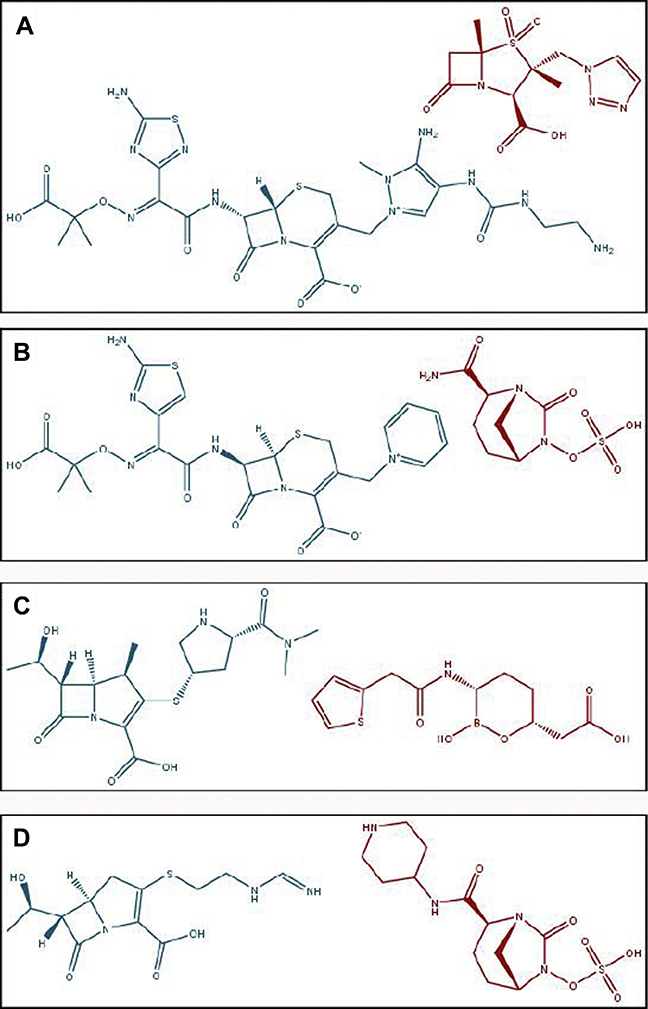
INTRODUCTION
As a class, the β-lactams are the most commonly prescribed and clinically dependable antimicrobials in the United States, representing more than 65% of injected antibiotic prescriptions from 2004 to 20141 and 45% of oral antibiotic prescriptions in 2016.2 Given their effectiveness, the development of resistance to β-lactam antibiotics creates a major concern for physicians, scientists, and policymakers around the world. This review focuses on the emergence of resistance to the 4 novel β-lactam–β-lactamase inhibitor combinations approved between 2014 and 2019 in the United States: ceftolozane-tazobactam, ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam (Fig. 1). The resistance mechanisms that have been reported to date are summarized and the implications of these findings highlighted.
Fig. 1.
Structures of (A) ceftolozane-tazobactam, (B) ceftazidime-avibactam, (C) meropenem-vaborbactam, and (D) imipenem-relebactam. In all panels, the β-lactamase inhibitor (red) is located to the right of the β-lactam partner (blue).
MECHANISM OF ACTION OF β-LACTAM ANTIBIOTICS
Cell wall biosynthesis is critical to bacterial cell division, and most bacteria require a cell wall for survival.3 Cell walls are made of peptidoglycan, long polymers of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) joined in alternating order by β-1,4 glycosidic linkages. Each NAM subunit is attached a short pentapeptide, which is cross-linked between peptidoglycan strands to create a meshlike structure that provides strength to the cell wall. These linkages, which occur between the penultimate D-alanine of 1 peptide and the lysine or diaminopimelic acid of another, are catalyzed by DD-transpeptidases, known as penicillin-binding proteins (PBPs).4,5
β-Lactam antibiotics enter the transpeptidase active site of PBPs and stereochemically mimic the terminal D-alanine residues of the peptide.5 When the active site serine of the PBP attacks the β-lactam ring rather than a peptide bond, it forms a covalent acyl-enzyme complex that deacylates very slowly, crippling the PBP and preventing the final step of cell wall biosynthesis.1 This leads to potentially endless cycles of futile synthesis and degradation of nonfunctional peptidoglycan, depleting cellular stores of precursors and amplifying cytotoxicity in the process by permitting the entry of water into the cell.6
β-LACTAMASES
β-Lactamases are bacterial enzymes that hydrolyze the β-lactam bond of β-lactam antibiotics, rendering them nonfunctional. β-Lactamases are divided into 4 molecular classes by mechanism, conserved residues, and sequence homology. Classes A, C, and D β-lactamases use a conserved serine-based mechanism to hydrolyze the β-lactam bond. Class B metallo-β-lactamases catalyze the hydrolysis of the β-lactam bond using a Zn2+-based mechanism.7 For purposes of this review, alterations to class A and class C β-lactamases are discussed using the standardized numbering scheme of Ambler and colleagues8 and Structural Alignment-based Numbering of class C β-lactamases, respectively.9
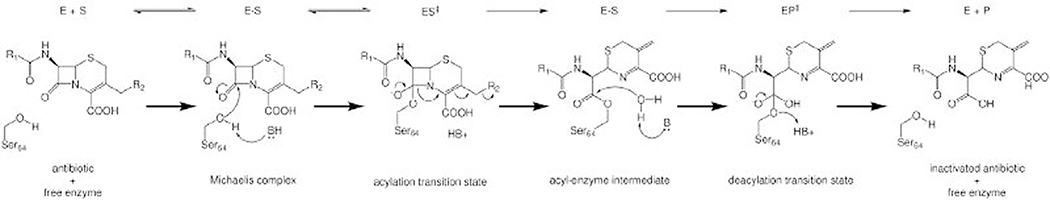
The basic mechanism used by class A and class C serine β-lactamases involves binding, acylation, and deacylation phases with 2 transition states (Fig. 2). Binding occurs when a substrate associates with the enzyme to form a reversible Michaelis complex. In the acylation phase of a class A or class C serine β-lactamase mechanism, a general base deprotonates the catalytic serine residue, permitting nucleophilic attack of the carbonyl carbon of the β-lactam ring, forming a high-energy transition state, which quickly collapses into the acyl-enzyme complex. Deacylation occurs when a water molecule is deprotonated and nucleophilically attacks the same carbon atom, creating a second high-energy transition state that collapses to restore the serine and release an inactive β-lactam (see Fig. 2).10–13
Fig. 2.
The class A and class C serine β-lactamase mechanism involves acylation and deacylation through high-energy tetrahedral transition states. In this example with a class C enzyme, B and HB+ represent a generic base and conjugate acid, respectively. The identity of the bases may vary by class, enzyme, and substrate.
THE Ω-LOOP OF β-LACTAMASES
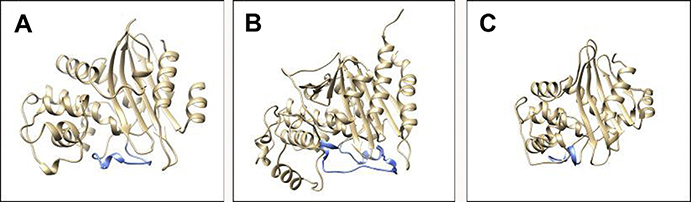
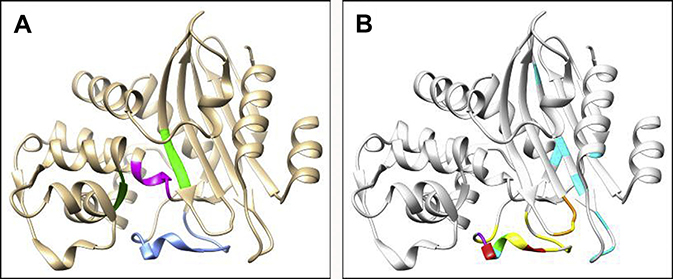
Named for its structural resemblance to the Greek letter Ω, the Ω-loop is a highly mobile and dynamic region in β-lactamases14 and is roughly defined as encompassing residues 164 to 179 in class A enzymes (15 amino acids), residues 188 through 221 (33 amino acids) in class C enzymes, and residues 143 through 173 (30 amino acids) in class D enzymes, although exact designations vary by research group and by family within a class (Fig. 3). The Ω-loop forms the floor of the active site and creates a wall that binds and positions the R1 group of β-lactams, helping to determine substrate specificity.15 In class A β-lactamases, the Ω-loop is believed to be rigid (rather than flexible) due to hydrogen bonding but remains mobile and able to move as a unit. Simulations suggest class C β-lactamases have a more balanced Ω-loop with both flexible and rigid characteristics and the ability to serve as a mechanical switch, meaning it is able to alternate between more flexible and more rigid states based on changes in the hydrogen bonding network.14 Amino acid substitutions in the Ω-loop of class A and class C β-lactamases were shown to expand the substrate spectrum of these enzymes toward oxyimino-cephalosporins.16–19 The precise details and mechanism by which this enhanced ability to hydrolyze these novel cephalosporins with complex R1 side chains occurs still are uncertain.
Fig. 3.
The location of the Ω-loop (blue) in (A) KPC-2 (PDB#: 2OV5), (B) PDC-1 (PDB#: 4GZB), and (C) OXA-48 (PDB#: 4S2P).
β-LACTAMASE INHIBITORS
An established approach to overcoming β-lactam resistance is to reduce the activity of β-lactamases, thus preserving the efficacy of penicillins, cephalosporins, and carbapenem antibiotics. Two approaches to inhibition have targeted β-lactamases successfully, using (1) a suicide, or mechanism-based, inhibitor, and (2) a reversible inhibitor. Suicide inhibitors (including clavulanic acid, sulbactam, and tazobactam) form stable acyl-enzyme complexes, which can undergo postacylation chemistry and fragment or deacylate very slowly. In some cases, these inhibitors permanently inactivate the enzyme in the process. In contrast, reversible inhibitors (including diazabicyclooctanes [DBOs] and boronates) are able to deacylate from the β-lactamase without being modified and proceed to inhibit another β-lactamase molecule.20–22 Current β-lactamase inhibitors fall into 1 of 3 chemical classes: the suicide inhibitors, which contain β-lactam rings but are less readily hydrolyzed (eg, clavulanic acid, sulbactam and tazobactam); the DBOs, which consist of an 8-membered ring partially analogous to the β-lactam bond (avibactam and relebactam); and the boronic acid transition state inhibitors, which are boronates that mimic transition states (vaborbactam).20,23
NEWER β-LACTAM AND β-LACTAMASE INHIBITOR COMBINATIONS: CEFTOLOZANE-TAZOBACTAM
Approved by the US Food and Drug Administration (FDA) in 2014, ceftolozane-tazobactam is indicated for use in complicated intra-abdominal infections (cIAIs), complicated urinary tract infections (cUTIs), hospital-acquired bacterial pneumonia (HABP), and ventilator-associated bacterial pneumonia (VABP) in adults.24 Ceftolozane-tazobactam additionally is being or has been investigated for use in adult patients with burns (NCT03002506), indwelling external ventricular drains (NCT03309657), and multidrug-resistant Pseudomonas aeruginosa infections (NCT03510351), and in pediatric patients for gram-negative infections or as perioperative prophylaxis (NCT02266706), for cUTIs (NCT03230838), and for cIAIs (NCT03217136). Ceftolozane-tazobactam is clinically effective against a wide variety of common gram-negative bacteria and some gram positives (Table 1).24
Table 1.
Clinical indications for the use of ceftolozane-tazobactam
| Indication | Indicated Bacteria | Patient Population | Clinical Trial |
|---|---|---|---|
| cIAIs | Enterobacter cloacae, Escherichia coli, K oxytoca, K pneumoniae, Proteus mirabilis, P aeruginosa, Bacteroides fragilis, Streptococcus anginosus, Streptococcus constellatus, and Streptococcus salivarius | Adults | ASPECT-cIAI25 NCT01445665 NCT01445678 NCT0273999726 |
| cUTIs, including pyelonephritis | E coli, K pneumoniae, Proteus mirabilis, and P aeruginosa | Adults | ASPECT-cUTI27 NCT01345929 NCT01345955 NCT0272808928 |
| HABP/VABP | Enterobacter cloacae, E coli, Haemophilus influenzae, K oxytoca, K pneumoniae, Proteus mirabilis, P aeruginosa, and S marcescens | Adults | ASPECT-NP29 NCT02070757 |
Data from Merck & Co., Inc. ZERBAXA (Ceftolozane and Tazobactam) for Injection, for Intravenous Use. Whitehouse Station, NJ 08889 USA; 2014.
Limitations for the combination against indicated organisms include P aeruginosa or Enterobacterales that carry class A and class B carbapenemases (eg, KPC, VIM, NDM, and IMP) or class A, class C, and class D extended-spectrum β-lactamases (ESBLs) (eg, GES-6, PER-1, FOX-4, and OXA-539) that are not readily inhibited by tazobactam.30–34 E coli–producing class A ESBLs are more susceptible to ceftolozane-tazobactam than K pneumoniae and Enterobacter cloacae–expressing class A ESBLs.35–38 Chromosomal and acquired blaAmpCs likely contribute to the former phenotype, because the hyperproduction of AmpCs or class A ESBLs was shown to reduce efficacy of ceftolozane-tazobactam.33,36,39–41
Ceftolozane is an expanded-spectrum cephalosporin that was developed with the intention of creating a novel, antipseudomonal β-lactam antibiotic that targets PBP3.42 Ceftolozane is modeled on the success of the closely related cephalosporin, ceftazidime, which is a first-line treatment of P aeruginosa infections. Specifically, ceftolozane was designed to be stable to the presence of Pseudomonas-derived cephalosporinase (PDC),43 the class C or AmpC β-lactamase of P aeruginosa.44 Unfortunately, as with other oxyimino-cephalosporins, ceftolozane is susceptible to hydrolysis by certain ESBLs (eg, PER-1) and carbapenemases (eg, KPCs) that often occur in conjunction with other ceftolozane-susceptible enzymes and overexpression of class C enzymes reduces its potency.45 Moreover, ceftolozane is readily hydrolyzed by class B metallo-β-lactamases (eg, VIM, IMP, and NDM) and activity against bacteria producing class D OXA β-lactamases is variable.46
Tazobactam is a penicillin-based sulfone derivative developed as a β-lactamase inhibitor47 that inactivates most class A β-lactamases. Tazobactam demonstrates variable activity against bacteria producing class A carbapenemases, class C β-lactamases, and class D β-lactamases.48,49 By using tazobactam in this combination, the goal was to inhibit class A ESBLs (eg, CTX-M) and tazobactam-susceptible class C β-lactamases (eg, CMY), thus extending the usefulness of the combination.33,35,45
REPORTS AND MECHANISMS OF RESISTANCE: CEFTOLOZANE-TAZOBACTAM
In Table 2, the reports of resistance to ceftolozane-tazobactam available to date are summarized. The most frequently reported cause of ceftolozane-tazobactam resistance in isolates for which it is indicated is alterations in PDC, the chromosomally encoded class C β-lactamase of P aeruginosa. Amino acid substitutions, insertions, and deletions in PDC were found in broad survey studies, individual case reports, and laboratory selection experiments, suggesting they can emerge in a variety of ways. Additional mechanisms including acquisition of rare class A β-lactamases as well as amino acid substitutions in OXA enzymes are described herein (see also above).
Table 2.
