Abstract
Synapses are dynamic connections that underlie essential functions of the nervous system. The addition, removal, and maintenance of synapses govern the flow of information in neural circuits throughout the lifetime of an animal. While extensive studies have elucidated many intrinsic mechanisms that neurons employ to modulate their connections, increasing evidence supports the roles of non-neuronal cells, such as glia, in synapse maintenance and circuit function. We previously showed that C. elegans epidermis regulates synapses through ZIG-10, a cell-adhesion protein of the immunoglobulin domain superfamily. Here we identified a member of the Pals1/MPP5 family, MAGU-2, that functions in the epidermis to modulate phagocytosis and the number of synapses by regulating ZIG-10 localization. Furthermore, we used light and electron microscopy to show that this epidermal mechanism removes neuronal membranes from the neuromuscular junction, dependent on the conserved phagocytic receptor CED-1. Together, our study shows that C. elegans epidermis constrains synaptic connectivity, in a manner similar to astrocytes and microglia in mammals, allowing optimized output of neural circuits.
Keywords: MAGUK, synapse elimination, glia, miniSOG, neuromuscular junction, ACR-2
Introduction
Synapses enable the transmission and integration of information within the nervous system. Proper synaptic connectivity is essential to govern nervous system functions such as sensory perception, learning, and coordinated movement. Aberrant synaptic connections have been associated with a variety of neurological disorders. Synapse formation, elimination, and maintenance work together to ensure precision and plasticity of neuronal circuits throughout the lifetime of an animal. Many neuronal-intrinsic mechanisms involve various classes of cell surface proteins and intracellular signaling pathways that establish synaptic connections (Cherra and Jin, 2015; de Wit and Ghosh, 2016; Sudhof, 2018). Additional work has shown how extrinsic mechanisms involving non-neuronal cells, such as astrocytes and microglia, cooperate with neurons to modulate neuronal circuits (Allen and Eroglu, 2017; Chung et al., 2015). It is now well established that astrocytes and microglia play an active role in the pruning of synaptic connections during development of the visual system in mice (Chung et al., 2013; Stevens et al., 2007). In Drosophila, glial cells also remove synapses and axonal components as a means to eliminate neuronal connections (Awasaki et al., 2006; Fuentes-Medel et al., 2009).
One common mechanism for non-neuronal cells to assist in the wiring and rewiring of circuits is through the phagocytosis pathway. The phagocytosis pathway is highly conserved throughout evolution and plays essential roles in the removal of cell corpses and cellular debris (Bangs et al., 2000; Mangahas and Zhou, 2005). While the core machinery that orchestrates engulfment has been widely studied, additional mechanisms that modulate the initiation or target specificity of this pathway still remain largely unknown. The C. elegans motor circuit provides a simple model to understand how non-neuronal cells modulate synapse elimination via the phagocytosis pathway. Our previous work uncovered a cell surface protein, ZIG-10, which mediates an interaction between the epidermis and neurons. The cell-cell interaction mediated by ZIG-10 enables the elimination of synapses through the activation of the phagocytosis pathway (Cherra and Jin, 2016).
In this study we have further investigated how the phagocytosis pathway maintains optimal synaptic connectivity in the motor circuit. We have discovered that a member of the MPP5/Pals1 membrane-associated guanylate kinase (MAGUK), MAGU-2, regulates synapse density. MAGU-2 functions in the epidermis and regulates ZIG-10 localization. Furthermore, we show that motor neuron membrane transfer to the epidermis is dependent on the phagocytosis receptor, CED-1. Our study highlights a new role for MAGUKs in regulating synaptic connectivity.
Results
Identification of MAGU-2 in ZIG-10 pathway
We have previously reported that maintenance of synaptic connectivity in the C. elegans motor circuit requires components of the phagocytosis pathway acting in the epidermis (Cherra and Jin, 2016). To further understand the regulation of phagocytosis-mediated synapse elimination, we screened for additional genes using a function-based assay. Ectopic expression of ZIG-10 in the ventral cord GABAergic motor neurons causes a reduction in GABAergic synapse number (Cherra and Jin, 2016) and enhances locomotor deficit caused by an acetylcholine receptor mutation, acr-2(n2420) (Jospin et al., 2009) (Supplemental Figure 1a–c). We employed this ectopic ZIG-10 expression induced phenotype to identify intracellular signaling molecules using RNAi against genes that contained SH3 domains (Cherra and Jin, 2016). One candidate corresponded to the magu-2 gene, knock-down of which decreased the locomotor defects in zig-10(tm6127); acr-2(n2420) animals that also ectopically expressed ZIG-10 in GABAergic neurons. The observed magu-2(RNAi) effect on locomotor defects was further verified using genetic null mutations in magu-2 (Supplementary Figure 1a). Additionally, the magu-2(gk218) null mutation restored the number of GABAergic synapses to wild type levels in animals ectopically expressing ZIG-10 (Supplementary Figure 1b–c). Since loss of function in magu-2 alone does not cause any defects in GABA synapse morphology (Supplementary Figure 1b–c), this suggests that MAGU-2 acts in a cellular context dependent on the ZIG-10 pathway.
