Abstract
Long noncoding RNAs (lncRNAs) are implicated in the autophagic-lysosomal pathway (ALP) and are closely linked to Parkinson’s disease (PD) pathology. β-Glucocerebrosidase (GCase) has also been reported to be correlated with α-synuclein (α-syn) proteostasis. However, lncRNAs and α-syn in neural-derived L1CAM exosomes and GCase activity in the plasma of PD patients have not been studied. This study used an ultrasensitive methodology, fluorescence nanoparticle tracking analysis (NTA), to measure plasma L1CAM exosomes and Quanterix Simoa to measure α-syn concentrations in L1CAM exosomes. Eighty-five healthy controls and 93 PD patients were enrolled, and several scales were used to rate the severity of PD. Receiver operating characteristic (ROC) curves were applied to map the diagnostic accuracy of categorizing PD patients and healthy subjects. We found increased Linc-POU3F3 and α-syn concentrations in L1CAM exosomes and decreased GCase activity in PD patients compared with controls. The three biomarkers displayed obvious differences among PD patients based on gender, H-Y stage, and UPDRS-III distribution. Interestingly, Linc-POU3F3 was significantly positively correlated with α-syn in L1CAM exosomes and inversely correlated with GCase activity in PD patients. Significant correlations were observed among L1CAM exosomal Linc-POU3F3 levels, GCase activity, and PD severity, including motor/cognitive dysfunction. Additionally, the combination of Linc-POU3F3 and α-syn in L1CAM exosomes and GCase activity could discriminate PD patients from controls. These results suggest that L1CAM exosomal Linc-POU3F3, L1CAM exosomal α-syn, and GCase activity may shed light on the mechanism underlying the autophagic-lysosomal system in the pathogenesis of PD and could be used to assess the severity of PD.
Electronic supplementary material
The online version of this article (10.1007/s13311-020-00842-5) contains supplementary material, which is available to authorized users.
Key words: Autophagic-lysosomal pathway, α-synuclein, L1CAM exosomes, Linc-POU3F3, Parkinson’s disease
Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, and disease-specific pathology will occur prior to its onset [1]. Imperative for the diagnosis of PD and for monitoring disease severity is the timely identification of potential biomarkers, which are currently lacking.
Mechanistically, α-synuclein (α-syn) plays a crucial role during PD pathogenesis [2, 3]. The latest study reported that α-syn is readily secreted out of cells and mostly exists in plasma, cerebrospinal fluid (CSF), and saliva [4, 5]. Although the mechanisms of α-syn secretion remain unknown, some reports suggest that exosomes are at least partially involved [5, 6].
Previous studies have shown that α-syn in neuronal-derived blood exosomes containing L1 cell adhesion molecules (L1CAM) is crucial for the pathogenesis and progression of synucleinopathies [4, 6], and L1CAM-containing exosomes isolated from human blood can be verified by fluorescence nanoparticle tracking analysis (NTA). Quanterix Simoa is a more sensitive methodology than traditional ELISA or Luminex to measure total plasma α-syn and L1CAM exosomal tau levels [7–9] but has yet to be used to examine L1CAM-exosomal α-syn levels. Moreover, increased secretion of exosomes has been associated with lysosomal dysfunction [10, 11]. The lysosomal enzyme β-glucocerebrosidase (GCase), encoded by the GBA1 gene, has been reported to be correlated with α-syn proteostasis and plays a crucial role in PD pathogenesis [12]. However, the association of L1CAM exosomal α-syn levels with plasma GCase activity and the severity of disease in PD patients has not been systemically investigated.
Long noncoding RNAs (lncRNAs) are noncoding RNAs with a length exceeding 200 nucleotides, which could regulate gene expression at epigenetic transcriptional and post-transcriptional levels [13]. Evidence suggests that the disordered regulation of lncRNAs is involved in the occurrence of human diseases, especially for nervous system diseases [14, 15]. Previous studies have proved the existence and stability of exosomal lncRNA in plasma [16, 17], which has been an emerging field for noninvasive diagnostic applications [17]. In this study, we screened LncRNAs in plasma L1CAM exosomes by Arraystar Human LncRNA array v3.0. Finally, we selected the Linc-POU3F3 as the candidate lncRNA. Recent studies have found that Linc-POU3F3 also plays a role in central nervous system (CNS) functions [18] as well as autophagic lysosome pathway [19]. In a previous study, Lang HL et al. provided proof of the existence and stability of exosomal Linc-POU3F3 in serum and reported its clinical significance [20]. However, the expression of L1CAM exosomal Linc-POU3F3 in the plasma of PD patients and the relationship of Linc-POU3F3 with α-syn in L1CAM exosomes and GCase activity have not been studied.
Therefore, investigating the levels of Linc-POU3F3 and α-syn in plasma L1CAM exosomes and GCase activity in PD patients would provide valuable insights. The primary aim of this study was to analyze the levels of Linc-POU3F3 and α-syn in plasma L1CAM exosomes as well as GCase activity for potential associations with PD and their suitability as biomarkers for PD. The secondary aim was to evaluate whether L1CAM exosomal Linc-POU3F3, L1CAM exosomal α-syn and GCase activity are associated with motor/nonmotor dysfunctions in PD. Third, we aimed to elucidate the relationship among plasma L1CAM exosomal Linc-POU3F3, L1CAM exosomal α-syn, and GCase activity in PD patients. Finally, we explored the diagnostic value of plasma L1CAM exosomal Linc-POU3F3, L1CAM exosomal α-syn, and GCase activity in PD patients.
Methods
Patients and Ethics Statement
A total of 85 healthy controls and 93 PD patients were recruited from the First Affiliated Hospital of Jinan University and the Third Affiliated Hospital of Sun Yat-sen University from November 2017 to March 2019 for this study. The diagnosis of PD was made according to the UK Parkinson’s Disease Society Brain Bank Criteria [21–23]. PD patients with H-Y stages 1 to 2 were included as early-stage patients, and those with H-Y stages 3 to 5 as advanced stage patients. This study was approved by the Local Ethics Committee of the First Affiliated Hospital of Jinan University and the Third Affiliated Hospital of Sun Yat-sen University. The exclusion criteria were as follows: 1) essential tremors, secondary parkinsonism, or Parkinson-plus syndrome; 2) severe craniocerebral injury; 3) tumor; 4) infectious, inflammatory, or autoimmune diseases of the peripheral and central nervous systems; 5) severe systemic diseases, such as anemia, liver disease, heart failure, lung disease, and chronic renal failure; and 6) other neurodegenerative diseases. We conducted this study in accordance with the principles outlined in the Helsinki declaration and the National Institutes of Health Human Subjects Policies and Guidance issued in January 1999 and on December 23, 1999, respectively. Written consent was provided for the investigation for all participants, who consented to have their plasma L1CAM exosomal Linc-POU3F3, plasma L1CAM exosomal α-syn and GCase activity levels measured. PD patients also completed additional disease-specific standard measurements in a blinded manner.
Blood Specimen Processing
Blood was collected in EDTA tubes to inhibit clotting, and plasma was separated within 2 h after blood collection. Blood cells were removed by centrifugation at 1000g for 20 min, and platelets and cell fragments were removed by centrifugation at 1600g for 20 min to prepare platelet-poor plasma. Blood samples were not exposed to excessive heat or cold to prevent the rupture of red blood cells. Hemolysis was evaluated quantitatively by free hemoglobin, and hemolytic samples were excluded to avoid bias before analysis; the plasma was stored at − 80 °C. When preparing peripheral blood lymphocytes (PBLs), we removed the plasma layer and then reconstituted the original volume with phosphate-buffered saline (PBS). The samples were then stratified with Histopaque-1077 (Sigma) and centrifuged at 800g for 15 min. The upper layer was removed, the PBL band was transferred to another tube, and PBS was added to reconstitute the original volume. PBL pellets were collected and stored at − 80 °C after discarding the supernatant.
Exosome Isolation
Exosomes containing L1CAM, possibly derived from the CNS, were isolated from human plasma using antibody-coated superparamagnetic microbeads according to previously published protocols [7, 24–26]. Briefly, 10 g of an anti-L1CAM antibody (Abcam, Cambridge, UK) (or a control of normal IgG (Santa Cruz Biotechnology, TX, USA)) was placed onto M-270 epoxy beads using a Dynabeads® Antibody Ligation Kit in accordance with the manufacturer’s instructions (Life Technologies, CA, USA). After rapid thawing at 37 °C, plasma samples (human > 300 μL) were sequentially centrifuged for 15 min at 2000g and for 30 min at 12,000g, and the supernatant was then diluted with PBS (pH 7.4) at 1:3. Single-antibody-coated magnetic beads and substitute plasma (Human plasma 900 L) were incubated for approximately 24 h at 4 °C under gentle rotation. The beads were then washed with 1 mL of 0.1% bovine serum albumin (BSA)/PBS three times and transferred to a new tube. Exosomes were eluted from the beads, and the beads were incubated in Triton X-100 with protease inhibitor mixture (Sigma, MO, USA) at room temperature for 1 h before immunoassay.
