Abstract
Medial temporal lobe (MTL) consists of hippocampal subfields and neighboring cortices. These heterogeneous structures are differentially involved in memory, cognitive and emotional functions, and present non-uniformly distributed atrophy contributing to cognitive disorders. The major goal of this study is to examine how genetics influences AD pathogenesis via MTL substructures by analyzing high-resolution MRI data. We performed GWAS to examine the associations between 565,373 SNPs and 14 MTL substructure volumes. A novel association with right Brodmann area 36 (R-BA36) volume was discovered in an ERC1 SNP (i.e., rs2968869). Further analyses on larger samples found rs2968869 associated with gray matter density and glucose metabolism measures in the right hippocampus, and disease status. Tissue-specific transcriptomics analysis identified the minor allele of rs2968869 (rs2968869-C) associated with reduced ERC1 expression in the hippocampus. All the findings indicated a protective role of rs2968869-C in AD. This study demonstrated the power of high-resolution MRI data and the promise of fine-grained MTL substructures for revealing the genetic basis of AD neurodegeneration biomarkers.
Keywords: Alzheimer’s disease, high-resolution T2-weighted MRI, medial temporal lobe, BA36, genome-wide association study (GWAS), ERC1
1. Introduction
Alzheimer’s disease (AD) is an irreversible neurodegenerative brain disease distinguished by progressive impairment of memory and decline in cognitive abilities. Based on the statistics from the 2019 Alzheimer’s disease facts and figures (Gaugler et al.;2019), AD is the 6th leading cause of death in the United States as it currently has no cure and is eventually fatal. AD is known as the most common type of age-related dementia, but the disease causes are unclear. Given neurodegeneration (i.e., N) as a major category of the amyloid-β deposition, pathologic tau, and neurodegeneration (ATN) classification for AD biomarkers (Jack et al.;2016), there is substantial research interest in AD neuroimaging studies with particular emphasis on critical memory structures. Although various imaging biomarkers have been identified to be related to disease status and progression, their genetic mechanisms remain unclear. Since human brain cognitive impairment diseases such as AD and Parkinson’s disease are strongly influenced by genetic, lifestyle and environmental factors, genetic analysis of brain imaging phenotypes is an important research topic. The goal is to reveal the genetic basis of brain phenotypes and contribute to the disease modeling and drug development.
Genome-wide association studies (GWAS) of quantitative endophenotypes have successfully identified a number of loci susceptible for AD (Saykin et al.;2015), and GWAS of whole hippocampal volume have been studied for late-onset AD (Horgusluoglu-Moloch et al.;2017,Nho et al.;2015, 2013). However, the critical subfields of the hippocampus and neighboring Medial temporal lobe (MTL) substructures are underexplored in genetic studies. Given that atrophy of these structures is not homogeneous (Foo et al.;2016), volume loss on specific MTL substructures has been identified as useful biomarkers in the existing AD studies (Cong et al.;2015). It becomes an increasingly important research topic to identify genetic susceptibility factors for hippocampal subfields and neighboring MTL substructures. Of note, hippocampal subfield volumes are highly heritable, and thus can be used as quantitative traits in genetic association and linkage studies (Greenspan et al.;2017). In recent GWAS findings, volume loss on hippocampal subfields was found to be associated with multiple genome-wide significant loci (Hibar et al.;2017,Morey et al.;2019, n.d.,Smeeth et al.;2019,Zhao et al.;2019), including novel genes which were not reported in prior genetic studies of the whole hippocampal volume (van der Meer et al.;2018). This suggests the promise of hippocampal subfields and neighboring MTL substructures as valuable quantitative traits in genetic association studies.
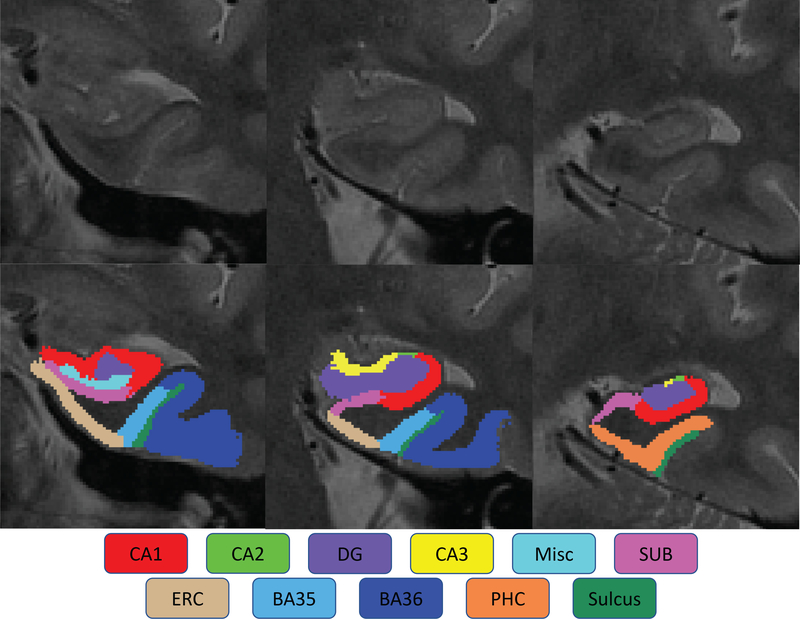
Most existing subfield-related GWAS are based on FreeSurfer (Iglesias et al.;2015,Van Leemput et al.;2009) segmentation results, which mainly focus on the hippocampal subfields and laminae. The adjacent cortical regions of the hippocampus are usually ignored, while some of those (e.g., perirhinal cortex) have been reported as the earliest affected regions in AD pathology (Sanchez-Mejias et al.;2019,Xie et al.;2017). To address this limitation, the regions of interest (ROIs) studied in this work are expanded from the hippocampus to the medial temporal lobe (MTL). The MTL in human brain mechanisms plays a prominent role in memory, cognitive, and emotional functions (Lech and Suchan;2013,Mulders et al.;2019). The MTL system is composed of a group of anatomically related structures that are essential for semantic memory and episodic memory. As shown in Figure 1, the MTL system consists of (1) hippocampal subfields including cornu ammonis (CA1/2/3), dentate gyrus (DG), and subiculum (SUB), and (2) adjacent neighboring regions such as perirhinal (PRC), entorhinal (ERC), and parahippocampal (PHC) cortices (Squire et al.;2004).
Figure 1. The medial temporal lobe system.
The medial temporal lobe (MTL) contains the hippocampus including cornu ammonis 1/2/3 (CA1, CA2 and CA3), dentate gyrus (DG), and subiculum (SUB); and medial temporal cortices including entorhinal cortex (ERC), perirhinal cortex ( PRC, including Brodmann Areas 35 and 36, or BA35 and BA36) and parahippocampal cortex (PHC).
Similar to hippocampal subfields, in AD progression, the neuron damage is not uniformly distributed across the entire MTL (Miller et al.;2013). Specialized functions of anatomically complex MTL sub-structures vary, and thus it is important to investigate not only the MTL as a whole but also each MTL substructure individually. The hippocampus is embedded at the end of a cortical processing hierarchy and recognized as fundamental for declarative memory formation, learning, and emotional processing. It serves as a content-independent hub (Schultz et al.;2019), while the areas of surrounding MTL cortices are thought to communicate with the hippocampus closely and comprehensively characterized in the aspects of spatial navigation, scene processing, and memory storage (Smith and Kosslyn;2013). The portions of PRC, ERC, and CA1 are involved in the onset of the neuropathological pathways of AD with evidence of remarkable neurofibrillary tangle (NFT) aggregation in the primitive stage of AD (Braak and Braak;1997,Carr et al.;2017,Maruszak and Thuret;2014), and the rest of the MTL regions are also affected with the development of AD.
