Pirfi3 | Horse | 56,455,805 | 10,992 | 1,463 | Haitjema et al. (2017) |
| Piromyces sp. | E2 | Elephant | 71,019,055 | 14,648 | 3,819 | Haitjema et al. (2017) |
ahttps://mycocosm.jgi.doe.gov (Grigoriev et al., 2013). bNot determined.
One striking feature of AF besides their large repertoire and diversity of CAZymes (Supplementary Table S1), is the ability of their CAZymes to form cellulosomes. Cellulosomes, first identified in anaerobic bacteria, are extracellular multi-enzyme complexes that tether together an assortment of cellulases and related accessory enzymes (Lamed et al., 1983). The assembly of these AF multi-protein complexes, in some cases with individual building blocks from different species, is facilitated by fungal dockerins, which can directly bind to plant cell wall components without the need for a scaffoldin (Fanutti et al., 1995). This is in stark contrast to bacterial cellulosomes, which are highly species-specific (Bayer et al., 2004).
Cellulosomes have been linked to the improved fitness and biomass-degradation phenotype of both anaerobic bacteria and AF (Bayer et al., 2004; Henske et al., 2018) by enabling the synergistic activity of the individual biomass-degrading enzymes. Cellulosomes have been shown to increase cellulolytic activity over free enzymes by up to 12-fold (Krauss et al., 2012). Synthetic cellulosomes, inspired by bacterial cellulosomes, have shown promise for industrial applications due to outperforming free cellulolytic enzymes when produced in recombinant systems (Gilmore et al., 2020). Although evidence of AF cellulosomes emerged about 20 years ago (Wilson and Wood, 1992), it is only recent work that has provided strong support for the hypothesis that these multi-enzyme machineries might hold the secret to the superior biomass-degrading capability of AF. However, cellulosomes from AF still remain poorly understood (Haitjema et al., 2017).
Horizontal gene transfer (HGT) between the different populations of rumen microbes appears to have played a significant role in the evolution of the Neocallimastigomycota (Murphy et al., 2019). HGT between rumen bacteria and rumen AF was proposed by Garcia-Vallvé et al. (2000) as a major mechanism that allowed rumen AF to acquire many of the CAZymes that have now been identified in their genomes (Supplementary Table S1). Since then, numerous other authors have supported the hypothesis that the acquisition of CAZymes from bacterial donors enabled the rumen AF, as well as rumen protozoa (Ricard et al., 2006), to successfully compete for plant carbohydrates in the herbivore gut (Duarte and Huynen, 2019; Wang et al., 2019). Despite the numerous CAZyme families that AF acquired via HGT, the cellulosome of AF appears to have unique attributes that distinguish it from those that are found in gut bacteria.
Cellulosomes, regardless of their origin, are typically composed of several enzymes (i.e., cellulases and hemicellulases) that contain an active site, one or several carbohydrate binding modules and a dockerin that facilitates the “docking” of the enzymatic multi-modular complex to one of the multiple cohesins that are displayed by a scaffoldin. In anaerobic bacteria, cellulosomes are anchored to the bacterial cell wall via dockerin-scaffoldin modules (Fontes and Gilbert, 2010). In contrast, AF cellulosomes are not necessarily anchored to the fungal cell wall, but can be also found as free multi-enzyme complexes that are released into the extracellular matrix (Haitjema et al., 2014). These free cellulosomes enable an increased concentration of enzymes that possess catalytic and carbohydrate binding sites without the need to have them linked to and displayed on the AF cell surface. More importantly, it has been suggested that these free cellulosomes are non-species specific and theoretically enable the synthesis of cellulosomes using components of different donors (Nagy et al., 2007).
Another aspect that distinguishes AF cellulosomes from their bacterial counterparts is the presence of modules that have sequence signatures which are conserved within members of glycoside hydrolase (GH) families GH3, GH6, and GH45 (Haitjema et al., 2017). The ability to expand the substrate repertoire of AF cellulosomes by incorporating these GH signatures might provide additional enzymatic capabilities not conveyed by bacterial cellulosomes, such as the conversion of cellulose to monosaccharides by the β-glucosidase activity of GH3.
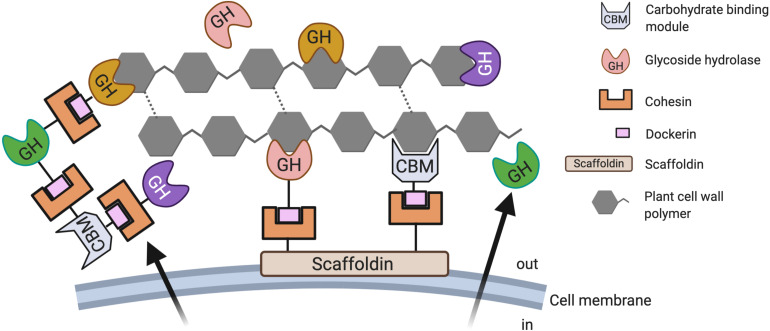
Besides cellulosomes, free CAZymes are also produced AF, which ultimately leaves the AF with several hydrolytic mechanisms to attack plant cell wall polymers from multiple directions (Haitjema et al., 2017) (Figure 2). The ability to produce secreted free CAZymes, cell-bound cellulosomes, as well as free cellulosomes might provide the AF with the competitive advantage over the CAZyme repertoire produced by the cellulolytic anaerobic bacteria also resident in the herbivore gut (Henske et al., 2017). It has also been proposed that the superior efficiency in biomass degradation of AF might be caused to a great extent by the ability to simultaneously employ a diverse set of cellulosome-bound as well as free (not cellulosome associated) CAZymes that also display a significant functional overlap (Couger et al., 2015).
FIGURE 2.
Carbohydrate-active enzymes employed by anaerobic fungi during biomass conversion. Anaerobic fungi (AF) deploy various strategies for the degradation of plant biomass. It has been suggested that their ability to produce secreted free CAZymes, cell-bound multi-enzyme complexes (cellulosomes), as well as free cellulosomes might provide the AF with the competitive advantage over the CAZyme repertoire produced by anaerobic bacteria (Henske et al., 2017).
This hypothesis, as well as the ability of AF to contribute a complementary set of CAZymes to the ones provided by bacteria was recently supported by genome-centric metaproteome and metatranscriptome approaches (Hagen et al., 2020). Shotgun metaproteome and metatranscriptome data were mapped back on previously assembled rumen genomes from prokaryotes and cultured AF, as well as on Metagenome Assembled Genomes to determine the origin of the different CAZymes detected in the microbiome that colonized recalcitrant plant material during rumen-incubation (Hagen et al., 2020). Results from this study revealed that the bacterial population contributed CAZymes mostly associated with the degradation of more readily degradable carbohydrates such as hemicellulose, whereas fungi provided CAZymes (e.g., enzymes belonging to the GH family GH5, GH6, GH8, and GH48) that targeted the more recalcitrant plant cell wall components. These data also provided clear evidence for the involvement of AF cellulosomes in the biomass degradation process. It can be assumed that a more detailed understanding of AF and their enzymatic repertoire, in the context of a highly diverse and synergistically working microbial ecosystem, will emerge as more of these complex and large-scale meta-omics data sets are generated and analyzed.
Anaerobic Fungi as Feed Additives to Promote Animal Health and Performance
The capability of AF to colonize and degrade otherwise recalcitrant plant structures via a set of highly efficient enzymes has led to an increased interest in the application of AF, and their enzymes, to boost the digestibility of low-quality feed and to increase the overall feed efficiency of herbivorous animals. Enabling the utilization of low-quality feed will be essential to enable the production of high-quality animal proteins, especially during a time when terrestrial areas for the production of high-quality feed will become scarce due to a changing climate and the rapid urbanization of areas that are currently being used to grow feed crops.
Several in vivo studies indicated that continuous dosing of ruminants with live culture of AF results in the changes of numerous parameters indicative of enhanced feed digestibility and feed efficiency, with benefits of AF dosing appearing to be more pronounced in young ruminants (Sehgal et al., 2008). Measurements performed to determine the animal’s ability to digest feedstuff in response to AF dosing, include dry matter (DM), neutral detergent fiber (NDF) and acid detergent fiber (ADF) digestibility, animal growth, milk yield and VFA concentrations (all parameters increased when AF were added) (Dey et al., 2004; Paul et al., 2004, 2011; Tripathi et al., 2007; Saxena et al., 2010).
Probiotic capabilities of live AF cultures for ruminants were indicated in the 1990s when living AF cultures were added to the rumen of cattle and sheep from which fungi had been previously removed. Forage intake by such fungus-free early weaned calves was 35% higher in those that had been dosed with Neocallimastix sp. R1 (Theodorou et al., 1990), and dosing of fungus-free sheep with Neocallimastix sp. SLl resulted in a 40% increase in intake of a straw based diet (Gordon and Phillips, 1993).
Despite the work that has been conducted to investigate how dosing the rumen ecosystem with AF cultures affected the abundance of the native fungal, bacterial and even the ciliate rumen populations (Lee et al., 2000a; Paul et al., 2004, 2011), there has been no evidence that suggests that dosing with AF, and therefore the increase of AF concentration in the rumen, resulted in a decrease of bacteria or ciliate protozoa nor in a drop of feed digestibility. In contrast to this, a positive correlation between fungal and bacterial concentrations, most likely due to the fact that the hyphae of AF physically open the plant tissue thereby increasing surface area available for colonization and nutrient access for other fibrolytic rumen microbes, has been reported (Bauchop, 1979; Akin and Borneman, 1990). This increased accessibility would also explain why counts of the holotrichs, the starch degrading protozoal population, increased in response to dosing with AF, while the overall count of the ciliate rumen protozoa remained stable (Paul et al., 2004).
Besides the lack of evidence that an increase in AF by dosing reduces the abundance of other rumen populations, it appears to be noteworthy that none of the studies centering on AF dosing has looked at potential changes of fiber-colonization by AF after the dosing event. Considering that dosing with AF cultures improved fiber digestion, VFA production and animal performance related parameters, suggesting that the altered microbiome is more efficient in the digestion of fibrous feeds than the original community, a closer investigation of the fiber colonization and deconstruction process post-dosing seems warranted.
