Abstract
In this study, which aims at early diagnosis of Covid-19 disease using X-ray images, the deep-learning approach, a state-of-the-art artificial intelligence method, was used, and automatic classification of images was performed using convolutional neural networks (CNN). In the first training-test data set used in the study, there were 230 X-ray images, of which 150 were Covid-19 and 80 were non-Covid-19, while in the second training-test data set there were 476 X-ray images, of which 150 were Covid-19 and 326 were non-Covid-19. Thus, classification results have been provided for two data sets, containing predominantly Covid-19 images and predominantly non-Covid-19 images, respectively. In the study, a 23-layer CNN architecture and a 54-layer CNN architecture were developed. Within the scope of the study, the results were obtained using chest X-ray images directly in the training-test procedures and the sub-band images obtained by applying dual tree complex wavelet transform (DT-CWT) to the above-mentioned images. The same experiments were repeated using images obtained by applying local binary pattern (LBP) to the chest X-ray images. Within the scope of the study, four new result generation pipeline algorithms having been put forward additionally, it was ensured that the experimental results were combined and the success of the study was improved. In the experiments carried out in this study, the training sessions were carried out using the k-fold cross validation method. Here the k value was chosen as 23 for the first and second training-test data sets. Considering the average highest results of the experiments performed within the scope of the study, the values of sensitivity, specificity, accuracy, F-1 score, and area under the receiver operating characteristic curve (AUC) for the first training-test data set were 0,9947, 0,9800, 0,9843, 0,9881 and 0,9990 respectively; while for the second training-test data set, they were 0,9920, 0,9939, 0,9891, 0,9828 and 0,9991; respectively. Within the scope of the study, finally, all the images were combined and the training and testing processes were repeated for a total of 556 X-ray images comprising 150 Covid-19 images and 406 non-Covid-19 images, by applying 2-fold cross. In this context, the average highest values of sensitivity, specificity, accuracy, F-1 score, and AUC for this last training-test data set were found to be 0,9760, 1,0000, 0,9906, 0,9823 and 0,9997; respectively.
Keywords: Covid-19, Corona 2019, Convolutional neural networks (CNN), Deep learning, Dual tree complex wavelet transform (DT-CWT), Local binary pattern (LBP), Chest X-ray classification
Introduction
In the last few months of 2019, a new type of virus, which is a member of the family Coronaviridae, emerged. The virus in question is considered to have had a zoonotic origin [1]. The virus that emerged in the city of Wuhan in Hubei province in China affected this region first and then spread all over the world in a short time. The virus generally affects the upper and lower respiratory tract, lungs, and, less frequently, the heart muscles [2]. While the virus generally affects young and middle-aged people and people who do not have any chronic diseases to a lesser extent, it can cause severe consequences, resulting in death, in people who suffer from diseases such as hypertension, cardiovascular disease, and diabetes [3]. The epidemic, which was declared to be a pandemic in March 2020 by the World Health Organization; as of the first week of October of the same year, had a number of cases approaching thirty-six million, while the death toll reached one million hundred thousand. Also, a modeling study carried out by Hernandez-Matamoros et al. [4] indicates that the effects of the epidemic will become more severe in the future.
In people suffering severely from the disease, the serious adverse effects are generally in the lungs [3]. In this context, many literature studies have been carried out in a short time in which these effects of the disease in the lungs were shown using CT scans of lungs and chest X-ray imaging. Literature studies indicate that radiological imaging, along with clinical symptoms, blood, and biochemical tests, is an effective and reliable diagnostic tool for the diagnosis of Covid-19 disease.
Many clinical studies in which X-ray images were examined [5–24] have shown that Covid-19 disease causes interstitial involvement, bilateral and irregular ground-glass opacities, parenchymal abnormalities, a unilobar reversed halo sign, and consolidation on the lungs. The recent review article published by Long and Ehrenfeld [25] highlighted the importance of using artificial intelligence methods to quickly diagnose Covid-19 disease and reduce the effects of the outbreak crisis. In this context, some literature studies have been carried out that diagnose Covid-19 disease (Covid-19 and non-Covid-19) through X-ray images and using deep learning methods. Table 1 contains some summary information about the number of images, study methods, and study results used in these literature studies.
Table 1.
Results of previous studies for Covid-19 and non-Covid-19 classification using X-ray images
| Study | Year | No. of Images | Methods | Test Methods | Results |
|---|---|---|---|---|---|
| Tuncer et al. [26] | 2020 | 321 images (87 Covid-19 and 234 Healthy) | Residual Exemplar Local Binary Pattern, Iterative Relief, Decision Tree, Linear Discriminant, Support Vector Machine, k-Nearest Neighborhood and Subspace Discriminant | 10-fold; 80% Train-20% Test; 50% Train-50% Test | Sen: 0,8755–0,9829/0,8297-0,9798/0,8149-1,0000; Spe: 0,9997-1,0000/0,9444-1,0000/0,9380-1,0000; Acc: 0,9663-0,9955/0,9130-0,9945/0,9049-0,9906 |
| Panwar et al. [27] | 2020 | 284 images (142 Covid-19 and 142 Healthy) | Convolutional Neural Network (nCOVnet) | 70% Train-30% Test | Sen: 0,9762; Spe: 0,7857; Acc: 0,881 |
| Ozturk et al. [28] | 2020 | 625 images (125 Covid-19 and 500 Healthy) | Convolutional Neural Network (DarkNet) | 5-fold | Sen: 0,9513; Spe: 0,953; Acc: 0,9808; F-1 Score: 0,9651 |
| Mohammed et al. [29] | 2020 | 50 images (25 Covid-19 and 25 Healthy) | Multi-Criteria Decision Making (Naive Bayes, Neural Network, Support Vector Machine, Radial Basis Function, k-Nearest Neighbors, Stochastic Gradient Descent, Random Forests, Decision Tree, AdaBoost, CN2 Rule Inducer Algorithm) | Unspecified | Sen: 0,706-0,974; Spe: 0,557-1,000; Acc: 0,620-0,987; F-1 Score: 0,555–0,987; AUC: 0,800-0,988; Time: 0,14–7,57 s. |
| Khan et al. [30] | 2020 | 594 images (284 Covid-19 and 310 Healthy) | Convolutional Neural Network (CoroNet (Xception)) | 4-fold | Sen: 0,993; Spe: 0,986; Acc: 0,990; F-1 Score: 0,985 |
| Apostolopoulos and Mpesiana [31] | 2020 | 728 images (224 Covid-19 and 504 Normal) | Transfer Learning with Convolutional Neural Networks (VGG19, MobileNet v2, Inception, Xception, Inception ResNet v2) | 10-fold | Sen: 0,9866; Spe: 0,9646; Acc: 0,9678 |
| Waheed et al. [32] | 2020 | 1.124 images (403 Covid-19 and 721 Healthy) | Convolutional Neural Network (VGG-16) and Synthetic Data Augmentation | Train: 932 (331 Covid-19 and 601 Healthy); Test: 192 (72 Covid-19 and 120 Healthy) | Sen: 0,69-0,90; Spe: 0,95-0,97; Acc: 0,85-0,95 |
| Mahmud et al. [33] | 2020 | 610 images (305 Covid-19 and 305 Healthy) | Transfer Learning with Convolutional Neural Networks (Stacked Multi-Resolution CovXNet) | 5-fold | Sen: 0,978; Spe: 0,947; Acc: 0,974; F-1 Score: 0,971; AUC: 0,969 |
| Vaid et al. [34] | 2020 | 545 images (181 Covid-19 and 364 Healthy) | Convolutional Neural Network (VGG-19) and Trainable Fully Connected Layers | Train: 348 (115 Covid-19 and 233 Healthy); Validation: 88 (32 Covid-19 and 56 Healthy); Test: 109 (34 Covid-19 and 75 Healthy) | Sen: 0,9863; Spe: 0,9166; Acc: 0,9633; F-1 Score: 0,9729 |
| Benbrahim et al. [35] | 2020 | 320 images (160 Covid-19 and 160 Healthy) | Transfer Learning with Convolutional Neural Networks (Inceptionv3 and ResNet50) | 70% Train-30% Test | Sen: 0,9803-0,9811; Acc: 0,9803-0,9901; F-1 Score: 0,9803-0,9901 |
| Elaziz et al. [36] | 2020 | Dataset-1: 1.891 images (216 Covid-19 and 1.675 Healthy); Dataset-2: 1.560 images (219 Covid-19 and 1.341 Healthy) | Fractional Multichannel Exponent Moments, Manta-Ray Foraging Optimization and KNN classifier | 80% Train-20% Test | Sen: 0,9875-0,9891; Acc: 0,9609-0,9809 |
| Martínez et al. [37] | 2020 | 240 images (120 Covid-19 and 120 Healthy) | Convolutional Neural Network (Neural Architecture Search Network (NASNet)) | 70% Train-30% Test | Sen: 0,97; Acc: 0,97; F-1 Score: 0,97 |
| Loey et al. [38] | 2020 | 148 images (69 Covid-19 and 79 Healthy) | Transfer Learning with Convolutional Neural Networks (Alexnet, Googlenet, and Resnet18) | Train: 130 (60 Covid-19 and 70 Healthy); Test: 18 (9 Covid-19 and 9 Healthy) | Sen: 1,000; Spe:1,000; Acc: 1,000 |
| Toraman et al. [39] | 2020 | 1.281 images (231 Covid-19 and 1.050 Healthy) | Convolutional Neural Network (CapsNet) | 10-fold | Sen: 0,28-0,9742; Spe:0,8095–0,98; Acc: 0,4914-0,9724; F-1 Score: 0,55-0,9724; Time: 16–500 s. (Note: The results show the average fold.) |
| Duran-Lopez et al. [40] | 2020 | 6.926 images (2.589 Covid-19 and 4.337 Healthy) | Convolutional Neural Network | 5-fold | Sen: 0,9253; Spe:0,9633; Acc: 0,9443; F-1 Score: 0,9314; AUC: 0,988 |
| Minaee et al. [41] | 2020 | 5.184 images (184 Covid-19 and 5.000 Healthy) | Transfer Learning with Convolutional Neural Networks (ResNet18, ResNet50, SqueezeNet, and DenseNet-121) | Train: 2.084 (84 Covid-19 and 2.000 Healthy); Test: 3.100 (100 Covid-19 and 3.000 Healthy) | Sen: 0,98; Spe:0,751-0,929 |
CT imaging generally contains more data than X-ray imaging. However, it has some disadvantages for the follow-up of all stages of the disease due to the excess amount of radiation that the patients are exposed to. For this reason, an artificial intelligence application using X-ray images was created and tested in the study.
In this study, which aims at early diagnosis of Covid-19 disease with the help of X-ray images, a deep learning approach, which is an artificial intelligence method applying the latest technology, was used. In this context, automatic classification of the images was carried out through the two different convolutional neural networks (CNNs). In the study, experiments were carried out for the use of images directly, using local binary pattern (LBP) as a pre-process and dual tree complex wavelet transform (DT-CWT) as a secondary operation, and the results of the automatic classification were calculated separately. Within the scope of the study, four new classification approaches that involve performing the experiments together and combining the results through a result generation algorithm, have been proposed and tested. The results of the study show that in the diagnosis of Covid-19 disease, the analysis of chest X-ray images using deep learning methods provides fast and highly accurate results.
Methods
Used Data
The chest X-ray images of patients with Covid-19 used in the study were obtained by combining metadata data sets that were made open access over GitHub after being created by Cohen et al. [42] and over Kaggle after being created by Dadario [43]. The images that these data sets contain in common and the clinical notes related to these images were combined and a mixed Covid-19 image data set consisting of 150 chest X-ray images was created. In the study, images obtained while the patients were facing the X-ray device directly were used. In the studies, the images taken from the same patient were obtained on different days of the course of the disease and therefore do not contain exactly the same content. The dimensions of the images in question vary between 255 px × 249 px and 4280 px × 3520 px (px is pixel abbreviation) and show a wide variety. Also, these images have different data formats such as png, jpg, jpeg and two different bit depths such as 8-bit (gray-level) and 24-bit (RGB). Standardization of the images is an essential process for use in this study. In this context, all of the images have been converted to 8-bit gray-level images. Then, to clarify the area of interest on the images, manual framing was performed so as to cover the chest area. After this process, all the images were rearranged to 448 px × 448 px and saved in png format.
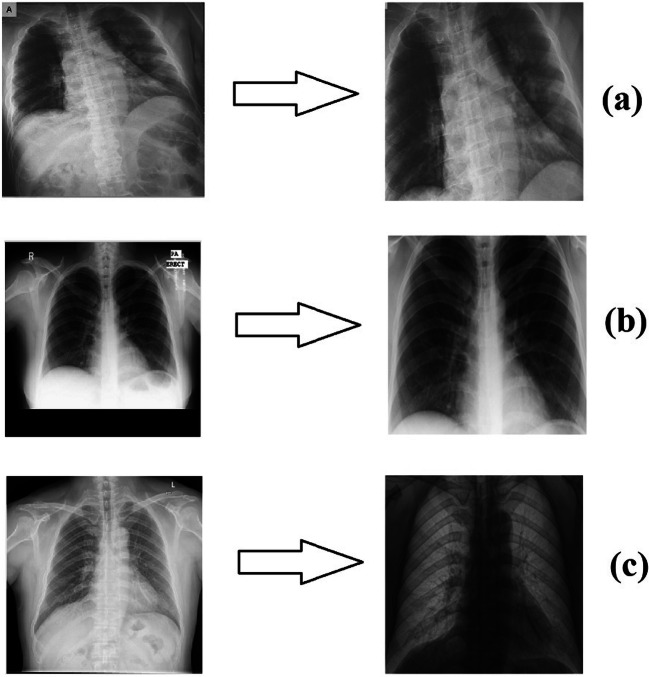
For the non-Covid-19 X-ray images in the study, two data sets, a Montgomery data set [44] and a Shenzhen data set [44], were used separately. These databases contain 80 and 326 non-Covid-19 X-ray images, respectively. The first training-test data set contains a total of 230 X-ray images, of which 150 are Covid-19 images and 80 are non-Covid-19 images, while the second training-test data set contains 476 X-ray images, of which 150 are Covid-19 images and 326 are non-Covid-19 images. Thus, it was ensured that classification results were obtained for the two data sets that contained predominantly Covid-19 images and predominantly non-Covid-19 images, respectively. The processes applied to the Covid-19 images were likewise applied to the non-Covid-19 images. In Fig. 1, original and edited versions of the X-ray images are shown; one belonging to a patient with Covid-19 and two belonging to people without Covid-19 (non-Covid-19 people).