Ignoring MTL neighboring regions of the hippocampus makes the genetic mechanisms related to MTL cortices such as ERC, PRC, and PHC underlying older adult neurogenesis in AD remaining underexplored. As a widely accepted fact, PRC and ERC are among the first regions of tau deposition. However, given the size, complexity, heterogeneity, and the large anatomical variability of the MTL cortices, in the existing subfield related GWAS, a major challenge of accurately capturing volumetric measures of the MTL substructures is imaging resolution. The conventional 3T T1-weighted magnetic resonance imaging (MRI) scans adopted in the existing GWAS usually have imaging resolution 1×1×1 mm3 or similar, while the 3T T2-weighted high-resolution MRI scans adopted in recent imaging studies (Cong et al.; 2018, 2016) have imaging resolution 0.4×0.4×2 mm3 or similar. Supplementary Figure 1 shows an example comparison between conventional 3T T1-weighted MRI and 3T T2-weighted high-resolution MRI on a same subject. With the higher MRI resolution, hippocampal subfield layers could be better distinguished from one another; thus, a more precise partition of MTL regions can be either manually or automatically obtained without requiring strong magnetic field strength during the process of MRI acquisition.
Recent studies (Sone et al.;2017,Wisse et al.;2018) adopt a strategy of using T2-weighted high-resolution MRI together with traditional T1-weighted MRI. By taking advantage of the more fine-grained MTL sub-regional measures, these studies have demonstrated that PRC atrophy occurred as the first affected region and was significantly correlated to early AD before the hippocampal subfields were influenced. Besides, these studies also illustrate the selectivity of the MTL atrophy by examining the volume changes of hippocampal subfields and neighboring cortical regions. As neurogenesis is essential for memory and cognition, volume loss (atrophy) on MTL sub-regions are found as a discriminative biomarker for preclinical detection of early AD. However, as mentioned before, the genetic mechanisms related to imaging biomarkers such as structural atrophy of MTL cortices underlying neurogenesis in AD remain underexplored. To bridge the above gap, this study is designed to investigate the genetic influences on the volumetric measures of both hippocampal subfields and the neighboring MTL regions using high-resolution MRI data, in order to explore MTL-specific regional neurogenetic mechanisms in AD.
2. Materials and methods
2.1. Alzheimer’s Disease Neuroimaging Initiative (ADNI)
Data used in the preparation of this article obtained from the ADNI database, which was initially launched in 2004 as a public-private partnership, and led by the Principal Investigator Michael W. Weiner, MD. One primary aim of ADNI has been to examine whether serial imaging biomarkers extracted from MRI, positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early AD. For up-to-date information, see www.adni-info.org.
2.2. Study participants
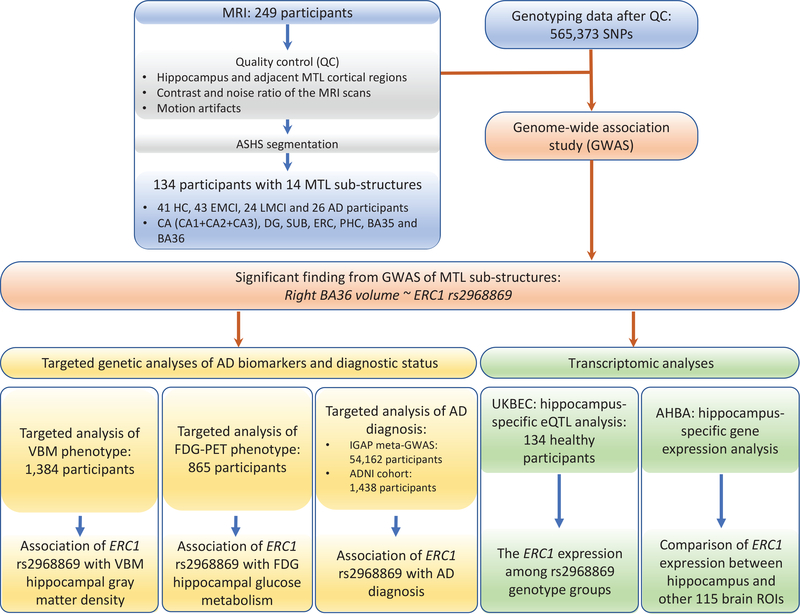
Participants included non-Hispanic Caucasian subjects from ADNI with both high-resolution MRI scans and genotype data available. The full inclusion and exclusion criteria for ADNI are described at www.adni-info.org. Detailed quality control (QC) steps for imaging and genotype data have been previously reported (Cong et al.;2018,Yao et al.;2019) and are briefly described below. Participants were restricted to non-Hispanic Caucasians to reduce the potential confounding effect of population stratification in the genetic analysis. Thus, the study analyzed a total of 134 non-Hispanic Caucasian subjects with high-resolution MRI data available and meeting all QC criteria described in (Cong et al.;2018), including 41 healthy control (HC), 43 early MCI (EMCI), 24 late MCI (LMCI) and 26 AD participants. Detailed characteristic information and the number of subjects in each sub-group are shown in Table 1. Besides participants included in the GWAS of MTL substructure volumes, non-overlapping samples with various AD endophenotypes were studied further for evaluating their associations with the resulting MTL genetic findings. The detailed information of study participants and flowchart are summarized in Figure 2. This study was approved by institutional review boards of all participating institutions, and written informed consent was obtained from all participants or authorized representatives.
Table 1.
Participant characteristics in the GWAS of MTL substructures.
| Diagnosis | HC | EMCI | LMCI | AD | p-value |
|---|---|---|---|---|---|
| Number | 41 | 43 | 24 | 26 | - |
| Gender (M/F) | 20/21 | 24/19 | 13/11 | 16/10 | 0.64 |
| Age | 76.10±6.88 | 73.98±7.21 | 72.71±8.38 | 74.42±7.62 | 0.32 |
| Education | 17.00±2.30 | 17.14±2.70 | 16.50±3.13 | 16.35±2.50 | 0.57 |
| ICV (103 cm3) | 1.51±0. 15 | 1.54±0.12 | 1.47±0.13 | 1.52±0.18 | 0.27 |
| CA_L (103 mm3) | 1.37±0.18 | 1.34±0.19 | 1.22±0.28 | 1.10±0.28 | 1.44E-05 |
| CA_R (103 mm3) | 1.43±0.21 | 1.41±0.21 | 1.19±0.29 | 1.08±0.28 | 1.07E-08 |
| DG_L (103 mm3) | 0.77±0.11 | 0.76±0.12 | 0.70±0.19 | 0.65±0.14 | 1.08E-03 |
| DG_R (103 mm3) | 0.84±0.15 | 0.81±0.13 | 0.70±0.17 | 0.63±0.16 | 7.28E-08 |
| SUB_L (103 mm3) | 0.46±0.08 | 0.48±0.06 | 0.41±0.08 | 0.39±0.08 | 1.04E-05 |
| SUB_R (103 mm3) | 0.45±0.07 | 0.48±0.06 | 0.40±0.07 | 0.38±0.09 | 9.04E-08 |
| ERC_L (103 mm3) | 0.45±0.09 | 0.46±0.08 | 0.43±0.09 | 0.39±0.07 | 1.50E-02 |
| ERC_R (103 mm3) | 0.44±0.09 | 0.45±0.09 | 0.39±0.10 | 0.39±0.09 | 1.80E-02 |
| PHC_L (103 mm3) | 0.88±0.15 | 0.90±0.17 | 0.80±0.16 | 0.77±0.22 | 8.90E-03 |
| PHC_R (103 mm3) | 0.94±0.15 | 0.98±0.20 | 0.88±0.12 | 0.85±0.16 | 7.60E-03 |
| BA35_L (103 mm3) | 0.55±0.11 | 0.52±0.10 | 0.49±0.10 | 0.45±0.11 | 1.93E-03 |
| BA36_L (103 mm3) | 1.77±0.33 | 1.70±0.28 | 1.65±0.35 | 1.52±0.30 | 1.60E-02 |
| BA35_R (103 mm3) | 0.51±0.09 | 0.50±0.09 | 0.45±0.09 | 0.43±0.11 | 1.30E-03 |
| BA36_R (103 mm3) | 1.68±0.32 | 1.67±0.25 | 1.47±0.30 | 1.39±0.39 | 2.33E-04 |
Note: P-values were assessed for significant differences among diagnostic groups, and were computed using one-way ANOVA (except for gender using chi-square test). The p-values < 0.05 are shown in bold. HC = Healthy Control; EMCI = Early Mild Cognitive Complaint; LMCI = Late Mild Cognitive Complaint; AD = Alzheimer’s disease; ICV = Intracranial Volume.
