Abstract
Neuropathic pain (NP) is directly caused by an injury or disease of the somatosensory nervous system. It is a serious type of chronic pain that is a burden to the economy and public health. Although recent studies have improved our understanding of NP, its pathogenesis has not been fully elucidated. Noncoding RNAs, including lncRNAs, circRNAs, and miRNAs, are involved in the pathological development of NP through many mechanisms. In addition, extensive evidence suggests that novel regulatory mechanisms among lncRNAs/circRNAs, miRNAs, and mRNAs play a crucial role in the pathophysiological process of NP. In this review, we comprehensively summarize the regulatory relationship among lncRNAs/circRNAs, miRNAs, and mRNAs and emphasize the important role of the lncRNA/circRNA–miRNA–mRNA axis in NP.
Electronic supplementary material
The online version of this article (10.1007/s13311-020-00881-y) contains supplementary material, which is available to authorized users.
Key Words: Noncoding RNA, neuropathic pain, function, mechanism, review.
Introduction
Neuropathic pain (NP) is a worldwide problem that can be caused by a lesion or disease of the somatosensory system [1, 2]. Based on a systematic review of NP epidemiological studies, estimates of NP prevalence range from 6.9 to 10% [3]. The 2 causes of NP [4, 5] have been confirmed to be central nerve injury (such as stroke, multiple sclerosis, and spinal cord injury [SCI]) and peripheral nerve injury (PNI) (such as diabetes mellitus, peripheral nerve compression, and postherpetic neuralgia). The prevalence of central NP and peripheral NP has increased because of the growing elderly population worldwide [1]. NP leads to serious pain, sleep disorders, impaired quality of life, anxiety, and depression [6], which can lead to economic burden, including direct expenses (such as healthcare expenditures) and indirect expenses (such as sick leave) [7, 8]. In Europe, the direct and indirect expenses (indirect expenses percentage in total expenses) of NP per patient were €9685 in the UK (57%), €10,597 in Spain (67%), €9305 in Italy (69%), €14,446 in Germany (78%), and €10,313 in France (69%) [8].
The mechanism of NP involves multiple organs and systems, such as the sciatic nerve, dorsal root ganglia (DRG), spinal cord, and brain [5, 9, 10]. Moreover, numerous NP mechanisms have been discovered in animal studies. Animal models of NP are conducted by surgical lesions of PNI and central nerve injury, such as the constriction, ligation, and transection of nerves [1]. The common animal models of NP are SCI, spared nerve injury (SNI), sciatic nerve chronic constriction injury (CCI), partial sciatic nerve ligation, chronic compression of the DRG, and spinal nerve ligation (SNL) [11–13]. Although some mechanisms of NP are well discovered, it remains difficult to prevent and treat NP without thoroughly illuminating the underlying mechanisms. Finding novel biomarkers for diagnosis and therapeutic targets of NP is important to develop more effective treatment programs. In recent decades, many studies have revealed the role of noncoding RNAs (ncRNAs) in the development and progression of NP [14, 15].
The discovery of ncRNAs brought novel landscapes for the diagnosis and treatment of NP. ncRNAs are not translated into proteins; they are functional small RNA molecules. Only 2% of the human genome consists of protein-coding genes, and approximately 98% of the rest are noncoding genes [16]. Currently, the ncRNAs mainly include microRNAs (miRNAs), long ncRNAs (lncRNAs), circular RNAs (circRNAs), and small nucleolar RNAs (snoRNAs) [17]. Numerous studies [18–20] have highlighted that miRNAs, lncRNAs, and circRNAs are pertinent to the pathological processes of NP, such as inflammation. In addition, many studies have focused on the interactions among lncRNAs/circRNAs, miRNAs, and messenger RNAs (mRNAs). The lncRNAs and circRNAs have a cross-talk with miRNAs and then regulate mRNA expression in the progression of NP. In this review, we elaborate the roles of miRNAs, lncRNAs, and circRNAs in NP and highlight the role of the lncRNA/circRNA–miRNA–mRNA axis in the processes of NP.
miRNAs and NP
miRNAs are small noncoding RNAs that were first reported in Caenorhabditis elegans in 1993. Lee and his colleagues revealed that the C. elegans-related gene lin-4 develops into 2 small RNAs instead of a protein [21, 22]. The length of miRNAs is approximately 21 to 25 nucleotides, and the target site of miRNAs is located in the 3′-untranslated region of mRNAs [23]. miRNAs are able to posttranscriptionally modulate gene expression via translation inhibition or mRNA degradation. When an miRNA is perfectly bound to the 3′-untranslated regions, the target mRNA degrades immediately; however, if the 2 species do not completely bind, then the miRNA will suppress the translation of the target mRNA [22, 24].
Under the development of bioinformatics, more than 1000 miRNAs have been detected in the human genome, and over 30% of the human genome may be modulated by these miRNAs [22, 25]. miRNAs participate in manifold biological regulation processes, including cell proliferation, differentiation, inflammation, and apoptosis [26–29]. Abundant evidence shows that miRNAs, as crucial regulators, participate in many pathological processes of human diseases, such as cancer [30], cardiac diseases [31], and diabetes [32]. Studies have shown the dysregulation of miRNAs in the animal models of NP, suggesting that miRNAs play a crucial part in the development of NP. Zhou et al. [15] used a sequencing technique to detect differences in the miRNA expression profiles in the spinal cord between an SNI group and a control group. Based on the sequencing results, the SNI group had 6 upregulated and 6 downregulated miRNAs at 14 days after injury compared with the control group. Among the 12 differential miRNAs, miR-344b-1-3p and miR-490-3p were verified by quantitative polymerase chain reaction (qPCR). Chang and his colleagues [33] assessed the expression of differential miRNAs in the DRG of rats via microarray analysis. Compared with the sham group, 83 differential miRNAs (49 upregulated and 35 downregulated) in the SNL group were observed 7 days after nerve injury. In addition, the authors validated the expression of miR-21, miR-31, miR-668, and miR-672 in the DRG through qPCR and predicted the potential signaling pathways regulated by the 4 miRNAs.
It has been reported that the reduction of 40 to 68% of the miR-183 family, including the miR-183/96/182 cluster, in the DRG is sufficient to significantly alter gene expression. Of the NP-induced pain genes, 80% are modulated by the miR-183 family, indicating that the miR-183 family plays a vital role in the maintenance of NP [34]. Several studies showed that the expression of miR-183 was significantly downregulated in CCI or SNL rat models, and the overexpression of miR-183 was able to relieve pain-like behavior by inhibiting the target genes of miR-183 [35–38]. MiR-96 and miR-182 were also decreased in the NP rat model. Cai et al. [39] found that miR-182 was significantly decreased in the DRG of SNI rats, and sodium channel 1.7 (Nav1.7) expression was elevated, causing mechanical hyperalgesia. The injection of miR-182 agomir reversed the overexpression of Nav1.7 and could restrain mechanical allodynia caused by NP. Similarly, several studies [34, 40] also reported that in NP animal mods, the expression of miR-96 was greatly downregulated in the DRG. Moreover, Cai et al. [41] reported that CCI caused miR-150 downregulation in the dorsal spinal cord at 7 days. AKT3 was directly targeted by miR-150 and upregulated in CCI rats. The overexpression of miR-150 could suppress the impacts of AKT3 and significantly relieve NP in CCI rats. Another study [42] found that miR-34a was significantly downregulated in the DRG at 12 days after CCI, with a significant reduction in the pain threshold. A luciferase assay was used to prove that vesicle-associated membrane protein 2 (VAMP-2) and voltage-gated sodium channel β2 subunit (SCN2B) were the target genes of miR-34a, which were confirmed by enzyme-linked immunosorbent assay. However, VAMP-2 expression was greatly enhanced, whereas SCN2B expression was only marginally altered, showing that the miR-34a/VAMP-2 pathway was possibly relevant to NP. Accumulating evidence has emphasized that the induction and maintenance of NP are accompanied by changes in the expression of miRNAs, and detailed information about these miRNAs is shown in Table 1.
Table 1.