Transmission Electron Microscopy
Isolated exosomes were mixed with 4% paraformaldehyde and 5% glutaraldehyde and then were absorbed on formvar/carbon-coated 300 mesh copper grids. Then dry them for 15 min at room temperature. After 2 min, grids were stained with 2% uranyl acetate for 20 min at room temperature. Samples were imaged on the FEI Tecnai G2 F20 X-TWIN (Thermo, CA, USA) transmission electron microscope and digital images were recorded by slowscan CCD Camera (Proscan, Scheuring, Germany).
Western Blot Analysis
The ProteinSimple capillary immunoassay (Wes) method was used for Western blot assay following the manuscript’s protocol and previous studies [27, 28]. The Exosome pellets were resuspended in lysis buffer containing diluted phosphatase and protease inhibitors. Protein concentration was measured by BCA assay (Pierce, Inc., IL, USA). Then the protein samples were mixed with the Master mix buffer from 12-230KDa Mouse Master kit (NO. PS-ST01-8, ProteinSimple, CA, USA), and then the Biotinylated Ladder was added to denature protein at 95 °C for 5 min. The protein samples were loaded on the pre-made gel from Wes 12-230 kDa Master kit with split Buffer (NO. SM-W002, ProteinSimple, CA, USA). Place the sample plate into Proteinsimple Wes instrument (NO. Wes S/N: WS-2494, ProteinSimple, CA, USA). For the information of parameters in the Assay program and primary antibodies, please refer to the Supplementary File.
Mass Spectrometry Analysis
Exosomes containing L1CAM were collected from approximately 3 mL of human plasma (negative control using normal mouse IgG) and lysed in 0.1% trifluoroacetic acid (TFA) without Triton X-100. Exosomal proteins were then digested with trypsin as previously described [29, 30]. We then analyzed peptides using a Q-Exactive Plus mass spectrometer (Thermo Scientific, Rockford, IL, USA) with data-dependent acquisition (DDA) or parallel reaction monitoring (PRM) targeting human α-syn peptides. We used C18 MicroSpin columns to clean up the digested samples in accordance with the manufacturer’s protocol. We performed liquid chromatography using NanoAcquity UPLC (Milford, MA, USA) and separated peptides with fused silica columns with a 2–80% acetonitrile/water gradient containing 0.1% formic acid. In the DDA approach, we performed a full mass spectrometry (MS) scan at 70,000 resolution, followed by a 20 MS/MS scan at 17,500 resolution, and the dynamic exclusion was set to 30 s. In the PRM method, 41 ion-mass α-syn peptides were targeted with an MS/MS scan resolution of 17,500. MS/MS spectral data were then converted to the MGF format, and MASCOT was used to search the UniProt Human database for the identification of peptides and proteins.
Fluorescence NTA
Platelet-free plasma from PD patients and healthy controls was removed in two centrifugation procedures (2000×g for 10 min, followed by 12,000×g for 25 min). Exosomes containing L1CAM were isolated from combined plasma samples using immunocapture technology, collected in 0.1 M glycine-HCl and neutralized with 1 M phosphate buffer. For penetration analysis, exosomes were fixed for 1 h in 0.25% paraformaldehyde/PBS at 4 °C and permeated for 15 min with 0.2% Tween-20/PBS at 37 °C. The samples were then washed in 1 mL of PBS (pH 7.4), centrifuged at hypervelocity (150,000g for 60 min) to precipitate exosomes, and resuspended in PBS. The permeated sample was then mixed with an anti-α-syn antibody (Thermo Fisher Scientific, Rockford, IL, USA) or a mouse IgG1 isotype control (EBioscience, San Diego, CA, USA) labeled with Alexa Fluor 488 according to the manufacturer’s instructions. For nonpermeabilization analysis, exosomes were labeled directly with Alexa Fluor 488-conjugated antibodies. We used 0.2% Tween-20/PBS (permeable samples) or PBS (nonpermeable samples) to wash immune-labeled samples and then resuspended the samples in PBS for fluorescence NTA analysis.
Microarray Analysis of lncRNAs
ExoRNeasy Midi Kit (Qiagen, Valencia, CA, USA) was used to extract plasma L1CAM exosomal RNA from plasma according to the manufacturer’s protocol. We then used NanoDrop ND-1000 (Thermo, CA, USA) to quantify the plasma L1CAM exosomal RNA of each sample and Agilent array platform for microarray analysis. Sample preparation and microarray hybridization were performed in accordance with the manufacturer’s instructions. And then, each sample was amplified and transcribed into fluorescence cRNA with entire transcription length by using random priming method. The labeled cRNAs were hybridized onto Human LncRNA array v3.0 (8 × 60 K, Arraystar). Then the arrays were scanned by Agilent-G2505C-Microarray Scanner (Agilent, California, USA).
RNA Isolation and Real-Time Quantitative PCR (QRT-PCR)
We used an exoRNeasy Midi Kit (Qiagen, Valencia, CA, USA) to extract plasma L1CAM exosomal RNA from plasma according to the manufacturer’s protocol. Next, cDNA was synthesized using a cDNA reverse transcription kit (Roche, Basel, Switzerland), and qRT-PCR was performed using the SYBR Green analysis method (Roche, Basel, Switzerland). LncRNA expression levels were standardized to those of GAPDH, and the 2-ΔCT method was used to calculate the relative expression of lncRNAs. The primer sequences are shown in Supplementary Table 1.
GCase Activity Assay
According to a previously described method [31], the enzymatic activity of GCase in PBLs was determined using the synthetic fluorescent substrate 4-methyl umbel-b-d-pyrans glucoside (4-MUG). PBL precipitates were lysed on ice in CellLytic lysis buffer (Sigma) for 30 min. We prepared and added the reaction mixture (0.15 M citrate/phosphate buffer, sodium taurocholate, and 1 nM 4-MUG) to each sample. A solution of 1 nM 4-methyl umbel ketone was added, and the reaction was terminated with 1 M glycine. Each sample was measured five times independently. Fluorescence was measured using a microplate reader. The final results were reported as nmol of substrate per microgram per hour.
Statistical Analyses
All continuous variables, including age, Unified Parkinson’s Disease Rating Scale (UPDRS) scores, Mini-Mental State Examination (MMSE) scores, Non-Motor Symptoms Scale (NMSS) scores, plasma L1CAM exosomal Linc-POU3F3 levels, plasma L1CAM exosomal α-syn levels, and GCase activity, are presented as the mean ± SD. The Mann–Whitney U test and Kruskal–Wallis test were used to evaluate the statistical significance of differences between the two groups when the data were not normally distributed. When the data were normally distributed, Student’s t test was used. One-way ANOVA followed by Tukey post hoc analysis was performed to compare L1CAM exosomal Linc-POU3F3, L1CAM exosomal α-syn, and GCase activity among normal subjects and PD patients, including subgroups based on UPDRS-III distribution. We assessed the correlations among different clinical parameters using Spearman’s rank correlation coefficients (rs). We performed receiver operating characteristic (ROC) analysis to assess the diagnostic criteria for PD to evaluate the performance of the clinical biomarkers. In addition, ROC curves were calculated for the combination of plasma L1CAM exosomal Linc-POU3F3, plasma L1CAM exosomal α-syn, and GCase activity as a potentially better prognostic tool. P < 0.05 was considered significant, and SPSS 13.0 was used for all statistical analyses (Chicago, Illinois, USA).
Results
Patient Characteristics
We enrolled 93 PD patients (53 males and 40 females) and 85 healthy subjects (48 males and 37 females) in this cross-sectional study. The mean years of age of the PD patients and healthy subjects were 66.93 ± 9.52 and 66.22 ± 10.30, respectively (Table 1). There was no significant difference in the age of PD patients and healthy subjects (Table 1). Then, PD patients were divided into two subgroups according to their H-Y stage. Fifty-one patients with H-Y stages 1 to 2 were included as early-stage patients, and the rest with H-Y stages 3 to 5 as advanced stage patients. When compared to PD patients in the early stage, advanced stage PD patients had higher UPDRS total (Table 1) and UPDRS-III scores (Table 1) and lower MMSE scores (Table 1). PD patients in the advanced stage also had a higher daily dose of L-dopa and longer disease duration than their early-stage counterparts. The demographic and clinical characteristics of all subjects are displayed in Table 1.
Table 1.
