Abstract
DNA damage-inducible transcript 4 (DDIT4) is known to participate in various cancers, including glioblastoma multiforme (GBM). However, contradictory roles of DDIT4 exist in inducing cell death and possessing anti-apoptotic functions against cancer progression. Herein, we investigated DDIT4 signaling in GBM and temozolomide (TMZ) drug resistance. We identified that TMZ induced DDIT4 upregulation, leading to desensitization against TMZ cytotoxicity in GBM cells. Higher DDIT4 levels were found in glioma cells and mesenchymal-type GBM patients, and these higher levels were positively correlated with mesenchymal markers. Furthermore, patients with lower DDIT4 levels, especially O-6-methylguanine-DNA methyltransferase (MGMT)-methylated patients, exhibited better TMZ therapeutic efficacy. We determined that higher levels of 5 DDIT4-associated downstream genes, including SLC2A3 (also known as glucose transporter 3 (GLUT3)), can be used to predict a poor prognosis. Among these 5 genes, only GLUT3 was upregulated in both TMZ-treated and DDIT4-overexpressing cells. DDIT4-mediated GLUT3 expression was also identified, and its expression decreased TMZ’s cytotoxicity. A significant correlation existed between DDIT4 and GLUT3. DDIT4 signaling was found to be involved in both glycolytic and autophagic pathways. However, GLUT3 only participated in the exhibition of DDIT4-mediated stemness, resulting from glycolytic regulation, but not in DDIT4-mediated autophagic signaling. Finally, we identified TMZ-upregulated activating transcription factor 4 (ATF4) as an upstream regulator of DDIT4-mediated GLUT3/stemness signaling and autophagy. Consequently, ATF4/DDIT4 signaling was connected to both autophagy and GLUT3-regulated stemness, which are involved in TMZ drug resistance and the poor prognoses of GBM patients. Targeting DDIT4/GLUT3 signaling might be a new direction for glioma therapy.
Electronic supplementary material
The online version of this article (10.1007/s13311-019-00826-0) contains supplementary material, which is available to authorized users.
Key Words: DNA damage-inducible transcript 4 (DDIT4), temozolomide (TMZ), SLC2A3 (GLUT3), autophagy, stemness, activating transcription factor 4 (ATF4)
Introduction
DNA damage-inducible transcript 4 (DDIT4) [1–3], also known as DNA damage response 1 (REDD1), is upregulated by different stressful conditions such as endoplasmic reticular (ER) stress [4], starvation [5], and hypoxia [6]. DDIT4 stabilizes the formation of tuberous sclerosis complex 1 (TSC1) and 2 (TSC2) to inhibit activation of the mammalian target of rapamycin (mTOR) complex (mTORC)-1, leading to cell death [7]. It seems that DDIT4 may have a tumor-suppressive role in cancer progression. In contrast, several studies suggested that DDIT4 possesses an anti-apoptotic function in several cancer types. Suppression of DDIT4 sensitizes cancer cells to chemotherapeutic drugs. For example, in gastric cancer cells, depleting DDIT4 enhanced 5-fluorouracil-induced apoptosis and cell-cycle arrest [8]. Silencing DDIT4 sensitized bladder urothelial carcinoma to paclitaxel through inhibiting autophagy [9]. Nevertheless, the roles of DDIT4 in glioma and chemoresistance are far less understood.
Glioblastoma multiforme (GBM), a grade IV glioma, is the most malignant type of brain tumor. More than 90% of GBM patients have a primary glioma which undergoes a de novo oncogenesis program [10]. Surgical resection is the first step in GBM therapy. However, the invasive nature and extensive infiltrating growth of glioma cells make it difficult to completely remove this malignant tumor. Following surgical resection, chemotherapy and radiation therapy are usually required [11]. Despite these multidisciplinary treatments, the median survival time of GBM patients is only ~ 12 to 15 months [11].
Temozolomide (TMZ), an alkylating agent of the imidazotetrazine series, is the first-line clinical chemotherapeutic drug for GBM patients [12]. Due to TMZ’s limited efficacy, GBM patients undergoing TMZ therapy still have poor prognoses. Therefore, investigating the molecular mechanisms associated with desensitization of the therapeutic efficacy of TMZ is a topic of urgent priority. Previous studies reported that TMZ-enhanced autophagy counteracts its own cytotoxicity [13]. Suppression of autophagy by chloroquine (CQ), an autophagy inhibitor, increased the efficacy of TMZ in randomized studies [14, 15]. In addition, cancer stem cells (CSCs) are a subpopulation of cancer cells that possesses multipotency and self-renewal properties. Other studies reported that CSCs are activated against therapeutic effects after chemotherapy [16] and are involved in the formation of resistance to chemotherapy and radiotherapy [17]. However, it is still unclear whether a key modulator upon TMZ treatment connects autophagy and cancer stemness in chemoresistance.
In our previous study, we identified TMZ-regulated transcriptome profiles in glioma cells [18]. Among the top 10 upregulated genes induced by TMZ, we found that DDIT4 expression was significantly enhanced. Moreover, using a meta-analysis of DDIT4 expression in multiple cohorts of glioma patients, DDIT4 was considered a poor prognostic indicator [3]. However, little is known about DDIT4’s role in gliomas. Previous study suggests that DDIT4 induction confers protection from radiotherapy and TMZ cytotoxicity in GBM through inhibition of mTORC1 complex [19]. This is the first study that indicates the role of DDIT4 in TMZ resistance of GBM. In the present study, to comprehensively investigate the role of DDIT4 in gliomas, we utilized transcriptome profiles from 3 public databases, including The Cancer Genome Atlas (TCGA), GSE13041, and GSE16011, to analyze downstream gene candidates and signaling pathways which were highly associated with DDIT4 expression. Further, the association between DDIT4 and TMZ efficacy was also evaluated using TCGA data. Finally, in vitro experiments were conducted to identify the functions and molecular mechanism of DDIT4 involved in TMZ-mediated cytotoxicity in GBM.
Materials and Methods
Chemicals and Reagents
We purchased various human glioblastoma cell lines, including U87-MG, Hs683, M059k, A172, and T98G cells from the Bioresource Collection and Research Center (Hsinchu City, Taiwan) and American Type Culture Collection (ATCC; Rockville, MD). Reagents for culturing cells were purchased from GIBCO-BRL (Grand Island, NY). Temozolomide (TMZ; cat. no. T2577) and chloroquine (CQ; cat. no. C6628) were purchased from Sigma-Aldrich (St. Louis, MO). Trizol reagent (cat. no. 15596026), Lipofectamine 3000 (cat. no. L3000015), secondary antibodies (cat. no. A16110), the MultiScribe reverse transcriptase kit (cat. no. 4311235), and SYBR Green PCR master mix (cat. no. 4309155) were purchased from Invitrogen (Carlsbad, CA). Primer sets were synthesized by Genomics BioSci & Tech (Xizhi, New Taipei City, Taiwan) and listed in Supplementary Table 1. The anti-glucose transporter 3 (GLUT3; cat. no. sc-74,399) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-DDIT4 (cat. no. GTX64553), activating transcription factor 4 (ATF4; cat. no. GTX101943), light chain 3B (LC3B; cat. no. GTX127375), p62 (cat. no. GTX100685), and β-actin (cat. no. GTX109639) were purchased from GeneTex (Hsinchu City, Taiwan). Caspase-3/CPP32 colorimetric assay kit (cat. no. k106) was purchased from BioVision, Inc. (Milpitas, CA). Scrambled short hairpin (sh) RNA, DDIT4 shRNA, and GLUT3 shRNA were purchased from the National RNAi Core Facility (Nankang, Taipei, Taiwan). Sequences of shRNAs are listed in Supplementary Table 1. Brain glioblastoma tissue arrays (cat. no. GL805) were purchased from US Biomax (Rockville, MD). Unless otherwise specified, all other reagents were of analytical grade.
Cell Culture, Gene Transfection, and Drug Treatment
Cells were cultured at 37 °C in a 5% CO2 incubator in Dulbecco’s modified Eagle’s medium (DMEM) with 2.5 mM GlutaMAX, 100 units/mL penicillin, 100 μg/mL streptomycin, 1 mM sodium pyruvate, and 10% fetal bovine serum (FBS; GIBCO-BRL). Lipofectamine 3000 (Invitrogen) was utilized in gene transfection experiments based on the manufacturer’s protocol. Two micrograms of empty pCDH plasmids, pCDH-DDIT4, pCDH-GLUT3, scrambled shRNA, DDIT4 shRNA, and GLUT3 shRNA were used in overexpression or knockdown experiments. All complementary (c) DNA constructs were established in our lab. For TMZ treatment, indicated concentrations or 200 μM of TMZ were added to overnight-cultured cells for 72 h. For transfection with TMZ treatment, cells were transfected with 2 μg cDNA or shRNA for 24 h. Then, 200 μM of TMZ were added for another 72 h.