Figure 2. Flowchart of MTL sub-structure GWAS design.
Abbreviations: ASHS, automatic segmentation of hippocampal subfields; SNPs, single nucleotide polymorphisms; MTL, medial temporal lobe; HC, healthy control; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; AD, Alzheimer’s disease; CA, cornu ammonis; DG, dentate gyrus; SUB, subiculum; ERC, entorhinal cortex; PHC, parahippocampal cortex; BA, Brodmann area; VBM, voxel-based morphometry; FDG-PET, Fludeoxyglucose - positron emission tomography; IGAP, International Genomics of Alzheimer’s Project; ADNI, Alzheimer’s Disease Neuroimaging Initiative; UKBEC, UK Brain Expression Consortium; AHBA, Allen human brain atlas; ROI, region of interest; ERC1: ELKS/RAB6-interacting/CAST family member 1.
2.3. MRI data acquisition and processing
The scanning protocols included a T1-weighted magnetization-prepared rapid acquisition gradient echo (MPRAGE) sequence with whole-brain coverage and a T2-weighted turbo spin-echo (TSE) sequence with partial-brain coverage and an oblique coronal slice orientation (positioned orthogonally to the main axis of the hippocampus). MRI data included T1-weighted MPRAGE scans with an acquisition matrix of 240×256×176 and voxel size 1.05×1.05×1.2 mm3 and T2-weighted scans containing 24 or 30 coronal slices with an acquisition matrix of 448×448 and voxel size 0.39×0.39×2 mm3. Image quality check was systematically performed on the segmentation results using ITK-SNAP 3.8.0 (Yushkevich et al.;2006) in the following aspects: 1) hippocampus and adjacent MTL cortices coverage, 2) contrast and noise ratio of the MRI scans, and 3) motion artifacts. As a result (see Figure 2), 134 out of 249 pairs of T1 and T2-weighted MRI scans from ADNI1 and ADNI2 passed the quality check.
2.4. Genotyping data acquisition and processing
Genotyping data were quality-controlled, imputed and combined as described in (Yao et al.;2019). Briefly, genotyping was performed on all ADNI participants following the manufacturer’s protocol using blood genomic DNA samples and Illumina GWAS arrays (610-Quad, OmniExpress, or HumanOmni2.5–4v1) (Saykin et al.;2010). Quality control was performed in PLINK v1.90 (Purcell et al.;2007) using the following criteria: 1) call rate per marker ≥ 95%, 2) minor allele frequency (MAF) ≥ 5%, 3) Hardy Weinberg Equilibrium (HWE) test P ≥ 1.0E-6, and 4) call rate per participant ≥ 95%. Significant relatedness pairs with PI_HAT > 0.45 were identified and thereafter one individual from each pair was randomly excluded (Ramanan et al.;2015). Participants were then checked for gender and identity-by-descent before imputation to identify the genotyping or coding error and to avoid the potential confounding effect due to the gender ambiguity or consanguinity such as sibling pairs. To restrict the studied participants to non-Hispanic Caucasians, we further performed population stratification using 988 subjects with known ancestry information from HapMap3 as reference data. We merged the ADNI and HapMap3 samples, and performed multidimensional scaling analysis using PLINK v1.90 with identity-by-state (IBS) pairwise distance matrix on the merged data to clustering samples in the principal component analysis space. ADNI participants were identified as non-Hispanic Caucasians if: 1) they were clustered with HapMap3 CEU or TSI subjects as well as had self-reported race/ethnicity as “non-Hispanic/white”, or 2) they were not clustered with any HapMap3 subjects while had self-reported race/ethnicity as “non-Hispanic/white”. Haplotype patterns from the 1,000 Genomes Project reference panel were then applied to impute the SNPs that were not directly genotyped from arrays. 5,574,300 SNPs were obtained for all subjects involved in this work.
Given the modest size of high-resolution imaging data, instead of using all the imputed SNPs, we focused on the analysis of the markers available on the ADNI1 610-Quad panel. In total, 565,373 SNPs were included in this GWAS of MTL substructure volumes (Figure 2). To appropriately control for population stratification, we used PLINK v1.90 to generate the top four principal components to be included as covariates in our genetic association analyses.
2.5. Segmentation of hippocampal subfields and neighboring MTL substructures
Automatic Segmentation of Hippocampal Subfields (ASHS) software (Yushkevich et al.;2015) was employed for segmenting hippocampal subfields and neighboring MTL substructures, using the atlas package provided by the Penn Memory Center at the University of Pennsylvania. The software has been validated by its authors using k-fold cross-validation against manually traced segmentation (Yushkevich et al.;2015), peer-reviewed (Mueller et al.;2018) and applied in recent neuroimaging studies (de Flores et al.;2015,Hindy et al.;2016). ASHS takes as inputs the conventional T1-weighted MRI scans and the corresponding high-resolution T2-weighted MRI scans, and performs multi-atlas segmentation by implementing the Joint Label Fusion method (Wang and Yushkevich;2013) and Corrective Learning (Wang et al.;2011). The segmentation results include the following hippocampal subfields and the neighboring MTL cortices: cornu ammonis 1/2/3 (CA1, CA2 and CA3), dentate gyrus (DG), subiculum (SUB), miscellaneous (MISC), entorhinal cortex (ERC), perirhinal cortex (PRC, including Brodmann Areas 35 and 36, or BA35 and BA36), parahippocampal cortex (PHC), and collateral sulcus (CS).
Given CA2 and CA3 as the smallest hippocampal subfields, we analyzed the entire CA region as a whole. The strategy of combining the CA regions could help increase the level of measurement stability, as shown in our prior study (Cong et al.;2018). As a result, in this study, we analyzed 14 MTL substructures extracted using the ASHS software, including seven regions on each hemisphere: CA (combining CA1, CA2, and CA3), DG, SUB, ERC, PHC, BA35 and BA36 (Figure 2).
2.6. Brain imaging genetic association analysis
GWAS on the volumetric measures of the 14 MTL substructures were performed using linear regression under an additive genetic model in PLINK v1.90 (Purcell et al.;2007). Age, gender, education, Intracranial Volume (ICV), diagnostic status and the top four principal components from population stratification analysis were included as covariates. Post-hoc analysis used Bonferroni correction for adjusting both the number of SNPs and the number of quantitative traits (i.e., significance threshold is 0.05 / 565,373 / 14 = 6.32E-09). Regional genetic association plot was generated using LocusZoom (Pruim et al.;2011). Gene-based association analysis was employed to gain collective statistical evidence of genetic findings at the gene- level (Figure 2). We used GATES (Li et al.;2011) to calculate a gene-level summary p-value for each gene by taking into account gene size, linkage disequilibrium (LD), and constituent SNP level p-values.