Compare of demographics and biomarkers between control subjects and PD
| PD | |||||||
|---|---|---|---|---|---|---|---|
| Control | Early | Advanced | Total | P1 | P2 | ||
| Number of cases | 85 | 51 | 42 | 93 | / | / | |
| Gender (n) | Male | 48 | 30 | 25 | 53 | 0.165 | 0.209 |
| Female | 37 | 22 | 18 | 40 | 0.213 | 0.227 | |
| Age (years) | 66.22 (10.30) | 64.71 (10.60) | 67.55 (8.15) | 66.93 (9.52) | 0.754 | 0.821 | |
| H&Y | / | 1.5 (0.5) | 3.0 (0.5) | 2.8 (0.5) | / | < 0.001 a*** | |
| MMSE | / | 28.72 (2.64) | 21.95 (2.57) | 24.28 (2.88) | < 0.001 b*** | ||
| UPDRS | / | 38.29 (20.21) | 54.7 (18.59) | 49.23 (18.13) | < 0.001a*** | ||
| UPDRS (I) | / | 3.35 (0.93) | 3.70 (0.86) | 3.52 (0.98) | 0.172 | ||
| UPDRS (II) | / | 11.28 (8.51) | 17.33 (9.29) | 15.17 (8.63) | 0.036b* | ||
| UPDRS (III) | / | 22.02 (18.53) | 31.41 (15.27) | 28.73 (16.04) | < 0.001a*** | ||
| UPDRS (IV) | / | 1.84 (2.50) | 2.26 (1.39) | 2.03 (1.84) | < 0.001a*** | ||
| NMSS (total) | / | ||||||
| Cardiovascular | 4.13 (2.35) | 4.60 (2.09) | 4.37 (2.81) | 0.063 | |||
| Sleep/fatigue | 16.25 (8.18) | 18.19 (8.18) | 17.03 (9.34) | 0.058 | |||
| Mood | 17.72 (13.72) | 19.94 (12.26) | 18.78 (10.65) | 0.055 | |||
| Perceptual problem | 2.86 (5.54) | 3.14 (4.20) | 3.05 (5.03) | 0.064 | |||
| Attention/memory | 10.16 (8.09) | 13.31 (9.36) | 12.66 (8.81) | 0.052 | |||
| Gastrointestina | 9.24 (6.33) | 10.61 (7.75) | 9.76 (7.52) | 0.881 | |||
| Urinar | 8.29 (8.46) | 9.35 (6.25) | 8.87 (6.60) | 0.923 | |||
| Sexual function | 5.44 (5.12) | 5.65 (6.23) | 5.61 (6.83) | 1.65 | |||
| Miscellaneous | 8.06 (9.03) | 9.38 (8.41) | 9.23 (8.49) | 0.058 | |||
| Daily dose of L-Dopa (mg) | 252 (58.52) | 398 (47.88) | 280 (51.76) | < 0.001b*** | |||
| Disease duration | 2.05 (3.40) | 5.33 (3.15) | 4.26 (2.52) | 0.003b** | |||
| L1CAM exosomal Linc-POU3F3 | 1.54 (0.59) | 2.24 (0.85) | 2.93 (0.63) | 2.61 (0.68) | 0.020a* | 0.041b* | |
| L1CAM exosomal α-syn (pg/ml) | 6.07 (1.35) | 7.19 (2.97) | 7.84 (2.46) | 7.62 (2.55) | 0.026a* | 0.033b* | |
| α-syn (ng/ml) | 3.01 (1.67) | 3.32 (1.83) | 3.51 (1.56) | 3.43 (1.61) | 0.723 | 0.693 | |
| Exo/total α-syn | 2.26 (1.52) | 3.01 (1.19) | 3.75 (1.36) | 3.30 (1.28) | 0.029a* | 0.040a* | |
| GCase activity (nmol/mg) | 10.43 (2.26) | 9.65 (2.79) | 8.06 (1.88) | 8.36 (2.05) | 0.038a* | 0.044b* | |
α-syn, α-synuclein; aMann–Whitney U test. bStudent’s t test; P1, compare between control subjects and PD; P2, compare between early stage PD and advanced stage PD. *P < 0.05, **P < 0.01, ***P < 0.001
Characterization of Exosomes Likely Derived from CNS in Human Plasma
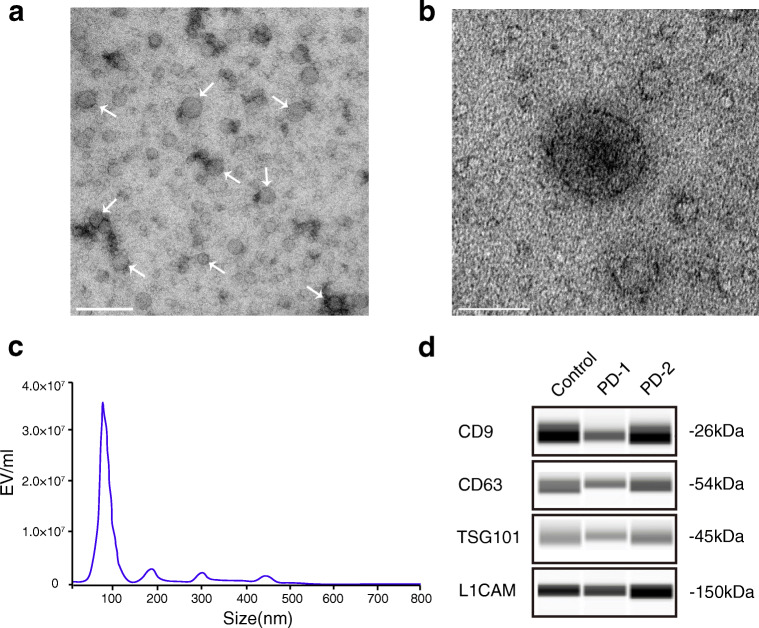
Exosomes isolated from the plasma of healthy subjects and patients were evaluated by TEM, Western blot and Nanosight to confirm the reliability of the isolation protocol and the purity of the exosome preparations. The ultracentrifugation pellet contained 40–120 nm membrane-bound vesicles (Fig. 1A, B), which displayed the neuronal-derived exosomal marker L1CAM and other general exosomal markers (CD9, CD63 TSG101) (Fig. 1D, Supplementary Figure 2). L1CAM is the founding member of a subfamily of cell adhesion molecules that are primarily expressed in the nervous system and is believed to be a surface marker of CNS-derived exosomes [7, 32, 33]. No differences in the expression of exosomal markers were found between PD patients and healthy subjects. Furthermore, NTA showed a particle size distribution ranging in diameter from 50 to 300 nm, with most particles detected in the size range consistent with exosome dimensions (Fig. 1C).
Fig. 1.
Characterization of exosomes likely derived from CNS in human plasma. (A, B) Representative transmission electron microscopy (TEM) images of isolated exosomes. Scale bar: (A) 200 nm and (B) 100 nm. (C) Size distribution and concentration of isolated L1CAM exosomes evaluated by NanoSight. (D) The neuronal-derived exosomal markers L1CAM and other general exosomal markers (CD9, CD63 TSG101) expression in neuronal-derived blood exosomes as evaluated with Western blot
Aberrant Expression of lncRNAs in L1CAM Exosomes from PD Patients and Controls
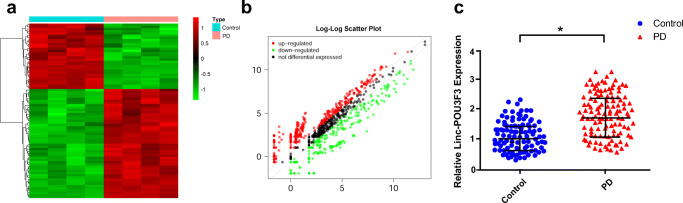
To determine whether lncRNAs were expressed in L1CAM exosomes in PD patients and controls, we applied a lncRNA microarray to detect the expression levels of various lncRNAs in L1CAM exosomes from the study subjects. As presented in Fig. 2A and B, significantly different expression profiles were observed between the two groups, which indicated that lncRNAs might be expressed in L1CAM exosomes among PD patients. We further ranked the differentially expressed lncRNAs according to both their fold change and their detection density, representing the detection abilities of the lncRNAs. Linc-POU3F3 presented the highest fold change value and the most stable detection density and was therefore selected as a candidate lncRNA. We then detected the expression level of Linc-POU3F3 in the larger samples mentioned above. QRT-PCR confirmed the upregulation of Linc-POU3F3 in plasma L1CAM exosomes in PD patients compared with healthy controls (Table 1 and Fig. 2C). In addition, we treated the plasma samples with prolonged room temperature exposure or several freeze-thaw cycles to explore the stability of plasma L1CAM exosomal Linc-POU3F3 in plasma, and the results showed that the treatment exerted no influence on the expression level of L1CAM exosomal Linc-POU3F3 (Supplementary Figure 1).
Fig. 2.
