Abstract
Studying the genetics of adaptation to new environments in ecologically and industrially important tree species is currently a major research line in the fields of plant science and genetic improvement for tolerance to abiotic stress. Specifically, exploring the genomic basis of local adaptation is imperative for assessing the conditions under which trees will successfully adapt in situ to global climate change. However, this knowledge has scarcely been used in conservation and forest tree improvement because woody perennials face major research limitations such as their outcrossing reproductive systems, long juvenile phase, and huge genome sizes. Therefore, in this review we discuss predictive genomic approaches that promise increasing adaptive selection accuracy and shortening generation intervals. They may also assist the detection of novel allelic variants from tree germplasm, and disclose the genomic potential of adaptation to different environments. For instance, natural populations of tree species invite using tools from the population genomics field to study the signatures of local adaptation. Conventional genetic markers and whole genome sequencing both help identifying genes and markers that diverge between local populations more than expected under neutrality, and that exhibit unique signatures of diversity indicative of “selective sweeps.” Ultimately, these efforts inform the conservation and breeding status capable of pivoting forest health, ecosystem services, and sustainable production. Key long-term perspectives include understanding how trees’ phylogeographic history may affect the adaptive relevant genetic variation available for adaptation to environmental change. Encouraging “big data” approaches (machine learning—ML) capable of comprehensively merging heterogeneous genomic and ecological datasets is becoming imperative, too.
Keywords: genomics of adaptation, genomic prediction, genome-wide association studies, genome-wide selection scans, assisted gene flow, machine learning, big data
Introduction
How trees will respond to climate change is a pressing question both in the contexts of natural forests and tree plantations (Kremer et al., 2014; Holliday et al., 2017; Isabel et al., 2020). Forests offer key ecological services, boosting significant resources of biodiversity in terms of species and habitats, while help mitigating the impact of excess air pollutants (Phillips et al., 2019; Pennisi, 2020). Trees also source natural renewable materials (i.e., wood itself, cellulose for the pulp industry, and lignin and hemicelluloses for energy production), likely to increase in the future as sustainable alternatives to fossil fuels (Carlson et al., 2014).
Yet, forest tree species are being threatened by climate change (Sullivan et al., 2020) due to fluctuations in the frequency and intensity of heat, drought, salinity (Naidoo et al., 2019), and the incidence of pathogens and pests (Naidoo et al., 2014; Christie et al., 2015). Hence, now more than ever it is essential to explore changing abiotic (Chakhchar et al., 2017; Alcaide et al., 2019b) and biotic (Meyer et al., 2016) interactions. Rampant phenotypic plasticity (Berlin et al., 2017; Hallingback et al., 2019) to climate gradients is presumed in trees, arguing resilience to variability throughout their long lives. Still, forests adaptability should also be assessed in the light of spatially varying local environmental selective pressures (Savolainen et al., 2013), and trees’ genetic and evolutionary potentials (Howe and Brunner, 2005). Both directly reflect and feedback overall adaptive genetic variation. Hence, understanding the genomic drivers that underpin adaptive trait variation becomes vital for conservation and industrial goals.
Developments in plant genomics (Brunner et al., 2007a; Neale and Kremer, 2011) have already disclosed the genetic basis of various useful traits (Khan and Korban, 2012; Tuskan et al., 2018). Yet, this information has limitedly been utilized in tree improvement and conservation (Flanagan et al., 2018), despite genetic gains (Figure 1) and optimized management are urgently required due to environmental issues (Scherer et al., 2020). Besides, breeding woody perennials is primarily bottlenecked by their outcrossing reproductive systems, prolonged juvenile phases (Grattapaglia et al., 2018), large genome sizes lacking elimination mechanisms of long-terminal transposons (Nystedt et al., 2013), and an excessive focus on productivity (Burdon and KlápšTě, 2019) that omits adaptive traits (Table 1; Li et al., 2019). Thus, here we discuss ways to side step these limitations by arguing how predictive genomics can increase selection accuracy and shorten generation intervals (Grattapaglia et al., 2018), assist the detection of exotic variants from tree germplasm (Migicovsky and Myles, 2017), and disclose the genomic potential of adaptation to different climates (Lind et al., 2018). These efforts will ultimately inform conservation and breeding to enhance forest health, ecosystem services, and sustainable production.
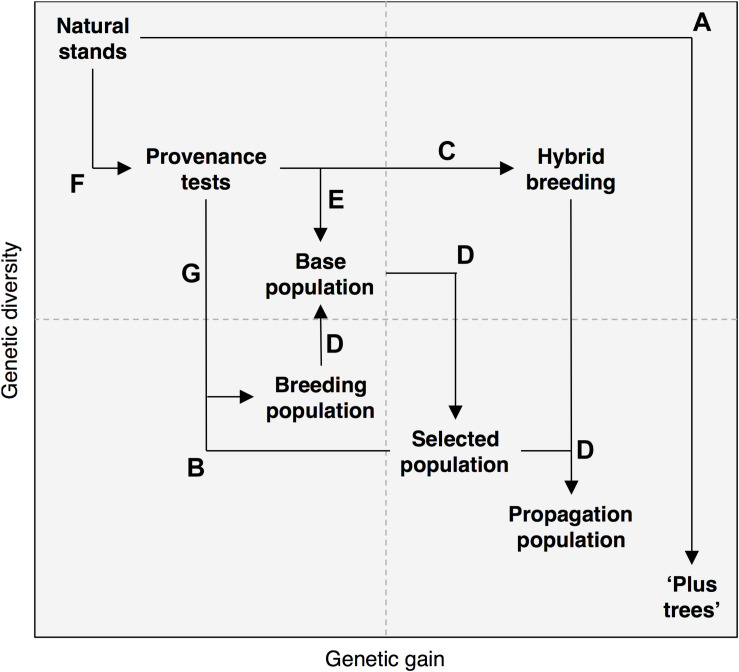
FIGURE 1.
Trans-disciplinary approaches (arrows) such as predictive breeding (GP) and machine learning (ML) promise supporting genome-wide marker-assisted (MAS) pre-breeding and breeding strategies for the selection of (A) “plus trees” in the wild, key (B) intra- and (C) inter- specific parental combinations, and (D) elite offspring from those parents. GP and ML should go beyond breeding and feedback (E) germplasm utilization and environmental niche classification (Cortés et al., 2013) and enviromics (Costa-Neto et al., 2020; Resende et al., 2020). Genomic-assisted characterizations, such as Genome-Wide Association Studies—GWAS (Neale and Savolainen, 2004), Genome–Environment Associations—GEA (Rellstab et al., 2015; Cortés and Blair, 2018; López-Hernández and Cortés, 2019) and Genome-Wide Selection Scans—GWSS (Zahn and Purnell, 2016), must also start considering more thoroughly (F) novel sources of local adaptation, (G) genetic-guided infusions and assisted gene flow (AGF), as well an overall systems genetics thinking (Ingvarsson et al., 2016; Myburg et al., 2019).
TABLE 1.
Predictive breeding (genomic prediction—GP, also known as genomic selection—GS) studies in forest tree species published during the last years.
| Species | Populations | Trait data | Genotyping data | GP algorithm | Key conclusions | References |
| Elaeis guineensis | 162 individuals from the Deli and Group B populations | Seven oil yield components | 262 SSRs | PBLUP, GBLUP | Genomic selection (GBLUP) calibrated according to conditions of the experiment showed higher trait precision when using pedigree-based model | Cros et al., 2015 |
| Elaeis guineensis | A × B hybrid progeny tests with almost 500 crosses for training and 200 crosses for independent validation | Seven oil yield components | (>5,000 GBS-derived SNPs | GBLUP, PBLUP | Preselection for yield components using GBS is the first possible application of GS in oil palm. | Cros et al., 2017 |
| Hevea brasilensis | 332 clones from the F1 cross PB 260 × RRIM 600 | Rubber production | 332 SSRs on site 1 and 296 SSRs on site 2 | RKHS, BLR_A, RR-BLUP-A, BLR_AD, RR-BLUP_AD | Mean between-site GS accuracy reached 0.561 when using the 125–200 SSRs with the highest Ho. The simulations showed that by applying a genomic preselection among 3,000 seedlings in the nursery there is a greater precision of selection of the genomic preselection compared to the phenotypic preselection. Statistical method had no effect on GS precision | Cros et al., 2019 |
| Eucalyptus grandis × E. urophylla hybrids | 999 individuals from 45 families | Cellulose content, composition of lignin monomer, total lignin, WD | 33,398 SNP | ABLUP, GBLUP, ssGBLUP | ssGBLUP is a tool with a great projection for the improvement of the precision and the bias of the classic GBLUP for the genomic evaluation in the improvement of Eucalyptus | Cappa et al., 2019 |
| Picea abies | 1,370 controlled-pollinated individuals from 46 unrelated parents | Quality features of solid wood, pilodyn penetration, acoustic speed | 116,765 SNP | ABLUP-A, ABLUP-AD, GBLUP-AD, GBLUD-ADE | GBLUP-AD is a model with great utility in production and propagation. Tree breeders can use it for seedling selection, or family and full-siblings selection | Chen et al., 2019 |
| Eucalyptus globulus | 646 individuals out of approximately 10 individuals per family | WD, branch quality, DBH, HT | 14,442 SNP | BRR, Bayes C, HAP, HAP-SNP | In general, the BRR and Bayes C methods had a higher predictive capacity for most of the traits. In particular, genomic models that included the haplotype effect (either HAP or HAP SNP) significantly increased the AP of traits with low heritability. | Ballesta et al., 2019 |
| Eucalyptus cladocalyx | 1,470 individuals from 49 families | DBH, HT, BHT, WD, STR, SLD, FI | 3.8 K Illumina Infinium EUChip60K SNPs | Bayes A, Bayes B, Bayes C, BRR | An GSq approach outperformed GS models in terms of predictive ability when the proportion of the variance explained by the significant marker-trait associations was higher than those explained by the polygenic background and non-significant markers | Ballesta et al., 2020 |
| Eucalyptus clones of E. urophylla× E. grandis | 1,130 clones of 69 full- sib families | Biomass production, WUE, wood properties | 3,303 SNPs | GBLUP | The inclusion of wood δ13C in the selection process may lead to Eucalyptus varieties adapted to marginal zones still presenting good performance for biomass and wood chemical traits | Bouvet et al., 2020 |
| Picea abies | 726 trees of 40 families of complete siblings from two localities | Density, microfiber angle, wood stiffness | 5,660 Infinium iSelect SNP matrix SNPs from exome capture and sequencing | Single-trait: GBLUP, BRR, GBLUP, TGBLUP, ABLUP. Multi-traits: GBLUP | Genomic prediction models showed similar results, but the multi-trait model stood out when weevil attacks were not available. Most of the results indicate that the weevil resistance genotypes were higher when there was a greater proportion of height to diameter and greater rigidity of the wood. | Lenz et al., 2020 |