Whereas ruminal fiber digestion improved in response to live AF, no shift in fermentation pattern was observed when AF-derived enzymes were added to the diet (Lee et al., 2000a). This highlights the importance of using viable cultures of AF as ruminant feed additives (Paul et al., 2011). In contrast to this, spent media containing AF enzymes seems to be effective in improving monogastric livestock production (Theodorou et al., 1996). Cell walls of cereals, major components of swine and poultry feed, contain difficult to digest non-starch polysaccharides, such as β-glucans in barley and wheat and arabinoxylans in rye and oats (Sánchez-Rodríguez et al., 2012). These polymers can have anti-nutritional effects due to their low digestibility and tendency to form high-molecular-weight aggregates that reduce passage rate, decrease diffusion of digestive enzymes, promote endogenous losses, and stimulate unwanted bacterial proliferation (Bedford and Schulze, 1998). Previous studies suggest that recombinant GHs from AF expressed in Lactobacillus reuteri maintain their fiber-degrading capability and their resistance to bile salt and acids, while L. reuteri itself still retained its high adhesion efficiency to mucin and mucus (Liu et al., 2005a,b, 2007; Cheng et al., 2014), which would explain the decomposition-promoting effect of these recombinant proteins in the monogastric animal. Despite these promising characteristics for recombinant AF-derived GHs, the ability to efficiently generate significant amounts of cheap, stable, and active recombinant enzymes will be essential for low-cost production and large-scale application of these enzymes as a feed additive for monogastrics.
Tannins present in many feeds and forages are inhibitory to rumen microbes, and are an anti-nutritional issue for ruminants. Likewise, upon anaerobic degradation of phenolic compounds present in fibrous feeds, different phenolic monomers (i.e., ferulic acid, p-coumaric acid, vanillic acid, vanillin, catechol etc.) are released into the rumen that are inhibitory to microbiota. Therefore, attempts were made to identify rumen microbes that had tannin or phenolic monomer tolerating or degrading capability, so that they could be utilized as direct fed microbials to mitigate adverse effects of these anti-nutritional factors.
The anaerobic rumen fungus Piromyces sp. FNG5, isolated from a wild herbivore, was found to be tolerant to phenolic monomers and its pure culture degraded p-coumaric acid (38.5–65.1%), 65.2–74.1% ferulic acid (65.2–74.1%) and vanillic acid (34.1–66.8%) after 14 days of incubation (Paul et al., 2003). McSweeney et al. (2001) reported that sheep fed with condensed tannins from Calliandra calothyrsus had reduced ruminal AF concentration, but the inhibitory effect was less prominent compared to rumen bacteria. Paul et al. (2006) reported that addition of Piromyces sp. FNG5 significantly increased in vitro degradation (12%) of condensed tannins and this AF isolate could tolerate tannic acid concentrations up to 20 g/L. This amount is higher than the theoretic tannic acid level expected in the rumen of animals fed a diet composed exclusively of high tannin content plants. Conversely, Kok et al. (2013) found that L. leucocephala hybrid-Bahru (containing condensed tannins) when fed to goats, significantly decreased ruminal AF concentration. Saminathan et al. (2019) reported that high MW fractions of condensed tannins had inhibitory effect on ruminal AF, but relative abundance of Piromyces 4 was increased indicating that this group of uncultured AF is likely to be tannin resistant. The mechanisms by which some AF species can overcome the growth inhibitory effects of condensed tannins or phenolic monomers is unknown. Whether AF produce tannase, or not, remains to be established; but many isolates of AF, especially those from wild ruminants adapted to tannin rich and fibrous diets, were shown to produce a variety of esterases capable of degrading phenolic compounds (Paul et al., 2003). It is possible that these esterases are directly linked the ability of these AF to overcome growth inhibition caused by phenolic compounds.
Although these studies highlight the potential of using AF as probiotics to enhance digestibility of highly fibrous or tanniferous feed, boosting ruminant livestock production, economic aspects such as the need to repeatedly administer oral-dosages of AF to maintain the desired response have to be considered when discussing AF as probiotics on a commercial scale (Ribeiro et al., 2016).
As well as positively impacting forage intake and feed digestibility, AF have the potential to contribute to the protein supply of the host animal. This is both indirectly through the production of proteolytic enzymes in the rumen and directly as a source of microbial protein. Unlike the cellulolytic rumen bacteria, many isolates of AF are protease positive and capable of penetrating the proteinaceous layer of feed particles (Wallace and Joblin, 1985; Asao et al., 1993; Michel et al., 1993; Yanke et al., 1993). In vitro studies with defined populations of both proteolytic and non-proteolytic rumen bacteria and a proteolytic Neocallimastix frontalis strain have further indicated that N. frontalis was able to contribute to rumen protein degradation, particularly when protein was associated with feed particles (Wallace and Munro, 1986).
Besides producing proteases, AF directly contribute to protein supply of the host in terms of being part of the microbial biomass that passes down to the intestines from rumen, for subsequent digestion and absorption. Gulati et al. (1989) showed that AF cells were composed of proteins with a well-balanced combination of amino acids that were highly available to the ruminant host. A high proportion of the protein components of three monocentric AF (i.e., Neocallimastix sp. LMI, Piromyces sp. SMI and Caecomyces sp. NMI) was digested and absorbed in the intestine of sheep, with digestibility factors of 0.91–0.98 (Gulati et al., 1988, 1989). These high in vivo digestibility values for AF protein compared favorably with a value of 0.77 for mixed rumen bacteria protein measured in a similar manner (Gulati et al., 1990). Although the amount of nutritional nitrogen derived directly from anaerobic rumen fungi might only amount to a small portion of the total nitrogen that is absorbed by the animal, its importance lies in its high quality and immediate availability.
Dietary Manipulation of Commensal Anaerobic Fungi
Rather than directly adding more AF to the ruminant animal, there have been numerous attempts to improve the concentration of AF in the rumen by providing animals with feed that increases their overall concentration. It was previously believed that increasing dietary recalcitrant fiber may increase fungal population in rumen, as some of the plant fiber breakdown products were shown to have a positive effect on zoosporogenesis and chemotactic effects on fungal zoospores. Few AF were seen in the rumen of animals that were fed lush pasture (i.e., legume or grass when green and leafy), and the number of AF increased when animals were fed the same pasture after it had matured and was more recalcitrant (Bauchop, 1989; Kostyukovsky et al., 1991). However, results later showed the opposite effect, with an increased AF count in ruminants fed a low-lignin diet reported compared to animals provided with more recalcitrant feed (Gordon and Phillips, 1998). To make matters even more complex, other studies suggested that there was no direct effect of the hay type (with different levels of lignin content) on AF populations (Sekine et al., 1995).
It has been suggested that an appropriate amount of starch or concentrate in diet may support ruminal AF growth and stimulate zoosporogenesis (Matsui et al., 1997), but in vivo study findings remain inconclusive (Ishaq et al., 2017). A possible explanation for these different responses to starch rich feed is that only some of the AF, namely species of the genera Neocallimastix, Piromyces and Orpinomyces, have been shown to produce amylases and, therefore, have the ability to ferment starch grains (Phillips and Gordon, 1988; McAllister et al., 1993; Yanke et al., 1993). More work in this area needs to be conducted in vivo before final conclusions can be made. Furthermore, feeding starch or concentrates tends to increase ruminal ciliate protozoal concentrations, and protozoa are known to predate on AF zoospores (Morgavi et al., 1994).
Beneficial effects of sulfur supplementation, specifically for low sulfur diets, on the number of AF in sheep and their relative contribution to fiber degradation was reported in the early 1980s (Akin et al., 1983). This beneficial effect was further confirmed in subsequent studies using alkali treated wheat straw (Gordon et al., 1983; Gulati et al., 1985; Weston et al., 1988) and other poor quality feeds (Morrison et al., 1990). Promkot and Wanapat (2009) suggested beneficial manipulation of AF concentrations was possible through provision of an appropriate dietary supplement containing sulfur. For a supplement of this type to be effective, it should ideally contain a single organic sulfur compound which is readily utilized by the rumen AF but not by other components of the rumen microbiota (i.e., bacteria, archaea, and protozoa).
Two organic sulfur nutrients, mercapto-1-propionic acid (MPA) and 3-mercapto-1-propanesulfonic acid, were tested in cattle trials and compared to an inorganic sulfur supplement. It was reported that the organic sulfur sources improved nitrogen utilization and microbial protein production, but surprisingly this was concluded to be due to a general improvement in the efficiency of microbial fermentation of lignocellulose and not from specific stimulation of ruminal AF (McSweeney and Denman, 2007). Conversely, in a patent (Gordon and Phillips, 2002) it was reported that administering an effective amount of a degradation resistant sulfur source (MPA or its functional equivalent) promoted the growth of AF in the rumen of animals fed low sulfur content diets. Within this patent, it was also demonstrated that ruminal MPA infusion increased AF zoospores concentrations, and had a strong, positive, response on the digestive performance of sheep. However, additional scientific literature in this field is scarce and a sulfur supplement specific for promoting anaerobic rumen fungi remains to be identified.
Influence of other dietary supplements on the rumen AF community has been less studied, but some interesting findings have been reported. Thiamine supplementation, used to attenuate rumen metabolic disorder caused by high concentrate diet through buffering the rumen pH, increased significantly the proportion of ruminal AF in dairy cows (Xue et al., 2018). Plant oils, which are attractive feed additives used to mitigate CH4 emissions, seem to have a negative effect on AF. The addition of soya oil significantly reduced ruminal AF diversity in steers (Boots et al., 2013). In dairy cows, sunflower oil addition decreased the concentration and diversity of AF. However, responses at the genus level were dependent on concentrate/forage ratios (Tapio et al., 2017). The addition of rapeseed oil led to a considerable decrease in the ruminal AF population, but the mechanism was not further investigated (Fonty and Grenet, 1994). Previously, Elliott et al. (1987) found that feeding a supplement of sunflower meal to sheep consuming a barley straw diet resulted in decrease of ruminal AF concentration to below detectable levels. In another study, the feeding of calcium salts of medium chain fatty acids (C6–Cl2) to sheep resulted in reduced numbers of AF zoospores in the rumen, whereas the salts of long chain fatty acids (C ≥ 14) had no effect on AF (Ushida et al., 1992). This indicates that the inhibitory effects of the long chain fatty acids common in oilseed meals can be alleviated, at least partly, by chemical pretreatment.