Aberrant expression of lncRNAs in L1CAM exosomes in PD patients and control subjects. (A, B) Different expression level of lncRNAs in L1CAM exosomes detected by lncRNA microarray presented as (A) heatmap and (B) scatter diagram (fold change > 2). (C) Upregulated Linc-POU3F3 in L1CAM exosomes was confirmed by qRT-PCR in 93 PD patients and 85 control subjects. *P < 0.05 by using the Mann–Whitney U test. The horizontal line and error bars represent the mean ± standard error
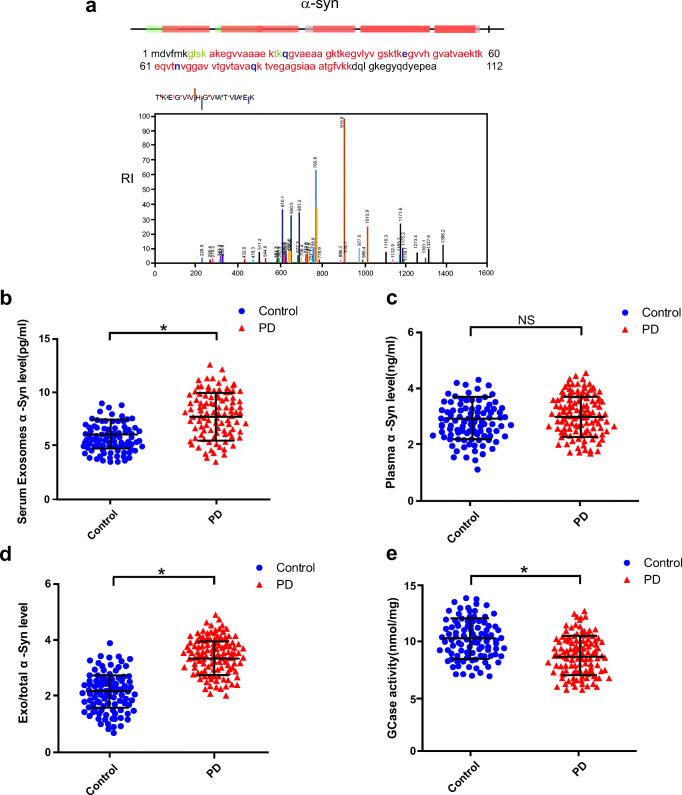
Evaluation of α-Syn in Plasma L1CAM Exosomes and GCase Activity in PD Patients and Healthy Subjects
We applied MS analysis and Quanterix Simoa to identify α-syn in L1CAM-containing exosomes isolated from human blood plasma (Fig. 3A), and used ELISAs to measure the plasma concentrations of total α-syn. The L1CAM exosomal α-syn concentration was significantly increased in PD patients compared with healthy controls (Table 1, Fig. 3B), but no significant difference in plasma α-syn was observed between PD patients and healthy controls (Table 1, Fig. 3C). The ratio of exosomal α-syn to total α-syn (exo/total) was also significantly higher in patients with PD than in the controls (Table 1, Fig. 3D). GCase activity was significantly decreased in PD patients compared to healthy controls (Table 1, Fig. 3E). The increased Linc-POU3F3 and α-syn levels in L1CAM exosomes, upregulated exo/total α-syn ratio and decreased GCase activity were especially prominent in male PD patients when compared with male healthy subjects (Supplementary Table 2).
Fig. 3.
Plasma L1CAM exosomal α-syn and GCase activity in PD patients and control subjects. (A) Mass spectrometry identification of α-syn in L1CAM-containing exosomes isolated from human blood plasma. (B) Plasma L1CAM exosomal α-syn concentrations were measured using Quanterix Simoa. (C) Plasma α-syn level. (D) Exosomal/total α-syn ratio. (E) GCase activity level in PD patients and control subjects; *P < 0.05 by using the Mann–Whitney U test. The horizontal line and error bars represent the mean ± standard error
Comparisons of Linc-POU3F3 and α-Syn Levels in Plasma L1CAM Exosomes and GCase Activity Among PD Patients According to PD Severity
For the evaluation of PD severity, PD patients were divided into two groups according to H-Y stage. The Linc-POU3F3 and α-syn levels in the L1CAM exosomes and the exo/total α-syn levels were upregulated in PD patients in the advanced stage compared with those in the early-stage (Table 1). Interestingly, GCase activity was downregulated in PD patients in the advanced stage compared with those in the early stage (Table 1).
Comparison of Linc-POU3F3 and α-Syn Levels in Plasma L1CAM Exosomes and GCase Activity among PD Patients According to Motor Severity
To evaluate motor symptom severity in PD, we classified PD patients into three subgroups according to UPDRS-III scores. The plasma L1CAM exosomal Linc-POU3F3/α-syn and exo/total α-syn levels were significantly higher in PD patients in the 31 < UPDRS-III < 50 subgroup than in the UPDRS-III ≤ 30 subgroup (Table 2). Similarly, significant increases were observed in plasma L1CAM exosomal Linc-POU3F3/α-syn and exo/total α-syn levels in PD patients in the UPDRS-III ≥ 50 subgroup when compared with the UPDRS-III ≤ 30 subgroup (Table 2). However, GCase activity was significantly downregulated in PD patients in the UPDRS-III ≥ 50 subgroup compared to the UPDRS-III ≤ 30 subgroup (Table 2).
Table 2.
Comparison of plasma L1CAM exosomal Linc-POU3F3, plasma L1CAM exosomal α-syn, and GCase activity between normal subjects and PD patients according to UPDRS (III) score
| Variable | PD UPDRS(III) ≤ 30 mean ± SD |
PD 31 < UPDRS(III) < 50 mean ± SD |
PD UPDRS(III) ≥ 50 mean ± SD |
P1 | P2 | P3 |
|---|---|---|---|---|---|---|
| L1CAM exosomal Linc-POU3F3 | 2.11 ± 0.62 | 2.62 ± 0.59 | 2.80 ± 0.85 | 0.025* | 0.91 | 0.019* |
| L1CAM exosomal α-syn (pg/ml) | 7.03 ± 1.46 | 7.87 ± 2.27 | 8.01 ± 1.09 | 0.014* | 0.083 | 0.008** |
| Plasma α-syn (ng/ml) | 3.32 ± 1.44 | 3.54 ± 0.95 | 3.61 ± 0.95 | 0.72 | 0.96 | 0.93 |
| Exo/total α-syn | 3.02 ± 1.19 | 3.75 ± 1.25 | 3.90 ± 1.77 | 0.021* | 0.65 | 0.017* |
| GCase activity (nmol/mg) | 9.54 ± 2.31 | 8.98 ± 2.17 | 8.13 ± 2.09 | 0.057 | 0.78 | 0.012* |
α-syn, α-synuclein; Kruskal-Wallis test for the comparison among three UPDRS(III) subgroups, Tukey’s post hoc analysis for the comparison in UPDRS(III) ≤ 30 vs. 31 < UPDRS(III) < 50 subgroup, 31 < UPDRS(III) < 50 vs. UPDRS(III) ≥ 50 subgroup, or UPDRS(III) ≤ 30 vs. UPDRS(III) ≥50 subgroup; P1, compare between UPDRS(III) ≤ 30 and 31 < UPDRS(III) < 50 subgroup; P2, compare between 31 < UPDRS(III) < 50 and UPDRS(III) ≥ 50 subgroup; P3, compare between UPDRS(III) ≤ 30 and UPDRS(III) ≥50 subgroup; *P < 0.05, **P < 0.01
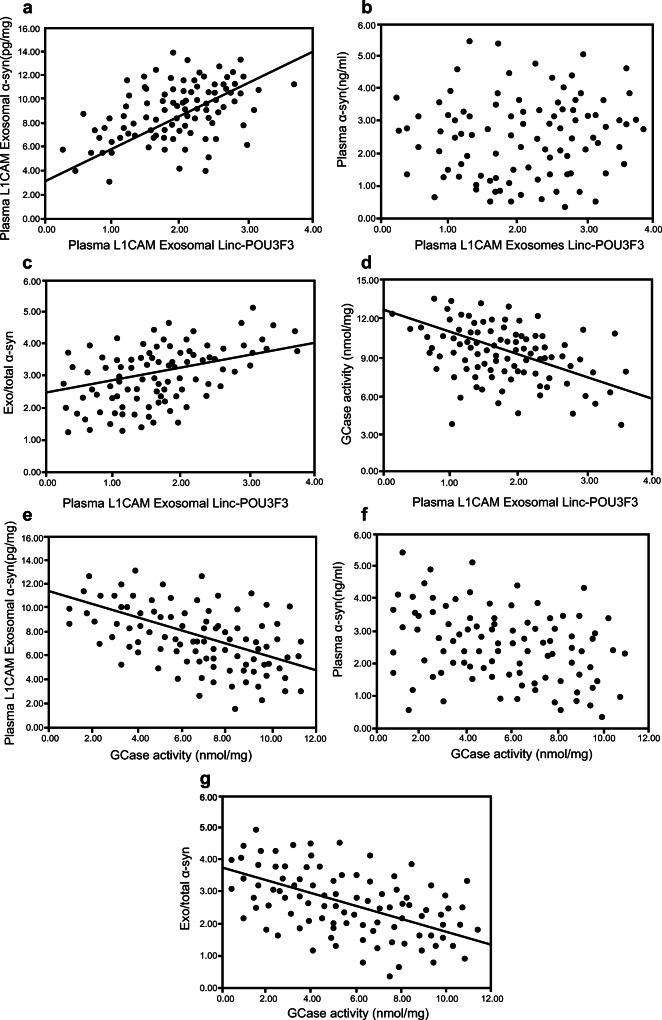
Correlation Analyses Between Plasma L1CAM Exosomal Linc-POU3F3 and L1CAM Exosomal α-Syn Levels in PD Patients
A profound positive correlation was observed between plasma L1CAM exosomal Linc-POU3F3 and exosomal α-syn levels in PD patients (r = 0.658, P = 0.009, Table 3, Fig. 4A). Similarly, a significant positive correlation between plasma L1CAM exosomal Linc-POU3F3 and exo/total α-syn levels was observed in PD patients (r = 0.429, P = 0.016, Table 3, Fig. 4C). However, no association was found between plasma L1CAM exosomal Linc-POU3F3 and plasma α-syn levels (r = 0.141, P = 0.521, Table 3, Fig. 4B). Strikingly, a significant negative correlation was observed between the plasma L1CAM exosomal Linc-POU3F3 levels and GCase activity in PD patients (r = −0.823, P = 0.005, Table 3, Fig. 4D).