Fig. 1.
a) X-ray image of a patient with Covid-19 (Phan et al. [23]) b) Non-Covid-19 X-ray image (Montgomery data set [44])) c) Non-Covid-19 X-ray image (Shenzhen data set [44]))
Local Binary Pattern (LBP)
Local binary pattern (LBP) is an approach that was proposed by Ojala et al. [45] to reveal local features. The method is basically based on comparing a pixel on the image to the neighboring pixels one by one, in terms of size.
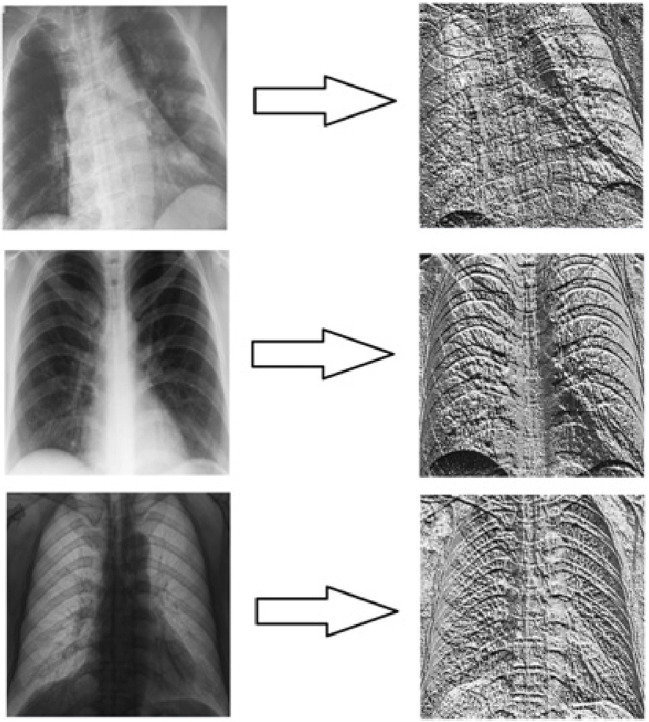
In Fig. 2, the images obtained by applying the LBP operation to the X-ray images given in Fig. 1 are included. The purpose of benefiting from LBP operation within the scope of this study is to observe the effects of using LBP images, which reflect the local features in the CNN input on the study results, rather than the original images. Additionally, the aim of the study is to increase the image feature depth used in the new result generation algorithm.
Fig. 2.
Images created by applying LBP and resizing the images in Fig. 1
Dual Tree Complex Wavelet Transform (DT-CWT)
Dual tree complex wavelet transform (DT-CWT) was first introduced by Kingsbury [46–48]. This method is generally similar to the Gabor wavelet transform. In the Gabor wavelet transform, low-pass and high-pass filters are applied to the rows and columns of the image horizontally and vertically. In this way, two different sub-band groups are formed in rows and columns as low (L) and high (H). Crossing is made during the conversion of the said one-dimensional bands into two dimensions. At the end of the process, a low sub-band, named LL, is obtained. In addition, three sub-bands containing high bands, LH, HL, and HH, are formed. Further sub-bands (such as LLL, LLH) can be obtained by applying the same operations to the LL sub-band.
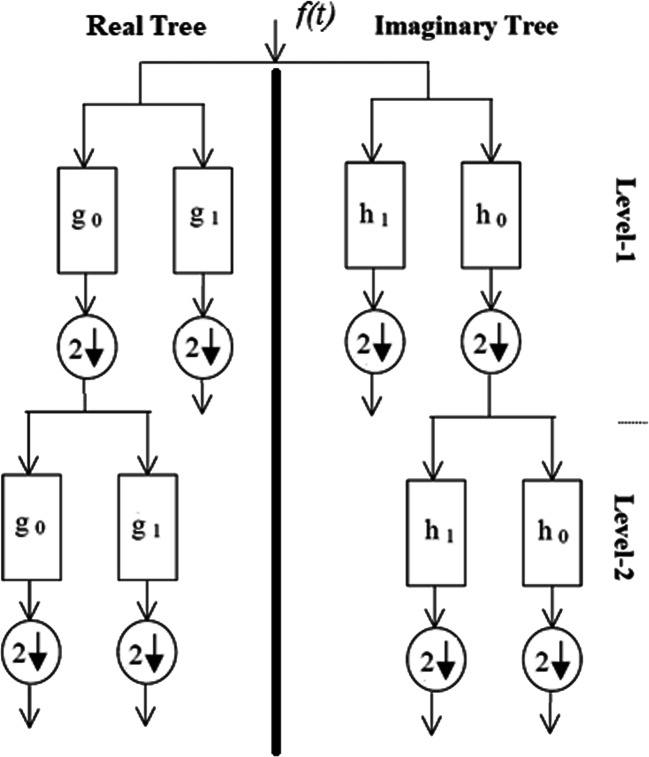
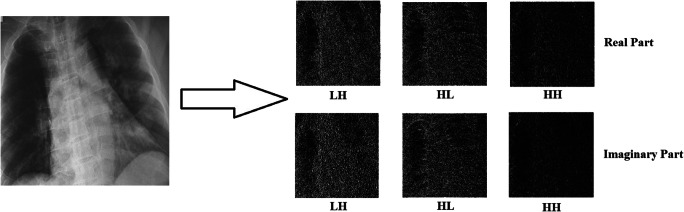
Unlike the Gabor wavelet transform, instead of a single filter, DT-CWT uses two filters that work in parallel. These two trees contain real and imaginary parts of complex numbers. That is, as a result of the DT-CWT process, a sub-band containing more directions than the Gabor wavelet transform is obtained. When DT-CWT is applied to an image, the processes are performed for six different directions, +15, −15, +45, −45, +75, and − 75 degrees. Three of these directions represent real sub-bands and the other three represent imaginary sub-bands. Figure 3 shows the DT-CWT decomposition tree. In Fig. 4, real and imaginary sub-band images obtained by applying the DT-CWT process (scale = 1) to the X-ray images given in Fig. 1, are shown. Within the scope of the study, the DT-CWT process was used with a scale (level) value of 1, and the dimensions of the sub-band images obtained were half the size of the original images. Since the complex wavelet transform has been successful in many studies [49–51] where medical images have previously been used, this conversion was preferred in the study.
Fig. 3.
Structure of the DT-CWT decomposition tree
Fig. 4.
Real and imaginary sub-band images obtained by applying DT-CWT to the X-ray Image (scale = 1)
Convolutional Neural Network (CNN)
Deep learning has come to the fore in recent years as an artificial intelligence approach that provides successful results in many image processing applications from image enhancement (such as [52]) to object identification (such as [53, 54]).
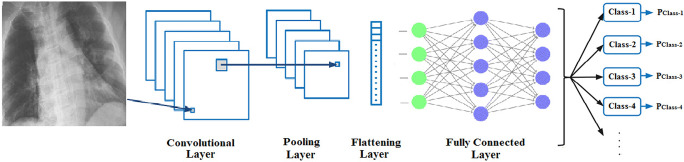
Convolutional neural network (CNN) has been the preferred deep learning model in image processing applications in recent years. The CNN classifier, in general, consists of a convolution layer, activation functions, a pooling layer, a flatten layer, and fully connected layer components. In this context, Fig. 5 describes the general operation of the CNN classifier. It is possible to examine more detailed information about the functions and operating modes of the layers in the CNN classifier from the studies [55–59].
Fig. 5.
General operation of the CNN classifier
Within the scope of the study, a CNN architecture with a total of 23 layers was designed. An effective design was aimed at, since increasing the number of layers in the CNN architecture leads to increased processing time in the training and classification processes. Table 2 contains details of the first CNN architecture used in the study. Also, a second CNN architecture was used to check whether the proposed pipeline approaches applied to other CNN architectures. In this context, an architecture modeled on VGG-16 CNN was used. However, to reduce the processing load, the number of filters and the fully connected layer sizes have been reduced. Additionally, normalization layers were added after the intermediate convolution layers. Details of this second CNN architecture used are given in Table 3.
Table 2.
First CNN architecture used within the scope of the study
| Layer | Layer Name | Layer Parameters (Matlab) |
|---|---|---|
| 1 | imageInputLayer | [448,448 1], [224,224 1], [224,224 2], [224,224 3] and [224,224 6] |
| 2 | convolution2dLayer | (3,4,'Padding’,'same’) |
| 3 | batchNormalizationLayer | default |
| 4 | reluLayer | default |
| 5 | maxPooling2dLayer | (2,'Stride’,2) |
| 6 | convolution2dLayer | (3,8,'Padding’,'same’) |
| 7 | batchNormalizationLayer | default |
| 8 | reluLayer | default |
| 9 | maxPooling2dLayer | (2,'Stride’,2) |
| 10 | convolution2dLayer | (3,16,'Padding’,'same’) |
| 11 | batchNormalizationLayer | default |
| 12 | reluLayer | default |
| 13 | maxPooling2dLayer | (2,'Stride’,2) |
| 14 | convolution2dLayer | (3,32,'Padding’,'same’) |
| 15 | batchNormalizationLayer | default |
| 16 | reluLayer | default |
| 17 | maxPooling2dLayer | (2,'Stride’,2) |
| 18 | convolution2dLayer | (3,64,'Padding’,'same’) |
| 19 | batchNormalizationLayer | default |
| 20 | reluLayer | default |
| 21 | fullyConnectedLayer | 2 |
| 22 | softmaxLayer | default |
| 23 | classificationLayer | default |
Table 3.
Second CNN architecture used within the scope of the study
| Layer | Layer Name | Layer Parameters (Matlab) |
|---|---|---|
| 1 | imageInputLayer | [448,448 1], [224,224 1], [224,224 2], [224,224 3] and [224,224 6] |
| 2 | convolution2dLayer | (3,8,'Padding’,'same’) |
| 3 | batchNormalizationLayer | default |
| 4 | reluLayer | default |
| 5 | convolution2dLayer | (3,8,'Padding’,'same’) |
| 6 | batchNormalizationLayer | default |
| 7 | reluLayer | default |
| 8 | maxPooling2dLayer | (2,'Stride’,2) |
| 9 | convolution2dLayer | (3,16,'Padding’,'same’) |
| 10 | batchNormalizationLayer | default |
| 11 | reluLayer | default |
| 12 | convolution2dLayer | (3,16,'Padding’,'same’) |
| 13 | batchNormalizationLayer | default |
| 14 | reluLayer | default |
| 15 | maxPooling2dLayer | (2,'Stride’,2) |
| 16 | convolution2dLayer | (3,32,'Padding’,'same’) |
| 17 | batchNormalizationLayer | default |
| 18 | reluLayer | default |
| 19 | convolution2dLayer | (3,32,'Padding’,'same’) |
| 20 | batchNormalizationLayer | default |
| 21 | reluLayer | default |
| 22 | convolution2dLayer | (3,32,'Padding’,'same’) |
| 23 | batchNormalizationLayer | default |
| 24 | reluLayer | default |
| 25 | maxPooling2dLayer | (2,'Stride’,2) |
| 26 | convolution2dLayer | (3,64,'Padding’,'same’) |
| 27 | batchNormalizationLayer | default |
| 28 | reluLayer | default |
| 29 | convolution2dLayer | (3,64,'Padding’,'same’) |
| 30 | batchNormalizationLayer | default |
| 31 | reluLayer | default |
| 32 | convolution2dLayer | (3,64,'Padding’,'same’) |
| 33 | batchNormalizationLayer | default |
| 34 | reluLayer | default |
| 35 | maxPooling2dLayer | (2,'Stride’,2) |
| 36 | convolution2dLayer | (3,64,'Padding’,'same’) |
| 37 | batchNormalizationLayer | default |
| 38 | reluLayer | default |
| 39 | convolution2dLayer | (3,64,'Padding’,'same’) |
| 40 | batchNormalizationLayer | default |
| 41 | reluLayer | default |
| 42 | convolution2dLayer | (3,64,'Padding’,'same’) |
| 43 | batchNormalizationLayer | default |
| 44 | reluLayer | default |
| 45 | maxPooling2dLayer | (2,'Stride’,2) |
| 46 | fullyConnectedLayer | 512 |
| 47 | reluLayer | default |
| 48 | dropoutLayer | 0,5 |
| 49 | fullyConnectedLayer | 512 |
| 50 | reluLayer | default |
| 51 | dropoutLayer | 0,5 |
| 52 | fullyConnectedLayer | 2 |
| 53 | softmaxLayer | default |
| 54 | classificationLayer | default |
In the context of the study, Matlab 2019a program was preferred as software. The layer names and parameters in Tables 2 and 3 are the names and parameters used directly in the software. In the study, more than one experiment was carried out and the sizes of the input images used in the experiments differ. For this reason, there are different sizes in the input layer in Tables 2 and 3. Those CNN architectures were used in all the experiments carried out within the scope of the study.