Mechanisms of resistance to ceftolozane-tazobactam
| Type | Organism | Mechanism(s) | Relevant Amino Acid Substitutions in β-Lactamase | Minimum Inhibitory Concentration of Isolatea | Minimum Inhibitory Concentration in a Susceptible Backgroundb | Reference |
|---|---|---|---|---|---|---|
| Case report | P aeruginosa | PDC | G156D | ≥64 | ≥64 | 57 |
| Case report | P aeruginosa | Overexpression of PDC, loss of oprD | T96I | >32 | 32 | 52 |
| Case report | P aeruginosa | Overexpression of PDC, loss of oprD | E219K | >32 | 16 | 52 |
| Case report | P aeruginosa | Overexpression of PDC, loss of oprD | ΔG202-E219 | 32 | 32 | 52 |
| Case report | P aeruginosa | Overexpression of PDC, loss of oprD, mutations in mexD, mexT, mexI and mexR | R52Q, T79 A, G156D | ≥256 | N/A | 61 |
| Case report | P aeruginosa | Overexpression of PDC | V211A | 64 | N/A | 50 |
| Case report | P aeruginosa | PDC, mutation in mexR | ΔP208-G214 | >128 | N/A | 50 |
| Case report | P aeruginosa | PDC, loss of oprD | G156D | 32–48 | N/A | 60 |
| Laboratory strain | P aeruginosa | Overexpression of PDC | G156D | 64 | 32 | 55 |
| Laboratory strain | P aeruginosa | Overexpression of PDC | E219K, V329I | 32 | 64 | 55 |
| Laboratory strain | P aeruginosa | Overexpression of PDC | F121L, Q130R, E219K, V329I | 128 | 128 | 55 |
| Survey | P aeruginosa | Overexpression of PDC, loss of oprD | Q128R, V211T, S279T | 256 | N/A | 54 |
| Survey | P aeruginosa | PDC, loss of oprD and mutations in mexT | A5V, V211A, G214R, G220S | 12 | N/A | 54 |
| Survey | P aeruginosa | Overexpression of PDC, loss of oprD | Q128R, V211A, G220S | 256 | N/A | 54 |
| Survey | P aeruginosa | Overexpression of PDC, loss of oprD | V211A | 256 | N/A | 54 |
| Survey | P aeruginosa | PDC | P153L | N/A | 8 | 51 |
| Survey | P aeruginosa | PDC | G214R | N/A | 8 | 51 |
| Survey | P aeruginosa | PDC | N346I | N/A | 4–8 | 51 |
| Survey | P aeruginosa | PDC | R100H, G214R | N/A | 16 | 51 |
| Survey | P aeruginosa | PDC | E219K | N/A | 64 to >64 | 51 |
| Survey | P aeruginosa | PDC | E219G | N/A | 32 | 51 |
| Survey | P aeruginosa | PDC | F121L, M174L | N/A | 16 | 51 |
| Survey | P aeruginosa | PDC | V211A, N346I | N/A | 16 | 51 |
| Survey | P aeruginosa | PDC | ΔT289-M291 | N/A | 4–8 | 51 |
| Survey | P aeruginosa | PDC | ΔT289-A292 | N/A | 16 | 51 |
| Case report | P aeruginosa | Overexpression of PDC, loss of oprD | ΔG202-E219 | 32 | N/A | 56 |
| Survey | P aeruginosa | Overexpression of PDC and/or MexAB-OprM or MexXY-OprM and/or loss of oprD | 8–16 | N/A | 68 | |
| Case report | P aeruginosa | PAC-1 | >128 | >128 | 74 | |
| Case report | P aeruginosa | FOX-4 | 16 | 16 | 34 | |
| Laboratory strain | P aeruginosa | GES-1 | N/A | 32 | 34 | |
| Laboratory strain | P aeruginosa | GES-2 | G170N | N/A | 16 | 34 |
| Laboratory strain | P aeruginosa | GES-5 | G170S | N/A | 8 | 34 |
| Laboratory Strain | P aeruginosa | GES-6 | E104K, G170S | N/A | 32 | 34 |
| Case report | P aeruginosa | GES-6 | E104K, G170S | 32 | 64 | 75 |
| Laboratory strain | P aeruginosa | PER-1 | N/A | 512 | 34 | |
| Laboratory strain | P aeruginosa | BEL-1 | N/A | 8 | 34 | |
| Laboratory strain | P aeruginosa | BEL-2 | N/A | 32 | 34 | |
| Survey | P aeruginosa | BEL-3 | P160S | 16 | N/A | 80 |
| Laboratory strain | P aeruginosa | VEB-1 | N/A | 64 | 34 | |
| Case report | P aeruginosa | OXA-2, loss of oprD | Duplication D149 | >32 | 16 | 78 |
| Case report | P aeruginosa | OXA-2, VIM-20, mutation of mexZ mexX, and mexB, loss of oprD | ΔI159 + E160 K in OXA-2 | >32 | 32 | 79 |
| Survey | P aeruginosa | OXA-2 | W159R | 16–32 | 16 | 68,80 |
| Survey | P aeruginosa | OXA-10 | N73S | 64 | N/A | 80 |
| Survey | P aeruginosa | OXA-101 | 32 | N/A | 80 | |
| Case report | P aeruginosa | OXA-10, loss of oprD | N146S | 32 | N/A | 52 |
| Laboratory strain | E coli | GES-1 | N/A | 8 | 31 | |
| Laboratory strain | E coli | GES-5 | G170S | N/A | 8 | 31 |
| Laboratory strain | E coli | GES-6 | E104K, G170S | N/A | 64 | 31 |
| Case report | E coli | GES-19, GES-26 | N/A | 48 | 77 | |
| Laboratory strain | E coli | PER-1 | N/A | 128 | 31 | |
| Laboratory strain | E coli | BEL-1 | N/A | 4 | 31 | |
| Laboratory strain | E coli | BEL-2 | N/A | 8 | 31 |
Tazobactam is maintained at 4 μg/mL when in combination with ceftolozane.81
P aeruginosa breakpoints: intermediate = 8 μg/mL; resistant ≥16 μg/mL.
Enterobacterales breakpoints: intermediate = 4 μg/mL; resistant ≥8 μg/mL.
Abbreviation: NA, not available.
The MIC values for the isolate represent the MIC values obtained for either a clinical isolate obtained from a patient in a case study or part of a surveillance study or a laboratory-selected strain.
The MIC values in a susceptible background represent the MIC value after the bla gene for the β-lactamase of interest was cloned and expressed in a susceptible strain.
Data from Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing. 30th ed.; 2020.
Pseudomonas-Derived Cephalosporinase Variants that Confer Ceftolozane-Tazobactam Resistance
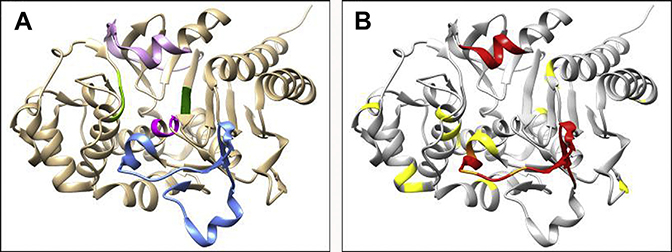
Not surprisingly, given the role it plays in β-lactamase function, the Ω-loop of PDC appears to be an important region for amino acid substitutions leading to ceftolozane-tazobactam resistance when these β-lactamases are expressed in bacteria, with V211A,50,51 G214R,51 E219G,51 E219K,51,52 and Y221H51 leading to varying levels of resistance (minimum inhibitory concentrations [MICs] range for clinical isolates: 32–>256 μg/mL) even as single amino acid substitutions (see Fig. 3, Table 2).53 Several of these substitutions also occur in tandem with others, including Q128R V211T S279T,54 A5V V211A G214R G220S,54 Q128R V211A G220S,54 E219K V329I,55 F121L Q130R E219K V329I,55 R100H G214R,51 and V211A N346I51 (see Fig. 3). Of these Ω-loop substitutions, the V211A, E219K, and E219G variants of PDC surfaced alone and E219K in conjunction with G154R substitution during treatment of patients.50,52,56
Outside the Ω-loop, substitutions are found in several regions of the enzyme but do not seem to cluster in a specific area. These substitutions include T96I,57,58 F121L,56,59 G156D alone55,57,60 and in combination with R52Q and T79A,61 P154L,51 L293P, N346I,51 and F121L M174L51 (Fig. 4). At this time, it is unknown if these substitutions outside the Ω loop serve to stabilize the protein (serve as a global suppressor) or specifically enhance catalytic activity. The T96I, F121L, and G156D variants of PDC emerged during treatment of patients for infections caused by P aeruginosa and ceftolozane-tazobactam MICs were elevated from 1 μg/mL to 4 μg/mL at the start of treatment to 32 μg/mL to 64 μg/mL during treatment.52,56,57,60,61 Moreover, multiple amino acid deletions leading to ceftolozane-tazobactam resistance were reported in PDC and are found both in the Ω-loop, deleting residues P208-G214,50,58,61 G202-E219,53,56 and G204-Y22158 as well as the R2 loop, deleting residues T289-P290, T289-M291, T289-A292, and L293-Q294.51 The R2 loop deletions tend to be associated with relatively low-level resistance when expressed in P aeruginosa 4098 (MIC range: 4–16 μg/mL)51 (see Fig. 4, Table 2). Of these loop deletions, to date, only Ω-loop deletion variants of PDC in P aeruginosa were reported to have surfaced in the clinic during treatment and resulted in ceftolozane-tazobactam MICs of 32 μg/mL to 256 μg/mL.50,52,56,58
Fig. 4.
PDC crystal structure highlighting the major active site motifs (blue: Ω-loop; magenta: S64X65X66K67 motif; dark green: K315T316G316 motif; light green: Y150X151K152 motif; and pink: R2 loop) (left) and the location of the amino acid substitutions, insertions, and deletions (red: deletion; yellow: substitution; and orange: deletion or substitution) that confer ceftolozane-tazobactam resistance (right).
The Molecular Basis of the Resistance Phenotype: PDC E219K, an Ω-Loop Variant
Among the best-characterized β-lactamase variants conferring ceftolozane-tazobactam resistance is the E219K variant of PDC. When the negatively charged glutamic acid (E) residue at 219 is changed to a positively charged lysine (K) and expressed in P aeruginosa PAO1 ΔblaPDC, the ceftolozane-tazobactam MIC increases from 0.5 μg/mL to 16 μg/mL.52 Steady-state kinetic characterization of the purified PDC-3 E219K variant with ceftolozane revealed a Km of 341 μM ± 64 μM and a kcat of 10 ± 1 s−1 compared with the wild-type PDC-3 enzyme, which had undetectable hydrolysis of ceftolozane and interacted with ceftolozane very poorly with a Ki app of 1300 μM.62 Moreover, electrospray-ionization mass spectrometry used to capture β-lactamase–ceftolozane adducts supported the kinetic observations, because, true to its design, ceftolozane was not detected bound to the wild-type Pseudomonas AmpC (PDC-3); conversely, ceftolozane was hydrolyzed by the PDC-3 E219K variant and acyl-enzyme complexes were not detected.62 Thermal stability assays revealed that the PDC-3 E219K variant possessed a lower Tm of 45°C compared with 52°C for PDC-3; these data suggest that the PDC-3 E219K variant is less stable.
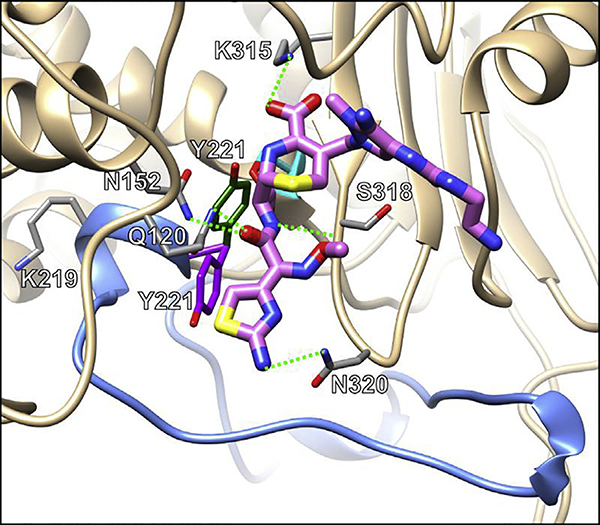
The molecular mechanism that allows the PDC-3 E219K variant to hydrolyze ceftolozane is perhaps best revealed by using classic atomistic molecular dynamics and well-tempered metadynamic simulations that model the interactions between the enzyme and substrate. The metadynamic simulations uncovered that the PDC-3 E219K variant was more conformationally flexible than the wild-type PDC-3. Moreover, the molecular dynamics showed that the flexibility of the PDC-3 E219K variant allows the nearby Y221 residue to rotate perpendicular to its usual position and open a hidden cavity adjacent to the active site (Fig. 5). This cavity is better able to accommodate the R1 group of ceftolozane (which normally faces a steric clash with Y221), allowing for better positioning of ceftolozane within the active site of the PDC-3 E219K variant to facilitate hydrolysis.
Fig. 5.
Ceftolozane (pink) docked in the active site of the PDC-3 E219K variant. Hydrogen bonds between ceftolozane and residues are indicated with green dashed lines, the catalytic S64 is in cyan, and the Ω-loop in blue. Two conformations of Y221 showcase the increased flexibility of the variant: purple represents a conformation found in both PDC-3 and the PDC-3 E219K variant whereas green represents a conformation found only in the PDC-3 E219K variant, the E219K substitution enables the movement between these conformations, allowing ceftolozane to enter the active site and bind while maintaining residues in catalytically favorable conformations.
Other Contributing Resistance Factors in Pseudomonas aeruginosa
Variants of PDC that result in ceftolozane-tazobactam resistance are found in strains that also have decreased expression of oprD and/or up-regulation of mexAB-oprM or mexXY-oprM efflux pumps and/or de-repression of blaPDC expression, which is normally expressed at a low basal level and induced by the presence of β-lactam antibiotics( Fig. 6, see Table 2).63–67 Regarding ceftolozane-tazobactam resistance, OprD does not appear to be necessary for entry of either compound and neither component is greatly impacted by hyperexpression of efflux pumps.43,68 Controlled (eg, ΔampD and ΔdacB) de-repression of wild-type blaPDC-1 in a P aeruginosa PAO1 background revealed that ceftolozane MICs are not impacted by hyperexpression of blaPDC-1.69 In conjunction with other factors (eg, blaPDC mutants and other bla genes), however, de-repression of blaPDC (eg, mutations in ampR, dacB, ampG, and ampD) was shown to elevate ceftolozane-tazobactam MICs.30,52,55 Importantly, high-level resistance due to overexpression of blaPDC variants was associated with the hypermutator background of P aeruginosa.55 Other potential contributors toward ceftolozane-tazobactam resistance in P aeruginosa are single amino acid substitutions in PBP3 (R504 C and F533 L); this may be an emerging phenotype due to selective pressure.30,59
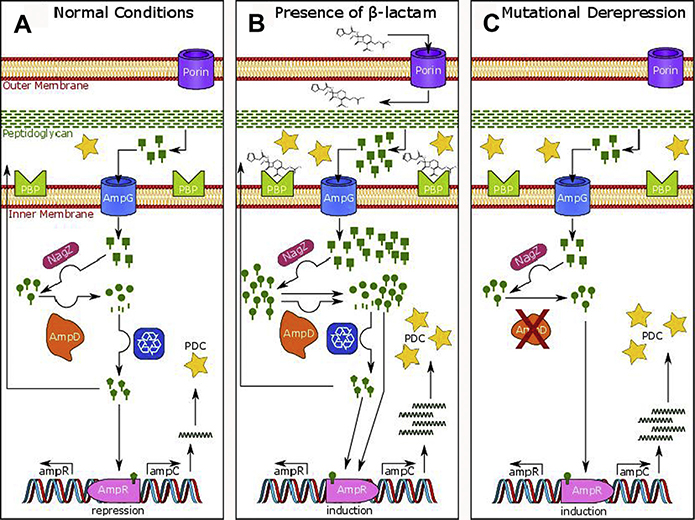
Fig. 6.
Regulation of blaPDC in P aeruginosa (61–64). (A) During normal cell growth, bacteria degrade approximately half their peptidoglycan and recycle approximately 90% of these degradation products or Glc-NAc-1,6-anhydro-MurNAc-peptides (green rectangular lollipops), which are transported into the cytoplasm via AmpG. In the cytosol, NagZ catalyzes the formation of 1,6-anhydro-MurNAc-peptides (green circular lollipops) that are activating peptides of AmpR, a LysR-type transcriptional regulator that controls expression of blaampC. AmpD, an N-acetylmuramyl-L-alanine amidase, cleaves the peptide (green stick) from the 1,6-anhydro-MurNAc (green circle) and the components enter the recycling pathway, keeping the cytoplasmic levels of activating 1,6-anhydro-MurNAc-peptides low and producing UDP-MurNAc-pentapeptides (green pentagonal lollipops) that are suppressing peptides that bind AmpR to repress the transcription of blaPDC. (B) In the presence of β-lactam antibiotics, the low molecular mass PBP4 (DacB) is inhibited (along with other PBPs), leading to an increase and shift in the composition of the Glc-NAc-1,6-anhydro-MurNAc-peptides entering the cytoplasm. This increase ultimately overpowers the capacity of AmpD to cleave the peptide from 1,6-anhydro-MurNAc, leading to a buildup of 1,6-anhydro-MurNAc-peptide in the cell. The 1,6-anhydro-MurNAc-peptide is then able to bind AmpR, activating transcription of blaampC. The AmpC β-lactamases are exported to the periplasm where they inactivate β-lactams. (C) Mutations in ampD are the most common cause of derepressed blaampC by severely crippling the production and/or activity of AmpD, levels of 1,6-anhydro-MurNAc-peptides greatly increase within the cell, bind to AmpR, and induce the production of high levels of blaampC.
The Impact of Other AmpCs
Resistance to ceftolozane-tazobactam in class C β-lactamases other than PDC is reported much less frequently or studied, but the Y221H substitution in CMY-2 was also shown to result in an elevated ceftolozane-tazobactam MIC (2.5 μg/mL).70 Other Ω-loop substitutions leading to ceftolozane-tazobactam resistance in P aeruginosa have been reported in other species producing class C β-lactamases, but in the context of nonsusceptibility to different substrates (eg, ceftazidime), including E219K in Citrobacter freundii AmpC71 and V211A combined with L239S in CMY-95.72
Additionally, SRT-1 of Serratia marcescens has lysine in the 219 position and exhibits better hydrolysis of several cephalosporins (eg, ceftazidime) than the closely related SST-1 with glutamic acid at 219.73 Additionally, 4 cases of P aeruginosa harboring blaPAC-1 were reported in patients repatriated from Mauritius and Afghanistan.74 PAC-1 is a unique class C β-lactamase with 47% sequence identity to PDC-1 and confers ceftolozane-tazobactam resistance; the introduction of blaPAC-1 into P aeruginosa PAO1 increased the ceftolozane-tazobactam MIC from less than or equal to 0.5 μg/mL to greater than 128 μg/mL. Recently, the FOX-4 cephamycinase was found responsible for elevated ceftolozane-tazobactam MICs (16 μg/mL) in a P aeruginosa clinical isolate.34 Much remains to be explored with non-PDC AmpCs and their involvement in ceftolozane-tazobactam resistance.