Table 3.
Spearman’s rank correlation coefficient (rs) and P values between plasma L1CAM exosomal Linc-POU3F3, plasma L1CAM exosomal α-syn, and GCase activity in PD patients
| Variable | PD | |
|---|---|---|
| Plasma L1CAM exosomal Linc-POU3F3 | ||
| r | P | |
| L1CAM exosomal α-syn (pg/ml) | 0.658** | 0.009 |
| α-syn (ng/ml) | 0.141 | 0.521 |
| Exo/total α-syn | 0.429* | 0.016 |
| GCase activity (nmol/mg) | −0.823** | 0.005 |
α-syn,α-synuclein; *P < 0.05,**P < 0.01, rs, Spearman’s rank correlation coefficient
Fig. 4.
Correlation analysis between L1CAM exosomal Linc-POU3F3, L1CAM exosomal α-syn, exosomal/total α-syn ratio and GCase activity in PD patients. (A, C, D) A significant correlation between (A) L1CAM exosomal Linc-POU3F3 and L1CAM exosomal α-syn in PD patients (rs = 0.658, P = 0.009); (C) L1CAM exosomal Linc-POU3F3 and exosomal/total α-syn ratio in PD patients (rs = 0.429, P = 0.016). (D) L1CAM exosomal Linc-POU3F3 and GCase activity in PD patients (rs = − 0.823, P = 0.005). (B) No significant correlation between L1CAM exosomal Linc-POU3F3 and plasma α-syn in PD patients (rs = 0.141, P = 0.521). (E, G) A significant negative correlation between e GCase activity and L1CAM exosomal α-syn in PD patients (rs = − 0.553, P = 0.011). (G) GCase activity and exosomal/total α-syn ratio in PD patients (rs = − 0.636, P = 0.008). (F) No significant correlation between GCase activity and plasma α-syn in PD patients (rs = − 0.072, P = 0.710)
Correlation Analyses Between GCase Activity and Plasma L1CAM Exosomal α-Syn Levels in PD Patients
A profound negative correlation was observed between GCase activity and plasma L1CAM exosomal α-syn levels in PD patients (r = − 0.553, P = 0.011, Table 4, Fig. 4E). Similarly, a significant negative correlation was observed between the GCase activity and exo/total α-syn levels in PD patients (r = − 0.636, P = 0.008, Table 4, Fig. 4G). No association was found between GCase activity and plasma α-syn levels (r = − 0.072, P = 0.710, Table 4, Fig. 4F).
Table 4.
Spearman’s rank correlation coefficient (rs) and P values between GCase activity and plasma L1CAM exosomal α-syn levels in PD patients
| Variable | PD | |
|---|---|---|
| GCase activity | ||
| r | P | |
| L1CAM exosomal α-syn (pg/ml) | − 0.553* | 0.011 |
| α-syn (ng/ml) | − 0.072 | 0.710 |
| Exo/total α-syn | − 0.636** | 0.008 |
α-syn, α-synuclein; *P < 0.05,**P < 0.01, rs, Spearman’s rank correlation coefficient
Correlations Among L1CAM Exosomal Linc-POU3F3/α-Syn, Exo/Total α-Syn Level, GCase Activity and Age, UPDRS Scores, H-Y Stages, MMSE Scores and NMSS (Total/Domain) Scores in PD Patients
Positive correlations were observed between plasma L1CAM exosomal Linc-POU3F3 level and UPDRS, UPDRS-III and H-Y staging, with a negative correlation with MMSE scores in PD patients (Table 5). Positive correlations were observed between plasma L1CAM exosomal Linc-POU3F3 levels and nonmotor symptom (NMS) burdens of mood and attention/memory in PD patients (Table 5). In addition, plasma L1CAM exosomal α-syn and exo/total α-syn levels were positively correlated with UPDRS, UPDRS-III, and H-Y staging in PD patients (Table 5).
Table 5.
Spearman’s rank correlation coefficient (rs) and P values between clinical variables and H&Y, MMSE, NMSS (total/domain) in PD patients
| Variable | L1CAM exosomal Linc-POU3F3 | L1CAM exosomal α-syn | Plasma α-syn | Exo/total α-syn | GCase activity | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | P | r | P | r | P | r | P | r | P | |
| Age | 0.198 | 0.271 | 0.132 | 0.553 | 0.165 | 0.431 | 0.074 | 0.587 | 0.096 | 0.551 |
| UPDRS | 0.325 | 0.030 | 0.304 | 0.043 | 0.119 | 0.613 | 0.495 | 0.011 | − 0.338 | 0.027 |
| Up(I) | 0.146 | 0.409 | 0.176 | 0.252 | 0.272 | 0.236 | 0.101 | 0.588 | − 0.185 | 0.393 |
| Up(II) | 0.091 | 0.541 | 0.161 | 0.267 | 0.131 | 0.368 | 0.258 | 0.269 | − 0.056 | 0.684 |
| Up(III) | 0.453 | 0.016 | 0.384 | 0.023 | 0.197 | 0.275 | 0.486 | 0.015 | − 0.567 | 0.011 |
| Up(IV) | 0.134 | 0.395 | 0.040 | 0.715 | 0.035 | 0.746 | 0.206 | 0.266 | −0.114 | 0.617 |
| H&Y | 0.412** | 0.005 | 0.321* | 0.033 | 0.144 | 0.413 | 0.351 | 0.028 | − 0.369 | 0.028 |
| MMSE | − 0.335* | 0.027 | − 0.064 | 0.639 | − 0.050 | 0.661 | − 0.110 | 0.573 | 0.551* | 0.015 |
| NMSS | 0.155 | 0.398 | 0.093 | 0.578 | 0.065 | 0.681 | 0.086 | 0.614 | 0.044 | 0.709 |
| Cardiovascular | 0.075 | 0.618, | 0.059 | 0.655 | 0.201 | 0.278 | 0.094 | 0.138 | − 0.105 | 0.581 |
| Sleep/fatigue | 0.07, | 0.713 | 0.417 | 0.064 | 0.048 | 0.705 | 0.026 | 0.831 | − 0.078 | 0.702 |
| Mood | 0.430* | 0.021 | 0.067 | 0.681 | 0.091 | 0.592 | 0.043 | 0.712 | − 0.451** | 0.004 |
| Perceptual problem | 0.126 | 0.432 | 0.080 | 0.653 | 0.185 | 0.105 | 0.076 | 0.606 | − 0.153 | 0.464 |
| Attention/memory | 0.483* | 0.018 | 0.109 | 0.576 | 0.241 | 0.376 | 0.143 | 0.420 | − 0.418** | 0.005 |
| Gastrointestina | 0.035 | 0.739 | 0.121 | 0.551 | 0.162 | 0.387 | 0.069 | 0.660 | − 0.092 | 0.611 |
| Urinar | 0.122 | 0.443 | 0.090 | 0.581 | 0.028 | 0.186 | 0.123 | 0.548 | − 0.025 | 0.833 |
| Sexual function | 0.095 | 0.607 | 0.033 | 0.806 | 0.197 | 0.454 | 0.083 | 0.651 | − 0.117 | 0.615 |
| Miscellaneous | 0.012 | 0.895 | − 0.107 | 0.579 | − 0.101 | 0.485 | − 0.411 | 0.029 | − 0.144 | 0.420 |
| Daily dose of L-Dopa (mg) | 0.164 | 0.483 | 0.138 | 0.427 | 0.060 | 0.668 | 0.058 | 0.658 | − 0.081 | 0.65 |
rs, Spearman’s rank correlation coefficient; UPDRS, Unified Parkinson’s disease rating scale; H&Y, the modified Hoehn and Yahr staging scale; MMSE, mini-mental state examination; NMSS, non-motor symptoms scale for Parkinson’s disease; *P < 0.05, **P < 0.01
Additionally, we found that GCase activity was negatively correlated with UPDRS, UPDRS-III, and H-Y staging and positively correlated with MMSE scores in PD patients (Table 5). Negative correlations were observed between GCase activity and the NMS burdens of mood and attention/memory in PD patients (Table 5). We also discovered that L-dopa treatment was not obviously correlated with plasma L1CAM exosomal Linc-POU3F3/α-syn, plasma α-syn or exo/total α-syn levels (Table 5). These results strongly indicated that L-dopa medication did not have an effect on the validity of these indicator measurements in PD patients.