Evaluation Criteria of the Classification Results
Within the scope of the study, confusion matrix and statistical parameters obtained from this matrix were used to evaluate the results. It is possible to examine detailed information about the confusion matrix, i.e., sensitivity (SEN), specificity (SPE), accuracy (ACC), and F-1 score (F-1), from the studies [60].
Receiver operating characteristic (ROC) analysis was also used to evaluate the results. In addition, the sizes of the areas under the ROC curve (Area Under Curve (AUC)) were calculated. ROC analysis basically reflects graphically the variation of sensitivity (SEN) (y-axis) relative to 1-SPE (x-axis) for the case that the threshold value is gradually changed with a certain precision between the minimum and maximum output predicted for the classification.
Pipeline Methodology
First of all, in the proposed pipeline algorithm, training and test procedures for images of size of 448 × 448 were performed and results were obtained.
Before the experiments after the first experiment were conducted, DT-CWT was applied to the images of size of 448 px × 448 px (scale = 1) and 224 px × 224 px sub-band images were obtained.
In the second experiment, results were obtained for the case of giving the real part of the LL sub-band image obtained by applying DT-CWT, as input to the CNN.
In the third experiment, training and testing procedures were carried out and results were obtained for the case of giving the imaginary part of the LL sub-band image obtained by applying DT-CWT, as input to the CNN.
In the fourth experiment, training and testing procedures were carried out and results were obtained for the case of giving the real parts of LL, LH and HL sub-band images obtained by applying DT-CWT as input to the CNN, together.
In the fifth experiment, training and test procedures were carried out and results were obtained for the case of giving the imaginary parts of the LL, LH and HL sub-band images obtained by applying DT-CWT as input to the CNN, together.
In the sixth experiment, results were obtained for the case of giving the real and imaginary parts of the LL sub-band image obtained by applying DT-CWT as input to the CNN, together.
In the seventh experiment, results were obtained for the case of giving the real and imaginary parts of the LL, LH, HL sub-band images obtained by applying DT-CWT, as input to the CNN, together.
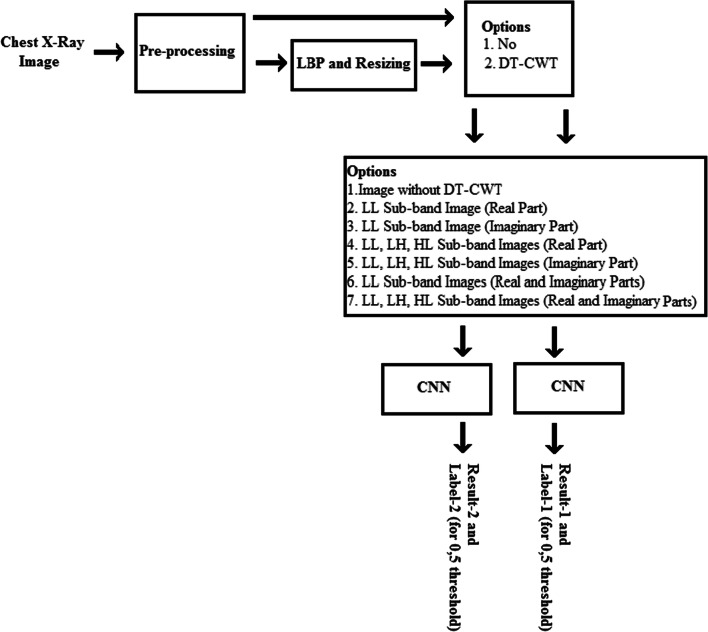
A block diagram of the experiments carried out in the study is shown in Fig. 6. The first seven experiments conducted were repeated using new images obtained by applying LBP to the X-ray images, and the first stage experiments were completed. Since the image size decreases after LBP processing, these images were rearranged as 448 px × 448 px in size.
Fig. 6.
Block diagram representation of the study of the experiments
In the ongoing part of the study, four pipeline classification algorithms were designed using the principle of parallel operation. These algorithms are based on combining the results of previous experiments to obtain new results. The first two pipeline classification algorithms mentioned above work as follows:
If the numbers of labeling (threshold value for 0,5) obtained in the experiments (with and without LBP) for an image are not equal to each other, the labeling result obtained in more than half of the experiments is considered to be the algorithm labeling result for Covid-19 or non-Covid-19. In this case, if the algorithm labeling result is Covid-19, the real experiment result is the closest to the number 0; if the algorithm labeling result is non-Covid-19, the closest to the number 1, respectively, is assigned as the algorithm result.
If the numbers of labeling (for threshold value 0,5) obtained in the experiments (with and without LBP) for an image are equal to each other, the actual test results obtained in the experiments conducted for the image are mixed (50%–50% and 75%–25%, respectively), and the result is accepted as the algorithm result. After that, the labeling of the image is realized as Covid-19 or non-Covid-19 (for the threshold value 0,5) according to this result obtained.
The basic coding of the first two pipeline classification approaches is included in Table 4. In the codes between Tables 4 and 6, Result-1 and Label-1 represent the actual test result and the label obtained without using LBP, while Result-2 and Label-2 represent the actual test result and the label obtained using LBP.
Table 4.
Basic coding of the pipeline algorithms (pipeline-1 and -2) proposed in the study
Table 6.
Basic coding of the pipeline algorithm (pipeline-4) proposed in the study
In the third and fourth pipeline algorithms, unlike the first two pipeline algorithms, if the tags obtained as a result of the classification experiment differ from each other, the result obtained without applying LBP has been taken into consideration with priority. Accordingly, in the case where the two classification tags are different from each other in the third pipeline algorithm, if the tag result obtained without applying LBP was abnormal, the result was considered abnormal. In the fourth pipeline algorithm, in the case of the two classification tags being different from each other, if the tag result obtained without applying LBP was normal, the result was considered normal. The other procedures are the same as for the first two pipeline algorithms. A mixing rate of 50% -50% was applied in the third and fourth pipeline algorithms. The basic coding of the third and fourth pipeline classification approaches is given in Tables 5 and 6.
Table 5.
Basic coding of the pipeline algorithm (pipeline-3) proposed in the study
Results
Experiments
In this study, which aims to detect Covid-19 disease early using X-ray images, the deep learning approach, which is the artificial intelligence method applying the latest technology, was used and automatic classification of the images was performed using CNN. In the first training-test data set used in the study, there were 230 X-ray images, of which 150 were Covid-19 and 80 were non-Covid-19, while in the second training-test data set there were 476 X-ray images, of which 150 were Covid-19 and 326 were non-Covid-19. Thus, it was ensured that the classification results were obtained separately from the two data sets containing predominantly abnormal images and predominantly normal images. The information from the training-test data sets is given in Table 7.
Table 7.
Information about the images used in the study
Within the scope of the study, chest X-ray images were manually framed to cover the lung region, primarily to determine the areas of interest on the image. Then, standardization was carried out since the images used were of very different sizes, formats, and bit depths. The areas of interest on the image were resized and the image sizes were arranged as 448 px × 448 px. After that, the images in question were saved in png format so as to be as gray-scale and 8-bit depth. These operations were applied to all the abnormal and normal images used in the study.
In the ongoing part of the study, a 23-layer CNN architecture and a 54-layer CNN architecture were designed and used, the details of which have been previously described. Those CNN architectures were used in all the experiments. Due to the fact that more than one experiment was performed within the scope of the study, only the images given to the CNN input differ in size.
In the experiments conducted in the study, the trainings were carried out with the k-fold cross validation method. In this context, the k value was chosen as 23. Since the first training-test data set consists of 230 images, 220 images, except for ten images at each stage (fold), were used for the training operations, and the remaining ten images were used for the testing operations. The second training-test data set consists of 476 images, and, in the same way, except 20/21 (16 groups consisting of 21 images and seven groups of 20 images) images, 456/455 images were used in the training operations, and the remaining 20/21 images were used in the testing operations. The test procedures were repeated 23 times and classification results were obtained for all the images.
Finally, within the scope of the study, all the images were combined and the training and testing procedures were repeated by applying a 2-fold cross for a total of 556 X-ray images comprising 150 Covid-19 images and 406 non-Covid-19 images. Considering the length of the study as well, the results that have been shared in the study are only for the input data that provided the best results for the first and second data sets.
In this part of the study, a total of 14 experiments were carried out. Some initial weights and parameters in the CNN are randomly assigned. To make the study results stable, each experiment was repeated five times in itself, and average results in the study are shown.
Within the scope of the study, the CPU time taken for an experiment to be completed entirely, including the training and testing, was divided by the total number of images processed, and the processing CPU time per image was measured. The experiments of this study were carried out using MATLAB 2019 (a) software running on a computer with 64 GB RAM and Intel(R) Xeon (R) CPU E5–2680 2.7 GHz (32 CPUs).
Results
In the first experimental group within the scope of the study, the training and testing procedures were first performed using the chest X-ray images, and the results were obtained. LBP operation was then applied to the images in question, and then the training and testing procedures were repeated and the results were calculated. Finally, the results were calculated using the pipeline classification algorithms, the details of which were previously described and proposed within the scope of the study. Due to the random assignment of some initial variables used in the internal structure of the CNN, each experiment group was repeated five times in order to make the results more stable. The image sizes given to the CNN as input for this experiment were 448 × 448 × 1. The results obtained from the experimental group are given in Table 8 (first training-test data set) and Table 9 (second training-test data set).
Table 8.
Results obtained directly using chest X-ray images (first training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 146,2 | 3,8 | 76,8 | 3,2 | 0,9747 | 0,9600 | 0,9696 | 0,9766 | 0,9961 | 18,964 |
| With LBP | 147,8 | 2,2 | 71,6 | 8,4 | 0,9853 | 0,8950 | 0,9539 | 0,9655 | 0,9939 | 18,226 | |
| Pipeline-1 | 148,6 | 1,4 | 76,2 | 3,8 | 0,9907 | 0,9525 | 0,9774 | 0,9828 | 0,9968 | 37,190 | |
| Pipeline-2 | 146,8 | 3,2 | 76,6 | 3,4 | 0,9787 | 0,9575 | 0,9713 | 0,9780 | 0,9968 | 37,190 | |
| Pipeline-3 | 148,8 | 1,2 | 75,8 | 4,2 | 0,9920 | 0,9475 | 0,9765 | 0,9822 | 0,9968 | 37,190 | |
| Pipeline-4 | 146,0 | 4,0 | 77,2 | 2,8 | 0,9733 | 0,9650 | 0,9704 | 0,9772 | 0,9969 | 37,190 | |
| Second CNN Architecture | Without LBP | 144,2 | 5,8 | 76,6 | 3,4 | 0,9613 | 0,9575 | 0,9600 | 0,9691 | 0,9949 | 33,105 |
| With LBP | 142,6 | 7,4 | 74,0 | 6,0 | 0,9507 | 0,9250 | 0,9417 | 0,9551 | 0,9815 | 36,433 | |
| Pipeline-1 | 147,2 | 2,8 | 78,4 | 1,6 | 0,9813 | 0,9800 | 0,9809 | 0,9853 | 0,9977 | 69,538 | |
| Pipeline-2 | 144,4 | 5,6 | 77,4 | 2,6 | 0,9627 | 0,9675 | 0,9643 | 0,9724 | 0,9973 | 69,538 | |
| Pipeline-3 | 148,0 | 2,0 | 76,6 | 3,4 | 0,9867 | 0,9575 | 0,9765 | 0,9821 | 0,9972 | 69,538 | |
| Pipeline-4 | 143,4 | 6,6 | 78,4 | 1,6 | 0,9560 | 0,9800 | 0,9643 | 0,9722 | 0,9958 | 69,538 |
Table 9.