Uncommon Class A β-Lactamases
The acquisition of several different class A β-lactamases (eg, GES, PER-1, BEL-1, BEL-2, and VEB-1) has been associated with the emergence of ceftolozane-tazobactam resistance.31,36,53,59,75,76 Depending on the strain background in which these β-lactamases are produced, P aeruginosa versus E coli, the ceftolozane-tazobactam MICs vary (eg, P aeruginosa with blaGES-1 MIC = 32 μg/mL vs E coli with blaGES-1 MIC = 8 μg/mL)31 (see Table 4). When the combination of blaGES-19 and blaGES-26 was expressed in E coli TG1, the MIC values for ceftolozane-tazobactam increased to 48 μg/mL compared with an MIC of greater than 256 μg/mL when in P aeruginosa.77 The production of PER-1 in P aeruginosa PA01 resulted in high level ceftolozane-tazobactam resistance (MIC: 512 μg/mL).31 Except for GES β-lactamases, rare class A carbapenemases (eg, IMI and SME) are poor cephalosporinases; thus, when expressed, these strains usually are susceptible to ceftolozane-tazobactam. 36
Table 4.
Mechanisms of resistance to ceftazidime-avibactam
| Type | Organism | Mechanism(s) | Relevant Amino Acid Substitutions in β-Lactamase | Minimum Inhibitory Concentration of Isolatea | Minimum Inhibitory Concentration in a Susceptible Backgroundb | Reference |
|---|---|---|---|---|---|---|
| Laboratory | E coli | KPC-2 | R164A | N/A | 16 | 102 |
| Laboratory | E coli | KPC-2 | R164P | N/A | 64 | 102 |
| Laboratory | E coli | KPC-2 | D179A | N/A | 64 | 102 |
| Laboratory | E coli | KPC-2 | D179Q | N/A | 32 | 102 |
| Laboratory | E coli | KPC-2 | D179N | N/A | 32 | 102 |
| Laboratory | E coli | KPC-2 | D179Y | N/A | 32 | 106 |
| Survey | K pneumoniae | KPC-2, SHV-12, TEM-1, and loss of ompK35, ompk36, and ompK37 | 16 | N/A | 166 | |
| Survey | E coli | KPC-2, TEM-1, 344_ins_TIPY_345_PBP3 | 8 | N/A | 153 | |
| Case | K pneumoniae | Overexpression of KPC-2, loss of ompK35 and ompK36 | 12 | N/A | 108 | |
| Laboratory | C freundii | KPC-2, overexpression of acrA, loss of ompF | D176Y | 32 | 8 | 110 |
| Laboratory | C freundii | KPC-2, overexpression of acrA, loss of ompF | R146S, P147L | 64 | 8 | 110 |
| Laboratory | C freundii | KPC-2, overexpression of acrA, loss of ompF | D179Y | 64 | 8 | 110 |
| Case | K pneumoniae | KPC-2, SHV-11, SHV-12, TEM-1, loss of ompK35, ompK36, and ompK37 | D179Y | >256 | 120 | |
| Survey | K pneumoniae | KPC-2 | E166_ins_EL_L167, V278_ins_ SEAV_A281 | 128 | 64 | 117 |
| Case | K pneumoniae | KPC-2 | L169P | 16 | 4 | 125 |
| Case | K pneumoniae | KPC-2, SHV-11 | L259_ins_AVYTRAPNKDDKHSE_V260 | >16 | 127 | |
| Laboratory | E coli | KPC-2 | ΔG242-T243 | N/A | 24 | 126 |
| Laboratory | K pneumoniae | KPC-3 | S181_ins_S_S182 | 64 | 103 | |
| Laboratory | K pneumoniae | KPC-3 | D179Y | 8–64 | 103 | |
| Laboratory | K pneumoniae | KPC-3 | D163G | 32 | 103 | |
| Laboratory | Enterobacter cloacae | KPC-3 | S181_ins_SS_S182 | 32 | 103 | |
| Laboratory | Enterobacter cloacae | KPC-3 | P174L | 8–16 | 103 | |
| Laboratory | Enterobacter cloacae | KPC-3 | T243P | 32 | 103 | |
| Laboratory | Enterobacter cloacae | KPC-3 | T265_ins_AR_R266 | 16 | 103 | |
| Laboratory | Enterobacter cloacae | KPC-3 | P183_ins-RAVTTSSP_R184 | 128 | 103 | |
| Case | K pneumoniae | KPC-3, SHV-11, TEM-1, OXA-9 and loss of ompk36 | D179Y | 64–256 | 8 | 104 |
| Case | K pneumoniae | KPC-3, SHV-11, TEM-1, OXA-9 and loss of ompk36 | D179Y, T243M | 256 | 64 | 104 |
| Case | K pneumoniae | KPC-3, SHV-11, TEM-1, OXA-9 | V240G | 32 | 2 | 104 |
| Case | K pneumoniae | KPC-3, SHV-11, TEM-1, OXA-9 and loss of ompk35 | A177E, D179Y | 128 | N/A | 107 |
| Case | K pneumoniae | Overexpression of KPC-3, loss of ompk35 and ompk35, SHV-11, SHV-12 | 32 | 145 | ||
| Survey | K pneumoniae | KPC-3 | ΔE166-L167 | 16–32 | 8–16 | 117 |
| Survey | K pneumoniae | KPC-3 | L7P, D179Y, T243M | 256 | N/A | 117 |
| Case | K pneumoniae | KPC-3, SHV-11, TEM-1A, OXA-9 loss of ompK35 and ompK36 | V240A | 16 | N/A | 118 |
| Laboratory | E coli | KPC-3 | ΔG242-T243 | N/A | 12 | 126 |
| Laboratory | K pneumoniae | KPC-3 | D163G | 32 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | R164S | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | E168_ins_EL_L169 | 32 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | L169_ins-KL_N170 | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | N170D | 32 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | N170D + ΔS171 | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | A172S | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | A172T + T243A | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | A172P | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | P174L | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | P174_ins_PGDARD_D179 | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | G175V | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | D176Y | 32 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | D179A | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | D179H | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | D179N | 32 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | D179Y | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | ΔV240 | 128 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | Y241N | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | T243M | 32 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | Y263_ins_YTRAPN_N269 | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | Y263_ins_YTRAPNKDDKYSEAV_V278 | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | R266_ins_RAS_P268 | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | R266_ins_RAPNKDDKYS_S275 | >256 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | P268_ins_PN_N269 | 64 | N/A | 109 |
| Laboratory | K pneumoniae | KPC-3 | K270_ins_ KD_D271 | 128 | N/A | 109 |
| Case | K pneumoniae | KPC-3 | K270_PNK_D271 | >128 | 128 | 128 |
| Case | K pneumoniae | KPC-3 | L169P, A172T | >16 | 122 | |
| Case | K pneumoniae | KPC-3 | A172T, T243A | >16 | 122 | |
| Case | K pneumoniae | KPC-3 | A172T | >16 | 122 | |
| Survey | K oxytoca | SHV-12, TEM-1 | 16 | 134 | ||
| Laboratory | E coli | CTX-M-15 | S130G, L169Q | N/A | 16 | 133 |
| Case | K pneumoniae | CTX-M-14, OXA-48 | CTX-M-14: P167S, T264I | 32 | 132 | |
| Laboratory | E coli | GES-5 | N/A | 0.5 | 31 | |
| Laboratory | E coli | PER-1 | N/A | 8 | 31 | |
| Laboratory | E coli | GES-19, GES-26 | N/A | 256 | 77 | |
| Case | K pneumoniae | VEB-1, TEM-1, 0XA-10 | K234R | 32–128 | 138 | |
| Survey | E coli | CMY-42, CTX-M-15, OXA-1, 333_ins_YRIK_334_PBP3 | 8 | N/A | 154 | |
| Survey | E coli | CMY-42, CMY-2, OXA-1, OXA-9, TEM-1, 333_ins_YRIK_334_PBP3 | 8 | N/A | 154 | |
| Survey | K pneumoniae | DHA-1, loss of ompK35 and ompK36 | 16 | N/A | 41 | |
| Survey | Enterobacter cloacae | AmpC | ΔS289-A294 | 64 | N/A | 140 |
| Laboratory | Enterobacter cloacae | AmpC | G156R | 64 | N/A | 139 |
| Laboratory | Enterobacter cloacae | AmpC | G156D | 16 | N/A | 139 |
| Laboratory | C freundii | AmpC | N346Y | 16 | N/A | 139 |
| Laboratory | C freundii | AmpC | R148P | 16 | N/A | 139 |
| Laboratory | C freundii | AmpC | R148H | 32 | N/A | 139 |
| Survey | K pneumoniae | OXA-48, SHV | 16 | N/A | 161 | |
| Survey | S marcescens | OXA-48, CTX-M-22, SHV | 64 | N/A | 161 | |
| Survey | P aeruginosa | GES-5 | 16 | N/A | 31 | |
| Case | P aeruginosa | GES-19, GES-26 | 128 to >256 | N/A | 77 | |
| Survey | P aeruginosa | PER-1 | 64 | N/A | 31 | |
| Laboratory | P aeruginosa | PDC | ΔR210-E219 | 64–256 | N/A | 141 |
| Laboratory | P aeruginosa | PDC | ΔK204a-G222 | 64 | N/A | 141 |
| Laboratory | P aeruginosa | PDC | ΔD217-Y221 | 256 | N/A | 141 |
| Laboratory/Case | P aeruginosa | PDC | G156D | 128 | >32 | 57,60,141,142 |
| Case | P aeruginosa | Overexpression of PDC, loss of oprD | T96I | 16–32 | 8 | 52 |
| Case | P aeruginosa | Overexpression of PDC, loss of oprD | E219K | >32 | 16 | 52 |
| Case | P aeruginosa | Overexpression of PDC, loss of oprD | ΔG202-E219 | 32 | 16 | 52 |
| Survey | P aeruginosa | Overexpression of PDC and MexAB-OprM, loss of oprD | 512 | N/A | 52 | |
| Case | P aeruginosa | Overexpression of PDC | V211A | >64 | N/A | 50 |
| Case | P aeruginosa | PDC, mutation in mexR | ΔP208-G214 | >64 | N/A | 50 |
| Survey | P aeruginosa | Overexpression of PDC, loss of oprD | Q128R, V211T, S279T | 256 | N/A | 54 |
| Survey | P aeruginosa | Overexpression of PDC, loss of oprD | Q128R, V211A, G220S | 256 | N/A | 54 |
| Survey | P aeruginosa | Overexpression of PDC, loss of oprD | V211A | 256 | N/A | 54 |
| Case | P aeruginosa | PAC-1 | >128 | >128 | 74 | |
| Case | P aeruginosa | FOX-4 | 32 | 32 | 34 | |
| Case | P aeruginosa | OXA-2, loss of oprD | Duplication D149 | >32 | 32 | 78 |
| Case | P aeruginosa | OXA-10, loss of oprD | N146S | 32 | N/A | 52 |
| Case | P aeruginosa | OXA-2, VIM-20, mutation of mexZ mexX, and mexB, loss of oprD | ΔI159 + E160K in OXA-2 | 32 | 32 | 79 |
Avibactam is maintained at 4 μg/mL when in combination with ceftazidime.81
P aeruginosa breakpoint: resistant greater than 8 μg/mL.
Enterobacterales breakpoint: resistant greater than 8 μg/mL.
Abbreviation: NA, not available.
The MIC values for the isolate represent the MIC values obtained for either a clinical isolate obtained from a patient in a case study or part of a surveillance study or a laboratory-selected strain.
The MIC values in a clean background represent the MIC value after the bla gene for the β-lactamase of interest was cloned and expressed in a susceptible strain.
Data from Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing. 30th ed.; 2020.
Resistance Observed in Class D β-Lactamases
Ceftolozane-tazobactam–resistant OXA variants (eg, OXA-14) also were identified in surveillance studies and have emerged during treatment of infections due to P aeruginosa52,56,68,78–80 (see Table 2). Many of these OXA variants acquired a single amino acid substitution, such as a strain of P aeruginosa producing the OXA-10 N146S variant possessed a ceftolozane-tazobactam MIC of 64 μg/mL.52 The continued evolution of narrow-spectrum oxacillinases in P aeruginosa (such as OXA-14) to ceftolozane-tazobactam resistant variants may represent an emerging challenge when using ceftolozane-tazobactam.
NEWER β-LACTAM AND β-LACTAMASE INHIBITOR COMBINATIONS: CEFTAZIDIME-AVIBACTAM
Ceftazidime-avibactam was approved by the FDA in 2015 for adult and pediatric use in cIAIs (with metronidazole) and cUTIs, including pyelonephritis, and for adult use in HABP and VABP.82 Ceftazidime-avibactam is being actively investigated for use in adult cystic fibrosis patients (NCT02504827), for nosocomial pneumonia in pediatric patients (NCT04040621), and in neonates and infants with gram-negative infections (NCT04126031).
Ceftazidime-avibactam is effective against a variety of gram-negative bacteria (Table 3) (Ceftazidime-Avibactam PI). Limitations for the combination against indicated organisms include P aeruginosa or Enterobacterales that carry class B carbapenemases (eg, VIM, NDM, and IMP)83,84 and non–OXA-48–like class D β-lactamases with ESBL activity (eg, OXA-2 variants).78,79,85,86
Table 3.
Clinical indications for the use of ceftazidime-avibactam
| Indication | Indicated Bacteria | Patient Population | |
|---|---|---|---|
| cIAI (with metronidazole) | E coli, K pneumoniae, Proteus mirabilis, Enterobacter cloacae, K oxytoca, C freundii complex, and P aeruginosa | Adult and pediatric (≥3 mo) | RECLAIM 1 and 287 NCT01499290 NCT01500239 REPRISE88 NCT01644643 NCT0247573389 |
| cUTI, including pyelonephritis | E coli, K pneumoniae, Enterobacter cloacae, C freundii complex, Proteus mirabilis, and P aeruginosa | Adult and pediatric (≥3 mo) | RECAPTURE90 NCT01595438 NCT01599806 REPRISE88 NCT01644643 NCT02497781 |
| HABP/VABP | K pneumoniae, Enterobacter cloacae, E coli, S marcescens, Proteus mirabilis, P aeruginosa, and Haemophilus influenzae | Adult | REPROVE91 NCT0180809292 |
Data from Allergan USA, Inc. AVYCAZ (Ceftazidime and Avibactam) for Injection, for Intravenous Use. Madison, NJ 07940 USA.; 2019.
Why is this combination so important? Ceftazidime is a broad-spectrum amino-thiazolyl cephalosporin originally approved by the FDA in July 1985. Ceftazidime demonstrates potent activity against a wide variety of gram-negative bacteria and some gram-positive bacteria, with particular strengths against P aeruginosa and Enterobacterales, including strains expressing many important β-lactamases.93,94 Unfortunately, resistance to ceftazidime rapidly emerged (presence of ESBLs in many enteric bacilli, such as E coli and K pneumoniae and the overexpression of class C β-lactamases among other mechanisms) and the drug became less attractive and use was heavily monitored.20,95,96 Ceftazidime-avibactam helped fill an important gap in the spectrum of other β-lactamases known and evolving at the time with its potent activity against P aeruginosa and ESBLs. In fact, an early article went so far as to describe it as “the most effective antibiotic thus far known against P. aeruginosa.”94
Avibactam is the first of a novel class of β-lactamase inhibitors known as the DBOs and has a wide spectrum of activity against class A, class C, and some class D β-lactamases. Unique among previously available β-lactamase inhibitors (clavulanic acid, tazobactam, and sulbactam), the DBOs do not contain a β-lactam group. Inhibition is accomplished by the formation of a covalent acyl-enzyme complex between the active-site serine of the β-lactamase and the 8-membered cyclooctane ring of the DBO. Interestingly, deacylation typically occurs through a reversible mechanism that regenerates an intact molecule of avibactam, allowing for inhibition of further enzymes. 22,97,98 KPC-2 and metallo-β-lactamases possess the ability to hydrolyze avibactam; however, the rate of hydrolysis is very slow.85,99
REPORTS AND MECHANISMS OF RESISTANCE: CEFTAZIDIME-AVIBACTAM
Table 4 lists the existing reports of resistance to ceftazidime-avibactam described to date. Phenotypic resistance to ceftazidime-avibactam appears to be driven largely by amino acid substitutions and deletions to the KPC carbapenemase found in Enterobacterales. These changes were reported mostly in case studies and laboratory selection experiments. Alarmingly, the first case report of ceftazidime-avibactam resistance in a K pneumoniae strain with blaKPC-3 was in the same year that ceftazidime-avibactam was released.100,101
Ceftazidime-Avibactam–Resistant KPC Variants
Substitutions in KPC that lead to ceftazidime-avibactam resistance tend to cluster into 1 of 2 regions of the enzyme: substitutions, insertions, and deletions in the Ω-loop, residues 164 to 179 in class A β-lactamases, or insertions in the B3–4 β-strands and adjacent helices (Fig. 7, see Table 4). The importance of the Ω-loop of KPC in ceftazidime-avibactam resistance was revealed first in the summer of 2015 shortly after the release of the combination.102 By exploiting the knowledge that evolution of the Ω-loop in β-lactamases is the Achilles heel for ceftazidime’s antimicrobial activity, 16–19 several Ω-loop variants (R164A, R164P, D179A, D179Q, and D179N) of KPC-2 were tested and found resistant to ceftazidime-avibactam (MIC range: 16–64 μg/mL). Additionally, in vitro selection experiments conducted using KPC-3-producing Enterobacterales resulted in the selection of the D179Y variant of KPC-3 among other alterations (see Table 4) that led to ceftazidime-avibactam resistance, further exposing the Ω-loop as a weakness for this combination.103 Subsequently, reports of ceftazidime-avibactam resistance began to emerge during treatment of patients with infections caused by carbapenem-resistant Enterobacterales carrying the D179Y variant of KPC-3, which elevated the ceftazidime-avibactam MICs from 2 μg/mL to 4 μg/mL prior to the start of treatment up to 64 μg/mL to greater than 256 μg/mL after treatment104,105 (see Table 4). Concomitant with the development of ceftazidime-avibactam resistance, bacteria producing the D179Y variant lose carbapenem resistance. Importantly, the tyrosine substitution at 179 was shown to revert back to aspartic acid when grown in the presence of a carbapenem; thus, KPC regains its ability to hydrolyze carbapenems when exposed to a carbapenem.106–109
Fig. 7.