ROC Curves for Plasma L1CAM Exosomal Linc-POU3F3/α-Syn and Plasma α-Syn Levels and GCase Activity in the Diagnosis of PD
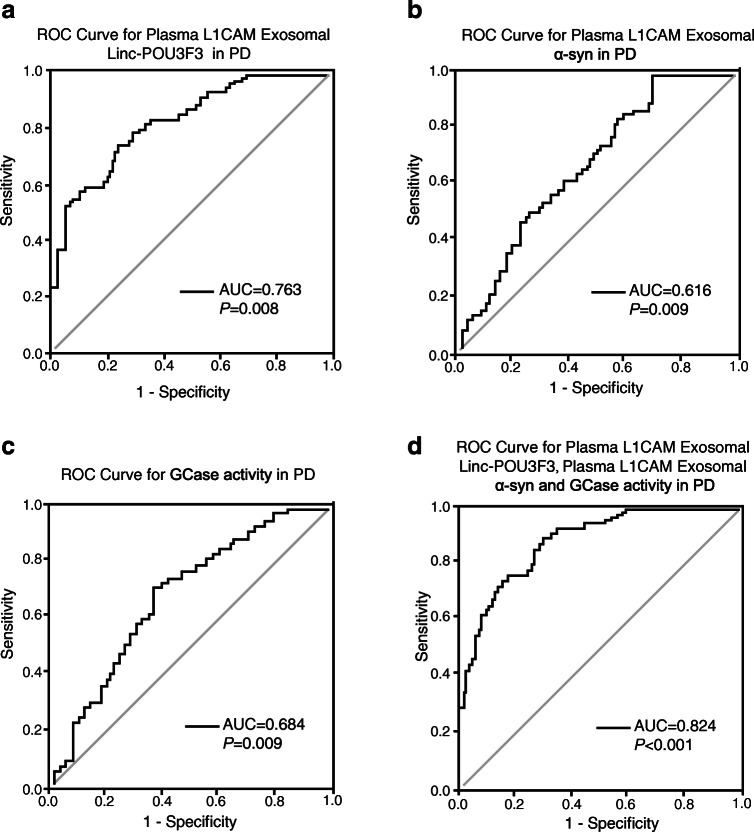
We conducted ROC curve analysis to detect whether plasma L1CAM exosomal Linc-POU3F3/α-syn, plasma α-syn, and GCase activity can credibly discriminate between PD patients and healthy subjects. ROC curves for plasma L1CAM exosomal Linc-POU3F3 showed that an area under the curve (AUC) value of 0.763 (Fig. 5A) was appropriate; the cutoff was 2.48, with a sensitivity of 68% and a specificity of 72%. The AUC for plasma L1CAM exosomal α-syn was 0.616 (Fig. 5B); the cutoff was 7.32 ng/mg, with a sensitivity of 55% and a specificity of 61%. In addition, the AUC for GCase activity was 0.684 (Fig. 5C); the cutoff was 8.85 nmol/mg, with a sensitivity of 64% and a specificity of 69%. Interestingly, when using the predicted risk algorithm, the AUC for the combination of plasma L1CAM exosomal Linc-POU3F3, plasma L1CAM exosomal α-syn and GCase activity was 0.824 (Fig. 5D), with a sensitivity of 71% and a specificity of 83% at a cutoff of 0.66. These results suggested that the combination variable was more credible than plasma L1CAM exosomal Linc-POU3F3, plasma L1CAM exosomal α-syn, or GCase activity alone in distinguishing PD patients from healthy subjects.
Fig. 5.
ROC curves to evaluate the utility of L1CAM exosomal Linc-POU3F3, L1CAM exosomal α-syn and GCase activity for the discrimination of PD patients from healthy controls. (A–D) The AUC of ROC curves for discrimination of PD patients from healthy controls (A) L1CAM exosomal Linc-POU3F3, (B) L1CAM exosomal α-syn, (C) GCase activity were 0.763 (95%CI: 0.650–0.809, P = 0.008), 0.616 (95%CI: 0.527–0.733, P = 0.009), and 0.684 (95%CI: 0.588–0.795, P = 0.009), respectively; The AUC of (D) L1CAM exosomal Linc-POU3F3+ L1CAM exosomal α-syn + GCase activity was 0.824 (95%CI: 0.739–0.915, P < 0.001)
Discussion
For the first time, we used NTA combined with QRT-PCR to measure L1CAM exosomal Linc-POU3F3 and NTA combined with Quanterix Simoa to measure L1CAM exosomal α-syn concentrations. This is also the first study to detect changes in Linc-POU3F3 and α-syn levels in L1CAM exosomes as well as GCase activity in PD patients and to assess the potential relationships between these biomarkers and the severity of PD. The interesting findings of the inverse correlation between Linc-POU3F3 and α-syn levels in plasma L1CAM exosomes and GCase activity may shed light on the mechanisms underlying PD pathogenesis.
Exosomes (50–150 nm in diameter), as cell-derived vesicles, are regarded as a new mode of cell-to-cell communication [34]. Exosomes also contain multiple cargos, including lipids, RNAs, proteins, and enzymes, and are present in all biological fluids; thus, they are believed to participate in multiple pathological conditions, including neurodegenerative diseases such as PD [35]. Exosomes provide a way for neurons to divert functionally important and disease-specific cargo from neurons into the CSF or peripheral blood through the blood–brain barrier (BBB) [36]. Furthermore, the double layer membrane of exosomes is an ideal shelter for protecting cargo from degradation, and the cargo stability is ideal for the use of noninvasive methods to diagnose neurodegenerative diseases [37]. NTA is a sensitive technique that can track the movement and size of individual vesicles and can provide an accurate count of extracellular vesicles (EVs) less than ~ 50 nm in diameter in biological fluids [38]. The purity of exosomal isolation and preparation is crucial. Our sample processing was optimized in accord with suggestions provided by the International Society for Extracellular Vesicles (ISEV) [39] and recent studies related to latest exosomes isolation and validation protocols and applications [24–26]. Plasma neural-derived exosomes (L1CAM-positive exosomes) are now regarded as potential sources from the CNS and could accurately reflect variations in the CNS [7, 8]. The L1CAM-containing exosomes isolated from the plasma of PD patients were verified by fluorescence NTA analysis. MS detection established α-syn in L1CAM-positive exosomes from human blood. Quanterix Simoa [40] is a more sensitive methodology than traditional ELISA or Luminex [41], and L1CAM-exosomal α-syn levels can thus be measured in the plasma range of sub pg/mL.
Exosomes are regarded as extracellular messengers that transfer lncRNAs from neurons into the blood circulation or CSF via the BBB [35]. In addition, exosomal lncRNAs are known as clinical biomarkers due to their stable characteristics in blood and their capacity to distinguish patients with disease from healthy subjects [42, 43]. A previous study provided proof of the existence and stability of exosomal Linc-POU3F3 in serum and reported its clinical significance. Linc-POU3F3 in A172 exosomes promotes human brain microvascular endothelial cell (HBMEC) migration, proliferation, and tubular-like structure formation both in vitro and in vivo [20]. However, the expression of Linc-POU3F3 in L1CAM-containing exosomes from PD blood samples has not yet been explored. In our study, the plasma L1CAM exosomal Linc-POU3F3 levels in patients with PD were significantly higher than those in healthy subjects, similar to findings from studies on other neurological diseases, such as glioma [18]. The upregulation of plasma L1CAM exosomal Linc-POU3F3 levels in PD patients compared with healthy controls suggests that Linc-POU3F3 may participate in the mechanisms underlying the pathophysiological process of PD. Moreover, plasma L1CAM exosomal Linc-POU3F3 levels were significantly higher in PD patients in the 31 < UPDRS-III < 50 and UPDRS-III > 50 subgroups than in the UPDRS-III ≤ 30 subgroup. Importantly, significant increases in the plasma L1CAM exosomal Linc-POU3F3 levels in PD patients were positively correlated with disease severity, as indexed by the H-Y score and motor dysfunction evaluated by the UPDRS-III score, even after considering confounder effects. These findings strongly imply that higher levels of L1CAM exosomal Linc-POU3F3 may contribute to the severity of PD and might involve in the pathophysiological process of PD. Therefore, L1CAM exosomal Linc-POU3F3 may be a potential useful diagnostic biomarker or/and therapeutic target in PD.
Plasma α-syn has been reported to be an inappropriate biomarker for PD diagnosis [44] due to considerations such as blood contamination in that erythrocyte-derived α-syn adulteration could affect the evaluation [45]. Recent studies found high α-syn levels in plasma neural-derived exosomes in PD patients by ELISA [46], similar to previous findings determined by Luminex assays [7]. However, there were still some limitations to these studies; for example, the sensitivity and specificity were not sufficiently high. In our study, plasma L1CAM exosomal α-syn levels were significantly upregulated in PD patients compared with healthy subjects as determined by fluorescence NTA analysis and Quanterix Simoa, which could indeed reflect α-syn content specifically derived from exosomes of neuronal origin among PD patients. Interestingly, we observed obvious upregulation of the plasma exosomal α-syn/total plasma α-syn ratio in PD patients, which may have contributed to equilibrium alterations between these forms of the α-syn protein. When referring to the H-Y stage subgroup of PD patients, the plasma L1CAM exosomal α-syn and exo/total α-syn levels were upregulated in PD patients in the advanced stage compared with those in the early stage. Moreover, the plasma L1CAM exosomal α-syn and exo/total α-syn levels were significantly higher in PD patients in the 31 < UPDRS-III < 50 and UPDRS-III > 50 subgroups than in the UPDRS-III ≤ 30 subgroup. Importantly, significant increases in the plasma L1CAM exosomal α-syn level and plasma exosomal α-syn/free α-syn ratio in PD patients were positively correlated with disease severity, as indexed by the H-Y score and motor dysfunction evaluated by the UPDRS-III score, even after considering confounder effects. This finding is in contrast to that reported in the study by Silvia Cerri et al., which showed that a significant increase in the plasma exosomal α-syn/free α-syn ratio in PD patients was negatively correlated with PD severity [47]. The possible reason for this discrepancy is that Silvia Cerri et al. detected the total pool of plasma exosomes and not merely neural-derived exosomes. Therefore, the exosomal α-syn levels in cells with different origins may have contributed to the different concentrations of exosomal α-syn. Similar to our study, Shi M et al. found that the plasma exosomal α-syn level was substantially higher in PD patients than in healthy controls and was significantly correlated with motor dysfunction and PD disease severity [7].