Results obtained directly using chest X-ray images (second training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 144,4 | 5,6 | 322,0 | 4,0 | 0,9627 | 0,9877 | 0,9798 | 0,9678 | 0,9975 | 24,878 |
| With LBP | 137,2 | 12,8 | 316,4 | 9,6 | 0,9147 | 0,9706 | 0,9529 | 0,9246 | 0,9899 | 24,995 | |
| Pipeline-1 | 146,0 | 4,0 | 322,0 | 4,0 | 0,9733 | 0,9877 | 0,9832 | 0,9733 | 0,9982 | 49,872 | |
| Pipeline-2 | 146,0 | 4,0 | 321,8 | 4,2 | 0,9733 | 0,9871 | 0,9828 | 0,9726 | 0,9983 | 49,872 | |
| Pipeline-3 | 147,0 | 3,0 | 321,0 | 5,0 | 0,9800 | 0,9847 | 0,9832 | 0,9735 | 0,9981 | 49,872 | |
| Pipeline-4 | 143,4 | 6,6 | 323,0 | 3,0 | 0,9560 | 0,9908 | 0,9798 | 0,9676 | 0,9979 | 49,872 | |
| Second CNN Architecture | Without LBP | 146,0 | 4,0 | 323,0 | 3,0 | 0,9733 | 0,9908 | 0,9853 | 0,9766 | 0,9977 | 51,817 |
| With LBP | 133,8 | 16,2 | 315,6 | 10,4 | 0,8920 | 0,9681 | 0,9441 | 0,9096 | 0,9885 | 54,239 | |
| Pipeline-1 | 148,0 | 2,0 | 322,6 | 3,4 | 0,9867 | 0,9896 | 0,9887 | 0,9821 | 0,9987 | 10,6056 | |
| Pipeline-2 | 147,0 | 3,0 | 323,2 | 2,8 | 0,9800 | 0,9914 | 0,9878 | 0,9806 | 0,9988 | 10,6056 | |
| Pipeline-3 | 148,8 | 1,2 | 322,0 | 4,0 | 0,9920 | 0,9877 | 0,9891 | 0,9828 | 0,9991 | 10,6056 | |
| Pipeline-4 | 145,2 | 4,8 | 323,6 | 2,4 | 0,9680 | 0,9926 | 0,9849 | 0,9758 | 0,9975 | 10,6056 |
In the second experimental group within the scope of the study, the training and testing procedures were performed using the real part of the LL sub-image obtained by applying DT-CWT to the chest X-ray images, and the results were obtained. Then, the training and testing procedures were performed using the real part of the LL sub-image obtained by applying the LBP and DT-CWT operations to the X-ray images, respectively. Finally, the results were calculated using the pipeline classification algorithms, the details of which were previously described and proposed within the scope of the study. The image sizes given to the CNN as input for this experiment were 224 × 224 × 1. The results obtained from the experimental group are given in Table 10 (first training-test data set) and Table 11 (second training-test data set).
Table 10.
Results obtained by using the LL real sub-band obtained by applying DT-CWT to the chest X-ray images (first training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 146,6 | 3,4 | 77,4 | 2,6 | 0,9773 | 0,9675 | 0,9739 | 0,9800 | 0,9983 | 0,6090 |
| With LBP | 146,0 | 4,0 | 72,6 | 7,4 | 0,9733 | 0,9075 | 0,9504 | 0,9624 | 0,9890 | 0,6252 | |
| Pipeline-1 | 149,0 | 1,0 | 77,4 | 2,6 | 0,9933 | 0,9675 | 0,9843 | 0,9881 | 0,9988 | 12,342 | |
| Pipeline-2 | 147,0 | 3,0 | 77,4 | 2,6 | 0,9800 | 0,9675 | 0,9757 | 0,9813 | 0,9989 | 12,342 | |
| Pipeline-3 | 149,2 | 0,8 | 76,6 | 3,4 | 0,9947 | 0,9575 | 0,9817 | 0,9862 | 0,9990 | 12,342 | |
| Pipeline-4 | 146,4 | 3,6 | 78,2 | 1,8 | 0,9760 | 0,9775 | 0,9765 | 0,9819 | 0,9986 | 12,342 | |
| Second CNN Architecture | Without LBP | 141,8 | 8,2 | 77,8 | 2,2 | 0,9453 | 0,9725 | 0,9548 | 0,9646 | 0,9934 | 12,146 |
| With LBP | 141,2 | 8,8 | 63,6 | 16,4 | 0,9413 | 0,7950 | 0,8904 | 0,9183 | 0,9580 | 12,339 | |
| Pipeline-1 | 146,6 | 3,4 | 77,6 | 2,4 | 0,9773 | 0,9700 | 0,9748 | 0,9806 | 0,9963 | 24,485 | |
| Pipeline-2 | 144,0 | 6,0 | 78,0 | 2,0 | 0,9600 | 0,9750 | 0,9652 | 0,9729 | 0,9962 | 24,485 | |
| Pipeline-3 | 147,4 | 2,6 | 77,2 | 2,8 | 0,9827 | 0,9650 | 0,9765 | 0,9820 | 0,9961 | 24,485 | |
| Pipeline-4 | 141,0 | 9,0 | 78,2 | 1,8 | 0,9400 | 0,9775 | 0,9530 | 0,9631 | 0,9944 | 24,485 |
Table 11.
Results obtained by using the LL real sub-band obtained by applying DT-CWT to the chest X-ray images (second training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 143,6 | 6,4 | 320,8 | 5,2 | 0,9573 | 0,9840 | 0,9756 | 0,9611 | 0,9973 | 0,7571 |
| With LBP | 135,2 | 14,8 | 311,2 | 14,8 | 0,9013 | 0,9546 | 0,9378 | 0,9013 | 0,9865 | 0,7569 | |
| Pipeline-1 | 146,4 | 3,6 | 322,2 | 3,8 | 0,9760 | 0,9883 | 0,9845 | 0,9753 | 0,9978 | 15,140 | |
| Pipeline-2 | 145,0 | 5,0 | 321,6 | 4,4 | 0,9667 | 0,9865 | 0,9803 | 0,9685 | 0,9979 | 15,140 | |
| Pipeline-3 | 146,8 | 3,2 | 320,4 | 5,6 | 0,9787 | 0,9828 | 0,9815 | 0,9709 | 0,9981 | 15,140 | |
| Pipeline-4 | 143,2 | 6,8 | 322,6 | 3,4 | 0,9547 | 0,9896 | 0,9786 | 0,9656 | 0,9973 | 15,140 | |
| Second CNN Architecture | Without LBP | 146,0 | 4,0 | 322,0 | 4,0 | 0,9733 | 0,9877 | 0,9832 | 0,9733 | 0,9983 | 15,029 |
| With LBP | 131,8 | 18,2 | 305,4 | 20,6 | 0,8787 | 0,9368 | 0,9185 | 0,8718 | 0,9770 | 18,120 | |
| Pipeline-1 | 147,6 | 2,4 | 321,6 | 4,4 | 0,9840 | 0,9865 | 0,9857 | 0,9775 | 0,9977 | 33,149 | |
| Pipeline-2 | 146,6 | 3,4 | 322,4 | 3,6 | 0,9773 | 0,9890 | 0,9853 | 0,9767 | 0,9980 | 33,149 | |
| Pipeline-3 | 148,2 | 1,8 | 320,8 | 5,2 | 0,9880 | 0,9840 | 0,9853 | 0,9770 | 0,9984 | 33,149 | |
| Pipeline-4 | 145,4 | 4,6 | 322,8 | 3,2 | 0,9693 | 0,9902 | 0,9836 | 0,9739 | 0,9975 | 33,149 |
In the third experimental group within the scope of the study, the training and testing procedures were performed using the imaginary part of the LL sub-image obtained by applying DT-CWT to the chest X-ray images, and the results were obtained. Then, the training and testing procedures were performed using the imaginary part of the LL sub-image obtained by applying the LBP and DT-CWT operations to the X-ray images, respectively. Finally, the results were calculated using the pipeline classification algorithms, the details of which were previously described and proposed within the scope of the study. The image sizes given to the CNN as input for this experiment were 224 × 224 × 1. The results obtained from the experimental group are given in Table 12 (first training-test data set) and Table 13 (second training-test data set).
Table 12.
Results obtained by using the LL imaginary sub-band obtained by applying DT-CWT to the chest X-ray images (first training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 146,4 | 3,6 | 77,4 | 2,6 | 0,9760 | 0,9675 | 0,9730 | 0,9792 | 0,9972 | 0,6033 |
| With LBP | 145,6 | 4,4 | 73,6 | 6,4 | 0,9707 | 0,9200 | 0,9530 | 0,9643 | 0,9889 | 0,5979 | |
| Pipeline-1 | 147,8 | 2,2 | 77,0 | 3,0 | 0,9853 | 0,9625 | 0,9774 | 0,9827 | 0,9980 | 12,012 | |
| Pipeline-2 | 146,6 | 3,4 | 77,4 | 2,6 | 0,9773 | 0,9675 | 0,9739 | 0,9799 | 0,9981 | 12,012 | |
| Pipeline-3 | 148,2 | 1,8 | 76,4 | 3,6 | 0,9880 | 0,9550 | 0,9765 | 0,9821 | 0,9985 | 12,012 | |
| Pipeline-4 | 146,0 | 4,0 | 78,0 | 2,0 | 0,9733 | 0,9750 | 0,9739 | 0,9799 | 0,9977 | 12,012 | |
| Second CNN Architecture | Without LBP | 141,0 | 9,0 | 77,2 | 2,8 | 0,9400 | 0,9650 | 0,9487 | 0,9599 | 0,9803 | 12,156 |
| With LBP | 143,6 | 6,4 | 60,2 | 19,8 | 0,9573 | 0,7525 | 0,8861 | 0,9167 | 0,9645 | 12,086 | |
| Pipeline-1 | 146,8 | 3,2 | 75,8 | 4,2 | 0,9787 | 0,9475 | 0,9678 | 0,9754 | 0,9923 | 24,243 | |
| Pipeline-2 | 143,0 | 7,0 | 77,4 | 2,6 | 0,9533 | 0,9675 | 0,9583 | 0,9675 | 0,9909 | 24,243 | |
| Pipeline-3 | 146,8 | 3,2 | 74,8 | 5,2 | 0,9787 | 0,9350 | 0,9635 | 0,9722 | 0,9898 | 24,243 | |
| Pipeline-4 | 141,0 | 9,0 | 78,2 | 1,8 | 0,9400 | 0,9775 | 0,9530 | 0,9631 | 0,9849 | 24,243 |
Table 13.
Results obtained by using the LL imaginary sub-band obtained by applying DT-CWT to the chest X-ray images (second training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 145,0 | 5,0 | 322,0 | 4,0 | 0,9667 | 0,9877 | 0,9811 | 0,9699 | 0,9970 | 0,7587 |
| With LBP | 134,8 | 15,2 | 312,0 | 14,0 | 0,8987 | 0,9571 | 0,9387 | 0,9022 | 0,9861 | 0,7578 | |
| Pipeline-1 | 147,2 | 2,8 | 322,0 | 4,0 | 0,9813 | 0,9877 | 0,9857 | 0,9774 | 0,9981 | 15,164 | |
| Pipeline-2 | 146,0 | 4,0 | 322,2 | 3,8 | 0,9733 | 0,9883 | 0,9836 | 0,9740 | 0,9982 | 15,164 | |
| Pipeline-3 | 147,6 | 2,4 | 321,0 | 5,0 | 0,9840 | 0,9847 | 0,9845 | 0,9755 | 0,9985 | 15,164 | |
| Pipeline-4 | 144,6 | 5,4 | 323,0 | 3,0 | 0,9640 | 0,9908 | 0,9824 | 0,9718 | 0,9969 | 15,164 | |
| Second CNN Architecture | Without LBP | 146,0 | 4,0 | 323,2 | 2,8 | 0,9733 | 0,9914 | 0,9857 | 0,9772 | 0,9981 | 15,022 |
| With LBP | 128,8 | 21,2 | 306,0 | 20,0 | 0,8587 | 0,9387 | 0,9134 | 0,8615 | 0,9773 | 16,851 | |
| Pipeline-1 | 147,4 | 2,6 | 322,6 | 3,4 | 0,9827 | 0,9896 | 0,9874 | 0,9800 | 0,9979 | 31,874 | |
| Pipeline-2 | 146,2 | 3,8 | 323,0 | 3,0 | 0,9747 | 0,9908 | 0,9857 | 0,9773 | 0,9984 | 31,874 | |
| Pipeline-3 | 147,8 | 2,2 | 322,0 | 4,0 | 0,9853 | 0,9877 | 0,9870 | 0,9794 | 0,9983 | 31,874 | |
| Pipeline-4 | 145,6 | 4,4 | 323,8 | 2,2 | 0,9707 | 0,9933 | 0,9861 | 0,9778 | 0,9982 | 31,874 |
In the fourth experimental group within the scope of the study, the training and testing procedures were performed using the real part of the LL, LH and HL sub-images obtained by applying DT-CWT to the chest X-ray images, and the results were obtained. Then, the training and testing procedures were performed using the real part of the LL, LH and HL sub-images obtained by applying the LBP and DT-CWT operations to the X-ray images, respectively. Finally, the results were calculated using the pipeline classification algorithms, the details of which were previously described and proposed within the scope of the study. The image sizes given to the CNN as input for this experiment were 224 × 224 × 3. The results obtained from the experimental group are given in Table 14 (first training-test data set) and Table 15 (second training-test data set).
Table 14.
Results obtained using the LL, LH, HL real sub-bands obtained by applying DT-CWT to the chest X-ray images (first training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 146,2 | 3,8 | 77,8 | 2,2 | 0,9747 | 0,9725 | 0,9739 | 0,9799 | 0,9976 | 10,596 |
| With LBP | 147,4 | 2,6 | 71,6 | 8,4 | 0,9827 | 0,8950 | 0,9522 | 0,9642 | 0,9926 | 10,598 | |
| Pipeline-1 | 148,4 | 1,6 | 77,4 | 2,6 | 0,9893 | 0,9675 | 0,9817 | 0,9860 | 0,9984 | 21,193 | |
| Pipeline-2 | 147,0 | 3,0 | 77,8 | 2,2 | 0,9800 | 0,9725 | 0,9774 | 0,9826 | 0,9984 | 21,193 | |
| Pipeline-3 | 148,4 | 1,6 | 77,0 | 3,0 | 0,9893 | 0,9625 | 0,9800 | 0,9847 | 0,9983 | 21,193 | |
| Pipeline-4 | 146,2 | 3,8 | 78,2 | 1,8 | 0,9747 | 0,9775 | 0,9757 | 0,9812 | 0,9983 | 21,193 | |
| Second CNN Architecture | Without LBP | 142,6 | 7,4 | 76,8 | 3,2 | 0,9507 | 0,9600 | 0,9539 | 0,9642 | 0,9876 | 16,868 |
| With LBP | 141,2 | 8,8 | 64,2 | 15,8 | 0,9413 | 0,8025 | 0,8930 | 0,9201 | 0,9664 | 16,890 | |
| Pipeline-1 | 146,8 | 3,2 | 76,4 | 3,6 | 0,9787 | 0,9550 | 0,9704 | 0,9774 | 0,9948 | 33,757 | |
| Pipeline-2 | 143,8 | 6,2 | 76,8 | 3,2 | 0,9587 | 0,9600 | 0,9591 | 0,9684 | 0,9944 | 33,757 | |
| Pipeline-3 | 147,4 | 2,6 | 75,8 | 4,2 | 0,9827 | 0,9475 | 0,9704 | 0,9775 | 0,9945 | 33,757 | |
| Pipeline-4 | 142,0 | 8,0 | 77,4 | 2,6 | 0,9467 | 0,9675 | 0,9539 | 0,9640 | 0,9903 | 33,757 |
Table 15.