The phenotypic variance explained by an identified genetic variant was evaluated using the linear regression after removing effects from covariates as described above. Both linear regression coefficient p-value and Cohen’s d statistic were used to illustrate the significance and effect size of the identified variant, and to facilitate the comparison among different genotyping groups.
Genetic findings of the MTL substructures were further examined in non-overlapping samples regarding their associations with hippocampal measures including voxel-based morphometry (VBM) gray matter density and Fludeoxyglucose-PET (FDG-PET) glucose metabolism. For both association tests, linear regression models were used. In particular, we applied additive genetic models implemented in PLINK v1.90 (Purcell et al.;2007), with age, gender, education and the top four principal components as covariates.
2.7. Targeted genetic analysis of VBM phenotypes
VBM, a computational approach for characterizing structural differences in brain disorders, has been widely applied to AD studies and reported the altered brain volume in a few brain regions including the hippocampus (Risacher et al.;2009, 2010). With this observation, we further evaluated our genetic findings of the MTL substructures for their associations with VBM gray matter density measure in the hippocampal region. The MRI data used in this study were obtained from the ADNI database. These preprocessed MRI data were co-registered to a T1-weighted template, segmented into grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF) compartments with bias correction, unmodulated normalized to Montreal Neurologic Institute (MNI) space as 1×1×1 mm3 voxels, and smoothed with an 8mm Gaussian kernel. ROI-level VBM GM density measurements were further extracted based on the MarsBaR automated anatomical labeling (AAL) atlas for 1,384 subjects who were not included in the GWAS of the MTL substructures. Subjects were treated as outliers and excluded if their VBM measures were greater or smaller than six standard deviations from the mean value. No subjects were removed under this criterion, as shown in Figure 2, 1,384 subjects with quality controlled VBM GM density measurements were analyzed (Supplementary Table 1).
2.8. Targeted genetic analysis of Fludeoxyglucose-PET (FDG-PET) phenotype
ROI-based FDG-PET studies have reported the altered hippocampal metabolism in MCI and AD compared with HC, showing the role of FDG hippocampal measure as a promising biomarker for AD (De Santi et al.;2001,Mosconi et al.;2005). With this observation, we examined the relationship between our MTL genetic findings and FDG measurements. Preprocessed FDG-PET scans were downloaded from the ADNI website (adni.loni.usc.edu) and processed as previously described in (Risacher et al.;2015,Yao et al.;2017a). FDG-PET scans were then aligned to the corresponding MRI scans and normalized to the MNI space as 2×2×2 mm3 voxels. ROI-level glucose metabolism measurements were further extracted based on the MarsBaR AAL atlas for 865 subjects who were not included in the GWAS of MTL substructures. Subjects were treated as outliers and excluded if their FDG-PET measures were greater or smaller than six standard deviations from the mean value. No subjects were removed under this criterion. As shown in Figure 2, finally 865 subjects with quality-controlled FDG-PET glucose metabolism measures were analyzed (Supplementary Table 2).
2.9. Targeted genetic association with AD status in IGAP
International Genomics of Alzheimer’s Project (IGAP) (Lambert et al.;2013) is a large two-stage study based upon GWAS on individuals of European ancestry. In Stage 1, IGAP employed genotyped and imputed data on 7,055,881 single nucleotide polymorphisms (SNPs) to meta-analyze four previously-published GWAS datasets consisting of 17,008 AD cases and 37,154 controls (the European Alzheimer’s disease Initiative (EADI), the Alzheimer Disease Genetics Consortium (ADGC), the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium (CHARGE), and the Genetic and Environmental Risk in AD consortium (GERAD)). In Stage 2, 11,632 SNPs were genotyped and tested for association in an independent set of 8,572 AD cases and 11,312 controls. Finally, a meta-analysis was performed by combining results from Stages 1 & 2. As illustrated in Figure 2, using meta-analysis summary statistics from IGAP stage 1, we checked the relationship between our MTL genetic findings with AD diagnostic status. We also checked the AD association of our findings in a more recent meta-GWAS of AD conducted by the IGAP (Kunkle et al.;2019), which analyzed 21,982 AD cases and 41,944 controls in Stage 1.
2.10. Targeted genetic association with AD status in ADNI
We analyzed the ADNI cohort to examine the direct AD association of our MTL genetic findings. As demonstrated in Figure 2, a total of 1,438 participants from the ADNI cohort (ADNI-1, ADNI-GO and ADNI-2) with corresponding baseline diagnosis (i.e., five values 1–5 indicating HC, SMC, EMCI, LMCI, and AD, respectively) were analyzed. Genetic association analysis with clinical diagnostic status was performed using linear regression under an additive genetic model in PLINK v1.90 (Purcell et al.;2007). Age, gender, education, and the top four principal components from population stratification analysis were included as covariates.
2.11. Tissue-specific eQTL analysis using UKBEC
In order to assess the potential role of the MTL genetic findings in regulating gene expression in hippocampus, we performed brain tissue-specific expression quantitative trait loci (eQTL) analysis. Specifically, we used brain tissue expression dataset available in BRAINEAC (http://www.braineac.org/), a web server for data from the UK Brain Expression Consortium (UKBEC) (Ramasamy et al.;2014). This dataset contains 12 brain tissues from 134 neuropathologically normal subjects (Figure 2). As the MTL substructures analyzed in our GWAS were located in the hippocampus and its neighboring regions, we performed the eQTL analysis using hippocampus-specific expression data. We examined the cis-effect of each identified SNP on the expression of genes located within ±100 kb from the SNP. This tissue-specific eQTL result can help provide novel insights into mechanisms of how the genetic variants affect brain structures via modulating regional gene expression levels to link genetics, transcriptomics, and brain phenomics.
2.12. Tissue-specific gene expression analysis using Allen human brain atlas
Allen human brain atlas (AHBA; http://human.brain-map.org/) includes the brain-wide genome-wide microarray-based gene expression data through systematic sampling of regional brain tissue. One goal of AHBA is to combine genomics with the neuroanatomy to better understand the connections between genes and brain functioning. Complete microarray datasets of six brains from healthy participants are available for download, including two full brains (H0351.2001 and H0351.2002) and four right hemispheres (H0351.1009, H0351.1012, H0351.1015 and H0351.1016). The datasets contain gene expression values normalized across all the brains. We downloaded the microarray data of all six brains, obtaining the expression measures of 58,692 probes in 3,703 brain samples. Then we merged probes to genes and mapped brain samples to MarsBaR AAL atlas using the mean statistics (Yao et al.;2017b). Finally, we obtained the expression data of 29,131 genes for 115 brain ROIs.
In this study, we explored the expression level of our top genetic finding in brain regions. Given that the genetic finding was associated with the hippocampus-relevant region, we compared its hippocampal expression with its expression in other 114 brain regions (Figure 2), to help provide valuable information for revealing the tissue-specific function of the identified gene.
3. Results
3.1. Participant characteristics
A total of 134 ADNI subjects were studied in the GWAS of volumetric measures of 14 MTL substructures (see Table 1 for their characteristics). Using one-way ANOVA or Chi-squared test, significant differences among diagnostic groups were observed for all MTL substructure volumes while not observed for age, gender, education or ICV.