RNA Isolation and Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
Cultured cells were used to extract total RNA using Trizol (Invitrogen) according to the manufacturer’s instructions. A260/A280 readings were used to check the RNA quality. Using a random primer and a MultiScribe Reverse Transcriptase kit (Invitrogen), cDNA was synthesized from 1 μg of total RNA, diluted 1:30 with PCR-grade water and stored at − 20 °C. Specific primers for detecting human DDIT4, GLUT3, and GAPDH levels for the real-time qPCR are listed in Supplementary Table 1. The Applied Biosystems StepOnePlus System (Thermo Fisher Scientific, Waltham, MA) with pre-optimized conditions was used to measure gene expression levels. Each PCR contained 5 μL 2× SYBR Green PCR master mix (Invitrogen), 0.2 μL primer sets, 1 μL cDNA, and 3.6 μL nucleotide-free H2O to yield a 10-μL reaction and was performed in triplicate. The normalized CT difference between the control and sample after adjusting for the amplification efficiency relative to the expression level of the GAPDH housekeeping gene was calculated as the expression rate.
Immunoblot Analysis
RIPA buffer (1% Nonidet P-40, 0.5% deoxycholate, and 0.1% sodium dodecylsulfate (SDS) in phosphate-buffered saline (PBS)) containing a protease inhibitor cocktail (Calbiochem, Billerica, MA) was used to lyse U87-MG cells. After centrifugation at 12,000 rpm for 10 min at 4 °C, the supernatant was collected. Buffers containing 2% SDS, 10 mM dithiothreitol, 60 mM Tris-hydrochloric acid (Tris-HCl, pH 6.8), and 0.1% bromophenol blue were used to denature cell lysates. Then, cell lysates (50 μg) were loaded onto a 10~15% polyacrylamide/SDS gel. After separated proteins were transferred onto a polyvinylidene difluoride membrane, the membrane was blocked for 1 h at room temperature in PBS containing 5% nonfat dry milk. The primary antibody was added to the membrane and incubated overnight at 4 °C. The anti-GLUT3 antibody, anti β-actin antibody, and other primary antibodies were respectively diluted with PBS containing 5% nonfat dry milk as 1:100, 1:10000, and 1:1000. After washing in PBS-T, the membrane was incubated with the secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature and then washed with PBS-T again. The dilution ratios of secondary antibodies for detecting GLUT3, β-actin, and other proteins were 1:500, 1:50000, and 1:3000, respectively. An enhanced chemiluminescence (ECL) nonradioactive detection system was used to detect the antibody–protein complexes.
Cell Viability Assay
Cells (104 cells/well) were seeded on a 96-well plate, followed by treatment with various concentrations of TMZ for 72 h. For transfection with TMZ treatment, cells were transfected with 2 μg cDNA or shRNA for 24 h. Then, 200 μM of TMZ were added for another 72 h. Then, 0.5 mg/mL MTT was added to each well for 4 h. After carefully aspirating the supernatants, formazan crystals were dissolved using dimethyl sulfoxide (DMSO). The absorbance was measured at 570 nm with a Thermo Varioskan Flash reader (Carlsbad, CA).
Caspase-3 Activity
The caspase-3 activity was measured by using caspase-3/CPP32 colorimetric assay kit (BioVision, Inc.; Milpitas, CA) according to the manufacturer’s instructions. The treated cells were lysed with 50 μL chilled cell lysis buffer on ice for 10 min. After centrifuging, supernatant was collected for protein concentration determination. One hundred micrograms of proteins were diluted to 50 μL cell lysis buffer for each assay, and then 50 μL of 2× Reaction Buffer (containing 10 mM DTT) was added to each sample. After adding 5 μL of the 4 mM DEVD-pNA substrate (200 μM final conc.) and incubating at 37 °C for 2 h, the absorbance was measured at 405 nm with a Thermo Varioskan Flash reader (Carlsbad, CA). Fold increase in caspase-3 activity was determined by comparing these results with the level of the untreated control.
Gene Expression Profile Analyses of Glioma Patients
Microarray data of GBM patients of TCGA were retrieved from the University of California, Santa Cruz (UCSC) cancer genomic browser (https://xena.ucsc.edu/welcome-to-ucsc-xena/). In the present analysis, we choose to use the level 3 microarray data of GBM patients (gdac.broadinstitute.org_GBM.mRNA_Preprocess_Median.Level_3), downloaded from broad institute Firehouse (http://gdac.broadinstitute.org/), which contained 539 GBM patients instead of using RNA sequencing data (n = 152). GSE13041 and GSE16011 microarray data were obtained from the GEO database. Expression profiles of each gene were normalized by the robust multichip average (RMA) method and were log2 transformed. Glioma patients were divided into groups with high and low DDIT4 expression levels based on the median cutoff of DDIT4 levels. A differential gene expression analysis was performed to identify differentially expressed genes (DEGs) associated with DDIT4 expression. DEGs were considered significant with a multiple of change of > 1.5 and a false detection rate (FDR) of < 0.01. To investigate associations of GBM mesenchymal markers with ATF4 and DDIT4 expressions, Pearson correlation analyses were conducted. For the association of DDIT4 and GLUT3 in all cancer types, we downloaded pan-cancer RNA sequencing (RNA-Seq) data from the UCSC cancer genomic browser (https://xena.ucsc.edu/welcome-to-ucsc-xena/) and conducted Pearson correlation analyses.
Survival and Pathway Analyses of Glioma Patients
To investigate the association between TMZ treatment and DDIT4 expression levels, we divided TCGA glioma patients into TMZ-treated and untreated groups. A log-rank test was conducted to investigate the survival benefit of receiving TMZ in groups with high and low DDIT4 expression. Furthermore, the O-6-methylguanine-DNA methyltransferase (MGMT)-methylated status of GBM patients was included for subgroup survival analyses. In order to explore important DDIT4-associated genes, a Cox regression analysis was conducted. Genes with a hazard ratio of > 1 and an FDR of < 0.1 were considered risk gene candidates. Using the beta coefficient of each risk gene candidate from TCGA, we established a risk score based on the formula:
Using this formula, we validated correlations of a glioma prognosis with DDIT4-associated genes in another independent dataset, GSE16011. A gene set enrichment analysis (GSEA) was conducted to investigate pathways associated with DDIT4 expression levels in 3 databases, including GSE13041, TCGA, and GSE16011. Gene candidates were ranked based on the log2 multiple of change between groups with high and low DDIT4 expression. A normalized enrichment score and FDR were generated after 1000 permutations. The glycolysis pathway was retrieved from the Hallmark database, and autophagy pathways were obtained from an in-house gene list.
Immunohistochemical (IHC) Analysis
For IHC staining, paraffin-embedded sections from 35 patients in a brain glioblastoma tissue array were baked at 60 °C overnight. These patients are all type IV glioblastoma. More detail information about age and gender of patients is listed in the website (https://www.biomax.us/tissue-arrays/Brain/GL805e). After dewaxing and hydration, 10 mmol/L sodium citrate buffer (pH 6) was used to heat sections in a microwave oven for 10 min. Then, sections were blocked with 10% horse serum in PBS for 30 min and incubated overnight at 4 °C with an anti-DDIT4 or anti-GLUT3 primary antibody at a dilution of 1:100. After washing, sections were incubated with biotinylated goat anti-rabbit immunoglobulin G (IgG) at a dilution of 1:1000 for 30 min. Immunoreactivity was detected with an avidin-biotin system using 3,3′-diaminobenzidine tetrahydrochloride as a chromogen and counterstaining with Mayer’s hematoxylin (Richard-Allan Scientific, Kalamazoo, MI).
Sphere Formation Assay
For tumor sphere formation, DMEM, B27 (1:50; Invitrogen, San Diego, CA), epidermal growth factor (EGF; 20 ng/mL, Invitrogen), basic fibroblast growth factor (bFGF; 20 ng/mL, Gibco), penicillin/streptomycin (Invitrogen), and L-glutamine (Invitrogen) were used to culture 3 × 104 cells in a Costar ultralow attachment multiple 6-well plate (Corning, Corning, NY). After day 4, spheres had formed. One third of the medium was replaced every 3 days. After 1 week, spheres were dissociated and replated in nonadherent plates to generate secondary spheres. After 2 weeks of culturing, spheres were collected for subsequent experiments.