Results obtained using the LL, LH, HL real sub-bands obtained by applying DT-CWT to the chest X-ray images (second training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 145,4 | 4,6 | 322,8 | 3,2 | 0,9693 | 0,9902 | 0,9836 | 0,9739 | 0,9974 | 13,986 |
| With LBP | 132,0 | 18,0 | 309,8 | 16,2 | 0,8800 | 0,9503 | 0,9282 | 0,8852 | 0,9807 | 14,043 | |
| Pipeline-1 | 146,6 | 3,4 | 320,8 | 5,2 | 0,9773 | 0,9840 | 0,9819 | 0,9715 | 0,9979 | 28,029 | |
| Pipeline-2 | 146,4 | 3,6 | 322,6 | 3,4 | 0,9760 | 0,9896 | 0,9853 | 0,9766 | 0,9982 | 28,029 | |
| Pipeline-3 | 147,8 | 2,2 | 320,6 | 5,4 | 0,9853 | 0,9834 | 0,9840 | 0,9749 | 0,9986 | 28,029 | |
| Pipeline-4 | 144,2 | 5,8 | 323,0 | 3,0 | 0,9613 | 0,9908 | 0,9815 | 0,9704 | 0,9970 | 28,029 | |
| Second CNN Architecture | Without LBP | 144,6 | 5,4 | 323,4 | 2,6 | 0,9640 | 0,9920 | 0,9832 | 0,9730 | 0,9985 | 21,551 |
| With LBP | 135,0 | 15,0 | 308,2 | 17,8 | 0,9000 | 0,9454 | 0,9311 | 0,8918 | 0,9828 | 21,720 | |
| Pipeline-1 | 146,6 | 3,4 | 323,0 | 3,0 | 0,9773 | 0,9908 | 0,9866 | 0,9786 | 0,9987 | 43,271 | |
| Pipeline-2 | 145,6 | 4,4 | 323,0 | 3,0 | 0,9707 | 0,9908 | 0,9845 | 0,9752 | 0,9989 | 43,271 | |
| Pipeline-3 | 147,2 | 2,8 | 322,4 | 3,6 | 0,9813 | 0,9890 | 0,9866 | 0,9787 | 0,9987 | 43,271 | |
| Pipeline-4 | 144,0 | 6,0 | 324,0 | 2,0 | 0,9600 | 0,9939 | 0,9832 | 0,9729 | 0,9987 | 43,271 |
In the fifth experimental group within the scope of the study, the training and testing procedures were performed using the imaginary part of the LL, LH and HL sub-images obtained by applying DT-CWT to the chest X-ray images, and the results were obtained. Then, the training and testing procedures were performed using the imaginary part of the LL, LH and HL sub-images obtained by applying the LBP and DT-CWT operations to the X-ray images, respectively. Finally, the results were calculated using the pipeline classification algorithms, the details of which were previously described and proposed within the scope of the study. The image sizes given to the CNN as input for this experiment were 224 × 224 × 3. The results obtained from the experimental group are given in Table 16 (first training-test data set) and Table 17 (second training-test data set).
Table 16.
Results obtained using the LL, LH, HL imaginary sub-bands obtained by applying DT-CWT to the chest X-ray images (first training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 145,2 | 4,8 | 77,4 | 2,6 | 0,9680 | 0,9675 | 0,9678 | 0,9751 | 0,9967 | 10,667 |
| With LBP | 146,8 | 3,2 | 71,8 | 8,2 | 0,9787 | 0,8975 | 0,9504 | 0,9628 | 0,9916 | 10,593 | |
| Pipeline-1 | 148,4 | 1,6 | 77,4 | 2,6 | 0,9893 | 0,9675 | 0,9817 | 0,9861 | 0,9982 | 21,261 | |
| Pipeline-2 | 145,8 | 4,2 | 77,4 | 2,6 | 0,9720 | 0,9675 | 0,9704 | 0,9772 | 0,9980 | 21,261 | |
| Pipeline-3 | 148,6 | 1,4 | 76,8 | 3,2 | 0,9907 | 0,9600 | 0,9800 | 0,9848 | 0,9983 | 21,261 | |
| Pipeline-4 | 145,0 | 5,0 | 78,0 | 2,0 | 0,9667 | 0,9750 | 0,9696 | 0,9764 | 0,9970 | 21,261 | |
| Second CNN Architecture | Without LBP | 143,6 | 6,4 | 76,2 | 3,8 | 0,9573 | 0,9525 | 0,9557 | 0,9657 | 0,9876 | 16,952 |
| With LBP | 140,4 | 9,6 | 66,8 | 13,2 | 0,9360 | 0,8350 | 0,9009 | 0,9252 | 0,9684 | 16,866 | |
| Pipeline-1 | 146,4 | 3,6 | 76,8 | 3,2 | 0,9760 | 0,9600 | 0,9704 | 0,9773 | 0,9940 | 33,818 | |
| Pipeline-2 | 144,2 | 5,8 | 76,0 | 4,0 | 0,9613 | 0,9500 | 0,9574 | 0,9671 | 0,9943 | 33,818 | |
| Pipeline-3 | 147,6 | 2,4 | 75,0 | 5,0 | 0,9840 | 0,9375 | 0,9678 | 0,9755 | 0,9936 | 33,818 | |
| Pipeline-4 | 142,4 | 7,6 | 78,0 | 2,0 | 0,9493 | 0,9750 | 0,9583 | 0,9674 | 0,9907 | 33,818 |
Table 17.
Results obtained using the LL, LH, HL imaginary sub-bands obtained by applying DT-CWT to the chest X-ray images (second training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 145,8 | 4,2 | 322,8 | 3,2 | 0,9720 | 0,9902 | 0,9845 | 0,9752 | 0,9980 | 14,026 |
| With LBP | 134,8 | 15,2 | 311,4 | 14,6 | 0,8987 | 0,9552 | 0,9374 | 0,9004 | 0,9839 | 14,067 | |
| Pipeline-1 | 146,2 | 3,8 | 322,0 | 4,0 | 0,9747 | 0,9877 | 0,9836 | 0,9740 | 0,9982 | 42,160 | |
| Pipeline-2 | 146,0 | 4,0 | 322,4 | 3,6 | 0,9733 | 0,9890 | 0,9840 | 0,9746 | 0,9983 | 70,253 | |
| Pipeline-3 | 147,4 | 2,6 | 321,2 | 4,8 | 0,9827 | 0,9853 | 0,9845 | 0,9755 | 0,9988 | 11,2414 | |
| Pipeline-4 | 144,6 | 5,4 | 323,6 | 2,4 | 0,9640 | 0,9926 | 0,9836 | 0,9737 | 0,9977 | 18,2667 | |
| Second CNN Architecture | Without LBP | 145,2 | 4,8 | 323,0 | 3,0 | 0,9680 | 0,9908 | 0,9836 | 0,9738 | 0,9987 | 21,541 |
| With LBP | 130,0 | 20,0 | 304,4 | 21,6 | 0,8667 | 0,9337 | 0,9126 | 0,8615 | 0,9733 | 21,514 | |
| Pipeline-1 | 147,2 | 2,8 | 321,8 | 4,2 | 0,9813 | 0,9871 | 0,9853 | 0,9768 | 0,9979 | 43,054 | |
| Pipeline-2 | 145,4 | 4,6 | 322,8 | 3,2 | 0,9693 | 0,9902 | 0,9836 | 0,9739 | 0,9984 | 43,054 | |
| Pipeline-3 | 147,8 | 2,2 | 321,6 | 4,4 | 0,9853 | 0,9865 | 0,9861 | 0,9782 | 0,9986 | 43,054 | |
| Pipeline-4 | 144,6 | 5,4 | 323,2 | 2,8 | 0,9640 | 0,9914 | 0,9828 | 0,9724 | 0,9982 | 43,054 |
In the sixth experimental group within the scope of the study, the training and testing procedures were performed using the real and imaginary parts of the LL sub-image obtained by applying DT-CWT to the chest X-ray images, and the results were obtained. Then, the training and testing procedures were performed using the real and imaginary parts of the LL sub-image obtained by applying the LBP and DT-CWT operations to the X-ray images, respectively. Finally, the results were calculated using the pipeline classification algorithms, the details of which were previously described and proposed within the scope of the study. The image sizes given to the CNN as input for this experiment were 224 × 224 × 2. The results obtained from the experimental group are given in Table 18 (first training-test data set) and Table 19 (second training-test data set).
Table 18.
Results obtained by using the LL real and imaginary sub-bands obtained by applying DT-CWT to the chest X-ray images (first training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 145,8 | 4,2 | 77,6 | 2,4 | 0,9720 | 0,9700 | 0,9713 | 0,9779 | 0,9970 | 0,8271 |
| With LBP | 145,8 | 4,2 | 68,8 | 11,2 | 0,9720 | 0,8600 | 0,9330 | 0,9500 | 0,9883 | 0,8273 | |
| Pipeline-1 | 148,2 | 1,8 | 77,2 | 2,8 | 0,9880 | 0,9650 | 0,9800 | 0,9847 | 0,9982 | 16,544 | |
| Pipeline-2 | 147,0 | 3,0 | 78,0 | 2,0 | 0,9800 | 0,9750 | 0,9783 | 0,9833 | 0,9983 | 16,544 | |
| Pipeline-3 | 148,4 | 1,6 | 76,4 | 3,6 | 0,9893 | 0,9550 | 0,9774 | 0,9828 | 0,9986 | 16,544 | |
| Pipeline-4 | 145,6 | 4,4 | 78,4 | 1,6 | 0,9707 | 0,9800 | 0,9739 | 0,9798 | 0,9970 | 16,544 | |
| Second CNN Architecture | Without LBP | 143,0 | 7,0 | 76,4 | 3,6 | 0,9533 | 0,9550 | 0,9539 | 0,9643 | 0,9902 | 14,416 |
| With LBP | 142,6 | 7,4 | 67,2 | 12,8 | 0,9507 | 0,8400 | 0,9122 | 0,9340 | 0,9738 | 14,399 | |
| Pipeline-1 | 148,0 | 2,0 | 77,4 | 2,6 | 0,9867 | 0,9675 | 0,9800 | 0,9847 | 0,9969 | 28,815 | |
| Pipeline-2 | 144,2 | 5,8 | 77,4 | 2,6 | 0,9613 | 0,9675 | 0,9635 | 0,9717 | 0,9957 | 28,815 | |
| Pipeline-3 | 148,0 | 2,0 | 75,6 | 4,4 | 0,9867 | 0,9450 | 0,9722 | 0,9789 | 0,9957 | 28,815 | |
| Pipeline-4 | 143,0 | 7,0 | 78,2 | 1,8 | 0,9533 | 0,9775 | 0,9617 | 0,9701 | 0,9922 | 28,815 |
Table 19.
Results obtained by using the LL real and imaginary sub-bands obtained by applying DT-CWT to the chest X-ray images (second training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 143,0 | 7,0 | 323,0 | 3,0 | 0,9533 | 0,9908 | 0,9790 | 0,9662 | 0,9980 | 10,808 |
| With LBP | 135,2 | 14,8 | 313,6 | 12,4 | 0,9013 | 0,9620 | 0,9429 | 0,9086 | 0,9863 | 10,820 | |
| Pipeline-1 | 146,8 | 3,2 | 321,8 | 4,2 | 0,9787 | 0,9871 | 0,9845 | 0,9754 | 0,9983 | 21,627 | |
| Pipeline-2 | 145,4 | 4,6 | 323,0 | 3,0 | 0,9693 | 0,9908 | 0,9840 | 0,9745 | 0,9983 | 21,627 | |
| Pipeline-3 | 147,2 | 2,8 | 321,4 | 4,6 | 0,9813 | 0,9859 | 0,9845 | 0,9755 | 0,9987 | 21,627 | |
| Pipeline-4 | 142,6 | 7,4 | 323,4 | 2,6 | 0,9507 | 0,9920 | 0,9790 | 0,9661 | 0,9977 | 21,627 | |
| Second CNN Architecture | Without LBP | 145,2 | 4,8 | 322,8 | 3,2 | 0,9680 | 0,9902 | 0,9832 | 0,9732 | 0,9982 | 18,172 |
| With LBP | 131,2 | 18,8 | 306,0 | 20,0 | 0,8747 | 0,9387 | 0,9185 | 0,8711 | 0,9797 | 18,135 | |
| Pipeline-1 | 147,4 | 2,6 | 323,0 | 3,0 | 0,9827 | 0,9908 | 0,9882 | 0,9813 | 0,9985 | 36,307 | |
| Pipeline-2 | 145,6 | 4,4 | 323,2 | 2,8 | 0,9707 | 0,9914 | 0,9849 | 0,9758 | 0,9986 | 36,307 | |
| Pipeline-3 | 148,0 | 2,0 | 321,8 | 4,2 | 0,9867 | 0,9871 | 0,9870 | 0,9795 | 0,9990 | 36,307 | |
| Pipeline-4 | 144,6 | 5,4 | 324,0 | 2,0 | 0,9640 | 0,9939 | 0,9845 | 0,9750 | 0,9980 | 36,307 |
In the seventh experimental group within the scope of the study, the training and testing procedures were performed using the real and imaginary parts of the LL, LH, HL sub-images obtained by applying DT-CWT to the chest X-ray images, and the results were obtained. Then, the training and testing procedures were performed using the real and imaginary parts of the LL, LH, HL sub-images obtained by applying the LBP and DT-CWT operations to the X-ray images, respectively. Finally, the results were calculated using the pipeline classification algorithms, the details of which were previously described and proposed within the scope of the study. The image sizes given to the CNN as input for this experiment were 224 × 224 × 6. The results obtained from he experimental group are given in Table 20 (first training-test data set) and Table 21 (second training-test data set).