3.2. GWAS of MTL substructures volumes
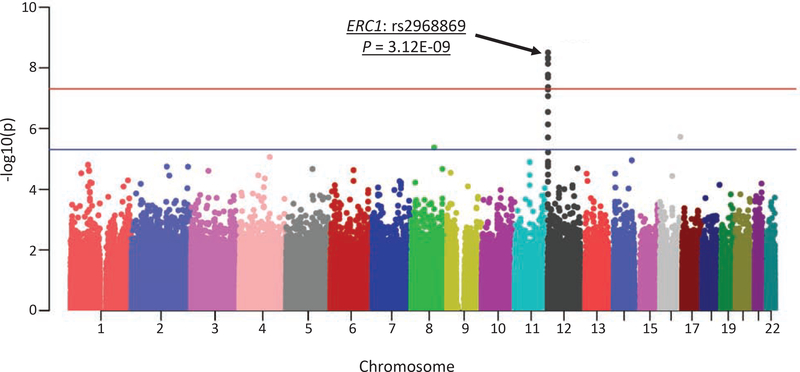
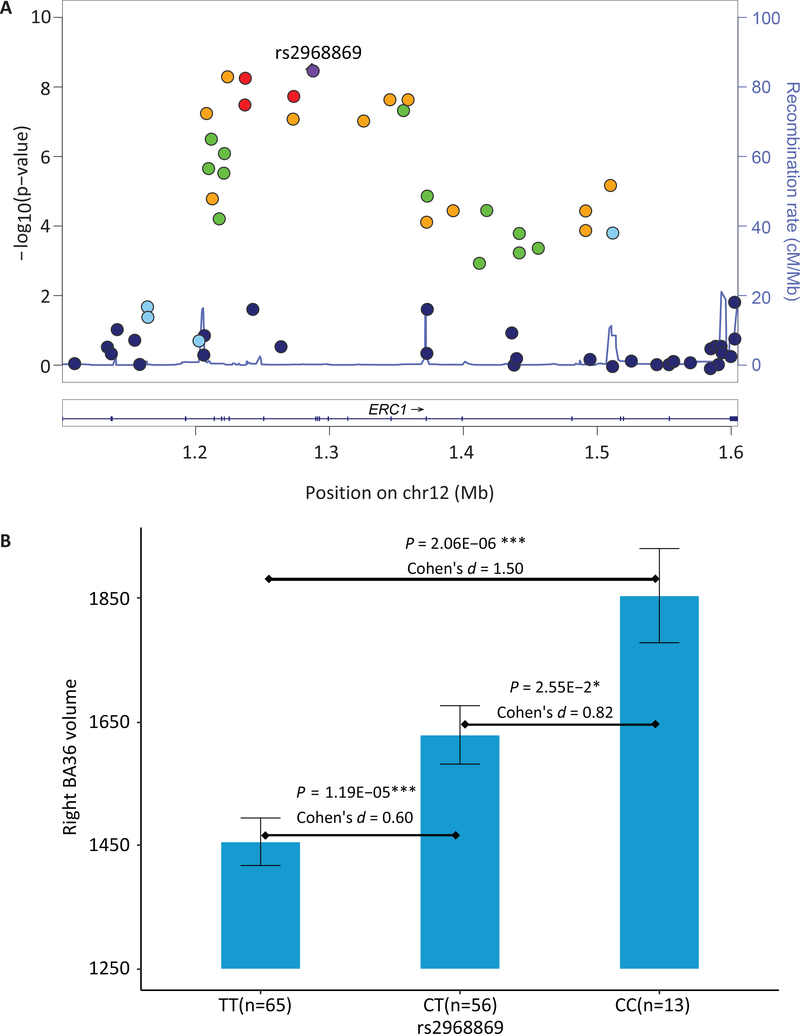
Genetic association between 565,373 SNPs and volumetric measures of 14 MTL substructures were assessed under the additive genetic model and controlled for age, gender, education, ICV, the top four principal components from population stratification, and diagnostic status. A novel significant association between rs2968869 and right BA36 (R-BA36) volume (p = 3.12E-09, corrected p = 3.12E-09 × 565,373 × 14 = 0.025) was identified after adjusting for both the number of SNPs and the number of phenotypes using the Bonferroni method (Figure 3). The minor allele C of rs2968869 (rs2968869-C) was associated with increased R-BA36 volume compared to its major allele T (Figure 4(B)), and the SNP accounted for 15.23% of the variance of the R-BA36 volume. As shown in Figure 4(B), significant differences of the R-BA36 volume exist among three rs2968869 genotype groups (i.e., p = 1.19E-05 between TT and CT, p = 2.06E-06 between TT and CC, and p = 2.55E-02 between CT and CC). In addition, ERC1 rs2968869-C was associated with increased R-BA36 volume across all diagnostic groups (Supplementary Figure 2).
Figure 3. Manhattan plot of GWAS results of right BA36 volume.
Blue and red lines correspond to the p-value of 5E-05 and 5E-07, respectively.
Figure 4. Association and effect of ERC1 rs2968869 on right BA36 volume.
(A) All SNPs within ERC1 are plotted based on their genetic association statistics -log10(P) values. NCBI build 37 genomic position and recombination rates are calculated from the 1,000 Genome Project reference data. The color scale of r2 values is used to label SNPs based on their degree of linkage disequilibrium with rs2968869. Genes in the region are labeled with arrows denoting 5’- to −3’ orientation. (B) Mean right BA36 volume with standard errors are plotted against rs2968869 genotype groups (TT, CT and CC). P value indicates the association significance of rs2968869 with right BA36 volume. P values are calculated from linear regression with age, gender, education, ICV, the top four principal components from population stratification analysis and diagnosis as covariates. Cohen’s d indicates the effect size of minor allele C (one copy or two copies) of rs2968869 on right BA36 volume, after being adjusted for age, gender, education, ICV, the top four principal components from population stratification analysis, and diagnosis. Presence of minor allele C of rs2968869 suggests an additive effect of increasing right BA36 volume and this SNP accounts for15.23% of the phenotypic variance.
3.3. Gene-based association analysis of MTL substructures volumes
An additional analysis was performed to test the gene-based association with R-BA36 volume. We used GATES (Li et al.;2011) to assess the gene-based association with the volume of right BA36. The protein-coding gene ERC1 was significantly associated with R-BA36 volume (corrected p-value = 3.97E-03), after Bonferroni correction for the number of genes.
3.4. Association of ERC1 rs2968869 with hippocampal gray matter density
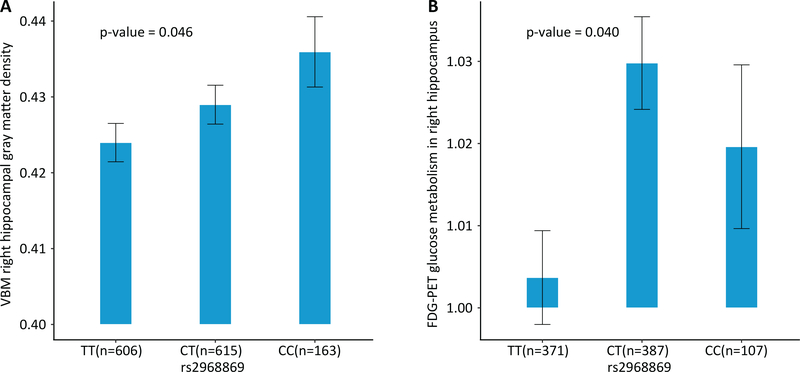
Given the effect of rs2968869 on R-BA36 volume, we further assessed the association of rs2968869 with the VBM gray matter density of the right hippocampus in 1,384 ADNI subjects (Supplementary Table 1) who were not included in the GWAS of MTL substructure volumes. The minor allele (C) of rs2968869 was significantly associated with increased right hippocampal gray matter density (p = 0.046; Figure 5(A)) under linear regression with age, gender, education and the top four principal components as covariates.
Figure 5. Association of ERC1 rs2968869 with VBM gray matter density and FDG metabolism in the right hippocampal region.