Statistical Analysis
Statistical analyses were carried out using Sigma Plot 12.5 (Systat Software, San Diego, CA). All data are presented as the mean ± standard deviation (SD). Significant differences among groups were determined using an unpaired t test. A value of p < 0.05 was taken as an indication of statistical significance. All figures shown in this article were obtained from at least 3 independent experiments with similar results.
Results
TMZ-Induced DDIT4 Upregulation Decreases the Therapeutic Efficacy of TMZ
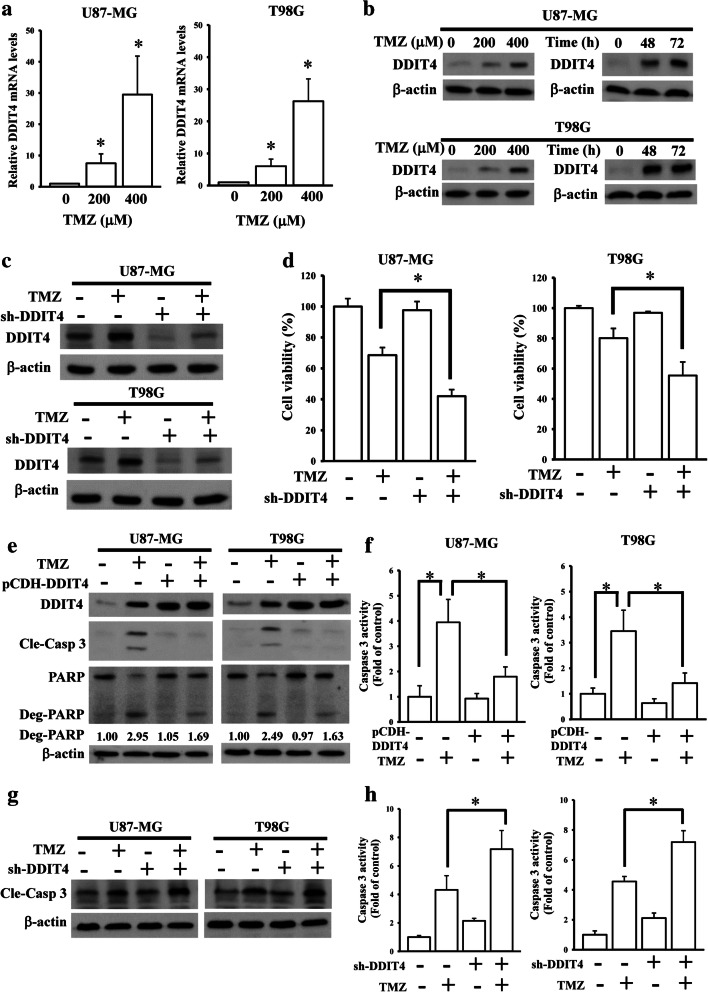
DDIT4, a TMZ-upregulated gene found in our previous microarray data (Supplementary Table 2) [18], was reported to contribute to chemotherapeutic resistance in several cancer types. By conducting qPCR and immunoblotting assays, we confirmed that TMZ upregulated both DDIT4 messenger (m)RNA and protein levels in a concentration-dependent manner (Fig. 1A, B). After using shRNA to deplete DDIT4 expression in glioma cells (Fig. 1C), increased TMZ cytotoxicity was observed (Fig. 1D). Depletion of DDIT4 expression reduced cell viabilities of both U87-MG and T98G cells. However, overexpression of DDIT4 in glioma cells did not significantly increase the cell growth or phenotypic changes. Overexpression and knockdown of DDIT4 influenced the TMZ-induced apoptotic death and caspase activity of glioma cells (Fig. 1E–H).
Fig. 1.
Upregulation of DNA damage-inducible transcript 4 (DDIT4) expression by temozolomide (TMZ) desensitizes its cytotoxicity towards gliomas. Enhancing effects of TMZ on DDIT4 mRNA (A) and protein levels (B). After being treated with the indicated doses of TMZ for 72 h, relative mRNA and protein levels of the DDIT4 gene were analyzed by real-time PCR and immunoblot assays. Data are the mean ± SD of 3 experiments. *p < 0.05. Depletion of endogenous DDIT4 expression reduced TMZ-upregulated DDIT4 levels (C), but enhanced TMZ cytotoxicity (D). (E, F) Overexpression of DDIT4 attenuated TMZ-induced cell apoptotic death and caspase-3 activity. (G, H) Knockdown of DDIT4 enhanced TMZ-induced caspase-3 activation. After cells were transfected with 2 μg cDNA or shRNA for 24 h. Then, 200 μM of TMZ were added for another 72 h. DDIT4 and cleavage caspase-3 (Cle-Casp 3) protein levels were detected by immunoblot assays. Caspase-3 activity was measured by using commercial kit according to the manufacturer’s instructions listed in method section. Cell viability was measured using an MTT assay. Data are the mean ± SD of 3 experiments. *p < 0.05
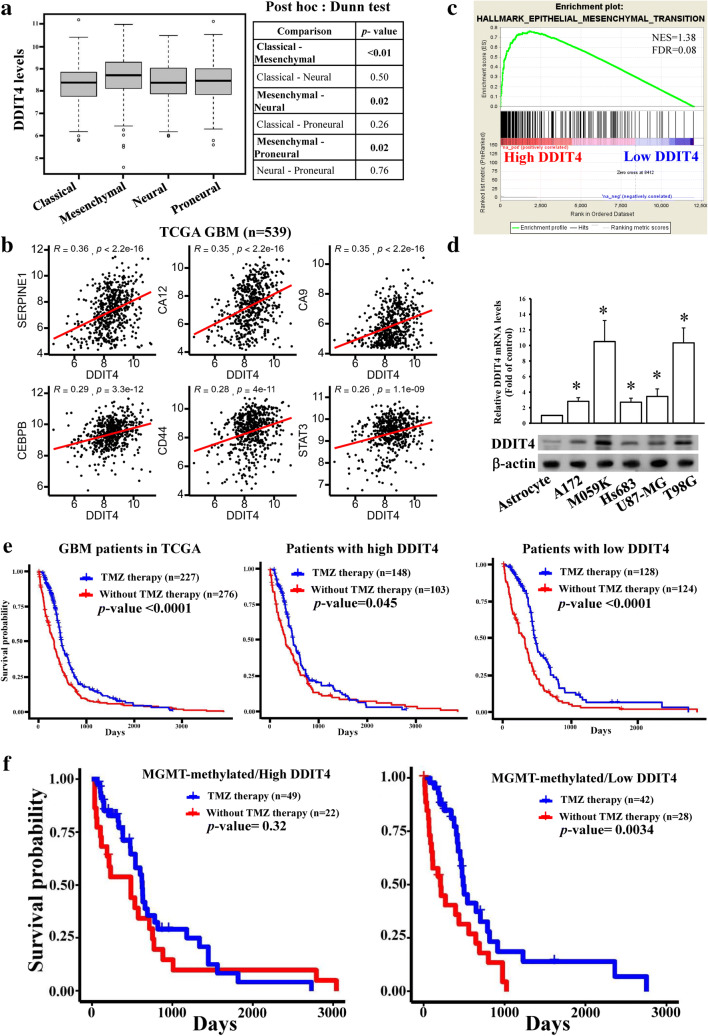
To further evaluate whether DDIT4 expression is associated with the therapeutic efficacy of TMZ, microarray data from TCGA GBM patients were analyzed. In a previous study [20], GBM was categorized into 4 different molecular types, and the mesenchymal type was highly associated with chemo-radiotherapeutic resistance. We found that DDIT4 exhibited the highest expression level in the mesenchymal type of GBM patients (Fig. 2A), and significant positive correlations were observed between DDIT4 expression and mesenchymal markers (Supplementary Table 3). The stemness markers we selected are based on Chow et al.’s study [21]. Top 6 of significantly correlated mesenchymal markers with DDIT4 levels are shown in Fig. 2B. Furthermore, by using GSEA analyses, we found that high DDIT4 expression was also significantly correlated with epithelial mesenchymal transition (EMT) signaling (Fig. 2C), suggesting that DDIT4 expression is associated with EMT in glioma. Higher mRNA and protein levels of DDITs existed in glioma cell lines compared to normal human astrocytes (Fig. 2D). Additionally, a better survival rate was observed in patients undergoing TMZ therapy compared to the non-TMZ therapy groups (Fig. 2E). Furthermore, the association between TMZ treatment and overall survival in high-DDIT4-expressing patients was less strong than that in patients with low DDIT4 levels (Fig. 2F). These data suggest that TMZ-enhanced DDIT4 expression dampened the therapeutic efficacy of TMZ against glioma cells.