| Pinus radiata | 457 POP2 descendants of 63 parents, and 524 POP3 descendants of 24 parents | Branching frequency, stem straightness, internal verification, and external bleeding | 1,371,123 exome sequencing capture SNPs | GBLUP, ABLUP | An efficient way to improve non-key traits is through genomic selection with a pedigree corrected using SNP information | Li et al., 2019 |
| Pseudotsuga menziesii | 13,615 individuals | HT, 13 environmental variables | 66,969 SNPs | ssGBLUP | GS-PA can be substantially improved using ECs to explain environmental heterogeneity and G × E effects. The ssGBLUP methodology allows historical genetic trials containing non-genotyped samples to contribute in genomic prediction, and, thus, effectively boosting training population size which is a critical step | Ratcliffe et al., 2019 |
| Shorea platyclados | 356 individuals from a half-sib progeny population | Seven important traits, including growth, branching quality, wood quality traits | 5,900 Illumina Hi-Seq X SNPs | rrBLUP | Selective breeding for these traits individually could be very effective, especially for increasing the diameter growth, branch diameter ratio and wood density simultaneously | Sawitri et al., 2020 |
| Hevea brasiliensis | 435 individual rubber trees at two sites. 252 F1 hybrids derived from a PR255 × PB217 cross, 146 F1 hybrids derived from a GT1 × RRIM701 cross, 37 genotypes from a GT1 × PB235 cross, and 4 testers (GT1, PB235, RRIM701, and RRIM600) | SC | 30,546 GBS-derived SNPs | BLUP, SM, MM, MDs, Mde | Multi-environment models were superior to the single-environment genomic models. Methods in which GS is incorporated resulted in a fivefold increase in response to selection for SC with multi-environment GS (MM, MDe, or MDs) | Souza et al., 2019 |
| Fraxinus excelsior | 1,250 individuals | Tree health, ash dieback resistance | 100–50,000 HiSeq X SNPs | RR-BLUP | Ash dieback resistance in F. excelsior is a polygenic trait that should respond well to both natural selection and breeding, which could be accelerated using genomic prediction | Stocks et al., 2019 |
| Eucalyptus nitens | 691 individuals | Solid wood production, height, DBH, stem straightness, WD, wood stiffness, wood shrinkage, growth strain | 12,236 Illumina EUChip60K SNPs | BLUP, GBLUP | The greatest improvement in genetic parameters was obtained for tangential air-dry wood shrinkage and growth strain | Suontama et al., 2019 |
| Pseudotsuga menziesii | A 38-year-old progeny test population (P1), selecting 37 of 165 families with complete siblings at random from 3 different settings. Validation population contained 247 descendants with controlled crosses from the 37 families | HT | Complete genotyping of exome capture | RR-BLUP, GRR, Byes-B | The validation of cross genomic selection of juvenile height in Douglas fir gave very similar results with the ABLUP predictive precision, but this precision may be linked to the relationship between training and validation conjugates | Thistlethwaite et al., 2019a |
| Pseudotsuga menziesii, Picea glauca, P. engelmannii | 1,321 Douglas-fir trees, representing 37 full-sib F1 families and 1,126 interior spruce trees, representing 25 open-pollinated (half-sib) families | Mid-rotation height, WD | 200–50,000 Illumina HiSeq 2000 SNPs | RR-BLUP | Reducing marker density cannot be recommended for carrying out GS in conifers. Significant LD between markers and putative causal variants was not detected using 50,000 SNPs | Thistlethwaite et al., 2020 |
| Pinus contorta | Half- and full- sibs represented by 57 base parents and 42 full-sib families with an calculated effective population size of 92 | Growth and wood quality | 51,213 Illumina HiSeq SNPs | Bayes C, Bayes B, BLUP, GBLUP, ABLUP | The predictions of Marker-based models had accuracies that were equal to or better than pedigree-based models (ABLUP) when using several cross-validation scenarios and were better at ranking trees within families | Ukrainetz and Mansfield, 2020 |
| Castanea dentate | 7,173 descendants of BC3F3 from 346 “Clapper” mothers and 198 “Serious” mothers. For the BC3F2 progeny, a total of 1,134 “Clapper” and 1,042 “Graves” were sampled | Cryphonectria parasitica fungus severity (BC3F3) or presence/absence data (BC3F2) | Sequencing of a C. dentata clone in the PacBio Sequel platform | HBLUP, ABLUP, Bayes C | By means of genomic prediction and estimation of hybrid indices, a trade-off is between resistance and a proportion of inherited genome. The results found show that the genetic architecture underlying the heritability of resistance to blight is complex | Westbrook et al., 2020 |
| Picea abies | 484 progeny trees from 62 half-sib families | WD, MOE, MFA | 130,269 Illumina HiSeq 2500 SNPs | ABLUP, GBLUP, rrBLUP, BayesB, RKHS | This study indicates standing tree-based measurements is a cost-effective alternative method for GS. Selection for density could be conducted at an earlier age than for MFA and MOE | Zhou et al. (2020) |
For a comprehensive summary of previous studies not included here see Grattapaglia et al. (2018). Detailed abbreviations are shown at the end of the table. WUE, water use efficiency; SC, stem circumference; WD, wood density; MOE, modulus of elasticity; MFA, microfibril angle; DBH, diameter at breast height; HT, total tree height; BHT, first bifurcation height; STR, stem straightness; SLD, slenderness index; FI, flowering intensity; SNP, single nucleotide polymorphism; SSR, simple sequence repeat; GBS, genotyping by sequencing.
Predictive Breeding Promises Boosting Forest Tree Genetic Improvement
The aim of forest tree breeding is rarely to develop new varieties, but instead advance gradual population improvement through recurrent selection and testing (Neale and Kremer, 2011). Because of the long generation times of forest trees, their breeding has traditionally relied on phenotypic selection from natural stands by choosing “plus-trees” (Figure 1A). Their superior phenotype (primarily productivity and tree architecture, and seldom adaptability) is often measured in situ or in provenance trials. This starting pool of preferred trees constitutes the base population, an arboretum from which further selection is carried out to build a selected population with elite seed/scion donors. Their estimated combinatory ability is gathered from genetic tests such as progeny trials, and parental re-selection (Figure 1B) from top families and single trees (White et al., 2007). After three steps of selection (from the natural, base, and selected populations), eroded genetic diversity may jeopardize overall population’s productivity and resilience due to inbreeding depression. In order to minimize this risk, a breeding population is established to increase genetic variability. Intermating may rely on infusions from external populations. Outbred multi-parental populations (Scott et al., 2020) hence become the base population of a second generation. A bottleneck of this approach is that each generation would last at least nine or 18 years, for seedling or elite clone identification, respectively, in a fast growing tree species such as Eucalyptus (Resende et al., 2012).
Shortcuts to speed up the traditional cycle of forest tree genetic improvement rely on hybrids and backcrossing. Hybrid breeding (Figure 1C) aims harnessing heterotic effects (hybrid vigor) due to dominance and over-dominance already existing in nature, capable of increasing yield and adaptability (Schilthuizen et al., 2004; Seehausen, 2004). Dominance refers to the masking of deleterious effects of recessive alleles as a consequence of the increased heterozygosity resulting from hybridization (i.e., an scape from inbreeding depression). On the other hand, over-dominance corresponds to the increase in aptitude as the result of the additive and epistatic effects of alleles that are naturally maintained by balancing selection and only coincide in hybrid genotypes. Hybrid breeding is nowadays widely used at operational plantations to maximize circumference at breast height (e.g., E. grandis× E. nitens and Pinus elliotti × P. oocarpa), height (e.g., P. caribaea × P. tecunumanii) and resistance to Fusarium spp. (i.e., P. patula × P. tecunumanii), among other potential uses (Burkhart et al., 2017). Backcrossing helps targeting the introgression of desired traits from exotic sources into elite populations, as has been done to transfer resistance to chestnut blight into American populations from Chinese wild donors (Cipollini et al., 2017).
Molecular breeding approaches (Badenes et al., 2016), in which genetic markers are used to assist selection, offer promising alternatives to speed up traditional tree breeding cycles, as well as hybrid and backcrossing schemes. Marker-Assisted Selection—MAS (Butcher and Southerton, 2007; Muranty et al., 2014) and Backcrossing—MAB (Herzog and Frisch, 2011) provide frameworks to pyramid target genetic variants of simple Mendelian traits, which are those regulated by few major genes (e.g., resistance to biotic stresses). Gene editing (Doudna and Charpentier, 2014; Dort et al., 2020) and transgenics (Campbell et al., 2003) can also transfer or silence allelic variants of major effects within a single generation (Pereira-Lorenzo et al., 2019). These may replicate the success of tolerant chestnuts (Alcaide et al., 2019a; Westbrook et al., 2019) and promote reproductive sterility (Meilan et al., 2001; Fritsche et al., 2018). Yet, molecular breeding via MAS, MAB and gene editing is often inefficient to trace quantitative traits as growth and adaptation to abiotic stresses. Adaptation is often polygenic (Cortés et al., 2018b; Barghi et al., 2020) due to many low-effect genes and their second-order interactions (Boyle et al., 2017).
