Abstract
Allogeneic hematopoietic stem cell transplantation (HCT) remains the only potentially curative option for myelodysplastic syndromes (MDS). Mortality after HCT is high, with deaths related to relapse or transplant-related complications. Thus, identifying patients who may or may not benefit from HCT is clinically important. We identified 1514 patients with MDS enrolled in the Center for International Blood and Marrow Transplant Research Registry and had their peripheral blood samples sequenced for the presence of 129 commonly mutated genes in myeloid malignancies. A random survival forest algorithm was used to build the model, and the accuracy of the proposed model was assessed by concordance index. The median age of the entire cohort was 59 years. The most commonly mutated genes were ASXL1(20%), TP53 (19%), DNMT3A (15%), and TET2 (12%). The algorithm identified the following variables prior to HCT that impacted overall survival: age, TP53 mutations, absolute neutrophils count, cytogenetics per International Prognostic Scoring System–Revised, Karnofsky performance status, conditioning regimen, donor age, WBC count, hemoglobin, diagnosis of therapy-related MDS, peripheral blast percentage, mutations in RAS pathway, JAK2 mutation, number of mutations/sample, ZRSR2, and CUX1 mutations. Different variables impacted the risk of relapse post-transplant. The new model can provide survival probability at different time points that are specific (personalized) for a given patient based on the clinical and mutational variables that are listed above. The outcomes’ probability at different time points may aid physicians and patients in their decision regarding HCT.
Keywords: MDS, Mutations, Genomic biomarkers
INTRODUCTION
The myelodysplastic syndromes (MDS) are clonal disorders characterized by the accumulation of complex molecular alterations and dysplastic features that yield to ineffective hematopoiesis, increased blasts presence, and risk of progression to acute myeloid leukemia [1–3]. The outcomes of patients with MDS are very heterogeneous and vary depending on each patient’s risk category and therapy. Allogeneic stem cell transplant (HCT) is the only potentially curative option for patients with MDS, although only 10% to 15% of patients with MDS undergo transplant [4]. This low number of patients undergoing HCT is due to several factors that include patient-related factors such as age and comorbidities and concerns regarding the higher risk of transplant-related morbidity and mortality.
The National Comprehensive Network Guidelines and other consensus guidelines recommend HCT for patients with MDS early in their disease if they have intermediate 2 or high-risk category per the International Prognostic Scoring System (IPSS) and later during their disease course (prior to acute myeloid leukemia progression) if they have lower-risk disease [5,6]. The rationale behind these recommendations is based on analyses that use Markov decision modeling to predict the benefit from transplants in patients with MDS receiving myeloablative and reduced-intensity transplant [5,6]. This type of modeling does not consider the extensive heterogeneity of patients with MDS and the inherent biases in our current prognostic scoring systems that overestimate or underestimate the actual risk of individual patients.
Several small- and large-scale genomic studies have shown that somatic mutations may have an impact on outcomes after HCT, although the prognostic impact of some of these mutations remained controversial except for TP53 mutations [7–9]. The differences in the results of these studies are likely related to differences in the patient cohort, sample size, number of mutations included in the genomic panel, and the statistical methods [7–9]. Although the outcomes of patients with MDS with TP53 mutations after HCT remain poor, with a median overall survival (OS) of 6 months, approximately 20% to 25% of these patients are alive at 24 months after transplant, suggesting that a small percentage of these patients still derives benefit from HCT compared to other available therapies [7–9]. Identifying these patients prior to HCT remains clinically important but a challenging task.
In this study, we took advantage of a large MDS cohort with fully annotated clinical and genomic data from the Center for Blood and Marrow Transplant Research (CIBMTR) registry and, using machine learning algorithms, built a personalized prediction model that can predict patient-specific outcomes after HCT.
METHODS
Patient Cohort
Patients with MDS diagnosed according to 2008 World Health Organization criteria and enrolled in the CIBMTR research database from 2004 to 2015 were included. Patients underwent HLA-matched, mismatched-donor, or umbilical cord transplant. Patients were excluded if blast percentage was >20% or had a diagnosis of overlap myelodysplastic-myeloproliferative neoplasms. IPSS, IPSS–Revised (IPSS-R), and CIBMTR MDS risk scores were calculated as described previously [10–12]. Cytogenetic karyotyping was stratified based on MDS Comprehensive Cytogenetic Scoring System and grouped based on IPSS and IPSS-R criteria [10,11,13].
All the patients provided written informed consent to participate in the CIBMTR research database and repository. Observational studies conducted by the CIBMTR are performed in compliance with all applicable federal regulations pertaining to the protection of human research participants. Protected health information used in the performance of such research is collected and maintained in the capacity of the CIBMTR as a public health authority under the Health Insurance Portability and Accountability Act Privacy Rule. More details regarding the data source and participating centers are described elsewhere [14].
DNA Sequencing Studies
Next-generation targeted deep sequencing of 129 genes that are known or suspected to be involved in myeloid malignancies was included. Annotation of the mutations was blinded in regard to clinical data [7]. The method of sequencing has been described previously [7]. Briefly, patient peripheral blood samples were collected prior to initiation of the preparative conditioning regimen and stored at the CIBMTR registry. Native genomic DNA was sheared and library constructed per manufacturer protocol (Agilent). Libraries were then quantified and pooled up to 24 samples per lane in equimolar amounts totaling 500 ng DNA. Fastq files were aligned to the hg19 version of the human genome with BWA 0.6.2. Single nucleotide and small insertion and deletion calling were performed with samtools-0.1.18 mpileup and Varscan 2.2.3. Variants were annotated to include information about cDNA and amino acid changes, sequence depth, number and percentage of reads supporting the variant allele, population allele frequency in 1000 Genomes release 2.2.2, the Exome Sequencing Project, Exome Aggregation Consortium, and presence in Catalogue of Somatic Mutations in Cancer (COSMIC), version 64.6. Variants were excluded if they had fewer than 15 total reads at the position, had fewer than 5 alternate reads, had variant allele fraction <2%, fell outside of the target coordinates, had excessive read strand bias, had an excessive number of calls in the local region, caused synonymous changes, or were recurrent small insertions/deletions at low variant allele fraction adjacent to homopolymer repeat regions. All clinical and genetic data are available for request through CIBMTR: https://www.cibmtr.org/Studies/Observational/ProposeStudy/pages/index.aspx.