Functional characterization of the miRNAs in NP
| miRNA | Expression | Target gene(s) | Model | Region | Functions | Reference |
|---|---|---|---|---|---|---|
| miR-21 | Up | CCL1, TIMP3 | CCI rat model | Spinal cord | Neuroinflammation | [43] |
| IL-1β | SNL/CCI rat model | DRG | Neuroinflammation | [44] | ||
| miR-195 | Up | Patched1 | CCI rat model | Cerebrospinal fluid | Neuroinflammation | [45] |
| ATG14 | SNL rat model | Spinal cord, microglia | Neuroinflammation | [46] | ||
| miR-155 | Up | SGK3 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [47] |
| SOCS1 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [48] | ||
| miR-34c-5p | Up | SIRT1, STAT3 | CCI rat model | Spinal cord, DRG | Neuroinflammation | [49] |
| miR-192-5p | Up | XIAP | SNI rat model | Spinal cord | Apoptosis | [50] |
| miR-15a/16 | Up | GRK2 | CCI rat model | Spinal cord | Neuroinflammation | [51] |
| miR-217 | Up | TLR5 | bCCI rat model | Spinal cord | Neuroinflammation | [52] |
| miR-32-5p | Up | Dusp5 | SNL rat model | Spinal cord, microglia | Neuroinflammation | [53] |
| miR-218 | Up | SOCS3 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [54] |
| miR-132-3p | Up | GluA1, GluA2 | SNI rat model | Spinal cord, DRG, microglia | Neuronal plasticity | [55] |
| miR-221 | Up | SOCS1 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [56] |
| miR-19a | Up | SOCS1 | CCI rat model | Spinal cord | Neuroinflammation | [57] |
| miR-183 | Down | MAP3K4 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [35] |
| mTOR/VEGF | CCI rat model | Spinal cord, PC12 cell | Neuroinflammation | [36] | ||
| TREK-1 | CCI rat model | DRG | Neuronal excitability | [37] | ||
| Nav1.3, BDNF | SNL rat model | DRG | Neuronal excitability | [38] | ||
| miR-182 | Down | Nav1.7 | SNI rat model | DRG | Neuronal excitability | [39] |
| miR-96 | Down | Nav1.3 | CCI rat model | DRG | Neuronal excitability | [40] |
| miR-96/182/183 | Down | – | SNL rat model | DRG | – | [34] |
| miR-150 | Down | AKT3 | CCI rat model | Spinal cord | Neuroinflammation | [41] |
| TLR5 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [58] | ||
| miR-7a | Down | NEFL, STAT3 | SNL rat model | DRG | Neuronal excitability | [59] |
| Scn2b | SNL rat model | DRG | Neuronal excitability | [60] | ||
| miR-206 | Down | HDAC4 | CCI rat model | DRG | Neuronal excitability | [61] |
| BDNF | CCI rat model | DRG, PC12 cell | Neuroinflammation | [62] | ||
| miR-30b | Down | Nav1.3 | SNL rat model | Spinal cord, DRG neuron | Neuronal excitability | [63] |
| Nav1.7 | SNI rat model | DRG, PC12 cell | Neuronal excitability | [64] | ||
| miR-194 | Down | FOXA1 | CCI rat model | Spinal cord | Neuroinflammation | [65] |
| miR-384-5p | Down | SCN3A | CCI rat model | DRG | Neuroinflammation | [66] |
| miR-15a | Down | AKT3 | CCI rat model | Spinal cord, PC12 cell | Autophagy | [67] |
| miR-144 | Down | RASA1 | CCI mice model | DRG | Neuroinflammation | [68] |
| miR-138 | Down | NF-κB | SNI mice model | Spinal cord | Neuroinflammation | [69] |
| miR-34a | Down | SCN2B, VAMP-2 | CCI rat model | DRG | Neuroinflammation | [42] |
| miR-340-5p | Down | Rap1A | CCI rat model | Spinal cord, microglia | Neuroinflammation | [70] |
| miR-202 | Down | Rap2A | CCI rat model | Spinal cord, PC12 cell | Neuronal plasticity | [71] |
| miR-98 | Down | HMGA2 | CCI rat model | Spinal cord | Neuroinflammation | [72] |
| miR-124-3p | Down | EZH2 | CCI rat model | Spinal cord | Neuroinflammation | [73] |
| miR-20b-5p | Down | AKT3 | CCI rat model | Spinal cord, PC12 cell | Neuroinflammation | [74] |
| miR-34c | Down | NLRP3 | CCI mice model | Spinal cord | Neuroinflammation | [75] |
| miR-20a | Down | PDZ-RhoGEF, RhoA, GAP43 | SDCL rat model | DRG | Sensory conduction | [76] |
| miR-152 | Down | MafB | PNI mice model | Spinal cord | Neuronal plasticity | [77] |
| miR-129-5p | Down | HMGB1 | CCI rat model | Spinal cord | Neuroinflammation | [78] |
| miR-214-3p | Down | DNMT3a, CSF1 | SNL rat model | DRG | Neuroinflammation | [79] |
| miR-362-3p | Down | PAX2 | SCI rat | Spinal cord | Neuroinflammation | [80] |
| miR-28-5p | Down | Zeb1 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [81] |
| miR-26a-5p | Down | MAPK6 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [82] |
| miR-93 | Down | STAT3 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [83] |
| miR-539 | Down | NR2B | CCI rat model | Brain | Neuronal plasticity | [84] |
| miR-449a | Down | TRPA1, KCNMA1, TPTE | SNI rat model | DRG | Neuronal excitability | [85] |
| miR-142-3p | Down | HMGB1 | SNL mice model | DRG, DRG neuron | Neuroinflammation | [86] |
| miR-200b/429 | Down | ZEB1 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [87] |
| miR-143 | Down | DNMT3a | SNL rat model | DRG | DNA methylation | [88] |
| miR-145 | Down | RREB1, p-AKT | CCI rat model | Spinal cord | Neuroinflammation | [89] |
| miR-141 | Down | HMGB1 | CCI rat model | DRG | Neuroinflammation | [90] |
| miR-1 | Down | BDNF, Cx43 | CCI rat model | Sciatic nerve | Neuroinflammation | [91] |
| miR-203 | Down | Rap1A | CCI rat model | Spinal cord, PC12 cell | Neuronal plasticity | [92] |
| miR-103 | Down | Cav1.2 | SNL rat model | Spinal cord, spinal neuron | Neuronal excitability | [93] |
CCI = sciatic nerve chronic constriction injury; SNL = sciatic nerve ligation; SNI = spared nerve injury; bCCI = bilateral sciatic nerve chronic constriction injury; SDCL = spinal cord dorsal column lesion; PNI = peripheral nerve injury; DRG = dorsal root ganglia; CCL1 = chemokines C-C motif ligand 1; TIMP3 = tissue inhibitor of metalloproteinase-3; IL-1β = interleukin-1β; SGK3 = serum and glucocorticoid–regulated protein kinase 3; SOCS = suppressor of cytokine signaling; SIRT1 = sirtuin-1; STAT3 = signal transducer and activator of transcription 3; XIAP = X-linked inhibitor of apoptosis protein; GRK2 = G protein-coupled receptor kinase 2; TLR5 = toll-like receptor 5; Dusp5 = dual-specificity phosphatase 5; SOCS3 = suppressor of cytokine signaling 3; GluA = AMPA receptor subunit; BDNF = brain-derived neurotrophic factor; NEFL = neurofilament light polypeptide; HDAC4 = histone deacetylase 4; FOXA1 = Forkhead box protein A1; RASA1 = RAS P21 Protein Activator 1; NF-κВ = nuclear factor-κВ; SCN2B = voltage-gated sodium channel β2 subunit; VAMP-2 = vesicle-associated membrane protein 2; Rap1A = Ras-related protein 1A; HMGA2 = high mobility group A2; NLRP3 = nucleotide binding domain-like receptor protein 3; HMGB1 = high mobility group protein B1; DNMT3a = DNA methyltransferase 3a; CSF1 = colony-stimulating factor-1; PAX2 = paired box gene 2; ZEB1 = zinc finger E-box-binding homeobox 1; MAPK = mitogen-activated protein kinases; TRPA1 = transient receptor potential cation channel subfamily A member 1; KCNMA1 = calcium-activated potassium channel subunit α-1; TPTE = transmembrane phosphatase with tension homology; RREB1 = ras responsive element binding protein 1; p-AKT = phosphorylated protein kinase B; Cx43 = Connexin 43
lncRNAs and NP
In the late 1980s, scientists discovered the first noncoding RNA, H19, but at that time, such RNA was considered mRNA [94]. Subsequently, another study reported that H19 was unusual compared with other mRNAs because the gene was not involved in translation, although it contained an open reading framework. Scientists suggested that H19 was an RNA molecule [95] until the early 1990s, when H19 was widely investigated as an lncRNA and then regarded as a prototype for manifold lncRNAs. lncRNAs are RNA molecules with greater than 200 nucleotides in length and that may or may not have a minor protein-coding function. On the basis of genomic location, lncRNAs could fall into 5 categories, namely, 1) sense, 2) anti-sense, 3) bidirectional, 4) intronic, and 5) intergenic. Recent research has shown that lncRNAs play crucial roles in biological processes, including cell growth, apoptosis, transcription, translation, cell differentiation, and immune responses, via various regulatory mechanisms, including the modulation of gene expression under specific conditions, DNA methylation, histone modification, sponging miRNAs, the modulation of mRNA stability, and the collection of transcription factors [96–99].
lncRNAs could play a crucial role in neuronal activity and injury through the control of nervous system development and synaptic plasticity and the deregulation of varying neurological and mental states [96]. Recently, considerable evidence has shown that lncRNAs are crucial players in NP processes. Microarray or RNA sequencing technologies have been applied to detect different lncRNA expression profiles in NP. Zhou and his colleagues [100] utilized second-generation RNA sequencing to detect lncRNA profiling in the SNI rat model and control rats. They discovered that the amount of differentially expressed lncRNAs peaked on the 7th day after SNI, and 25 lncRNAs were found to be upregulated and 101 downregulated in SNI rats compared with control rats at 7 days. Subsequently, 4 upregulated lncRNAs (XLOC_041439, Mlxipl, XLOC_022312 and LOC100911498) and 3 downregulated lncRNAs (Rn50_X_0739.1, XLOC_001451, and XLOC_026060) were validated by qPCR. Du et al. [101] found that compared with the control group, 1481 differential lncRNAs (1026 upregulated and 455 downregulated) and 1096 differential mRNAs (463 upregulated and 633 downregulated) were discovered via microarray analysis in the spinal cord of diabetic mice. Four of the lncRNAs (ENSMUST00000134111, NR_003513, ENSMUST00000150952, and AK047066) were verified by qPCR, and the results were in accordance with the sequencing results. Moreover, through the analysis of these differentially expressed genes, Du et al. detected 346 related lncRNA–mRNA pairs that participate in the process of NP.
On the basis of the increasing discovery of the functional properties of differentially expressed lncRNAs in NP, several studies have further discussed the functions and molecular mechanisms of certain lncRNAs acting on NP (Table 2). The lncRNA X-inactive specific transcript (XIST) is 1 of the lncRNAs that plays a key role in the regulation of X chromosome inactivation. Notably, several studies have proven that XIST is upregulated in NP animal models, including the CCI rat model, SCI rat model, and complete Freund’s adjuvant (CFA) rat model. The silencing of XIST could reduce neuroinflammation and relieve pain-like behavior in NP [18, 102–106]. Several studies have indicated that lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) significantly increased in the spinal cord or Schwann cells. Reducing the expression of MALAT1 could improve pain-like behavior and repress the progression of inflammation, proliferation, and migration by regulating target genes [107–110]. The lncRNAs uc.48+ [111–113], BC168687 [114–116], and NONRATT021972 [117–119] were also upregulated in the NP animal models. They relieved hyperalgesia and decreased neuroinflammation by modulating the P2X3 receptor or P2X7 receptor. In addition to the lncRNAs reported above, some lncRNAs have also been reported to be associated with NP [19, 120–138]. For more detailed information, see Table 2.
Table 2.