A last-generation predictive breeding (Figure 1D) approach designed for quantitative polygenic traits is known as Genomic Prediction—GP (Desta and Ortiz, 2014; Crossa et al., 2017; Grattapaglia et al., 2018). GP standardizes infinitesimal marker-based additive predictive models by relying on historical phenotypic data (Meuwissen et al., 2001; Gianola et al., 2006; de los Campos et al., 2013). Trait data must be in Linkage Disequilibrium—LD or genetic auto-correlation (e.g., Kelleher et al., 2012), with the molecular markers or with the samples’ genetic co-ancestry. GP utility has been demonstrated (Table 1) in model forest tree species such as Eucalyptus (Resende et al., 2012; Suontama et al., 2019), and conifers as Pinus (Resende M. F. et al., 2012; Li et al., 2019) and Douglas-fir (Thistlethwaite et al., 2017, 2019b), but also in non-model perennial crops such as coffee (Sousa et al., 2018), rubber (Cros et al., 2019; Souza et al., 2019) and oil palm (Cros et al., 2015). GP may even fit epigenetics (Roudbar et al., 2020), as well as multi-trait genomic models as was recently confirmed in Norway spruce for growth, wood quality and weevil resistance traits (Lenz et al., 2020). GP could also be coupled with somatic embryo-genesis for clonal propagation of elite genotypes by selecting elite zygotic embryos based on their genomic breeding value (Grattapaglia et al., 2018). GP has the potential to predict untested hybrid genotypes (Technow et al., 2014) in woody perennials (Cros et al., 2017; Tan et al., 2017) by genotyping potential parental lines and phenotyping few F1 hybrids. Prioritizing inter-specific combinations for field trials can speed up hybrid breeding. Meanwhile, like already envision for chestnut (Westbrook et al., 2020), Genomic-Assisted Backcrossing (GABC) will replace MAB as the strategy to assist introgression breeding into elite populations from exotic germplasm.
Assisting Genomic Characterization of Tree Germplasm to Capture Novel Variants
Exploiting tree wild populations for genomics-assisted breeding (Figure 1E) is key to broaden the genetic basis of woody perennial breeding programs (Migicovsky and Myles, 2017). Specifically, diverse seed bank collections and novel tree provenances might source (Ulian et al., 2020) exotic variation (e.g., unique wood quality properties). They also help avoiding genetic erosion (e.g., via infusions) and increasing long-term adaptability to climate change (e.g., making forests more tolerant to abiotic stresses such as drought and heat). For example, genomic diversity analyses helped capturing rare variants in P. trichocarpa germplasm (Piot et al., 2019) often missed by Genome-Wide Association Studies (GWAS) in the related species P. tremula (Khan and Korban, 2012). Expanded phylogenomic (Wang M. et al., 2020) and species (Wang et al., 2020) diversity may source novel alleles to support selective breeding, as in wood quality traits for improved bioenergy feedstock. In turn, GP might go beyond breeding, the focus of the previous section, and feedback seed bank characterization (Hickey et al., 2017)—e.g., by predicting seed traits (Kehel et al., 2020) and overall yield (Crossa et al., 2007, 2016) in diverse accessions that otherwise could not have been tested at once in genetic field trials. Although the use of GP for germplasm characterization is latent, it has not been fully explored in forest tree species, a main research gap to be filled in the oncoming years.
Tree species rich in evolutionary diversity (Shang et al., 2020) could leverage breeding. Hybridization (Nieto Feliner et al., 2020), introgression (Burgarella et al., 2019), and polyploidy (Mason and Wendel, 2020) have already pumped morphological novelty by testing more genetic compatibilities than humans ever will. Yet, genomics of adaptive radiations (Seehausen, 2004; Madriñán et al., 2013; Cortés et al., 2018a; Marques et al., 2019) are challenging (Schilthuizen et al., 2004; de la Harpe et al., 2017). Long-living oaks—Quercus (Plomion et al., 2018; Leroy et al., 2020b; Plomion and Martin, 2020) are a classical syngameon (Cannon and Petit, 2020) – a promiscuous network of weakly isolated species that has driven peerless historical (Crowl et al., 2020; Hipp et al., 2020; Leroy et al., 2020c) and current (Leroy et al., 2020a) adaptive introgression (Kremer and Hipp, 2020).
In short, marker-assisted schemes are liable to be implemented at various stages during pre-breeding—e.g., in the selection of “plus trees” from the wild (De Dato et al., 2018), of target parental pairs (Blair et al., 2013), and of superior offspring (Galeano et al., 2012). These approaches also aid conservation (Martín et al., 2012; Mattioni et al., 2017) and germplasm tracing (Cortés et al., 2011; Blair et al., 2012; Chiocchini et al., 2016). Still, genomic-assisted studies of germplasm may risk focusing on productive traits and disregard locally adapted trait variation.
Genomics of Adaptation to Different Environments
Local genetic adaptation (Figure 1F) may prove useful in the reaction of forests to climate change (Savolainen et al., 2013; Lascoux et al., 2016), for instance via gene swamping of pre-adapted alleles (Kremer et al., 2014; de Visser et al., 2018). Nowadays there is a wide portfolio of genomic tools that appeal to environmental variables in order to infer the genetic basis of adaptation to abiotic stresses. Specifically, Genome-Wide Selection Scans—GWSS (Zahn and Purnell, 2016) and Genome–Environment Associations – GEA (Rellstab et al., 2015) aim detecting signatures of selection across environmental gradients by pinpointing sections in the genomes that correlate with habitat heterogeneity (Forester et al., 2016). These approaches have successfully been used to assess variation in bud-break phenology (McKown et al., 2018) and stomata patterning (McKown et al., 2014) as potential responses to climate warming in natural populations of P. trichocarpa. They have also allow comparing the likelihoods of adaptive reactions at continental (Holliday et al., 2011; Evans et al., 2014; Zhou et al., 2014; Stölting et al., 2015) and regional scales (Eckert et al., 2010; Holliday et al., 2016; Pluess et al., 2016; Ingvarsson and Bernhardsson, 2020) across phylogenetically diverse taxa (Yeaman et al., 2016). Currently there are even multi-scale approaches to detect widespread divergent selection in non-model tree species experiencing population decline (Mayol et al., 2020).
Local adaptation to climate change can be further enhanced (Figure 1G) via assisted gene flow—AGF (Aitken and Whitlock, 2013). AGF aims minimizing endogenous negative, while maximizing exogenous positive, selection by trans-locating pre-adapted individuals to facilitate adaptation of planted forests to climate change (Aitken and Bemmels, 2016). Management of local adaptation in a changing climate was recently examined in populations from lodgepole pine (P. contorta) across western Canada (Mahony et al., 2020). Yet, operational uses of genomic data to guide seed transfer or AGF are still lacking. Alternatively, genetic containment may be desired for transgenic trees (Brunner et al., 2007b; Klocko et al., 2016). The utility of these approaches in tropical forests remains to be explored. Tropical trees are more at risk from warming because they are closer to upper thermal limits (Freeman et al., 2020; Sentinella et al., 2020), as in montane (Cortés and Wheeler, 2018; Feeley et al., 2020; Tito et al., 2020) and alpine (Wheeler et al., 2014, 2016; Valencia et al., 2020) habitats. Disclosing the genetic, pan-genomic (Bayer et al., 2020), and epigenetic (Brautigam et al., 2013; Sow et al., 2018; Barrera-Redondo et al., 2020) bases of traits underlying adaptive responses in tree species will assist AGF, industrial milestones, and conservation priorities (Isabel et al., 2020) across meta-populations (Gonzalez et al., 2020), and even micro-habitats (Cortés et al., 2014; Abdelaziz et al., 2020).
Concluding Remarks
A major question in the interface between forests and their environments that genomics have the potential to assist is whether tree adaptation to the fast pace of climate change can happen despite their long generation times (Holliday et al., 2017). Specifically, GP offers a feasible way to predict adaptation from allele frequencies in many genes of low effects underlying polygenic traits (Isabel et al., 2020). This way, the role of adaptive responses can be balanced in relation with range shifts (i.e., migration) and extinction as possible climate change outcomes for tree populations (Aitken et al., 2008; Alberto et al., 2013). This question is equally insightful for domesticated and wild stands of forest trees, and must be coupled with reflections regarding the best propagation and conservation schemes. For instance, the factual consequences on genetic diversity of clonal and seedling forestry (Ingvarsson and Dahlberg, 2018), and of assisted gene flow (Aitken and Whitlock, 2013; Aitken and Bemmels, 2016), must be compiled.
Forest genomics tends focusing on economically important species. Yet, the power of population genomics must be further extended to comprehend neutral and adaptive processes in non-commercial species of ecological value in order to advance not just productivity, but also climate adaptation, forest health and conservation (Isabel et al., 2020). In this sense, GP is starting to permeate novel non-key traits other than growth and wood density, but still of interest for breeding, such as branching, stem straightness and external resin bleeding (Li et al., 2019). GP is also predicting adaptive trait variation for abiotic (Eckert et al., 2010) and biotic (Westbrook et al., 2020) stresses. In parallel to an enrichment of target traits, emerging genomic technologies might unlock woody plant trait diversity beyond the model tree species poplar, eucalyptus, willow, oak, chestnut and pecan (Tuskan et al., 2018).
There is currently a rich mosaic of alternative genetic methods to carry out both explicit (direct) and implied (indirect) selection on economic- (Burdon and KlápšTě, 2019) and ecological-worth (Holliday et al., 2017; Isabel et al., 2020) functions. These different traits can enlighten our understanding of the consequences of genetic divergence on the reaction of tree populations to climate change (Kremer et al., 2014). However, novel methodological developments should target more comprehensively complex trait–environment relationships (Bruelheide et al., 2018). They should also mingle between adaptive (Cortés et al., 2015b; Sedlacek et al., 2016) and range shift (Sedlacek et al., 2014; Wheeler et al., 2015) responses across altitudinal (Lenoir et al., 2008; Steinbauer et al., 2018), latitudinal (Chen et al., 2011) and micro-habitat (Sedlacek et al., 2015; Little et al., 2016) gradients.
Perspectives
Exploring natural adaptation to changing climate and genetic breeding for tolerance to abiotic stress in forest tree species has traditionally been assisted by GWAS, GWSS, GEA (Cortés et al., 2020), and AGF techniques. These approaches have allowed identifying and utilizing naturally available, locally adapted, variants. More recently, major developments in the field of predictive breeding (i.e., GP) promise to speed up selection from natural sources, as well as within the breeding cycle, by shortening the generation intervals and increasing the selection accuracy prior field trials. We have already identified and discussed major improvements in this line, such as multi-trait GP models (Lenz et al., 2020), coupled with integrative selection scores (Burdon and KlápšTě, 2019) on novel non-key (Li et al., 2019) and ecological-worth (Holliday et al., 2017; Isabel et al., 2020) traits. These innovations can capture multi-scale trait–environment relationships (Bruelheide et al., 2018) in non-model tree species (Mayol et al., 2020). Given the complexity and heterogeneity of trans-disciplinary data sources, Machine Learning (ML) offers a timely predictive and synthetizing approach capable of merging the highlights of the GWAS, GWSS, GEA, AGF and GP techniques.