Statistical Analysis
A total of 1514 patients were included in the final analysis and divided randomly into a training cohort comprising 70% (n = 1059) and a validation cohort including the remaining 30% (n = 455). These patients were treated at 130 different transplant centers that participate in the CIBMTR database and provide a very diverse patient cohort, including different institutional treatment practices. The training cohort was used to build the new model, and the validation set was used to assess the prognostic accuracy of the proposed model. Overall survival was defined as the time from date of transplant to date of death from any cause or until censoring at the last date of follow-up.
To build the model in the training cohort, a random survival forest (RSF) algorithm was used. This algorithm allows an efficient nonparametric analysis of time-to-event data. The algorithm randomly chooses variables to produce the desired outcome, thus overcoming the shortcoming of the stepwise selection approach using Cox regression models. Further details regarding the application of this algorithm for survival analysis are summarized in the supplementary materials. To identify the most important variables that impacted the algorithm decisions, a variable importance analysis was used, and variables were ranked from the most important to the least important. The final proposed model was then rebuilt using only the variables that were included in the variable importance analysis. Concordance indices were calculated for the proposed prognostic scoring system, IPSS, IPSS-R, and the CIBMTR MDS risk score models using the validation cohort as previously described [12–14]. All analyses were conducted using R statistical environment version 3.3.
RESULTS
Patient Cohort
A total of 1514 patients with MDS were included. The clinical characteristics are summarized in Table 1 and were described previously [7]. The median follow-up of survivors was 47 months. The percentage of patients for whom complete follow-up data was available at 1, 3, and 5 years was 98%, 90%, and 88%, respectively. The median age of the entire cohort was 59 years (range, 0.4 to 77). Donors were 8/8 HLA-matched unrelated donors (n = 863, 57%), HLA-identical siblings (n = 181, 12%), <8/8 HLA-matched unrelated donors (n = 296, 20%), or umbilical cord blood (n = 174, 11%). Conditioning regimens included myeloablative (n = 789, 52%), reduced intensity (n = 582, 38%), and nonmyeloablative (n = 130, 9%). Hematopoietic stem cell sources included 221 (15%) from bone marrow, 1114 (74%) from peripheral blood, 168 (11%) from umbilical cord, and 11 (1%) from combined sources. For cytogenetic analysis by IPSS-R criteria, 579 (38%) were very good/ good, 269 (18%) were intermediate, 287 (19%) were poor, 125 (8%) were very poor, and 254 (17%) were missing (Table 1). Seventy-nine percent of the patient cohort had primary MDS and 21% had therapy-related MDS (t-MDS) (Table 1). Risk stratification per IPSS-R at HCT included 26% with very low/low risk, 22% intermediate risk, 14% high risk, 11% very high risk, and 24% missing (unable to calculate due to missing data).
Table 1.
Patient Characteristics
| Variable | No. (%) |
|---|---|
| Number of patients | 1514 |
| Number of centers | 130 |
| HLA matching out of 8 loci | |
| < or = 6/8 | 116 (10) |
| 7/8 | 177 (15) |
| 8/8 | 870 (75) |
| Unknown | 351 (N/A) |
| Cytogenetic category per IPSS system criteria | |
| Favorable | 471 (31) |
| Intermediate | 215 (14) |
| Poor | 504 (33) |
| Unknown | 324 (21) |
| Cytogenetic category for IPSS-R system criteria | |
| Very good | 9 (1) |
| Good | 542 (36) |
| Intermediate | 253 (17) |
| Poor | 265 (18) |
| Very poor | 121 (8) |
| Unknown | 324 (21) |
| Recipient age at transplant, yr | |
| 0–39 | 241 (16) |
| 40–49 | 167 (11) |
| 50–59 | 387 (26) |
| 60 and older | 719 (47) |
| Median (range) | 59 (0–77) |
| Ethnicity | |
| Caucasian | 1397 (93) |
| African American | 57 (4) |
| Asian | 37 (2) |
| Other | 16 (1) |
| Unknown | 25 (N/A) |
| Recipient sex | |
| Male | 912 (60) |
| Female | 602 (40) |
| Karnofsky performance score | |
| 10–80 | 419(28) |
| 90–100 | 817(54) |
| Missing | 278(18) |
| Disease status at transplant | |
| Early | 576 (38) |
| Advanced | 796 (53) |
| Other | 142 (9) |
| Stem cell source | |
| Marrow | 221 (15) |
| PBSC | 1114 (74) |
| UCB | 168 (11) |
| Other | 11 (<1) |
| Donor source | |
| Unrelated donor | 1165 (77) |
| Related donor | 181 (12) |
| Cord blood | 168 (11) |
| Donor group | |
| BM | 221 (15) |
| Unrelated | 209 (14) |
| Related | 12 (1) |
| PBSC | 1114 (74) |
| Unrelated | 947 (63) |
| Related | 167 (11) |
| UCB | 168 (11) |
| Missing | 11 (1) |
| Conditioning regimen intensity | |
| Myeloablative | 789 (52) |
| RIC | 582 (38) |
| Nonmyeloablative | 130 (9) |
| Unknown | 13 (1) |
| In vivo T cell depletion | |
| No | 843 (56) |
| Yes | 605 (40) |
| Unknown | 66 (4) |
| Donor/recipient sex matching | |
| Male/male | 610 (43) |
| Male/female | 362 (25) |
| Female/male | 250 (18) |
| Female/female | 204 (14) |
| Unknown | 88 (N/A) |
| Donor/recipient CMV matching | |
| Negative/negative | 439 (29) |
| Negative/positive | 459 (30) |
| Positive/negative | 162 (11) |
| Positive/positive | 379 (25) |
| Unknown | 75 (5) |
| Donor age at donation, yr | |
| Median (range) | 29 (0–77) |
| Year of transplant | |
| 2004–2007 | 300 (20) |
| 2008–2011 | 652 (43) |
| 2012–2015 | 562 (37) |
PBSC indicates peripheral blood stem cell; UCB, umbilical cord blood; BM, bone marrow; RIC, reduced-intensity conditioning; N/A, not applicable; CMV, cytomegalovirus.