MAGU-2 belongs to the membrane-associated guanylate kinase (MAGUK) family that contains a PDZ domain, an SH3 domain, and a guanylate kinase domain in its carboxy-terminus (Supplementary Figure 2a) (Hobert, 2013; Zhu et al., 2016). The family of MAGUKs modulate various forms of intercellular junctions by regulating protein localization (Zhu et al., 2016). In mammals, CASK and PSD95 modulate intercellular junctions in the nervous system through their regulation of synaptic transmission and neurotransmitter receptor clustering. Based on homology, MAGU-2 belongs to the MAGUK subfamily of membrane palmitoylated proteins (MPP) and displays highest percent of total identity (~32%) with Mpp5/Pals1 (Supplementary Figure 2b). In the developing mouse nervous system, Mpp5/Pals1 is expressed in Schwann cells and Muller glia and is essential for establishing cell polarity (Ozcelik et al., 2010; van Rossum et al., 2006). Additionally, Mpp5/Pals1 regulates the maintenance of cerebellar progenitors (Park et al., 2016) and axon sorting in the peripheral nervous system (Zollinger et al., 2015). Retinal ganglion cell-specific Mpp5/Pals1 knockout mice display neuronal degeneration early during development, mimicking the clinical degeneration observed in Leber congenital amaurosis patients (Cho et al., 2012; Park et al., 2011). The Drosophila homolog, Stardust, acts as a scaffolding protein at epithelial intercellular junctions to establish cell polarity (Tepass, 2012). However, it is unclear whether the MPP subfamily also regulates synaptic connectivity.
MAGU-2 acts in the epidermis to regulate the synapse number of cholinergic motor neurons
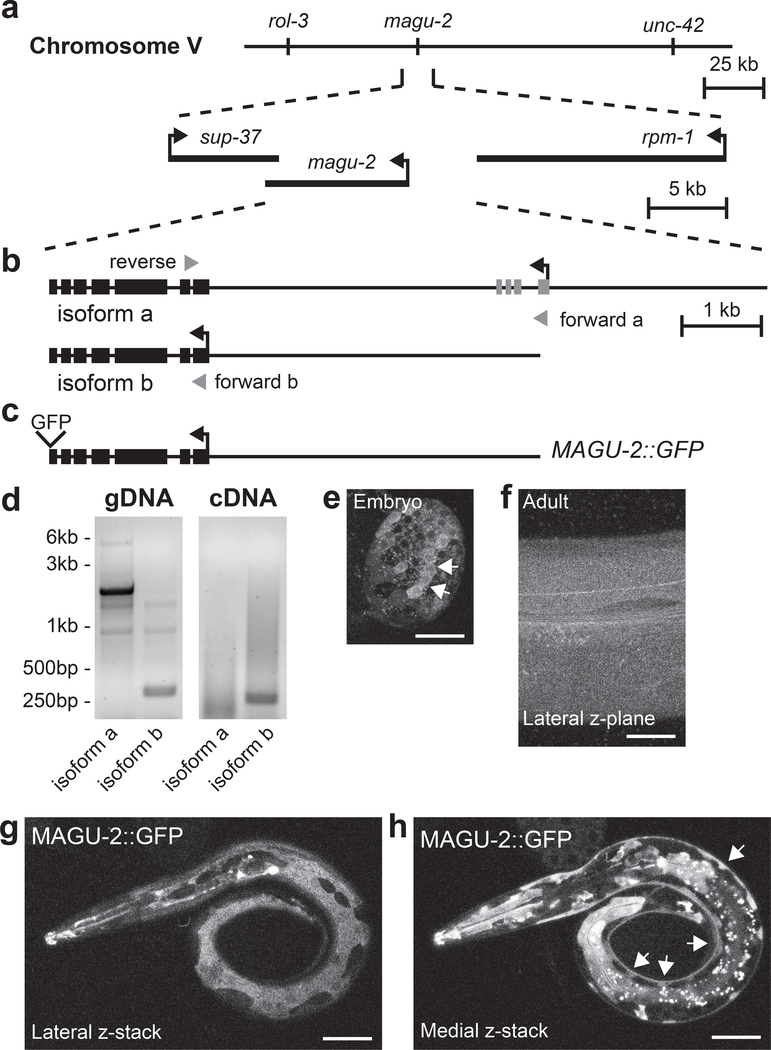
To determine where MAGU-2 functions to modulate synaptic connectivity, we first investigated the magu-2 gene structure to identify its promoter based on the information from AceView and Wormbase (Lee et al., 2018; Thierry-Mieg and Thierry-Mieg, 2006). Both sources predicted magu-2 to span more than 10kb, producing two potential isoforms (Figure 1a–b), with the long isoform a being supported by a recent dataset based on single-cell RNA-seq (Cao et al., 2017). We isolated RNA from a mixed population of wild type N2 animals. We then used isoform-specific forward primers and a common reverse primer to amplify each putative isoform. Using genomic DNA as a control, we found that only the shorter isoform b was detected in our assay as a 270bp-band, whereas 748bp-band indicative of the longer isoform was not detected (Figure 1d). This analysis does not exclude the possibility that isoform a may be expressed at lower levels or in a very limited subset of tissues.
Figure 1. MAGU-2 is expressed in the epidermis.
(a) Diagram of the magu-2 genetic locus on chromosome V. The magu-2 gene is flanked by the neighboring genes, sup-37 and rpm-1. (b) Diagram of magu-2 gene structure representing the predicted a and b isoforms. Gray arrowheads indicate primers used for isoform analysis. (c) Diagram of MAGU-2::GFP transgene. (d) Images of PCR products using magu-2a and magu-2b specific primers after gel electrophoresis. Genomic DNA (gDNA) from N2 was used as a PCR reaction control. Messenger RNAs from N2 were reverse transcribed into cDNA. PCR of magu-2a is expected to produce a 5.7kb band from gDNA and 748bp band from cDNA; magu-2b is expected to produce a 323bp band from gDNA and a 270bp band from cDNA. (e) MAGU-2::GFP expression in an embryo, between 150 and 300 minutes after fertilization. Arrows indicate cluster of cells expressing MAGU-2::GFP. (f) MAGU-2::GFP expression in a z-plane through the lateral epidermis of an adult animal. (g-h) Representative fluorescence images of L1 larva expressing MAGU-2::GFP. Scale bars are 20 μm. (g) Maximum projection through superficial layers of lateral aspect of animal showing MAGU-2 expression in epidermis, but not in neuronal commissures. (h) Maximum projection through medial layers around nerve cord showing MAGU-2 expression in epidermal ridge, but not in neuronal cell bodies. Arrows indicate epidermal ridge.