Functional characterization of the lncRNAs in NP
| LncRNA | Expression | Target genes | Related genes | Model | Region | Functions | Reference |
|---|---|---|---|---|---|---|---|
| XIST | Up | miR-150 | ZEB1 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [102] |
| miR-137 | TNFAIP1 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [103] | ||
| miR-154-5p | TLR5 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [104] | ||
| miR-494 | STAT3 | SCI rat model | Spinal cord | Apoptosis | [105] | ||
| miR-544 | STAT3 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [18] | ||
| miR-146a | Nav1.7 | CFA rat model | DRG | Neuroinflammation | [106] | ||
| MALAT1 | Up | miR-154-5p | AQP9 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [107] |
| miR-129-5p | BDNF | PNI mice model | Schwann cell | Proliferation and migration | [108] | ||
| miR-129-5p | HMGB1 | CCI rat model | Spinal cord, microglia | Neuroinflammation | [109] | ||
| miR-206 | ZEB2 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [110] | ||
| uc.48+ | Up | P2X7 receptor | p-ERK1/2 | TN rat model | Trigeminal ganglia | Phosphorylation | [111] |
| – | Diabetic rat model | DRG | Neuroinflammation | [112] | |||
| p-ERK1/2 | Diabetic rat model | Superior cervical ganglia | Autonomic neuropathy | [113] | |||
| BC168687 | Up | P2X7 receptor | – | Diabetic rat model | DRG | Neuroinflammation | [114] |
| TRPV1 | TNF-α, IL-1β, ERK, MAPK | Diabetic rat model | DRG | Neuroinflammation | [115] | ||
| P2X7, TRPV1 receptor | – | Diabetic rat model | DRG | Neuronal excitability | [116] | ||
| NONRATT021972 | Up | P2X3 receptor | ERK1/2, p-ERK | Diabetic rat model | DRG | Neuroinflammation | [117] |
| P2X7 receptor | – | Diabetic rat model | Superior cervical ganglion, PC12 cell | Neuroinflammation | [118] | ||
| P2X7 receptor | – | Diabetic rat model | DRG | Neuroinflammation | [119] | ||
| BC088259 | Up | Vimentin | – | SNI rat model | Schwann cell | Migration | [120] |
| Linc00657 | Up | miR-136 | ZEB1 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [121] |
| CRNDE | Up | miR-136 | IL6R | CCI rat model | Spinal cord, microglial | Neuroinflammation | [122] |
| NEAT1 | Up | miR-381 | HMGB1 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [19] |
| uc.153 | Up | miR-182-5p | EphB1-NMDA receptors | CCI mice model | Spinal cord | Neuronal excitability | [123] |
| Linc00052 | Up | miR-448 | JAK1 | SNL rat model | Spinal cord, PC12 cell | Neuroinflammation | [124] |
| Linc00311, AK141205 | Up | STAT3 | CCL-2, COX-2, IL-1β, IL-6, and TNF-α | CCI rat model | Microglia | Neuroinflammation | [125] |
| SNHG5 | Up | miR-154-5p | CXCL13 | CCI rat model | DRG, microglia | Astrocyte inhibition, microglia activation | [126] |
| H19 | Up | – | – | SNL rat model | DRG | Neuroinflammation | [127] |
| H19 | Up | – | – | SNL mice model | DRG, DRG cell | Neuronal excitability | [128] |
| PKIA-AS1 | Up | – | CDK6 | SNL rat model | Spinal cord | Neuroinflammation | [129] |
| BC088327 | Up | – | Heregulin-1β | PNI rat model | Sciatic nerve | Proliferation | [130] |
| MRAK009713 | Up | P2X3 receptor | – | CCI rat model | DRG | Neuronal excitability | [131] |
| Kcna2 AS RNA | Up | Kcna2 mRNA | G9a | CCI/SNL mice model | DRG | Neuronal excitability | [132] |
| Kcna2 mRNA | – | CCI/SNL rat model | DRG | Neuronal excitability | [133] | ||
| DGCR5 | Down | miR-330-3p | PDCD4 | CCI rat model | Spinal cord, microglial | Neuroinflammation | [134] |
| TNXA-PS1 | Down | miR-24-3p/miR-152-3p | Dusp1 | SNI rat model | DRG | Migration | [135] |
| CCAT1 | Down | miR-155 | SGK3 | CCI rat model | DRG, spinal cord, hippocampus, anterior cingulate cortex, PC12 cell | Neuronal excitability | [136] |
| uc.217 | Down | – | – | SNI rat model | DRG | Neurite outgrowth | [137] |
| BC089918 | Down | – | – | SNI rat model | DRG | Nerve regeneration | [138] |
CCI = sciatic nerve chronic constriction injury; SCI = spinal cord injury; CFA = complete Freund’s adjuvant; PNI = peripheral nerve injury; TN = trigeminal neuralgia; DRG = dorsal root ganglia; XIST = X-inactive specific transcript; ZEB1 = zinc finger E-box-binding homeobox 1; TNFAIP1 = tumor necrosis factor alpha-induced protein 1; TLR5 = toll-like receptor 5; STAT3 = signal transducer and activator of transcription 3; MALAT1 = metastasis-associated lung adenocarcinoma transcript 1; AQP9 = aquaporin 9; BDNF = brain-derived neurotrophic factor; HMGB1 = high mobility group protein B1; TRPV1 = transient receptor potential vanilloid type 1; IL = interleukin; TNF-α = tumor necrosis factor-α; ERK = extracellular regulated protein kinases; MAPK = mitogen-activated protein kinases; CRNDE = Colorectal Neoplasia Differentially Expressed; JAK1 = Janus kinase 1; CCL-2 = chemokine CC motif ligand 2; COX-2 = cyclooxygenase 2; SNHG5 = small nucleolar RNA host gene 5; CXCL13 = C-X-C motif chemokine 13; CDK6 = cyclin-dependent kinases 6; NF-κВ = nuclear factor-κВ; Dusp 1 = Dual-specificity phosphatase 1; CCAT1 = colon cancer–associated transcript-1; SGK3 = Serum and glucocorticoid–regulated protein kinase 3
circRNAs and NP
The circRNAs were first reported in viroids in 1976, and they are closed circular RNA molecules without 3′ polyadenylated tails or terminal 5′ caps [139, 140]. Abundant circRNAs were found with the advancement of RNA sequencing analyses, beginning in approximately 2010. More than 10,000 circRNAs have been found in various fungi, plants, and animals [141]. At present, 4 kinds of circRNAs have been identified: intergenic circRNAs, exon–intron circRNAs (EIciRNAs), and circRNAs from introns and exonic circRNAs (ecircRNAs) [142]. In addition, many circRNAs showed dynamic expression in various physiological and pathological scenarios, including cell apoptosis and cancer [143, 144]. Given that circRNAs have circular structures and are not influenced by RNA exonuclease, the expression of circRNAs could be more stable than the expression of linear RNAs [145–147]. The mechanism and function of circRNAs could not be fully identified; however, an increasing number of studies have revealed that circRNAs play a critical role in the diagnosis and treatment of different diseases.
Expression profiles of circRNAs were revealed in NP induced by CCI of the sciatic nerve and SNI [15, 148]. Cao et al. [149] used a circRNA microarray to detect the differential expression of circRNAs between CCI NP model rats and sham CCI rats. A total of 469 differentially expressed circRNAs could be found in the spinal dorsal horn between sham and CCI rats (fold change ≥ 2). In the CCI group, 106 of 469 circRNAs were statistically downregulated, and the other 363 were upregulated. The expression of 3 circRNAs (circRNA_003724, circRNA_008008, and circRNA_013779) was more than 10 times higher in the CCI group than in the sham group. The expression levels of 4 downregulated circRNAs (circRNA_011111, circRNA_007419, circRNA_007512, circRNA_010913) and 4 upregulated circRNAs (circRNA_008973, circRNA_013779, circRNA_008646, circRNA_35215) were validated by qPCR in the CCI group. Another study [15] used RNA sequencing analyses to predict the expression patterns of circRNAs between the SNI NP rat model and sham SNI rats. Compared with the sham group, Zou et al. [15] found that 188 circRNAs (120 downregulated and 68 upregulated) were abnormally expressed in the spinal cord of rats in the SNI group at 14 days after surgery. The expression of 2 selected circRNAs (circ 0004058 and circ 0005854) was verified by qPCR.
Currently, although few studies have revealed the functional roles of circRNAs in NP, several studies have reported the mechanism and function of circRNAs in the development and progression of NP (Table 3). Zhang and colleagues [20] found that the expression of circAnks1a was significantly upregulated in the spinal cord of SNL rats after 3, 7, 10, and 14 days compared with sham SNL rats. Notably, the pain-like behavior of SNL rats could be relieved through the downregulation of circAnks1a by siRNA. circAnks1a alleviated NP by regulating the vascular endothelial growth factor B (VEGFB) expression to decrease the excitability of the spinal cord in SNL rats. Pan and colleagues [149] demonstrated that the expression of the circRNA filamin A interacting protein 1-like (Filip1l) was significantly increased in spinal neurons of CCI mice and CFA-induced chronic inflammatory pain mice. The downregulation of circRNA Filip1l could alleviate the pain-like behavior in CFA mice by the injection of anti-Filip1l. The circRNA Filip1l regulated chronic pain by targeting ubiquitin protein ligase E3 component n-recognin 5 (Ubr5). Mao and colleagues [153] found that the circRNA ankyrin repeat and in-between Ring finger (IBR) domain containing 1 (Ankib1) was downregulated at 1, 4, 7, and 14 days after PNI in the sciatic nerve, which targeted cytochrome P450, family 26, subfamily B, polypeptide 1 (Cyp26b1) to regulate Schwann cell proliferation in the sciatic nerve. Zhou and colleagues [152] reported that, compared with sham PNI rats, circRNA-2837 was inhibited in the spinal cord of PNI rats and targeted LC3-II and p62 to regulate neuronal autophagy. Wang and colleagues [150] revealed that the expression of circHIPK3 was significantly upregulated in the DRG of NP-induced diabetic rats. In addition, the circHIPK3 regulated IL-1b, IL-6, IL-12, and tumor necrosis factor (TNF)-α to alleviate NP by inhibiting inflammation in diabetic NP rats. Cai and colleagues [151] demonstrated that ciRS-7 was significantly increased and that the degree of inflammation and autophagy was also upregulated in the spinal cord of CCI rats.
Table 3.
Functional characterization of the circRNAs in NP
| circRNA | Expression | Target genes | Related genes | Model | Region | Functions | Reference |
|---|---|---|---|---|---|---|---|
| circ-Filip1l | Up | miRNA-1224 | Ago2, Ubr5 | CCI mice model | Spinal cord, astrocytes, and microglia | Neuroinflammation | [149] |
| circHIPK3 | Up | miR-124 | IL-1b, IL-6, IL-12, and TNF-α | Diabetic rat model | DRG, PC12 cell | Neuroinflammation | [150] |
| ciRS-7 | Up | miR-135a-5p | IL-6, IL-12, TNF-α, Iba1 and Beclin-1, p62, LC3-I, LC3-II | CCI rat model | Spinal cord | Neuroinflammation, autophagy | [151] |
| circRNA.2837 | Down | miR-34a | LC3-II, p62 | SNI rat model | Spinal cord, sciatic nerve | Neuronal autophagy | [152] |
| circ-Ankib1 | Down | miR-423-5p/485-5p/666-3p | DHX9, Cyp26b1 | Sciatic nerve crush rat model | Schwann cell | Proliferation | [153] |
| circAnks1a | Up | miR-324-3p | VEGFB, YBX1 | SNL rat model | Spinal cord | Central sensitization | [20] |
CCI = sciatic nerve chronic constriction injury; SNI = spared nerve injury; SNL = sciatic nerve ligation; DRG = dorsal root ganglia; Ago2 = Argonaute-2; Ubr5 = ubiquitin protein ligase E3 component n-recognin 5; IL = interleukin; TNF-α = tumor necrosis factor-α; Ankib1 = ankyrin repeat and in-between Ring finger (IBR) domain containing 1; DHX9 = DEx/H-box helicase 9; Cyp26b1 = cytochrome P450, family 26, subfamily B, polypeptide 1
Interactions Among lncRNAs, miRNAs, and mRNAs in NP
Recently, extensive evidence suggests that the lncRNA–miRNA–mRNA axis plays a critical role in the physiology and pathology of many diseases, including cancer, osteoarthritis, and cardiovascular diseases [98, 154, 155]. The potential mechanisms of lncRNA, miRNA, and mRNA interactions are as follows: 1) lncRNA acts as a competitive endogenous RNA (ceRNA) to sponge miRNA. lncRNAs have the effect of keeping miRNA away from mRNAs. These lncRNAs are called ceRNAs. They act as sponges for miRNAs, reducing the number of available miRNAs and helping to improve the translation of target mRNAs. For example, Cheng and his colleagues found that the lncRNAs CRNDE and MALAT1 may sponge miRNAs related to sepsis, participating in the regulation of sepsis modules [156]. Another study showed that the lncRNA TPTE pseudogene 1 (TPTEP1) could suppress the process of proliferation in lung cancer by sponging miR-328-5p [157]. 2) miRNA leads to the degradation of lncRNA. miRNAs can change the abundance of lncRNAs by reducing the stability of their target lncRNAs, thus affecting diverse cellular processes. For example, miR-145-5p was a target gene of lncRNA-RoR in human embryonic stem cells, whereas increasing the concentration of miR-145-5p reduced the activity of lncRNA-RoR [158]. 3) lncRNA combines with mRNA by competing with miRNA. lncRNAs can directly bind to complementary mRNAs at the miRNA–mRNA binding site region, thereby removing the regulation of miRNA on mRNA. For example, the tumor-related lncRNA noncoding Nras functional RNA (ncNRFR) decreases the function of let-7 by competing with let-7 for target mRNAs [159]. 4) lncRNA generates miRNA. For example, it has been reported that lncRN-MD1 could generate miR-206 and miR-133, playing roles in muscle differentiation and dystrophy [160]. However, it has been reported that the reciprocity of ncRNAs is involved in the pathological process of NP, including neuroinflammation, cell migration, and cell apoptosis. The potential mechanism of lncRNA, miRNA, and mRNA acting on NP is that lncRNAs serve as ceRNAs, thereby sponging miRNAs and inhibiting their downstream target genes (Fig. 1).