A total of 168 patients in our patient cohort received a transplant from a cord graft. Compared to other graft types, the median OS for patients who received cord graft was 13.2 months (range, 8.9 to 23.6) compared to bone marrow graft (19.8; range, 15.3 to 35.1) and peripheral blood graft (21.9; range, 17.9 to 26.6), P= .64, Supplementary Figure S1. The median relapse-free survival was 8.8 months (range, 5.8 to 12.9), 12 months (range, 8.5 to 19.3), and 12.9 months (range, 10.9 to 17.5), respectively, P= .95, Supplementary Figure S2.
Genetic Characteristics and Mutational Landscape
At least 1196 patients (79%) had 1 or more mutations in their blood samples prior to transplant with a median of 2 mutations per sample (range, 0 to 15). Only 31 mutated genes were present in at least 20 patients in the study cohort (Figure 1). The prognostic impact of each of these mutations on OS and relapse-free survival in univariate and multivariate analysis was reported previously [7]. Briefly, only mutations in TP53 (hazard ratio [HR] for death, 1.96), PPM1D (HR, 1.64), and JAK2 mutations (HR, 1.77) were associated with shorter OS, while no mutations predicted prolonged survival. In multivariate analyses and after adjustment for recipient age, donor group, IPSS-R score prior to HCT, Karnofsky score, t-MDS status, donor-recipient sex match, and year of HCT, mutations in TP53 (HR, 1.72), JAK 2 (HR, 1.67), and RAS pathway mutations (HR, 1.25) independently impacted OS15. In multivariate analyses, TP53 mutations and RAS pathway mutations were associated with an increased risk of relapse [7].
Figure 1.
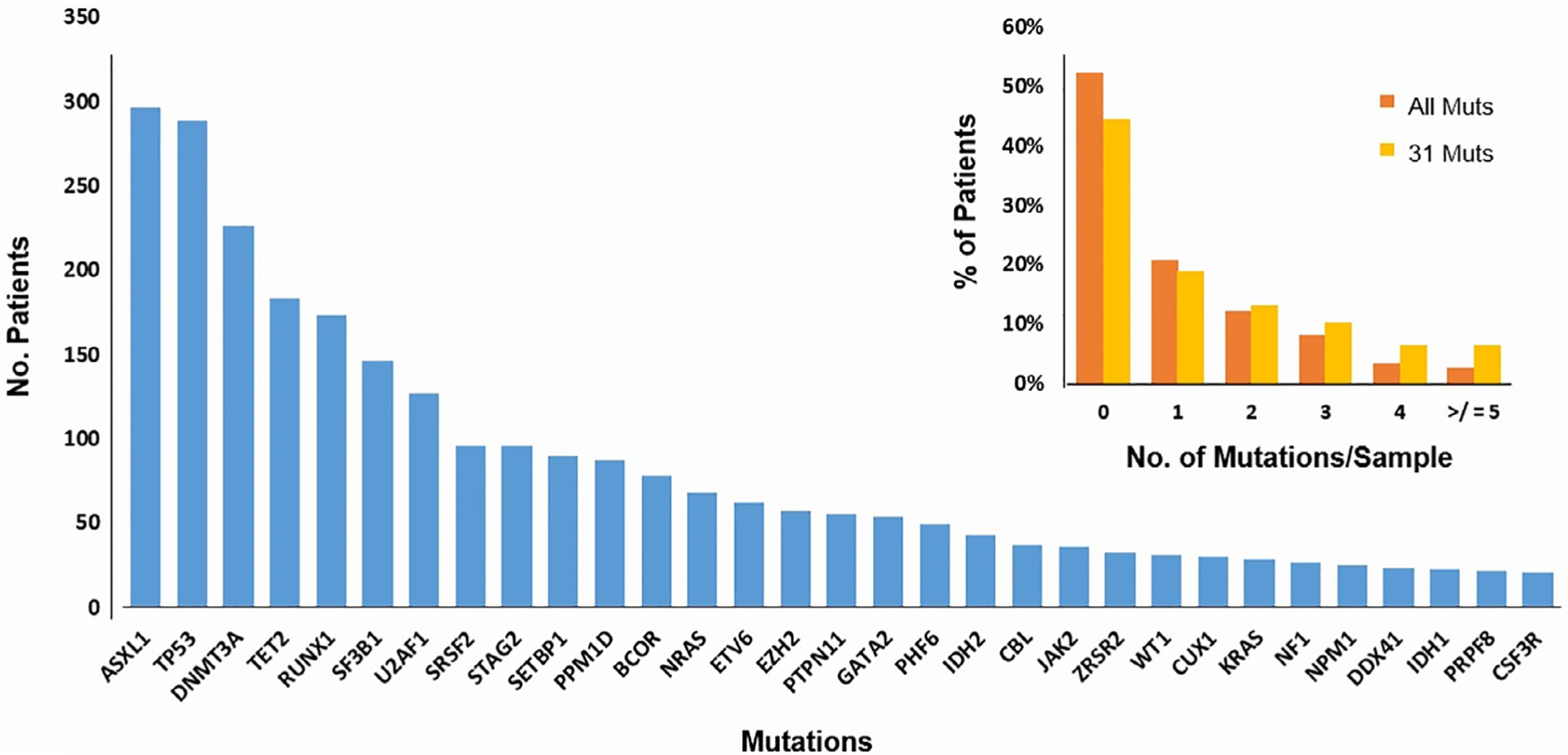
Most commonly mutated genes in the study cohort. (A) Frequency of mutations in the study cohort included the 31 genes that are present in 20 or more patients. The panel also shows the number of mutations per sample using all genes (129 genes) in orange and the 31 commonly mutated genes in yellow. Mut, mutations. Panel B numbr of mutations per sample given all mutations versus the 31 genes included in the final analysis.