Fig. 2.
DNA damage-inducible transcript 4 (DDIT4) levels are significantly correlated with a mesenchymal phenotype and temozolomide (TMZ)’s therapeutic effects in glioblastoma multiforme (GBM) patients. DDIT4 was enriched in mesenchymal-type patients (A) and was positively associated with mesenchymal markers (B) according an analysis of GBM patient data of The Cancer Genome Atlas. (C) GSEA analyses showed high DDIT4 levels were positively correlated with epithelial mesenchymal transition. (D) Comparison of endogenous DDIT4 levels in different cell lines. DDIT4 protein and mRNA levels were respectively detected by immunoblot assays and real-time PCR. Data are the mean ± SD of 3 experiments. *p < 0.05. Lower DDIT4 levels in patients (E) and methyl guanine methyl transferase (MGMT)-methylated patients (F) were highly correlated with better TMZ efficacy
DDIT4 Is Correlated with TMZ Therapy
Methylation of the MGMT promoter is a recognized predictor for evaluating TMZ responses. However, not all MGMT-methylated patients have a good response to TMZ therapy. Hence, we asked whether expression levels of DDIT4 could be used to stratify MGMT-methylated patients into good and poor TMZ responders. First, we found that treatment with TMZ was not associated with glioma prognosis in patients with unmethylated MGMT (Supplementary Fig. 1A). In contrast, methylated MGMT patients undergoing TMZ treatment had better prognoses (Supplementary Fig. 1B), suggesting that patients with distinct MGMT levels possessed different TMZ therapeutic responses. More importantly, when we divided MGMT-methylated patients into high- and low-DDIT4-expressing groups, TMZ treatment was correlated with a favorable prognosis only in the low-DDIT4-expressing group and not in the high-DDIT4-expressing group (Fig. 2E). Taken together, we concluded that DDIT4 is significantly correlated with TMZ therapeutic responses in MGMT-methylated GBM patients.
The DDIT4-Associated Gene Signature Is Correlated with a Poor Prognosis
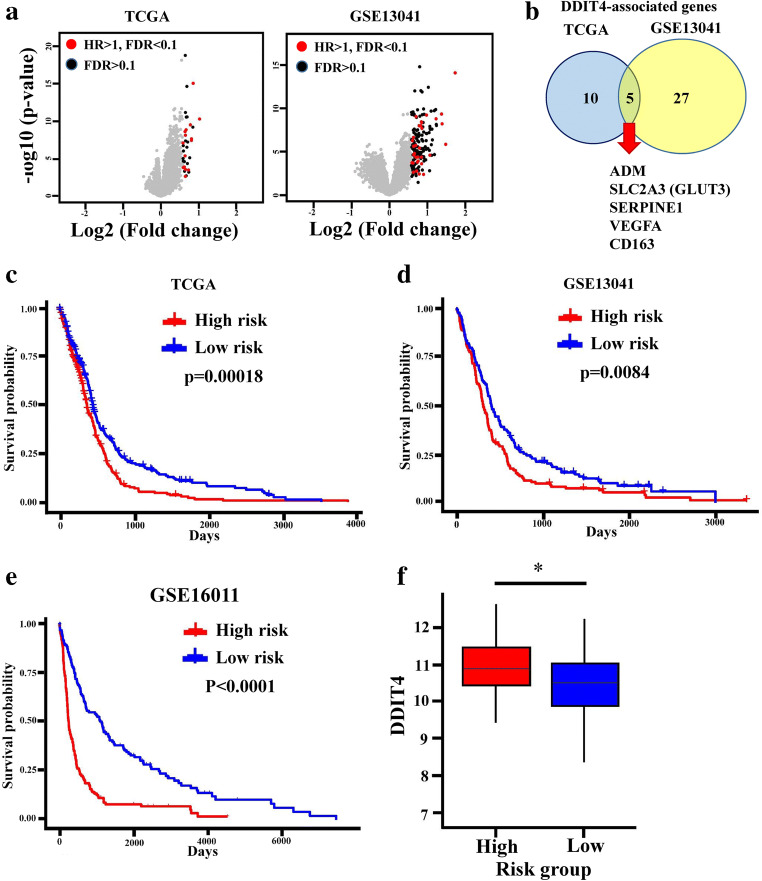
To comprehensively investigate DDIT4-associated gene candidates in glioma patients, differential gene expression analyses between high- and low-DDIT4-expressing groups based on a medium cutoff value were performed using TCGA and GSE13041 microarray data (Fig. 3A and Supplementary Table 4). Most of the upregulated gene candidates in the high-DDIT4-expressing groups were associated with a poor prognosis in glioma patients (red dots in Fig. 3A). Poor prognosis-related gene candidates in both TCGA and GSE13041 intersected, including adrenomedullin (ADM), vascular endothelial growth factor A (VEGFA), solute carrier family 2 member 3 (SLC2A3), serpin family E member 1 (SERPINE1), and cluster of differentiation 163 (CD163) (Fig. 3B). Further, we established a risk score for each patient based on the coefficient of these 5 gene candidates derived from a Cox regression analysis in TCGA. The high-risk group was associated with a poor prognosis in both TCGA (Fig. 3C) and GSE13041 databases (Fig. 3D). Next, we utilized this genetic signature-based risk score to analyze another independent dataset, GSE16011. The high-risk score also predicted a poor prognosis in those glioma patients (Fig. 3E) and was correlated with higher expression levels of DDIT4 (Fig. 3F). These findings identified that the DDIT4-associated gene signature is correlated with poor prognoses in GBM.
Fig. 3.
Five DNA damage-inducible transcript 4 (DDIT4)-associated genes predict poor patient prognoses. (A) Volcano plots showing risk genes among DDIT4-related genes. Red and black dots respectively indicate risk genes (hazard ratio (HR) > 1) and nonrisk genes among genes highly associated with DDIT4 using The Cancer Genome Atlas (TCGA) and GSE13041 analyses. (B) Five DDIT4-associated genes existed in both TCGA and GSE13041 data. A higher risk score created by the 5 DDIT4-associated genes predicted a poor prognosis in TCGA (C), GSE13041 (D), and GSE16011 (E). (F) Higher DDIT4 levels were seen in patients with high-risk scores. *p < 0.05
TMZ-Induced DDIT4/SLC2A3 Signaling Reduces TMZ Cytotoxicity
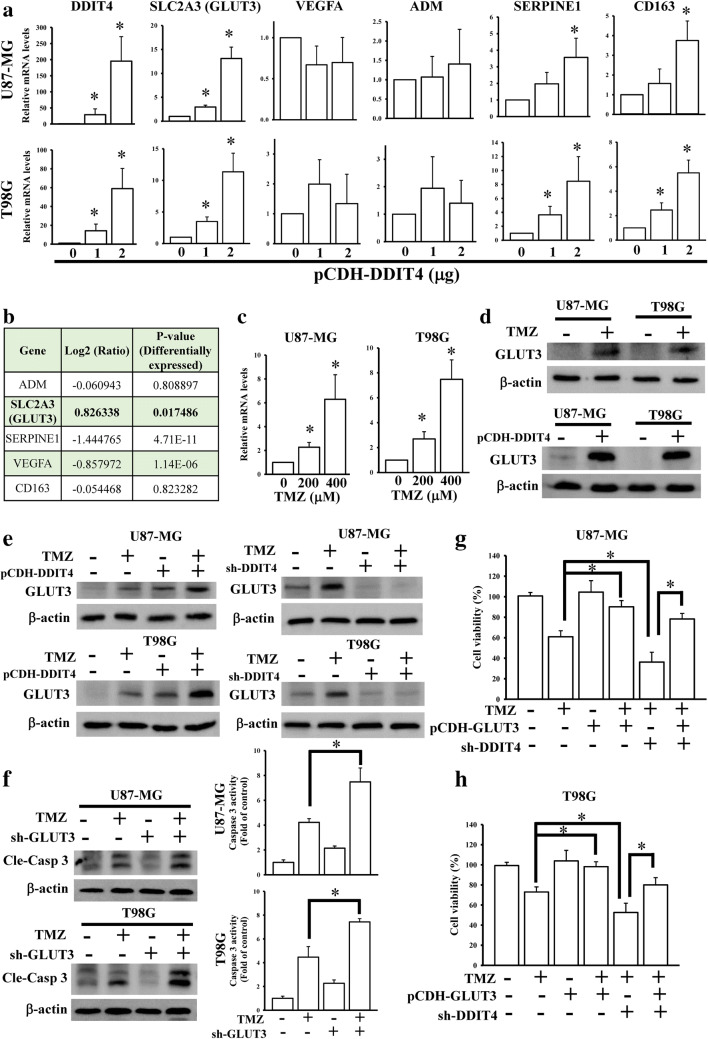
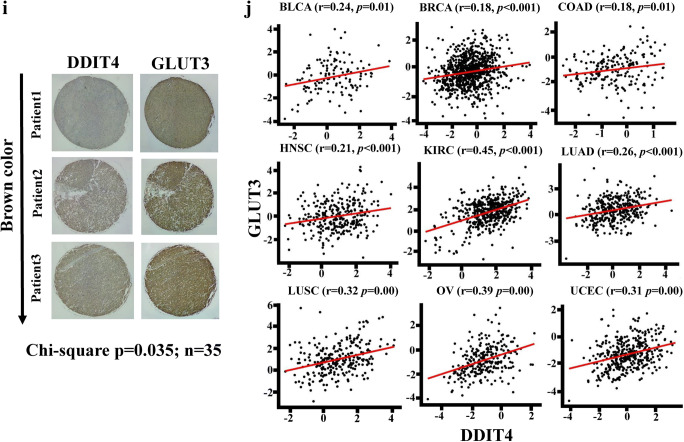