Table 20.
Results obtained by using the LL, LH, HL real and imaginary sub-bands obtained by applying DT-CWT to the chest X-ray images (first training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 147,0 | 3,0 | 77,6 | 2,4 | 0,9800 | 0,9700 | 0,9765 | 0,9819 | 0,9975 | 17,203 |
| With LBP | 145,4 | 4,6 | 74,2 | 5,8 | 0,9693 | 0,9275 | 0,9548 | 0,9655 | 0,9902 | 17,311 | |
| Pipeline-1 | 148,2 | 1,8 | 77,4 | 2,6 | 0,9880 | 0,9675 | 0,9809 | 0,9854 | 0,9984 | 34,513 | |
| Pipeline-2 | 147,2 | 2,8 | 77,8 | 2,2 | 0,9813 | 0,9725 | 0,9783 | 0,9833 | 0,9984 | 34,513 | |
| Pipeline-3 | 148,8 | 1,2 | 77,2 | 2,8 | 0,9920 | 0,9650 | 0,9826 | 0,9867 | 0,9986 | 34,513 | |
| Pipeline-4 | 146,4 | 3,6 | 77,8 | 2,2 | 0,9760 | 0,9725 | 0,9748 | 0,9806 | 0,9981 | 34,513 | |
| Second CNN Architecture | Without LBP | 142,8 | 7,2 | 76,8 | 3,2 | 0,9520 | 0,9600 | 0,9548 | 0,9648 | 0,9884 | 24,206 |
| With LBP | 142,0 | 8,0 | 70,4 | 9,6 | 0,9467 | 0,8800 | 0,9235 | 0,9417 | 0,9763 | 24,210 | |
| Pipeline-1 | 146,6 | 3,4 | 77,6 | 2,4 | 0,9773 | 0,9700 | 0,9748 | 0,9806 | 0,9956 | 48,417 | |
| Pipeline-2 | 144,6 | 5,4 | 77,4 | 2,6 | 0,9640 | 0,9675 | 0,9652 | 0,9731 | 0,9948 | 48,417 | |
| Pipeline-3 | 147,8 | 2,2 | 76,4 | 3,6 | 0,9853 | 0,9550 | 0,9748 | 0,9808 | 0,9953 | 48,417 | |
| Pipeline-4 | 141,6 | 8,4 | 78,0 | 2,0 | 0,9440 | 0,9750 | 0,9548 | 0,9646 | 0,9909 | 48,417 |
Table 21.
Results obtained by using the LL, LH, HL real and imaginary sub-bands obtained by applying DT-CWT to the chest X-ray images (second training-test data set)
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 144,8 | 5,2 | 323,0 | 3,0 | 0,9653 | 0,9908 | 0,9828 | 0,9724 | 0,9982 | 23,185 |
| With LBP | 129,2 | 20,8 | 308,6 | 17,4 | 0,8613 | 0,9466 | 0,9197 | 0,8710 | 0,9786 | 23,401 | |
| Pipeline-1 | 147,0 | 3,0 | 321,6 | 4,4 | 0,9800 | 0,9865 | 0,9845 | 0,9755 | 0,9974 | 46,586 | |
| Pipeline-2 | 145,2 | 4,8 | 322,8 | 3,2 | 0,9680 | 0,9902 | 0,9832 | 0,9732 | 0,9978 | 46,586 | |
| Pipeline-3 | 147,6 | 2,4 | 321,4 | 4,6 | 0,9840 | 0,9859 | 0,9853 | 0,9768 | 0,9984 | 46,586 | |
| Pipeline-4 | 144,2 | 5,8 | 323,2 | 2,8 | 0,9613 | 0,9914 | 0,9819 | 0,9710 | 0,9975 | 46,586 | |
| Second CNN Architecture | Without LBP | 144,6 | 5,4 | 323,4 | 2,6 | 0,9640 | 0,9920 | 0,9832 | 0,9731 | 0,9984 | 31,573 |
| With LBP | 131,4 | 18,6 | 304,8 | 21,2 | 0,8760 | 0,9350 | 0,9164 | 0,8686 | 0,9806 | 31,369 | |
| Pipeline-1 | 147,4 | 2,6 | 322,4 | 3,6 | 0,9827 | 0,9890 | 0,9870 | 0,9794 | 0,9982 | 62,942 | |
| Pipeline-2 | 145,0 | 5,0 | 322,6 | 3,4 | 0,9667 | 0,9896 | 0,9824 | 0,9719 | 0,9986 | 62,942 | |
| Pipeline-3 | 148,2 | 1,8 | 321,8 | 4,2 | 0,9880 | 0,9871 | 0,9874 | 0,9802 | 0,9986 | 62,942 | |
| Pipeline-4 | 143,8 | 6,2 | 324,0 | 2,0 | 0,9587 | 0,9939 | 0,9828 | 0,9723 | 0,9983 | 62,942 |
Finally, all the training-test data sets were combined to test the performance of the proposed method and the pipeline approaches. In this context, a collective training-test data set containing a total of 556 X-ray images comprising 150 Covid-19 and 406 non-Covid-19 images was created. Then the k value was determined as 2 (cross training and testing for 75 Covid-19 and 203 non-Covid-19 images). The training and testing processes were realized for the input images (original image and the LL (real sub-band)), ensuring the best results in the first and second training-test data sets. The results obtained are given in Tables 22 and 23.
Table 22.
Results obtained directly using chest X-ray images (k = 2 and a total of 556 images (150 Covid-19 and 406 non-Covid-19 images))
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 138,8 | 11,2 | 401,6 | 4,4 | 0,9253 | 0,9892 | 0,9719 | 0,9468 | 0,9937 | 0,1460 |
| With LBP | 133,4 | 16,6 | 400,8 | 5,2 | 0,8893 | 0,9872 | 0,9608 | 0,9241 | 0,9932 | 0,1401 | |
| Pipeline-1 | 144,8 | 5,2 | 406,0 | 0,0 | 0,9653 | 1,0000 | 0,9906 | 0,9823 | 0,9997 | 0,2861 | |
| Pipeline-2 | 139,8 | 10,2 | 402,4 | 3,6 | 0,9320 | 0,9911 | 0,9752 | 0,9529 | 0,9991 | 0,2861 | |
| Pipeline-3 | 146,4 | 3,6 | 401,6 | 4,4 | 0,9760 | 0,9892 | 0,9856 | 0,9734 | 0,9984 | 0,2861 | |
| Pipeline-4 | 137,2 | 12,8 | 406,0 | 0,0 | 0,9147 | 1,0000 | 0,9770 | 0,9554 | 0,9977 | 0,2861 | |
| Second CNN Architecture | Without LBP | 138,6 | 11,4 | 403,2 | 2,8 | 0,9240 | 0,9931 | 0,9745 | 0,9511 | 0,9949 | 0,2853 |
| With LBP | 120,6 | 29,4 | 395,0 | 11,0 | 0,8040 | 0,9729 | 0,9273 | 0,8565 | 0,9786 | 0,2811 | |
| Pipeline-1 | 143,0 | 7,0 | 405,4 | 0,6 | 0,9533 | 0,9985 | 0,9863 | 0,9740 | 0,9994 | 0,5664 | |
| Pipeline-2 | 141,0 | 9,0 | 404,6 | 1,4 | 0,9400 | 0,9966 | 0,9813 | 0,9644 | 0,9992 | 0,5664 | |
| Pipeline-3 | 145,6 | 4,4 | 403,0 | 3,0 | 0,9707 | 0,9926 | 0,9867 | 0,9752 | 0,9991 | 0,5664 | |
| Pipeline-4 | 136,0 | 14,0 | 405,6 | 0,4 | 0,9067 | 0,9990 | 0,9741 | 0,9495 | 0,9964 | 0,5664 |
Table 23.
Results obtained using the LL real sub-band obtained by applying DT-CWT to the chest X-ray images (k = 2 and a total of 556 images (150 Covid-19 and 406 non-Covid-19 images))
| CNN Type | Method | TP | FN | TN | FP | SEN | SPE | ACC | F-1 | AUC | CPU Time |
|---|---|---|---|---|---|---|---|---|---|---|---|
| First CNN Architecture | Without LBP | 136,2 | 13,8 | 401,4 | 4,6 | 0,9080 | 0,9887 | 0,9669 | 0,9366 | 0,9939 | 0,0496 |
| With LBP | 121,6 | 28,4 | 396,0 | 10,0 | 0,8107 | 0,9754 | 0,9309 | 0,8634 | 0,9798 | 0,0492 | |
| Pipeline-1 | 140,8 | 9,2 | 405,4 | 0,6 | 0,9387 | 0,9985 | 0,9824 | 0,9663 | 0,9987 | 0,0987 | |
| Pipeline-2 | 138,8 | 11,2 | 403,2 | 2,8 | 0,9253 | 0,9931 | 0,9748 | 0,9519 | 0,9985 | 0,0987 | |
| Pipeline-3 | 142,8 | 7,2 | 401,4 | 4,6 | 0,9520 | 0,9887 | 0,9788 | 0,9603 | 0,9981 | 0,0987 | |
| Pipeline-4 | 134,2 | 15,8 | 405,4 | 0,6 | 0,8947 | 0,9985 | 0,9705 | 0,9423 | 0,9965 | 0,0987 | |
| Second CNN Architecture | Without LBP | 137,8 | 12,2 | 403,4 | 2,6 | 0,9187 | 0,9936 | 0,9734 | 0,9486 | 0,9975 | 0,0932 |
| With LBP | 99,2 | 50,8 | 395,0 | 11,0 | 0,6613 | 0,9729 | 0,8888 | 0,7580 | 0,9553 | 0,0920 | |
| Pipeline-1 | 136,4 | 13,6 | 406,0 | 0,0 | 0,9093 | 1,0000 | 0,9755 | 0,9523 | 0,9988 | 0,1852 | |
| Pipeline-2 | 139,2 | 10,8 | 404,6 | 1,4 | 0,9280 | 0,9966 | 0,9781 | 0,9577 | 0,9987 | 0,1852 | |
| Pipeline-3 | 141,0 | 9,0 | 403,4 | 2,6 | 0,9400 | 0,9936 | 0,9791 | 0,9603 | 0,9983 | 0,1852 | |
| Pipeline-4 | 133,2 | 16,8 | 406,0 | 0,0 | 0,8880 | 1,0000 | 0,9698 | 0,9401 | 0,9987 | 0,1852 |
Conclusion
In this section, first of all, the results that were obtained without using pipeline algorithms are compared. When the results of the study given between Tables 8 and 23 are examined within the scope of the study, it can be seen that the results of the study obtained without using LBP are generally better than the results of the study using LBP, for the same input image. In this context, it is understood that there are exceptions for the sensitivity parameter of some results obtained using the first CNN architecture for the first training-test data set. Within the scope of the study, the highest mean sensitivity, specificity, accuracy, F-1 score, and AUC values obtained without using the pipeline algorithms were, respectively; 0,9853, 0,9725, 0,9765, 0,9819, 0,9983 for the first training-test data set and the first CNN architecture, 0,9613, 0,9725, 0,9600, 0,9691, 0,9949 for the first training-test data set and the second CNN architecture, 0,9720, 0,9908, 0,9845, 0,9752, 0,9982 for the second training-test data set and the first CNN architecture, and 0,9733, 0,9920, 0,9857, 0,9772, 0,9987 for the second training-test data set and the second CNN architecture. In this context, it can be seen that the achievements of the first and second CNN architectures are generally close to each other. However, when a comparison is made in terms of CPU run-time, it is understood that the second CNN architecture is two times slower than the first CNN architecture in terms of CPU run-time. The main reason for this is that the number of layers in the second CNN architecture is approximately twice that as high as in the first CNN architecture. A similar situation arose in the experiments performed by combining all the data and using 2-fold cross. For these experiments, the highest mean sensitivity, specificity, accuracy, F-1 score, and AUC values obtained without using the pipeline algorithms are respectively; 0,9253, 0,9892, 0,9719, 0,9468, 0,9939 for the first CNN architecture and 0,9240, 0,9936, 0,9745, 0,9511, 0,9975 for the second CNN architecture.