Anaerobic fungi are also sensitive to a shortage of nitrogen. A low protein diet decreased rumen AF concentration in dairy cows compared to a high protein diet (Belanche et al., 2012). However, the AF community composition was modified by the level of dietary protein only when cows consumed the starch-rich diet, but not the fiber-rich diet (Belanche et al., 2012). This highlights the potential for further complexities when trying to determine the effects of individual dietary components on ruminal AF.
Future Perspectives and Opportunities for Anaerobic Fungi Based Applications in Animal Production and Health
With consumer demand for affordable high-quality animal products increasing and with a decline in natural resources, such as farmable land area, it will be essential to create new and refine existing strategies to improve the utilization of low-quality forages for animal feed. Such strategies will rely heavily on approaches that render the recalcitrant fraction of the plant material more accessible to the fermenting microorganisms that are indigenous to the herbivore gut. AF with their ability to break open recalcitrant plant structures will play a significant role in these new approaches.
Whilst AF are clearly beneficial for the ruminant host, the underlying mechanisms to boost indigenous AF populations through standard feed components such as fiber, starch, nitrogen and lipid are inherently complex and are still not well understood. As such, there still remains great interest in developing a reliable and reproducible feed-based strategy to increase AF in order to improve animal performance and health. However, whilst the benefits of AF for ruminants has been well established, their value for hindgut herbivores remains to be confirmed. The observed increase in feed intake with AF supplemented ruminants is thought to be due to the AF causing more rapid clearance of digesta from the rumen, due to their physical and enzymatic disruption of fibrous plant particles (Gordon and Phillips, 1998). If this is also the case in equines, development of an equine AF probiotic may enable replacement of some of the energy dense concentrates used in horse feeds with more bulky fibrous feeds. This will contribute to reducing the risk of colic and dysbiosis of hindgut microbiota, which is commonly observed in working and/or performance equines fed high grain/concentrate diets in order to meet their higher energy requirements (Shirazi-Beechey, 2008; Durham, 2013; Julliand and Grimm, 2017).
Most of the studies focused on AF-based strategies for improving animal production and health have relied on the repeated oral-dosing of AF. This approach can only become economically feasible if the cost of industrial scale production of an AF probiotic is significantly less than the economic return gained by livestock producers. Considering the significant benefits that have been reported for live AF supplementation to date for ruminants, the probiotic use of AF is likely to have a significant return on investment for ruminant livestock producers. One possibility to produce an AF probiotic could be the use of encapsulated cultures. However, methods for large-scale production of encapsulated AF are currently not available, and would have to be developed before this approach could become commercially feasible. An alternative approach could be the use of AF resting structures, with their subsequent revival into active hydrolytic AF occurring within the host. Fundamental understanding of these structures is, however, currently too limited for this to be practically realized in the near future. Advances in understanding of the biology of the AF resting phase would not only facilitate the utilization of AF in livestock production, but also their application in other areas where lignocellulosic plant material is used to produce biofuels and platform chemicals. As such, characterization of the resting phase of AF should be a high priority research area for development.
The resulting increased efficiency of livestock production through the application of AF will undoubtedly have beneficial impacts in terms of the environmental footprint and sustainability of livestock production. The impact of AF based strategies on ruminant derived methane production remains to be determined, however, it is clear that increased efficiency of lignocellulosic plant material utilization will also decrease the need for arable land for animal feed production. Recent advances in molecular techniques enable a detailed understanding of role of the AF and how it affects the performance and health of its herbivorous host. Research based on state-of-the-art methods will allow the development of more advanced and holistic approaches to manipulate the composition and function of the gut microbiome and ultimately the health and performance of the host animal. Such progress will facilitate a more sustainable livestock industry to provide affordable high-quality animal products for a growing global population.
Conclusion
Although AF were first observed more than hundred years ago, it was not until Colin Orpin’s ground-breaking work in the mid 1970s that AF were correctly identified, overturning the paradigm of the time that all fungi required oxygen. Considering AF have only been effectively studied for ∼50 years, advances made during this time have been significant. Whereas initial work required the refinement of isolation and cultivation techniques and was mostly driven by morphological observations and phenotypic characterization, more recent insights have been enabled by novel molecular approaches. These molecular techniques have already greatly advanced understanding of the complexity and diversity of the AF, however, our full understanding is still far from complete. With the ability to sequence long genomic regions such as the entire rrn operon, it is inevitable that a more accurate and complete understanding of the AF phylogeny will soon emerge. Understanding the phylogenetic relationship of individual AF will be essential to increasing understanding of their evolutionary history, and factors that drive their niche specialization within and between different types of host. However, to practically develop AF for application in livestock production, as well as other industries, functional systems microbiology approaches will be key. Technologies such as (meta)genomics, (meta)transcriptomics and proteomics will enable the pinpointing of specific genes, proteins and reactions that are employed by AF in response to extrinsic conditions (e.g., host genotype) and changes therein (e.g., host health status and dietary composition). Despite the significant progress made to date, the ecological role of AF and their quantitative contribution to host function and health still remains to be clarified in the full range of mammalian herbivores where they naturally reside. Furthermore, a key phase of the AF life cycle, the resting phase, is an area of very limited knowledge that urgently needs to be researched. Together, newly obtained knowledge in these areas will enable utilization of AF and their enzymes to transform the sustainability and environmental footprint of livestock agriculture, as well as revolutionizing biotechnological processes involving plant-based feedstocks. Whilst we increasingly understand more about the evolution, biology and ecology of AF, there still remains many key “why” questions to be answered: “Why” are AF sensitive to oxygen? “Why” do AF have the lowest GC-content among all known microorganisms? “Why” are AF the only fungi with polyflagellate zoospores, hydrogenosomes, and cellulosomes? Considering that the AF phylum is currently composed of just one family, are we only looking at the tip of an iceberg? Or have we missed something crucial when classifying these microorganisms? Whatever the answers are, we know for certain that many fundamental questions still remain to be answered before the true potential of this highly valuable and paradigm shifting phylum of microorganisms is fully understood and can be harnessed.
Author Contributions
MH designed the manuscript, coordinated the co-author contributions, and wrote the plant cell-wall degradation section. KF wrote the habitat, life cycle, morphology and taxonomy section and contributed to other sections. SP and AP contributed to the feed additive section. MG contributed to the methanogenesis section. CS co-authored the plant cell wall degradation section. JE wrote the section on hindgut herbivores and contributed to other sections. All authors reviewed the manuscript, offered critical feedback, and approved the final version.
Conflict of Interest
During the development of the manuscript, JE changed employment from Wageningen University & Research to the company Palital Feed Additives, Netherlands. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.584893/full#supplementary-material
References
- Akin D. E., Borneman W. S. (1990). Role of rumen fungi in fiber degradation. J. Dairy Sci. 73 3023–3032. 10.3168/jds.s0022-0302(90)78989-8 [DOI] [PubMed] [Google Scholar]
- Akin D. E., Gordon G. L., Hogan J. P. (1983). Rumen bacterial and fungal degradation of Digitaria pentzii grown with or without sulfur. Appl. Environ. Microbiol. 46 738–748. 10.1128/aem.46.3.738-748.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asao N., Ushida K., Kojima Y. (1993). Proteolytic activity of rumen fungi belonging to the genera Neocallimastix and Piromyces. Lett. Appl. Microbiol. 16 247–250. 10.1111/j.1472-765x.1993.tb01410.x [DOI] [Google Scholar]