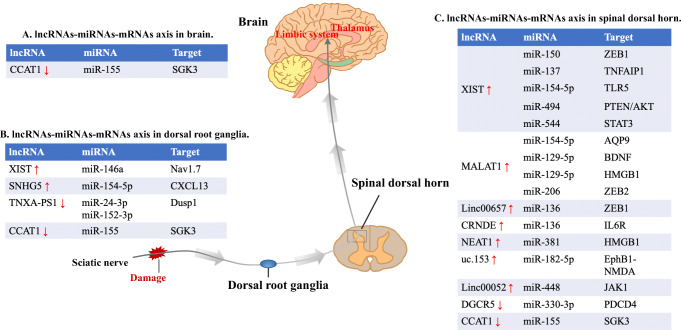
Fig. 1.
lncRNA–miRNA–mRNA axis in NP. The noxious stimulus reaches the spinal dorsal horn via the afferent nerve fibers. Spinal dorsal horn is the center responsible for integrating and processing input information. The output from the spinal cord network is then sent to higher cortical centers. The thalamus and limbic system are important parts of the brain for NP expression. The roles of the lncRNA–miRNA–mRNAs axis in NP was reflected in the DRG, spinal dorsal horn, and brain regions. CCAT1 = colon cancer–associated transcript-1; SGK3 = serum and glucocorticoid–regulated protein kinase 3; XIST = X-inactive specific transcript; SNHG5 = small nucleolar RNA host gene 5; CXCL13 = C-X-C motif chemokine 13; Dusp 1 = Dual-specificity phosphatase 1; ZEB = zinc finger E-box-binding homeobox; TNFAIP1 = tumor necrosis factor alpha-induced protein 1; TLR5 = toll-like receptor 5; STAT3 = signal transducer and activator of transcription 3; AQP9 = aquaporin 9; BDNF = brain-derived neurotrophic factor; HMGB1 = high mobility group protein B1; IL6R = interleukin 6R, JAK1 = Janus kinase 1
Several studies have proven that the lncRNA XIST was significantly upregulated in the CCI or CFA animal model, and XIST could induce the neuroinflammation of NP by regulating the target mRNAs as sponges of miR-150, miR-137, miR-154-5p, miR-544, and miR-146a [18, 102–104, 106]. Another study showed that the knockdown of XIST could relieve pain-like behavior and apoptosis in the spinal cord of SCI rats by competitively sponging miR-494, thereby negatively modulating the expression of AKT [105]. In addition, there were articles reporting that the lncRNA MALAT1 participated in the process of NP by sponging miRNA-154-5p, miR-129-5p, and miR-206. The downregulation of MALAT1 or its target mRNAs helps relieve pain and repress inflammation, proliferation, and migration [107–110]. Yao et al. demonstrated that the lncRNA TNXA-PS1 was significantly decreased in the DRG of SNI rats. Furthermore, they indicated that TNXA-PS1 could competitively bind with miR-24-3p/miR-152-3p, thereby modulating migration in the DRG by controlling the expression of Dusp1 [135]. Chen and colleagues [126] clarified that lncRNA SNHG5 could sponge miR-154-5p to modulate C-X-C motif chemokine 13 (CXCL13) in CCI rats and that the knockdown of SNHG5 could relieve pain and suppress the sensitization of astrocytes and microglia. Shen et al. and Zhang et al. [121, 122] found that lncRNA00657 and the lncRNA CRNDE were significantly increased in the CCI rat model, and they could regulate ZEB1 and interleukin 6 receptor (IL6R) by sponging miR-136. Based on the study of Xia et al. [19], the lncRNA NEAT1 was overexpressed in the CCI rat model. The NEAT1 modulated the expression of HMGB1 to relieve pain and decrease inflammation by sponging miR-381 in the spinal cord and microglia of CCI rats. Zhang and colleagues [123] found that the lncRNA uc.153 played regulatory roles in NP via competitive combination of miR-182-5p and then the regulation of EphB1-NMDA receptors. Wang et al. suggested that lnc00052 [124] could induce pain-like behavior and inflammation in the spinal cord of SNL rats by sponging miR-448 and modulating JAK1. Compared with sham CCI, the lncRNA DGCR5 was downregulated in the spinal cord of CCI rats. DGCR5 could negatively regulate the expression of miR-330-3p and suppress neuroinflammation and hyperalgesia by sponging miR-330-3p and modulating the downstream target PDCD4 [134]. Dou and colleagues [136] showed that the lncRNA CCAT1 was significantly decreased in the DRG, spinal cord, hippocampus, and anterior cingulate cortex of CCI rats. CCAT1 could inhibit pain-like behaviors by sponging miR-155 and then regulating the expression level of SGK3. The abovementioned results showed that lncRNAs could serve as miRNA sponges in the interactions among lncRNAs, miRNAs, and mRNAs in NP.
Interactions Among circRNAs, miRNAs, and mRNAs in NP
At present, many studies focus on the novel mechanism underlying the interactions among circRNAs, miRNAs, and mRNAs. Cross-talk was found between circRNAs and miRNAs, which regulated the relevant mRNA expression in physiologic processes and pathological mechanisms [161–163]. The interactions among circRNAs, miRNAs, and mRNAs could participate in the pathological mechanisms of NP, such as neuroinflammation [149, 150], neuronal excitability [20], cell proliferation [153], and neuronal autophagy [152] (Fig. 2). The relative interaction mechanisms among circRNAs, miRNAs, and mRNAs are debated; however, 2 kinds of mechanisms could be identified: 1) circRNAs sponging miRNAs. Song et al. found that circHMCU, as a sponger of the let-7 family, played a crucial role in the proliferation and metastasis of breast cancer [164]. Another study showed that the circRNA MAN2B2 was abnormally expressed in hepatocellular carcinoma tissues. The knockdown of the circRNA MAN2B2 could suppress the process of cell proliferation in the hepatocellular carcinoma cells by sponging miR-217 [165]. 2) miRNAs mediating circRNAs. Hansen and his colleagues found that miR-671 mediated the cleavage of the circRNA cerebellar degeneration-related protein 1 (CDR1) in the mouse brain [166].
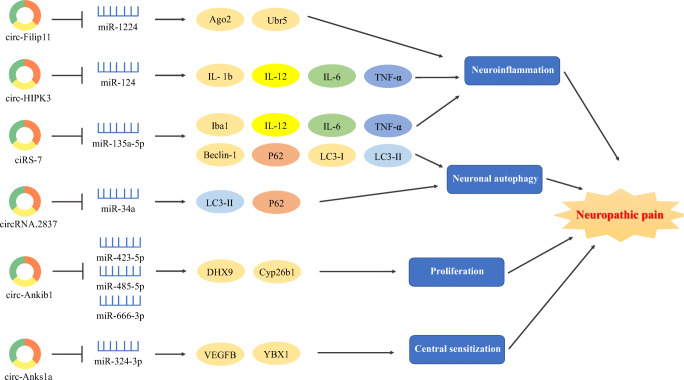
Fig. 2.
circRNA–miRNA–mRNA axis in NP. circRNAs could regulate the target genes by sponging miRNAs, leading to the emergence and development of NP. Ago2 = Argonaute-2; Ubr5 = ubiquitin protein ligase E3 component n-recognin 5; IL = interleukin; TNF-α = tumor necrosis factor-α; Ankib1 = ankyrin repeat and in-between Ring finger (IBR) domain containing 1; DHX9 = DEx/H-box helicase 9; Cyp26b1 = cytochrome P450, family 26, subfamily B, polypeptide 1
The novel mechanism of “circRNAs sponging miRNAs” is frequent in the cross-talk among circRNAs, miRNAs, and mRNAs in NP. Compared with sham SNL, circAnks1a was overexpressed in the SNL model of NP. circAnks1a regulated the VEGFB expression to reduce the excitability of the spinal cord by sponging miR-324-3p in the spinal cord of SNL rats [20]. Based on the research of Mao et al. [153], their results showed that circ-Ankib1 was verified to sponge miR-423-5p, miR-485-5p, and miR-666-3p and then regulate Schwann cell proliferation by targeting the Cyp26b1 protein in PNI. As shown in the research of Zhou et al. [152], circRNA-2837 could sponge the miR-34 family (miR-34a, miR-34b, and miR-34c) to regulate LC3-II and p62 in SNI rats and then protect neurons by decreasing neuronal autophagy. Wang and colleagues [150] found that circHIPK3 could alleviate pain by sponging miR-124, which induced inflammation by regulating IL-1b, IL-6, IL-12, and TNF-α in diabetic NP rats. Cai and colleagues [151] showed that ciRS-7 was involved in regulating inflammation and autophagy in NP progression by sponging miR-135-5p. These results demonstrated the novel mechanism of “circRNAs sponging miRNAs” in NP.
Few studies have revealed the novel mechanism of “miRNAs mediating circRNAs” in the cross-talk among circRNAs, miRNAs, and mRNAs in NP. Based on the research of Pan et al. [149], miR-1224 expression was downregulated in the spinal cord at 1, 3, and 7 days after CFA-induced chronic inflammatory pain, and the expression of spinal circRNA-Filip1l increased because of the reduction of miR-1224 expression through the binding and splicing of precursor of circRNA-Filip1l in an Argonaute-2-dependent manner. The knockdown of miR-1224 resulted in thermal hyperalgesia and mechanical allodynia in mice. They found that the ubiquitin protein ligase E3 component n-recognin 5 (Ubr5) was a target gene of circRNA-Filip1l and played vital roles in the modulation of nociception. Pan and colleagues [149] revealed that miR-1224 could mediate circRNA-Filip1l expression by regulating the Ubr5 in the spinal cord of CFA rats. Their results proved the novel mechanism of “miRNAs mediating circRNAs” for the interactions among circRNAs, miRNAs, and mRNAs in chronic pain.