Within the scope of the study, DT-CWT was used to reduce the image dimensions. In this way, DT-CWT tolerated the increase in result-producing time due to the use of the pipeline algorithm. In this context, when the results obtained using the original images and the ones obtained using DT-CWT are compared, it can be seen that there is no serious decrease in the results, in general. Using DT-CWT, the image sizes were reduced successfully and a reduction in the result-producing times was achieved, in the study.
The pipeline algorithms proposed within the scope of the study are based on combining the results obtained without using LBP and with using LBP, as detailed previously. After this stage, the study results obtained by using the pipeline algorithms were analyzed. With the introduction of the pipeline algorithms, improvements were achieved in all the parameters obtained by using both training-test data sets and the CNN architectures. In this context, an improvement was achieved in general, according to the highest results obtained without LBP and with using LBP, in terms of percentage ranging between 0,67% and 3,73% for the sensitivity parameter, between 0,06% and 2,25% for the specificity parameter, between 0% to 2,61% for the accuracy parameter, between 0,03% and 2,04% for the F-1 score parameter, and between 0% and 1,20% for the AUC parameter.
It was also observed that similar improvements were achieved for the experiments performed by combining all data and using 2-fold cross. In this context, according to the highest results obtained without LBP and with using LBP, an improvement was achieved generally in terms of percentage ranging between 2,13% and 5,07% for the sensitivity parameter, between 0,59% and 1,08% for the specificity parameter, between 0,58% and 1,87% for the accuracy parameter, between 1,18% and 3,55% for the F-1 score parameter, and between 0,13% and 0,59% for the AUC parameter.
When comparing the success of pipeline algorithms in improving the results in general, it can be seen that the algorithms of pipeline-1 and pipeline-3 obtain the highest sensitivity values; pipeline-4 obtains the highest specificity values; pipeline-1 and pipeline-3 obtain the highest accuracy values; pipeline-1 and pipeline-3 obtain the highest F-1 scores values; and pipeline-1, pipeline-2 and pipeline-3 algorithms successfully obtained the highest AUC values.
When the input data with the best results obtained by using the pipeline algorithms are examined, it can be seen that using the real part of the LL sub-image band for the first training-test data set and using the original images for the second training-test data set provided the best results. Experiments performed using the 2-fold cross by combining all the data also confirm this situation. For this reason, only the results of the experiments mentioned were included in the study, in consideration of the length of the study.
The highest mean sensitivity, specificity, accuracy, F-1 score, and AUC values obtained using the study pipeline algorithms are as follows, respectively; 0,9947, 0,9800, 0,9843, 0,9881, 0,9990 for the first training-test data set and the first CNN architecture; 0,9867, 0,9800, 0,9809, 0,9853, 0,9977 for the first training-test data set and the second CNN architecture; 0,9853, 0,9926, 0,9857, 0,9774, 0,9988 for the second training-test data set and the first CNN architecture; and 0,9920, 0,9939, 0,9891, 0,9828, 0,9991 for the second training-test and the second CNN architecture.
The highest mean sensitivity, specificity, accuracy, F-1 score and AUC values obtained in the experiments performed by combining all data and using the 2-fold cross were respectively; 0,9760, 1,0000, 0,9906, 0,9823, 0,9997 for the first CNN architecture; and 0,9707, 1,0000, 0,9867, 0,9752, 0,9994 for the second CNN architecture.
Within the scope of the study, the best results obtained before and after using the pipeline algorithm and the comparison of these results with the recent literature studies are given in Table 24.
Table 24.
Comparison of the results obtained, within the scope of the study, with previous studies
| Study | SEN | SPE | ACC | F-1 | AUC |
|---|---|---|---|---|---|
| Tuncer et al. [26] | 0,8149-1,0000 | 0,9380-1,0000 | 0,9049-0,9955 | X | X |
| Panwar et al. [27] | 0,9762 | 0,7857 | 0,881 | X | X |
| Ozturk et al. [28] | 0,9513 | 0,953 | 0,9808 | 0,9651 | X |
| Mohammed et al. [29] | 0,706-0,974 | 0,557-1,000 | 0,620-0,987 | 0,555–0,987 | 0,800-0,988 |
| Khan et al. [30] | 0,993 | 0,986 | 0,990 | 0,985 | X |
| Apostolopoulos and Mpesiana [31] | 0,9866 | 0,9646 | 0,9678 | X | X |
| Waheed et al. [32] | 0,69-0,90 | 0,95-0,97 | 0,85-0,95 | X | X |
| Mahmud et al. [33] | 0,978 | 0,947 | 0,974 | 0,971 | 0,969 |
| Vaid et al. [34] | 0,9863 | 0,9166 | 0,9633 | 0,9729 | X |
| Benbrahim et al. [35] | 0,9803-0,9811 | X | 0,9803-0,9901 | 0,9803-0,9901 | X |
| Elaziz et al. [36] | 0,9875-0,9891 | X | 0,9609-0,9809 | X | X |
| Martínez et al. [37] | 0,97 | X | 0,97 | 0,97 | X |
| Loey et al. [38] | 1,0000 | 1,0000 | 1,0000 | X | X |
| Toraman et al. [39] | 0,28-0,9742 | 0,8095–0,98 | 0,4914-0,9724 | 0,55-0,9724 | X |
| Duran-Lopez et al. [40] | 0,9253 | 0,9633 | 0,9443 | 0,9314 | 0,988 |
| Minaee et al. [41] | 0,98 | 0,751-0,929 | X | X | X |
| Our Study (Before Pipeline-First data set) | 0,9853 | 0,8950 | 0,9539 | 0,9655 | 0,9939 |
| Our Study (Before Pipeline-First data set) | 0,9747 | 0,9725 | 0,9739 | 0,9799 | 0,9976 |
| Our Study (Before Pipeline-First data set) | 0,9800 | 0,9700 | 0,9765 | 0,9819 | 0,9975 |
| Our Study (Before Pipeline-First data set) | 0,9773 | 0,9675 | 0,9739 | 0,9800 | 0,9983 |
| Our Study (Before Pipeline-Second data set) | 0,9733 | 0,9914 | 0,9857 | 0,9772 | 0,9981 |
| Our Study (Before Pipeline-Second data set) | 0,9640 | 0,9920 | 0,9832 | 0,9730 | 0,9985 |
| Our Study (Before Pipeline-Second data set) | 0,9680 | 0,9902 | 0,9832 | 0,9732 | 0,9982 |
| Our Study (Before Pipeline-Combined data set) | 0,9253 | 0,9892 | 0,9719 | 0,9468 | 0,9937 |
| Our Study (Before Pipeline-Combined data set) | 0,9187 | 0,9936 | 0,9734 | 0,9486 | 0,9975 |
| Our Study (Before Pipeline-Combined data set) | 0,9240 | 0,9931 | 0,9745 | 0,9511 | 0,9949 |
| Our Study (After Pipeline-First data set) | 0,9947 | 0,9575 | 0,9817 | 0,9862 | 0,9990 |
| Our Study (After Pipeline-First data set) | 0,9813 | 0,9800 | 0,9809 | 0,9853 | 0,9977 |
| Our Study (After Pipeline-First data set) | 0,9933 | 0,9675 | 0,9843 | 0,9881 | 0,9988 |
| Our Study (After Pipeline-Second data set) | 0,9920 | 0,9877 | 0,9891 | 0,9828 | 0,9991 |
| Our Study (After Pipeline-Second data set) | 0,9640 | 0,9939 | 0,9845 | 0,9750 | 0,9980 |
| Our Study (After Pipeline-Combined data set) | 0,9760 | 0,9892 | 0,9856 | 0,9734 | 0,9984 |
| Our Study (After Pipeline-Combined data set) | 0,9653 | 1,0000 | 0,9906 | 0,9823 | 0,9997 |
Discussion
As a result of our study on the automatic classification of chest X-ray images and using one of the deep learning methods, the CNN, some important and comprehensive test results were obtained for early diagnosis of Covid-19 disease. When the results obtained within the scope of the study are compared with the literature studies detailed in Tables 1 and 24, the results of the study were found to be better than the 14 out of the 16 studies in which this value was calculated for the sensitivity parameter, than all the 13 studies in which this value was calculated for the specificity parameter, than the 13 out of the 15 studies in which this value was calculated for the accuracy parameter, than the eight out of the nine studies in which this value was calculated for the F-1 score parameter, and than all the 3 studies in which this value was calculated for the AUC parameter. Moreover, if it is necessary to make a comparison in terms of run-times, it was found that it produced a result at least three times faster in terms of run-time than the result was obtained in the study conducted by Mohammed et al. [29]. This study is the only study in which this parameter was calculated. Also, it is at least ten times faster than the study conducted by Toraman et al. [39]. These two studies were studies in which the run-times were shared. No information was given about run-times in the other previous studies.
Overall, the results obtained within the scope of the study lagged behind the results obtained in studies conducted by Tuncer et al. [26], Benbrahim et al. [35], and Loey et al. [38]. However, in order to make a more detailed comparison, the number of images used in these studies should be compared with the number of images used in our study. The number of images used in our study is higher than the number of images used in these studies. In particular, the number of images used in our study is almost three times the number of images used by Loey et al. [38]. Another important issue is the procedure for training and testing. There was no cross validation in the studies by Benbrahim et al. [35] and Loet et al. [38]. In our study, cross-validation in the training-test processes is one of the important measures taken against the overfitting problem that occurs during the training of the network. However, it is known that cross validation improves the reliability of the study results while balancing the study results. In this context, these issues should be taken into consideration when making a comparison.
In the context of the study, if an evaluation should be based on the differentiation made between giving the images to the CNN as input directly and after the LBP was applied, it can be seen that the images obtained by applying the LBP produced worse results than the original images. However, the pipeline classification algorithm presented in the context of this study enabled the results obtained to be improved by combining the original and LBP-applied images. In this context, a significant part of the best results obtained in the study was provided using the pipeline classification algorithm. In this sense, it can be seen that the results of the study support some other literature studies [61–66] where the CNN and LBP methods are used together and use of the LBP was shown to increase the success of the relevant study.
The success achieved through the pipeline approaches in the study is due to the fact that some classification results that could not be revealed without using the LBP alone and with using the LBP alone were revealed by using the two methods together. Feeding the results from the two sources in the pipeline approaches results in an increase in running time. However, the results obtained within the scope of the study show that this time cost can be eliminated by using DT-CWT. In this way, it has been observed that working success can be increased significantly without time cost. It is considered that this model is within the scope of the study and can be used in many other deep learning studies.
It was evaluated that another important factor in achieving the successful results in this study was the framing process, which included the chest region and clarified the area of interest before the training and test procedures started. Hence, thanks to this pre-process carried out in this context, the parts lacking medical diagnostic information were removed from the images and only the relevant areas on the images were used in the procedures.
As the size of the inputs given to the CNN increases, the time taken for the training and testing increases. The DT-CWT transformation used in the study reduces the size of the image by half. Although the image sizes are reduced by half, there is no serious adverse effect on the study results. By contrast, some of the best results achieved in the study were obtained using the DT-CWT. In this context, although the pipeline classification algorithms proposed in the study increase the time to produce the results for the image, the times in question are less than half the time required for the images to be used directly without applying LBP and DT-CWT. Also, all the training and test procedures provided in the study reflect the amount per image. However, approximately 98% of these periods are spent on the training procedures. In this context, in the case where the results obtained by the transfer learning approach are used with the pipeline classification algorithm proposed in the study, the periods mentioned will decrease accordingly.
The pipeline algorithms revealed within the scope of the study were tested for data sets with different weights in terms of the number of Covid-19 and non-Covid-19 images, for different training-test ratios and different CNN architectures. The pipeline algorithms were successful for all these situations that may have affected the results. This shows that the proposed pipeline algorithms are not partial but are general solutions. From this point of view, it is obvious that if the pipeline algorithms mentioned above are added to the algorithms used in other literature studies, this would increase the success of these studies.
The results of the study show that analyzing chest X-ray images in the diagnosis of Covid-19 disease using deep learning methods will speed up the diagnosis and significantly reduce the burden on healthcare personnel. To further improve the results of the study, increasing the number of images in the training set, i.e., the creation of databases in which the clinical data of patients with Covid-19 that are accessible to the public, is of prime importance.
After this stage, it is aimed to realize applications using CT images of the lungs an important diagnostic tool, such as chest X-ray images, in Covid-19 disease diagnosis. In addition, it is planned to analyze the effects of using the results obtained, through direct transfer learning in pipeline classification algorithms, on the study results. This is evaluated as another important application to classify the complex-valued sub-bands of images obtained by applying DT-CWT, with the help of using the complex-valued CNN directly.
Biographies
Huseyin Yasar
received his B.Sc. and M.Sc. degrees in electrical and electronics engineering from the Selcuk University, Konya, Turkey, in 2010, and 2015, respectively. He has been working electrical and electronics engineering at the Ministry of Health of Republic of Turkey since 2012. His current research interests include image processing, medical image processing, biomedical engineering, and artificial intelligence. Information about his academic studies can be obtained from the link below. https://scholar.google.com.tr/citations?user=auLebQEAAAAJ&hl=tr
Murat Ceylan
received B.Sc. degree in 2000, M.Sc. degree in 2004 and Ph.D. degree in 2009 from Electrical–Electronics Engineering Department of Selçuk University, Konya, Turkey. His current research interests are digital image processing, biomedical systems, artificial intelligence, deep learning, transfer learning and thermal imaging and hyperspectral imaging for biomedical applications.