- Baldwin R. L., Allison M. J. (1983). Rumen metabolism. J. Anim. Sci. 57 461–477. [PubMed] [Google Scholar]
- Barr D. J. S. (1980). An outline for the reclassification of the Chytridiales, and for a new order, the Spizellomycetales. Can. J. Bot. 58 2380–2394. 10.1139/b80-276 [DOI] [Google Scholar]
- Barr D. J. S. (1988). How modern systematics relates to the rumen fungi. Biosystems 21 351–356. 10.1016/0303-2647(88)90032-9 [DOI] [PubMed] [Google Scholar]
- Barr D. J. S., Kudo H., Jakober K. D., Cheng K. J. (1989). Morphology and development of rumen fungi: Neocallimastix sp., Piromyces communis, and Orpinomyces bovis gen.nov., sp.nov. Can. J. Bot. 67 2815–2824. 10.1139/b89-361 [DOI] [Google Scholar]
- Bauchop T. (1979). Rumen anaerobic fungi of cattle and sheep. Appl. Environ. Microbiol. 38 148–158. 10.1128/aem.38.1.148-158.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauchop T. (1989). Biology of gut anaerobic fungi. Biosystems 23 53–64. 10.1016/0303-2647(89)90008-7 [DOI] [PubMed] [Google Scholar]
- Bauchop T., Mountfort D. O. (1981). Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl. Environ. Microbiol. 42 1103–1110. 10.1128/aem.42.6.1103-1110.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58 521–554. 10.1146/annurev.micro.57.030502.091022 [DOI] [PubMed] [Google Scholar]
- Bedford M. R., Schulze H. (1998). Exogenous enzymes for pigs and poultry. Nutr. Res. Rev. 11 91–114. 10.1079/nrr19980007 [DOI] [PubMed] [Google Scholar]
- Belanche A., Doreau M., Edwards J. E., Moorby J. M., Pinloche E., Newbold C. J. (2012). Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation. J. Nutr. 142 1684–1692. 10.3945/jn.112.159574 [DOI] [PubMed] [Google Scholar]
- Boots B., Lillis L., Clipson N., Petrie K., Kenny D. A., Boland T. M., et al. (2013). Responses of anaerobic rumen fungal diversity (phylum Neocallimastigomycota) to changes in bovine diet. J. Appl. Microbiol. 114 626–635. 10.1111/jam.12067 [DOI] [PubMed] [Google Scholar]
- Borrel G., O’Toole P. W., Harris H. M. B., Peyret P., Brugère J.-F., Gribaldo S. (2013). Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol. Evol. 5 1769–1780. 10.1093/gbe/evt128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braune R. A. (1913). Untersuchungen über die im wiederkäuermagen vorkommenden protozoen. Arch. Protistenk. 32 111–170. [Google Scholar]
- Breton A., Bernalier A., Dusser M., Fonty G., Gaillard-Martinie B., Guillot J. (1990). Anaeromyces mucronatus nov. gen., nov. sp. A new strictly anaerobic rumen fungus with polycentric thallus. FEMS Microbiol. Lett. 58 177–182. 10.1111/j.1574-6968.1990.tb13974.x [DOI] [PubMed] [Google Scholar]
- Brookman J. L., Ozkose E., Rogers S., Trinci A. P. J., Theodorou M. K. (2000). Identification of spores in the polycentric anaerobic gut fungi which enhance their ability to survive. FEMS Microbiol. Ecol. 31 261–267. 10.1111/j.1574-6941.2000.tb00692.x [DOI] [PubMed] [Google Scholar]
- Callaghan T. M., Podmirseg S. M., Hohlweck D., Edwards J. E., Puniya A. K., Dagar S. S., et al. (2015). Buwchfawromyces eastonii gen. nov., sp. nov.: a new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. Mycokeys 9 11–28. 10.3897/mycokeys.9.9032 [DOI] [Google Scholar]
- Capper J. L., Bauman D. E. (2013). The role of productivity in improving the environmental sustainability of ruminant production systems. Annu. Rev. Anim. Biosci. 1 469–489. 10.1146/annurev-animal-031412-103727 [DOI] [PubMed] [Google Scholar]
- Cheng H. L., Hu C. Y., Lin S. H., Wang J. Y., Liu J. R., Chen Y. C. (2014). Characterization of two truncated forms of xylanase recombinantly expressed by Lactobacillus reuteri with an introduced rumen fungal xylanase gene. Enzyme Microb. Technol. 64–65 6–10. 10.1016/j.enzmictec.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Cheng Y. F., Edwards J. E., Allison G. G., Zhu W. Y., Theodorou M. K. (2009). Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour. Technol. 100 4821–4828. 10.1016/j.biortech.2009.04.031 [DOI] [PubMed] [Google Scholar]
- Clauss M., Dittmann M. T., Vendl C., Hagen K. B., Frei S., Ortmann S., et al. (2020). Review: comparative methane production in mammalian herbivores. Animal 14 s113–s123. [DOI] [PubMed] [Google Scholar]
- Couger M. B., Youssef N. H., Struchtemeyer C. G., Liggenstoffer A. S., Elshahed M. S. (2015). Transcriptomic analysis of lignocellulosic biomass degradation by the anaerobic fungal isolate Orpinomyces sp. strain C1A. Biotechnol. Biofuels 8:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagar S. S., Kumar S., Griffith G. W., Edwards J. E., Callaghan T. M., Singh R., et al. (2015). A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol. 119 731–737. 10.1016/j.funbio.2015.04.005 [DOI] [PubMed] [Google Scholar]
- Dagar S. S., Kumar S., Mudgil P., Singh R., Puniya A. K. (2011). D1/D2 domain of large-subunit ribosomal DNA for differentiation of Orpinomyces spp. Appl. Environ. Microbiol. 77 6722–6725. 10.1128/aem.05441-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E. A., van der Giezen M., Horner D. S., Embley T. M., Howe C. J. (2002). An [Fe] hydrogenase from the anaerobic hydrogenosome-containing fungus Neocallimastix frontalis L2. Gene 296 45–52. 10.1016/s0378-1119(02)00873-9 [DOI] [PubMed] [Google Scholar]
- Davies D. R., Theodorou M. K., Brooks A. E., Trinci A. P. (1993). Influence of drying on the survival of anaerobic fungi in rumen digesta and faeces of cattle. FEMS Microbiol. Lett. 106 59–63. 10.1111/j.1574-6968.1993.tb05935.x [DOI] [PubMed] [Google Scholar]
- Dearing M. D., Kohl K. D. (2017). Beyond fermentation: other important services provided to endothermic herbivores by their gut microbiota. Integr. Comp. Biol. 57 723–731. 10.1093/icb/icx020 [DOI] [PubMed] [Google Scholar]
- Dehority B. A. (2002). Gastrointestinal tracts of herbivores, particularly the ruminant: anatomy, physiology and microbial digestion of plants. J. Appl. Anim. Res. 21 145–160. 10.1080/09712119.2002.9706367 [DOI] [Google Scholar]
- Denman S. E., McSweeney C. S. (2006). Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 58 572–582. 10.1111/j.1574-6941.2006.00190.x [DOI] [PubMed] [Google Scholar]
- Dey A., Sehgal J. P., Puniya A. K., Singh K. (2004). Influence of an anaerobic fungal culture (Orpinomyces sp.) administration on growth rate, ruminal fermentation and nutrient digestion in calves. Asian Australas. J. Anim. Sci. 17 820–824. 10.5713/ajas.2004.820 [DOI] [Google Scholar]
- Dollhofer V., Callaghan T. M., Dorn-In S., Bauer J., Lebuhn M. (2016). Development of three specific PCR-based tools to determine quantity, cellulolytic transcriptional activity and phylogeny of anaerobic fungi. J. Microbiol. Methods 127 28–40. 10.1016/j.mimet.2016.05.017 [DOI] [PubMed] [Google Scholar]
- Dollhofer V., Callaghan T. M., Griffith G. W., Lebuhn M., Bauer J. (2017). Presence and transcriptional activity of anaerobic fungi in agricultural biogas plants. Bioresour. Technol. 235 131–139. 10.1016/j.biortech.2017.03.116 [DOI] [PubMed] [Google Scholar]
- Duarte I., Huynen M. A. (2019). Contribution of lateral gene transfer to the evolution of the eukaryotic fungus Piromyces sp. E2: massive bacterial transfer of genes involved in carbohydrate metabolism. bioRxiv [Preprint] 10.1101/514042 [DOI] [Google Scholar]
- Durham A. E. (2013). “Intestinal disease,” in Equine Applied and Clinical Nutrition, eds Geor R. J., Harris P. A., Coenen M. (Philadelphia, PA: W.B. Saunders; ), 568–581. [Google Scholar]
- Ebersberger I., de Matos Simoes R., Kupczok A., Gube M., Kothe E., Voigt K., et al. (2012). A consistent phylogenetic backbone for the fungi. Mol. Biol. Evol. 29 1319–1334. 10.1093/molbev/msr285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. E., Forster R. J., Callaghan T. M., Dollhofer V., Dagar S. S., Cheng Y., et al. (2017). PCR and omics based techniques to study the diversity, ecology and biology of anaerobic fungi: insights, challenges and opportunities. Front. Microbiol. 8:1657. 10.3389/fmicb.2017.01657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. E., Hermes G. D. A., Kittelmann S., Nijsse B., Smidt H. (2019). Assessment of the accuracy of high-throughput sequencing of the ITS1 region of Neocallimastigomycota for community composition analysis. Front. Microbiol. 10:2370. 10.3389/fmicb.2019.02370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. E., Kingston-Smith A. H., Jimenez H. R., Huws S. A., Skot K. P., Griffith G. W., et al. (2008). Dynamics of initial colonization of nonconserved perennial ryegrass by anaerobic fungi in the bovine rumen. FEMS Microbiol. Ecol. 66 537–545. 10.1111/j.1574-6941.2008.00563.x [DOI] [PubMed] [Google Scholar]
- Edwards J. E., Schennink A., Burden F., Long S., van Doorn D. A., Pellikaan W. F., et al. (2020a). Domesticated equine species and their derived hybrids differ in their fecal microbiota. Anim. Microbiome 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. E., Shetty S. A., van den Berg P., Burden F., van Doorn D. A., Pellikaan W. F., et al. (2020b). Multi-kingdom characterization of the core equine fecal microbiota based on multiple equine (sub)species. Anim. Microbiome 2:6 10.1111/eve.03_12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elekwachi C. O., Wang Z., Wu X., Rabee A., Forster R. J. (2017). Total rRNA-Seq analysis gives insight into bacterial, fungal, protozoal and archaeal communities in the rumen using an optimized RNA isolation method. Front. Microbiol. 8:1814. 10.3389/fmicb.2017.01814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R., Ash A. J., Calderon-Cortes F., Norton B. W., Bauchop T. (1987). The influence of anaerobic fungi on rumen volatile fatty acid concentrations in vivo. J. Agric. Sci. 109 13–17. 10.1017/s0021859600080928 [DOI] [Google Scholar]