Clinical Implications
Recently, it has been widely reported that aberrant expression of ncRNAs exists after NP injury, and these differentially expressed ncRNAs are considered to be potential biomarkers for the diagnosis, assessment, treatment, prediction, and prognosis of NP. For example, Heyn et al. showed that miR-124a and miR-155 were significantly increased in patients with NP compared with healthy volunteers. However, the aberrant expression of these miRNAs repressed the expression of sirtuin1 (SIRT1), a direct target gene of miR-124a and miR-155, resulting in the overexpression of anti-inflammatory differentiation of regulatory T cells, thereby limiting the development of inflammation and relieving the pain [167]. Another study demonstrated that the level of the lncRNA NONRATT021972 was upregulated in the blood of patients with diabetic neuropathic pain, which was correlated with TNF-α pathways. Furthermore, in animal experiments, they further demonstrated that lncRNA NONRATT021972 siRNA could suppress inflammation and alleviate NP by decreasing TNF-α [168]. All these studies indicate that ncRNAs may act as potential biomarkers for clinical utility in patients with NP. However, because most of the existing studies are focused on animal NP models, these data are difficult to apply to humans. Therefore, further studies need to explore more clinical trials to provide more validated evidence that ncRNAs can be used in the diagnosis, treatment, and prognosis of NP.
Conclusion and Future Prospects
Recently, studies on ncRNAs in NP have become increasingly extensive. The role of miRNAs in NP has been studied the most, but its mechanism in NP is still unclear because of its wide range of target genes and different signaling pathways. Considering the improvement of high-throughput sequencing technologies, such as microarray and RNA sequencing, a large number of lncRNAs and circRNAs have been discovered, thereby elucidating transcriptome complexity and demonstrating the dysregulation of the expression of many lncRNAs and circRNAs in NP. Therefore, these findings suggest the importance of explorations of the potential use of ncRNAs as biomarkers and potential therapeutic targets for NP. However, the annotated ncRNAs that we have discussed may be just the tip of the iceberg. More broadly, future research is also needed to determine more valuable functions of ncRNAs in the process of NP.
lncRNAs and circRNAs have been successively added to the miRNA network and can be used as “miRNA sponges” to regulate miRNA expression in NP. However, because the study was limited to the obtained clinical specimens, it was difficult to further explore the network mechanism of the lncRNA/circRNA–miRNA–mRNA axis. In future investigations, subsequent in-depth studies are required to verify clinical diagnosis and treatment protocols and to utilize the lncRNA/circRNA–miRNA–mRNA axis as a treatment target in NP.
Electronic Supplementary Material
(PDF 1224 kb)
Author Contributions
G S, Z Y, JB G, YL Z, X S, and XQ W searched the literature and wrote the manuscript.
Funding Information
This work was supported by the National Natural Science Foundation of China (81871844), the Shuguang Program supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission (18SG48), the Shanghai Municipal Commission of Health and Family Planning (201840346), and Fok Ying-Tong Education Foundation of China (161092).
Compliance with Ethical Standards
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ge Song and Zheng Yang are the co-first authors. These authors contributed equally to this work.
References
- 1.Colloca L, Ludman T, Bouhassira D, et al. Neuropathic pain. Nat Rev Dis Primers. 2017;3:17002. doi: 10.1038/nrdp.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen TS, Baron R, Haanpaa M, et al. A new definition of neuropathic pain. Pain. 2011;152(10):2204–5. doi: 10.1016/j.pain.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 3.van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155(4):654–62. doi: 10.1016/j.pain.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Scholz J, Finnerup NB, Attal N, et al. The IASP classification of chronic pain for ICD-11: chronic neuropathic pain. Pain. 2019;160(1):53–9. doi: 10.1097/j.pain.0000000000001365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang S, Zhou J, Jing H, et al. Functional roles of lncRNAs and its potential mechanisms in neuropathic pain. Clinical Epigenetics. 2019;11(1):78. doi: 10.1186/s13148-019-0671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Failde I, Duenas M, Ribera MV, et al. Prevalence of central and peripheral neuropathic pain in patients attending pain clinics in Spain: factors related to intensity of pain and quality of life. Journal of Pain Research. 2018;11:1835–47. doi: 10.2147/JPR.S159729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blyth FM. Global burden of neuropathic pain. Pain. 2018;159(3):614–7. doi: 10.1097/j.pain.0000000000001127. [DOI] [PubMed] [Google Scholar]
- 8.Liedgens H, Obradovic M, De Courcy J, Holbrook T, Jakubanis R. A burden of illness study for neuropathic pain in Europe. Clinicoecon Outcomes Res. 2016;8:113–26. doi: 10.2147/CEOR.S81396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Wei H, Piirainen S, et al. Spinal versus brain microglial and macrophage activation traits determine the differential neuroinflammatory responses and analgesic effect of minocycline in chronic neuropathic pain. Brain, behavior, and immunity. 2016;58:107–17. doi: 10.1016/j.bbi.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Tramullas M, Frances R, de la Fuente R, et al. MicroRNA-30c-5p modulates neuropathic pain in rodents. Science Translational Medicine. 2018;10(453). [DOI] [PubMed]
- 11.Guo JB, Zhu Y, Chen BL, et al. Network and pathway-based analysis of microRNA role in neuropathic pain in rat models. Journal of Cellular and Molecular Medicine. 2019;23(7):4534–44. doi: 10.1111/jcmm.14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaggi AS, Jain V, Singh N. Animal models of neuropathic pain. Fundam Clin Pharmacol. 2011;25(1):1–28. doi: 10.1111/j.1472-8206.2009.00801.x. [DOI] [PubMed] [Google Scholar]
- 13.Kumar A, Kaur H, Singh A. Neuropathic Pain models caused by damage to central or peripheral nervous system. Pharmacol Rep. 2018;70(2):206–16. doi: 10.1016/j.pharep.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Raju HB, Tsinoremas NF, Capobianco E. Emerging putative associations between non-coding RNAs and protein-coding genes in neuropathic pain: added value from reusing microarray data. Front Neurol. 2016;7:168. doi: 10.3389/fneur.2016.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Xiong Q, Chen H, Yang C, Fan Y. Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Frontiers in Molecular Neuroscience. 2017;10:91. doi: 10.3389/fnmol.2017.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. The Journal of Clinical Investigation. 2017;127(3):761–71. doi: 10.1172/JCI84424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, Wang Y, Shu S, Cai J, Tang C, Dong Z. Non-coding RNAs in kidney injury and repair. American Journal of Physiology Cell Physiology. 2019;317(2):C177–C88. doi: 10.1152/ajpcell.00048.2019. [DOI] [PubMed] [Google Scholar]
- 18.Jin H, Du XJ, Zhao Y, Xia DL. XIST/miR-544 axis induces neuropathic pain by activating STAT3 in a rat model. Journal of Cellular Physiology. 2018;233(8):5847–55. doi: 10.1002/jcp.26376. [DOI] [PubMed] [Google Scholar]
- 19.Xia LX, Ke C, Lu JM. NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. Journal of Cellular Physiology. 2018;233(9):7103–11. doi: 10.1002/jcp.26526. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SB, Lin SY, Liu M, et al. CircAnks1a in the spinal cord regulates hypersensitivity in a rodent model of neuropathic pain. Nature Communications. 2019;10(1):4119. doi: 10.1038/s41467-019-12049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Zhen S, Liu H, et al. Effects of miR-26a-5p on neuropathic pain development by targeting MAPK6 in in CCI rat models. Biomed. Pharmacother. 2018;107:644–649. doi: 10.1016/j.biopha.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5(4):316–23. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 25.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Reviews Genetics. 2011:540. [DOI] [PubMed]
- 26.Huang X, Huang M, Kong L, Li Y. miR-372 suppresses tumour proliferation and invasion by targeting IGF2BP1 in renal cell carcinoma. Cell proliferation. 2015;48(5):593–9. doi: 10.1111/cpr.12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin H, He H, Shen X, et al. miR-9-5p inhibits skeletal muscle satellite cell proliferation and differentiation by targeting IGF2BP3 through the IGF2-PI3K/Akt signaling pathway. International Journal of Molecular Sciences. 2020;21(5):1665. doi: 10.3390/ijms21051655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y, Cao DL, Jiang BC, Yang T, Gao YJ. MicroRNA-146a-5p attenuates neuropathic pain via suppressing TRAF6 signaling in the spinal cord. Brain, Behavior, and Immunity. 2015;49:119–29. doi: 10.1016/j.bbi.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 29.Yao B, Wan X, Zheng X, et al. Critical roles of microRNA-141-3p and CHD8 in hypoxia/reoxygenation-induced cardiomyocyte apoptosis. Cell & Bioscience. 2020;10:20. doi: 10.1186/s13578-020-00384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raeisi F, Mahmoudi E, Dehghani-Samani M, et al. Differential expression profile of miR-27b, miR-29a, and miR-155 in chronic lymphocytic leukemia and breast cancer patients. Molecular Therapy Oncolytics. 2020;16:230–7. doi: 10.1016/j.omto.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yvan D, Mélanie V, Emeline G, et al. Use of circulating microRNAs to diagnose acute myocardial infarction. Clinical Chemistry. 2020;3:3. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Fan J, Chen N. A novel regulator of type II diabetes: microRNA-143. Trends in Endocrinology & Metabolism. 2018;29(6):380–388. doi: 10.1016/j.tem.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Chang HL, Wang HC, Chunag YT, et al. miRNA expression change in dorsal root ganglia after peripheral nerve injury. Journal of Molecular Neuroscience. 2017;61(2):169–77. doi: 10.1007/s12031-016-0876-7. [DOI] [PubMed] [Google Scholar]
- 34.Aldrich BT, Frakes EP, Kasuya J, Hammond DL, Kitamoto T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience. 2009;164(2):711–23. doi: 10.1016/j.neuroscience.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L, Wang L. Upregulation of miR-183 represses neuropathic pain through inhibiton of MAP3K4 in CCI rat models. Journal of Cellular Physiology. 2020;235(4):3815–22. doi: 10.1002/jcp.29276. [DOI] [PubMed] [Google Scholar]