Number of Mutated Genes per Sample as a Prognostic Marker
Mutation number per sample can carry important prognostic value, although the actual number can vary depending on the number of genes included in the sequencing panel and the level of detection. Overall, the median number of mutations per sample was 2 (range, 0 to 15) when 129 genes were considered, and it was 2 (range, 0 to 8) when the analysis was focused only on the 31 mutations in >20 patients (Figure 1). When 129 genes were considered, the median OS for patients with zero mutations was 43.2 months, compared to 20, 15, 14, 25, and 19 months for patients with 1, 2, 3, 4, and ≥5 mutations per sample, P < .001, Figure 2A. Similar median OS was observed when the analysis was restricted to 31 commonly mutated genes with a median OS of 42.3 months for patients with zero mutations compared to 18, 13, 24, 27, and 13 months, respectively, P < .001, Figure 2B. In univariate analysis, mutation number/sample impacted OS when all mutations were accounted for (HR, 1.07; 95% confidence interval, 1.03 to 1.11; P < .001) and when the analysis was restricted to 31 genes (HR, 1.08; 95% CI, 1.03 to 1.13; P < .001). However, this significant impact was lost after adjustment for other clinical variables such as age, donor group, t-MDS versus de novo, IPSS-R prior to transplant, conditioning regimen, and year of transplant (Supplementary Tables S1 and S2). We also observed a statistically significant but weak correlation between the number of mutations/sample and the bone marrow blasts percentage (correlation coefficient for number of mutations versus bone marrow percentage is 0.19; P < .001), Supplementary Figure S1.
Figure 2.
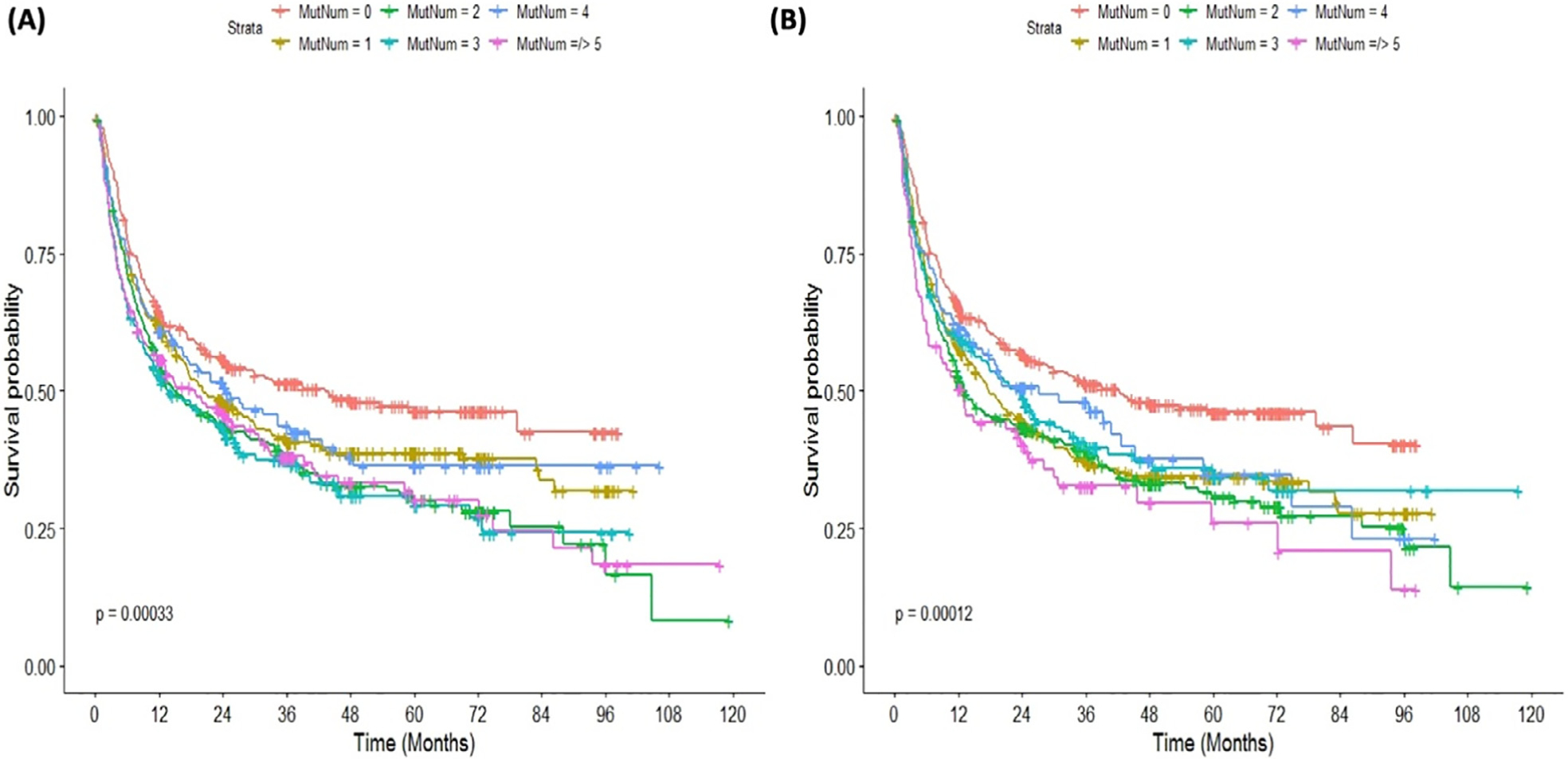
Kaplan-Meier curves for overall survival according to mutation number per sample. (A) The survival curves based on 129 genes included. (B) The overall survival curves based on 31 (commonly mutated) genes.
Machine Learning Model Building and Validation
The new model was built in the training cohort using the RSF algorithm by introducing all variables (clinical, transplant related, and molecular data). The algorithm randomly choses variables to produce the desired outcomes (OS and time to relapse), thus overcoming the shortcoming of the stepwise selection approach using Cox regression models. After building the model, a variable importance analysis was conducted to extract the most important variables that affected the outcome. The variables are summarized from the most important to the least important for OS and time to relapse in Figure 3. As expected, age and TP53 mutations were very important variables for OS while TP53 was the most important variable for relapse. Mutation number along with CUX-1 mutations and PPM1D mutation was also important for OS and time to leukemia transformation while TET2 mutation and mutations in the RAS pathway were among important variables for time to relapse (Figure 3). When mutation characteristics such as variant allele frequency were added to the random survival algorithm, no improvement in the performance of the model was observed. Thus, we focused the analysis on whether the mutations were present or absent. The final model was developed using variables highlighted in Figure 3.