In our study, GCase activity was decreased significantly in PD patients, especially in those in the advanced stage and in the UPDRS-III > 50 subgroup, when compared to healthy controls. Moreover, GCase activity was negatively correlated with UPDRS, UPDRS-III and H-Y staging in PD patients. These results imply that lysosomal enzymes were increasingly dysfunctional as PD advances, which was correlated with manifested enhanced motor dysfunction. In a murine PD model study, inhibition of GCase activity in PD transgenic mice affected their motor function at late time points, and low levels of GCase activity correlated with the development of fine motor deficits [48]. Specifically, GCase activity was negatively associated with the NMS burdens of mood and attention/memory and positively associated with MMSE scores in PD patients, further demonstrating that attention and cognitive dysfunctions may be important targets for disease evaluation using GCase activity. These results are consistent with those of previous studies showing that GCase activity reduction is attributed to decreased hippocampal memory function in a PD transgenic mouse model [48]. However, other studies report discrepant findings. Han-Joon Kim et al. found that leukocyte GCase activity was not different in patients with sporadic PD (sPD), PD with PARK2 mutations, and SCA17 with parkinsonism from those of the controls [31]. Papagiannakis et al. also demonstrated that GCase protein levels were decreased in only the genetic PD groups, whereas increased mRNA levels and decreased activity were detected in only the GCase mutation group [49]. Silvia Cerri et al. also indicated that GCase activity in peripheral blood mononuclear cells was unchanged between PD patients without GBA1 mutations and healthy subjects [47]. Interestingly, Niimi Y et al. found that serum β-galactosidase activity was significantly higher in sPD patients without GBA1 mutations and parkinsonian syndrome patients than in the negative control group [50]. Some possible explanations for this discrepancy include 1) different races (85 healthy controls and 93 PD patients in the Chinese population were recruited in our study), 2) genetic factors, and 3) different sources of GCase activity in blood.
To our knowledge, our study is the first to correlate GCase activity with L1CAM exosomal lncRNA and L1CAM exosomal α-syn levels in blood. The autophagy-lysosomal system is crucial for α-syn degradation and PD pathogenesis [51, 52]. If this system is impaired, α-syn is released from cells via exosomes as a sort of compensation [53]. We first found that GCase activity was negatively correlated with plasma L1CAM exosomal α-syn levels among PD patients. The negative correlation supports the theory that GCase deficiency could lead to extracellular α-syn release by inhibiting the autophagy-lysosomal pathway [4, 54–56]. Additionally, Ting-Ting Du et al. indicated that GCase deficiency can inhibit autophagy by downregulating the expression of LC3-II (autophagosome marker), LAMP1 (lysosome marker), or the autophagy pathway component Beclin-1 [57]. However, the mechanism by which GCase deficiency regulates the expression of the abovementioned autophagic-related proteins remains unknown. Interestingly, we also discovered that decreased GCase activity levels were also negatively correlated with increased Linc-POU3F3 levels in exosomal L1CAM of PD patients. Previous study has shown that Linc-POU3F3 could also inhibit autophagy by downregulating LC3-II, ATG5 and Beclin 1 expression, which counteracts the effects of GCase activity [19]. We believe that GCase deficiency may decrease the expression of autophagy-related proteins by upregulating Linc-POU3F3 levels, subsequently inhibiting α-syn degradation and finally leading to compensatory release of α-syn from cells via exosomes. The excessive Linc-POU3F3 could also be released, accompanied by α-syn, via exosomes. Most interestingly, multiple lines of evidence show that the increased release of α-syn in exosomes can also increase α-syn transmission to recipient cells, which further promotes a-syn aggregation in neurons. Then, the increased α-syn levels will in turn inhibit GCase activity and exacerbate autophagic lysosome dysfunction in a self-perpetuating manner [58, 59]. Previous research has demonstrated that lncRNAs could also regulate α-syn transmission or aggregation as well as GCase activity [60]. It is also possible that Linc-POU3F3 in exosomes may mediate α-syn transmission or aggregation or even inhibit GCase activity together with α-syn in exosomes. Therefore, further investigation of the causal relationships and mechanisms underlying Linc-POU3F3 and α-syn levels in L1CAM exosomes and GCase activity in PD is warranted.
There are several limitations of this study: 1) it was small in size (85 healthy subjects and 93 PD patients); 2) longitudinal cohort studies will be necessary in the future to detect the three biomarkers regarding PD progression; and 3) genetic factors such as the GBA1 genotype were not considered. Therefore, longitudinal cohort studies with large populations is necessitated in the future.
In conclusion, for the first time, we used NTA combined with QRT-PCR to measure L1CAM exosomal Linc-POU3F3 concentrations and NTA combined with Quanterix Simoa to measure L1CAM exosomal α-syn concentrations. This is the first clinical study to explore the potential relationships between Linc-POU3F3 and α-syn levels in L1CAM exosomes and GCase activity in PD patients and to assess the potential relationships between these biomarkers and motor/nonmotor dysfunction and PD severity. These results suggest that the Linc-POU3F3 and α-syn levels in L1CAM exosomes and GCase activity may shed light on the mechanism underlying PD pathogenesis and could be used to assess the severity of PD.
Electronic Supplementary Material
(DOCX 389 kb)
(PDF 1225 kb)
Authors’ Contribution
JZ, YG, LW, and ADX designed the study, analyzed the data, and wrote the manuscript. LW, FY, BY, and ADX helped with data collection and analysis. LW and ADX review of the manuscript.
Funding Information
This study was supported by the Chinese Postdoctoral Science Foundation (2019M653293 to JZ) and the Postdoctoral Fund of the First Affiliated Hospital, Jinan University (809003 to JZ).
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no conflict of interest.
Ethical Approval
The Local Ethics Committee of the First Affiliated Hospital of Jinan University and the Third Affiliated Hospital of Sun Yat-sen University.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Zou and Yue Guo contributed equally to this work.
References
- 1.Conte A, Khan N, Defazio G, Rothwell JC, Berardelli A. Pathophysiology of somatosensory abnormalities in Parkinson disease. Nat Rev Neurol. 2013;9:687–697. doi: 10.1038/nrneurol.2013.224. [DOI] [PubMed] [Google Scholar]
- 2.Lesage S, Brice A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 3.Zhou H, Shao M, Guo B et al. Tetramethylpyrazine Analogue T-006 Promotes the Clearance of Alpha-synuclein by Enhancing Proteasome Activity in Parkinson’s Disease Models. Neurotherapeutics. 2019. 10.1007/s13311-019-00759-8 [DOI] [PMC free article] [PubMed]
- 4.Papadopoulos VE, Nikolopoulou G, Antoniadou I, et al. Modulation of beta-glucocerebrosidase increases alpha-synuclein secretion and exosome release in mouse models of Parkinson’s disease. Hum Mol Genet. 2018;27:1696–1710. doi: 10.1093/hmg/ddy075. [DOI] [PubMed] [Google Scholar]
- 5.Stuendl A, Kunadt M, Kruse N, et al. Induction of alpha-synuclein aggregate formation by CSF exosomes from patients with Parkinson’s disease and dementia with Lewy bodies. Brain. 2016;139:481–494. doi: 10.1093/brain/awv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngolab J, Trinh I, Rockenstein E, et al. Brain-derived exosomes from dementia with Lewy bodies propagate alpha-synuclein pathology. Acta Neuropathol Commun. 2017;5:46. doi: 10.1186/s40478-017-0445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi M, Liu C, Cook TJ, et al. Plasma exosomal alpha-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol. 2014;128:639–650. doi: 10.1007/s00401-014-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi M, Kovac A, Korff A, et al. CNS tau efflux via exosomes is likely increased in Parkinson’s disease but not in Alzheimer’s disease. Alzheimers Dement. 2016;12:1125–1131. doi: 10.1016/j.jalz.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng ASL, Tan YJ, Lu Z, et al. Plasma alpha-synuclein detected by single molecule array is increased in PD. Ann Clin Transl Neurol. 2019;6:615–619. doi: 10.1002/acn3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia Y, Zhang G, Han C, et al. Microglia as modulators of exosomal alpha-synuclein transmission. Cell Death Dis. 2019;10:174. doi: 10.1038/s41419-019-1404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riva P, Battaglia C, Venturin M. Emerging Role of Genetic Alterations Affecting Exosome Biology in Neurodegenerative Diseases. Int J Mol Sci. 2019; 20. 10.3390/ijms20174113 [DOI] [PMC free article] [PubMed]
- 12.Parnetti L, Paciotti S, Eusebi P, et al. Cerebrospinal fluid beta-glucocerebrosidase activity is reduced in parkinson’s disease patients. Mov Disord. 2017;32:1423–1431. doi: 10.1002/mds.27136. [DOI] [PubMed] [Google Scholar]