- 36.Xie X, Ma L, Xi K, Zhang W, Fan D. MicroRNA-183 suppresses neuropathic pain and expression of AMPA receptors by targeting mTOR/VEGF signaling pathway. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2017;41(1):181–92. doi: 10.1159/000455987. [DOI] [PubMed] [Google Scholar]
- 37.Shi DN, Yuan YT, Ye D, Kang LM, Wen J, Chen HP. MiR-183-5p alleviates chronic constriction injury-induced neuropathic pain through inhibition of TREK-1. Neurochemical Research. 2018;43(6):1143–9. doi: 10.1007/s11064-018-2529-4. [DOI] [PubMed] [Google Scholar]
- 38.Lin CR, Chen KH, Yang CH, Huang HW, Sheen-Chen SM. Intrathecal miR-183 delivery suppresses mechanical allodynia in mononeuropathic rats. The European Journal of Neuroscience. 2014;39(10):1682–9. doi: 10.1111/ejn.12522. [DOI] [PubMed] [Google Scholar]
- 39.Cai W, Zhao Q, Shao J, et al. MicroRNA-182 alleviates neuropathic pain by regulating Nav1.7 following spared nerve injury in rats. Scientific Reports. 2018;8(1):16750. doi: 10.1038/s41598-018-34755-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen HP, Zhou W, Kang LM, et al. Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury. Neurochemical Research. 2014;39(1):76–83. doi: 10.1007/s11064-013-1192-z. [DOI] [PubMed] [Google Scholar]
- 41.Cai W, Zhang Y, Liu Y, Liu H, Zhang Z, Su Z. Effects of miR-150 on neuropathic pain process via targeting AKT3. Biochemical and Biophysical Research Communications. 2019;517(3):532–7. doi: 10.1016/j.bbrc.2019.07.061. [DOI] [PubMed] [Google Scholar]
- 42.Brandenburger T, Johannsen L, Prassek V, et al. MiR-34a is differentially expressed in dorsal root ganglia in a rat model of chronic neuropathic pain. Neuroscience Letters. 2019;708:134365. doi: 10.1016/j.neulet.2019.134365. [DOI] [PubMed] [Google Scholar]
- 43.Zhong L, Xiao W, Wang F, Liu J, Zhi LJ. miR-21-5p inhibits neuropathic pain development via directly targeting C-C motif ligand 1 and tissue inhibitor of metalloproteinase-3. Journal of Cellular Biochemistry. 2019;120(10):16614–23. doi: 10.1002/jcb.28920. [DOI] [PubMed] [Google Scholar]
- 44.Sakai A, Suzuki H. Nerve injury-induced upregulation of miR-21 in the primary sensory neurons contributes to neuropathic pain in rats. Biochemical and Biophysical Research Communications. 2013;435(2):176–81. doi: 10.1016/j.bbrc.2013.04.089. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Wang H, Zhang T, et al. Inhibition of microRNA-195 alleviates neuropathic pain by targeting Patched1 and inhibiting SHH signaling pathway activation. Neurochemical Research. 2019;44(7):1690–702. doi: 10.1007/s11064-019-02797-2. [DOI] [PubMed] [Google Scholar]
- 46.Shi G, Shi J, Liu K, et al. Increased miR-195 aggravates neuropathic pain by inhibiting autophagy following peripheral nerve injury. Glia. 2013;61(4):504–12. doi: 10.1002/glia.22451. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Zhu B, Sun Y, Xie X. MiR-155 modulates the progression of neuropathic pain through targeting SGK3. International Journal of Clinical and Experimental Pathology. 2015;8(11):14374–82. [PMC free article] [PubMed] [Google Scholar]
- 48.Tan Y, Yang J, Xiang K, Tan Q, Guo Q. Suppression of microRNA-155 attenuates neuropathic pain by regulating SOCS1 signalling pathway. Neurochemical Research. 2015;40(3):550–60. doi: 10.1007/s11064-014-1500-2. [DOI] [PubMed] [Google Scholar]
- 49.Mo Y, Liu B, Qiu S, et al. Down-regulation of microRNA-34c-5p alleviates neuropathic pain via the SIRT1/STAT3 signaling pathway in rat models of chronic constriction injury of sciatic nerve. J Neurochem. 2020;00:1–15. doi: 10.1111/jnc.14998. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Cui X, Guan G, Dong Y, Zhang Z. microRNA-192-5p is involved in nerve repair in rats with peripheral nerve injury by regulating XIAP. Cell Cycle. 2020;19(3):326–38. doi: 10.1080/15384101.2019.1710916. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Li T, Wan Y, Sun L, et al. Inhibition of microRNA-15a/16 expression alleviates neuropathic pain development through upregulation of G protein-coupled receptor kinase 2. Biomolecules & Therapeutics. 2019;27(4):414–22. doi: 10.4062/biomolther.2018.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang W, Wang Q, Yu X, Lu T, Zhang P. MicroRNA-217 relieved neuropathic pain through targeting toll-like receptor 5 expression. Journal of Cellular Biochemistry. 2019;120(3):3009–17. doi: 10.1002/jcb.27269. [DOI] [PubMed] [Google Scholar]
- 53.Yan T, Zhang F, Sun C, et al. miR-32-5p-mediated Dusp5 downregulation contributes to neuropathic pain. Biochemical and Biophysical Research Communications. 2018;495(1):506–11. doi: 10.1016/j.bbrc.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Zhao G. Downregulation of microRNA-218 relieves neuropathic pain by regulating suppressor of cytokine signaling 3. International Journal of Molecular Medicine. 2016;37(3):851–8. doi: 10.3892/ijmm.2016.2455. [DOI] [PubMed] [Google Scholar]
- 55.Leinders M, Uceyler N, Pritchard RA, Sommer C, Sorkin LS. Increased miR-132-3p expression is associated with chronic neuropathic pain. Experimental Neurology. 2016;283(Pt A):276–86. doi: 10.1016/j.expneurol.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xia L, Zhang Y, Dong T. Inhibition of microRNA-221 alleviates neuropathic pain through targeting suppressor of cytokine signaling 1. Journal of Molecular Neuroscience. 2016;59(3):411–20. doi: 10.1007/s12031-016-0748-1. [DOI] [PubMed] [Google Scholar]
- 57.Wang C, Jiang Q, Wang M, Li D. MiR-19a targets suppressor of cytokine signaling 1 to modulate the progression of neuropathic pain. International Journal of Clinical and Experimental Pathology. 2015;8(9):10901–7. [PMC free article] [PubMed] [Google Scholar]
- 58.Ji LJ, Shi J, Lu JM, Huang QM. MiR-150 alleviates neuropathic pain via inhibiting toll-like receptor 5. Journal of Cellular Biochemistry. 2018;119(1):1017–26. doi: 10.1002/jcb.26269. [DOI] [PubMed] [Google Scholar]
- 59.Yang FR, Chen J, Yi H, Peng LY, Hu XL, Guo QL. MicroRNA-7a ameliorates neuropathic pain in a rat model of spinal nerve ligation via the neurofilament light polypeptide-dependent signal transducer and activator of transcription signaling pathway. Molecular Pain. 2019;15:1744806919842464. doi: 10.1177/1744806919842464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H. miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain: a Journal of Neurology. 2013;136(Pt 9):2738–50. doi: 10.1093/brain/awt191. [DOI] [PubMed] [Google Scholar]
- 61.Wen J, He T, Qi F, Chen H. MiR-206-3p alleviates chronic constriction injury-induced neuropathic pain through targeting HDAC4. Experimental Animals. 2019;68(2):213–20. doi: 10.1538/expanim.18-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun W, Zhang L, Li R. Overexpression of miR-206 ameliorates chronic constriction injury-induced neuropathic pain in rats via the MEK/ERK pathway by targeting brain-derived neurotrophic factor. Neuroscience Letters. 2017;646:68–74. doi: 10.1016/j.neulet.2016.12.047. [DOI] [PubMed] [Google Scholar]
- 63.Su S, Shao J, Zhao Q, et al. MiR-30b attenuates neuropathic pain by regulating voltage-gated sodium channel Nav1.3 in rats. Frontiers in Molecular Neuroscience. 2017;10:126. doi: 10.3389/fnmol.2017.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao J, Cao J, Wang J, et al. MicroRNA-30b regulates expression of the sodium channel Nav1.7 in nerve injury-induced neuropathic pain in the rat. Molecular Pain. 2016;12. [DOI] [PMC free article] [PubMed]
- 65.Zhang X, Chen Q, Shen J, Wang L, Cai Y, Zhu KR. miR-194 relieve neuropathic pain and prevent neuroinflammation via targeting FOXA1. Journal of Cellular Biochemistry. 2020;121(5-6):3278–85. doi: 10.1002/jcb.29598. [DOI] [PubMed] [Google Scholar]
- 66.Ye G, Zhang Y, Zhao J, et al. miR-384-5p ameliorates neuropathic pain by targeting SCN3A in a rat model of chronic constriction injury. Neurol Res. 2020;42(4):299–307. doi: 10.1080/01616412.2020.1723313. [DOI] [PubMed] [Google Scholar]
- 67.Cai L, Liu X, Guo Q, Huang Q, Zhang Q, Cao Z. MiR-15a attenuates peripheral nerve injury-induced neuropathic pain by targeting AKT3 to regulate autophagy. Genes & Genomics. 2020;42(1):77–85. doi: 10.1007/s13258-019-00881-z. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, Guo H, Xie A, Liao O, Ju F, Zhou Y. MicroRNA-144 relieves chronic constriction injury-induced neuropathic pain via targeting RASA1. Biotechnol Appl Biochem. 2019;18. [DOI] [PubMed]
- 69.Zhu B, Gao J, Ouyang Y, Hu Z, Chen X. Overexpression of miR138 ameliorates spared sciatic nerve injury-induced neuropathic pain through the anti-inflammatory response in mice. Journal of Pain Research. 2019;12:3135–45. doi: 10.2147/JPR.S219462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao L, Pu X, Huang Y, Huang J. MicroRNA-340-5p relieved chronic constriction injury-induced neuropathic pain by targeting Rap1A in rat model. Genes & Genomics. 2019;41(6):713–21. doi: 10.1007/s13258-019-00802-0. [DOI] [PubMed] [Google Scholar]
- 71.Fang B, Wei L, Dong K, Niu X, Sui X, Zhang H. miR-202 modulates the progression of neuropathic pain through targeting RAP1A. Journal of Cellular Biochemistry. 2019;120(3):2973–82. doi: 10.1002/jcb.27025. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, Su Z, An LJ, et al. miR-98 acts as an inhibitor in chronic constriction injury-induced neuropathic pain via downregulation of high-mobility group AT-hook 2. Journal of Cellular Biochemistry. 2019;120(6):10363–9. doi: 10.1002/jcb.28320. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y, Liu HL, An LJ, et al. miR-124-3p attenuates neuropathic pain induced by chronic sciatic nerve injury in rats via targeting EZH2. Journal of Cellular Biochemistry. 2019;120(4):5747–55. doi: 10.1002/jcb.27861. [DOI] [PubMed] [Google Scholar]
- 74.You H, Zhang L, Chen Z, Liu W, Wang H, He H. MiR-20b-5p relieves neuropathic pain by targeting Akt3 in a chronic constriction injury rat model. Synapse (New York, NY) 2019;73(12):e22125. doi: 10.1002/syn.22125. [DOI] [PubMed] [Google Scholar]
- 75.Xu L, Wang Q, Jiang W, Yu S, Zhang S. MiR-34c ameliorates neuropathic pain by targeting NLRP3 in a mouse model of chronic constriction injury. Neuroscience. 2019;399:125–34. doi: 10.1016/j.neuroscience.2018.12.030. [DOI] [PubMed] [Google Scholar]