“Supervised” ML typically utilizes “labeled” training datasets in order to cross-validate the “recall” rate of a target classification (e.g., selection). ML powerfully handles high-dimensional inputs of heterogeneous “features” without a joint probability distribution (Schrider and Kern, 2018). This way, algorithmically generated non-parametric models that avoid rejection sampling sidestep the “curse of dimensionality” and offer new ways to reveal complex systems (Myburg et al., 2019). ML has historically been utilized in functional genomics (Libbrecht and Noble, 2015) and ecological niche modeling (Phillips et al., 2017). Yet, it is now transitioning into GWAS-coupled MAS (Cortés et al., 2015a), GP (Crossa et al., 2019; Abdollahiarpanahi et al., 2020), GWSS (Schrider and Kern, 2018), and demographics—as when coupled with Approximate Bayesian Computation (Elleouet and Aitken, 2018; Liu et al., 2019).
We anticipate that ML techniques will brace GP predictions for various traits in multi-environment trials that aim disentangling the additive genetic variance and the genotype × environment components. Novel developments in the field of ML will further allow building more accurate predictions by merging environmental variables, microhabitat diversity, and genome-wide divergence, all within a tree-breeding context to pivot “plus tree” selection, hybrid breeding and GABC schemes, as well as in terms of adaptation to climate change in natural forests. Integrative assessments (Ingvarsson et al., 2016) via ML promise harnessing adaptive trait variation in forest tree species.
Author Contributions
AC conceived this review. MR-M and LB-C collected literature and prepared summary tables. AC wrote the first draft of the review with later edits made by MR-M and LB-C.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This review homages C. Lexer R.I.P. (Fay and Palma-Silva, 2020; Karrenberg et al., 2020; Schloötterer, 2020) for his visionary contributions to the fields of forest tree genetics and population genomics, and for his exceptional enthusiasm while mentoring and welcoming pupils and colleagues at his affable research group. In particular, AC enormously appreciates his stimulating and supportive role as an inspiring doctoral co-advisor (2011–2015), and gratefully remembers his hospitality in Fribourg (Switzerland) during 2011–2013 through countless discussions, refreshing Fondue mealtimes, hikes and Pétanque contests. C. Lexer is also thanked for making possible exciting joint field trips with S. Humbert and Y. Naciri to Col du Sanetsch, Lac de Moiry and Lac des Autannes (Valais, Switzerland) in August 2011, and with S. Humbert and A. Tribsch to Hohe Tauern and Niedere Tauern (Austria) in July 2013, as well as for encouraging thought-provoking scientific discussions during the Swiss National Science Foundation (SNSF) Sinergia Salix Kickoff Meeting held in April 2011 at Davos (Graubünden, Switzerland), the European Society for Evolutionary Biology Congress held in August 2011 at Tubingen (Germany), the Swiss National Science Foundation (SNSF) Sinergia Salix Closure Meeting held in February 2013 at Fribourg (Switzerland), and the European Molecular Biology Organization (EMBO) workshop on Mechanisms of Plant Speciation (Lafon-Placette et al., 2016) held in June 2015 at Åkersberga (Sweden). Special recognition is additionally granted to S. Arenas, J.P. Jaramillo-Correa, F. López-Hernández and M.J. Torres-Urrego for debates while writing this review. Topic editors and reviewers are acknowledged for conceiving and pushing through a timely Research Topic on “Forests and Their Interactions with the Environment”.
Footnotes
Funding. Supported to AC during the early phases of this work was made through the grants 4.1-2016-00418 and BS2017-0036 from Vetenskapsrådet (VR) and from Kungliga Vetenskapsakademien (KVA), respectively. The editorial fund from the Colombian Corporation for Agricultural Research (AGROSAVIA) waged the publication of this review.
References
- Abdelaziz M., Anderson J. T., Rochford M. E., Bemmels J. B., Jameel M. I., Denney D. A. (2020). Small spaces, big impacts: contributions of micro-environmental variation to population persistence under climate change. AoB PLANTS 12:plaa005. 10.1093/aobpla/plaa005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahiarpanahi R., Gianola D., Peñagaricano F. (2020). Deep learning versus parametric and ensemble methods for genomic prediction of complex phenotypes. Genet. Sel. Evol. 52:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken S. N., Bemmels J. B. (2016). Time to get moving: assisted gene flow of forest trees. Evol. Appl. 9 271–290. 10.1111/eva.12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken S. N., Whitlock M. C. (2013). Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 44 367–388. 10.1146/annurev-ecolsys-110512-135747 [DOI] [Google Scholar]
- Aitken S. N., Yeaman S., Holliday J. A., Wang T., Curtis-Mclane S. (2008). Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1 95–111. 10.1111/j.1752-4571.2007.00013.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberto F. J., Aitken S. N., Alia R., Gonzalez-Martinez S. C., Hanninen H., Kremer A., et al. (2013). Potential for evolutionary responses to climate change - evidence from tree populations. Glob. Chang. Biol. 19 1645–1661. 10.1111/gcb.12181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide F., Solla A., Cherubini M., Mattioni C., Cuenca B., Camisón Á., et al. (2019a). Adaptive evolution of chestnut forests to the impact of ink disease in Spain. J. Syst. Evol. 58 504–516. 10.1111/jse.12551 [DOI] [Google Scholar]
- Alcaide F., Solla A., Mattioni C., Castellana S., Martín M. A. (2019b). Adaptive Diversity and drought tolerance in Castanea Sativa assessed through genic markers Est-Ssr. Forestry 92 287–296. 10.1093/forestry/cpz007 [DOI] [Google Scholar]
- Badenes M. L., Fernandez I. M. A., Rios G., Rubio-Cabetas M. J. (2016). Application of genomic technologies to the breeding of trees. Front. Genet. 7:198. 10.3389/fgene.2016.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesta P., Bush D., Silva F. F., Mora F. (2020). Genomic predictions using low-density Snp markers, Pedigree and Gwas information: a case study with the non-model species Eucalyptus Cladocalyx. Plants 9:99. 10.3390/plants9010099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesta P., Maldonado C., Perez-Rodriguez P., Mora F. (2019). Snp and Haplotype-based genomic selection of quantitative traits in Eucalyptus Globulus. Plants 8:331. 10.3390/plants8090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barghi N., Hermisson J., SchloöTterer C. (2020). Polygenic adaptation: a unifying framework to understand positive selection. Nat. Rev. Genet. 10.1038/s41576-020-0276-2 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Barrera-Redondo J., Pinero D., Eguiarte L. E. (2020). Genomic, transcriptomic and epigenomic tools to study the domestication of plants and animals: a field guide for beginners. Front. Genet. 11:742. 10.3389/fgene.2020.00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer P. E., Golicz A. A., Scheben A., Batley J., Edwards D. (2020). Plant pan-genomes are the new reference. Nat. Plants 6 914–920. 10.1038/s41477-020-0733-0 [DOI] [PubMed] [Google Scholar]
- Berlin S., Hallingbäck H. R., Beyer F., Nordh N. E., Weih M., Rönnberg-Wästljung A. C. (2017). Genetics of phenotypic plasticity and biomass traits in hybrid willows across contrasting environments and years. Ann. Bot. 120 87–100. 10.1093/aob/mcx029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair M. W., Cortés A. J., Penmetsa R. V., Farmer A., Carrasquilla-Garcia N., Cook D. R. (2013). A high-throughput Snp marker system for parental polymorphism screening, and diversity analysis in common bean (Phaseolus Vulgaris L.). Theor. Appl. Genet. 126 535–548. 10.1007/s00122-012-1999-z [DOI] [PubMed] [Google Scholar]
- Blair M. W., Soler A., Cortés A. J. (2012). Diversification and Population Structure in Common Beans (Phaseolus vulgaris L.). PLoS One 7:e49488. 10.1371/journal.pone.0049488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet J.-M., Makouanzi Ekomono C. G., Brendel O., Laclau J.-P., Bouillet J.-P., Epron D. (2020). Selecting for water use efficiency, wood chemical traits and biomass with genomic selection in a Eucalyptus breeding program. For. Ecol. Manage. 465:118092 10.1016/j.foreco.2020.118092 [DOI] [Google Scholar]
- Boyle E. A., Li Y. I., Pritchard J. K. (2017). An expanded view of complex traits: from polygenic to omnigenic. Cell 169 1177–1186. 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam K., Vining K. J., Lafon-Placette C., Fossdal C. G., Mirouze M., Marcos J. G., et al. (2013). Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol. Evol. 3 399–415. 10.1002/ece3.461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruelheide H., Dengler J., Purschke O., Lenoir J., Jiménez-Alfaro B., Hennekens S. M., et al. (2018). Global trait–environment relationships of plant communities. Nat. Ecol. Evol. 2 1906–1917. [DOI] [PubMed] [Google Scholar]
- Brunner A. M., Difazio S. P., Groover A. T. (2007a). Forest genomics grows up and branches out. New Phytol. 174 707–710. [DOI] [PubMed] [Google Scholar]
- Brunner A. M., Li J., Difazio S. P., Shevchenko O., Montgomery B. E., Mohamed R., et al. (2007b). Genetic containment of forest plantations. Tree Genet. Genomes 3 75–100. 10.1007/s11295-006-0067-8 [DOI] [Google Scholar]
- Burdon R. D., KlápšTě J. (2019). Alternative selection methods and explicit or implied economic-worth functions for different traits in tree breeding. Tree Genet. Genomes 15:79. [Google Scholar]
- Burgarella C., Barnaud A., Kane N. A., Jankowski F., Scarcelli N., Billot C., et al. (2019). Adaptive introgression: an untapped evolutionary mechanism for crop adaptation. Front. Plant Sci. 10:4. 10.3389/fpls.2019.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart H. E., Brunner A. M., Stanton B. J., Shuren R. A., Amateis R. L., Creighton J. L. (2017). An assessment of potential of hybrid poplar for planting in the Virginia Piedmont. New Forests 48 479–490. 10.1007/s11056-017-9576-6 [DOI] [Google Scholar]