(A) Mean right hippocampal gray matter density with standard errors is plotted against the rs2968869 genotype groups (i.e., TT, CT and CC). P value indicates the significance of the association between rs2968869 and mean right hippocampal gray matter density, with age, gender, education and the top four principal components from population stratification analysis as covariates. The C allele of rs2968869 is associated with increased right hippocampal gray matter density. (B) Mean right hippocampal FDG-PET glucose metabolism with standard errors is plotted against the rs2968869 genotype groups (i.e., i.e., TT, CT and CC). The C allele of rs2968869 is associated with increased FDG-PET measured glucose metabolism in right hippocampus.
3.5. Association of ERC1 rs2968869 with FDG-PET glucose metabolism
Altered hippocampal metabolism have been reported in AD compared to HC. In this study, we explored whether our volumetric genetic finding could also be associated with glucose metabolism in the same region. We examined the association of rs2968869 with right hippocampal metabolism in 865 ADNI subjects (Supplementary Table 2) who were not included in the GWAS of MTL substructure volumes. We observed that rs2968869-C was significantly associated with increased glucose metabolism in the right hippocampus (p = 0.040; Figure 5(B)). The heterozygous group showed higher metabolism compared with the homozygous groups. Of note, these two AD endophenotypes (i.e., gray matter density and glucose metabolism of right hippocampus) were highly correlated with each other (correlation coefficient = 0.6). Thus we reported the original p-values without correcting for multiple comparison.
3.6. Association of ERC1 rs2968869 with AD
The hippocampus has been widely studied as one of the earliest affected brain regions in the progression of AD, and various imaging measures of the hippocampus are associated with AD status. With this observation, we further examined whether the genetic finding from our imaging GWAS was directly associated with AD by leveraging the results from IGAP, a large meta-analysis of AD (Lambert et al.;2013). According to the summary statistics of IGAP stage 1, rs2968869 showed significant association with AD (p = 0.045, N = 54,162), with effect size of −0.035 indicating the protective role of rs2968869-C for AD. However, in a more recent IGAP study (Kunkle et al.;2019), no significant AD association (p = 0.113, N = 63,926) was identified for rs2968869.
We also examined the rs2968869 association with AD in the ADNI cohort, and observed that rs2968869 was significantly associated with AD diagnostic status (p = 0.0069, N = 1,438) with effect size of −0.072 indicating the protective role of rs2968869-C for AD. Both AD association findings from IGAP and ADNI data aligned well with our imaging genetic result.
3.7. Hippocampus-specific eQTL analysis of rs2968869
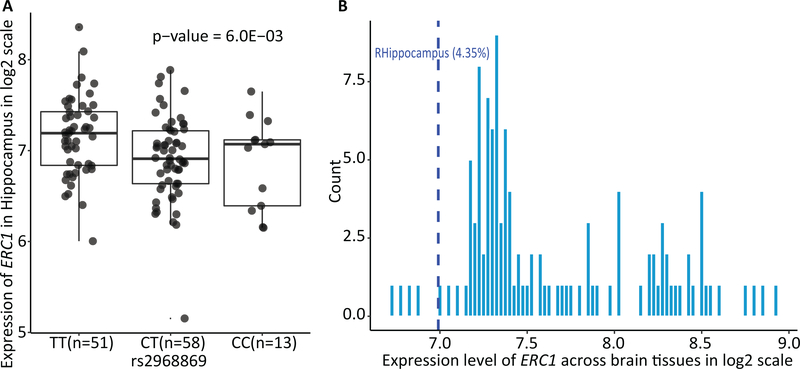
Hippocampus-specific cis-eQTL analysis identified that rs2968869-C was significantly associated with downregulated expression of ERC1 in the hippocampus (p = 6.00E-03). Figure 6(A) shows the differential expression level of ERC1 among rs2968869 genotype groups. We observed that the heterozygous group of rs2968869 exhibited the lowest level ERC1 expression. To further validate if rs2968869 was the lead eQTL for ERC1 in hippocampus, we examined if other eQTLs for ERC1 in the hippocampus were in high LD with rs2968869. From the ADNI data, there were a total of 117 SNPs located within ±100kb of ERC1, among which 18 SNPs (including rs2968869) were eQTLs (uncorrected p < 0.05) for ERC1 in the UKBEC. Thus, we evaluated the LD (D’ and r2) between rs2968869 with each of 17 eQTLs using the ADNI genotyping data. Only one SNP rs2906109 was in high LD (D’ > 0.8, r2 > 0.8) with the iQTL SNP rs2968869. Both rs2968869 and rs2906109 are significantly associated with the ERC1 gene expression (p = 6.00E-03 and 3.00E-03, respectively) and R-BA36 imaging phenotype (p = 3.12E-09 and 1.70E-08, respectively). It appears that rs2906109 shows a slightly stronger association with the gene expression while rs2968869 shows a slightly stronger association with the R-BA36 imaging phenotype.
Figure 6. Hippocampus-specific expression of ERC1.
(A) illustrates the UKBEC hippocampus-specific cis-eQTL results. It shows the expression level of ERC1 in the hippocampus stratified by rs2968869 groups (TT, CT and CC). P-value indicates the association significance of rs2968869 with the expression level of ERC1 in the hippocampus. P-value is calculated from linear regression with gender as covariate. Presence of minor allele C of rs2968869 suggests the effect of decreased expression of ERC1. (B) illustrates the expression of ERC1 in the right hippocampus compared with other brain regions in AHBA. Histogram shows the distribution of ERC1 expression across 115 brain AAL ROIs. Blue line illustrates the expression level of ERC1 in the right hippocampus at the 4.35th percentile, indicating the ERC1 expression in the right hippocampus is higher than 4.35% brain regions.
We also conducted a summary Mendelian Randomization (SMR) analysis (Zhu et al.;2016) which used Wald estimator to evaluate the association between gene expression and the trait due to either causality or pleiotropy. The SMR analysis was performed using the GWAS results of the R-BA36 volume and the eQTL analysis result of the hippocampus. The SMR p-value is not significant (p > 0.05). Of note, the statistical power of SMR analysis on our data appears substantially limited by two factors: 1) SMR requires GWAS with very large sample size (N > 10,000; (Teumer;2018)) while our GWAS included only 134 participants; and 2) our GWAS analysis was performed on the R-BA36 volume while the eQTL analysis was performed on the hippocampus. We hope to further investigate the mechanisms among the lead SNP, ERC1 expression and R-BA36 volume when more relevant data become available (e.g., (1) large GWAS data coupled with the R-BA36 volumetric phenotype and (2) ERC1 expression in the R-BA36 region coupled with the lead SNP information).
3.8. Hippocampus-specific gene expression analysis of ERC1
The R-BA36 associated genetic finding rs2968869 is located in ERC1. Therefore, we assessed the hippocampus-specific expression of ERC1 by comparing it with its expression in other brain re,gions. Figure 6(B) illustrated the distribution of ERC1 expression across all 115 brain ROIs, from which the expression of ERC1 in the hippocampus was lower than 95.65% other brain regions. This suggests the low expression level of ERC1 in the normal hippocampus. Further discussion on the effect of ERC1 expression level on brain function and disease is available in the next section.