- Embley T. M., Finlay B. J. (1994). The use of small subunit rRNA sequences to unravel the relationships between anaerobic ciliates and their methanogen endosymbionts. Microbiology 140(Pt 2), 225–235. 10.1099/13500872-140-2-225 [DOI] [PubMed] [Google Scholar]
- Fanutti C., Ponyi T., Black G. W., Hazlewood G. P., Gilbert H. J. (1995). The conserved noncatalytic 40-residue sequence in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J. Biol. Chem. 270 29314–29322. 10.1074/jbc.270.49.29314 [DOI] [PubMed] [Google Scholar]
- Ferry J. G. (2010). How to make a living by exhaling methane. Annu. Rev. Microbiol. 64 453–473. 10.1146/annurev.micro.112408.134051 [DOI] [PubMed] [Google Scholar]
- Finlay B. J., Esteban G., Clarke K. J., Williams A. G., Embley T. M., Hirt R. P. (1994). Some rumen ciliates have endosymbiotic methanogens. FEMS Microbiol. Lett. 117 157–161. 10.1111/j.1574-6968.1994.tb06758.x [DOI] [PubMed] [Google Scholar]
- Fontes C. M., Gilbert H. J. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79 655–681. 10.1146/annurev-biochem-091208-085603 [DOI] [PubMed] [Google Scholar]
- Fonty G., Grenet E. (1994). “Effects of diet on the fungal population of the digestive tract of ruminants,” in Anaerobic Fungi: Biology, Ecology and Function, eds Mountfort D. O., Orpin C. G. (New York, NY: Marcel Dekker; ), 229–239. 10.1201/9781003067085-8 [DOI] [Google Scholar]
- France J., Theodorou M. K., Davies D. (1990). Use of zoospore concentrations and life cycle parameters in determining the population of anaerobic fungi in the rumen ecosystem. J. Theor. Biol. 147 413–422. 10.1016/s0022-5193(05)80496-5 [DOI] [PubMed] [Google Scholar]
- Garcia-Vallvé S., Romeu A., Palau J. (2000). Horizontal gene transfer of glycosyl hydrolases of the rumen fungi. Mol. Biol. Evol. 17 352–361. 10.1093/oxfordjournals.molbev.a026315 [DOI] [PubMed] [Google Scholar]
- Gay L. (1991). Chitin content and chitin synthase activity as indicators of the growth of three different anaerobic rumen fungi. FEMS Microbiol. Lett. 64 99–102. 10.1111/j.1574-6968.1991.tb04643.x [DOI] [PubMed] [Google Scholar]
- Gilmore S. P., Lankiewicz T. S., Wilken S. E., Brown J. L., Sexton J. A., Henske J. K., et al. (2019). Top-down enrichment guides in formation of synthetic microbial consortia for biomass degradation. ACS Synth. Biol. 8 2174–2185. 10.1021/acssynbio.9b00271 [DOI] [PubMed] [Google Scholar]
- Gilmore S. P., Lillington S. P., Haitjema C. H., de Groot R., O’Malley M. A. (2020). Designing chimeric enzymes inspired by fungal cellulosomes. Synth. Syst. Biotechnol. 5 23–32. 10.1016/j.synbio.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold J. J., Heath I. B., Bauchop T. (1988). Ultrastructural description of a new chytrid genus of caecum anaerobe, Caecomyces equi gen. nov., sp. nov., assigned to the Neocallimasticaceae. Biosystems 21 403–415. 10.1016/0303-2647(88)90039-1 [DOI] [PubMed] [Google Scholar]
- Gordon G. L., Phillips M. W. (1998). The role of anaerobic gut fungi in ruminants. Nutr. Res. Rev. 11 133–168. 10.1079/nrr19980009 [DOI] [PubMed] [Google Scholar]
- Gordon G. L., Phillips M. W. (2002). Fungal sulphur source and methods of using the same. U.S. Patent No 645,858,0B1. Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Gordon G. L. R., Gulati S. K., Ashes J. R. (1983). Influence of low-sulphur straw on anaerobic fungal numbers in a sheep rumen. Proc. Nutr. Soc. Aust. 8:188. [Google Scholar]
- Gordon G. L. R., Phillips M. W. (1993). Removal of anaerobic fungi from the rumen of sheep by chemical treatment and the effect on feed consumption and in vivo fibre digestion. Lett. Appl. Microbiol. 17 220–223. 10.1111/j.1472-765x.1993.tb01451.x [DOI] [Google Scholar]
- Griffith G. W., Ozkose E., Theodorou M. K., Davies D. R. (2009). Diversity of anaerobic fungal populations in cattle revealed by selective enrichment culture using different carbon sources. Fungal Ecol. 2 87–97. 10.1016/j.funeco.2009.01.005 [DOI] [Google Scholar]
- Grigoriev I. V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., et al. (2013). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42 D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati S. K., Ashes J. R., Gordon G. L. (1988). Digestibility of sulphur amino acids in rumen fungal species. Proc. Nutr. Soc. Aust. 13:133. [Google Scholar]
- Gulati S. K., Ashes J. R., Gordon G. L. R. (1990). Comparative digestibility of sulphur amino acids in rumen bacteria and fungal proteins by sheep. Proc. Nutr. Aust. 15 128–131. [Google Scholar]
- Gulati S. K., Ashes J. R., Gordon G. L. R., Connell P. J., Rogers P. L. (1989). Nutritional availability of amino acids from the rumen anaerobic fungus Neocallimastix sp. LM1 in sheep. J. Agric. Sci. 113 383–387. 10.1017/s002185960007009x [DOI] [Google Scholar]
- Gulati S. K., Ashes J. R., Gordon G. L. R., Phillips M. W. (1985). Possible contribution of rumen fungi to fibre digestion in sheep. Proc. Nutr. Soc. Aust. 10:96. [Google Scholar]
- Hagen L. H., Brooke C. G., Shaw C., Norbeck A. D., Piao H., Arntzen M. Ø., et al. (2020). Proteome specialization of anaerobic fungi during ruminal degradation of recalcitrant plant fiber. ISME J. 10.1038/s41396-020-00769-x [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitjema C. H., Gilmore S. P., Henske J. K., Solomon K. V., de Groot R., Kuo A., et al. (2017). A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2:17087. [DOI] [PubMed] [Google Scholar]
- Haitjema C. H., Solomon K. V., Henske J. K., Theodorou M. K., O’Malley M. A. (2014). Anaerobic gut fungi: advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol. Bioeng. 111 1471–1482. 10.1002/bit.25264 [DOI] [PubMed] [Google Scholar]
- Hanafy R. A., Elshahed M. S., Liggenstoffer A. S., Griffith G. W., Youssef N. H. (2017). Pecoramyces ruminantium, gen. nov., sp. nov., an anaerobic gut fungus from the feces of cattle and sheep. Mycologia 109 231–243. 10.1080/00275514.2017.1317190 [DOI] [PubMed] [Google Scholar]
- Hanafy R. A., Elshahed M. S., Youssef N. H. (2018). Feramyces austinii, gen. nov., sp. nov., an anaerobic gut fungus from rumen and fecal samples of wild Barbary sheep and fallow deer. Mycologia 110 513–525. 10.1080/00275514.2018.1466610 [DOI] [PubMed] [Google Scholar]
- Hanafy R. A., Lanjekar V. B., Dhakephalkar P. K., Callaghan T. M., Dagar S. S., Griffith G. W., et al. (2020). Seven new Neocallimastigomycota genera from wild, zoo-housed, and domesticated herbivores greatly expand the taxonomic diversity of the phylum. Mycologia 10.1080/00275514.2019.1696619 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Heath I. B., Bauchop T., Skipp R. A. (1983). Assignment of the rumen anaerobe Neocallimastix frontalis to the Spizellomycetales (Chytridiomycetes) on the basis of its polyflagellate zoospore ultrastructure. Can. J. Bot. 61 295–307. 10.1139/b83-033 [DOI] [Google Scholar]
- Heath I. B., Kaminskyj S. G., Bauchop T. (1986). Basal body loss during fungal zoospore encystment: evidence against centriole autonomy. J. Cell Sci. 83 135–140. [DOI] [PubMed] [Google Scholar]
- Hegarty R., Gerdes R. (1999). Hydrogen production and transfer in the rumen. Recent Adv. Anim. Nutr. Aust. 12 37–44. [Google Scholar]
- Henderson G., Cox F., Ganesh S., Jonker A., Young W., Global Rumen Census C., et al. (2015). Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henske J. K., Gilmore S. P., Knop D., Cunningham F. J., Sexton J. A., Smallwood C. R., et al. (2017). Transcriptomic characterization of Caecomyces churrovis: a novel, non-rhizoid-forming lignocellulolytic anaerobic fungus. Biotechnol. Biofuels 10:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henske J. K., Wilken S. E., Solomon K. V., Smallwood C. R., Shutthanandan V., Evans J. E., et al. (2018). Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose. Biotechnol. Bioeng. 115 874–884. 10.1002/bit.26515 [DOI] [PubMed] [Google Scholar]
- Hess M., Sczyrba A., Egan R., Kim T., Chokhawala H., Schroth G., et al. (2011). Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331 463–467. 10.1126/science.1200387 [DOI] [PubMed] [Google Scholar]
- Hibbett D. S., Binder M., Bischoff J. F., Blackwell M., Cannon P. F., Eriksson O. E., et al. (2007). A higher-level phylogenetic classification of the Fungi. Mycol. Res. 111 509–547. [DOI] [PubMed] [Google Scholar]
- Ho Y. W., Abdullah N., Jalaludin S. (1988). Penetrating structures of anaerobic rumen fungi in cattle and swamp buffalo. Microbiology 134 177–181. 10.1099/00221287-134-1-177 [DOI] [Google Scholar]
- Ho Y. W., Bauchop T. (1991). Morphology of three polycentric rumen fungi and description of a procedure for the induction of zoosporogenesis and release of zoospores in cultures. J. Gen. Microbiol. 137 213–217. 10.1099/00221287-137-1-213 [DOI] [PubMed] [Google Scholar]
- Huws S. A., Creevey C. J., Oyama L. B., Mizrahi I., Denman S. E., Popova M., et al. (2018). Addressing global ruminant agricultural challenges through understanding the rumen microbiome: past, present, and future. Front. Microbiol. 9:2161. 10.3389/fmicb.2018.02161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irbis C., Ushida K. (2004). Detection of methanogens and proteobacteria from a single cell of rumen ciliate protozoa. J. Gen. Appl. Microbiol. 50 203–212. 10.2323/jgam.50.203 [DOI] [PubMed] [Google Scholar]