KPC-2 crystal structure highlighting the major active site motifs (blue: Ω-loop, R164-D179; magenta: S70X71X72K73 motif; dark green: S130D131N132 loop; and bright green: K234T235G236 motif) (left) and the location of the amino acid substitutions, insertions, and deletions (cyan: insertion after residue; yellow: substitution; orange: deletion or substitution; red: deletion; green: insertion or substitution; and purple deletion or insertion) that confer ceftazidime-avibactam resistance (right).
Another group found this reversion phenotype with other Ω-loop amino acid substitutions (D176Y, P174L, and R164S) in KPC that also caused elevated ceftazidime-avibactam MICs110 In 1 case, the 179 substitution reverted to aspartic acid and ceftazidime-avibactam resistance was maintained (MIC: 12 μg/mL) through amplification of blaKPC-2 and loss of OmpK35 and OmpK36 when treating the patient with meropenem/polymyxin B.108 Moreover, the addition of a polymyxin, colistin, to ceftazidime-avibactam does not prevent the emergence of resistance to ceftazidime-avibactam by KPC-producing Enterobacterales.111 Another study assessed population diversity and found that wild-type KPC-3 and KPC-3 D179Y coexisted as a mixed population.112 Based on these observations, a case can be made that carbapenem monotherapy should not be considered as a therapeutic regimen against ceftazidime-avibactam–resistant KPC-producing Enterobacterales despite their carbapenem-susceptible phenotype.113 The clinical risk factors associated with the potential for KPC-producing Enterobacterales to acquire ceftazidime-avibactam resistance during treatment include pneumonia and renal replacement therapy.114 Likely, the concentration of the drug combination at the infection site does not remain above the MIC for a sufficient time and resistant variants are selected; therapeutic drug monitoring and proper dosage selection may be helpful in these cases.115,116 This leads to speculation that perhaps the dosing of ceftazidime avibactam should be modified in certain cases.
In addition to the D179Y variant of KPC-3, other variants, D179Y T243M, V240G, A177E D179Y, Δ166–167, L7P D179Y T243M, and V240A, also began to emerge in the clinic.104,117,118 The D179Y variant,103,104,106,109,110,117,119–123 however, continues to be the most commonly reported substitution leading to ceftazidime-avibactam resistance and also has emerged in KPC-2.108 The KPC-2 D179Y variant also was identified in a P aeruginosa strain in Chile, where ceftazidime-avibactam was never used.124 Importantly, this study was evaluating a rapid immunochromatographic test for detection of KPC and the variant was not identified by this test or Carba-NP testing.124 Also, in the Ω-loop, L169P occurred in KPC-2 during the treatment of a patient for VABP; the cloned KPC-2 L169P variant expressed in E coli DH5α possessed an MIC of 4 μg/mL, while the parent clinical strain’s MIC was 16 μg/mL.125 Subsequently, another L169P variant in conjunction with an A172T substitution emerged in KPC-3 during the treatment of an IAI caused by K pneumoniae; in the same study, the patient also was infected with 2 ceftazidime-avibactam–resistant K pneumoniae carrying the KPC-3 D179Y variant and a KPC-3 A172T variant and became colonized by a fourth ceftazidime-avibactam–resistant KPC-3 A172T T243A variant.122 The rapidly converting and changing phenotypes of these KPC-3 variants is concerning. Other KPC-2 variants that surfaced during treatment, include 1 that acquired 2 insertions (E and L) between Ω-loop residues 166–167 and S-E-A-V between the C-terminal α-helix residues 278–281 that resulted in a ceftazidime-avibactam MIC of 128 μg/mL.117 A deletion of residues G242-T243 in the B4 β-strand of KPC-2 and KPC-3 (KPC-14 and KPC-28, respectively) resulted in low-level resistance to ceftazidime-avibactam (MICs: 12–24 μg/mL) due to increased catalytic efficiency toward ceftazidime mediated by lower Km values; inhibition kinetics revealed that avibactam possessed similar IC50 values (range: 107–586 nM) against all variants tested.126
Two laboratory studies selected for ceftazidime-avibactam–resistant mutants using various Enterobacterales parent strains producing KPC-3 and found many substitutions that conferred resistance to ceftazidime-avibactam.103,109 The D179Y, D163G (a location before the Ω loop), and P174L substitutions were identified in both studies; however, only D179Y emerged in the clinic. One study further examined if imipenem susceptibility was affected by the acquisition of ceftazidime-avibactam resistance.109 The KPC-3 D163G, R164S, N170D, A172S, A172T, A172P, P174L, G175V, Y241N, and T243M variants, along with a KPC-3 ΔV240 variant, and several KPC-3 variants with different insertions in the B5 β-strand and subsequent loop residues (263–278) maintained imipenem resistance (MICs: ≥32 μg/mL), while correspondingly acquiring ceftazidime-avibactam resistance (range: 32 to >256 μg/mL).109 These data further exemplify the need to screen KPC-producers against carbapenems and ceftazidime-avibactam.
Unfortunately, these gain-of-function observations were not limited to the laboratory. The B5 β-strand seems capable of absorbing large changes leading to increases in ceftazidime-avibactam MICs, with an insertion of NH2-A-V-Y-T-R-A-P-N-K-D-D-K-H-S-E-CO2 in the B5 β-strand between residues 261 and 262 of KPC-2 raising MICs from 1 μg/mL to greater than 16 μg/mL; this KPC-2 variant surfaced during treatment of a patient for bacteremia due to K pneumoniae.127 In addition, another insertion in the B5 β-strand of PNK between K270 and D271 of KPC-3 in a K pneumoniae strain was obtained from a rectal swab of a patient and resulted in a ceftazidime-avibactam MIC of greater than 128 μg/mL.128 This KPC-3 variant was purified for kinetic characterization and the results implicated a lower Kd for the variant with ceftazidime as the primary driver for resistance, as conversely the Ki for avibactam increased 6-fold from KPC-3.128
The Mechanism Behind the Resistance: KPC D179N, an Ω-Loop Variant
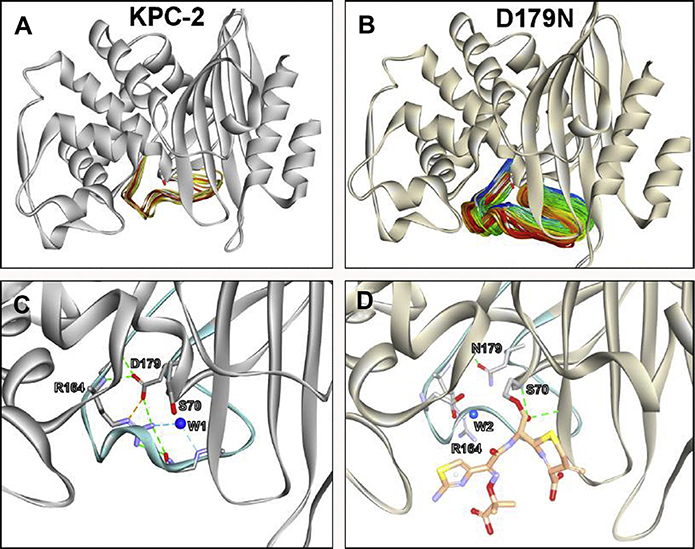
Among the most-studied β-lactamase variants leading ceftazidime-avibactam resistance is the D179N variant of KPC-2. The D179 residue forms a salt bridge with R164 in wild-type KPC-2 (Fig. 8). When the negatively charged aspartic acid residue at 179 (D) is changed to a polar asparagine (N) and expressed in E coli DH10 B, the ceftazidime-avibactam MIC increases from 1 μg/mL to 16 μg/mL.129 Steady-state kinetic characterization of the purified KPC-2 D179N variant revealed that ceftazidime is hydrolyzed at a slower rate with the variant compared with wild-type KPC-2. The apparent Km value for ceftazidime with the KPC-2 D179N variant, however, was 130 μM compared with 3500 μM with wild-type KPC-2. These kinetic observations suggest that the KPC-2 D179N variant forms more favorable interactions with ceftazidime than wild-type KPC-2 does. The inhibition by avibactam of the KPC-2 D179N variant was not significantly altered compared with wild-type KPC-2 (acylation rates: 38,000 M−1s−1 vs 17,000 M−1s−1, respectively).102 Electrospray-ionization mass spectrometry used to capture β-lactamase adducts revealed the unique trapping phenotype of the KPC-2 D179N variant. Acyl-enzyme adducts were detected when all β-lactams (eg, ceftazidime, ceftolozane, and imipenem) were incubated with the KPC-2 D179N variant but not with wild-type KPC-2, which presumably hydrolyzed these substrates. Moreover, when the β-lactamases were incubated with equimolar concentrations of β-lactam and avibactam, the KPC-2 D179N variant preferentially bound the β-lactam, whereas KPC-2 favored avibactam. Molecular modeling revealed that the flexibility and mobility of the Ω-loop were increased in the KPC-2 D179N variant due to disruption of the salt bridge with R164; this mobility likely allows ceftazidime to interact more favorably with the active site of the variant (see Fig. 8A, B). In addition, the catalytic residue S70 and the general base E166 were repositioned, thus allowing for a longer-lasting acyl-enzyme complex with the variant and the observed trapping phenotype (see Fig. 8C, D).129
Fig. 8.
Molecular modeling and 500-ns molecular dynamic simulation revealed the flexibility and mobility of the Ω-loop was increased in the (B) KPC-2 D179N variant due to disruption of the salt bridge with R164; mobility is RMSD of (A) 2 Å in KPC-2 versus (B) 10 Å for D179N variant. (C) In KPC-2, the R164 residue forms a salt bridge with D179 and hydrogen bonding network with a water molecule (W1). (D) The substitution D179N disrupts the salt bridge, and the nucleophilic S70 and the general base, E166 are repositioned, which results in the repositioning of the catalytic water (W2) and the formation of a longer-lasting acyl-enzyme complex with the variant and ceftazidime.
In addition to the analysis of the KPC-2 D179N variant, the KPC-2 D179Y variant was investigated by another group and they found the variant to have a greater than 43-fold decrease in Km toward ceftazidime and a greater than 1000-fold decrease in kcat compared with wild-type KPC-2.106 These data suggest that the variant can form favorable interactions with ceftazidime more readily than wild-type but is not able to hydrolyze ceftazidime; this is similar to the observation with the KPC-2 D179N variant.106 Conversely, the acylation rate of avibactam toward the KPC-2 D179Y variant was decreased significantly compared with wild-type (0.4 M−1s−1 vs 29,000 M−1s−1, respectively). The KPC-2 D179Y variant also appears to trap ceftazidime but is not effectively inhibited by avibactam.
Avibactam-Resistant Variants of KPC
Oxapenem, sulfones, and DBO β-lactamase inhibitors follow a similar reaction pathway toward acyl-enzyme formation. Indeed, substitutions (eg, S130G, K234R, and R220M) that have an impact on inhibition by traditional inhibitors, such as clavulanic acid, also effect the ability of avibactam to inhibit β-lactamases.130 The S130G substitution in KPC-2 resulted in the inability of avibactam to effectively acylate the enzyme. A subsequent report revealed the importance of the N132 residue in KPC-2 for acylation by avibactam; the N132G mutant was also unable to be acylated by avibactam.131 Fortunately, these substitutions also reduced KPC-2’s hydrolysis of β-lactams; thus, the contribution toward ceftazidime-avibactam resistance was limited. As KPC enzymes continue to evolve, however, the impact of these inhibitor-resistant substitutions may emerge.
Contributions of Other Class A Enzymes
Amino acid substitution of P167S in the Ω-loop of CTX-M-14 occurring in combination with T264I and OXA-48 resulted in a ceftazidime-avibactam MIC of 32 μg/mL for K pneumoniae.132 The role of the Ω-loop in expanding the spectrum of CTX-M-15 enzymes against ceftazidime-avibactam was assessed and the combination of L169Q (Ω-loop) and S130G (SDN loop) in CTX-M-15 resulted in an MIC of 16 μg/mL, when expressed in E coli; the purified CTX-M-15 S130 G L169Q variant hydrolyzed ceftazidime efficiently and was not inhibited by avibactam (IC50 >50 mM).133 K oxytoca with blaTEM-1 and blaSHV-12 tested nonsusceptible to ceftazidime-avibactam in a large surveillance study.134 In other large surveillance studies, P aeruginosa isolates producing class A PER, GES, or VEB β-lactamases demonstrated reduced susceptibility to ceftazidime-avibactam76,135–137; however, some of these isolates were not evaluated for other potentially contributing mechanisms.135 Another study found that the production of GES-5 and PER-1 in P aeruginosa did result in ceftazidime-avibactam resistance; however, when these bla genes were cloned and expressed in E coli TOP10, the MICs were lowered to 0.5 μg/mL and 16 μg/mL, respectively.31
Similarly, the P aeruginosa background causes elevated MICs against these agents, as was seen with ceftolozane-tazobactam. Low-level resistance in E coli is amplified when both GES-19 and GES-26 are introduced in E coli TG1; the ceftazidime-avibactam MIC increased from 0.5 μg/mL to 256 μg/mL, which was comparable to the parent P aeruginosa MICs of 128 to greater than 256 μg/mL.77 Recently, 2 different patients in Greece acquired K pneumoniae producing a VEB-1 K234R variant that demonstrated resistance to ceftazidime-avibactam (MICs: 32–128 μg/mL).138
Resistance in Enterobacterales AmpCs
Enterobacterales AmpCs can acquire resistance to ceftazidime-avibactam. Single amino acid substitutions were selected for in Enterobacter cloacae AmpC (G156R and G156D) and C freundii AmpC (R148H, R148P, and N346Y) that raised MICs to ceftazidime-avibactam from 0.5 μg/mL for the parent strains to 16 μg/mL to 32 μg/mL for the selected isolates.139 Moreover, a deletion of 289–294 in the Enterobacter cloacae AmpC resulted in a ceftazidime-avibactam MIC of 64 μg/mL.140 Evidence suggests that deletions in the vicinity of residue 290 are the result of enlargement in the R2 binding pocket allowing for β-lactams with larger R2 groups, such as ceftazidime, to be better accommodated.140
Ceftazidime-Avibactam Resistance of Pseudomonas-Derived Cephalosporinase Variants
Laboratory selection experiments in P aeruginosa identified several changes in PDC (ΔR210-E219, ΔK204a-G222, and ΔD217-Y221) that resulted in ceftazidime-avibactam resistance.141 The ΔD217-Y221 in PDC increased the baseline MIC from 8 μg/mL to 256 μg/mL for ceftazidime-avibactam when expressed in P aeruginosa.141 Purification of the wild-type and variant PDCs revealed that the ΔD217-Y221 variant’s kcat for ceftazidime increased by 650-fold and IC50 value for avibactam increased by 25-fold. Thus, resistance in this variant was due to increased turnover of ceftazidime as well as reduced inhibition by avibactam. Subsequently, during the selection of ceftolozane-tazobactam–resistant variants, cross-resistance to ceftazidime-avibactam was revealed. Several of the same substitutions in PDC T96I,52 G156D,57,60,141,142 including Ω-loop substitutions V211A50,54 and E219K,52 combinations Q128R V211T S279T, and Q128R V211A G220S,54 and ΔP208-G21450 and ΔG202-E219,52 that result in resistance to ceftolozane-tazobactam have been reported to also lead to ceftazidime-avibactam resistance in P aeruginosa. Cross-resistance in P aeruginosa to ceftazidime-avibactam and ceftolozane-tazobactam is highly alarming. A large surveillance study also revealed many PDC variants present in P aeruginosa led to resistance to ceftazidime-avibactam; however, the contribution of these variants toward this resistance was not validated.136 Acquisition of novel non-PDC AmpCs in P aeruginosa, PAC-1, or FOX-4 also resulted in resistance to ceftazidime-avibactam.34,74 Moreover, the ability of P aeruginosa to become ceftazidime-avibactam–resistant was found more pronounced in a hypermutator background.142
Other Considerations
In Enterobacterales, loss of outer membrane proteins (porins) did not result in resistance to ceftazidime-avibactam even in KPC-producing, AmpC-producing, and/or ESBL-producing strains.143,144 Strains carrying KPC-2, ESBLs (ie, TEM, SHV, or CTX-M), and ompK36 porin mutations, however, demonstrated statistically significant higher MICs toward ceftazidime-avibactam.143 Moreover, overexpression of blaKPC in conjunction with either loss of OmpK35 and OmpK36 and/or production of ESBLs has been reported to contribute to ceftazidime-avibactam resistance119,123,145–148; in a patient who failed on ceftazidime-avibactam treatment, the ceftazidime-avibactam MICs increased from 4 μg/mL to 32 μg/mL after therapy. Loss of OmpK35 and OmpK36 in concurrence with the production of the DHA-1 class C β-lactamase in K pneumoniae also elevated ceftazidime-avibactam MICs to 16 μg/mL,41 as did loss of porins and production of CTX-M-15 and OXA-1 in K pneumoniae.149
In 3 large surveillance studies of 10,998 Klebsiella species, 6209 Enterobacterales, and 36,380 Enterobacterales, small subsets of isolates (n = 16, n = 5, and n = 14, respectively) were resistant to ceftazidime-avibactam and the mechanisms for most of these resistant strains could not be determined.150–152 The investigators of 1 study proposed that potentially these isolates may have novel modifications of β-lactamases or PBP sequences or changes in drug efflux levels.150 Indeed, 2 different 4–amino acid insertions in PBP3 found in 3 different E coli isolates carrying various bla genes possessed elevated ceftazidime-avibactam MICs of 8 μg/mL.153,154 Contrary to Enterobacterales, out of 7062 P aeruginosa tested in a surveillance study, 272 isolates were resistant to ceftazidime-avibactam also with undefined resistance mechanisms; thus, a higher proportion of P aeruginosa are resistant to ceftazidime-avibactam compared with Enterobacterales.155 In P aeruginosa, overexpression of blaPDC as well as efflux and permeability of ceftazidime-avibactam appear to play a role in resistance to the combination.63,136,156–159 In a P aeruginosa PAO1 background, however, controlled (eg, ΔampD and ΔdacB) de-repression of wildtype blaPDC-1, loss of oprD, and/or hyperexpression of efflux pumps (eg, ΔmexR and ΔmexZ) did not have a a significant impact on ceftazidime-avibactam MICs (range: 1–4 μg/mL).156 In vitro selection experiments using P aeruginosa strain PA14 revealed the mutations in dnaJ, pepA, ctpA, glnD, flgF, pcm, spoT, and genes encoding an unidentified 2-component system and efflux pump component also effected resistance to ceftazidime-avibactam (MICs: ≥256 μg/mL); the exact mechanisms are not well understood.160
The Impact of Class D Oxacillinases
Except for OXA-48, most class D β-lactamases are not inhibited well by avibactam; however, as long as these enzymes are poor ceftazidimases, ceftazidime-avibactam will restore susceptibility, with ceftazidime doing the heavy lifting. Mutations in genes encoding OXA enzymes that extend their profile to ceftazidime have been reported to cause ceftazidime-avibactam resistance.52,78,79 When the D149 residue in OXA-2 was duplicated, the ceftazidime-avibactam MIC for P aeruginosa PAO1 expressing OXA-2 versus the variant enzyme increased from 1 μg/mL to 32 μg/mL.78 The expression of other bla genes (eg, blaSHV and blaCTX-M) also has been shown to result in elevated ceftazidime-avibactam MICs (16–64 μg/mL) in Enterobacterales producing OXA-48.161 In vitro laboratory selection on ceftazidime-avibactam using E coli MG1655 expressing blaOXA-48 revealed that substitutions of P68A and Y211S in OXA-48 elevated ceftazidime-MICs and the OXA-48 P68A Y211S variant possessed an approximately 6-fold increase in Ki for avibactam.162
Overcoming the Limitation Incurred from Class B Metallo-β-Lactamases
Not surprising, given that they fall outside the target activity of avibactam, the class B metallo-β-lactamases commonly are associated with high-level resistance to ceftazidime-avibactam. Notably, clinical case reports77 and laboratory testing163–165 suggest that the addition of aztreonamto ceftazidime-avibactammay be able to overcome resistance caused by the coproduction of metallo-β-lactamases and serine β-lactamases.