Figure 3.
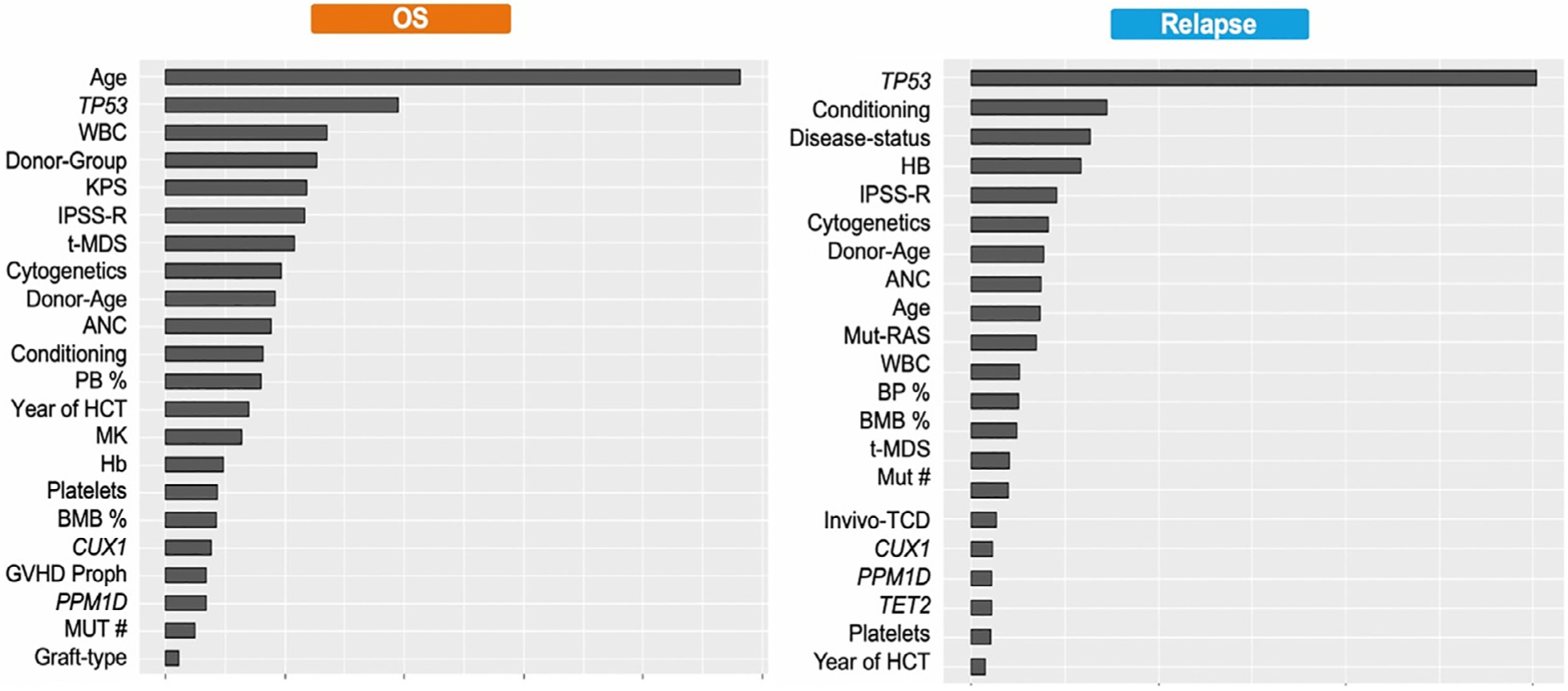
Important variables that impacted overall survival and risk of relapse based on the machine learning algorithm. The variables are ranked from the most important to the least important and excluded variables with no impact on either endpoint. (More details regarding variable importance analysis are included in supplementary materials.) KPS indicates Karnofsky performance score; Conditioning, transplant conditioning regimen; ANC, absolute neutrophil count; MK, monosomal karyotype; hb, hemoglobin; BMB %, bone marrow blast percentage; proph, prophylaxis; MUT #, mutation number.
To further understand the prognostic impact of the clinical and transplant-related variables on outcomes among patients with higher-risk disease such as older age and patients with TP53 mutations, we reran the RSF algorithm in each subgroup and then highlighted the most important variables (ranked from the most to the least important) using variable importance analysis. A total of 289 patients had TP53 mutations in our patient cohort. When we focused the analysis on these patients only, the clinical and transplant-related factors impact changed for OS time to relapse, as shown in Supplementary Figures S4 and S5. Interestingly, some of the transplant-related variables such as graft type and the graft-versus-host disease (GVHD) prophylaxis did not impact OS while graft type, conditioning regimen, donor age, Karnofsky performance status, and GVHD prophylaxis did not impact time to relapse (Supplementary Figures S4 and S5).
We also identified 730 patients in our patient cohort who were ≥60 years old. When we focused the analysis on this patient subgroup, the clinical and transplant-related factors impact changed for OS and time to relapse as shown in Supplementary Figures S6 and S7. Interestingly, transplant year, graft type, and the GVHD prophylaxis did not impact OS while only graft type did not impact time to relapse (Supplementary Figures S6 and S7).
Predictability of the Proposed Model Compared to IPSS-R and CIBMTR MDS Models and Its Clinical Application
To investigate whether the proposed system was superior compared to IPSS-R and CIBMTR model, concordance indices were measured for each model prior to transplant. The C-index for the proposed model was .62 for OS and .68 for time to relapse compared to .55/.58 for IPSS-R and .57/.61 for the CIBMTR MDS model, respectively (Figure 4). When we built the model based only on the top 4 variables that impacted the OS and time to relapse outcomes (age, TP53 mutations, conditioning regimen, and donor group), we observed a significant drop in the c-index for OS (.57) and time to relapse (.58) outcomes, suggesting that all variables that were included in our final model are needed to boost the model performance. Although a simpler model is always preferred to ease the implementation and adaptation of any model in clinical practice, the performance of the final model should be at least comparable to a more complex model.