To explore whether DDIT4 influenced ADM, SLC2A3, SERPINE1, and CD163 expressions, DDIT4 cDNA was cloned into a pCDH vector and transfected into glioma cells. We found that SLC2A3, SERPINE1, and CD163, but not VEGFA or ADM, were upregulated in DDIT4-overexpressing cells (Fig. 4A). Further, by examining microarray data of TMZ-treated glioma cells [18], SLC2A3, also known as GLUT3, was enhanced by TMZ treatment (Fig. 4B). TMZ-upregulated GLUT3 expression was also confirmed by a qPCR and immunoblot assay (Fig. 4C, D). Overexpression of DDIT4 also enhanced GLUT3 protein levels in glioma cells (Fig. 4D). By conducting overexpression and knockdown of DDIT4 experiments in TMZ-treated cells, we found that upregulation of DDIT4 further increased TMZ-induced GLUT3 levels. However, DDIT4 depletion attenuated the effects of TMZ on enhancing GLUT3 expression (Fig. 4E). Knockdown of GLUT3 enhanced TMZ-induced cell apoptotic death (Fig. 4F). Additionally, using cell viability assays, we found that overexpression of GLUT3 significantly decreased the cytotoxicity of TMZ (Fig. 4G, H). Furthermore, the increased GLUT3 expression rescued the cell viability of DDIT4-depleted glioma cells after TMZ treatment. To further investigate the clinical significance of DDIT4-mediated GLUT3 signaling, we conducted analyses of a tissue array containing 35 GBM patients (Fig. 4I). DDIT4 protein expression was positively correlated with GLUT3 protein levels. Moreover, by performing a pan-cancer analysis from TCGA RNA sequencing data, DDIT4 expression was also positively correlated with GLUT3 levels in multiple cancer types (Fig. 4J). These results indicated that DDIT4-regulated GLUT3 signaling reduces the therapeutic efficacy of TMZ, and the levels of these 2 molecules are tightly associated with each other in clinical patients.
Fig. 4.
SLC2A3 (glucose transporter 3 (GLUT3)) is a downstream gene in temozolomide (TMZ)-induced DNA damage-inducible transcript 4 (DDIT4) signaling. (A) Effects of DDIT4 overexpression on mRNA levels of 5 DDIT4-associated genes. (B) Only GLUT3 upregulation in microarray data of TMZ-treated cells. (C) TMZ treatment increased GLUT3 mRNA levels in a dose-dependent manner. (D) TMZ treatment and overexpression of DDIT4 enhanced GLUT3 protein levels. (E) Overexpression and knockdown of DDIT4 influenced TMZ-induced GLUT3 expression. (F) Knockdown of GLUT3 enhanced TMZ-induced caspase-3 activation. Overexpression of GLUT3 attenuated the effects of DDIT4 depletion of enhancing TMZ cytotoxicity in U87-MG (G) and T98G cells (H). After transfection with the indicated dose or 2 μg of DDIT4 plasmids for 24 h, indicated dose or 200 μM TMZ was added for another 72 h. Relative mRNA and protein levels, and cell viability were respectively measured by real-time PCR, immunoblot, and MTT assays. Caspase-3 activity was measured by using commercial kit according to the manufacturer’s instructions listed in the “Materials and Methods” section. Data are the mean ± SD of 3 experiments. *p < 0.05. A significant positive correlation existed between DDIT4 and GLUT3 in glioblastoma multiforme (GBM) tissue arrays (I) and The Cancer Genome Atlas (TCGA) pan-cancer analyses (J). BLCA = urothelial bladder carcinoma; BRCA = breast invasive carcinoma; COAD = colon adenocarcinoma; HNSC = head-neck squamous cell carcinoma; KIRC = kidney renal clear cell carcinoma; LUAD = lung adenocarcinoma; LUSC = lung squamous cell carcinoma; OV = ovarian cancer; UCEC = uterine corpus endometrial carcinoma
DDIT4 Mediates GLUT3 Signaling to Increase Glioma Stemness
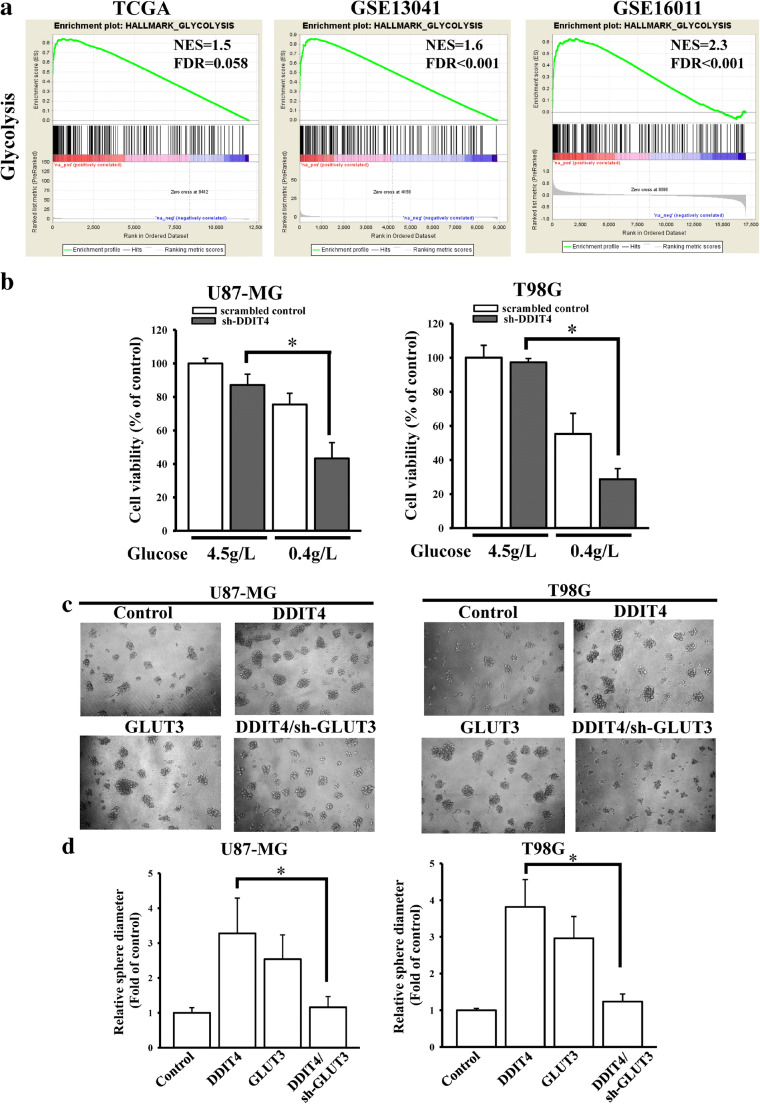
A previous study reported that GLUT3-mediated glycolysis signaling is critical for glioma stem cell maintenance, and cancer stem cells are highly correlated with chemotherapeutic resistance. Therefore, we tried to explore whether DDIT4 could promote cancer stemness through GLUT3 signaling. By conducting a GSEA with TCGA, GSE13041, and GSE16011 datasets, we found that glycolysis signaling was significantly enriched in high-DDIT4 glioma patients (Fig. 5A). According to a previous study [22], inhibition of GLUT3 reduced cell viability in a glucose-restricted condition (0.4 g/L glucose). Similarly, DDIT4 depletion also reduced cell viability with glucose restriction (Fig. 5B), demonstrating that DDIT4 participates in GLUT3-mediated glucose metabolism. Next, we tested the effects of DDIT4 on GLUT3-mediated stemness. Overexpression of DDIT4 or GLUT3 enhanced the sphere formation size of glioma cells (Fig. 5C, D). GLUT3 depletion reduced DDIT4-promoted sphere formation. Taken together, these findings showed that enhanced DDIT4 expression may decrease TMZ efficacy through GLUT3-mediated glioma stemness via glycolysis metabolism.