NEWER β-LACTAM AND β-LACTAMASE INHIBITOR COMBINATIONS: MEROPENEM-VABORBACTAM
In 2017, the FDA approved meropenem-vaborbactam (vaborbactam previously was known as RPX-7009) for the treatment of adult patients with cUTI, including pyelonephritis. 167 The combination also is undergoing clinical testing for use in pediatric patients with severe infections (NCT02687906) as well as in patients with HABP/VABP (NCT03006679).
Meropenem-vaborbactam demonstrates antimicrobial activity against E coli, K pneumoniae, a n d Enterobacter cloacae complex (Table 5) (Meropenem-Vaborbactam PI). Limitations for this combination against indicated organisms include those that carry class B metallo-carbapenemases (eg, VIM, NDM, and IMP) and/or class D OXA β-lactamases that are not susceptible to inhibition by meropenem or vaborbactam.
Table 5.
Clinical indications for the use of meropenem-vaborbactam
| Indication | Indicated Bacteria | Patient Population | Clinical Trials |
|---|---|---|---|
| cUTI, including pyelonephritis | E coli, K pneumoniae, and Enterobacter cloacae species complex | Adult | TANGO I168 NCT02166476 |
Data from Melinta Therapeutics, Inc. VABOMERE (Meropenemand and Vaborbactam) for Injection, for Intravenous Use. Lincolnshire, IL 60069 USA; 2019.
Meropenem has been in use in the United States since 1996 and is a carbapenem β-lactam antibiotic noted for its stability in the presence of human dehydropeptidase I (an enzyme which quickly metabolizes imipenem).169 Unfortunately, meropenem is susceptible to hydrolysis by class A carbapenemases, such as KPC; class B metallo-β-lactamases, such as NDM, VIM, and IMP; and class D OXA carbapenemases (eg, OXA-48). In addition, loss of outer membrane proteins (eg, OmpK35 and OmpK36 in K pneumoniae) and increases in efflux pump production (eg, AcrAB-TolC) effect its penetration.170
Vaborbactam is a cyclic boronate-based β-lactamase inhibitor designed to be selective for β-lactamases over other serine hydrolase enzymes. Intended to have a carbapenem partner from early development, the focus was placed on inhibiting the class A carbapenemase, KPC.171 Vaborbactam is a potent inhibitor of KPC (Ki app = 69 nM) and other class A β-lactamases (CTX-M, SHV, and TEM) as well as class C β-lactamases (eg, CMY-2, P99) but not class D serine or class B metallo-β-lactamases.172
REPORTS AND MECHANISMS OF RESISTANCE: MEROPENEM-VABORBACTAM
In Table 6, the reports of resistance to meropenem-vaborbactam available to date are presented. Despite 2 years on the market, clinical case reports of meropenem-vaborbactam resistance remain elusive in the literature. Whether this is indicative of the properties of the combination itself or the result of limited use and careful screening for susceptibility on the part of clinicians remains to be seen.
Table 6.
Mechanisms of resistance to meropenem-vaborbactam
| Type | Organism | Mechanism(s) | Minimum Inhibitory Concentraiton of Isolatea | Reference |
|---|---|---|---|---|
| Survey | K pneumoniae | KPC, loss of ompK37 and up-regulation of AcrAB-TolC | 16 | 177 |
| Survey | K pneumoniae | KPC-2, TEM-181, SHV-11, loss of ompK35 and ompK36 | 16 | 173 |
| Survey | K pneumoniae | KPC-2, TEM-1, SHV-11, SHV-12, loss of ompK35 and ompK36 | 32 | 173 |
| Survey | K pneumoniae | KPC, loss of ompK35 and ompK36 | 32 | 174 |
| Survey | K pneumoniae | KPC-2, SHV, TEM, OXA-10, loss of ompK35 and ompK36 | 64 | 178 |
| Survey | K pneumoniae | KPC-3, SHV-11, SHV-12, loss of ompK35 and ompK36 | 16 | 178 |
| Survey | K pneumoniae | KPC-3, SHV-11, loss of ompK35, ompK36, and ompK37 | 256 | 123 |
| Survey | K pneumoniae | KPC-3, SHV-11, TEM1a, OXA-9, loss of ompK35, ompK36, and ompK37 | 256 | 123 |
Vaborbactam is maintained at 8 μg/mL when in combination with meropenem
E coli, K pneumoniae, and Enterobacter cloacae complex breakpoint: resistant ≥16 μg/mL.81
The MIC values for the isolate represent the MIC values obtained for either a clinical isolate obtained from a patient in a case study or part of a surveillance study or a laboratory-selected strain.
Data from Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing. 30th ed.; 2020.
Enterobacterales Resistant to Meropenem-Vaborbactam
Unlike ceftolozane-tazobactam and ceftazidime-avibactam, reports of strains producing β-lactamase variants resistant to meropenem-vaborbactam are only now beginning to be reported. Moreover, meropenem-vaborbactam was shown to be effective against a case of bacteremia caused by a ceftazidime-avibactam–resistant K pneumoniae producing a KPC-2 D179Y variant.120 Likely, the change from a cephalosporin β-lactam partner to a carbapenem β-lactam partner is a significant factor for the differences observed in the resistance patterns. The use of a carbapenem partner in a β-lactam–β-lactamase inhibitor combination, however, also presents its own challenges as resistance due to increased efflux and decreased permeability of carbapenems is more problematic than with cephalosporins in gram negatives.170 In addition to carbapenems using porins for bacterial cell entry, vaborbactam also was found to traverse OmpK35 and OmpK36 in K pneumoniae.172 Thus, permeability is likely the largest hurdle for meropenem-vaborbactam efficacy. Resistance to meropenem-vaborbactam largely was reported in strains of K pneumoniae producing KPC with loss of expression of OmpK35, OmpK36, and/or OmpK37—mutations in porin genes that result in the production of partially functioning porins (eg, duplication of GD at positions 134 and 135 in OmpK36) also elevated meropenem-vaborbactam MICs.123,173–178 Increased expression of acrAB and/or blaKPC additionally was reported. 123,176,177 In vitro selection of K pneumoniae producing KPC-2 on meropenem-vaborbactam revealed that the primary resistance mechanisms toward meropenem-vaborbactam were loss of OmpK36 as well as increased copy number of blaKPC.173 One report revealed that the emergence of meropenem-vaborbactam nonsusceptibility (MIC: 8 μg/mL) due to loss of ompK36 during treatment of a patient for bacteremia due to K pneumoniae producing KPC-3.179 On a promising note, at least 1 study demonstrated synergy between meropenem-vaborbactam and aztreonam in the treatment of Enterobacterales carrying a metallo-β-lactamases.165
NEWER β-LACTAM AND β-LACTAMASE INHIBITOR COMBINATIONS: IMIPENEM-CILASTATIN-RELEBACTAM
Approved by the FDA in 2019, imipenem-cilastatin-relebactam is indicated for adult use in treating cIAIs and cUTIs, including pyelonephritis. The combination also is being evaluated for use in severe gram-negative infections in pediatric patients (NCT03969901 and NCT03230916) and for HABP/VABP in adults (NCT03583333) and was studied for bacterial pneumonia more broadly (NCT02493764). The combination demonstrates antimicrobial activity against a variety of gram-negative pathogens, including the anaerobes Bacteroides spp (Table 7).180 Limitations of imipenem-cilastatin-relebactam against organisms for which it is approved to treat include Enterobacterales or P aeruginosa that carry class B metallo-carbapenemases (eg, VIM, NDM, and IMP) or class D OXA β-lactamases that are not susceptible to inhibition by imipenem or relebactam.181
Table 7.
Clinical indications for the use of imipenem-cilastatin-relebactam
| Indication | Indicated Bacteria | Patient Population | Clinical Trials |
|---|---|---|---|
| cIAI | Enterobacter cloacae, E coli, K aerogenes, K pneumoniae, and P aeruginosa | Adult | RESTORE- IMI 1182 NCT02452047 |
| cUTI, including pyelonephritis | Bacteroides caccae, Bacteroides fragilis, Bacteroides ovatus, Bacteroides stercoris, Bacteroides thetaiotaomicron, Bacteroides uniformis, Bacteroides vulgatus, C freundii, Enterobacter cloacae, E coli, Fusobacterium nucleatum, K aerogenes, K oxytoca, K pneumoniae, Parabacteroides distasonis, and P aeruginosa | Adult | RESTORE-IMI 1182 NCT02452047 |
Data from Merck & Co., Inc. RECARBRIO (Imipenem, Cilastatin, and Relebactam) for Injection, for Intravenous Use. Whitehouse Station, NJ 08889 USA.; 2019.
Imipenem-cilastatin originally approved by the FDA in November 1985 and was the first carbapenem in clinical use. The combination brings together a potent carbapenem antibiotic with an inhibitor of human renal dehydropeptidase (cilastatin), reducing renal metabolism of imipenem.180 Unfortunately, imipenem is susceptible to hydrolysis by class A carbapenemases (KPC), class B metallo-β-lactamases (eg, VIM, NDM, and IMP), and class D carbapenemases (OXA-48). Moreover, decreased permeability due to loss of outer membrane porins (OmpK35, OmpK36, and OprD) and/or increases in efflux pump production (AcrAB-TolC and MexAB-OprM) effect its activity.170
Relebactam is a DBO β-lactamase inhibitor that was chosen from among many similar candidate compounds in a search for inhibitors to potentiate imipenem activity. It was selected for having particularly strong inhibitory activity against both class A and class C β-lactamases, demonstrating highly compatible pharmacokinetics with imipenem, effectiveness in mouse models of imipenem-resistant P aeruginosa and K pneumoniae strains, and favorable results in safety testing.183
REPORTS AND MECHANISMS OF RESISTANCE: IMIPENEM-RELEBACTAM
Table 8 lists all the reports of resistance to imipenem-relebactam described to date. On the market in the United States for less than 6 months at the time of this writing, clinical case reports of resistance to imipenem-cilastatin-relebactam are not present in the literature. As with meropenem-vaborbactam, reports of strains producing β-lactamase variants resistant to imipenem-relebactam have not been described. Moreover, the imipenem, carbapenem partner, is susceptible to the same mechanisms of resistance as meropenem; increased efflux and decreased permeability are major barriers for carbapenem activity.170 Being new to the market, a reasonable assumption is that further studies of resistance mechanisms and clinical case reports of emerging resistance and treatment failure of imipenem-relebactam will begin to emerge in the coming year, but until that time, data remains sparse and care should be taken to not draw any speculative conclusions.
Table 8.
Mechanisms of resistance to imipenem-relebactam
| Type | Organism | Mechanism(s) | Minimum Inhibitory Concentration of Isolatea | Reference |
|---|---|---|---|---|
| Survey | K pneumoniae | KPC, loss of ompK36 | 8 | 184 |
| Survey | K pneumoniae | Overexpression of KPC-2, TEM-1, SHV-12, loss of ompK35 and ompK36 | 512 | 186 |
| Survey | K pneumoniae | KPC-3 TEM-1/SHV-11, loss of ompK35 and ompK36 | 8 | 186 |
| Survey | S marcescens | SME-1 | 4 | 190 |
| Survey | P aeruginosa | PDC, loss of oprD | 8 | 184 |
| Survey | P aeruginosa | Hyperexpression of PDC, loss of oprD | 8 | 184 |
| Survey | P aeruginosa | GES-5 | >8 | 53 |
Relebactam is maintained at 4 μg/mL when in combination with imipenem.81
P aeruginosa breakpoint: resistant ≥8 μg/mL.
Enterobacterales breakpoint: resistant ≥4 μg/mL.
The MIC values for the isolate represent the MIC values obtained for either a clinical isolate obtained from a patient in a case study or part of a surveillance study or a laboratory-selected strain.
Data from Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing. 30th ed.; 2020.
Enterobacterales Resistant to Imipenem-Relebactam
Resistance to imipenem-relebactam was reported mostly in Enterobacterales due to loss of OmpK35/OmpF and OmpK36/OmpC as well as hyperexpression of blaKPC.184–188 For β-lactamase–mediated resistance, to date, 1 isolate of K pneumoniae was resistant to imipenem-relebactam and produced a GES-20,189 whereas 2 isolates of S marcescens with SME also were reported as resistant.32,190 The contribution (eg, lack of inhibition by relebactam vs enhance hydrolysis of imipenem) of these β-lactamases toward imipenem-relebactam resistance remains to be established.
Pseudomonas aeruginosa Resistant to Imipenem-Relebactam
Contrary to Enterobacterales, most oprD mutants in P aeruginosa were more susceptible to imipenem-relebactam, despite imipenem using OprD for entry into P aeruginosa. 181,191,192 A previous study revealed that when oprD is not expressed, blaPDC must be expressed,193 and because blaPDC is inhibited by relebactam the imipenem-relebactam combination is effective against many oprD mutants even when blaPDC is overexpressed.53,183,194 Some P aeruginosa strains with decreased expression of oprD and either wild-type or overexpressed levels of PDC, however, were resistant to imipenem-relebactam (MIC: 8 μg/mL).184 These somewhat contradictory data may be due to the fact that the baseline MICs of oprD mutants toward imipenem and imipenem-relebactam were higher compared with wild-type P aeruginosa strains181,184; thus, a fine line between susceptibility and resistance exists in these oprD mutants. Efflux was found to not have an impact on the activity of imipenem-relebactam; an oprD mutant strain of P aeruginosa overexpressing MexAB, MexCD, MexXY, and MexJK possessed imipenem-relebactam MICs between 0.125 μg/mL and 1 μg/mL.53,195 In 2 surveillance studies, of 17 of 589 and 5 of 42, P aeruginosa were found resistant to imipenem-relebactam (MIC range: 8–32 μg/mL) and the mechanism was not defined.194,196 As with Enterobacterales, the presence of GES carbapenemases (eg, GES-5 and GES-6) in P aeruginosa was found to result in resistance to imipenem-relebactam.197,198 Indeed, 11% of imipenem-relebactam–resistant P aeruginosa isolates carried GES β-lactamases; imipenem-relebactam MICs ranged from 8 μg/mL to 32 μg/mL for these isolates.195
SUMMARY
Although the combination therapies covered in this review are highly effective against large collections of clinical isolates, none is perfect, and all have shortcomings. KPC and PDC, the major resistant determinants in Enterobacterales and P aeruginosa, respectively, are evolving at an unprecedented rate. Perhaps the most important takeaway from this review on the development of resistance to some of the most promising advancements in β-lactam antibiotics from the past decade is a reminder: humanity is locked in a constant battle with bacteria that have a huge evolutionary advantage in the fight. Although every new antibiotic or inhibitor that makes it to market provides new tools for physicians to treat otherwise untreatable infections, resistance seemingly remains inevitable. The release of promising new drugs should be heralded, but everyone from doctors and scientists to pharmaceutical companies to policymakers and to the general public needs to realize and remember to not become complacent and that continued research into resistance mechanisms, stewardship and conservation of existing drugs, and development of novel treatments remain essential in the great war between humans and microbe. Judicious use and extensive laboratory testing are needed to prevent further spread especially as new agents enter the armamentarium.