- Butcher P., Southerton S. (2007). “Marker-Assisted Selection in Forestry Species,” in Marker-Assisted Selection – Current Status and Future Perspectives in Crops, Livestock, Forestry and Fish, eds Guimarães E., Ruane J., Scherf B., Sonnino A., Dargie J. (Rome: FAO; ). [Google Scholar]
- Campbell M. M., Brunner A. M., Jones H. M., Strauss S. H. (2003). Forestry’s fertile crescent: the application of biotechnology to forest trees. Plant Biotechnol. J. 1 141–154. 10.1046/j.1467-7652.2003.00020.x [DOI] [PubMed] [Google Scholar]
- Cannon C. H., Petit R. J. (2020). The oak syngameon: more than the sum of its parts. New Phytol. 226 978–983. 10.1111/nph.16091 [DOI] [PubMed] [Google Scholar]
- Cappa E. P., De Lima B. M., Da Silva-Junior O. B., Garcia C. C., Mansfield S. D., Grattapaglia D. (2019). Improving genomic prediction of growth and wood traits in Eucalyptus using phenotypes from non-genotyped trees by single-step Gblup. Plant Sci. 284 9–15. 10.1016/j.plantsci.2019.03.017 [DOI] [PubMed] [Google Scholar]
- Carlson C. H., Gouker F. E., Serapiglia M. J., Tang H., Krishnakumar V., Town C. D., et al. (2014). “Annotation of the Salix purpurea L. genome and gene families important for biomass production,” in Proceedings of the Plant and Animal Genetics Conference XXII, San Diego, CA. [Google Scholar]
- Chakhchar A., Haworth M., El Modafar C., Lauteri M., Mattioni C., Wahbi S., et al. (2017). An assessment of genetic diversity and drought tolerance in Argan tree (Argania Spinosa) populations: potential for the development of improved drought tolerance. Front. Plant Sci. 8:276. 10.3389/fpls.2017.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. C., Hill J. K., Ohlemuller R., Roy D. B., Thomas C. D. (2011). Rapid range shifts of species associated with high levels of climate warming. Science 333 1024–1026. 10.1126/science.1206432 [DOI] [PubMed] [Google Scholar]
- Chen Z. Q., Baison J., Pan J., Westin J., Gil M. R. G., Wu H. X. (2019). Increased prediction ability in Norway Spruce Trials marker X environment interaction and non-additive genomic selection model. J. Hered. 110 830–843. 10.1093/jhered/esz061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchini F., Mattioni C., Pollegioni P., Lusini I., Martín M. A., Cherubini M., et al. (2016). Mapping the genetic diversity of Castanea Sativa: exploiting spatial analysis for biogeography and conservation studies. J. Geogr. Information Syst. 08 248–259. 10.4236/jgis.2016.82022 [DOI] [Google Scholar]
- Christie N., Tobias P. A., Naidoo S., Kulheim C. (2015). The Eucalyptus Grandis Nbs-Lrr gene family: physical clustering and expression hotspots. Front. Plant Sci. 6:1238. 10.3389/fpls.2015.01238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollini M., Dingley N. R., Felch P., Maddox C. (2017). Evaluation of phenotypic traits and blight-resistance in an American chestnut backcross orchard in Georgia. Glob. Ecol. Conserv. 10 1–8. 10.1016/j.gecco.2017.01.004 [DOI] [Google Scholar]
- Cortés A. J., Blair M. W. (2018). Genotyping by sequencing and genome – environment associations in wild common bean predict widespread divergent adaptation to drought. Front. Plant Sci. 9:128. 10.3389/fpls.2018.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Chavarro M. C., Blair M. W. (2011). Snp marker diversity in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 123 827–845. 10.1007/s00122-011-1630-8 [DOI] [PubMed] [Google Scholar]
- Cortés A. J., Garzón L. N., Valencia J. B., Madriñán S. (2018a). On the causes of rapid diversification in the Páramos: isolation by ecology and genomic divergence in Espeletia. Front. Plant Sci. 9:1700. 10.3389/fpls.2018.01700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Liu X., Sedlacek J., Wheeler J. A., Lexer C., Karrenberg S. (2015a). Maintenance of Female-Bias in a Polygenic Sex Determination System is Consistent with Genomic Conflict. On the Big Challenges of a Small Shrub: Ecological Genetics of Salix Herbacea L. (Uppsala: Acta Universitatis Upsaliensis; ). [Google Scholar]
- Cortés A. J., Monserrate F., Ramírez-Villegas J., Madriñán S., Blair M. W. (2013). Drought tolerance in wild plant populations: the case of common beans (Phaseolus vulgaris L.). PLoS One 8:e62898. 10.1371/journal.pone.0062898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Skeen P., Blair M. W., Chacón-Sánchez M. I. (2018b). Does the genomic landscape of species divergence in phaseolus beans coerce parallel signatures of adaptation and domestication? Front. Plant Sci. 9:1816. 10.3389/fpls.2018.01816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Waeber S., Lexer C., Sedlacek J., Wheeler J. A., Van Kleunen M., et al. (2014). Small-scale patterns in snowmelt timing affect gene flow and the distribution of genetic diversity in the alpine dwarf shrub Salix Herbacea. Heredity 113 233–239. 10.1038/hdy.2014.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortés A. J., Wheeler J. A. (2018). “The environmental heterogeneity of mountains at a fine scale in a changing world,” in Mountains, Climate, and Biodiversity, eds Hoorn C., Perrigo A., Antonelli A. (New York, NY: Wiley; ). [Google Scholar]
- Cortés A. J., Wheeler J. A., Sedlacek J., Lexer C., Karrenberg S. (2015b). Genome-Wide Patterns of Microhabitat-Driven Divergence in the Alpine Dwarf Shrub Salix Herbacea L. On the Big Challenges of a Small Shrub: Ecological Genetics of Salix Herbacea L. (Uppsala: Acta Universitatis Upsaliensis; ). [Google Scholar]
- Cortés A. J., López-Hernández F., Osorio-Rodriguez D. (2020). Predicting thermal adaptation by looking into populations’ genomic past. Front. Genet. 11:564515 10.3389/fgene.2020.564515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Neto G., Fritsche-Neto R., Crossa J. (2020). Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials. Heredity 10.1038/s41437-020-00353-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros D., Bocs S., Riou V., Ortega-Abboud E., Tisné S., Argout X., et al. (2017). Genomic preselection with genotyping-by- sequencing increases performance of commercial oil palm hybrid crosses. BMC Genomics 18:839. 10.1186/s12864-017-4179-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros D., Denis M., SaìNchez L., Cochard B., Flori A., Durand-Gasselin T., et al. (2015). Genomic selection prediction accuracy in a perennial crop: case study of oil palm (Elaeis Guineensis Jacq.). Theor. Appl. Genet. 128 397–410. 10.1007/s00122-014-2439-z [DOI] [PubMed] [Google Scholar]
- Cros D., Mbo-Nkoulou L., Bell J. M., Oum J., Masson A., Soumahoro M., et al. (2019). Within-family genomic selection in rubber tree (Hevea brasiliensis) increases genetic gain for rubber production. Ind. Crops Prod. 138:111464 10.1016/j.indcrop.2019.111464 [DOI] [Google Scholar]
- Crossa J., Burgueno J., Dreisigacker S., Vargas M., Herrera-Foessel S. A., Lillemo M., et al. (2007). Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177 1889–1913. 10.1534/genetics.107.078659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Jarquin D., Franco J., Perez-Rodriguez P., Burgueno J., Saint-Pierre C., et al. (2016). Genomic prediction of gene bank wheat landraces. G3 6 1819–1834. 10.1534/g3.116.029637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Martini J. W. R., Gianola D., Perez-Rodriguez P., Jarquin D., Juliana P., et al. (2019). Deep kernel and deep learning for genome-based prediction of single traits in multienvironment breeding trials. Front. Genet. 10:1168. 10.3389/fgene.2019.01168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Perez-Rodriguez P., Cuevas J., Montesinos-Lopez O., Jarquin D., De Los Campos G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22 961–975. [DOI] [PubMed] [Google Scholar]
- Crowl A. A., Manos P. S., Mcvay J. D., Lemmon A. R., Lemmon E. M., Hipp A. L. (2020). Uncovering the genomic signature of ancient introgression between white oak lineages (Quercus). New Phytol. 226 1158–1170. 10.1111/nph.15842 [DOI] [PubMed] [Google Scholar]
- De Dato G., Teani A., Mattioni C., Marchi M., Monteverdi M. C., Ducci F. (2018). Delineation of seed collection zones based on environmental and genetic characteristics for Quercus Suber L. in Sardinia, Italy. iForest 11 651–659. 10.3832/ifor2572-011 17959540 [DOI] [Google Scholar]
- de la Harpe M., Paris M., Karger D. N., Rolland J., Kessler M., Salamin N., et al. (2017). Molecular ecology studies of species radiations: current research gaps, opportunities and challenges. Mol. Ecol. 26 2608–2611. 10.1111/mec.14110 [DOI] [PubMed] [Google Scholar]
- de los Campos G., Hickey J. M., Pong-Wong R., Daetwyler H. D., Calus M. P. (2013). Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 193 327–345. 10.1534/genetics.112.143313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desta Z. A., Ortiz R. (2014). Genomic selection: genome-wide prediction in plant improvement. Trends Plant Sci. 19 592–601. 10.1016/j.tplants.2014.05.006 [DOI] [PubMed] [Google Scholar]
- de Visser J. A. G. M., Elena S. F., Fragata I. S., Matuszewski S. (2018). The utility of fitness landscapes and big data for predicting evolution. Heredity 121 401–405. 10.1038/s41437-018-0128-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dort E. N., Tanguay P., Hamelin R. C. (2020). Crispr/Cas9 gene editing: an unexplored frontier for forest pathology. Front. Plant Sci. 11:1126. 10.3389/fpls.2020.01126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). Genome editing. the new frontier of genome engineering with Crispr-Cas9. Science 346:1258096. [DOI] [PubMed] [Google Scholar]