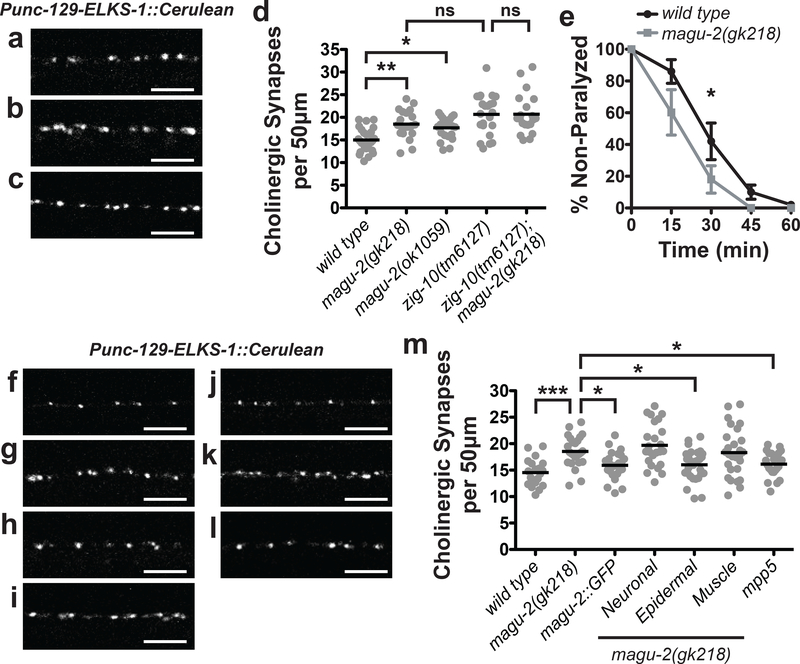
Based on our transcript analysis, we created a MAGU-2::GFP fusion protein driven by the 4kb DNA sequence upstream of magu-2 isoform b to determine where MAGU-2 was expressed (Figure 1c). We detected MAGU-2::GFP expression from embryo to adulthood (Figure 1e–f). MAGU-2::GFP was visible as a diffuse signal throughout the epidermis and in various cells in the head, pharyngeal muscle, and posterior intestine, but was not observed in neurons or muscle (Figure 1g–h). Since ZIG-10 acts in both epidermis and cholinergic neurons to regulate cholinergic synapse density (Cherra and Jin, 2016), we investigated whether magu-2 affects cholinergic synapses. We analyzed two magu-2 deletion mutants, ok1059 and gk218, which both remove most of the gene, and therefore likely are null alleles (Figure 2a–d; Supplementary Figure 2a). Both alleles displayed an increase in cholinergic synapses, similar to zig-10(tm6127) animals (Figure 2d). We observed that zig-10(tm6127); magu-2(gk218) double mutants showed no further increase in cholinergic synapses (Figure 2d). To assess whether the additional cholinergic synapses in magu-2 mutants formed functional postsynaptic compartments, we assayed the animals’ sensitivity to the cholinergic agonist, levamisole. We have previously shown that the excessive synapses observed in the zig-10(tm6127) animals caused hypersensitivity to levamisole, leading to more rapid paralysis than wild type animals (Cherra and Jin, 2016). The magu-2(gk218) mutants showed an increased sensitivity to levamisole similar to zig-10(tm6127) mutants (Figure 2e). Overall, these observations are consistent with MAGU-2 and ZIG-10 functioning in the same pathway.
Figure 2. MAGU-2 functions in the epidermis to regulate cholinergic synapse density.
Representative images of cholinergic synapses labeled by ELKS-1::Cerulean in wild type (a), magu-2(gk218) (b), and magu-2(ok1059) (c) animals. Scale bars are 5 μm. (d) Quantification of synapses from the indicated genotypes of animals expressing ELKS-1::Cerulean in cholinergic neurons. Gray dots represent individual animals; the black lines indicate the means. *p<0.05; **p<0.01; ns= not significant. (e) Quantification of paralysis over time in the presence of 1 mM levamisole; *p<0.05. Representative images of cholinergic synapses labeled by ELKS-1::Cerulean in wild type (f), magu-2(gk218) (g), and magu-2(gk218); MAGU-2::GFP (h), magu-2(gk218); Neuronal::MAGU-2 (i), magu-2(gk218); Epidermal::MAGU-2 (j), magu-2(gk218); Muscle::MAGU-2 (k), and magu-2(gk218); Epidermal::mpp5 (l) animals. Scale bars are 5 μm. (m) Quantification of cholinergic synapses labeled by ELKS-1::Cerulean in wild type or magu-2(0) animals. Transgenic expression of MAGU-2 was achieved using its endogenous promoter, rgef-1 promoter for neurons, col-10 promoter for epidermis, or myo-3 promoter for muscle. Mouse mpp5 cDNA was expressed using the col-10 promoter. Gray dots represent individual animals; the black lines indicate the means; *p<0.05; ***p<0.001.
Expression of MAGU-2::GFP in magu-2(gk218) animals was sufficient to restore cholinergic synapse density to wild type levels (Figure 2h and 2m), demonstrating that isoform b was sufficient to replace MAGU-2 function in the gk218 mutant. We further addressed in which cells MAGU-2 was functioning to regulate synapse density using tissue-specific promoters to express MAGU-2 isoform b cDNA. Expression of MAGU-2 cDNA in the epidermis of magu-2(gk218) animals restored the density of cholinergic synapses to wild type levels (Figure 2j and 2m). However, expression of MAGU-2 in the nervous system or muscles was not sufficient to restore cholinergic synapse density (Figure 2i, 2k, and 2m). Moreover, expressing mouse Mpp5 cDNA in the epidermis sufficiently restored synapse number to wild type levels (Figure 2l–m), indicating that mpp5 can function similarly to MAGU-2. Together, these data indicate that MAGU-2 is expressed and functions solely in the epidermis to modulate synaptic connectivity.