KEY POINTS.
The novel β-lactam–β-lactamase inhibitor combinations (ceftazidime-avibactam, ceftolozane-tazobactam, meropenem-vaborbactam, and imipenem-relebactam) are a significant advance in the therapeutic armamentarium against multidrug-resistant gram-negative pathogens. Unfortunately, resistance to these very powerful agents is emerging rapidly in clinics.
Resistance to ceftolozane-tazobactam is mediated largely by amino acid substitutions, insertions, and/or deletions in the chromosomal AmpC, Pseudomonas-derived cephalosporinase (PDC), of P aeruginosa.
Mutations in blaKPC are the source of most reports of ceftazidime-avibactam resistance in gram negatives.
A majority of ceftolozane-tazobactam–resistant variants of PDC are cross-resistant to ceftazidime-avibactam.
The cephalosporin partners in ceftolozane-tazobactam and ceftazidime-avibactam are the major evolutionary drivers toward resistance in these combinations.
Permeability and efflux are the primary basis for resistance to meropenem-vaborbactam and imipenem-relebactam.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) to R.A. Bonomo under Award Numbers R01AI100560, R01AI063517, and R01AI072219. This study also was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, Award Numbers 1I01BX002872 to K.M. Papp-Wallace and 1I01BX001974 to R.A. Bonomo. from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, and from the Geriatric Research Education and Clinical Center VISN 10. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Department of Veterans Affairs.
DISCLOSURE
K.M. Papp-Wallace and R.A. Bonomo are funded in part by research grants from VenatoRx, Entasis, and Merck. K.M. Papp-Wallace and R.A. Bonomo are participating or have participated in collaborative research projects with Allecra, Allergan, Entasis, Merck, Roche, VenatoRx, and Wockhardt.
REFERENCES
- 1.Bush K, Bradford PA. β-lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med 2016;6(8):a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Outpatient Antibiotic prescriptions — United States, 2016 2018. Available at: https://www.cdc.gov/antibiotic-use/community/programs-measurement/state-local-activities/outpatient-antibioticprescriptions-US-2016.html Accessed November 25, 2018.
- 3.Errington J L-form bacteria, cell walls and the origins of life. Open Biol 2013; 3(1):120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborn MJ. Structure and Biosynthesis of the Bacterial Cell Wall. Annu Rev Biochem 1969;38(1):501–38. [DOI] [PubMed] [Google Scholar]
- 5.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A 1965;54(4):1133–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 2014;159(6):1300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K The ABCD’s of β-lactamase nomenclature. J Infect Chemother 2013; 19(4):549–59. [DOI] [PubMed] [Google Scholar]
- 8.Ambler RP, Coulson AF, Frère JM, et al. A standard numbering scheme for the class A β-lactamases. Biochem J 1991;276(Pt 1):269–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack AR, Barnes MD, Taracila MA, et al. A standard numbering scheme for class C β-lactamases. Antimicrob Agents Chemother 2019. 10.1128/AAC.01841-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oefner C, D’Arcy A, Daly JJ, et al. Refined crystal structure of β-lactamase from Citrobacter freundii indicates a mechanism for β-lactam hydrolysis. Nature 1990;343(6255):284–8. [DOI] [PubMed] [Google Scholar]
- 11.Galleni M, Lamotte-Brasseur J, Raquet X, et al. The enigmatic catalytic mechanism of active-site serine β-lactamases. Biochem Pharmacol 1995;49(9): 1171–8. [DOI] [PubMed] [Google Scholar]
- 12.Beadle BM, Trehan I, Focia PJ, et al. Structural milestones in the reaction pathway of an amide hydrolase: substrate, acyl, and product complexes of cephalothin with ampc β-lactamase. Structure 2002;10(3):413–24. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, McReynolds A, Shoichet BK. Re-examining the role of Lys67 in class C β-lactamase catalysis. Protein Sci 2009. 10.1002/pro.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown JR, Livesay DR. Flexibility correlation between active site regions is conserved across four AmpC β-lactamase enzymes. PLoS One 2015;10(5): e0125832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev 2009;22(1):161–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palzkill T, Le Q-Q, Venkatachalam KV, et al. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol Microbiol 1994;12(2):217–29. [DOI] [PubMed] [Google Scholar]
- 17.Banerjee S, Pieper U, Kapadia G, et al. Role of the Ω-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry 1998; 37(10):3286–96. [DOI] [PubMed] [Google Scholar]
- 18.Nukaga M, Taniguchi K, Washio Y, et al. Effect of an amino acid insertion into the omega loop region of a class C β-lactamase on its substrate specificity. Biochemistry 1998;37(29):10461–8. [DOI] [PubMed] [Google Scholar]
- 19.Crichlow GV, Kuzin AP, Nukaga M, et al. Structure of the extended-spectrum class C β-Lactamase of Enterobacter cloacae GC1, a natural mutant with a tandem tripeptide insertion. Biochemistry 1999;38(32):10256–61. [DOI] [PubMed] [Google Scholar]
- 20.Drawz SM, Bonomo RA. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 2010;23(1):160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush K β-lactamase inhibitors from laboratory to clinic. Clin Microbiol Rev 1988; 1(1):109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehmann DE, Jahić H, Ross PL, et al. Avibactam is a covalent, reversible, non–β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A 2012;109(29):11663–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papp-Wallace KM, Bonomo RA. New β-lactamase inhibitors in the clinic. Infect Dis Clin North Am 2016;30(2):441–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merck & Co., Inc. ZERBAXA (Ceftolozane and Tazobactam) for injection, for intravenous use. Whitehouse Station (NJ); 2014.
- 25.Solomkin J, Hershberger E, Miller B, et al. Ceftolozane/tazobactam plus metronidazole for complicated intra-abdominal infections in an era of multidrug resistance: results from a randomized, double-blind, phase 3 trial (ASPECT-cIAI). Clin Infect Dis 2015;60(10):1462–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikamo H, Monden K, Miyasaka Y, et al. The efficacy and safety of tazobactam/ceftolozane in combination with metronidazole in Japanese patients with complicated intra-abdominal infections. J Infect Chemother 2019;25(2):111–6. [DOI] [PubMed] [Google Scholar]
- 27.Huntington JA, Sakoulas G, Umeh O, et al. Efficacy of ceftolozane/tazobactam versus levofloxacin in the treatment of complicated urinary tract infections (cUTIs) caused by levofloxacin-resistant pathogens: results from the ASPECT-cUTI trial. J Antimicrob Chemother 2016;71(7):2014–21. [DOI] [PubMed] [Google Scholar]
- 28.Arakawa S, Kawahara K, Kawahara M, et al. The efficacy and safety of tazobactam/ceftolozane in Japanese patients with uncomplicated pyelonephritis and complicated urinary tract infection. J Infect Chemother 2019;25(2):104–10. [DOI] [PubMed] [Google Scholar]
- 29.Kollef MH, Nováček M, Kivistik Ü, et al. Ceftolozane-tazobactam versus meropenem for treatment of nosocomial pneumonia (ASPECT-NP): a randomised, controlled, double-blind, phase 3, non-inferiority trial. Lancet Infect Dis 2019; 19(12):1299–311. [DOI] [PubMed] [Google Scholar]
- 30.Del Barrio-Tofiño E, López-Causapé C, Cabot G, et al. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa Isolates from Spain. Antimicrob Agents Chemother 2017;61(11). 10.1128/AAC.01589-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortiz de la Rosa J-M, Nordmann P, Poirel L. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother 2019;74(7):1934–9. [DOI] [PubMed] [Google Scholar]
- 32.Senchyna F, Gaur RL, Sandlund J, et al. Diversity of resistance mechanisms in carbapenem-resistant Enterobacteriaceae at a health care system in Northern California, from 2013 to 2016. Diagn Microbiol Infect Dis 2019;93(3):250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt-Malan SM, Mishra AJ, Mushtaq A, et al. In vitro activity of imipenem-relebactam and ceftolozane-tazobactam against resistant gram-negative bacilli. Antimicrob Agents Chemother 2018;62(8). 10.1128/AAC.00533-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraile-Ribot PA, Del Rosario-Quintana C, López-Causapé C, et al. Emergence of resistance to novel β-lactam-β-lactamase inhibitor combinations due to horizontally acquired AmpC (FOX-4) in Pseudomonas aeruginosa Sequence Type 308. Antimicrob Agents Chemother 2019;64(1). 10.1128/AAC.02112-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castanheira M, Doyle TB, Mendes RE, et al. Comparative activities of ceftazidime-avibactam and ceftolozane-tazobactam against enterobacteriaceae isolates producing extended-spectrum β-lactamases from U.S. Hospitals. Antimicrob Agents Chemother 2019;63(7). 10.1128/AAC.00160-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livermore DM, Mushtaq S, Meunier D, et al. Activity of ceftolozane/tazobactam against surveillance and “problem” Enterobacteriaceae, Pseudomonas aeruginosa and non-fermenters from the British Isles. J Antimicrob Chemother 2017; 72(8):2278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farrell DJ, Flamm RK, Sader HS, et al. Antimicrobial activity of ceftolozane-tazobactam tested against Enterobacteriaceae and Pseudomonas aeruginosa with various resistance patterns isolated in U.S. Hospitals (2011–2012). Antimicrob Agents Chemother 2013;57(12):6305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tato M, García-Castillo M, Bofarull AM, et al. , CENIT Study Group. In vitro activity of ceftolozane/tazobactam against clinical isolates of Pseudomonas aeruginosa and Enterobacteriaceae recovered in Spanish medical centres: Results of the CENIT study. Int J Antimicrob Agents 2015;46(5):502–10. [DOI] [PubMed] [Google Scholar]
- 39.Castanheira M, Duncan LR, Mendes RE, et al. Activity of Ceftolozane-Tazobactam against Pseudomonas aeruginosa and Enterobacteriaceae Isolates Collected from Respiratory Tract Specimens of Hospitalized Patients in the United States during 2013 to 2015. Antimicrob Agents Chemother 2018; 62(3). 10.1128/AAC.02125-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robin F, Auzou M, Bonnet R, et al. In Vitro activity of ceftolozane-tazobactam against enterobacter cloacae complex clinical isolates with different β-lactam resistance phenotypes. Antimicrob Agents Chemother 2018;62(9). 10.1128/AAC.00675-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas-Chanoine M-H, Mayer N, Guyot K, et al. Interplay between membrane permeability and enzymatic barrier leads to antibiotic-dependent resistance in Klebsiella Pneumoniae. Front Microbiol 2018;9:1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moyá B, Beceiro A, Cabot G, et al. Pan-β-lactam resistance development in Pseudomonas aeruginosa clinical strains: molecular mechanisms, penicillin-binding protein profiles, and binding affinities. Antimicrob Agents Chemother 2012;56(9):4771–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda S, Nakai T, Wakai Y, et al. In vitro and in vivo activities of a new cephalosporin, FR264205, against Pseudomonas aeruginosa. Antimicrob Agents Chemother 2007;51(3):826–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodríguez-Martínez J-M, Poirel L, Nordmann P. Extended-spectrum cephalo-sporinases in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009; 53(5):1766–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livermore DM, Mushtaq S, Ge Y. Chequerboard titration of cephalosporin CXA-101 (FR264205) and tazobactam versus β-lactamase-producing Enterobacteriaceae. J Antimicrob Chemother 2010;65(9):1972–4. [DOI] [PubMed] [Google Scholar]
- 46.Giacobbe DR, Bassetti M, De Rosa FG, et al. Ceftolozane/tazobactam: place in therapy. Expert Rev Anti Infect Ther 2018;16(4):307–20. [DOI] [PubMed] [Google Scholar]
- 47.Aronoff SC, Jacobs MR, Johenning S, et al. Comparative activities of the β-lactamase inhibitors YTR 830, sodium clavulanate, and sulbactam combined with amoxicillin or ampicillin. Antimicrob Agents Chemother 1984;26(4):580–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 2010;54(3):969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bush K, Macalintal C, Rasmussen BA, et al. Kinetic interactions of tazobactam with β-lactamases from all major structural classes. Antimicrob Agents Chemother 1993;37(4):851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skoglund E, Abodakpi H, Rios R, et al. In Vivo resistance to ceftolozane/tazobactam in Pseudomonas aeruginosa Arising by AmpC- and Non-AmpC-mediated pathways. Case Rep Infect Dis 2018. 10.1155/2018/9095203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berrazeg M, Jeannot K, Enguéné VYN, et al. Mutations in β-lactamase AmpC increase resistance of Pseudomonas aeruginosa isolates to antipseudomonal cephalosporins. Antimicrob Agents Chemother 2015;59(10):6248–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraile-Ribot PA, Cabot G, Mulet X, et al. Mechanisms leading to in vivo ceftolozane/tazobactam resistance development during the treatment of infections caused by MDR Pseudomonas aeruginosa. J Antimicrob Chemother 2018; 73(3):658–63. [DOI] [PubMed] [Google Scholar]
- 53.Fraile-Ribot P, Zamorano L, Orellana R, et al. Activity of imipenem/relebactam against a large collection of Pseudomonas aeruginosa clinical isolates and isogenic β-lactam resistant mutants. Antimicrob Agents Chemother 2019. 10.1128/AAC.02165-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zamudio R, Hijazi K, Joshi C, et al. Phylogenetic analysis of resistance to ceftazidime/avibactam, ceftolozane/tazobactam and carbapenems in piperacillin/tazobactam-resistant Pseudomonas aeruginosa from cystic fibrosis patients. Int J Antimicrob Agents 2019;53(6):774–80. [DOI] [PubMed] [Google Scholar]
- 55.Cabot G, Bruchmann S, Mulet X, et al. Pseudomonas aeruginosa ceftolozane-tazobactam resistance development requires multiple Mutations leading to overexpression and structural modification of AmpC. Antimicrob Agents Chemother 2014;58(6):3091–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Díaz-Cañestro M, Periañez L, Mulet X, et al. Ceftolozane/tazobactam for the treatment of multidrug resistant Pseudomonas aeruginosa: experience from the Balearic Islands. Eur J Clin Microbiol Infect Dis 2018;37(11):2191–200. [DOI] [PubMed] [Google Scholar]
- 57.MacVane SH, Pandey R, Steed LL, et al. Emergence of ceftolozane-tazobactam-resistant Pseudomonas aeruginosa during treatment is mediated by a single AmpC structural mutation. Antimicrob Agents Chemother 2017; 61(12). e01183–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haidar G, Philips NJ, Shields RK, et al. Ceftolozane-tazobactam for the treatment of multidrug-resistant Pseudomonas aeruginosa infections: clinical effectiveness and evolution of resistance. Clin Infect Dis 2017;65(1):110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Del Barrio-Tofiño E, Zamorano L, Cortes-Lara S, et al. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J Antimicrob Chemother 2019;74(7):1825–35. [DOI] [PubMed] [Google Scholar]
- 60.Boulant T, Jousset AB, Bonnin RA, et al. A 2.5-years within-patient evolution of a Pseudomonas aeruginosa with in vivo acquisition of ceftolozane-tazobactam and ceftazidime-avibactam resistance upon treatment. Antimicrob Agents Chemother 2019. 10.1128/AAC.01637-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.So W, Shurko J, Galega R, et al. Mechanisms of high-level ceftolozane/tazobactam resistance in Pseudomonas aeruginosa from a severely neutropenic patient and treatment success from synergy with tobramycin. J Antimicrob Chemother 2019. 10.1093/jac/dky393. [DOI] [PubMed] [Google Scholar]
- 62.Barnes MD, Taracila MA, Rutter JD, et al. Deciphering the Evolution of Cephalosporin Resistance to Ceftolozane-Tazobactam in Pseudomonas aeruginosa. mBio 2018;9(6). 10.1128/mBio.02085-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wi YM, Greenwood-Quaintance KE, Schuetz AN, et al. Activity of ceftolozane-tazobactam against carbapenem-resistant, non-carbapenemase-producing Pseudomonas aeruginosa and associated resistance mechanisms. Antimicrob Agents Chemother 2018;62(1). 10.1128/AAC.01970-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009;22(4):582–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juan C, Torrens G, González-Nicolau M, et al. Diversity and regulation of intrinsic β-lactamases from non-fermenting and other Gram-negative opportunistic pathogens. FEMS Microbiol Rev 2017;41(6):781–815. [DOI] [PubMed] [Google Scholar]
- 66.Johnson JW, Fisher JF, Mobashery S. Bacterial cell-wall recycling. Ann N Y Acad Sci 2013;1277:54–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dik DA, Fisher JF, Mobashery S. Cell-wall recycling of the gram-negative bacteria and the nexus to antibiotic resistance. Chem Rev 2018;118(12):5952–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castanheira M, Mills JC, Farrell DJ, et al. Mutation-driven β-lactam resistance mechanisms among contemporary ceftazidime-nonsusceptible Pseudomonas aeruginosa isolates from U.S. hospitals. Antimicrob Agents Chemother 2014; 58(11):6844–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moya B, Zamorano L, Juan C, et al. Activity of a new cephalosporin, CXA-101 (FR264205), against β-lactam-resistant Pseudomonas aeruginosa mutants selected in vitro and after antipseudomonal treatment of intensive care unit patients. Antimicrob Agents Chemother 2010;54(3):1213–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zavala A, Retailleau P, Elisée E, et al. Genetic, biochemical, and structural characterization of CMY-136 β-lactamase, a peculiar CMY-2 variant. ACS Infect Dis 2019;5(4):528–38. [DOI] [PubMed] [Google Scholar]
- 71.Tsukamoto K, Ohno R, Sawai T. Extension of the substrate spectrum by an amino acid substitution at residue 219 in the Citrobacter freundii cephalosporinase. J Bacteriol 1990;172(8):4348–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crémet L, Caroff N, Giraudeau C, et al. Detection of clonally related Escherichia coli isolates producing different CMY β-lactamases from a cystic fibrosis patient. J Antimicrob Chemother 2013;68(5):1032–5. [DOI] [PubMed] [Google Scholar]
- 73.Matsumura N, Minami S, Mitsuhashi S. Sequences of homologous β-lactamases from clinical isolates of Serratia marcescens with different substrate specificities. Antimicrob Agents Chemother 1998;42(1):176–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bour M, Fournier D, Jové T, et al. Acquisition of class C β-lactamase PAC-1 by ST664 strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2019. 10.1128/AAC.01375-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poirel L, Ortiz De La Rosa J-M, Kieffer N, et al. Acquisition of Extended-Spectrum β-Lactamase GES-6 Leading to Resistance to Ceftolozane-Tazobactam Combination in Pseudomonas aeruginosa. Antimicrob Agents Chemother 2019;63(1). 10.1128/AAC.01809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sid Ahmed MA, Abdel Hadi H, Hassan AAI, et al. Evaluation of in vitro activity of ceftazidime/avibactam and ceftolozane/tazobactam against MDR Pseudomonas aeruginosa isolates from Qatar. J Antimicrob Chemother 2019;74(12): 3497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khan A, Tran TT, Rios R, et al. Extensively Drug-Resistant Pseudomonas aeruginosa ST309 harboring tandem guiana extended spectrum β-lactamase enzymes: a newly emerging threat in the United States. Open Forum Infect Dis 2019;6(7):ofz273 10.1093/ofid/ofz273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fraile-Ribot PA, Mulet X, Cabot G, et al. In vivo emergence of resistance to novel cephalosporin-β-lactamase inhibitor combinations through the duplication of amino acid D149 from OXA-2 β-Lactamase (OXA-539) in Sequence Type 235 Pseudomonas aeruginosa. Antimicrob Agents Chemother 2017;61(9). 10.1128/AAC.01117-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arca-Suárez J, Fraile-Ribot P, Vázquez-Ucha JC, et al. Challenging antimicrobial susceptibility and evolution of resistance (OXA-681) during treatment of a long-term nosocomial infection caused by a Pseudomonas aeruginosa ST175 Clone. Antimicrob Agents Chemother 2019;63(10). 10.1128/AAC.01110-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Juan C, Zamorano L, Pérez JL, et al. Activity of a new antipseudomonal cephalosporin, CXA-101 (FR264205), against carbapenem-resistant and multidrug-resistant Pseudomonas aeruginosa clinical strains. Antimicrob Agents Chemother 2010;54(2):846–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptibility Testing. 30th edition. 2020.