4. Discussion
We performed GWAS on volumetric measures of 14 MTL substructures, and identified a novel association between ERC1 SNP rs2968869 and R-BA36 volume in 134 ADNI subjects. To the best of our knowledge, this is among the first GWAS of hippocampal subfields and neighboring MTL substructures extracted from the high-resolution MRI data in AD-related study. The minor allele C of rs2968869 is associated with increased R-BA36 volume, demonstrating a protective effect. The neuroprotective role of rs2968869-C is further confirmed by several post-hoc analyses. First, rs2968869-C carriers exhibited higher gray matter density and higher glucose metabolism in the right hippocampus in larger and independent ADNI samples. Second, a large-scale landmark meta GWAS in AD (N = 54,162) also indicated the protective effect of rs2968869-C. Third, a tissue-specific cis-eQTL analysis identified an association between rs2968869-C and lower ERC1 expression in the hippocampus. Finally, brain-wide genome-wide expression data in healthy samples also demonstrated the lower expression of ERC1 in the right hippocampus compared with that in a majority of other brain regions. Below we discuss the functions and involvements of ERC1 and BA36 in brain regions and AD. We also explore possible underlying molecular mechanisms of AD by linking our genetic finding with phenotype via hippocampus-specific transcriptome eQTL analysis of ERC1 rs2968869.
ERC1 (also known as ELKS) encoding protein ELKS/RAB6-interacting/CAST family member 1, which is a family of Rab3-interacting molecule (RIM)-binding proteins, has been widely studied in brain disorders given that it is highly enriched in the active zone of the brain, especially in the hippocampal region. In the active zone, ERC1 encoded protein controls the presynaptic Ca2+ signal to regulate neurotransmitter release, which typically is impaired in complex brain disorders like AD. A number of molecular studies have investigated the important role of ERC1 in functions and organizations of the active zone. For example, ERC1 protein has shown its positive regulation effects on both neurotransmitter release at synapses and Ca2+ influx in nerve terminals in the study of mouse hippocampus (Dong et al.;2018,Liu et al.;2014). These findings indicate the functions of ERC1 in the active zone. ERC1 is also implicated in the NF-kappaB signaling pathway which participates in the regulation of neuroinflammation. This might suggest additional aspects for the involvement of ERC1 in neurodegenerative diseases (Liu et al.;2017). Given the critical role of ERC1 in brain disorders and hippocampal region, our reported variant rs2968869, may indicate a possible function for AD by modulating the transcription of ERC1 in the medial temporal region.
Pathologically, NFT of the MTL substructure has been reported for the involvement in the hippocampus and extrahippocampal cortical regions in early AD (Didic et al.;2011,Nelson et al.;2012). Studies discover that the PRC is indeed the first region displaying NFT pathology in early stage of AD (Mason et al.;2017,Wolk et al.;2017), instead of any hippocampal subfields as one of the earliest affected regions in AD. Another early AD study showed that regional tau deposition was associated with atrophy in PRC but not in hippocampal subfields (Sone et al.;2017). PRC is comprised of BA35 and BA36, of which BA35 has been widely studied for its association with early NFT deposition in AD pathology. However, in recent studies, BA36 demonstrates a similar or even better performance than BA35 for discriminating preclinical AD from normal aging (Wisse et al.;2018,Wolk et al.;2017). Accordingly, the BA36 associated genetic finding can help provide deeper insights into the understanding of molecular mechanisms of AD pathogenesis, especially for the early stage of AD.
Our imaging genetic analysis identified the association between ERC1 rs2968869 and R-BA36. To understand the underlying molecular basis, it is important to examine the influence of rs2968869 on gene expression in this particular brain region. Our cis-eQTL analysis detected the down-regulation effect of minor allele (C) of rs2968869 on ERC1 expression in the hippocampus. Thus, the protect role of rs2968869 suggests relatively high ERC1 expression in the hippocampus in AD. This hypothesis has been supported in a mouse model study, which analyzed the expression level of hippocampal ERC1 in AD and control samples, and reported significantly higher expression of ERC1 in the AD hippocampus compared with the control hippocampus (4.45 fold; p < 0.05) (Acquaah-Mensah et al.;2015). The brain-wide gene expression data on healthy brains also show relatively low expression of ERC1 in the hippocampus compared with most of other brain regions. We acknowledge that although we did not identify casual or pleiotropic association between ERC1 expression and R-BA36 volume, it warrants further investigation to explore the underlying mechanisms among rs2968869, ERC1 expression and R-BA36 volume when more data become available.
In conclusion, we have revealed a novel association between the minor allele (C) of ERC1 rs2968869 and increased right BA36 volume. This genetic finding has been found to be associated with multiple AD phenotypes including right hippocampal gray matter density and glucose metabolism as well as AD diagnostic status, all indicating the protective effect of rs2968869-C in AD. We also checked the associations between rs2968869 with brain imaging phenotypes available in the ENIGMA study (http://enigma.ini.usc.edu/) and other AD phenotypes including CSF biomarkers (ABeta, Tau and pTau) and MMSE in the ADNI study, from which we identified one nominally significant association between rs2968869 with the caudate volume (p = 0.032). An additional cis-eQTL analysis on brain transcriptome data has connected rs2968869-C with lower ERC1 expression in the hippocampus. It warrants further investigation of the molecular mechanism of the identified ERC1 rs2968869 in the hippocampus, including the understanding of possible molecular pathway on how the variant modulates the ERC1 expression in the hippocampus and subsequently affects the neurotransmitter release at the hippocampal active zone.
The advances of high-resolution MRI technology allow the extraction of improved measures from the MTL substructures for studying their role in AD progression and their genetic basis, which provides enormous opportunities to gain deeper insights into the molecular mechanisms of AD pathogenesis. One limitation of this work is the modest sample size. Some ADNI-1/GO/2 subjects have relatively low image quality due to either incomplete coverage of the hippocampus or low image contrast, and thus are not included in the study. We anticipate the newly collected ADNI-3 data to have improved image quality. An interesting future topic is to perform a replication study on the ADNI-3 data.
Supplementary Material
Highlights.
MTL structures are promising indicators for early detection of AD
Fine-grained MTL substructures are valuable traits for studying AD genetics
An ERC1 locus is significantly associated with the right Brodmann area 36 volume
The identified genetic variation is also associated with several AD biomarkers
The finding can help understand AD genetic and molecular mechanism
Acknowledgements
This work was supported in part by NIH R01 EB022574, R01 LM011360, RF1 AG063481, R01 AG060054, U19 AG024904, R01 AG019771, P30 AG010133, R01 LM012535, and R03 AG054936, and by NSF IIS 1837964. See ADNI, UKBEC and IGAP Acknowledgements in Supplementary Materials.