- Ishaq S. L., AlZahal O., Walker N., McBride B. (2017). An investigation into rumen fungal and protozoal diversity in three rumen fractions, during high-fiber or grain-induced sub-acute ruminal acidosis conditions, with or without active dry yeast supplementation. Front. Microbiol. 8:1943. 10.3389/fmicb.2017.01943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen P. H. (2010). Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 160 1–22. 10.1016/j.anifeedsci.2010.07.002 [DOI] [Google Scholar]
- Jeyanathan J., Martin C., Morgavi D. P. (2014). The use of direct-fed microbials for mitigation of ruminant methane emissions: a review. Animal 8 250–261. 10.1017/s1751731113002085 [DOI] [PubMed] [Google Scholar]
- Jin W., Cheng Y. F., Mao S. Y., Zhu W. Y. (2011). Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour. Technol. 102 7925–7931. 10.1016/j.biortech.2011.06.026 [DOI] [PubMed] [Google Scholar]
- Joblin K. N., Campbell G. P., Richardson A. J., Stewart C. S. (1989). Fermentation of barley straw by anaerobic rumen bacteria and fungi in axenic culture and in co-culture with methanogens. Lett. Appl. Microbiol. 9 195–197. 10.1111/j.1472-765x.1989.tb00323.x [DOI] [Google Scholar]
- Joshi A., Lanjekar V. B., Dhakephalkar P. K., Callaghan T. M., Griffith G. W., Dagar S. S. (2018). Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. Mycokeys 40 89–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julliand V., Grimm P. (2017). The impact of diet on the hindgut microbiome. J. Equine Vet. Sci. 52 23–28. 10.1016/j.jevs.2017.03.002 [DOI] [Google Scholar]
- Julliand V., Riondet C., de Vaux A., Alcaraz G., Fonty G. (1998). Comparison of metabolic activities between Piromyces citronii, an equine fungal species, and Piromyces communis, a ruminal species. Anim. Feed Sci. Technol. 70 161–168. 10.1016/s0377-8401(97)00043-6 [DOI] [Google Scholar]
- Kittelmann S., Naylor G. E., Koolaard J. P., Janssen P. H. (2012). A proposed taxonomy of anaerobic fungi (class neocallimastigomycetes) suitable for large-scale sequence-based community structure analysis. PLoS One 7:e36866. 10.1371/journal.pone.0036866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelmann S., Pinares-Patino C. S., Seedorf H., Kirk M. R., Ganesh S., McEwan J. C., et al. (2014). Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9:e103171. 10.1371/journal.pone.0103171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittelmann S., Seedorf H., Walters W. A., Clemente J. C., Knight R., Gordon J. I., et al. (2013). Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. 10.1371/journal.pone.0047879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetschan C., Kittelmann S., Lu J., Al-Halbouni D., Jarvis G. N., Müller T., et al. (2014). Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota). PLoS One 9:e91928. 10.1371/journal.pone.0091928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok C. M., Sieo C. C., Tan H. Y., Saad W. Z., Liang J. B., Ho Y. W. (2013). Anaerobic cellulolytic rumen fungal populations in goats fed with and without Leucaena leucocephala hybrid, as determined by real-time PCR. J. Microbiol. 51 700–703. 10.1007/s12275-013-2540-z [DOI] [PubMed] [Google Scholar]
- Kostyukovsky V. A., Okunev O. N., Tarakanov B. V. (1991). Description of two anaerobic fungal strains from the bovine rumen and influence of diet on the fungal population in vivo. J. Gen. Microbiol. 137 1759–1764. 10.1099/00221287-137-7-1759 [DOI] [PubMed] [Google Scholar]
- Krauss J., Zverlov V. V., Schwarz W. H. (2012). In vitro reconstitution of the complete Clostridium thermocellum cellulosome and synergistic activity on crystalline cellulose. Appl. Environ. Microbiol. 78 4301–4307. 10.1128/aem.07959-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamed R., Setter E., Bayer E. A. (1983). Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156 828–836. 10.1128/jb.156.2.828-836.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S., Ha J. K., Cheng K. J. (2000a). Influence of an anaerobic fungal culture administration on in vivo ruminal fermentation and nutrient digestion. Anim. Feed Sci. Technol. 88 201–217. 10.1016/s0377-8401(00)00216-9 [DOI] [Google Scholar]
- Lee S. S., Ha J. K., Cheng K. J. (2000b). Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl. Environ. Microbiol. 66 3807–3813. 10.1128/aem.66.9.3807-3813.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Jin W., Mu C., Cheng Y., Zhu W. (2017). Indigenously associated methanogens intensified the metabolism in hydrogenosomes of anaerobic fungi with xylose as substrate. J. Basic Microbiol. 57 933–940. 10.1002/jobm.201700132 [DOI] [PubMed] [Google Scholar]
- Li Y., Li Y., Jin W., Sharpton T. J., Mackie R. I., Cann I., et al. (2019). Combined genomic, transcriptomic, proteomic, and physiological characterization of the growth of Pecoramyces sp. F1 in monoculture and co-culture with a syntrophic methanogen. Front. Microbiol. 10:435. 10.3389/fmicb.2019.00435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebetanz E. (1910). Die parasitischen protozoen des wiederkauermagens. Arch. Protistenkunde 19 19–90. [Google Scholar]
- Liggenstoffer A. S., Youssef N. H., Couger M. B., Elshahed M. S. (2010). Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. ISME J. 4 1225–1235. 10.1038/ismej.2010.49 [DOI] [PubMed] [Google Scholar]
- Liu C., Meng Q., Chen Y., Xu M., Shen M., Gao R., et al. (2017). Role of age-related shifts in rumen bacteria and methanogens in methane production in cattle. Front. Microbiol. 8:1563. 10.3389/fmicb.2017.01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. R., Yu B., Lin S. H., Cheng K. J., Chen Y. C. (2005a). Direct cloning of a xylanase gene from the mixed genomic DNA of rumen fungi and its expression in intestinal Lactobacillus reuteri. FEMS Microbiol. Lett. 251 233–241. 10.1016/j.femsle.2005.08.008 [DOI] [PubMed] [Google Scholar]
- Liu J. R., Yu B., Liu F. H., Cheng K. J., Zhao X. (2005b). Expression of rumen microbial fibrolytic enzyme genes in probiotic Lactobacillus reuteri. Appl. Environ. Microbiol. 71 6769–6775. 10.1128/aem.71.11.6769-6775.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. R., Yu B., Zhao X., Cheng K. J. (2007). Coexpression of rumen microbial beta-glucanase and xylanase genes in Lactobacillus reuteri. Appl. Microbiol. Biotechnol. 77 117–124. 10.1007/s00253-007-1123-5 [DOI] [PubMed] [Google Scholar]
- Ljungdahl L. G. (2008). The cellulase/hemicellulase system of the anaerobic fungus Orpinomyces PC-2 and aspects of its applied use. Ann. N. Y. Acad. Sci. 1125 308–321. 10.1196/annals.1419.030 [DOI] [PubMed] [Google Scholar]
- Lowe S. E., Grifith G. G., Milne A., Theodorou M. K., Trinci A. P. J. (1987a). The life cycle and growth kinetics of an anaerobic rumen fungus. J. Gen. Microbiol. 133 1815–1827. 10.1099/00221287-133-7-1815 [DOI] [Google Scholar]
- Lowe S. E., Theodorou M. K., Trinci A. P. J. (1987b). Isolation of anaerobic fungi from saliva and faeces of sheep. Microbiology 133 1829–1834. 10.1099/00221287-133-7-1829 [DOI] [Google Scholar]
- Lyu Z., Shao N., Akinyemi T., Whitman W. B. (2018). Methanogenesis. Curr. Biol. 28 R727–R732. [DOI] [PubMed] [Google Scholar]
- Marinier S. L., Alexander A. J. (1995). Coprophagy as an avenue for foals of the domestic horse to learn food preferences from their dams. J. Theor. Biol. 173 121–124. 10.1006/jtbi.1995.0049 [DOI] [Google Scholar]
- Marvin-Sikkema F. D., Pedro Gomes T. M., Grivet J.-P., Gottschal J. C., Prins R. A. (1993). Characterization of hydrogenosomes and their role in glucose metabolism of Neocallimastix sp. L2. Arch. Microbiol. 160 388–396. [DOI] [PubMed] [Google Scholar]
- Marvin-Sikkema F. D., Richardson A. J., Stewart C. S., Gottschal J. C., Prins R. A. (1990). Influence of hydrogen-consuming bacteria on cellulose degradation by anaerobic fungi. Appl. Environ. Microbiol. 56 3793–3797. 10.1128/aem.56.12.3793-3797.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H., Ushida K., Kojima Y. (1997). Effect of dietary concentrate on fungal zoosporogenesis in sheep rumen. Asian Australas. J. Anim. Sci. 10 599–602. 10.5713/ajas.1997.599 [DOI] [Google Scholar]
- McAllister T. A., Dong Y., Yanke L. J., Bae H. D., Cheng K. J., Costerton J. W. (1993). Cereal grain digestion by selected strains of ruminal fungi. Can. J. Microbiol. 39 367–376. 10.1139/m93-054 [DOI] [PubMed] [Google Scholar]
- McGranaghan P., Davies J. C., Griffith G. W., Davies D. R., Theodorou M. K. (1999). The survival of anaerobic fungi in cattle faeces. FEMS Microbiol. Ecol. 29 293–300. 10.1111/j.1574-6941.1999.tb00620.x [DOI] [Google Scholar]
- McSweeney C. S., Denman S. E. (2007). Effect of sulfur supplements on cellulolytic rumen micro-organisms and microbial protein synthesis in cattle fed a high fibre diet. J. Appl. Microbiol. 103 1757–1765. 10.1111/j.1365-2672.2007.03408.x [DOI] [PubMed] [Google Scholar]
- McSweeney C. S., Palmer B., Bunch R., Krause D. O. (2001). Effect of the tropical forage calliandra on microbial protein synthesis and ecology in the rumen. J. Appl. Microbiol. 90 78–88. 10.1046/j.1365-2672.2001.01220.x [DOI] [PubMed] [Google Scholar]
- Michel V., Fonty G., Millet L., Bonnemoy F., Gouet P. (1993). In vitro study of the proteolytic activity of rumen anaerobic fungi. FEMS Microbiol. Lett. 110 5–9. 10.1111/j.1574-6968.1993.tb06287.x [DOI] [PubMed] [Google Scholar]
- Milne A., Theodorou M. K., Jordan M. G. C., Kingspooner C., Trinci A. P. J. (1989). Survival of anaerobic fungi in feces, in saliva, and in pure culture. Exp. Mycol. 13 27–37. 10.1016/0147-5975(89)90005-4 [DOI] [Google Scholar]