- 82.Allergan USA, Inc. AVYCAZ (Ceftazidime and Avibactam) for Injection, for Intravenous Use. Madison (NJ): 2019.
- 83.Castanheira M, Farrell SE, Krause KM, et al. Contemporary diversity of β-lactamases among Enterobacteriaceae in the nine U.S. census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent β-lactamase groups. Antimicrob Agents Chemother 2014;58(2):833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Jonge BLM, Karlowsky JA, Kazmierczak KM, et al. In vitro susceptibility to ceftazidime-avibactam of carbapenem-nonsusceptible enterobacteriaceae isolates collected during the INFORM Global Surveillance Study (2012 to 2014). Antimicrob Agents Chemother 2016;60(5):3163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ehmann DE, Jahic H, Ross PL, et al. Kinetics of avibactam inhibition against Class A, C, and D β-lactamases. J Biol Chem 2013;288(39):27960–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stone GG, Bradford PA, Yates K, et al. In vitro activity of ceftazidime/avibactam against urinary isolates from patients in a Phase 3 clinical trial programme for the treatment of complicated urinary tract infections. J Antimicrob Chemother 2017;72(5):1396–9. [DOI] [PubMed] [Google Scholar]
- 87.Mazuski JE, Gasink LB, Armstrong J, et al. Efficacy and safety of ceftazidime-avibactam plus metronidazole versus meropenem in the treatment of complicated intra-abdominal infection: results from a randomized, controlled, double-blind, phase 3 program. Clin Infect Dis 2016;62(11):1380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carmeli Y, Armstrong J, Laud PJ, et al. Ceftazidime-avibactam or best available therapy in patients with ceftazidime-resistant Enterobacteriaceae and Pseudomonas aeruginosa complicated urinary tract infections or complicated intraabdominal infections (REPRISE): a randomised, pathogen-directed, phase 3 study. Lancet Infect Dis 2016;16(6):661–73. [DOI] [PubMed] [Google Scholar]
- 89.Bradley JS, Broadhurst H, Cheng K, et al. Safety and efficacy of ceftazidime-avibactam plus metronidazole in the treatment of children ≥3 months to <18 years with complicated intra-abdominal infection: results from a phase 2, randomized, controlled trial. Pediatr Infect Dis J 2019;38(8):816–24. [DOI] [PubMed] [Google Scholar]
- 90.Wagenlehner FM, Sobel JD, Newell P, et al. Ceftazidime-avibactam versus doripenem for the treatment of complicated urinary tract infections, including acute pyelonephritis: RECAPTURE, a phase 3 randomized trial program. Clin Infect Dis 2016;63(6):754–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Torres A, Zhong N, Pachl J, et al. Ceftazidime-avibactam versus meropenem in nosocomial pneumonia, including ventilator-associated pneumonia (REPROVE): a randomised, double-blind, phase 3 non-inferiority trial. Lancet Infect Dis 2018; 18(3):285–95. [DOI] [PubMed] [Google Scholar]
- 92.Bradley JS, Roilides E, Broadhurst H, et al. Safety and efficacy of ceftazidime-avibactam in the treatment of children ≥3 months to <18 years with complicated urinary tract infection: results from a phase 2 randomized, controlled trial. Pediatr Infect Dis J 2019;38(9):920–8. [DOI] [PubMed] [Google Scholar]
- 93.O’Callaghan CH, Acred P, Harper PB, et al. GR 20263, a new broad-spectrum cephalosporin with anti-pseudomonal activity. Antimicrob Agents Chemother 1980;17(5):876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Verbist L, Verhaegen J. GR-20263: a new aminothiazolyl cephalosporin with high activity against Pseudomonas and Enterobacteriaceae. Antimicrob Agents Chemother 1980;17(5):807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mushtaq S, Warner M, Livermore DM. In vitro activity of ceftazidime1NXL104 against Pseudomonas aeruginosa and other non-fermenters. J Antimicrob Chemother 2010;65(11):2376–81. [DOI] [PubMed] [Google Scholar]
- 96.Endimiani A, Perez F, Bonomo RA. Cefepime: a reappraisal in an era of increasing antimicrobial resistance. Expert Rev Anti Infect Ther 2008;6(6): 805–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lahiri SD, Mangani S, Durand-Reville T, et al. Structural Insight into Potent Broad-Spectrum Inhibition with Reversible Recyclization Mechanism: Avibactam in Complex with CTX-M-15 and Pseudomonas aeruginosa AmpC β-Lactamases. Antimicrob Agents Chemother 2013;57(6):2496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lahiri SD, Johnstone MR, Ross PL, et al. Avibactam and class C β-lactamases: mechanism of inhibition, conservation of the binding pocket, and implications for resistance. Antimicrob Agents Chemother 2014;58(10):5704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lohans CT, Brem J, Schofield CJ. New Delhi metallo-β-lactamase 1 catalyzes avibactam and aztreonam hydrolysis. Antimicrob Agents Chemother 2017; 61(12). 10.1128/AAC.01224-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Humphries RM, Yang S, Hemarajata P, et al. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae Isolate. Antimicrob Agents Chemother 2015;59(10):6605–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spellberg B, Bonomo RA. Editorial commentary: ceftazidime-avibactam and carbapenem-resistant enterobacteriaceae: “we’re gonna need a bigger boat.” Clin Infect Dis 2016;63(12):1619–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Winkler ML, Papp-Wallace KM, Bonomo RA. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β-lactamases with single amino acid substitutions in the Ω-loop. J Antimicrob Chemother 2015;70(8):2279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Livermore DM, Warner M, Jamrozy D, et al. In vitro selection of ceftazidime-avibactam resistance in Enterobacteriaceae with KPC-3 carbapenemase. Antimicrob Agents Chemother 2015;59(9):5324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shields RK, Chen L, Cheng S, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blakpc-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 2017; 61(3). 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shields RK, Potoski BA, Haidar G, et al. Clinical outcomes, drug toxicity, and emergence of ceftazidime-avibactam resistance among patients treated for carbapenem-resistant enterobacteriaceae infections. Clin Infect Dis 2016; 63(12):1615–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Compain F, Arthur M. Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D179Y substitution in the KPC-2 β-lactamase. Antimicrob Agents Chemother 2017;61(7). 10.1128/AAC.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shields RK, Nguyen MH, Press EG, et al. In vitro selection of meropenem resistance among ceftazidime-avibactam-resistant, meropenem-susceptible Klebsiella pneumoniae isolates with variant KPC-3 carbapenemases. Antimicrob Agents Chemother 2017;61(5). 10.1128/AAC.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Giddins MJ, Macesic N, Annavajhala MK, et al. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-Harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 2018;62(3). 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Göttig S, Frank D, Mungo E, et al. Emergence of ceftazidime/avibactam resistance in KPC-3-producing Klebsiella pneumoniae in vivo. J Antimicrob Chemother 2019;74(11):3211–6. [DOI] [PubMed] [Google Scholar]
- 110.Castanheira M, Arends SJR, Davis AP, et al. Analyses of a ceftazidime-avibactam-resistant Citrobacter freundii Isolate Carrying blaKPC-2 Reveals a Heterogenous Population and Reversible Genotype. mSphere 2018;3(5). 10.1128/mSphere.00408-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shields RK, Nguyen MH, Hao B, et al. Colistin does not potentiate ceftazidime-avibactam killing of carbapenem-resistant enterobacteriaceae in vitro or suppress emergence of ceftazidime-avibactam resistance. Antimicrob Agents Chemother 2018;62(8). 10.1128/AAC.01018-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gaibani P, Campoli C, Lewis RE, et al. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J Antimicrob Chemother 2018;73(6):1525–9. [DOI] [PubMed] [Google Scholar]
- 113.Shields RK, Nguyen MH, Press EG, et al. Emergence of ceftazidime-avibactam resistance and restoration of carbapenem susceptibility in klebsiella pneumoniae carbapenemase-producing K pneumoniae: a case report and review of literature. Open Forum Infect Dis 2017;4(3):ofx101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shields RK, Nguyen MH, Chen L, et al. Pneumonia and renal replacement therapy are risk factors for ceftazidime-avibactam treatment failures and resistance among patients with carbapenem-resistant enterobacteriaceae infections. Antimicrob Agents Chemother 2018;62(5). 10.1128/AAC.02497-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bidell MR, Lodise TP. Suboptimal clinical response rates with newer antibiotics among patients with moderate renal impairment: review of the literature and potential pharmacokinetic and pharmacodynamic considerations for observed findings. Pharmacotherapy 2018;38(12):1205–15. [DOI] [PubMed] [Google Scholar]
- 116.Yasmin M, Fouts DE, Jacobs MR, et al. Monitoring Ceftazidime-Avibactam (CAZ-AVI) and Aztreonam (ATM) Concentrations in the Treatment of a Bloodstream Infection Caused by a Multidrug-Resistant Enterobacter sp. Carrying both KPC-4 and NDM-1 carbapenemases. Clin Infect Dis 2019. 10.1093/cid/ciz1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilson WR, Kline EG, Jones CE, et al. Effects of KPC Variant and Porin Genotype on the In Vitro activity of meropenem-vaborbactam against carbapenem-resistant enterobacteriaceae. Antimicrob Agents Chemother 2019;63(3). 10.1128/AAC.02048-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Galani I, Antoniadou A, Karaiskos I, et al. Genomic characterization of a KPC-23-producing Klebsiella pneumoniae ST258 clinical isolate resistant to ceftazidime-avibactam. Clin Microbiol Infect 2019;25(6):763.e5–e8. [DOI] [PubMed] [Google Scholar]
- 119.Zhang P, Shi Q, Hu H, et al. Emergence of ceftazidime/avibactam resistance in carbapenem-resistant Klebsiella pneumoniae in China. Clin Microbiol Infect 2019. 10.1016/j.cmi.2019.08.020. [DOI] [PubMed] [Google Scholar]
- 120.Athans V, Neuner EA, Hassouna H, et al. Meropenem-vaborbactam as salvage therapy for ceftazidime-avibactam-resistant Klebsiella pneumoniae bacteremia and abscess in a liver transplant recipient. Antimicrob Agents Chemother 2019;63(1). 10.1128/AAC.01551-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Venditti C, Nisii C, D’Arezzo S, et al. Molecular and phenotypical characterization of two cases of antibiotic-driven ceftazidime-avibactam resistance in blaKPC-3-harboring Klebsiella pneumoniae. Infect Drug Resist 2019;12:1935–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cano Á, Guzmán-Puche J, García-Gutiérrez M, et al. Use of carbapenems in the combined treatment of emerging ceftazidime/avibactam-resistant and carbapenem-susceptible KPC-producing Klebsiella pneumoniae infections: report of a case and review of the literature. J Glob Antimicrob Resist 2019. 10.1016/j.jgar.2019.11.007. [DOI] [PubMed] [Google Scholar]
- 123.Gaibani P, Carla Re M, Campoli C, et al. Bloodstream infection caused by KPC-producing Klebsiella pneumoniae resistant to ceftazidime/avibactam: Epidemiology and genomic characterization. Clin Microbiol Infect 2019. 10.1016/j.cmi.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 124.Wozniak A, Paillavil B, Legarraga P, et al. Evaluation of a rapid immunochromatographic test for detection of KPC in clinical isolates of Enterobacteriaceae and Pseudomonas species. Diagn Microbiol Infect Dis 2019;95(2):131–3. [DOI] [PubMed] [Google Scholar]
- 125.Hemarajata P, Humphries RM. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J Antimicrob Chemother 2019;74(5):1241–3. [DOI] [PubMed] [Google Scholar]
- 126.Oueslati S, Iorga BI, Tlili L, et al. Unravelling ceftazidime/avibactam resistance of KPC-28, a KPC-2 variant lacking carbapenemase activity. J Antimicrob Chemother 2019;74(8):2239–46. [DOI] [PubMed] [Google Scholar]
- 127.Räisänen K, Koivula I, Ilmavirta H, et al. Emergence of ceftazidime-avibactamresistant Klebsiella pneumoniae during treatment, Finland, 2018. Euro Surveill 2019;24(19). 10.2807/1560-7917.ES.2019.24.19.1900256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mueller L, Masseron A, Prod’Hom G, et al. Phenotypic, biochemical and genetic analysis of KPC-41, a KPC-3 variant conferring resistance to ceftazidime-avibactam and exhibiting reduced carbapenemase activity. Antimicrob Agents Chemother 2019. 10.1128/AAC.01111-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Barnes MD, Winkler ML, Taracila MA, et al. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at ambler position asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 2017;8(5). 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Papp-Wallace KM, Winkler ML, Taracila MA, et al. Variants of β-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother 2015;59(7):3710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ourghanlian C, Soroka D, Arthur M. Inhibition by Avibactam and Clavulanate of the β-Lactamases KPC-2 and CTX-M-15 Harboring the Substitution N132G in the Conserved SDN Motif. Antimicrob Agents Chemother 2017;61(3). 10.1128/AAC.02510-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Both A, Büttner H, Huang J, et al. Emergence of ceftazidime/avibactam nonsusceptibility in an MDR Klebsiella pneumoniae isolate. J Antimicrob Chemother 2017;72(9):2483–8. [DOI] [PubMed] [Google Scholar]
- 133.Compain F, Dorchène D, Arthur M. Combination of amino acid substitutions leading to CTX-M-15-mediated resistance to the ceftazidime-avibactam combination. Antimicrob Agents Chemother 2018;62(9). 10.1128/AAC.00357-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karlowsky JA, Biedenbach DJ, Kazmierczak KM, et al. Activity of Ceftazidime-Avibactam against Extended-Spectrum- and AmpC β-Lactamase-Producing Enterobacteriaceae Collected in the INFORM Global Surveillance Study from 2012 to 2014. Antimicrob Agents Chemother 2016;60(5):2849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Karlowsky JA, Kazmierczak KM, Bouchillon SK, et al. In Vitro Activity of Ceftazidime-Avibactam against Clinical Isolates of Enterobacteriaceae and Pseudomonas aeruginosa Collected in Latin American Countries: Results from the INFORM Global Surveillance Program, 2012 to 2015. Antimicrob Agents Chemother 2019;63(4). 10.1128/AAC.01814-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Castanheira M, Doyle TB, Smith CJ, et al. Combination of MexAB-OprM overexpression and mutations in efflux regulators, PBPs and chaperone proteins is responsible for ceftazidime/avibactam resistance in Pseudomonas aeruginosa clinical isolates from US hospitals. J Antimicrob Chemother 2019;74(9):2588–95. [DOI] [PubMed] [Google Scholar]
- 137.Stone GG, Smayevsky J, Kazmierczak K. Longitudinal analysis of the in vitro activity of ceftazidime-avibactam vs. Pseudomonas aeruginosa, 2012–2016. Diagn Microbiol Infect Dis 2020;96(1):114835. [DOI] [PubMed] [Google Scholar]
- 138.Voulgari E, Kotsakis SD, Giannopoulou P, et al. Detection in two hospitals of transferable ceftazidime-avibactam resistance in Klebsiella pneumoniae due to a novel VEB β-lactamase variant with a Lys234Arg substitution, Greece, 2019. Euro Surveill 2020;25(2). 10.2807/1560-7917.ES.2020.25.2.1900766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Livermore DM, Mushtaq S, Doumith M, et al. Selection of mutants with resistance or diminished susceptibility to ceftazidime/avibactam from ESBL- and AmpC-producing Enterobacteriaceae. J Antimicrob Chemother 2018;73(12): 3336–45. [DOI] [PubMed] [Google Scholar]
- 140.Lahiri SD, Giacobbe RA, Johnstone MR, et al. Activity of avibactam against Enterobacter cloacae producing an extended-spectrum class C β-lactamase enzyme. J Antimicrob Chemother 2014;69(11):2942–6. [DOI] [PubMed] [Google Scholar]