MAGU-2 affects phagocytosis in epidermis
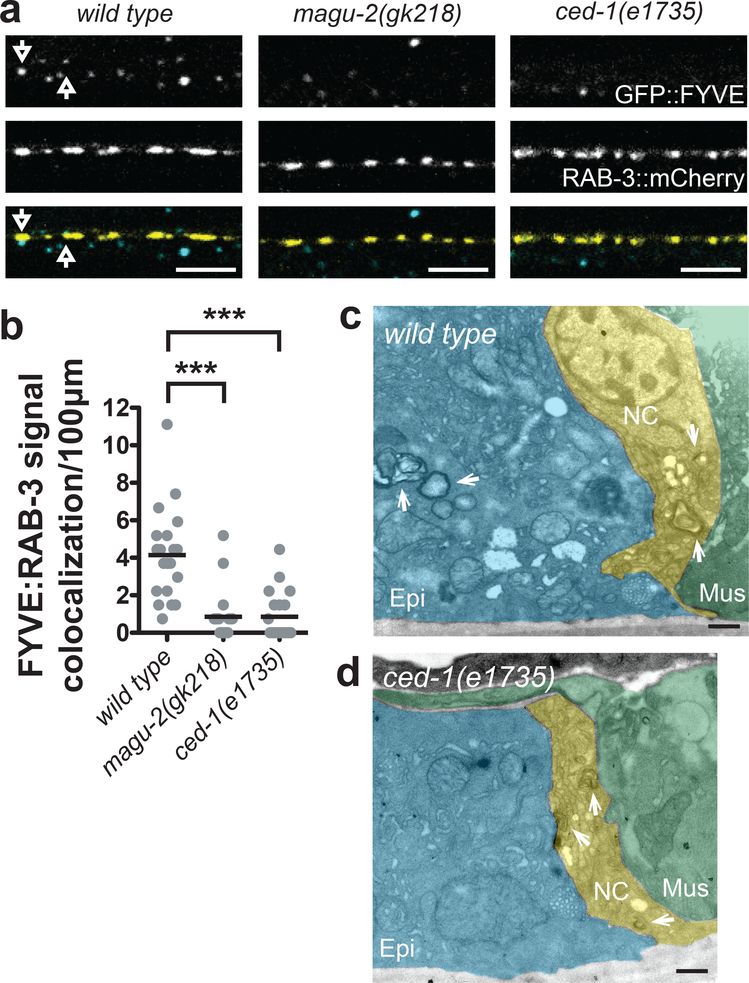
To determine if MAGU-2 regulates epidermal phagocytosis, we investigated whether magu-2(gk218) animals displayed changes in the epidermal structures marked by GFP::FYVE. FYVE domains associate with the membranes of endosomes and phagosomes in developing embryos of C. elegans (Yu et al., 2008). We have reported that both fluorescently-labeled FYVE domain and CED-1 expressed in the epidermis show colocalization with puncta labeled for presynaptic proteins (Cherra and Jin, 2016). Here, we analyzed the colocalization between GFP::FYVE structures in the epidermis and cholinergic synapses labeled by the presynaptic marker, mCherry::RAB-3. We observed that wild type animals displayed ~4 colocalization events per 100 micrometers where GFP::FYVE-labeled structures were associated with mCherry::RAB-3 expressed by cholinergic motor neurons (Figure 3a–b). In magu-2(gk218) animals, we found a significant decrease in the number of phagosomes near cholinergic synapses, similar to that of ced-1(e1735) animals (Figure 3a–b). These results indicate that MAGU-2 regulates epidermal phagocytosis.
Figure 3. MAGU-2 modulates epidermal phagocytosis of neuronal membranes.
(a) Representative single, 0.5μm-plane fluorescence images of wild type, magu-2(gk218), and ced-1(e1735) animals expressing GFP::FYVE to label epidermal phagosomes and mCherry::RAB-3 to label cholinergic synapses. Arrows indicate GFP::FYVE spots that colocalize with mCherry::RAB-3. Scale bars are 5 μm. (b) Quantification of colocalization between epidermal phagosomes and cholinergic synapses. Gray dots represent individual animals; the black lines indicate the means; ***<0.001. (c-d) Representative EM images of the ventral nerve cord in animals expressing miniSOG::ZIG-10 in cholinergic neurons. Scale bars are 500 nm. Cholinergic miniSOG-labeled structures are indicated by white arrows. Green shading indicates the muscle, blue indicates the epidermis, and yellow indicates the nerve cord. (c) Representative image of the ventral nerve cord flanked by muscle and epidermis. Cholinergic membranous material labeled by miniSOG::ZIG-10 is visible in the nerve cord and inside the epidermis in a wild type animal. (d) Representative image of the ventral cord in ced-1(e1735) animals, which show no miniSOG labeled structures inside the epidermis.
To further evaluate the phagocytosis of neuronal materials by the epidermis, we performed electron microscopy analysis using the mini singlet oxygen generator (miniSOG) to label specific tissues. When stimulated by blue light, miniSOG produces singlet oxygen, and in the presence of diaminobenzadine (DAB), miniSOG oxidizes DAB to generate electron-dense osmiophilic polymers within nanometers of miniSOG localization (Shu et al., 2011). We expressed miniSOG fused to ZIG-10 in cholinergic neurons to determine if cholinergic membranes were engulfed by the epidermis. On electron micrographs of the nerve cord, miniSOG::ZIG-10 signals were detected in a subset of neuronal processes, whose positions were consistent with being cholinergic axons and dendrites (Figure 3c–d). Importantly, in addition to the miniSOG signals found in the nerve cord, we also observed miniSOG-containing vesicles within the epidermis (Figure 3c). This suggested that the epidermis engulfs portions of cholinergic neuronal membranes. As further support for this idea, we analyzed cholinergic-driven miniSOG::ZIG-10 in the phagocytotic receptor ced-1(e1735) animals, which are deficient in executing phagocytosis (Hedgecock et al., 1983; Zhou et al., 2001). In over 300 EM sections from two wild type animals, we found more than twenty miniSOG-labeled vesicles derived from cholinergic neurons in the epidermis. In the same number of sections from three ced-1(e1735) animals, we only observed miniSOG-labeled cholinergic axons or dendrites (Figure 3d). Thus, these data from miniSOG-mediated correlated-light-electron-microscopy analyses show that neuronal membranes are transferred to epidermis through the phagocytosis pathway.