Figure 4.
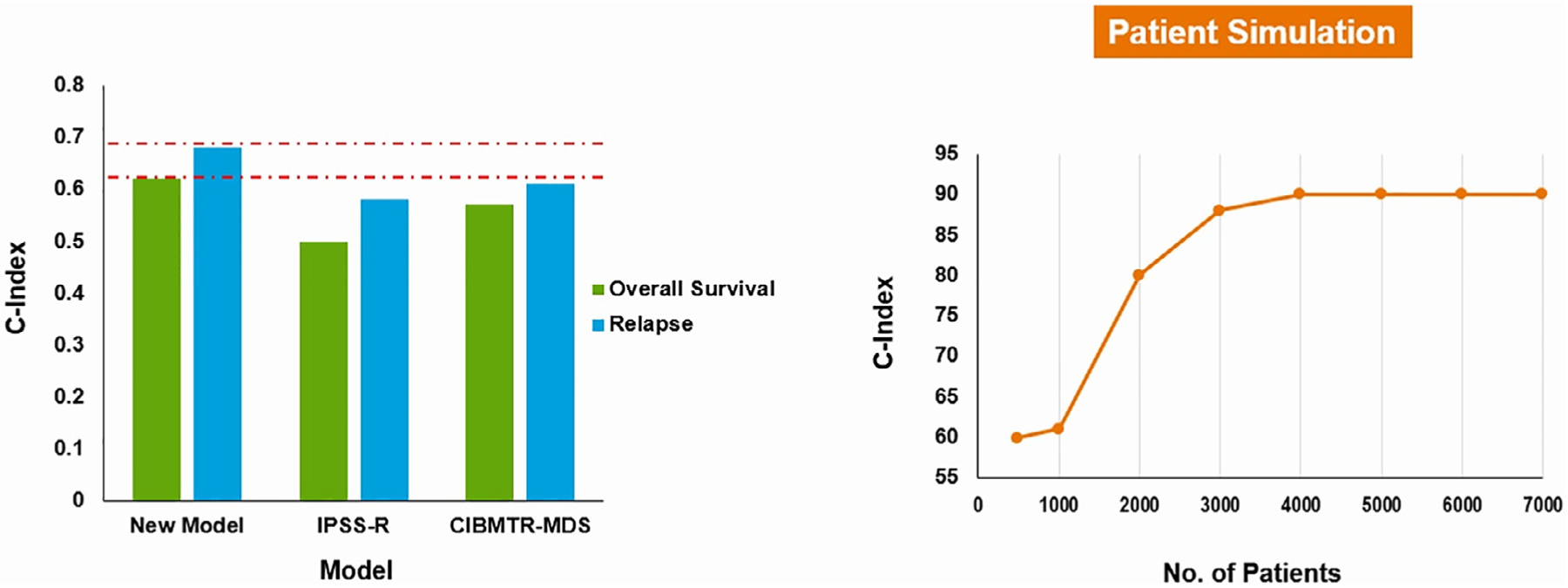
Summary of the c-index of the proposed model compared to IPSS-R and CIBMTR models. The panel on the right shows the increase in the correlation between c-index and sample size. This suggests that increasing our patient cohort could improve the c-index of the proposed model.
The modest improvement in c-index could be related to the sample size. When our patient cohort was oversampled to a larger number, a significant increase in the c-index was observed (Figure 4).
Further, our model can provide survival probability at 6, 12, and 24 months after transplant that is specific for a given patient based on his or her clinical and mutational data. These probabilities can aid physicians and patients in their decision whether to proceed or not with the transplant (Figure 5). To ease the translation of this model into the clinic, a web application that input the patient clinical and mutational data and output the survival probability at different time points is currently under construction on the CIBMTR public portal (Figure 5). A preliminary version of the web application can be accessed at https://azizn38.shinyapps.io/aziz/ (please copy the link to your web browser or hold the control key to open the link and refresh the website every time you change the variables).
Figure 5.
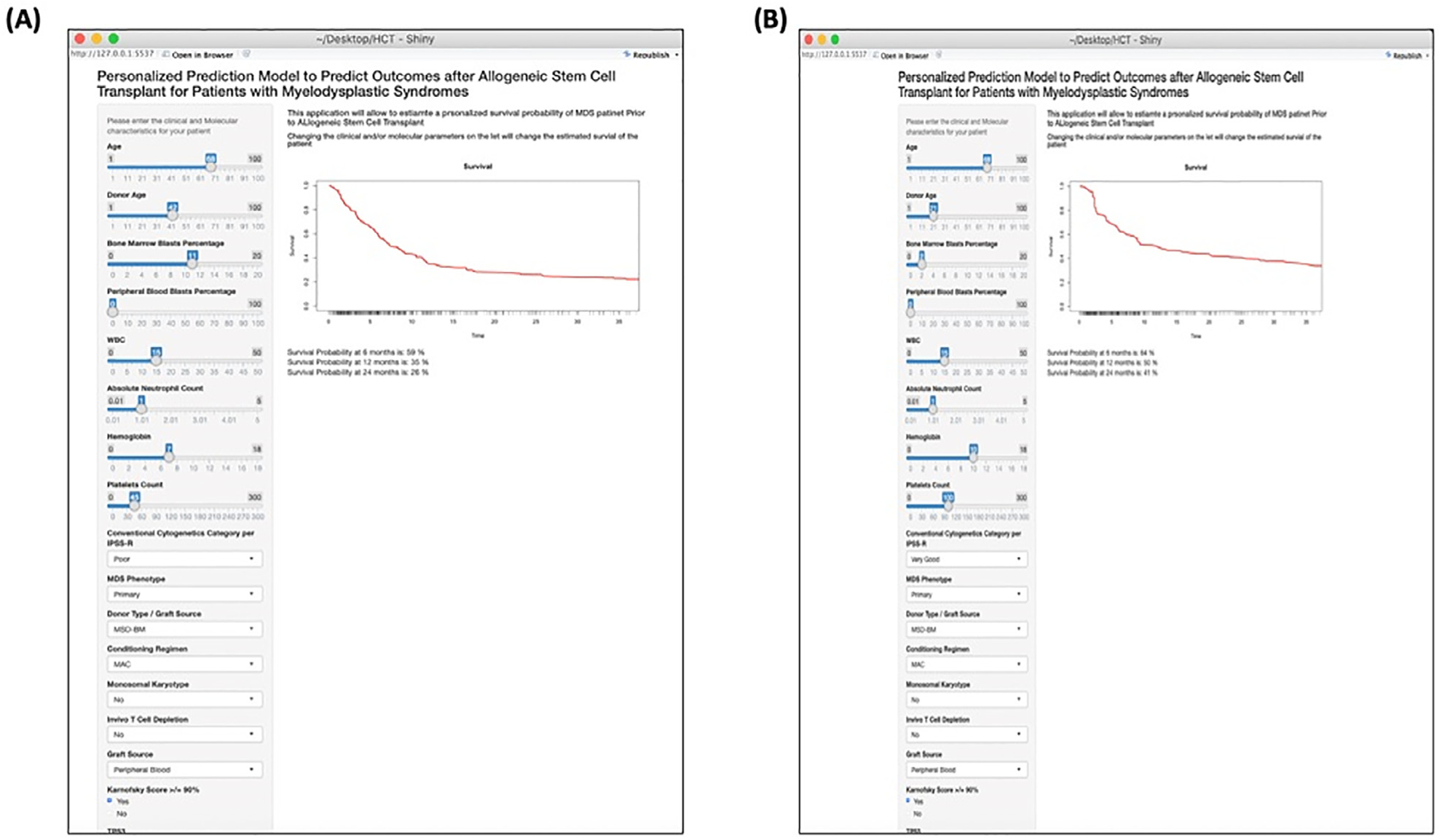
A snapshot from the web application for 2 different patients. The patients (A and B) have different clinical and mutational variables. On the right is the survival curve that is personalized for the patient with survival probability at different time points post-transplant. Please note that the variables are not included.