- Mizrahi I. (2013). “Rumen symbioses,” in The Prokaryotes: Prokaryotic Biology and Symbiotic Associations, eds Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Berlin: Springer; ), 533–544. 10.1007/978-3-642-30194-0_1 [DOI] [Google Scholar]
- Morais S., Mizrahi I. (2019). Islands in the stream: from individual to communal fiber degradation in the rumen ecosystem. FEMS Microbiol. Rev. 43 362–379. 10.1093/femsre/fuz007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgavi D. P., Kelly W. J., Janssen P. H., Attwood G. T. (2013). Rumen microbial (meta)genomics and its application to ruminant production. Animal 7 (Suppl. 1), 184–201. 10.1017/s1751731112000419 [DOI] [PubMed] [Google Scholar]
- Morgavi D. P., Sakurada M., Mizokami M., Tomita Y., Onodera R. (1994). Effects of ruminal protozoa on cellulose degradation and the growth of an anaerobic ruminal fungus, Piromyces sp. strain OTS1, in vitro. Appl. Environ. Microbiol. 60 3718–3723. 10.1128/aem.60.10.3718-3723.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M., Murray R. M., Boniface A. N. (1990). Nutrient metabolism and rumen micro-organisms in sheep fed a poor-quality tropical grass hay supplemented with sulphate. J. Agric. Sci. 115 269–275. 10.1017/s0021859600075237 [DOI] [Google Scholar]
- Müller M. (1993). The hydrogenosome. J. Gen. Microbiol. 139 2879–2889. [DOI] [PubMed] [Google Scholar]
- Mura E., Edwards J., Kittelmann S., Kaerger K., Voigt K., Mrazek J., et al. (2019). Anaerobic fungal communities differ along the horse digestive tract. Fungal Biol. 123 240–246. 10.1016/j.funbio.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Murphy C. L., Youssef N. H., Hanafy R. A., Couger M. B., Stajich J. E., Wang Y., et al. (2019). Horizontal gene transfer as an indispensable driver for evolution of Neocallimastigomycota into a distinct gut-dwelling fungal lineage. Appl. Environ. Microbiol. 85:e00988-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja T. G. (2016). “Microbiology of the rumen,” in Rumenology, eds Millen D. D., De Beni Arrigoni M., Lauritano Pacheco R. D. (Cham: Springer International Publishing; ), 39–61. [Google Scholar]
- Nagler M., Kozjek K., Etemadi M., Insam H., Podmirseg S. M. (2019). Simple yet effective: microbial and biotechnological benefits of rumen liquid addition to lignocellulose-degrading biogas plants. J. Biotechnol. 300 1–10. 10.1016/j.jbiotec.2019.05.004 [DOI] [PubMed] [Google Scholar]
- Nagler M., Podmirseg S. M., Griffith G. W., Insam H., Ascher-Jenull J. (2018). The use of extracellular DNA as a proxy for specific microbial activity. Appl. Microbiol. Biotechnol. 102 2885–2898. 10.1007/s00253-018-8786-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy T., Tunnicliffe R. B., Higgins L. D., Walters C., Gilbert H. J., Williamson M. P. (2007). Characterization of a double dockerin from the cellulosome of the anaerobic fungus Piromyces equi. J. Mol. Biol. 373 612–622. 10.1016/j.jmb.2007.08.007 [DOI] [PubMed] [Google Scholar]
- Obispo N. E., Dehority B. A. (1992). A most probable number method for enumeration of rumen fungi with studies on factors affecting their concentration in the rumen. J. Microbiol. Methods 16 259–270. 10.1016/0167-7012(92)90016-w [DOI] [Google Scholar]
- Offre P., Spang A., Schleper C. (2013). Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 67 437–457. 10.1146/annurev-micro-092412-155614 [DOI] [PubMed] [Google Scholar]
- Orpin C. G. (1975). Studies on the rumen flagellate Neocallimastix frontalis. J. Gen. Microbiol. 91 249–262. 10.1099/00221287-91-2-249 [DOI] [PubMed] [Google Scholar]
- Orpin C. G. (1976). Studies on the rumen flagellate Sphaeromonas communis. J. Gen. Microbiol. 94 270–280. 10.1099/00221287-94-2-270 [DOI] [PubMed] [Google Scholar]
- Orpin C. G. (1977a). The occurrence of chitin in the cell walls of the rumen organisms Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J. Gen. Microbiol. 99 215–218. 10.1099/00221287-99-1-215 [DOI] [PubMed] [Google Scholar]
- Orpin C. G. (1977b). The rumen flagellate Piromonas communis: its life-history and invasion of plant material in the rumen. J. Gen. Microbiol. 99 107–117. 10.1099/00221287-99-1-107 [DOI] [PubMed] [Google Scholar]
- Orpin C. G. (1981). Isolation of cellulolytic phycomycete fungi from the caecum of the horse. J. Gen. Microbiol. 123 287–296. 10.1099/00221287-123-2-287 [DOI] [PubMed] [Google Scholar]
- Orpin C. G. (1994). “Anaerobic fungi: taxonomy, biology and distribution in nature,” in Anaerobic Fungi, Vol. 12 eds Mountfort D. O., Orpin C. G. (New York, NY: Dekker; ), 1–45. 10.1201/9781003067085-1 [DOI] [Google Scholar]
- Orpin C. G., Bountiff L. (1978). Zoospore chemotaxis in the rumen phycomycete Neocallimaslix frontalis. J. Gen. Microbiol. 104 113–122. 10.1099/00221287-104-1-113 [DOI] [PubMed] [Google Scholar]
- Orpin C. G., Greenwood Y. (1986). The role of haems and related compounds in the nutrition and zoosporogenesis of the rumen chytridiomycete Neocallimastix frontalis H8. Microbiology 132 2179–2185. 10.1099/00221287-132-8-2179 [DOI] [Google Scholar]
- Ozkose E., Thomas B. J., Davies D. R., Griffith G. W., Theodorou M. K. (2001). Cyllamyces aberensis gen.nov. sp.nov., a new anaerobic gut fungus with branched sporangiophores isolated from cattle. Can. J. Bot. 79 666–673. 10.1139/cjb-79-6-666 [DOI] [Google Scholar]
- Partida-Martinez L. P., Monajembashi S., Greulich K. O., Hertweck C. (2007). Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 17 773–777. 10.1016/j.cub.2007.03.039 [DOI] [PubMed] [Google Scholar]
- Paul S. S., Bu D., Xu J., Hyde K. D., Yu Z. (2018). A phylogenetic census of global diversity of gut anaerobic fungi and a new taxonomic framework. Fungal Divers. 89 253–266. 10.1007/s13225-018-0396-6 [DOI] [Google Scholar]
- Paul S. S., Deb S. M., Punia B. S., Das K. S., Singh G., Ashar M. N., et al. (2011). Effect of feeding isolates of anaerobic fungus Neocallimastix sp. CF 17 on growth rate and fibre digestion in buffalo calves. Arch. Anim. Nutr. 65 215–228. 10.1080/1745039x.2011.559722 [DOI] [PubMed] [Google Scholar]
- Paul S. S., Kamra D. N., Sastry V. R., Sahu N. P. (2006). Effect of adding an anaerobic fungal culture isolated from a wild blue bull (Boselophus tragocamelus) to rumen fluid from buffaloes on in vitro fibrolytic enzyme activity, fermentation and degradation of tannins and tannin-containing Kachnar tree (Bauhinia variegata) leaves and wheat straw. J. Sci. Food Agric. 86 258–270. 10.1002/jsfa.2303 [DOI] [Google Scholar]
- Paul S. S., Kamra D. N., Sastry V. R., Sahu N. P., Kumar A. (2003). Effect of phenolic monomers on biomass and hydrolytic enzyme activities of an anaerobic fungus isolated from wild nil gai (Baselophus tragocamelus). Lett. Appl. Microbiol. 36 377–381. 10.1046/j.1472-765x.2003.01331.x [DOI] [PubMed] [Google Scholar]
- Paul S. S., Kamra D. N., Sastry V. R. B., Sahu N. P., Agarwal N. (2004). Effect of administration of an anaerobic gut fungus isolated from wild blue bull (Boselaphus tragocamelus) to buffaloes (Bubalus bubalis) on in vivo ruminal fermentation and digestion of nutrients. Anim. Feed Sci. Technol. 115 143–157. 10.1016/j.anifeedsci.2004.01.010 [DOI] [Google Scholar]
- Peng X. N., Gilmore S. P., O’Malley M. A. (2016). Microbial communities for bioprocessing: lessons learned from nature. Curr. Opin. Chem. Eng. 14 103–109. 10.1016/j.coche.2016.09.003 [DOI] [Google Scholar]
- Phillips M. W., Gordon G. L. R. (1988). Sugar and polysaccharide fermentation by rumen anaerobic fungi from Australia, Britain and New Zealand. Biosystems 21 377–383. 10.1016/0303-2647(88)90036-6 [DOI] [PubMed] [Google Scholar]
- Poulsen M., Schwab C., Borg Jensen B., Engberg R. M., Spang A., Canibe N., et al. (2013). Methylotrophic methanogenic Thermoplasmata implicated in reduced methane emissions from bovine rumen. Nat. Commun. 4:1428. [DOI] [PubMed] [Google Scholar]
- Promkot C., Wanapat M. (2009). Effect of elemental sulfur supplementation on rumen environment parameters and utilization efficiency of fresh cassava foliage and cassava hay in dairy cattle. Asian Australas. J. Anim. Sci. 22 1366–1376. 10.5713/ajas.2009.90141 [DOI] [Google Scholar]
- Reay D. S., Smith P., Christensen T. R., James R. H., Clark H. (2018). Methane and global environmental change. Annu. Rev. Environ. Resour. 43 165–192. [Google Scholar]
- Rezaeian M., Beakes G. W., Parker D. S. (2004a). Distribution and estimation of anaerobic zoosporic fungi along the digestive tracts of sheep. Mycol. Res. 108(Pt 10), 1227–1233. 10.1017/s0953756204000929 [DOI] [PubMed] [Google Scholar]
- Rezaeian M., Beakes G. W., Parker D. S. (2004b). Methods for the isolation, culture and assessment of the status of anaerobic rumen chytrids in both in vitro and in vivo systems. Mycol. Res. 108 1215–1226. 10.1017/s0953756204000917 [DOI] [PubMed] [Google Scholar]
- Ribeiro G. O., Gruninger R. J., Badhan A., McAllister T. A. (2016). Mining the rumen for fibrolytic feed enzymes. Anim. Front. 6 20–26. 10.2527/af.2016-0019 32704858 [DOI] [Google Scholar]
- Ricard G., McEwan N. R., Dutilh B. E., Jouany J.-P., Macheboeuf D., Mitsumori M., et al. (2006). Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7:22. 10.1186/1471-2164-7-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehe R., Dewhurst R. J., Duthie C. A., Rooke J. A., McKain N., Ross D. W., et al. (2016). Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet. 12:e1005846. 10.1371/journal.pgen.1005846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotterová J., Salomaki E., Pánek T., Bourland W., Žihala D., Táborskı P., et al. (2020). Genomics of new ciliate lineages provides insight into the evolution of obligate anaerobiosis. Curr. Biol. 30 2037–2050.e6. [DOI] [PubMed] [Google Scholar]