ZIG-10 localization depends on magu-2
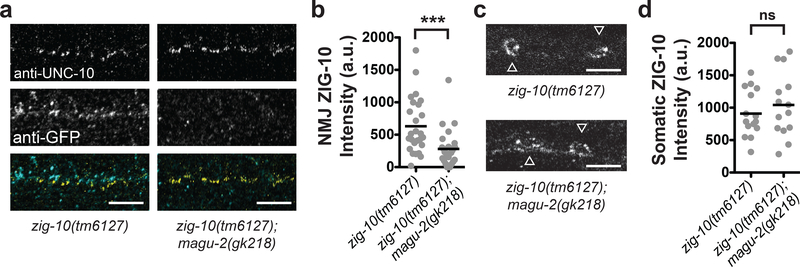
MAGUKs regulate localization and surface expression of ion channels (El-Husseini et al., 2000), glutamate receptors (Mi et al., 2004), and N-cadherins (Wang et al., 2014). Mutations in the extracellular Ig domain of ZIG-10 or RNAi-mediated knockdown displayed a similar phenotype to the zig-10(tm6127) mutation, suggesting that extracellular interactions and cell surface expression levels of ZIG-10 were essential to its function (Cherra and Jin, 2016). We hypothesized that MAGU-2 may regulate ZIG-10 expression or localization, similar to the interactions between cell adhesion proteins and other MAGUKs. To test this possibility, we generated a single-copy insertion of a functional GFP::ZIG-10 (Supplemental Figure 3). We then co-stained for GFP::ZIG-10 and the presynaptic active zone protein, UNC-10 in wild type and magu-2(gk218) animals harboring the GFP::ZIG-10 transgene. In magu-2(gk218) animals, there was a significant decrease in GFP::ZIG-10 near UNC-10-labeled synapses (Figure 4a–b). However, there was no change in GFP::ZIG-10 intensity in neuronal cell bodies (Figure 4c–d). These data suggest that MAGU-2 regulates ZIG-10 localization near synapses but does not alter ZIG-10 expression levels.
Figure 4. MAGU-2 regulates ZIG-10 localization.
(a) Representative single, 0.5μm-plane fluorescence images of the dorsal nerve cord in zig-10(tm6127) and zig-10(tm6127); magu-2(gk218) animals expressing a single-copy transgene of GFP::ZIG-10. Animals were stained for UNC-10 to label synapses and GFP to identify ZIG-10 localization. Scale bars are 5 μm. (b) Quantification of fluorescence intensity of GFP::ZIG-10 near synapses. Gray dots represent individual animals; the black lines indicate the means; ***p<0.001. (c) Representative single, 0.5μm-plane fluorescence images of neuronal cell bodies from zig-10(tm6127) and zig-10(tm6127); magu-2(gk218) animals stained for GFP::ZIG-10. Scale bars are 5 μm. White arrowheads indicate cell bodies. (d) Quantification of fluorescence intensity of GFP::ZIG-10 in neuronal cell bodies. Gray dots represent individual animals; the black lines indicate the means; ns = not significant.
Discussion
The regulation of synaptic connections plays an essential role in promoting circuit function and robust signaling. Cell-cell signaling is a common mechanism to modulate synapse formation and maintenance. In mammals, Drosophila, and C. elegans non-neuronal cells play essential roles in controlling synapse number through multiple mechanisms (Allen, 2013; Cherra and Jin, 2015; Corty and Freeman, 2013). Phagocytosis-mediated synapse pruning requires cell-cell interactions that enable glia or other non-neuronal cells to remove synapses (Cherra and Jin, 2016; Chung et al., 2013; MacDonald et al., 2006). Here, we have presented a new approach for investigating cellular interactions by electron microscopy, using genetically encoded miniSOG enzyme that enables the labeling of specific proteins or cellular compartments for electron microscopic analysis (Shu et al., 2011). MiniSOG can be expressed in a tissue-specific or temporal manner to enable the analysis of discrete interactions, such as between the epidermis and neurons. This approach provides a complementary method for immuno-EM analysis of protein localization or cell-cell interactions, including the analysis of phagocytosis.
Within the nervous system, phagocytosis by glia or epidermal cells plays multiple roles in removing dead cells or debris, degrading axons after injury, and pruning axons or synapses to modulate neural circuits (Awasaki et al., 2006; Cherra and Jin, 2016; MacDonald et al., 2006; Rasmussen et al., 2015; Stevens et al., 2007). While the removal of apoptotic cells by phagocytes has been widely studied, the recognition and engulfment of synapses is not well understood. In addition to phagocyte-corpse interactions mediated by a phagocytosis receptor, CED-1/Draper/Megf, other cell surface proteins further modulate the phagocytosis process, such as integrin and the immunoglobulin domain superfamily member, ZIG-10 (Albert et al., 2000; Cherra and Jin, 2016). Here, we uncovered a role for the Pals1/Mpp5 homolog, MAGU-2, in regulating non-neuronal phagocytosis and synapse density at the neuromuscular junction. MPP members establish cell polarity, participate in axon sorting, and reduce or prevent neuronal degeneration through the maintenance of cell-cell interactions (Cho et al., 2012; van Rossum et al., 2006; Zollinger et al., 2015). Our data suggest that cell polarity and MPPs may also modulate synaptic connectivity.