DISCUSSION
Outcomes of patients with MDS who undergo HCT are very heterogeneous, and our current models do not provide risk prediction tailored to each patient’s characteristics.
In this study, we took advantage of a large registry-based fully annotated cohort of patients with MDS to develop a personalized prediction model that uses clinical and mutational data and predicts OS and relapse risk that is specific to each patient. We found that a panel of 31 most commonly mutated genes in our cohort showed a similar impact on outcomes compared to the whole panel of 129 genes, suggesting that only a panel of approximately 25 to 30 genes is needed to improve risk stratification prior to transplant.
Our machine learning algorithm identified several clinical factors that impacted OS and risk of relapse after HCT along with 3 mutations (TP53, CUX-1, and PP1MD) for OS and (RAS pathway and TET2 mutations) for relapse risk. The differences between the variables included in our final model and the clinical and mutational variables that have been shown to impact survival in prior studies are related to the differences in the analytic approach. The traditional model building typically starts with univariate analyses of variables and then stepwise multivariate analyses that include only significant variables. Such an approach can ignore the significance of some variables that are significant only in the context of other variables. Further, risk stratification tools that divide the patient cohort into 4 or 5 risk categories undermine the significant heterogeneity in outcomes of patients in the same risk category.
Several studies have investigated the impact of somatic mutations on transplant outcomes with some conflicting results [7,9]. In a large cohort of 1514 patients with MDS who were reported in the CIBMTR Repository between 2005 and 2014, TP53 mutations independently predicted shorter OS and a higher risk of relapse even after adjustment for known clinical risk factors [7]. In another study from the Japanese bone marrow transplant registry, mutations in TP53, NRAS, CBL, and complex karyotype were independently associated with shorter OS after adjusting for clinical variables [9]. More important, the negative impact of TP53 mutations was mainly seen in patients who had a complex karyotype [9]. Patients with TP53 mutations without complex karyotype (CK) had a significantly better survival post-transplant (73% were alive at 60 months) [9]. The differences in the results of these studies and others are mainly related to differences in the patient cohorts, different analytic methods, and the clinical and mutational variables that were included in the final model. Most important, these studies did not compare the impact of mutations on the outcome in patients who did not receive a transplant and treated with other available therapies alone such as hypomethylating agents. Such analysis is important as it is evident that a subset of patients with TP53 mutations can have prolonged OS with HCT that cannot be achieved by other therapies.
Our study also has some limitations. Although the study cohort is relatively large, a larger number is needed to improve the c-index of the model. In an attempt to oversample our patient cohort using the machine learning algorithm, a cohort of approximately 5000 patients can potentially improve the accuracy of the model. Although our model was built on data from patients with MDS from different transplant centers across the United States and validated in a randomly selected patient cohort from the same database, an independent validation from an external cohort is still needed to ensure the reproducibility of the model.
In conclusion, we built a personalized prediction model that uses clinical, molecular, and transplant-related data and can provide MDS patient-specific survival outcomes post-transplant. The new model identified several clinical and molecular variables that impacted OS and the cause-specific hazard of relapse. The OS probability at different time points may aid physicians and patients in their decision of whether to proceed to HCT.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge all participating centers and researchers from the CIBMTR.