- 76.Wang T, Li B, Yuan X, et al. MiR-20a plays a key regulatory role in the repair of spinal cord dorsal column lesion via PDZ-RhoGEF/RhoA/GAP43 axis in rat. Cellular and Molecular Neurobiology. 2019;39(1):87–98. doi: 10.1007/s10571-018-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tozaki-Saitoh H, Masuda J, Kawada R, et al. Transcription factor MafB contributes to the activation of spinal microglia underlying neuropathic pain development. Glia. 2019;67(4):729–40. doi: 10.1002/glia.23570. [DOI] [PubMed] [Google Scholar]
- 78.Tian J, Song T, Wang W, Wang H, Zhang Z. miR-129-5p alleviates neuropathic pain through regulating HMGB1 expression in CCI rat models. Journal of Molecular Neuroscience : MN. 2020;70(1):84–93. doi: 10.1007/s12031-019-01403-y. [DOI] [PubMed] [Google Scholar]
- 79.Liu L, Xu D, Wang T, et al. Epigenetic reduction of miR-214-3p upregulates astrocytic colony-stimulating factor-1 and contributes to neuropathic pain induced by nerve injury. Pain. 2020;161(1):96–108. doi: 10.1097/j.pain.0000000000001681. [DOI] [PubMed] [Google Scholar]
- 80.Hu Y, Liu Q, Zhang M, Yan Y, Yu H, Ge L. MicroRNA-362-3p attenuates motor deficit following spinal cord injury via targeting paired box gene 2. Journal of Integrative Neuroscience. 2019;18(1):57–64. doi: 10.31083/j.jin.2019.01.12. [DOI] [PubMed] [Google Scholar]
- 81.Bao Y, Wang S, Xie Y, Jin K, Bai Y, Shan S. MiR-28-5p relieves neuropathic pain by targeting Zeb1 in CCI rat models. Journal of Cellular Biochemistry. 2018;119(10):8555–63. doi: 10.1002/jcb.27096. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Su Z, Liu HL, et al. Effects of miR-26a-5p on neuropathic pain development by targeting MAPK6 in in CCI rat models. Biomedicine & Pharmacotherapy = Biomedecine & pharmacotherapie. 2018;107:644–9. doi: 10.1016/j.biopha.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 83.Yan XT, Ji LJ, Wang Z, et al. MicroRNA-93 alleviates neuropathic pain through targeting signal transducer and activator of transcription 3. International Immunopharmacology. 2017;46:156–62. doi: 10.1016/j.intimp.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 84.Ding M, Shen W, Hu Y. The role of miR-539 in the anterior cingulate cortex in chronic neuropathic pain. Pain Medicine (Malden, Mass) 2017;18(12):2433–42. doi: 10.1093/pm/pnx004. [DOI] [PubMed] [Google Scholar]
- 85.Lu S, Ma S, Wang Y, Huang T, Zhu Z, Zhao G. Mus musculus-microRNA-449a ameliorates neuropathic pain by decreasing the level of KCNMA1 and TRPA1, and increasing the level of TPTE. Molecular Medicine Reports. 2017;16(1):353–60. doi: 10.3892/mmr.2017.6559. [DOI] [PubMed] [Google Scholar]
- 86.Zhang Y, Mou J, Cao L, Zhen S, Huang H, Bao H. MicroRNA-142-3p relieves neuropathic pain by targeting high mobility group box 1. International Journal of Molecular Medicine. 2018;41(1):501–10. doi: 10.3892/ijmm.2017.3222. [DOI] [PubMed] [Google Scholar]
- 87.Yan XT, Zhao Y, Cheng XL, et al. Inhibition of miR-200b/miR-429 contributes to neuropathic pain development through targeting zinc finger E box binding protein-1. Journal of Cellular Physiology. 2018;233(6):4815–24. doi: 10.1002/jcp.26284. [DOI] [PubMed] [Google Scholar]
- 88.Xu B, Cao J, Zhang J, et al. Role of microRNA-143 in nerve injury-induced upregulation of Dnmt3a expression in primary sensory neurons. Frontiers in Molecular Neuroscience. 2017;10:350. doi: 10.3389/fnmol.2017.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pang X, Tang Y, Zhang D. Role of miR-145 in chronic constriction injury in rats. Experimental and Therapeutic Medicine. 2016;12(6):4121–7. doi: 10.3892/etm.2016.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang J, Zhang H, Zi T. Overexpression of microRNA-141 relieves chronic constriction injury-induced neuropathic pain via targeting high-mobility group box 1. International Journal of Molecular Medicine. 2015;36(5):1433–9. doi: 10.3892/ijmm.2015.2342. [DOI] [PubMed] [Google Scholar]
- 91.Neumann E, Hermanns H, Barthel F, Werdehausen R, Brandenburger T. Expression changes of microRNA-1 and its targets Connexin 43 and brain-derived neurotrophic factor in the peripheral nervous system of chronic neuropathic rats. Molecular Pain. 2015;11:39. doi: 10.1186/s12990-015-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Huang Y, Ma C, Yu X, Zhang Z, Shen L. MiR-203 involves in neuropathic pain development and represses Rap1a expression in nerve growth factor differentiated neuronal PC12 cells. The Clinical Journal of Pain. 2015;31(1):36–43. doi: 10.1097/AJP.0000000000000070. [DOI] [PubMed] [Google Scholar]
- 93.Favereaux A, Thoumine O, Bouali-Benazzouz R, et al. Bidirectional integrative regulation of Cav1.2 calcium channel by microRNA miR-103: role in pain. The EMBO Journal. 2011;30(18):3830–41. doi: 10.1038/emboj.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pachnis V, Belayew A, Tilghman SM. Locus unlinked to alpha-fetoprotein under the control of the murine raf and Rif genes. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(17):5523–7. doi: 10.1073/pnas.81.17.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10(1):28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Z, Li X, Chen X, et al. Emerging roles of long non-coding RNAs in neuropathic pain. Cell Proliferation. 2019;52(1):e12528. doi: 10.1111/cpr.12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Z, Li X, Chen C, et al. Long non-coding RNAs in nucleus pulposus cell function and intervertebral disc degeneration. Cell Proliferation. 2018;51(5):e12483. doi: 10.1111/cpr.12483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang SD, Lu J, Deng ZH, Li YS, Lei GH. Long noncoding RNAs in osteoarthritis. Joint, Bone, Spine : revue du rhumatisme. 2017;84(5):553–6. doi: 10.1016/j.jbspin.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 99.Jiang SD, Lu J, Deng ZH, Li YS, Lei GH. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Proliferation. 2016;49(4):471–5. doi: 10.1111/cpr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou J, Fan Y, Chen H. Analyses of long non-coding RNA and mRNA profiles in the spinal cord of rats using RNA sequencing during the progression of neuropathic pain in an SNI model. RNA Biology. 2017;1:1. doi: 10.1080/15476286.2017.1371400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Du H, Liu Z, Tan X, Ma Y, Gong Q. Identification of the genome-wide expression patterns of long non-coding RNAs and mRNAs in mice with streptozotocin-induced diabetic neuropathic pain. Neuroscience. 2019;402:90–103. doi: 10.1016/j.neuroscience.2018.12.040. [DOI] [PubMed] [Google Scholar]
- 102.Yan XT, Lu JM, Wang Y, et al. XIST accelerates neuropathic pain progression through regulation of miR-150 and ZEB1 in CCI rat models. Journal of Cellular Physiology. 2018;233(8):6098–106. doi: 10.1002/jcp.26453. [DOI] [PubMed] [Google Scholar]
- 103.Zhao Y, Li S, Xia N, Shi Y, Zhao CM. Effects of XIST/miR-137 axis on neuropathic pain by targeting TNFAIP1 in a rat model. Journal of Cellular Physiology. 2018;233(5):4307–16. doi: 10.1002/jcp.26254. [DOI] [PubMed] [Google Scholar]
- 104.Wei M, Li L, Zhang Y, Zhang ZJ, Liu HL, Bao HG. LncRNA X inactive specific transcript contributes to neuropathic pain development by sponging miR-154-5p via inducing toll-like receptor 5 in CCI rat models. Journal of Cellular Biochemistry. 2018. [DOI] [PubMed]
- 105.Gu S, Xie R, Liu X, Shou J, Gu W, Che X. Long coding RNA XIST contributes to neuronal apoptosis through the downregulation of AKT phosphorylation and is negatively regulated by miR-494 in rat spinal cord injury. International Journal of Molecular Sciences. 2017;18(4). [DOI] [PMC free article] [PubMed] [Retracted]
- 106.Sun W, Ma M, Yu H, Yu H. Inhibition of lncRNA X inactivate-specific transcript ameliorates inflammatory pain by suppressing satellite glial cell activation and inflammation by acting as a sponge of miR-146a to inhibit Nav 1.7. Journal of Cellular Biochemistry. 2018;119(12):9888–98. doi: 10.1002/jcb.27310. [DOI] [PubMed] [Google Scholar]
- 107.Wu J, Wang C, Ding H. LncRNA MALAT1 promotes neuropathic pain progression through the miR1545p/AQP9 axis in CCI rat models. Molecular Medicine Reports. 2020;21(1):291–303. doi: 10.3892/mmr.2019.10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu G, Li X, Li M, Zhang Z. Long non-coding RNA MALAT1 promotes the proliferation and migration of Schwann cells by elevating BDNF through sponging miR-129-5p. Experimental Cell Research. 2020;390(1):111937. doi: 10.1016/j.yexcr.2020.111937. [DOI] [PubMed] [Google Scholar]
- 109.Ma X, Wang H, Song T, Wang W, Zhang Z. lncRNA MALAT1 contributes to neuropathic pain development through regulating miR-129-5p/HMGB1 axis in a rat model of chronic constriction injury. Int J Neurosci. 2020:1-10. [DOI] [PubMed]
- 110.Chen ZL, Liu JY, Wang F, Jing X. Suppression of MALAT1 ameliorates chronic constriction injury-induced neuropathic pain in rats via modulating miR-206 and ZEB2. Journal of Cellular Physiology. 2019:1-7. [DOI] [PubMed]
- 111.Xiong W, Tan M, Tong Z, et al. Effects of long non-coding RNA uc.48+ on pain transmission in trigeminal neuralgia. Brain Research Bulletin. 2019;147:92–100. doi: 10.1016/j.brainresbull.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 112.Wang S, Xu H, Zou L, et al. LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signalling. 2016;12(1):139–48. doi: 10.1007/s11302-015-9488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu B, Zhang C, Zou L, et al. LncRNA uc.48+ siRNA improved diabetic sympathetic neuropathy in type 2 diabetic rats mediated by P2X7 receptor in SCG. Auton Neurosci. 2016;197:14–8. doi: 10.1016/j.autneu.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 114.Liu CL, Deng ZY, Du ER, Xu CS. Long noncoding RNA BC168687 small interfering RNA reduces high glucose and high free fatty acidinduced expression of P2X7 receptors in satellite glial cells. Molecular Medicine Reports. 2018;17(4):5851–9. doi: 10.3892/mmr.2018.8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu C, Li C, Deng Z, Du E, Xu C. Long non-coding RNA BC168687 is involved in TRPV1-mediated diabetic neuropathic pain in rats. Neuroscience. 2018;374:214–22. doi: 10.1016/j.neuroscience.2018.01.049. [DOI] [PubMed] [Google Scholar]
- 116.Liu C, Tao J, Wu H, et al. Effects of lncRNA BC168687 siRNA on diabetic neuropathic pain mediated by P2X7 receptor on SGCs in DRG of rats. BioMed Research International. 2017;2017:7831251. doi: 10.1155/2017/7831251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peng H, Zou L, Xie J, et al. lncRNA NONRATT021972 siRNA decreases diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root ganglia. Molecular Neurobiology. 2017;54(1):511–23. doi: 10.1007/s12035-015-9632-1. [DOI] [PubMed] [Google Scholar]