- Eckert A. J., Van Heerwaarden J., Wegrzyn J. L., Nelson C. D., Ross-Ibarra J., Gonzalez-Martinez S. C., et al. (2010). Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus Taeda L., Pinaceae). Genetics 185 969–982. 10.1534/genetics.110.115543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elleouet J. S., Aitken S. N. (2018). Exploring approximate bayesian computation for inferring recent demographic history with genomic markers in nonmodel species. Mol. Ecol. Resour. 18 525–540. 10.1111/1755-0998.12758 [DOI] [PubMed] [Google Scholar]
- Evans L. M., Slavov G. T., Rodgers-Melnick E., Martin J., Ranjan P., Muchero W., et al. (2014). Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 46 1089–1096. 10.1038/ng.3075 [DOI] [PubMed] [Google Scholar]
- Fay M. F., Palma-Silva C. (2020). Professor Christian Lexer (23.05.1971-15.12.2019). Bot. J. Linn. Soc. 192 589–591. 10.1093/botlinnean/boaa006 [DOI] [Google Scholar]
- Feeley K., Martinez-Villa J., Perez T., Silva Duque A., Triviño Gonzalez D., Duque A. (2020). The thermal tolerances, distributions, and performances of tropical montane tree species. Front. For. Glob. Change 3:25 10.3389/ffgc.2020.00025 [DOI] [Google Scholar]
- Flanagan S. P., Forester B. R., Latch E. K., Aitken S. N., Hoban S. (2018). Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evol. Appl. 11 1035–1052. 10.1111/eva.12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forester B. R., Jones M. R., Joost S., Landguth E. L., Lasky J. R. (2016). Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol. Ecol. 25 104–120. 10.1111/mec.13476 [DOI] [PubMed] [Google Scholar]
- Freeman B. G., Song Y., Feeley K. J., Zhu K. (2020). Montane species and communities track recent warming more closely in the tropics. bioRxiv [Preprint]. 10.1101/2020.05.18.102848 [DOI] [Google Scholar]
- Fritsche S., Klocko A. L., Boron A., Brunner A. M., Thorlby G. (2018). Strategies for engineering reproductive sterility in plantation forests. Front. Plant Sci. 9:1671. 10.3389/fpls.2018.01671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeano C. H., Cortés A. J., Fernandez A. C., Soler A., Franco-Herrera N., Makunde G., et al. (2012). Gene-based single nucleotide polymorphism markers for genetic and association mapping in common bean. BMC Genet. 13:48. 10.1186/1471-2156-13-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianola D., Fernando R. L., Stella A. (2006). Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 173 1761–1776. 10.1534/genetics.105.049510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A., Germain R. M., Srivastava D. S., Filotas E., Dee L. E., Gravel D., et al. (2020). Scaling-up biodiversity-ecosystem functioning research. Ecol. Lett. 23 757–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattapaglia D., Silva-Junior O. B., Resende R. T., Cappa E. P., Muller B. S. F., Tan B., et al. (2018). Quantitative genetics and genomics converge to accelerate forest tree breeding. Front. Plant Sci. 9:1693. 10.3389/fpls.2018.01693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallingback H. R., Berlin S., Nordh N. E., Weih M., Ronnberg-Wastljung A. C. (2019). Genome wide associations of growth, phenology, and plasticity traits in willow [Salix Viminalis (L.)]. Front. Plant Sci. 10:753. 10.3389/fpls.2019.00753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog E., Frisch M. (2011). Selection strategies for marker-assisted backcrossing with high-throughput marker systems. Theor. Appl. Genet. 123 251–260. 10.1007/s00122-011-1581-0 [DOI] [PubMed] [Google Scholar]
- Hickey J. M., Chiurugwi T., Mackay I., Powell W. Implementing Genomic Selection in Cgiar Breeding Programs Workshop Participants (2017). Genomic prediction unifies animal and plant breeding programs to form platforms for biological discovery. Nat. Genet. 49 1297–1303. 10.1038/ng.3920 [DOI] [PubMed] [Google Scholar]
- Hipp A. L., Manos P. S., Hahn M., Avishai M., Bodenes C., Cavender-Bares J., et al. (2020). Genomic landscape of the global oak phylogeny. New Phytol. 226 1198–1212. 10.1111/nph.16162 [DOI] [PubMed] [Google Scholar]
- Holliday J. A., Aitken S. N., Cooke J. E., Fady B., González-Martínez S. C., Heuertz M., et al. (2017). Advances in ecological genomics in forest trees and applications to genetic resources conservation and breeding. Mol. Ecol. 26 706–717. 10.1111/mec.13963 [DOI] [PubMed] [Google Scholar]
- Holliday J. A., Suren H., Aitken S. N. (2011). Divergent selection and heterogeneous migration rates across the range of sitka spruce (Picea Sitchensis). Proc. Biol. Sci. 279 1675–1683. 10.1098/rspb.2011.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday J. A., Zhou L., Bawa R., Zhang M., Oubida R. W. (2016). Evidence for extensive parallelism but divergent genomic architecture of adaptation along altitudinal and latitudinal gradients in Populus trichocarpa. New Phytol. 209 1240–1251. 10.1111/nph.13643 [DOI] [PubMed] [Google Scholar]
- Howe G. T., Brunner A. M. (2005). An evolving approach to understanding plant adaptation. New Phytol. 167 1–5. 10.1111/j.1469-8137.2005.01469.x [DOI] [PubMed] [Google Scholar]
- Ingvarsson P. K., Bernhardsson C. (2020). Genome-wide signatures of environmental adaptation in European Aspen (Populus Tremula) under current and future climate conditions. Evol. Appl. 13 132–142. 10.1111/eva.12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvarsson P. K., Dahlberg H. (2018). The effects of clonal forestry on genetic diversity in wild and domesticated stands of forest trees. Scand. J. For. Res. 34 370–379. 10.1080/02827581.2018.1469665 [DOI] [Google Scholar]
- Ingvarsson P. K., Hvidsten T. R., Street N. R. (2016). Towards integration of population and comparative genomics in forest trees. New Phytol. 212 338–344. 10.1111/nph.14153 [DOI] [PubMed] [Google Scholar]
- Isabel N., Holliday J. A., Aitken S. N. (2020). Forest genomics: advancing climate adaptation, forest health, productivity, and conservation. Evol. Appl. 13 3–10. 10.1111/eva.12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrenberg S., Buerkle C. A., Field D. L., Savolainen V. (2020). Dedication: Christian Lexer (1971-2019). Philos. Trans. R. Soc. Lond. B Biol. Sci. 375:20200232. 10.1098/rstb.2020.0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehel Z., Sanchez-Garcia M., El Baouchi A., Aberkane H., Tsivelikas A., Charles C., et al. (2020). Predictive characterization for seed morphometric traits for genebank accessions using genomic selection. Front. Ecol. Evol. 8:32 10.3389/fevo.2020.00032 [DOI] [Google Scholar]
- Kelleher C. T., Wilkin J., Zhuang J., Cortés A. J., Quintero ÁL. P., Gallagher T. F., et al. (2012). Snp discovery, gene diversity, and linkage disequilibrium in wild populations of Populus tremuloides. Tree Genet. Genomes 821–829. 10.1007/s11295-012-0467-x [DOI] [Google Scholar]
- Khan M. A., Korban S. S. (2012). Association mapping in forest trees and fruit crops. J. Exp. Bot. 63 4045–4060. 10.1093/jxb/ers105 [DOI] [PubMed] [Google Scholar]
- Klocko A. L., Brunner A. M., Huang J., Meilan R., Lu H., Ma C., et al. (2016). Containment of transgenic trees by suppression of leafy. Nat. Biotechnol. 34 918–922. 10.1038/nbt.3636 [DOI] [PubMed] [Google Scholar]
- Kremer A., Hipp A. L. (2020). Oaks: an evolutionary success story. New Phytol. 226 987–1011. 10.1111/nph.16274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A., Potts B. M., Delzon S., Bailey J. (2014). Genetic divergence in forest trees: understanding the consequences of climate change. Funct. Ecol. 28 22–36. 10.1111/1365-2435.12169 [DOI] [Google Scholar]
- Lafon-Placette C., Vallejo-Marín M., Parisod C., Abbott R. J., Köhler C. (2016). Current plant speciation research: unravelling the processes and mechanisms behind the evolution of reproductive isolation barriers. New Phytol. 209 29–33. 10.1111/nph.13756 [DOI] [PubMed] [Google Scholar]
- Lascoux M., Glémin S., Savolainen O. (2016). Local adaptation in plants. Encycl. Life Sci. 0025270 1–7. 10.1002/9780470015902.a0025270 [DOI] [Google Scholar]
- Lenoir J., Gegout J. C., Marquet P. A., De Ruffray P., Brisse H. (2008). A significant upward shift in plant species optimum elevation during the 20th century. Science 320 1768–1771. 10.1126/science.1156831 [DOI] [PubMed] [Google Scholar]
- Lenz P. R. N., Nadeau S., Mottet M. J., Perron M., Isabel N., Beaulieu J., et al. (2020). Multi-trait genomic selection for weevil resistance, growth, and wood quality in Norway Spruce. Evol. Appl. 13 76–94. 10.1111/eva.12823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T., Louvet J. M., Lalanne C., Le Provost G., Labadie K., Aury J. M., et al. (2020a). Adaptive introgression as a driver of local adaptation to climate in European white oaks. New Phytol. 226 1171–1182. 10.1111/nph.16095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T., Plomion C., Kremer A. (2020b). Oak symbolism in the light of genomics. New Phytol. 226 1012–1017. 10.1111/nph.15987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy T., Rougemont Q., Dupouey J. L., Bodenes C., Lalanne C., Belser C., et al. (2020c). Massive postglacial gene flow between European white oaks uncovered genes underlying species barriers. New Phytol. 226 1183–1197. 10.1111/nph.16039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Klápště J., Telfer E., Wilcox P., Graham N., Macdonald L., et al. (2019). Genomic selection for non-key traits in radiata pine when the documented pedigree is corrected using DNA marker information. BMC Genomics 20:1026. 10.1186/s12864-019-6420-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht M. W., Noble W. S. (2015). Machine learning applications in genetics and genomics. Nat. Rev. Genet. 16 321–332. 10.1038/nrg3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind B. M., Menon M., Bolte C. E., Faske T. M., Eckert A. J. (2018). The genomics of local adaptation in trees: are we out of the woods yet? Tree Genet. Genomes 14:29. [Google Scholar]
- Little C. J., Wheeler J. A., Sedlacek J., Cortés A. J., Rixen C. (2016). Small-scale drivers: the importance of nutrient availability and snowmelt timing on performance of the alpine shrub Salix Herbacea. Oecologia 180 1015–1024. 10.1007/s00442-015-3394-3 [DOI] [PubMed] [Google Scholar]