Footnotes
Disclosure statement
The authors have no actual or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acquaah-Mensah GK et al. , 2015. A regulatory role for the insulin- and BDNF-Linked RORA in the hippocampus: Implications for Alzheimer’s disease. J. Alzheimer’s Dis. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, 1997. Staging of Alzheimer-related cortical destruction. Int. psychogeriatrics 9 Suppl 1,257–61; discussion 269–72. [PubMed] [Google Scholar]
- Carr VA et al. , 2017. Individual differences in associative memory among older adults explained by hippocampal subfield structure and function. Proc. Natl. Acad. Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong S et al. , 2018. Volumetric comparison of hippocampal subfields extracted from 4-minute accelerated vs. 8-minute high-resolution T2-weighted 3T MRI scans. Brain Imaging Behav. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong S et al. , 2016. Building a surface atlas of hippocampal subfields from high resolution T2-weighted MRI scans using landmark-free surface registration, in: 2016 IEEE 59th International Midwest Symposium on Circuits and Systems (MWSCAS) pp. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong S et al. , 2015. Surface-based morphometric analysis of hippocampal subfields in mild cognitive impairment and Alzheimer’s disease, in: Proceedings of the 58th International Midwest Symposium on Circuits and Systems (MWSCAS) pp. 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Flores R et al. , 2015. Structural imaging of hippocampal subfields in healthy aging and Alzheimer’s disease. Neuroscience 309, 29–50. [DOI] [PubMed] [Google Scholar]
- De Santi S et al. , 2001. Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiol. Aging. [DOI] [PubMed] [Google Scholar]
- Didic M et al. , 2011. Which memory system is impaired first in alzheimer’s disease? J. Alzheimer’s Dis. 27, 11–22. [DOI] [PubMed] [Google Scholar]
- Dong W et al. , 2018. CAST/ELKS Proteins Control Voltage-Gated Ca2+ Channel Density and Synaptic Release Probability at a Mammalian Central Synapse. Cell Rep. 24, 284–293.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo H et al. , 2016. Associations of hippocampal subfields in the progression of cognitive decline related to Parkinson’s disease. NeuroImage Clin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler J et al. , 2019. 2019 Alzheimer’s disease facts and figures. ALZHEIMERS Dement. 15, 321–387. [Google Scholar]
- Greenspan KS et al. , 2017. Heritability of Hippocampal Formation Sub-region Volumes. J. Neurol. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP et al. , 2017. Novel genetic loci associated with hippocampal volume. Nat. Commun. 8, 13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindy NC et al. , 2016. Linking pattern completion in the hippocampus to predictive coding in visual cortex. Nat Neurosci 19, 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgusluoglu-Moloch E et al. , 2017. Targeted neurogenesis pathway-based gene analysis identifies ADORA2A associated with hippocampal volume in mild cognitive impairment and Alzheimer’s disease. Neurobiol. Aging 60, 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE et al. , 2015. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage 115, 117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR et al. , 2016. A/T/N: An unbiased descriptive classification scheme for Alzheimer disease biomarkers. Neurology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW et al. , 2019. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Ap, tau, immunity and lipid processing. Nat. Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC et al. , 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lech RK, Suchan B, 2013. The medial temporal lobe: Memory and beyond. Behav. Brain Res. [DOI] [PubMed] [Google Scholar]
- Li MX et al. , 2011. GATES: A rapid and powerful gene-based association test using extended Simes procedure. Am. J. Hum. Genet. 88, 283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C et al. , 2014. The active zone protein family ELKS supports Ca 2+ influx at nerve terminals of inhibitory hippocampal neurons. J. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T et al. , 2017. NF-kB signaling in inflammation. Signal Transduct. Target. Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruszak A, Thuret S, 2014. Why looking at the whole hippocampus is not enough—a critical role for anteroposterior axis, subfield and activation analyses to enhance predictive value of hippocampal changes for Alzheimer’s disease diagnosis. Front. Cell. Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason EJ et al. , 2017. Family History of Alzheimer’s Disease is Associated with Impaired Perceptual Discrimination of Novel Objects. J. Alzheimer’s Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI et al. , 2013. The diffeomorphometry of temporal lobe structures in preclinical Alzheimer’s disease. Neuroimage Clin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey R et al. , 2019. Neuroimaging Phenotypes Implicated For GWAS of PTSD Through The PGC And ENIGMA Worldwide Consortia. Eur. Neuropsychopharmacol. [Google Scholar]
- Morey RA et al. , n.d. Hippocampal subfield GWAS in PTSD Genome wide association study of hippocampal subfield volume in PTSD cases and trauma-exposed controls fThe VA Mid-Atlantic MIRECC Workgroup includes the following contributors. [Google Scholar]
- Mosconi L et al. , 2005. Reduced hippocampal metabolism in MCI and AD: Automated FDG-PET image analysis. Neurology. [DOI] [PubMed] [Google Scholar]
- Mueller SG et al. , 2018. Systematic comparison of different techniques to measure hippocampal subfield volumes in ADNI2. NeuroImage Clin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders P et al. , 2019. Electroconvulsive Therapy for Depression: Neurobiological Mechanisms, in: Neurobiology of Depression. [Google Scholar]
- Nelson PT et al. , 2012. Correlation of alzheimer disease neuropathologic changes with cognitive status: A review of the literature. J. Neuropathol. Exp. Neurol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho K et al. , 2015. Protective variant for hippocampal atrophy identified by whole exome sequencing. Ann. Neurol. 77, 547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nho K et al. , 2013. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol. Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ et al. , 2011. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 27, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S et al. , 2007. PLINK: A Tool Set for Whole-Genome Association and Population- Based Linkage Analyses. Am. J. Hum. Genet. 81,559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK et al. , 2015. GWAS of longitudinal amyloid accumulation on 18 F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A et al. , 2014. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat. Neurosci. 17, 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher S et al. , 2009. Baseline MRI Predictors of Conversion from MCI to Probable AD in the ADNI Cohort. Curr. Alzheimer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL et al. , 2015. APOE effect on Alzheimer’s disease biomarkers in older adults with significant memory concern. Alzheimer’s Dement. 11, 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risacher SL et al. , 2010. Longitudinal MRI atrophy biomarkers: Relationship to conversion in the ADNI cohort. Neurobiol. Aging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mejias E et al. , 2019. Distinct disease-sensitive GABAergic neurons in the perirhinal cortex of Alzheimer’s mice and patients. Brain Pathol. bpa.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ et al. , 2015. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimer’s Dement. 11, 792–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saykin AJ et al. , 2010. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer’s Dement. 6, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz H et al. , 2019. Content tuning in the medial temporal lobe cortex: Voxels that perceive, retrieve. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeeth DM et al. , 2019. Polygenic risk for circulating reproductive hormone levels and their influence on hippocampal volume and depression susceptibility. Psychoneuroendocrinology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EE, Kosslyn SM, 2013. Cognitive psychology: mind and brain. Pearson Higher Ed. [Google Scholar]
- Sone D et al. , 2017. Regional tau deposition and subregion atrophy of medial temporal structures in early Alzheimer’s disease: A combined positron emission tomography/magnetic resonance imaging study. Alzheimer’s Dement. Diagnosis, Assess. Dis. Monit. 9, 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire LR et al. , 2004. THE MEDIAL TEMPORAL LOBE. Annu. Rev. Neurosci. [DOI] [PubMed] [Google Scholar]
- Teumer A, 2018. Common Methods for Performing Mendelian Randomization. Front. Cardiovasc. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer D et al. , 2018. Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Mol. Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leemput K et al. , 2009. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus 19, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H et al. , 2011. A learning-based wrapper method to correct systematic errors in automatic image segmentation: consistently improved performance in hippocampus, cortex and brain segmentation. Neuroimage 55, 968–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yushkevich PA, 2013. Multi-atlas segmentation without registration: a supervoxel-based approach, in: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse L et al. , 2018. MEDIAL TEMPORAL LOBE SUBREGIONAL ATROPHY PATTERN IN SEMANTIC VARIANT PRIMARY PROGRESSIVE APHASIA AND ASSOCIATIONS WITH COGNITIVE IMPAIRMENTS. Alzheimer’s Dement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk DA et al. , 2017. Medial temporal lobe subregional morphometry using high resolution MRI in Alzheimer’s disease. Neurobiol. Aging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L et al. , 2017. Multi-template analysis of human perirhinal cortex in brain MRI: Explicitly accounting for anatomical variability. Neuroimage 144, 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X et al. , 2019. Targeted genetic analysis of cerebral blood flow imaging phenotypes implicates the INPP5D gene. Neurobiol. Aging 81, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X et al. , 2017a. Tissue-specific network-based genome wide study of amygdala imaging phenotypes to identify functional interaction modules. Bioinformatics 33, 3250–3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X et al. , 2017b. Two-dimensional enrichment analysis for mining high-level imaging genetic associations. Brain Informatics 4, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA et al. , 2015. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum. Brain Mapp. 36, 258–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA et al. , 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116–28. [DOI] [PubMed] [Google Scholar]
- Zhao B et al. , 2019. GWAS of 19,629 individuals identifies novel genetic variants for regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z et al. , 2016. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.