We propose that MAGU-2 functions in the epidermis to maintain cellular interactions between the epidermis and neurons by regulating the localization of ZIG-10. In the presence of MAGU-2, ZIG-10 localizes near synapses and enables the execution of phagocytosis to reduce the number of cholinergic synapses at the neuromuscular junction. Since MPPs are expressed in brain regions where glial-mediated synaptic pruning occurs (Clarke et al., 2018; Zhang et al., 2014), this raises the possibility that a conserved mechanism involving MPPs and phagocytosis may underlie synaptic pruning in mammals. Together these results provide new insights into how the phagocytosis pathway modulates synaptic connectivity.
Materials and Methods
Strains and Transgenes
All strains were maintained at 20°C as previously described (Brenner, 1974). Transgenic animals were created by microinjection as previously described (Mello et al., 1991). Single-copy insertion of GFP::ZIG-10 at cxTi10882 on chromosome IV was generated using CRISPR/Cas9 and modified plasmids (Frokjaer-Jensen et al., 2012; Takayanagi-Kiya et al., 2016). For further strain and transgene information, see Table S1 and Supplementary Figure 3.
mRNA Analysis for magu-2
To determine if magu-2 produced both predicted isoforms, we isolated RNA from mixed stage worms using Trizol according to manufacturer’s specifications. SuperScript First Strand Synthesis kit (Invitrogen) was used to generate cDNA. We performed RT-PCR using primers: 5’-TCGATACCACAGGCACTGTCC-3’ as magu-2a forward, 5’-CATTGCTGCAACATCTGGACC-3’ as magu-2b forward, and 5’-TTTCTTCAACTTCAGCGAGTGG-3’ as the common magu-2 reverse primer.
DNA Plasmid Construction
MAGU-2::GFP was constructed by PCR amplifying the 4 kb sequence upstream of magu-2 b as its promoter and cloning it into pCR8 (Invitrogen) to generate pCZGY3355. The genomic magu-2 coding sequence was amplified by PCR from wild type worms and Gibson assembly was used to fuse GFP to the C-terminus of magu-2 to generate MAGU-2::GFP (pCZGY3358). For tissue-specific rescue constructs, magu-2 cDNA was cloned from wild type animals using SuperScript III First-Strand Synthesis System (Invitrogen). The splice leader 1 sequence was used with gene-specific primers that recognized the longest predicted mRNA to generate magu-2 cDNA, which then was cloned into pCR8 (Invitrogen) to produce pCZGY3350. Mouse mpp5 cDNA was amplified from a mouse brain cDNA library and cloned into pCR8 to produce pCZGY3359. Generation of tissue-specific rescue constructs was performed using LR reactions (Invitrogen) with destination vectors containing tissue-specific promoters: rgef-1, col-10, unc-17b and myo-3 (Altun-Gultekin et al., 2001; Cherra and Jin, 2016; Okkema et al., 1993; Rand, 2007). For single-copy insertion, Gibson assembly was used to clone Pzig-10-gfp::zig-10, Prps-0-hygromycin resistance gene, and homology arms around the cxTi10882 locus on chromosome IV (pCZGY3354). See Table S2 for more information.
Whole-mount immunocytochemistry
To visualize GFP::ZIG-10 at near endogenous levels, CRISPR technology was used to insert into chromosome IV a single-copy of GFP::ZIG-10, which encodes the genomic locus of ZIG-10 with GFP fused directly after the signal peptide. The localization of GFP::ZIG-10 (juSi333) in the zig-10(tm6127) background in the presence or absence of the magu-2(gk218) allele was analyzed. Adult animals were washed 3 times in M9, followed by one 30-minute wash in water. Animals were then placed on poly-L-lysine coated slides for freeze-crack and fixation with methanol and acetone as previously described (Duerr et al., 1999). Following fixation, slides were washed in PBST and blocked with 5% goat serum. Samples were then incubated overnight with antibodies against GFP (1:500, A1112, RRID:AB_10073917, Invitrogen) and against UNC-10 (1:50, RRID:AB_10570332, Developmental Studies Hybridoma Bank). Samples were washed and incubated with secondary antibodies: goat anti-mouse Alexa 594 (RRID:AB_141372, Invitrogen) or goat anti-rabbit Alexa 488 (RRID:AB_143165, Invitrogen). After washing, samples were mounted with Vectashield mounting media (RRID:AB_2336789, Vector Labs).
MiniSOG Photo-oxidation and Electron Microscopy
To investigate the cellular interactions between the epidermis and the nerve cord, we used miniSOG to label cholinergic neurons using tissue-specific promoters. miniSOG-labeled cellular membranes were visualized by EM after fixation and photo-oxidation. Adult animals were placed in 2% glutaraldehyde, 2% paraformaldehyde solution in 100 mM sodium cacodylate buffer. After the animals stopped moving, they were cut in half and incubated for 1 hour on ice. The samples were washed in 100 mM sodium cacodylate buffer and then blocked in 50 mM glycine, 10 mM potassium cyanide, 20 mM aminotriazole in 100 mM sodium cacodylate buffer for 2 hours on ice. The samples were placed in a MatTek culture dish containing ice-cold 2.5 mM oxygentated diaminobenzadine and 10 mM HCl in 100 mM sodium cacodylate buffer and illuminated with blue light using a Leica SPEII confocal microscope for 20 minutes. The samples were then washed in 100 mM sodium cacodylate buffer and post fixed overnight at 4°C in 2% osmium tetroxide in 100 mM sodium cacodylate buffer. The following day samples were rinsed with ice-cold ddH2O, dehydrated with ethanol and acetone, embedded in Durcupan, and baked for 3 days at 60°C.