- 118.Xu H, He L, Liu C, et al. LncRNA NONRATT021972 siRNA attenuates P2X7 receptor expression and inflammatory cytokine production induced by combined high glucose and free fatty acids in PC12 cells. Purinergic Signalling. 2016;12(2):259–68. doi: 10.1007/s11302-016-9500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu S, Zou L, Xie J, et al. LncRNA NONRATT021972 siRNA regulates neuropathic pain behaviors in type 2 diabetic rats through the P2X7 receptor in dorsal root ganglia. Molecular Brain. 2016;9:44. doi: 10.1186/s13041-016-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yao C, Chen Y, Wang J, et al. LncRNA BC088259 promotes Schwann cell migration through vimentin following peripheral nerve injury. Glia. 2020;68(3):670–9. doi: 10.1002/glia.23749. [DOI] [PubMed] [Google Scholar]
- 121.Shen F, Zheng H, Zhou L, Li W, Zhang Y, Xu X. LINC00657 expedites neuropathic pain development by modulating miR-136/ZEB1 axis in a rat model. Journal of Cellular Biochemistry. 2019;120(1):1000–10. doi: 10.1002/jcb.27466. [DOI] [PubMed] [Google Scholar]
- 122.Zhang D, Mou JY, Wang F, Liu J, Hu X. CRNDE enhances neuropathic pain via modulating miR-136/IL6R axis in CCI rat models. Journal of Cellular Physiology. 2019;234(12):22234–41. doi: 10.1002/jcp.28790. [DOI] [PubMed] [Google Scholar]
- 123.Zhang C, Peng Y, Wang Y, Xu H, Zhou X. Transcribed ultraconserved noncoding RNA uc.153 is a new player in neuropathic pain [in press]. Pain. 2020. [DOI] [PubMed]
- 124.Wang L, Zhu K, Yang B, Cai Y. Knockdown of Linc00052 alleviated spinal nerve ligation-triggered neuropathic pain through regulating miR-448 and JAK1 [in press]. Journal of Cellular Physiology. 2020. [DOI] [PubMed]
- 125.Pang H, Ren Y, Li H, Chen C, Zheng X. LncRNAs linc00311 and AK141205 are identified as new regulators in STAT3-mediated neuropathic pain in bCCI rats. European Journal of Pharmacology. 2020;868:172880. doi: 10.1016/j.ejphar.2019.172880. [DOI] [PubMed] [Google Scholar]
- 126.Chen M, Yang Y, Zhang W, et al. Long noncoding RNA SNHG5 knockdown alleviates neuropathic pain by targeting the miR-154-5p/CXCL13 axis. Neurochemical Research. 2020. [DOI] [PubMed]
- 127.Iwasaki H, Sakai A, Maruyama M, Ito T, Sakamoto A, Suzuki H. Increased H19 long non-coding RNA expression in schwann cells in peripheral neuropathic pain. Journal of Nippon Medical School = Nippon Ika Daigaku zasshi. 2019;86(4):215–21. doi: 10.1272/jnms.JNMS.2018_86-402. [DOI] [PubMed] [Google Scholar]
- 128.Wen J, Yang Y, Wu S, et al. Long noncoding RNA H19 in the injured dorsal root ganglion contributes to peripheral nerve injury-induced pain hypersensitivity. Transl Perioper Pain Med. 2020;7(2):176–84. [PMC free article] [PubMed] [Google Scholar]
- 129.Hu JZ, Rong ZJ, Li M, et al. Silencing of lncRNA PKIA-AS1 attenuates spinal nerve ligation-induced neuropathic pain through epigenetic downregulation of CDK6 expression. Frontiers in Cellular Neuroscience. 2019;13:50. doi: 10.3389/fncel.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang H, Wu J, Zhang X, Ding L, Zeng Q. Microarray analysis of the expression profile of lncRNAs reveals the key role of lncRNA BC088327 as an agonist to heregulin1betainduced cell proliferation in peripheral nerve injury. International Journal of Molecular Medicine. 2018;41(6):3477–84. doi: 10.3892/ijmm.2018.3571. [DOI] [PubMed] [Google Scholar]
- 131.Li G, Jiang H, Zheng C, et al. Long noncoding RNA MRAK009713 is a novel regulator of neuropathic pain in rats. Pain. 2017;158(10):2042–52. doi: 10.1097/j.pain.0000000000001013. [DOI] [PubMed] [Google Scholar]
- 132.Liang L, Gu X, Zhao JY, et al. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Scientific Reports. 2016;6:37704. doi: 10.1038/srep37704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao X, Tang Z, Zhang H, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nature Neuroscience. 2013;16(8):1024–31. doi: 10.1038/nn.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Peng C, Zhang C, Su Z, Lin D. DGCR5 attenuates neuropathic pain through sponging miR-330-3p and regulating PDCD4 in CCI rat models. Journal of Cellular Physiology. 2019;234(5):7292–300. doi: 10.1002/jcp.27487. [DOI] [PubMed] [Google Scholar]
- 135.Yao C, Wang Y, Zhang H, et al. lncRNA TNXA-PS1 modulates Schwann cells by functioning as a competing endogenous RNA following nerve injury. The Journal of Neuroscience : the official journal of the Society for Neuroscience. 2018;38(29):6574–85. doi: 10.1523/JNEUROSCI.3790-16.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dou L, Lin H, Wang K, et al. Long non-coding RNA CCAT1 modulates neuropathic pain progression through sponging miR-155. Oncotarget. 2017;8(52):89949–57. doi: 10.18632/oncotarget.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yao C, Wang J, Zhang H, et al. Long non-coding RNA uc.217 regulates neurite outgrowth in dorsal root ganglion neurons following peripheral nerve injury. The European Journal of Neuroscience. 2015;42(1):1718–25. doi: 10.1111/ejn.12966. [DOI] [PubMed] [Google Scholar]
- 138.Yu B, Zhou S, Hu W, et al. Altered long noncoding RNA expressions in dorsal root ganglion after rat sciatic nerve injury. Neuroscience Letters. 2013;534:117–22. doi: 10.1016/j.neulet.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 139.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73(11):3852–6. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. The EMBO Journal. 2019;38(16):e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Molecular Cell. 2018;71(3):428–42. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 142.Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cheng X, Zhang L, Zhang K, et al. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Annals of the Rheumatic Diseases. 2018;77(5):770–9. doi: 10.1136/annrheumdis-2017-212056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–81. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xie L, Mao M, Xiong K, Jiang B. Circular RNAs: a novel player in development and disease of the central nervous system. Frontiers in Cellular Neuroscience. 2017;11:354. doi: 10.3389/fncel.2017.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen X, Yang T, Wang W, et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9(2):588–607. doi: 10.7150/thno.29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Molecular Therapy Nucleic Acids. 2019;16:118–29. doi: 10.1016/j.omtn.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cao S, Deng W, Li Y, et al. Chronic constriction injury of sciatic nerve changes circular RNA expression in rat spinal dorsal horn. Journal of Pain Research. 2017;10:1687–96. doi: 10.2147/JPR.S139592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pan Z, Li GF, Sun ML, et al. MicroRNA-1224 splicing circular RNA-Filip1l in an Ago2-dependent manner regulates chronic inflammatory pain via targeting Ubr5. The Journal of Neuroscience : the official journal of the Society for Neuroscience. 2019;39(11):2125–43. doi: 10.1523/JNEUROSCI.1631-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang L, Luo T, Bao Z, Li Y, Bu W. Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic rats. Biochemical and Biophysical Research Communications. 2018;505(3):644–50. doi: 10.1016/j.bbrc.2018.09.158. [DOI] [PubMed] [Google Scholar]
- 151.Cai W, Zhang Y, Su Z. ciRS-7 targeting miR-135a-5p promotes neuropathic pain in CCI rats via inflammation and autophagy. Gene. 2020;736:144386. doi: 10.1016/j.gene.2020.144386. [DOI] [PubMed] [Google Scholar]
- 152.Zhou ZB, Niu YL, Huang GX, Lu JJ, Chen A, Zhu L. Silencing of circRNA.2837 plays a protective role in sciatic nerve injury by sponging the miR-34 family via regulating neuronal autophagy. Molecular Therapy Nucleic Acids. 2018;12:718–29. doi: 10.1016/j.omtn.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mao S, Zhang S, Zhou S, et al. A Schwann cell-enriched circular RNA circ-Ankib1 regulates Schwann cell proliferation following peripheral nerve injury. FASEB Journal : official publication of the Federation of American Societies for Experimental Biology. 2019:fj201900965R. [DOI] [PMC free article] [PubMed]
- 154.Deng K, Wang H, Guo X, Xia J. The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta biochimica et biophysica Sinica. 2016;48(2):111–6. doi: 10.1093/abbs/gmv120. [DOI] [PubMed] [Google Scholar]
- 155.Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. Journal of Cellular and Molecular Medicine. 2018;22(12):5768–75. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cheng L, Nan C, Kang L, et al. Whole blood transcriptomic investigation identifies long non-coding RNAs as regulators in sepsis. Journal of Translational Medicine. 2020;18(1):217. doi: 10.1186/s12967-020-02372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cao F, Wang Z, Feng Y, et al. lncRNA TPTEP1 competitively sponges miR-328-5p to inhibit the proliferation of non-small cell lung cancer cells. Oncology Reports. 2020;43(5):1606–18. doi: 10.3892/or.2020.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Franklin JL, Rankin CR, Levy S, et al. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochemical and Biophysical Research Communications. 2013;440(1):99–104. doi: 10.1016/j.bbrc.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019. [DOI] [PubMed]
- 162.Kleaveland B, Shi CY, Stefano J, Bartel DP. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174(2):350–62. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 164.Song X, Liang Y, Sang Y, et al. circHMCU promotes proliferation and metastasis of breast cancer by sponging the let-7 family. Molecular Therapy Nucleic Acids. 2020;20:518–33. doi: 10.1016/j.omtn.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fu X, Zhang J, He X, et al. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J Cancer. 2020;11(11):3318–26. doi: 10.7150/jca.36500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. The EMBO Journal. 2011;30(21):4414–22. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Heyn J, Luchting B, Hinske LC, Hübner M, Azad SC, Kreth S. miR-124a and miR-155 enhance differentiation of regulatory T cells in patients with neuropathic pain. Journal of Neuroinflammation. 2016;13(1):248. doi: 10.1186/s12974-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yu W, Zhao GQ, Cao RJ, Zhu ZH, Li K. LncRNA NONRATT021972 was associated with neuropathic pain scoring in patients with type 2 diabetes. Behavioural Neurology. 2017;2017:2941297. doi: 10.1155/2017/2941297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)