- Saminathan M., Kumari Ramiah S., Gan H. M., Abdullah N., Wong C. M. V. L., Ho Y. W., et al. (2019). In vitro study on the effects of condensed tannins of different molecular weights on bovine rumen fungal population and diversity. Ital. J. Anim. Sci. 18 1451–1462. 10.1080/1828051x.2019.1681304 [DOI] [Google Scholar]
- Sánchez-Rodríguez C., Bauer S., Hématy K., Saxe F., Ibáñez A. B., Vodermaier V., et al. (2012). Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24 589–607. 10.1105/tpc.111.094672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S., Sehgal J., Puniya A., Singh K. (2010). Effect of administration of rumen fungi on production performance of lactating buffaloes. Benef. Microbes 1 183–188. 10.3920/bm2009.0018 [DOI] [PubMed] [Google Scholar]
- Sehgal J. P., Jit D., Puniya A. K., Singh K. (2008). Influence of anaerobic fungal administration on growth, rumen fermentation and nutrient digestion in female buffalo calves. J. Anim. Feed Sci. 17 510–518. 10.22358/jafs/66678/2008 [DOI] [Google Scholar]
- Sekine J., Shinoda M., Kamel H., Oura R. (1995). Effect of kinds of bay on population densities of rumen anaerobic fungi. Indian J. Anim. Sci. 65 1352–1355. [Google Scholar]
- Seshadri R., Leahy S. C., Attwood G. T., Teh K. H., Lambie S. C., Cookson A. L., et al. (2018). Cultivation and sequencing of rumen microbiome members from the Hungate1000 collection. Nat. Biotechnol. 36 359–367. 10.1038/nbt.4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Moon C. D., Leahy S. C., Kang D., Froula J., Kittelmann S., et al. (2014). Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 24 1517–1525. 10.1101/gr.168245.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirazi-Beechey S. P. (2008). Molecular insights into dietary induced colic in the horse. Equine Vet. J. 40 414–421. 10.2746/042516408x314075 [DOI] [PubMed] [Google Scholar]
- Sijtsma L., Tan B. (1996). Degradation of perennial ryegrass leaf and stem cell walls by the anaerobic fungus Neocallimastix sp. strain CS3b. Appl. Environ. Microbiol. 62 1437–1440. 10.1128/aem.62.4.1437-1440.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon K. V., Haitjema C. H., Henske J. K., Gilmore S. P., Borges-Rivera D., Lipzen A., et al. (2016). Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351 1192–1195. 10.1126/science.aad1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R. D., Auffret M. D., Warr A., Walker A. W., Roehe R., Watson M. (2019). Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 37 953–961. 10.1038/s41587-019-0202-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapio I., Fischer D., Blasco L., Tapio M., Wallace R. J., Bayat A. R., et al. (2017). Taxon abundance, diversity, co-occurrence and network analysis of the ruminal microbiota in response to dietary changes in dairy cows. PLoS One 12:e0180260. 10.1371/journal.pone.0180260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L., Sánchez-Ramírez S., Kõljalg U., Bahram M., Döring M., Schigel D., et al. (2018). High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 90 135–159. 10.1007/s13225-018-0401-0 [DOI] [Google Scholar]
- Theodorou M. K., Gill M., King-Spooner C., Beever D. E. (1990). Enumeration of anaerobic chytridiomycetes as thallus-forming units: novel method for quantification of fibrolytic fungal populations from the digestive tract ecosystem. Appl. Environ. Microbiol. 56 1073–1078. 10.1128/aem.56.4.1073-1078.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorou M. K., Zhu W. Y., Rickers A., Nielsen B. B., Gull K., Trinci A. P. J. (1996). “Biochemistry and ecology of anaerobic fungi,” in Human and Animal Relationships, eds Howard D. H., Miller J. D. (Berlin: Springer; ), 265–295. [Google Scholar]
- Trinci A. P. J., Davies D. R., Gull K., Lawrence M. I., Nielsen B. B., Rickers A., et al. (1994). Anaerobic fungi in herbivorous animals. Mycol. Res. 98 129–152. 10.1016/s0953-7562(09)80178-0 [DOI] [Google Scholar]
- Tripathi V. K., Sehgal J. P., Puniya A. K., Singh K. (2007). Effect of administration of anaerobic fungi isolated from cattle and wild blue bull (Boselaphus tragocamelus) on growth rate and fibre utilization in buffalo calves. Arch. Anim. Nutr. 61 416–423. 10.1080/17450390701556759 [DOI] [PubMed] [Google Scholar]
- Ushida K., Tanaka H., Kojima Y. (1989). A simple in situ method for estimating fungal population size in the rumen. Lett. Appl. Microbiol. 9 109–111. 10.1111/j.1472-765x.1989.tb00302.x [DOI] [Google Scholar]
- Ushida K., Umeda M., Kishigami N., Kojima Y. (1992). Effect of medium-chain and long-chain fatty acid calcium salts on rumen microorganisms and fiber digestion in sheep. Nihon Chikusan Gakkaiho 63 591–597. 10.2508/chikusan.63.591 [DOI] [Google Scholar]
- Valle E. R., Henderson G., Janssen P. H., Cox F., Alexander T. W., McAllister T. A. (2015). Considerations in the use of fluorescence in situ hybridization (FISH) and confocal laser scanning microscopy to characterize rumen methanogens and define their spatial distributions. Can. J. Microbiol. 61 417–428. 10.1139/cjm-2014-0873 [DOI] [PubMed] [Google Scholar]
- van der Giezen M. (2009). Hydrogenosomes and mitosomes: conservation and evolution of functions. J. Eukaryot. Microbiol. 56 221–231. 10.1111/j.1550-7408.2009.00407.x [DOI] [PubMed] [Google Scholar]
- Vogels G. D., Hoppe W. F., Stumm C. K. (1980). Association of methanogenic bacteria with rumen ciliates. Appl. Environ. Microbiol. 40 608–612. 10.1128/aem.40.3.608-612.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. J., Joblin K. N. (1985). Proteolytic activity of a rumen anaerobic fungus. FEMS Microbiol. Lett. 29 19–25. 10.1111/j.1574-6968.1985.tb00828.x [DOI] [Google Scholar]
- Wallace R. J., Munro C. A. (1986). Influence of the rumen anaerobic fungus Neocallimastix frontalis on the proteolytic activity of a defined mixture of rumen bacteria growing on a solid substrate. Lett. Appl. Microbiol. 3 23–26. 10.1111/j.1472-765x.1986.tb01539.x [DOI] [Google Scholar]
- Wallace R. J., Sasson G., Garnsworthy P. C., Tapio I., Gregson E., Bani P., et al. (2019). A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 5:eaav8391. 10.1126/sciadv.aav8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu X., Groenewald J. Z. (2017). Phylogeny of anaerobic fungi (phylum Neocallimastigomycota), with contributions from yak in China. Antonie Van Leeuwenhoek 110 87–103. 10.1007/s10482-016-0779-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Youssef N. H., Couger M. B., Hanafy R. A., Elshahed M. S., Stajich J. E. (2019). Molecular dating of the emergence of anaerobic rumen fungi and the impact of laterally acquired genes. mSystems 4:e00247-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston R. H., Lindsay J. R., Purser D. B., Gordon G. L. R., Davis P. (1988). Feed intake and digestion responses in sheep to the addition of inorganic sulfur to a herbage diet of low sulfur content. Aust. J. Agric. Res. 39 1107–1119. 10.1071/ar9881107 [DOI] [Google Scholar]
- Wilson C. A., Wood T. M. (1992). The anaerobic fungus Neocallimastix frontalis: isolation and properties of a cellulosome-type enzyme fraction with the capacity to solubilize hydrogen-bond-ordered cellulose. Appl. Microbiol. Biotechnol. 37 125–129. [Google Scholar]
- Woese C. R., Fox G. E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U.S.A. 74 5088–5090. 10.1073/pnas.74.11.5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubah D. A., Fuller M. S., Akin D. E. (1991). Resistant body formation in Neocallimastix sp., an anaerobic fungus from the rumen of cow. Mycologia 83 40–47. 10.2307/3759831 [DOI] [Google Scholar]
- Wubah D. A., Kim D. S. H. (1996). Chemoattraction of anaerobic ruminai fungi zoospores to selected phenolic acids. Microbiol. Res. 151 257–262. 10.1016/s0944-5013(96)80022-x [DOI] [PubMed] [Google Scholar]
- Wurzbacher C., Larsson E., Bengtsson-Palme J., Van den Wyngaert S., Svantesson S., Kristiansson E., et al. (2019). Introducing ribosomal tandem repeat barcoding for fungi. Mol. Ecol. Resour. 19 118–127. 10.1111/1755-0998.12944 [DOI] [PubMed] [Google Scholar]
- Xue F., Nan X., Sun F., Pan X., Guo Y., Jiang L., et al. (2018). Metagenome sequencing to analyze the impacts of thiamine supplementation on ruminal fungi in dairy cows fed high-concentrate diets. AMB Express 8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanke L. J., Dong Y., McAllister T. A., Bae H. D., Cheng K. J. (1993). Comparison of amylolytic and proteolytic activities of ruminal fungi grown on cereal grains. Can. J. Microbiol. 39 817–820. 10.1139/m93-121 [DOI] [PubMed] [Google Scholar]
- Yarlett N., Orpin C. G., Munn E. A., Yarlett N. C., Greenwood C. A. (1986). Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem. J. 236 729–739. 10.1042/bj2360729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef N. H., Couger M. B., Struchtemeyer C. G., Liggenstoffer A. S., Prade R. A., Najar F. Z., et al. (2013). The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 79 4620–4634. 10.1128/aem.00821-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Difford G., Sahana G., Løvendahl P., Lassen J., Lund M. S., et al. (2020). Bayesian modeling reveals host genetics associated with rumen microbiota jointly influence methane emission in dairy cows. ISME J. 14 2019–2033. 10.1038/s41396-020-0663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.