- Liu S., Cornille A., Decroocq S., Tricon D., Chague A., Eyquard J. P., et al. (2019). The complex evolutionary history of apricots: species divergence, gene flow and multiple domestication events. Mol. Ecol. 28 5299–5314. 10.1111/mec.15296 [DOI] [PubMed] [Google Scholar]
- López-Hernández F., Cortés A. J. (2019). Last-generation genome–environment associations reveal the genetic basis of heat tolerance in common bean (Phaseolus vulgaris L.). Front. Genet. 10:22. 10.3389/fgene.2019.00954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madriñán S., Cortés A. J., Richardson J. E. (2013). Páramo is the world’s fastest evolving and coolest biodiversity hotspot. Front. Genet. 4:192. 10.3389/fgene.2013.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony C. R., Maclachlan I. R., Lind B. M., Yoder J. B., Wang T., Aitken S. N. (2020). Evaluating genomic data for management of local adaptation in a changing climate: a lodgepole pine case study. Evol. Appl. 13 116–131. 10.1111/eva.12871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques D. A., Meier J. I., Seehausen O. (2019). A combinatorial view on speciation and adaptive radiation. Trends Ecol. Evol. 34 531–544. 10.1016/j.tree.2019.02.008 [DOI] [PubMed] [Google Scholar]
- Martín M. A., Herrera M. A., MartiìN L. M. (2012). In situ conservation and landscape genetics in forest species. J. Nat. Resour. Dev. 2 1–5. [Google Scholar]
- Mason A. S., Wendel J. F. (2020). Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 11:1014. 10.3389/fgene.2020.01014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioni C., Martin M. A., Chiocchini F., Gaudet M., Pollegioni P., Velichkov I., et al. (2017). Landscape genetics structure of european sweet chestnut (Castanea Sativa Mill): indications for conservation priorities. Tree Genet. Genomes 13:39. [Google Scholar]
- Mayol M., Riba M., Cavers S., Grivet D., Vincenot L., Cattonaro F., et al. (2020). A multiscale approach to detect selection in nonmodel tree species: widespread adaptation despite population decline in Taxus baccata L. Evol. Appl. 13 143–160. 10.1111/eva.12838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown A. D., Guy R. D., Quamme L., Klapste J., La Mantia J., Constabel C. P., et al. (2014). Association genetics, geography and ecophysiology link stomatal patterning in Populus trichocarpa with carbon gain and disease resistance trade-offs. Mol. Ecol. 23 5771–5790. 10.1111/mec.12969 [DOI] [PubMed] [Google Scholar]
- McKown A. D., Klapste J., Guy R. D., El-Kassaby Y. A., Mansfield S. D. (2018). Ecological genomics of variation in bud-break phenology and mechanisms of response to climate warming in Populus trichocarpa. New Phytol. 220 300–316. 10.1111/nph.15273 [DOI] [PubMed] [Google Scholar]
- Meilan R., Brunner A. M., Skinnera J. S., Strauss S. H. (2001). Modification of flowering in transgenic trees. Prog. Biotechnol. 18 247–256. [Google Scholar]
- Meuwissen T. H. E., Hayes B. J., Goddard M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. E., Shuey L. S., Naidoo S., Mamni T., Berger D. K., Myburg A. A., et al. (2016). Dual Rna-sequencing of Eucalyptus nitens during Phytophthora cinnamomi challenge reveals pathogen and host factors influencing compatibility. Front. Plant Sci. 7:191. 10.3389/fpls.2016.00191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migicovsky Z., Myles S. (2017). Exploiting wild relatives for genomics-assisted breeding of perennial crops. Front. Plant Sci. 8:460. 10.3389/fpls.2017.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranty H., Jorge V., Bastien C., Lepoittevin C., Bouffier L., Sanchez L. (2014). Potential for marker-assisted selection for forest tree breeding: lessons from 20 years of mas in crops. Tree Genet. Genomes 10 1491–1510. 10.1007/s11295-014-0790-5 [DOI] [Google Scholar]
- Myburg A. A., Hussey S. G., Wang J. P., Street N. R., Mizrachi E. (2019). Systems and synthetic biology of forest trees: a bioengineering paradigm for Woody biomass feedstocks. Front. Plant Sci. 10:775. 10.3389/fpls.2019.00775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo S., Külheim C., Zwart L., Mangwanda R., Oates C. N., Visser E. A., et al. (2014). Uncovering the defence responses of Eucalyptus to pests and pathogens in the genomics age. Tree Physiol. 34 931–943. 10.1093/treephys/tpu075 [DOI] [PubMed] [Google Scholar]
- Naidoo S., Slippers B., Plett J. M., Coles D., Oates C. N. (2019). The road to resistance in forest trees. Front. Plant Sci. 10:273. 10.3389/fpls.2019.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale D. B., Kremer A. (2011). Forest tree genomics: growing resources and applications. Nat. Rev. Genet. 12 111–122. 10.1038/nrg2931 [DOI] [PubMed] [Google Scholar]
- Neale D. B., Savolainen O. (2004). Association genetics of complex traits in conifers. Trends Plant Sci. 9 325–330. 10.1016/j.tplants.2004.05.006 [DOI] [PubMed] [Google Scholar]
- Nieto Feliner G., Casacuberta J., Wendel J. F. (2020). Genomics of evolutionary novelty in hybrids and polyploids. Front. Genet. 11:792. 10.3389/fgene.2020.00792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B., Street N. R., Wetterbom A., Zuccolo A., Lin Y.-C., Scofield D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497 579–584. [DOI] [PubMed] [Google Scholar]
- Pennisi E. (2020). Tropical forests store carbon despite warming. Science 368:813. 10.1126/science.368.6493.813 [DOI] [PubMed] [Google Scholar]
- Pereira-Lorenzo S., Ramos-Cabrer A. M., Barreneche T., Mattioni C., Villani F., Díaz-Hernández B., et al. (2019). Instant domestication process of European chestnut cultivars. Ann. Appl. Biol. 174 74–85. 10.1111/aab.12474 [DOI] [Google Scholar]
- Phillips J., Ramirez S., Wayson C., Duque A. (2019). Differences in carbon stocks along an elevational gradient in tropical mountain forests of Colombia. Biotropica 51 490–499. 10.1111/btp.12675 [DOI] [Google Scholar]
- Phillips S. J., Anderson R. P., Dudík M., Schapire R. E., Blair M. E. (2017). Opening the black box: an open-source release of maxent. Ecography 40 887–893. 10.1111/ecog.03049 [DOI] [Google Scholar]
- Piot A., Prunier J., Isabel N., Klapste J., El-Kassaby Y. A., Villarreal Aguilar J. C., et al. (2019). Genomic diversity evaluation of Populus trichocarpa germplasm for rare variant genetic association studies. Front. Genet. 10:1384. 10.3389/fgene.2019.01384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomion C., Aury J. M., Amselem J., Leroy T., Murat F., Duplessis S., et al. (2018). Oak genome reveals facets of long lifespan. Nat. Plants 4 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomion C., Martin F. (2020). Oak genomics is proving its worth. New Phytol. 226 943–946. 10.1111/nph.16560 [DOI] [PubMed] [Google Scholar]
- Pluess A. R., Frank A., Heiri C., Lalague H., Vendramin G. G., Oddou-Muratorio S. (2016). Genome-environment association study suggests local adaptation to climate at the regional scale in Fagus sylvatica. New Phytol. 210 589–601. 10.1111/nph.13809 [DOI] [PubMed] [Google Scholar]
- Ratcliffe B., Thistlethwaite F., El-Dien O. G., Cappa E. P., Porth I., Klápštì J., et al. (2019). Inter- and intra-generation genomic predictions for Douglas-Fir growth in unobserved environments. bioRxiv [Preprint]. 10.1101/540765 [DOI] [Google Scholar]
- Rellstab C., Gugerli F., Eckert A. J., Hancock A. M., Holderegger R. (2015). A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 24 4348–4370. 10.1111/mec.13322 [DOI] [PubMed] [Google Scholar]
- Resende M. D. V., Resende M. F. R., Sansaloni C. P., Petroli C. D., Missiaggia A. A., Aguiar A. M. (2012). Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol. 194 116–128. 10.1111/j.1469-8137.2011.04038.x [DOI] [PubMed] [Google Scholar]
- Resende M. F., Muñoz P., Resende M. D., Garrick D. J., Fernando R. L., Davis J. M., et al. (2012). Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 190 1503–1510. 10.1534/genetics.111.137026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende R. T., Piepho H. P., Rosa G. J. M., Silva-Junior O. B., Silva F. F., Resende M. D. V., et al. (2020). Enviromics in breeding: applications and perspectives on envirotypic-assisted selection. Theor. Appl. Genet. 10.1007/s00122-020-03684-z [DOI] [PubMed] [Google Scholar]
- Roudbar M. A., Momen M., Mousavi S. F., Ardestani S. S., Lopes F. B., Gianola D., et al. (2020). Genome-wide methylation prediction of biological age using reproducing Kernel Hilbert spaces and Bayesian ridge regressions. bioRxiv [Preprint]. 10.1101/2020.08.25.266924 [DOI] [Google Scholar]
- Savolainen O., Lascoux M., Merilä J. (2013). Ecological genomics of local adaptation. Nat. Rev. Genet. 14 807–820. 10.1038/nrg3522 [DOI] [PubMed] [Google Scholar]
- Sawitri S., Tani N., Na’iem M., Widiyatno, Indrioko S., Uchiyama K., et al. (2020). Potential of genome-wide association studies and genomic selection to improve productivity and quality of commercial timber species in tropical rainforest, a case study of Shorea platyclados. Forests 11:239 10.3390/f11020239 [DOI] [Google Scholar]
- Scherer L., Svenning J. C., Huang J., Seymour C. L., Sandel B., Mueller N., et al. (2020). Global priorities of environmental issues to combat food insecurity and biodiversity loss. Sci. Total Environ. 730:139096. 10.1016/j.scitotenv.2020.139096 [DOI] [PubMed] [Google Scholar]
- Schilthuizen M., Hoekstra R. F., Gittenberger E. (2004). Hybridization, rare alleles and adaptive radiation. Trends Ecol. Evol. 19 404–405. 10.1016/j.tree.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Schloötterer C. (2020). Christian Lexer: a lifelong passion for trees. Mol. Ecol. 29 443–444. 10.1111/mec.15363 [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Kern A. D. (2018). Supervised machine learning for population genetics: a new paradigm. Trends Genet. 34 301–312. 10.1016/j.tig.2017.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. F., Ladejobi O., Amer S., Bentley A. R., Biernaskie J., Boden S. A., et al. (2020). Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding. Heredity. 10.1038/s41437-020-0336-6 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlacek J., Bossdorf O., Cortés A. J., Wheeler J. A., Van-Kleunen M. (2014). What role do plant-soil interactions play in the habitat suitability and potential range expansion of the alpine dwarf shrub Salix herbacea? Basic Appl. Ecol. 15 305–315. 10.1016/j.baae.2014.05.006 [DOI] [Google Scholar]