After embedding, 60-nm-thick serial sections were prepared for analysis. MiniSOG-labeled structures were identified by visual inspection of electron micrographs as vesicular structures appearing darker than background and that persist through at least two consecutive serial sections. For miniSOG::ZIG-10 expressed in cholinergic neurons, the number of miniSOG-labeled structures was counted in the nerve cord and in the epidermis by an individual blinded to the sample genotypes.
Fluorescent Microscopy
All images were captured at 63X magnification using an LSM710 confocal microscope (Zeiss) using identical settings for each fluorescent protein marker. L4 animals were immobilized in M9 buffer by rolling animals such that their dorsal or ventral surface contacted the coverslip. All image analysis was performed using the Fiji distribution of NIH ImageJ.
For synapse number, dorsal synapses posterior to the vulva were imaged, and a single z-plane of 0.5-μm thickness was analyzed. Synapse density was determined using the ImageJ Analyze Particles function to count the number of synaptic puncta larger than 0.05 μm2 over a defined length of nerve cord.
For colocalization of phagosomes with dorsal synapses posterior to the vulva, confocal images were manually inspected for colocalization between GFP::FYVE and mCherry::RAB-3, as defined by particles containing pixels from both the red and green channels. The number of colocalization events per length of nerve cord analyzed was then determined.
For quantifying ZIG-10 near the neuromuscular junctions, synapses were identified by eye as bright spots labeled by the UNC-10 antibody. GFP::ZIG-10 intensity was quantified by drawing a 2-μm-wide rectangle adjacent to the UNC-10-labeled nerve cord using ImageJ. The integrated density from the green channel was then measured inside of the rectangle. The rectangle was then moved 10 μm perpendicular to the nerve cord to measure the background integrated density. The background signal was subtracted from the GFP::ZIG-10 signal adjacent to the nerve cord to produce the measured intensity of ZIG-10 at the neuromuscular junction.
Levamisole Sensitivity Assay
L4 animals were moved to a fresh plate. The following day young adult animals were placed on plates containing 1 mM levamisole (Sigma). Animals were gently touched every 15 minutes for one hour to assess paralysis; animals that did not move after three touches were considered paralyzed.
Statistical Analysis
All quantitative data are displayed as mean with individual data points represented as circles. For comparisons between two groups with normal distributions, Student’s t-test was used. A Mann-Whitney test was used for comparisons between two groups that did not display Gaussian distribution. For comparisons between multiple groups, an ANOVA was used followed by post-hoc t-tests using a Bonferroni correction for multiple comparisons. A p-value < 0.05 was considered statistically significant. Power analysis was performed to ensure that data was collected from a large enough sample size to provide a beta error ≤ 0.2.
Supplementary Material
Acknowledgments
We thank the members of the Jin laboratory for constructive comments. We thank J.S. Dittman for reporter lines. Some strains were provided by National BioResource Project (Dr. S. Mitani) and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40-OD010440). We appreciate Wormbase for genetic and genomic information. The electron micrographs were taken in the UCSD Cellular and Molecular Medicine Electron microscopy core facility, which is supported in part by National Institutes of Health Award (S10-OD023527). This work was supported by grants from the National Institutes of Health to S.J.C. (K99-NS097638), D.B. (R01-GM086197), M.E. (P41-GM103412) and Y.J. (R01 and R37-NS035546).
Footnotes
Conflict of interest
None of the authors declare any conflicts of interest.
Supplemental material
The supplemental material includes three figures. Supplementary Figure 1 displays the quantification of convulsions per minute in magu-2 and zig-10 deletion mutants in the acr-2(gf) background. Supplementary Figure 2 illustrates the functional domains of MAGU-2 and the portions deleted in the ok1059 and gk218 mutations. Supplementary Figure 2 also includes a phylogenetic tree of the MPP subfamily of proteins. Supplementary Figure 3 contains diagrams of the coding sequences of plasmids used for the creation of transgenic animals in this study.
References
- Albert ML, Kim JI, and Birge RB. 2000. alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol. 2:899–905. [DOI] [PubMed] [Google Scholar]
- Allen NJ 2013. Role of glia in developmental synapse formation. Curr Opin Neurobiol. 23:1027–1033. [DOI] [PubMed] [Google Scholar]
- Allen NJ, and Eroglu C. 2017. Cell Biology of Astrocyte-Synapse Interactions. Neuron. 96:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun-Gultekin Z, Andachi Y, Tsalik EL, Pilgrim D, Kohara Y, and Hobert O. 2001. A regulatory cascade of three homeobox genes, ceh-10, ttx-3 and ceh-23, controls cell fate specification of a defined interneuron class in C. elegans. Development. 128:1951–1969. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, and Ito K. 2006. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 50:855–867. [DOI] [PubMed] [Google Scholar]
- Bangs P, Franc N, and White K. 2000. Molecular mechanisms of cell death and phagocytosis in Drosophila. Cell Death Differ. 7:1027–1034. [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, and Shendure J. 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 357:661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ 3rd, and Jin Y. 2015. Advances in synapse formation: forging connections in the worm. Wiley Interdiscip Rev Dev Biol. 4:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ 3rd, and Jin Y. 2016. A Two-Immunoglobulin-Domain Transmembrane Protein Mediates an Epidermal-Neuronal Interaction to Maintain Synapse Density. Neuron. 89:325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Kim JY, Simons DL, Song JY, Le JH, Swindell EC, Jamrich M, Wu SM, and Kim S. 2012. Genetic ablation of Pals1 in retinal progenitor cells models the retinal pathology of Leber congenital amaurosis. Hum Mol Genet. 21:2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Allen NJ, and Eroglu C. 2015. Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb Perspect Biol. 7:a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, Smith SJ, and Barres BA. 2013. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 504:394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Liddelow SA, Chakraborty C, Munch AE, Heiman M, and Barres BA. 2018. Normal aging induces A1-like astrocyte reactivity. Proc Natl Acad Sci U S A. 115:E1896–E1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty MM, and Freeman MR. 2013. Cell biology in neuroscience: Architects in neural circuit design: glia control neuron numbers and connectivity. J Cell Biol. 203:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, and Ghosh A. 2016. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 17:22–35. [DOI] [PubMed] [Google Scholar]