Compliance with ethical standards
Conflict of interest
Dr. Ceylan declares that he has no conflict of interest. Mr. Yasar declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J Pediatrics. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez-Matamoros A, Fujita H, Hayashi T, Perez-Meana H. Forecasting of COVID19 per regions using ARIMA models and polynomial functions. Appl Soft Comput. 2020;96:106610. doi: 10.1016/j.asoc.2020.106610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albarello F, Pianura E, Stefanoa FD, Cristofaro M, Petrone A, Marchioni L, Palazzolo C, Schininà V, Nicastri E, Petrosillo N, Campioni P, Eskild P, Zumla A, Ippolito G. 2019-novel coronavirus severe adult respiratory distress syndrome in two cases in Italy: an uncommon radiological presentation. Int J Infect Dis. 2020;93:192–197. doi: 10.1016/j.ijid.2020.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsih WH, Cheng MY, Ho MW, Chou CH, Lin PC, Chi CY, Liao WC, Chen CY, Leong LY, Tien N, Lai HC, Lai YC, Lu MC. Featuring COVID-19 cases via screening symptomatic patients with epidemiologic link during flu season in a medical center of Central Taiwan. J Microbiol Immunol Infect. 2020;53:459–466. doi: 10.1016/j.jmii.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, Akdis CA, Gao YD. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan. China Allergy. 2020;75:1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 8.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee NY, Li CW, Tsai HP, Chen PL, Syue LS, Li MC, Tsai CS, Lo CL, Hsueh PR, Ko WC. A case of COVID-19 and pneumonia returning from Macau in Taiwan: clinical course and anti-SARS-CoV-2 IgG dynamic. J Microbiol Immunol Infect. 2020;53:485–487. doi: 10.1016/j.jmii.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu YC, Liao CH, Chang CF, Chou CC, Lin YR. A locally transmitted case of SARS-CoV-2 infection in Taiwan. N Engl J Med. 2020;382(11):1070–1072. doi: 10.1056/NEJMc2001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thevarajan I, Nguyen TH, Koutsakos M, Druce J, Caly L, van de Sandt CE, Jia X, Nicholson S, Catton M, Cowie B, Tong SYC, Lewin SR, Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26:450–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim J, Jeon S, Shin HY, Kim MJ, Seong YM, Lee WJ, Choe KW, Kang YM, Lee B, Park SJ. Case of the index patient who caused tertiary transmission of COVID-19 infection in Korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J Korean Med Sci. 2020;35(6):e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong W, Agarwal PP. Chest imaging appearance of COVID-19 infection. Radiol: Cardiothoracic Imaging. 2020;2(1):e200028. doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon SH, Lee KH, Kim JY, Lee YK, Ko H, Kim KH, Park CM, Kim YH. Chest radiographic and ct findings of the 2019 novel coronavirus disease (Covid-19): analysis of nine patients treated in Korea. Korean J Radiol. 2020;21(4):494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Liu J, Zhao X, Liu C, Wang W, Wang D, Xu W, Zhang C, Yu J, Jiang B, Cao H, Li L. Clinical characteristics of imported cases of coronavirus disease 2019 (COVID-19) in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020;71:706–712. doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zu ZY, Jiang MD, Xu PP, Chen W, Ni QQ, Lu GM, Zhang LJ. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;200490:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song F, Shi N, Shan F, Zhang Z, Shen J, Lu H, Ling Y, Jiang Y, Shi Y. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1):210–217. doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng SC, Chang YC, Chiang YLF, Chien YC, Cheng M, Yang CH, Huang CH, Hsu YN. First case of coronavirus disease 2019 (COVID-19) pneumonia in Taiwan. J Formos Med Assoc. 2020;119(3):747–751. doi: 10.1016/j.jfma.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng MY, Lee EY, Yang J, Yang F, Li X, Wang H, Lui MM, Lo CSY, Leung B, Khong PL, Hui CKM, Yuen KY, Kuo MD. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol: Cardiothoracic Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H, Han X, Zheng C. Evolution of CT manifestations in a patient recovered from 2019 novel coronavirus (2019-nCoV) pneumonia in Wuhan, China. Radiology. 2020;295(1):20–20. doi: 10.1148/radiol.2020200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverstein WK, Stroud L, Cleghorn GE, Leis JA. First imported case of 2019 novel coronavirus in Canada, presenting as mild pneumonia. Lancet. 2020;395(10225):734. doi: 10.1016/S0140-6736(20)30370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen HT, Le HQ, Nguyen TT, Cao TM, Pham QD. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giang HTN, Shah J, Hung TH, Reda A, Truong LN, Huy NT (2020) The first Vietnamese case of COVID-19 acquired from China. Lancet Infect Dis 20(4):408–409.10.1016/S1473-3099(20)30111-0 [DOI] [PMC free article] [PubMed]
- 25.Long JB, Ehrenfeld JM. The role of augmented intelligence (ai) in detecting and preventing the spread of novel coronavirus. J Med Syst. 2020;44:59. doi: 10.1007/s10916-020-1536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tuncer T, Dogan S, Ozyurt F. An automated residual exemplar local binary pattern and iterative ReliefF based corona detection method using lung X-ray image. Chemom Intell Lab Syst. 2020;203:104054. doi: 10.1016/j.chemolab.2020.104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos, Solitons Fractals. 2020;138:109944. doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammed MA, Abdulkareem KH, Al-Waisy AS, Mostafa SA, Al-Fahdawi S, Dinar AM, Arbaiy N. Benchmarking methodology for selection of optimal COVID-19 diagnostic model based on entropy and TOPSIS methods. IEEE Access. 2020;8:99115–99131. doi: 10.1109/ACCESS.2020.2995597. [DOI] [Google Scholar]
- 30.Khan AI, Shah JL, Bhat MM. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Prog Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. Covidgan: data augmentation using auxiliary classifier Gan for improved covid-19 detection. IEEE Access. 2020;8:91916–91923. doi: 10.1109/ACCESS.2020.2994762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahmud T, Rahman MA, Fattah SA. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. 2020;122:103869. doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaid S, Kalantar R, Bhandari M. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int Orthop. 2020;44:1539–1542. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benbrahim H, Hachimi H, Amine A. Deep transfer learning with apache spark to detect COVID-19 in chest X-ray images. Romanian J Inform Sci Technol. 2020;23:S117–S129. [Google Scholar]
- 36.Elaziz MA, Hosny KM, Salah A, Darwish MM, Lu S, Sahlol AT. New machine learning method for image-based diagnosis of COVID-19. PLoS One. 2020;15(6):e0235187. doi: 10.1371/journal.pone.0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez F, Martínez F, Jacinto E. Performance evaluation of the NASNet convolutional network in the automatic identification of COVID-19. Int J Adv Sci Eng Inform Technol. 2020;10(2):662. doi: 10.18517/ijaseit.10.2.11446. [DOI] [Google Scholar]
- 38.Loey M, Smarandache F, Khalifa NEM. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry. 2020;12(4):651. doi: 10.3390/sym12040651. [DOI] [Google Scholar]
- 39.Toraman S, Alakus TB, Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fract. 2020;140:110122. doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duran-Lopez L, Dominguez-Morales JP, Corral-Jaime J, Vicente-Diaz S, Linares-Barranco A. COVID-XNet: a custom deep learning system to diagnose and locate COVID-19 in chest X-ray images. Appl Sci. 2020;10(16):5683. doi: 10.3390/app10165683. [DOI] [Google Scholar]
- 41.Minaee S, Kafieh R, Sonka M, Yazdani S, Soufi GJ. Deep-covid: predicting covid-19 from chest x-ray images using deep transfer learning. Med Image Anal. 2020;65:101794. doi: 10.1016/j.media.2020.101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection, arXiv. https://github.com/ieee8023/covid-chestxray-dataset (access time :11 may 2020)
- 43.Dadario AMV (2020) COVID-19 X rays. Kaggle. 10.34740/KAGGLE/DSV/1019469
- 44.Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. 2014;4(6):475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ojala T, Pietikäinen M, Harwood D. A comparative study of texture measures with classification based on featured distributions. Pattern Recogn. 1996;29(1):51–59. doi: 10.1016/0031-3203(95)00067-4. [DOI] [Google Scholar]
- 46.Selesnick IW, Baraniuk RG, Kingsbury NC. The dual-tree complex wavelet transform. IEEE Signal Process Mag. 2005;22(6):123–151. doi: 10.1109/MSP.2005.1550194. [DOI] [Google Scholar]
- 47.Kingsbury N (1998) The dual-tree complex wavelet transform: a new efficient tool for image restoration and enhancement. In Signal Process Conference:1-4
- 48.Kingsbury N. Shift invariant properties of the dual-tree complex wavelet transform. Int Confer Acoustics Speech Signal Process. 1999;3:1221–1224. doi: 10.1109/ICASSP.1999.756198. [DOI] [Google Scholar]
- 49.Iqbal MZ, Ghafoor A, Siddiqui AM, Riaz MM, Khalid U. Dual-tree complex wavelet transform and SVD based medical image resolution enhancement. Signal Process. 2014;105:430–437. doi: 10.1016/j.sigpro.2014.05.011. [DOI] [Google Scholar]
- 50.Ceylan M, Ozbay Y, Ucan ON, Yildirim E. A novel method for lung segmentation on chest CT images: complex-valued artificial neural network with complex wavelet transform. Turk J Electr Eng Comput Sci. 2010;18(4):613–624. doi: 10.3906/elk-0908-137. [DOI] [Google Scholar]
- 51.Ceylan M, Yasar H (2013) Blood vessel extraction from retinal images using complex wavelet transform and complex-valued artificial neural network. Int Conference Telecommun Signal Process:822-825.10.1109/TSP.2013.6614053
- 52.Zhang H, Zhang T, Pedrycz W, Zhao C, Miao D. Improved adaptive image retrieval with the use of shadowed sets. Pattern Recogn. 2019;90:390–403. doi: 10.1016/j.patcog.2019.01.029. [DOI] [Google Scholar]
- 53.Zhao CR, Chen K, Zang D, Zhang Z, Zuo W, Miao D. Uncertainty-optimized deep learning model for small-scale person re-identification. Science China Inf Sci. 2019;62(12):220102. doi: 10.1007/s11432-019-2675-3. [DOI] [Google Scholar]
- 54.Fu J, Zhao C, Xia Y, Liu W. Vehicle and wheel detection: a novel SSD-based approach and associated large-scale benchmark dataset. Multimed Tools Appl. 2020;79:12615–12634. doi: 10.1007/s11042-019-08523-y. [DOI] [Google Scholar]
- 55.Cui Y, Zhou F, Wang J, Liu X, Lin Y, Belongie S (2017) Kernel pooling for convolutional neural networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp 2921–2930. 10.1109/CVPR.2017.325
- 56.Wang J, Zhou F, Wen S, Liu X, Lin Y (2017) Deep metric learning with angular loss. In: Proceedings of the IEEE International Conference on Computer Vision. pp 2593–2601. 10.1109/ICCV.2017.283
- 57.Pagola M, Forcen JI, Barrenechea E, Fernández J, Bustince H (2017) A study on the cardinality of ordered average pooling in visual recognition. In: Iberian Conference on Pattern Recognition and Image Analysis. pp 437–444. 10.1007/978-3-319-58838-4_48
- 58.Wang F, Zhong SH, Peng J, Jiang J, Liu Y (2018) Data augmentation for eeg-based emotion recognition with deep convolutional neural networks. In: International Conference on Multimedia Modeling. pp 82–93. 10.1007/978-3-319-73600-6_8
- 59.Jiao L, Zhang S, Li L, Liu F, Ma W. A modified convolutional neural network for face sketch synthesis. Pattern Recogn. 2018;76:125–136. doi: 10.1016/j.patcog.2017.10.025. [DOI] [Google Scholar]
- 60.Yasar H, Ceylan M (2020) A novel comparative study for detection of Covid-19 on CT lung images using texture analysis, machine learning, and deep learning methods. Multimed Tools Appl. 10.1007/s11042-020-09894-3 [DOI] [PMC free article] [PubMed]
- 61.Hardalac F, Yasar H, Akyel A, Kutbay U. A novel comparative study using multi-resolution transforms and convolutional neural network (CNN) for contactless palm print verification and identification. Multimed Tools Appl. 2020;79:22929–22963. doi: 10.1007/s11042-020-09005-2. [DOI] [Google Scholar]
- 62.Zhang H, Qu Z, Yuan L, Li G (2017) A face recognition method based on LBP feature for CNN. In advanced information technology, electronic and automation control conference (IAEAC). pp 544-547. 10.1109/IAEAC.2017.8054074
- 63.Yang X, Li M, Zhao S (2017) Facial expression recognition algorithm basedon CNN and LBP feature fusion. In: International Conference on Robotics and Artificial Intelligence. pp 33-38. 10.1145/3175603.3175615
- 64.Juefei-Xu F, Naresh Boddeti V, Savvides M (2017) Local binary convolutional neural networks. In: Conference on Computer Vision and Pattern Recognition. pp 19–28
- 65.Ke P, Cai M, Wang H, Chen J (2018) A novel face recognition algorithm based on the combination of LBP and CNN. In international conference on signal processing (ICSP). pp 539-543. 10.1109/ICSP.2018.8652477
- 66.Touahri R, AzizI N, Hammami NE, Aldwairi M, Benaida F (2019) Automated breast tumor diagnosis using local binary patterns (LBP) based on deep learning classification. In international conference on computer and information sciences (ICCIS). pp 1-5. 10.1109/ICCISci.2019.8716428