Financial disclosure: The CIBMTR is supported primarily by Public Health Service grant/cooperative agreement U24CA076518 with the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); grant/cooperative agreement U24HL138660 with NHLBI and NCI; grant U24CA233032 from the NCI; grants OT3HL147741, R21HL140314, and U01HL128568 from the NHLBI; contract HHSH250201700006C with Health Resources and Services Administration (HRSA); grants N00014-18-1-2888 and N00014-17-1-2850 from the Office of Naval Research; suba-ward from prime contract award SC1MC31881-01-00 with HRSA; subawards from prime grant awards R01HL131731 and R01HL126589 from NHLBI; subawards from prime grant awards 5P01CA111412, 5R01HL129472, R01CA152108, 1R01HL131731, 1U01AI126612, and 1R01CA231141 from the NIH; and industry funds from Actinium Pharmaceuticals, Inc.; Adaptive Biotechnologies; Allovir, Inc.; Amgen, Inc.; anonymous donation to the Medical College of Wisconsin; Anthem, Inc.; Astellas Pharma US; Atara Biotherapeutics, Inc.; BARDA; Be the Match Foundation; bluebird bio, Inc.; Boston Children’s Hospital; Bristol Myers Squibb Co.; Celgene Corp.; Children’s Hospital of Los Angeles; Chimerix, Inc.; City of Hope Medical Center; CSL Behring; CytoSen Therapeutics, Inc.; Daiichi Sankyo Co., Ltd.; Dana Farber Cancer Institute; Enterprise Science and Computing, Inc.; Fred Hutchinson Cancer Research Center; Gamida-Cell, Ltd.; Genzyme; Gilead Sciences, Inc.; GlaxoS-mithKline (GSK); HistoGenetics, Inc.; Immucor; Incyte Corporation; Janssen Biotech, Inc.; Janssen Pharmaceuticals, Inc.; Janssen Research & Development, LLC; Janssen Scientific Affairs, LLC; Japan Hematopoietic Cell Transplantation Data Center; Jazz Pharmaceuticals, Inc.; Karius, Inc.; Karyopharm Therapeutics, Inc.; Kite, a Gilead Company; Kyowa Kirin; Magenta Therapeutics; Mayo Clinic and Foundation Rochester; Medac GmbH; Mediware; Memorial Sloan Kettering Cancer Center; Merck & Company, Inc.; Mesoblast; MesoScale Diagnostics, Inc.; Millennium, Takeda Oncology Co.; Miltenyi Biotec, Inc.; Mundipharma EDO; National Marrow Donor Program; Novartis Oncology; Novartis Pharmaceuticals Corporation; Omeros Corporation; Oncoimmune, Inc.; OptumHealth; Orca Biosystems, Inc.; PCORI; Pfizer, Inc.; Phamacyclics, LLC; PIRCHE AG; Regeneron Pharmaceuticals, Inc.; REGiMMUNE Corp.; Sanofi Genzyme; Seattle Genetics; Shire; Sobi, Inc.; Spectrum Pharmaceuticals, Inc.; St. Baldrick’s Foundation; Swedish Orphan Biovitrum, Inc.; Takeda Oncology; The Medical College of Wisconsin; University of Minnesota; University of Pittsburgh; University of Texas MD Anderson; University of Wisconsin, Madison; Viracor Eurofins; and Xenikos BV. The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, HRSA, or any other agency of the US government.
Footnotes
Conflict of interest statement: A.N.: Incyte, Novartis, Karyopharma, Tolero, MEI, Jazz Pharma, Daiichi Sankyo. J.C.: Jazz Pharma, Daiichi Sankyo, Incyte. Robert Peter Gale: Celgene. U. G.: Astellas, Incyte, Merck, Jazz Pharma. M.R.G.: Agios, Abbvie, Amgen, Cardinal Health, Celgene, Incyte, Merck, Pfizer, Trava-gene, Daiichi Sankyo, Medtrnoic, Forma Theraputics, Amgen, Genentech, Janssen, Novartis. D.R.: Abbvie, Agios, Jazz Pharma, Novartis, Sanofi, Teva, AROG, Bayer, Pfizer, Celgene, Gilead, Stemline. G.H.: Sangamo Bioscience, Axim Biotechnology, Juno Theraputics, Kite Pharma, Novartis, insys Theraputics, Abbvie, GW Pharmaceuticals, Cardina health, Immunomedics, Endocyte, Clovis Oncology, Aetna, CSV health, Bleubird Bio, BMD, Crispr therapeutics, IDEXX laboratoris, Johnson & Johnson, Pfizer, Procter & Gamble, Vertex, Bayer, Scott-Miracle, Pfizer, Kite Pharma, Incyte, Jazz Pharmaceuticals, Astellas. D.V.: Celgene, Novartis, Jazz Pharma, Amgen, GSK, Takeda, Astellas Pharma.
SUPPLEMENTARY MATERIALS
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.bbmt.2020.08.003.
REFERENCES
- 1.Tefferi A, Vardiman JW. Myelodysplastic syndromes. N Engl J Med. 2009;361:1872–1885. [DOI] [PubMed] [Google Scholar]
- 2.Nazha A The MDS genomics-prognosis symbiosis. Hematology Am Soc Hematol Educ Program. 2018;2018:270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ades L, Itzykson R, Fenaux P. Myelodysplastic syndromes. Lancet. 2014;383:2239–2252. [DOI] [PubMed] [Google Scholar]
- 4.Bartenstein M, Deeg HJ. Hematopoietic stem cell transplantation for MDS. Hematol Oncol Clin North Am. 2010;24:407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutler CS, Lee SJ, Greenberg P, et al. A decision analysis of allogeneic bone marrow transplantation for the myelodysplastic syndromes: delayed transplantation for low-risk myelodysplasia is associated with improved outcome. Blood. 2004;104:579–585. [DOI] [PubMed] [Google Scholar]
- 6.Koreth J, Pidala J, Perez WS, et al. Role of reduced-intensity conditioning allogeneic hematopoietic stem-cell transplantation in older patients with de novo myelodysplastic syndromes: an international collaborative decision analysis. J Clin Oncol. 2013;31:2662–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376:536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Della Porta MG, Galli A, Bacigalupo A, et al. Clinical effects of driver somatic mutations on the outcomes of patients with myelodysplastic syndromes treated with allogeneic hematopoietic stem-cell transplantation. J Clin Oncol. 2016;34:3627–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshizato T, Nannya Y, Atsuta Y, et al. Impact of genetic alterations in stem-cell transplantation for myelodysplasia and secondary acute myeloid leukemia. Blood. 2017. 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg P, Cox C, LeBeau MM, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 11.Greenberg PL, Tuechler H, Schanz J, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaffer BC, Ahn KW, Hu Z-H, et al. Scoring system prognostic of outcome in patients undergoing allogeneic hematopoietic cell transplantation for myelodysplastic syndrome. J Clin Oncol. 2016;34:1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schanz J, Tuchler H, Sole F, et al. New comprehensive cytogenetic scoring system for primary myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia after MDS derived from an international database merge. J Clin Oncol. 2012;30:820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horowitz M The role of registries in facilitating clinical research in BMT: examples from the Center for International Blood and Marrow Transplant Research. Bone Marrow Transplant. 2008;42((suppl 1)):S1–S2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