- Duerr JS, Frisby DL, Gaskin J, Duke A, Asermely K, Huddleston D, Eiden LE, and Rand JB. 1999. The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J Neurosci. 19:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Topinka JR, Lehrer-Graiwer JE, Firestein BL, Craven SE, Aoki C, and Bredt DS. 2000. Ion channel clustering by membrane-associated guanylate kinases. Differential regulation by N-terminal lipid and metal binding motifs. J Biol Chem. 275:23904–23910. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, and Jorgensen EM. 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 9:117–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Medel Y, Logan MA, Ashley J, Ataman B, Budnik V, and Freeman MR. 2009. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol. 7:e1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock EM, Sulston JE, and Thomson JN. 1983. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science. 220:1277–1279. [DOI] [PubMed] [Google Scholar]
- Hobert O 2013. The neuronal genome of Caenorhabditis elegans. WormBook:1–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jospin M, Qi YB, Stawicki TM, Boulin T, Schuske KR, Horvitz HR, Bessereau JL, Jorgensen EM, and Jin Y. 2009. A neuronal acetylcholine receptor regulates the balance of muscle excitation and inhibition in Caenorhabditis elegans. PLoS Biol. 7:e1000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RYN, Howe KL, Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Davis P, Gao S, Grove C, Kishore R, Muller HM, Nakamura C, Nuin P, Paulini M, Raciti D, Rodgers F, Russell M, Schindelman G, Tuli MA, Van Auken K, Wang Q, Williams G, Wright A, Yook K, Berriman M, Kersey P, Schedl T, Stein L, and Sternberg PW. 2018. WormBase 2017: molting into a new stage. Nucleic Acids Res. 46:D869–D874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, and Ambros V. 1991. Alternative temporal control systems for hypodermal cell differentiation in Caenorhabditis elegans. Nature. 350:162–165. [DOI] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, and Freeman MR. 2006. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 50:869–881. [DOI] [PubMed] [Google Scholar]
- Mangahas PM, and Zhou Z. 2005. Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol. 16:295–306. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, and Ambros V. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. The EMBO journal. 10:3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi R, Sia GM, Rosen K, Tang X, Moghekar A, Black JL, McEnery M, Huganir RL, and O’Brien RJ. 2004. AMPA receptor-dependent clustering of synaptic NMDA receptors is mediated by Stargazin and NR2A/B in spinal neurons and hippocampal interneurons. Neuron. 44:335–349. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, and Fire A. 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 135:385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcelik M, Cotter L, Jacob C, Pereira JA, Relvas JB, Suter U, and Tricaud N. 2010. Pals1 is a major regulator of the epithelial-like polarization and the extension of the myelin sheath in peripheral nerves. J Neurosci. 30:4120–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Alves CH, Lundvig DM, Tanimoto N, Beck SC, Huber G, Richard F, Klooster J, Andlauer TF, Swindell EC, Jamrich M, Le Bivic A, Seeliger MW, and Wijnholds J. 2011. PALS1 is essential for retinal pigment epithelium structure and neural retina stratification. J Neurosci. 31:17230–17241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Hughes LJ, Moon UY, Park R, Kim SB, Tran K, Lee JS, Cho SH, and Kim S. 2016. The apical complex protein Pals1 is required to maintain cerebellar progenitor cells in a proliferative state. Development. 143:133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JB 2007. Acetylcholine. WormBook:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JP, Sack GS, Martin SM, and Sagasti A. 2015. Vertebrate epidermal cells are broad-specificity phagocytes that clear sensory axon debris. J Neurosci. 35:559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, and Tsien RY. 2011. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 9:e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AM, Lambris JD, Smith SJ, John SW, and Barres BA. 2007. The classical complement cascade mediates CNS synapse elimination. Cell. 131:1164–1178. [DOI] [PubMed] [Google Scholar]
- Sudhof TC 2018. Towards an Understanding of Synapse Formation. Neuron. 100:276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi-Kiya S, Zhou K, and Jin Y. 2016. Release-dependent feedback inhibition by a presynaptically localized ligand-gated anion channel. Elife. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepass U 2012. The apical polarity protein network in Drosophila epithelial cells: regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu Rev Cell Dev Biol. 28:655–685. [DOI] [PubMed] [Google Scholar]
- Thierry-Mieg D, and Thierry-Mieg J. 2006. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 7 Suppl 1:S12 11–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum AG, Aartsen WM, Meuleman J, Klooster J, Malysheva A, Versteeg I, Arsanto JP, Le Bivic A, and Wijnholds J. 2006. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Muller glia cells. Hum Mol Genet. 15:2659–2672. [DOI] [PubMed] [Google Scholar]
- Wang SH, Celic I, Choi SY, Riccomagno M, Wang Q, Sun LO, Mitchell SP, Vasioukhin V, Huganir RL, and Kolodkin AL. 2014. Dlg5 regulates dendritic spine formation and synaptogenesis by controlling subcellular N-cadherin localization. J Neurosci. 34:12745–12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lu N, and Zhou Z. 2008. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 6:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, and Wu JQ. 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 34:11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, and Horvitz HR. 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 104:43–56. [DOI] [PubMed] [Google Scholar]
- Zhu J, Shang Y, and Zhang M. 2016. Mechanistic basis of MAGUK-organized complexes in synaptic development and signalling. Nat Rev Neurosci. 17:209–223. [DOI] [PubMed] [Google Scholar]
- Zollinger DR, Chang KJ, Baalman K, Kim S, and Rasband MN. 2015. The Polarity Protein Pals1 Regulates Radial Sorting of Axons. J Neurosci. 35:10474–10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.