Fig. 5.
DNA damage-inducible transcript 4 (DDIT4) enhances stemness properties through glucose transporter 3 (GLUT3)-mediated glycolysis pathways. (A) Genes positively correlated with DDIT4 were found to be enriched in glycolysis signaling using gene set enrichment analyses (GSEAs). (B) Inhibition of DDIT4 influenced cell viability in a glucose-restricted condition. Cell viability was measured using an MTT assay. Data are the mean ± SD of 3 experiments. *p < 0.05. (C) The effects of DDIT4-GLUT3 signaling on sphere formation. (D) Quantitative data according to measurements of sphere diameters from tumorsphere formation assays. After transfection with 2 μg cDNA or shRNA for 24 h, cells were collected for glucose-restricted or tumorsphere formation assays
GLUT3 Signaling Is Not Involved in DDIT4-Promoted Autophagy Against TMZ Cytotoxicity
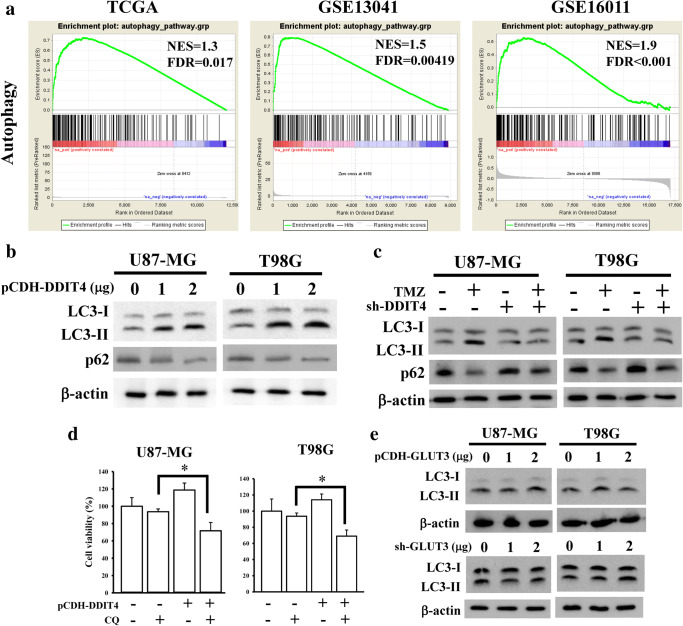
TMZ was reported to promote autophagy, leading to a decrease in its therapeutic efficacy. Additionally, other studies also indicated that DDIT4 can increase autophagy signaling and contribute to chemotherapeutic resistance. Hence, we attempted to clarify whether DDIT4 is involved in TMZ-promoted autophagy of gliomas. The autophagy gene list was obtained from human autophagy database [23]. The gene candidates in this database have been reported to directly or indirectly involve in autophagy. Through a GSEA, autophagy was found to be significantly enriched in the high-DDIT4-expression group (Fig. 6A). The ectopic expression of DDIT4 in glioma cells increased the ratio of LC3-I/LC3-II and decreased p62 protein levels (Fig. 6B), indicating that DDIT4 can induce autophagy. Furthermore, knockdown of DDIT4 decreased the TMZ-promoted autophagy of glioma cells (Fig. 6C). By treating DDIT4-overexpressing glioma cells with the autophagy inhibitor, CQ, DDIT4 promoted protective autophagy in glioma cells (Fig. 6D). Finally, we tested the effects of GLUT3 on DDIT4-mediated autophagy signaling. However, overexpression or knockdown of GLUT3 did not influence autophagy generation in glioma cells (Fig. 6E), suggesting that DDIT4-mediated autophagy is independent of GLUT3 signaling.
Fig. 6.
DNA damage-inducible transcript 4 (DDIT4) induces protective autophagy against temozolomide (TMZ) cytotoxicity, but not through glucose transporter 3 (GLUT3) signaling. (A) Genes positively correlated with DDIT4 were found to be enriched in autophagy signaling using gene set enrichment analyses (GSEAs). (B) DDIT4 induced autophagy formation. After cells were transfected with 2 μg of DDIT4 plasmids for 24 h, LC3 levels from cell lysates were detected by immunoblot assays. (C) Depletion of DDIT4 reduced TMZ-enhanced autophagy formation. After transfection with 2 μg of DDIT4 shRNAs for 24 h, 200 μM TMZ was added for another 72 h. LC3 and p62 levels from cell lysates were detected by immunoblot assays. (D) Inhibition of autophagy by chloroquine (CQ) treatment reduced cell viability in DDIT4-overecpressing cells. After transfection with 2 μg of DDIT4 plasmids for 24 h, 10 μM CQ was added for another 24 h. Cell viability was measured by MTT assays. Data are the mean ± SD of 3 experiments. *p < 0.05. (E) GLUT3 showed no effects on autophagy formation. After transfection with 2 μg of GLUT3 shRNAs or cDNAs for 24 h, LC3 and p62 levels from cell lysates were detected by immunoblot assays
TMZ-Enhanced ATF4 Is Responsible for Upregulation of DDIT4-Mediated GLUT3 Signaling
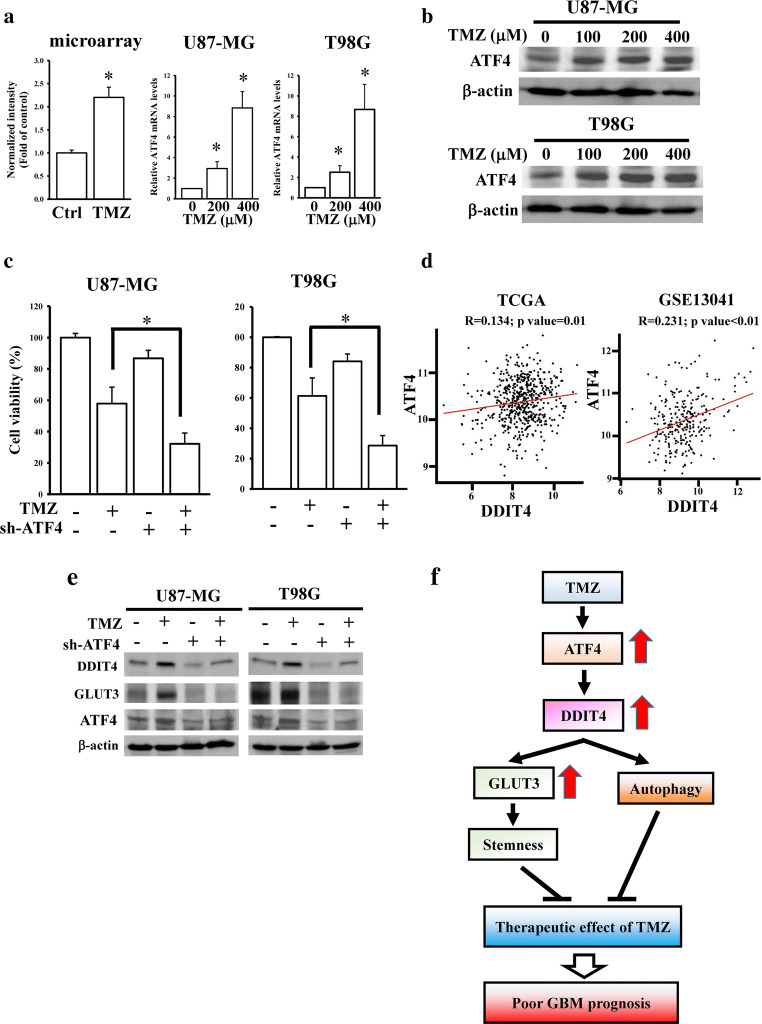
ER stress was reported to promote DDIT4 upregulation through increasing ATF4 expression. Interestingly, we found that ATF4 was upregulated in microarray data of TMZ-treated glioma cells [18]. By conducting qPCR and immunoblot assays, we confirmed that TMZ increased ATF4 mRNA and protein expressions in concentration-dependent manners (Fig. 7A, B and Supplementary Fig. 2). ATF4 depletion enhanced TMZ-mediated cytotoxicity in glioma cells (Fig. 7C). Next, we hypothesized that upregulated ATF4 expression is responsible for TMZ-mediated DDIT4 signaling. By analyzing the transcriptome profiles of TCGA and GSE13041 datasets, we found that ATF4 expression was positively correlated with DDIT4 levels (Fig. 7D). Knockdown of ATF4 abolished TMZ-induced DDIT4 and GLUT3 protein expressions (Fig. 7E). Taken together, we identified that TMZ-induced DDIT4/GLUT3 signaling decreased the therapeutic efficacy of TMZ in GBM progression through ATF4 upregulation.