- 141.Lahiri SD, Walkup GK, Whiteaker JD, et al. Selection and molecular characterization of ceftazidime/avibactam-resistant mutants in Pseudomonas aeruginosa strains containing derepressed AmpC. J Antimicrob Chemother 2015;70(6): 1650–8. [DOI] [PubMed] [Google Scholar]
- 142.Khil PP, Dulanto Chiang A, Ho J, et al. Dynamic emergence of mismatch repair deficiency facilitates rapid evolution of ceftazidime-avibactam resistance in Pseudomonas aeruginosa Acute infection. mBio 2019;10(5). 10.1128/mBio.01822-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shields RK, Clancy CJ, Hao B, et al. Effects of Klebsiella pneumoniae carbapenemase subtypes, extended-spectrum β-lactamases, and porin mutations on the in vitro activity of ceftazidime-avibactam against carbapenem-resistant K. pneumoniae. Antimicrob Agents Chemother 2015;59(9):5793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.López-Hernández I, Alonso N, Fernández-Martínez M, et al. Activity of ceftazidime-avibactam against multidrug-resistance Enterobacteriaceae expressing combined mechanisms of resistance. Enferm Infecc Microbiol Clin 2017;35(8):499–504. [DOI] [PubMed] [Google Scholar]
- 145.Humphries RM, Hemarajata P. Resistance to ceftazidime-avibactam in Klebsiella pneumoniae due to porin mutations and the increased expression of KPC-3. Antimicrob Agents Chemother 2017;61(6). 10.1128/AAC.00537-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nelson K, Hemarajata P, Sun D, et al. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother 2017;61(10). 10.1128/AAC.00989-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Giani T, Antonelli A, Sennati S, et al. Results of the Italian infection-Carbapenem Resistance Evaluation Surveillance Trial (iCREST-IT): activity of ceftazidime/avibactam against Enterobacterales isolated from urine. J Antimicrob Chemother 2020. 10.1093/jac/dkz547. [DOI] [PubMed] [Google Scholar]
- 148.Coppi M, Di Pilato V, Monaco F, et al. Ceftazidime-avibactam resistance associated with increased blaKPC-3 gene copy number mediated by pKpQIL plasmid derivatives in ST258 Klebsiella pneumoniae. Antimicrob Agents Chemother 2020. 10.1128/AAC.01816-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Viala B, Zaidi FZ, Bastide M, et al. Assessment of the In Vitro activities of ceftolozane/tazobactam and ceftazidime/avibactam in a collection of beta-lactam-resistant enterobacteriaceae and Pseudomonas aeruginosa Clinical Isolates at Montpellier University Hospital, France. Microb Drug Resist 2019;25(9): 1325–9. [DOI] [PubMed] [Google Scholar]
- 150.Hackel M, Kazmierczak KM, Hoban DJ, et al. Assessment of the In Vitro Activity of Ceftazidime-Avibactam against Multidrug-Resistant Klebsiella spp. Collected in the INFORM Global Surveillance Study, 2012 to 2014. Antimicrob Agents Chemother 2016;60(8):4677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sader HS, Castanheira M, Flamm RK. Antimicrobial activity of ceftazidime-avibactam against gram-negative bacteria isolated from patients hospitalized with pneumonia in U.S. Medical Centers, 2011 to 2015. Antimicrob Agents Chemother 2017;61(4). 10.1128/AAC.02083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sader HS, Castanheira M, Shortridge D, et al. Antimicrobial activity of ceftazidime-avibactam tested against multidrug-resistant enterobacteriaceae and Pseudomonas aeruginosa Isolates from U.S. Medical Centers, 2013 to 2016. Antimicrob Agents Chemother 2017;61(11). 10.1128/AAC.01045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang Y, Kashikar A, Brown CA, et al. Unusual Escherichia coli PBP 3 Insertion Sequence Identified from a Collection of Carbapenem-Resistant Enterobacteriaceae Tested In Vitro with a Combination of Ceftazidime-, Ceftaroline-, or Aztreonam-Avibactam. Antimicrob Agents Chemother 2017;61(8). 10.1128/AAC.00389-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alm RA, Johnstone MR, Lahiri SD. Characterization of Escherichia coli NDM isolates with decreased susceptibility to aztreonam/avibactam: role of a novel insertion in PBP3. J Antimicrob Chemother 2015;70(5):1420–8. [DOI] [PubMed] [Google Scholar]
- 155.Nichols WW, de Jonge BLM, Kazmierczak KM, et al. In Vitro Susceptibility of Global Surveillance Isolates of Pseudomonas aeruginosa to Ceftazidime-Avibactam (INFORM 2012 to 2014). Antimicrob Agents Chemother 2016; 60(8):4743–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Torrens G, Cabot G, Ocampo-Sosa AA, et al. Activity of Ceftazidime-Avibactam against Clinical and Isogenic Laboratory Pseudomonas aeruginosa Isolates Expressing Combinations of Most Relevant β-Lactam Resistance Mechanisms. Antimicrob Agents Chemother 2016;60(10):6407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Winkler ML, Papp-Wallace KM, Hujer AM, et al. Unexpected challenges in treating multidrug-resistant gram-negative bacteria: resistance to ceftazidime-avibactam in archived isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2015;59(2):1020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chalhoub H, Sáenz Y, Nichols WW, et al. Loss of activity of ceftazidime-avibactam due to MexAB-OprM efflux and overproduction of AmpC cephalosporinase in Pseudomonas aeruginosa isolated from patients suffering from cystic fibrosis. Int J Antimicrob Agents 2018;52(5):697–701. [DOI] [PubMed] [Google Scholar]
- 159.Atkin SD, Abid S, Foster M, et al. Multidrug-resistant Pseudomonas aeruginosa from sputum of patients with cystic fibrosis demonstrates a high rate of susceptibility to ceftazidime-avibactam. Infect Drug Resist 2018;11:1499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sanz-García F, Hernando-Amado S, Martínez JL. Mutation-driven evolution of Pseudomonas aeruginosa in the presence of either ceftazidime or ceftazidime-avibactam. Antimicrob Agents Chemother 2018;62(10). 10.1128/AAC.01379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kazmierczak KM, Bradford PA, Stone GG, et al. In vitro activity of ceftazidime-avibactam and aztreonam-avibactam against OXA-48-Carrying Enterobacteriaceae Isolated as Part of the International Network for Optimal Resistance Monitoring (INFORM) Global Surveillance Program from 2012 to 2015. Antimicrob Agents Chemother 2018;62(12). 10.1128/AAC.00592-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fröhlich C, Sørum V, Thomassen AM, et al. OXA-48-mediated ceftazidime-avibactam resistance is associated with evolutionary trade-offs. mSphere 2019;4(2). e00024–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Marshall S, Hujer AM, Rojas LJ, et al. Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in enterobacteriaceae? Antimicrob Agents Chemother 2017;61(4). 10.1128/AAC.02243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Davido B, Fellous L, Lawrence C, et al. Ceftazidime-avibactam and aztreonam, an interesting strategy to overcome β-lactam resistance conferred by metallo-β-lactamases in enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 2017;61(9). 10.1128/AAC.01008-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Biagi M, Wu T, Lee M, et al. Searching for the optimal treatment for metallo- and serine-β-lactamase producing enterobacteriaceae: aztreonam in combination with ceftazidime-avibactam or meropenem-vaborbactam. Antimicrob Agents Chemother 2019. 10.1128/AAC.01426-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Castanheira M, Mendes RE, Sader HS. Low Frequency of Ceftazidime-Avibactam Resistance among Enterobacteriaceae Isolates Carrying blaKPC Collected in U.S. Hospitals from 2012 to 2015. Antimicrob Agents Chemother 2017;61(3). 10.1128/AAC.02369-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Melinta Therapeutics, Inc. VABOMERE (Meropenem and Vaborbactam) for Injection, for Intravenous Use. Lincolnshire (IL): 2019.
- 168.Kaye KS, Bhowmick T, Metallidis S, et al. Effect of meropenem-vaborbactam vs piperacillin-tazobactam on clinical cure or improvement and microbial eradication in complicated urinary tract infection: the TANGO I randomized clinical trial. JAMA 2018;319(8):788–99. 10.1001/jama.2018.0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Edwards JR, Turner PJ, Wannop C, et al. In vitro antibacterial activity of SM-7338, a carbapenem antibiotic with stability to dehydropeptidase I. Antimicrob Agents Chemother 1989;33(2):215–22. 10.1128/aac.33.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Papp-Wallace KM, Endimiani A, Taracila MA, et al. Carbapenems: past, present, and future. Antimicrob Agents Chemother 2011;55(11):4943–60. 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hecker SJ, Reddy KR, Totrov M, et al. Discovery of a Cyclic Boronic Acid β-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases. J Med Chem 2015;58(9):3682–92. 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- 172.Lomovskaya O, Sun D, Rubio-Aparicio D, et al. Vaborbactam: Spectrum of Beta-Lactamase Inhibition and Impact of Resistance Mechanisms on Activity in Enterobacteriaceae. Antimicrob Agents Chemother 2017;61(11). 10.1128/AAC.01443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sun D, Rubio-Aparicio D, Nelson K, et al. Meropenem-Vaborbactam Resistance Selection, Resistance Prevention, and Molecular Mechanisms in Mutants of KPC-Producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2017; 61(12). 10.1128/AAC.01694-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pfaller MA, Huband MD, Mendes RE, et al. In vitro activity of meropenem/vaborbactam and characterisation of carbapenem resistance mechanisms among carbapenem-resistant Enterobacteriaceae from the 2015 meropenem/vaborbactam surveillance programme. Int J Antimicrob Agents 2018;52(2):144–50. 10.1016/j.ijantimicag.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 175.Lapuebla A, Abdallah M, Olafisoye O, et al. Activity of Meropenem Combined with RPX7009, a Novel β-Lactamase Inhibitor, against Gram-Negative Clinical Isolates in New York City. Antimicrob Agents Chemother 2015;59(8):4856–60. 10.1128/AAC.00843-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhou M, Yang Q, Lomovskaya O, et al. In vitro activity of meropenem combined with vaborbactam against KPC-producing Enterobacteriaceae in China. J Antimicrob Chemother 2018;73(10):2789–96. 10.1093/jac/dky251. [DOI] [PubMed] [Google Scholar]
- 177.Castanheira M, Rhomberg PR, Flamm RK, et al. Effect of the β-Lactamase Inhibitor Vaborbactam Combined with Meropenem against Serine Carbapenemase-Producing Enterobacteriaceae. Antimicrob Agents Chemother 2016;60(9): 5454–8. 10.1128/AAC.00711-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Griffith DC, Sabet M, Tarazi Z, et al. Pharmacokinetics/Pharmacodynamics of Vaborbactam, a Novel Beta-Lactamase Inhibitor, in Combination with Meropenem. Antimicrob Agents Chemother 2019;63(1). 10.1128/AAC.01659-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shields RK, McCreary EK, Marini RV, et al. Early experience with meropenem-vaborbactam for treatment of carbapenem-resistant Enterobacteriaceae infections. Clin Infect Dis 2019. 10.1093/cid/ciz1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Merck & Co., Inc. RECARBRIO (imipenem, cilastatin, and relebactam) for injection, for intravenous Use. Whitehouse Station (NJ); 2019.
- 181.Livermore DM, Warner M, Mushtaq S. Activity of MK-7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa. J Antimicrob Chemother 2013;68(10):2286–90. 10.1093/jac/dkt178. [DOI] [PubMed] [Google Scholar]
- 182.Motsch J, Murta de Oliveira C, Stus V, et al. RESTORE-IMI 1: A Multicenter, Randomized, Double-blind Trial Comparing Efficacy and Safety of Imipenem/Relebactam vs Colistin Plus Imipenem in Patients With Imipenem-nonsusceptible Bacterial Infections. Clin Infect Dis 2019. 10.1093/cid/ciz530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Blizzard TA, Chen H, Kim S, et al. Discovery of MK-7655, a β-lactamase inhibitor for combination with Primaxin®. Bioorg Med Chem Lett 2014;24(3):780–5. 10.1016/j.bmcl.2013.12.101. [DOI] [PubMed] [Google Scholar]
- 184.Lapuebla A, Abdallah M, Olafisoye O, et al. Activity of Imipenem with Relebactam against Gram-Negative Pathogens from New York City. Antimicrob Agents Chemother 2015;59(8):5029–31. 10.1128/AAC.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Haidar G, Clancy CJ, Chen L, et al. Identifying Spectra of Activity and Therapeutic Niches for Ceftazidime-Avibactam and Imipenem-Relebactam against Carbapenem-Resistant Enterobacteriaceae. Antimicrob Agents Chemother 2017;61(9). 10.1128/AAC.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Balabanian G, Rose M, Manning N, et al. Effect of porins and blakpc expression on activity of imipenem with Relebactam in Klebsiella pneumoniae: can antibiotic combinations overcome resistance? Microb Drug Resist 2018;24(7): 877–81. [DOI] [PubMed] [Google Scholar]
- 187.Gomez-Simmonds A, Stump S, Giddins MJ, et al. Clonal Background, Resistance Gene Profile, and Porin Gene Mutations Modulate In Vitro Susceptibility to Imipenem-Relebactam in Diverse Enterobacteriaceae. Antimicrob Agents Chemother 2018;62(8). 10.1128/AAC.00573-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Galani I, Souli M, Nafplioti K, et al. In vitro activity of imipenem-relebactam against non-MBL carbapenemase-producing Klebsiella pneumoniae isolated in Greek hospitals in 2015–2016. Eur J Clin Microbiol Infect Dis 2019;38(6): 1143–50. [DOI] [PubMed] [Google Scholar]
- 189.Lob SH, Hackel MA, Kazmierczak KM, et al. In Vitro Activity of imipenem-relebactam against gram-negative ESKAPE pathogens isolated by clinical laboratories in the united states in 2015 (results from the SMART global surveillance program). Antimicrob Agents Chemother 2017;61(6). 10.1128/AAC.02209-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Carpenter J, Neidig N, Campbell A, et al. Activity of imipenem/relebactam against carbapenemase-producing Enterobacteriaceae with high colistin resistance. J Antimicrob Chemother 2019;74(11):3260–3. [DOI] [PubMed] [Google Scholar]
- 191.Hirsch EB, Ledesma KR, Chang K-T, et al. In vitro activity of MK-7655, a novel β-lactamase inhibitor, in combination with imipenem against carbapenem-resistant Gram-negative bacteria. Antimicrob Agents Chemother 2012;56(7): 3753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Horner C, Mushtaq S, Livermore DM. BSAC Resistance Surveillance Standing Committee. Potentiation of imipenem by relebactam for Pseudomonas aeruginosa from bacteraemia and respiratory infections. J Antimicrob Chemother 2019;74(7):1940–4. [DOI] [PubMed] [Google Scholar]
- 193.Livermore DM. Interplay of impermeability and chromosomal β-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 1992;36(9):2046–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Barnes MD, Bethel CR, Alsop J, et al. Inactivation of the Pseudomonas-Derived Cephalosporinase-3 (PDC-3) by Relebactam. Antimicrob Agents Chemother 2018;62(5). 10.1128/AAC.02406-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Young K, Painter RE, Raghoobar SL, et al. In vitro studies evaluating the activity of imipenem in combination with relebactam against Pseudomonas aeruginosa. BMC Microbiol 2019;19(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lob SH, Hackel MA, Kazmierczak KM, et al. In vitro activity of imipenem-relebactam against gram-negative bacilli isolated from patients with lower respiratory tract infections in the United States in 2015 - Results from the SMART global surveillance program. Diagn Microbiol Infect Dis 2017;88(2):171–6. [DOI] [PubMed] [Google Scholar]
- 197.Karlowsky JA, Lob SH, Kazmierczak KM, et al. In vitro activity of imipenem/relebactam against Gram-negative ESKAPE pathogens isolated in 17 European countries: 2015 SMART surveillance programme. J Antimicrob Chemother 2018;73(7):1872–9. [DOI] [PubMed] [Google Scholar]
- 198.Karlowsky JA, Lob SH, Kazmierczak KM, et al. In vitro activity of imipenem-relebactam against Enterobacteriaceae and Pseudomonas aeruginosa isolated from intraabdominal and urinary tract infection samples - SMART Surveillance United States 2015–2017. J Glob Antimicrob Resist 2019. 10.1016/j.jgar.2019.10.028. [DOI] [PubMed] [Google Scholar]