- Sedlacek J., Cortés A. J., Wheeler J. A., Bossdorf O., Hoch G., Klapste J., et al. (2016). Evolutionary potential in the alpine: trait heritabilities and performance variation of the dwarf willow Salix herbacea from different elevations and microhabitats. Ecol. Evol. 6 3940–3952. 10.1002/ece3.2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlacek J., Wheeler J. A., Cortés A. J., Bossdorf O., Hoch G., Lexer C., et al. (2015). The response of the alpine dwarf shrub Salix herbacea to altered snowmelt timing: lessons from a multi-site transplant experiment. PLoS One 10:e0122395. 10.1371/journal.pone.0122395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. (2004). Hybridization and adaptive radiation. Trends Ecol. Evol. 19 198–207. 10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- Sentinella A. T., Warton D. I., Sherwin W. B., Offord C. A., Moles A. T., Wang Z. (2020). Tropical plants do not have narrower temperature tolerances, but are more at risk from warming because they are close to their upper thermal limits. Glob. Ecol. Biogeogr. 29 1387–1398. 10.1111/geb.13117 [DOI] [Google Scholar]
- Shang H., Hess J., Pickup M., Field D. L., Ingvarsson P. K., Liu J., et al. (2020). Evolution of strong reproductive isolation in plants: broad-scale patterns and lessons from a perennial model group. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375:20190544. 10.1098/rstb.2019.0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa T. V., Caixeta E. T., Alkimim E. R., Oliveira A. C. B., Pereira A. A., Sakiyama N. S., et al. (2018). Early selection enabled by the implementation of genomic selection in Coffea arabica breeding. Front. Plant Sci. 9:1934. 10.3389/fpls.2018.01934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza L. M., Francisco F. R., Goncalves P. S., Scaloppi Junior E. J., Le Guen V., Fritsche-Neto R., et al. (2019). Genomic selection in rubber tree breeding: a comparison of models and methods for managing GxE interactions. Front. Plant Sci. 10:1353. 10.3389/fpls.2019.01353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sow M. D., Allona I., Ambroise C., Conde D., Fichot R., Gribkova S., et al. (2018). Epigenetics in forest trees: state of the art and potential implications for breeding and management in a context of climate change. Adv. Bot. Res. 88 387–453. 10.1016/bs.abr.2018.09.003 [DOI] [Google Scholar]
- Steinbauer M. J., Grytnes J. A., Jurasinski G., Kulonen A., Lenoir J., Pauli H., et al. (2018). Accelerated increase in plant species richness on mountain summits is linked to warming. Nature 556 231–234. [DOI] [PubMed] [Google Scholar]
- Stocks J. J., Metheringham C. L., Plumb W. J., Lee S. J., Kelly L. J., Nichols R. A., et al. (2019). Genomic basis of European ash tree resistance to ash dieback fungus. Nat. Ecol. Evol. 3 1686–1696. 10.1038/s41559-019-1036-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stölting K. N., Paris M., Meier C., Heinze B., Castiglione S., Bartha D., et al. (2015). Genome-wide patterns of differentiation and spatially varying selection between postglacial recolonization lineages of Populus Alba (Salicaceae), a widespread forest tree. New Phytol. 207 723–734. 10.1111/nph.13392 [DOI] [PubMed] [Google Scholar]
- Sullivan M., Lewis S. L., Affum-Baffoe K., Castilho C., Costa F., Sanchez A. C., et al. (2020). Long-term thermal sensitivity of earth’s tropical forests. Science 368 869–874. [DOI] [PubMed] [Google Scholar]
- Suontama M., Klápště J., Telfer E., Graham N., Stovold T., Low C., et al. (2019). Efficiency of genomic prediction across two Eucalyptus nitens seed orchards with different selection histories. Heredity 122 370–379. 10.1038/s41437-018-0119-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B., Grattapaglia D., Martins G. S., Ferreira K. Z., Sundberg B. R., Ingvarsson P. R. K. (2017). Evaluating the accuracy of genomic prediction of growth and wood traits in two Eucalyptus species and their F1 hybrids. BMC Plant Biol. 17:110. 10.1186/s12870-017-1059-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Technow F., Schrag T. A., Schipprack W., Bauer E., Simianer H., Melchinger A. E. (2014). Genome properties and prospects of genomic prediction of hybrid performance in a breeding program of maize. Genetics 197 1343–1355. 10.1534/genetics.114.165860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistlethwaite F. R., Gamal El-Dien O., Ratcliffe B., Klapste J., Porth I., Chen C., et al. (2020). Linkage Disequilibrium Vs. Pedigree: genomic selection prediction accuracy in conifer species. PLoS One 15:e0232201. 10.1371/journal.pone.0232201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistlethwaite F. R., Ratcliffe B., Klápštì J., Porth I., Chen C., Stoehr M. U., et al. (2017). Genomic prediction accuracies in space and time for height and wood density of Douglas-Fir using exome capture as the genotyping platform. BMC Genomics 18:930. 10.1186/s12864-017-4258-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistlethwaite F. R., Ratcliffe B., Klápštì J., Porth I., Chen C., Stoehr M. U., et al. (2019a). Genomic selection of juvenile height across a single-generational gap in Douglas-Fir. Heredity 122 848–863. 10.1038/s41437-018-0172-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistlethwaite F. R., Ratcliffe B., Klápštì J., Porth I., Chen C., Stoehr M. U., et al. (2019b). Genomic selection of juvenile height across a single-generational gap in Douglas-Fir. Heredity 122 848–863. 10.1038/s41437-018-0172-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tito R., Vasconcelos H. L., Feeley K. J. (2020). Mountain ecosystems as natural laboratories for climate change experiments. Front. For. Glob. Change 3:38 10.3389/ffgc.2020.00038 [DOI] [Google Scholar]
- Tuskan G. A., Groover A. T., Schmutz J., Difazio S. P., Myburg A., Grattapaglia D., et al. (2018). Hardwood tree genomics: unlocking woody plant biology. Front. Plant Sci. 9:1799. 10.3389/fpls.2018.01799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukrainetz N. K., Mansfield S. D. (2020). Assessing the sensitivities of genomic selection for growth and wood quality traits in lodgepole pine using Bayesian models. Tree Genet. Genomes 16:14. [Google Scholar]
- Ulian T., Diazgranados M., Pironon S., Padulosi S., Liu U., Davies L., et al. (2020). Unlocking plant resources to support food security and promote sustainable agriculture. Plants People Planet 2 421–445. 10.1002/ppp3.10145 [DOI] [Google Scholar]
- Valencia J. B., Mesa J., León J. G., Madriñán S., Cortés A. J. (2020). Climate vulnerability assessment of the Espeletia complex on PaìRamo sky islands in the Northern Andes. Front. Ecol. Evol. 8:565708 10.3389/fevo.2020.565708 [DOI] [Google Scholar]
- Wang J., Street N. R., Park E. J., Liu J., Ingvarsson P. K. (2020). Evidence for widespread selection in shaping the genomic landscape during speciation of Populus. Mol. Ecol. 29 1120–1136. 10.1111/mec.15388 [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang L., Zhang Z., Li M., Wang D., Zhang X., et al. (2020). Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol. 225 1370–1382. 10.1111/nph.16215 [DOI] [PubMed] [Google Scholar]
- Westbrook J. W., Holliday J. A., Newhouse A. E., Powell W. A. (2019). A plan to diversify a transgenic blight-tolerant American chestnut population using citizen science. Plants People Planet 2 84–95. 10.1002/ppp3.10061 [DOI] [Google Scholar]
- Westbrook J. W., Zhang Q., Mandal M. K., Jenkins E. V., Barth L. E., Jenkins J. W., et al. (2020). Optimizing genomic selection for blight resistance in American chestnut backcross populations: a trade-off with American chestnut ancestry implies resistance is polygenic. Evol. Appl. 13 31–47. 10.1111/eva.12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler J. A., Cortés A. J., Sedlacek J., Karrenberg S., Van Kleunen M., Wipf S., et al. (2016). The snow and the willows: accelerated spring snowmelt reduces performance in the low-lying alpine shrub Salix herbacea. J. Ecol. 104 1041–1050. 10.1111/1365-2745.12579 [DOI] [Google Scholar]
- Wheeler J. A., Hoch G., Cortés A. J., Sedlacek J., Wipf S., Rixen C. (2014). Increased spring freezing vulnerability for alpine shrubs under early snowmelt. Oecologia 175 219–229. 10.1007/s00442-013-2872-8 [DOI] [PubMed] [Google Scholar]
- Wheeler J. A., Schnider F., Sedlacek J., Cortés A. J., Wipf S., Hoch G., et al. (2015). With a little help from my friends: community facilitation increases performance in the dwarf shrub Salix herbacea. Basic Appl. Ecol. 16 202–209. 10.1016/j.baae.2015.02.004 [DOI] [Google Scholar]
- White T., Adams W., Neale D. (2007). Forest Genetics. New York, NY: CSIRO-CABI Publishing. [Google Scholar]
- Yeaman S., Hodgins K. A., Lotterhos K. E., Suren H., Nadeau S., Degner J. C., et al. (2016). Convergent local adaptation to climate in distantly related conifers. Science 353 1431–1433. 10.1126/science.aaf7812 [DOI] [PubMed] [Google Scholar]
- Zahn L. M., Purnell B. A. (2016). Genes under pressure. Science 354:52. 10.1126/science.354.6308.52 [DOI] [PubMed] [Google Scholar]
- Zhou L., Bawa R., Holliday J. A. (2014). Exome resequencing reveals signatures of demographic and adaptive processes across the genome and range of black cottonwood (Populus trichocarpa). Mol. Ecol. 23 2486–2499. 10.1111/mec.12752 [DOI] [PubMed] [Google Scholar]
- Zhou L., Chen Z., Olsson L., Grahn T., Karlsson B., Wu H. X., et al. (2020). Effect of number of annual rings and tree ages on genomic predictive ability for solid wood properties of Norway spruce. BMC Genomics 21:323. 10.1186/s12864-020-6737-3 [DOI] [PMC free article] [PubMed] [Google Scholar]