Fig. 7.
Activating transcription factor 4 (ATF4) is involved in temozolomide (TMZ)-induced DNA damage-inducible transcript 4 (DDIT4) signaling. Effects of TMZ on ATF4 mRNA (A) and protein (B) levels. After treating with indicated doses of TMZ for 72 h, relative ATF4 mRNA and protein levels were respectively measured by real-time PCR and immunoblot assays. Data are the mean ± SD of 3 experiments. *p < 0.05. (C) Knockdown of ATF4 enhanced TMZ cytotoxicity. (D) A positive correlation existed between DDIT4 and ATF4. Depletion of ATF4 influenced TMZ/DDIT4/glucose transporter 3 (GLUT3) signaling (E). After transfection with the 2 μg ATF4 shRNAs for 24 h, 200 μM TMZ was added for another 72 h. ATF4 protein levels were measured by immunoblot assays. Cell viability was measured by MTT assays. Data are the mean ± SD of 3 experiments. *p < 0.05. (F) ATF4-mediated DDIT4 signaling desensitizes TMZ cytotoxicity through autophagy and GLUT3-regulated stemness. GBM patients with higher DDIT4 levels exhibited lower TMZ therapeutic efficacy and poorer prognoses
Discussion
In this study, we identified that DDIT4 is significantly upregulated in TMZ-treated glioma cells. Further, by conducting an in silico analysis, elevated DDIT4 expression was identified to be associated with poor TMZ efficacy in GBM patients (Fig. 7F). It is worth noting that MGMT-methylated GBM patients who supposedly would receive greater benefits from TMZ therapy could be further categorized into good and bad TMZ responders based on DDIT4 expression. By mining 3 public databases, including TCGA, GSE13041, and GSE16011, genes positively associated with DDIT4 were found to be correlated with a poor prognosis. These genes are involved in both an autophagy pathway and glycolysis signaling which plays an important role in maintaining glioma stemness. By intersecting the microarray data in TMZ-treated glioma cells with DDIT4-correlated genes, we determined that GLUT3 upregulation by DDIT4 after TMZ stimulation is the key molecule. TMZ-increased DDIT4/GLUT3 signaling through inducing ATF4 signaling was also validated, leading to promotion of TMZ intrinsic resistance. Taken together, our results suggest that DDIT4 decreases TMZ efficacy in gliomas through multiple mechanisms including autophagy activation and GLUT3-mediated cancer stemness.
It was reported that elevated DDIT4 levels are induced under different stressful conditions including chemo-radiotherapy [19]. Similarly, in our study, we found that DDIT4 was one of the top 10 upregulated genes after TMZ treatment according to microarray analyses. Increasing mRNA and protein levels of DDIT4 were both observed in TMZ-treated cells, which emphasizes that DDIT4 may have a critical role in TMZ-regulated downstream signaling. Additionally, through mTORC1 inhibition, DDIT4 was identified to be involved in TMZ and radiotherapy resistance by GBM [19]. However, in addition to being an mTORC1 inhibitor, whether DDIT4 possesses multiple functions in regulating GBM progression and TMZ resistance remains unclear. Hence, we comprehensively analyzed transcriptome profiles of glioma patients using 3 public databases. We identified 5 candidate genes, including ADM, SLC2A3, SERPINE1, VEGFA, and CD163, that were consistently associated with high DDIT4 expression and worse prognoses. Interestingly, among these 5 candidate genes, only SLC2A3, also known as GLUT3, was significantly upregulated in TMZ-treated cells. Mounting evidence suggests that GLUT3 is critical for maintaining cancer stemness [24–26]. However, GLUT3 is also abundantly expressed by neurons [27], which makes it less than an ideal target for drug development for GBM. This issue highlights the importance of identifying upstream and dysregulated modulators for activating GLUT3 signaling in GBM. Herein, we identified that DDIT4 upregulation enhanced tumor sphere formation. Moreover, knockdown of GLUT3 in DDIT4-overxpressing cells reduced glioma stemness. Through analyses from TCGA pan-cancer data and GBM tissue arrays, DDIT4 was found to be positively correlated with GLUT3 at both the RNA and protein levels. This evidence suggests that DDIT4 is a novel regulator in promoting GLUT3-mediated cancer stemness.
Autophagy, a cellular process that is activated in response to stressful stimuli such as hypoxia, starvation, and exposure to chemotherapy, is critical for cell survival in harsh microenvironments. In response to the chemotherapeutic drug, TMZ, glioma cells activate a reversible autophagic process as a protective mechanism against cell death [28]. A previous study reported that DDIT4 suppresses the phosphorylation of mTOR, contributing to induction of autophagy upon encountering stressful conditions [29]. DDIT4-activating autophagy through mTOR-EE2K signaling also confers intrinsic resistance of bladder urothelial carcinoma to paclitaxel treatment [9]. In our study, we validated that DDIT4 gene expression was positively associated with autophagy signaling in glioma patients. We also identified that DDIT4 activates a protective autophagic mechanism in desensitizing TMZ cytotoxicity to glioma cells. In addition, a previous study reported that heat stress induced autophagy activation accompanied by GLUT3 upregulation [30]. Similar findings also exist in TMZ-treated glioma cells. Furthermore, we suggest that DDIT4 plays a key role in connecting these 2 physiological phenomena to restrain TMZ cytotoxicity. As a consequence, DDIT4-mediated autophagy independent of GLUT3 upregulation elicits the intrinsic TMZ resistance.
Recently, several studies identified that autophagy is highly associated with maintenance of cancer stemness properties in a variety of cancers including breast cancer [31], liver cancer [32], and glioblastomas [33]. As for GBM, autophagy was identified as a self-defense mechanism for glioblastoma stem cells to resist ferroptosis formation, which sensitizes them to TMZ cytotoxicity [33]. However, linkage of the maintenance of glioma stemness with autophagy induction is still poorly understood. Herein, we identified the key molecule, DDIT4, that respectively promotes glioma stemness and autophagy. Still, some limitations exist in this study. In vivo experiments and additional GBM patient tissues are needed for data validation. Elucidating the molecular mechanisms in DDIT4’s regulation of GLUT3 expression still requires further investigation. Elevated DDIT4 levels desensitize TMZ’s efficacy, leading to the poor prognosis of GBM patients. In addition, constitutive overexpression of DDIT4 enhances HSP27 and HSP70 induction, leading to ionizing radiation resistance of lung cancer [34]. DDIT4 overexpression protects bone marrow mesenchymal stromal cells (BMSCs) from injury induced by ionizing radiation exposure [35], suggesting that DDIT4 also plays a critical role in ionizing radiation resistance against cancer therapy. Because both ionizing radiation and chemotherapy are important adjuvant therapies for glioma, targeting DDIT4 signaling may provide novel strategies in future glioma therapies.
Electronic Supplementary Material
(PDF 670 kb)
Acknowledgments:
This study was sponsored by grants from the Ministry of Science and Technology, Taiwan (MOST 106-2320-B-038-051-MY3) and the Ministry of Education, Taiwan (DP2-107-21121-01-NK and DP2-108-21121-01-NK). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kuo-Hao Ho and Peng-Hsu Chen contributed equally to this work.