- 13.Qian X, Zhao J, Yeung PY, Zhang QC, Kwok CK. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem Sci. 2019;44:33–52. doi: 10.1016/j.tibs.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qureshi IA, Mehler MF. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics. 2013;10:632–646. doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68. doi: 10.1186/s12943-018-0817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu T, Zhang X, Gao S, et al. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo H, Wu L, Yang Q, Ye M, Zhu X. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene. 2015;554:114–119. doi: 10.1016/j.gene.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 19.Shan TD, Xu JH, Yu T, et al. Knockdown of linc-POU3F3 suppresses the proliferation, apoptosis, and migration resistance of colorectal cancer. Oncotarget. 2016;7:961–975. doi: 10.18632/oncotarget.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang HL, Hu GW, Chen Y, et al. Glioma cells promote angiogenesis through the release of exosomes containing long non-coding RNA POU3F3. Eur Rev Med Pharmacol Sci. 2017;21:959–972. [PubMed] [Google Scholar]
- 21.Gelb DJ, Oliver E, Gilman S. Diagnostic criteria for Parkinson disease. Arch Neurol. 1999;56:33–39. doi: 10.1001/archneur.56.1.33. [DOI] [PubMed] [Google Scholar]
- 22.Postuma RB, Berg D, Stern M, et al. MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord. 2015;30:1591–1601. doi: 10.1002/mds.26424. [DOI] [PubMed] [Google Scholar]
- 23.Postuma RB, Poewe W, Litvan I, et al. Validation of the MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord. 2018;33:1601–1608. doi: 10.1002/mds.27362. [DOI] [PubMed] [Google Scholar]
- 24.Manna I, Iaccino E, Dattilo V, et al. Exosome-associated miRNA profile as a prognostic tool for therapy response monitoring in multiple sclerosis patients. Faseb j. 2018;32:4241–4246. doi: 10.1096/fj.201701533R. [DOI] [PubMed] [Google Scholar]
- 25.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in Exosome Isolation Techniques. Theranostics. 2017;7:789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manier S, Liu CJ, Avet-Loiseau H, et al. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood. 2017;129:2429–2436. doi: 10.1182/blood-2016-09-742296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin Z, Balimunkwe RM, Quan T. Age-related reduction of dermal fibroblast size upregulates multiple matrix metalloproteinases as observed in aged human skin in vivo. Br J Dermatol. 2017;177:1337–1348. doi: 10.1111/bjd.15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beekman C, Janson AA, Baghat A, van Deutekom JC, Datson NA. Use of capillary Western immunoassay (Wes) for quantification of dystrophin levels in skeletal muscle of healthy controls and individuals with Becker and Duchenne muscular dystrophy. PLoS One. 2018;13:e0195850. doi: 10.1371/journal.pone.0195850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devic I, Hwang H, Edgar JS, et al. Salivary alpha-synuclein and DJ-1: potential biomarkers for Parkinson’s disease. Brain. 2011;134:e178. doi: 10.1093/brain/awr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin X, Cook TJ, Zabetian CP, et al. DJ-1 isoforms in whole blood as potential biomarkers of Parkinson disease. Scientific Reports. 2012;2:954. doi: 10.1038/srep00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HJ, Jeon B, Song J, et al. Leukocyte glucocerebrosidase and beta-hexosaminidase activity in sporadic and genetic Parkinson disease. Parkinsonism Relat Disord. 2016;23:99–101. doi: 10.1016/j.parkreldis.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Kenwrick S, Watkins A, De Angelis E. Neural cell recognition molecule L1: relating biological complexity to human disease mutations. Hum Mol Genet. 2000;9:879–886. doi: 10.1093/hmg/9.6.879. [DOI] [PubMed] [Google Scholar]
- 33.Lachenal G, Pernet-Gallay K, Chivet M, et al. Release of exosomes from differentiated neurons and its regulation by synaptic glutamatergic activity. Mol Cell Neurosci. 2011;46:409–418. doi: 10.1016/j.mcn.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Baldwin S, Deighan C, Bandeira E, et al. Analyzing the miRNA content of extracellular vesicles by fluorescence nanoparticle tracking. Nanomedicine. 2017;13:765–770. doi: 10.1016/j.nano.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caruso Bavisotto C, Scalia F, Marino Gammazza A et al. Extracellular Vesicle-Mediated Cell(−)Cell Communication in the Nervous System: Focus on Neurological Diseases. Int J Mol Sci. 2019; 20. 10.3390/ijms20020434 [DOI] [PMC free article] [PubMed]
- 37.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Dragovic RA, Gardiner C, Brooks AS, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witwer KW, Buzas EI, Bemis LT et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013; 2. 10.3402/jev.v2i0.20360 [DOI] [PMC free article] [PubMed]
- 40.Rissin DM, Kan CW, Campbell TG, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zetterberg H, Wilson D, Andreasson U, et al. Plasma tau levels in Alzheimer’s disease. Alzheimers Res Ther. 2013;5:9. doi: 10.1186/alzrt163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z, Yang S, Zhou Q, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17:82. doi: 10.1186/s12943-018-0831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng R, Du M, Wang X, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. 2018;17:143. doi: 10.1186/s12943-018-0880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong Z, Shi M, Chung KA, et al. DJ-1 and alpha-synuclein in human cerebrospinal fluid as biomarkers of Parkinson’s disease. Brain. 2010;133:713–726. doi: 10.1093/brain/awq008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barbour R, Kling K, Anderson JP, et al. Red blood cells are the major source of alpha-synuclein in blood. Neurodegener Dis. 2008;5:55–59. doi: 10.1159/000112832. [DOI] [PubMed] [Google Scholar]
- 46.Zhao ZH, Chen ZT, Zhou RL, et al. Increased DJ-1 and alpha-Synuclein in Plasma Neural-Derived Exosomes as Potential Markers for Parkinson’s Disease. Front Aging Neurosci. 2018;10:438. doi: 10.3389/fnagi.2018.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cerri S, Ghezzi C, Sampieri M, et al. The Exosomal/Total alpha-Synuclein Ratio in Plasma Is Associated With Glucocerebrosidase Activity and Correlates With Measures of Disease Severity in PD Patients. Front Cell Neurosci. 2018;12:125. doi: 10.3389/fncel.2018.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rockenstein E, Clarke J, Viel C, et al. Glucocerebrosidase modulates cognitive and motor activities in murine models of Parkinson’s disease. Hum Mol Genet. 2016;25:2645–2660. doi: 10.1093/hmg/ddw124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papagiannakis N, Xilouri M, Koros C, et al. Lysosomal alterations in peripheral blood mononuclear cells of Parkinson’s disease patients. Mov Disord. 2015;30:1830–1834. doi: 10.1002/mds.26433. [DOI] [PubMed] [Google Scholar]
- 50.Niimi Y, Ito S, Mizutani Y, et al. Altered regulation of serum lysosomal acid hydrolase activities in Parkinson’s disease: A potential peripheral biomarker? Parkinsonism Relat Disord. 2019;61:132–137. doi: 10.1016/j.parkreldis.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 51.Pan T, Kondo S, Le W, Jankovic J. The role of autophagy-lysosome pathway in neurodegeneration associated with Parkinson’s disease. Brain. 2008;131:1969–1978. doi: 10.1093/brain/awm318. [DOI] [PubMed] [Google Scholar]
- 52.Bellomo G, Paciotti S, Gatticchi L, Parnetti L. The vicious cycle between alpha-synuclein aggregation and autophagic-lysosomal dysfunction. Mov Disord. 2019. 10.1002/mds.27895 [DOI] [PubMed]
- 53.Fussi N, Hollerhage M, Chakroun T, et al. Exosomal secretion of alpha-synuclein as protective mechanism after upstream blockage of macroautophagy. Cell Death Dis. 2018;9:757. doi: 10.1038/s41419-018-0816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bae EJ, Yang NY, Lee C, et al. Loss of glucocerebrosidase 1 activity causes lysosomal dysfunction and alpha-synuclein aggregation. Exp Mol Med. 2015;47:e153. doi: 10.1038/emm.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siebert M, Sidransky E, Westbroek W. Glucocerebrosidase is shaking up the synucleinopathies. Brain. 2014;137:1304–1322. doi: 10.1093/brain/awu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bae EJ, Yang NY, Song M, et al. Glucocerebrosidase depletion enhances cell-to-cell transmission of alpha-synuclein. Nat Commun. 2014;5:4755. doi: 10.1038/ncomms5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du TT, Wang L, Duan CL, et al. GBA deficiency promotes SNCA/alpha-synuclein accumulation through autophagic inhibition by inactivated PPP2A. Autophagy. 2015;11:1803–1820. doi: 10.1080/15548627.2015.1086055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alvarez-Erviti L, Seow Y, Schapira AH, et al. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol Dis. 2011;42:360–367. doi: 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Danzer KM, Kranich LR, Ruf WP, et al. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Straniero L, Rimoldi V, Samarani M, et al. The GBAP1 pseudogene acts as a ceRNA for the glucocerebrosidase gene GBA by sponging miR-22-3p. Sci Rep. 2017;7:12702. doi: 10.1038/s41598-017-12973-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 389 kb)
(PDF 1225 kb)