References
- 1.Jia W, Chang B, Sun L, Zhu H, Pang L, Tao L, et al. REDD1 and p-AKT over-expression may predict poor prognosis in ovarian cancer. Int J Clin Exp Pathol. 2014;7(9):5940–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Molitoris JK, McColl KS, Swerdlow S, Matsuyama M, Lam M, Finkel TH, et al. Glucocorticoid elevation of dexamethasone-induced gene 2 (Dig2/RTP801/REDD1) protein mediates autophagy in lymphocytes. J Biol Chem. 2011;286(34):30181–9. doi: 10.1074/jbc.M111.245423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto JA, Rolfo C, Raez LE, Prado A, Araujo JM, Bravo L, et al. In silico evaluation of DNA Damage Inducible Transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies. Sci Rep. 2017;7(1):1526. doi: 10.1038/s41598-017-01207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitney ML, Jefferson LS, Kimball SR. ATF4 is necessary and sufficient for ER stress-induced upregulation of REDD1 expression. Biochem Biophys Res Commun. 2009;379(2):451–5. doi: 10.1016/j.bbrc.2008.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schupp M, Chen F, Briggs ER, Rao S, Pelzmann HJ, Pessentheiner AR, et al. Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues. BMC Genomics. 2013;14:758. doi: 10.1186/1471-2164-14-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22(2):239–51. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du F, Sun L, Chu Y, Li T, Lei C, Wang X, et al. DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways. Cancer Commun (Lond) 2018;38(1):45. doi: 10.1186/s40880-018-0315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng Q, Liu J, Cao P, Li J, Liu X, Fan X, et al. Inhibition of REDD1 sensitizes bladder urothelial carcinoma to paclitaxel by inhibiting autophagy. Clin Cancer Res. 2018;24(2):445–59. doi: 10.1158/1078-0432.CCR-17-0419. [DOI] [PubMed] [Google Scholar]
- 10.Ohgaki H, Burger P, Kleihues P. Definition of primary and secondary glioblastoma—response. Clin Cancer Res. 2014;20(7):2013. doi: 10.1158/1078-0432.CCR-14-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis ME. Glioblastoma: overview of disease and treatment. Clin J Oncol Nurs. 2016;20(5 Suppl):S2–8. doi: 10.1188/16.CJON.S1.2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komotar RJ, Otten ML, Moise G, Connolly ES., Jr Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma—a critical review. Clin Med Oncol. 2008;2:421–2. doi: 10.4137/cmo.s390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wurstle S, Schneider F, Ringel F, Gempt J, Lammer F, Delbridge C, et al. Temozolomide induces autophagy in primary and established glioblastoma cells in an EGFR independent manner. Oncol Lett. 2017;14(1):322–8. doi: 10.3892/ol.2017.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briceno E, Reyes S, Sotelo J. Therapy of glioblastoma multiforme improved by the antimutagenic chloroquine. Neurosurg Focus. 2003;14(2):e3. doi: 10.3171/foc.2003.14.2.4. [DOI] [PubMed] [Google Scholar]
- 15.Sotelo J, Briceno E, Lopez-Gonzalez MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144(5):337–43. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 16.Wei Z, Deng X, Hong M, Su Q, Liu A, Huang Y, et al. Icariin has synergistic effects with methylprednisolone to ameliorate EAE via modulating HPA function, promoting anti-inflammatory and anti-apoptotic effects. Int J Clin Exp Med. 2015;8(11):20188–97. [PMC free article] [PubMed] [Google Scholar]
- 17.Prieto-Vila M, Takahashi RU, Usuba W, Kohama I, Ochiya T. Drug resistance driven by cancer stem cells and their niche. Int J Mol Sci. 2017;18(12). [DOI] [PMC free article] [PubMed]
- 18.Chen PH, Shen WL, Shih CM, Ho KH, Cheng CH, Lin CW, et al. The CHAC1-inhibited Notch3 pathway is involved in temozolomide-induced glioma cytotoxicity. Neuropharmacology. 2017;116:300–14. doi: 10.1016/j.neuropharm.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Foltyn M, Luger AL, Lorenz NI, Sauer B, Mittelbronn M, Harter PN, et al. The physiological mTOR complex 1 inhibitor DDIT4 mediates therapy resistance in glioblastoma. Br J Cancer. 2019;120(5):481–7. doi: 10.1038/s41416-018-0368-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chow KH, Park HJ, George J, Yamamoto K, Gallup AD, Graber JH, et al. S100A4 is a biomarker and regulator of glioma stem cells that is critical for mesenchymal transition in glioblastoma. Cancer Res. 2017;77(19):5360–73. doi: 10.1158/0008-5472.CAN-17-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosset E, Ilmjarv S, Dutoit V, Elliott K, von Schalscha T, Camargo MF, et al. Glut3 addiction is a druggable vulnerability for a molecularly defined subpopulation of glioblastoma. Cancer Cell. 2017;32(6):856–68. doi: 10.1016/j.ccell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, et al. The acquisition of resistance to TNFalpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy. 2011;7(7):760–70. doi: 10.4161/auto.7.7.15454. [DOI] [PubMed] [Google Scholar]
- 24.Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–82. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu M, Tanaka N. IL-8-induced O-GlcNAc modification via GLUT3 and GFAT regulates cancer stem cell-like properties in colon and lung cancer cells. Oncogene. 2019;38(9):1520–33. doi: 10.1038/s41388-018-0533-4. [DOI] [PubMed] [Google Scholar]
- 26.Chang CW, Chen YS, Chou SH, Han CL, Chen YJ, Yang CC, et al. Distinct subpopulations of head and neck cancer cells with different levels of intracellular reactive oxygen species exhibit diverse stemness, proliferation, and chemosensitivity. Cancer Res. 2014;74(21):6291–305. doi: 10.1158/0008-5472.CAN-14-0626. [DOI] [PubMed] [Google Scholar]
- 27.Maher F, Davies-Hill TM, Lysko PG, Henneberry RC, Simpson IA. Expression of two glucose transporters, GLUT1 and GLUT3, in cultured cerebellar neurons: evidence for neuron-specific expression of GLUT3. Mol Cell Neurosci. 1991;2(4):351–60. doi: 10.1016/1044-7431(91)90066-W. [DOI] [PubMed] [Google Scholar]
- 28.Kanzawa T, Germano IM, Komata T, Ito H, Kondo Y, Kondo S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004;11(4):448–57. doi: 10.1038/sj.cdd.4401359. [DOI] [PubMed] [Google Scholar]
- 29.Chen R, Wang B, Chen L, Cai D, Li B, Chen C, et al. DNA damage-inducible transcript 4 (DDIT4) mediates methamphetamine-induced autophagy and apoptosis through mTOR signaling pathway in cardiomyocytes. Toxicol Appl Pharmacol. 2016;295:1–11. doi: 10.1016/j.taap.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 30.Bao ZQ, Liao TT, Yang WR, Wang Y, Luo HY, Wang XZ. Heat stress-induced autophagy promotes lactate secretion in cultured immature boar Sertoli cells by inhibiting apoptosis and driving SLC2A3, LDHA, and SLC16A1 expression. Theriogenology. 2017;87:339–48. doi: 10.1016/j.theriogenology.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 31.Gong C, Bauvy C, Tonelli G, Yue W, Delomenie C, Nicolas V, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32(18):2261–72, 72e 1–11. [DOI] [PMC free article] [PubMed]
- 32.Song YJ, Zhang SS, Guo XL, Sun K, Han ZP, Li R, et al. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient-deprived tumor microenvironment. Cancer Lett. 2013;339(1):70–81. doi: 10.1016/j.canlet.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 33.Buccarelli M, Marconi M, Pacioni S, De Pascalis I, D'Alessandris QG, Martini M, et al. Inhibition of autophagy increases susceptibility of glioblastoma stem cells to temozolomide by igniting ferroptosis. Cell Death Dis. 2018;9(8):841. doi: 10.1038/s41419-018-0864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin HO, Hong SE, Kim JY, Kim MR, Chang YH, Hong YJ, et al. Induction of HSP27 and HSP70 by constitutive overexpression of Redd1 confers resistance of lung cancer cells to ionizing radiation. Oncol Rep. 2019;41(5):3119–26. doi: 10.3892/or.2019.7036. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z, Li T, Zhu F, Deng S, Li X, He Y. Regulatory roles of miR-22/Redd1-mediated mitochondrial ROS and cellular autophagy in ionizing radiation-induced BMSC injury. Cell Death Dis. 2019;10(3):227. doi: 10.1038/s41419-019-1373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 670 kb)