ABSTRACT
This study investigated the genetic basis of multidrug resistance in two strains of Achromobacter xylosoxidans isolated from patients attending a hospital in Thailand in 2012. These isolates were highly resistant to cephalosporins, aminoglycosides, fluoroquinolones, co-trimoxazole and carbapenems. Whole genome sequencing revealed that the two isolates were not clonally related and identified a carbapenem resistance gene-habouring integron (In687), residing in a novel genomic island, AcGI1. This In687 shares 100% identical nucleotide sequence with ones found in Acinetobacter baumannii Aci 16, isolated from the same hospital in 2007. We report the first analysis of multidrug-resistant A. xylosoxidans isolated in Thailand, and the first example of this island in A. xylosoxidans. Our data support the idea that resistance has spread in Thailand via horizontal gene transfer between species and suggest the possibility of A. xylosoxidans may serve as a reservoir of antibiotic resistance, especially in hospital setting.
Keywords: antibiotic resistance, Achromobacter xylosoxidans, class 1 integron, In687, horizontal gene transfer, AcGI1
The first identification of a novel genomic island, AcGI1, containing multiple antimicrobial resistance genes in A. xylosoxidans.
INTRODUCTION
Members of the genus Achromobacter are Gram-negative bacteria of growing importance as opportunistic pathogens, particularly in relation to patients with Cystic Fibrosis (CF) (Hansen et al. 2013) and immunocompromised patients (Bellissimo et al. 2013). Amongst Achromobacter isolated from chronic lung infections of CF patients, Achromobacter xylosoxidans is the most abundant species (40–60%) (Spilker, Vandamme and LiPuma 2012; Barrado et al. 2013; Hansen et al. 2013; Coward et al. 2015). In part the clinical problems associated with these bacteria are due to the fact that a number of isolates display high levels of antibiotic resistance. All A. xylosoxidans isolates show high levels of innate resistances to cephalosporin and aminoglycoside antibiotics and some of the genes associated with this innate resistance have been described (Doi et al. 2008; Bador et al. 2011, 2013). Additionally, there is evidence that a number of resistance genes have been obtained by horizontal transfer (Yamamoto et al. 2012).
A number of different metallo-β-lactamases, carried in class 1 integrons, have been described in A. xylosoxidans. These include VIM-type (Sofianou et al. 2005; Di Pilato, Pollini and Rossolini 2014) and IMP-type β-lactamases (Yamamoto et al. 2012). Also, extended-spectrum β-lactamases, such as Vietnamese extended-spectrum β-lactamase (VEB)-type (Neuwirth et al. 2006), are found in A. xylosoxidans. There are currently more than 50 types of IMP β-lactamases, based on amino acid sequence variations(Zhao and Hu 2011; Pournaras et al. 2013; Shakibaie, Azizi and Shahcheraghi 2017). In addition to β-lactam resistance, genes associated with the resistance to fosfomycin, aminoglycosides and sulfamethoxazole/trimethoprim have been identified previously from the complete genome sequence data of A. xylosoxidans (Jakobsen et al. 2013).
In this study, we investigated two multidrug resistant isolates of A. xylosoxidans from Thailand, using whole genome sequencing to reveal the presence and localization of genes responsible for carbapenem resistance, within a novel genomic island called Achromobacter genomic island 1 (AcGI 1).
MATERIALS AND METHODS
Bacterial strains
Multidrug-resistant A. xylosoxidans strains R4 and R8 were isolated from an emergency unit between 2010 and 2012, at Ramathibodi Hospital in Bangkok, Thailand. Strain R4 was isolated from a urine sample and strain R8 from a peripheral blood culture. The collection of these clinical isolates was approved by the ethical review committee, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Thailand (MURA 2009/1396/S8; ID 03-52-93). The A. xylosoxidans reference strain, NCIMB11015 (Braker, Fesefeldt and Witzel 1998), was purchased from NCIMB, UK, and used as a non-clinical isolate control. The relationship of the isolates to previously reported strains of A. xylosoxidans was determined using multilocus sequence typing (MLST) assignment (Jolley, Bray and Maiden 2018) (http://pubmlst.org/achromobacter/).
Antimicrobial susceptibility testing
Antimicrobial susceptibility was determined by disk diffusion assays, using guidelines of the European Society Committee on Antimicrobial Susceptibility Testing (The European Committee on Antimicrobial Susceptibility Testing 2020). All isolates were grown on Iso-Sensitest agar (Oxoid, UK). All antimicrobial disks were purchased from Thermo Scientific Inc., Waltham, MA, USA. The minimum inhibitory concentrations (MIC) were determined using the Sensititre (Trek diagnostic systems Ltd, UK). The antibiotics used in this study were cefuroxime (CXM), cefotaxime (CTX), ceftazidime (CAZ), ceftriaxone (CRO), meropenem (MEM), imipenem (IPM), ertapenem (ETP), ciprofloxacin (CIP), levofloxacin (LVX), gentamicin (GEN), amikacin (AMK), co-trimoxazole (SXT) and Piperacillin/tazobactam (TZP). Metallo-β-lactamase detection assay was performed using disk diffusion with 10 µg MEM + 750 µg EDTA disk (Thermo Scientific Inc.). β-lactamase activity was assessed using a nitrocefin broth method (O'Callaghan et al. 1972). Briefly, nitrocefin was added into mid-log culture to reach a final concentration of 51.6 µg/mL. Optical density (OD) at 486 nm and 390 nm was measured over 30 min, when incubated at 37°C. Activity was expressed as the ratio of OD at 486 nm to OD at 390 nm (OD486/OD390 ratio). Assays were conducted in triplicate. The significance of differences in the OD486/OD390 ratio between three isolates was determined by one-way ANOVA with Tukey's test. For the comparison of OD486/OD390 ratio of each isolate between 0 and 30 min, the significance of differences in the OD486/OD390 ratio was evaluated using paired t-test. Statistical significance was defined as a P-value of < 0.05.
Whole genome sequencing
Achromobacter xylosoxidans strains R4, R8 and NCIMB11015 were grown in LB broth at 37°C overnight. Genomic DNA from all A. xylosoxidans strains was extracted using DNeasy Blood and Tissue kit (QIAGEN, UK). Whole genome sequencing was performed using the Pacific Bioscience® RS II platform by the Centre for Genomic Research, University of Liverpool. Genome assemblies of the isolates were obtained using the HGAP software (Chin et al. 2013) and annotated using Prokka 1.13 (Seemann 2014). Comparative genomic analysis between sequenced genomes and a reference genome NH44784-1996 (Jakobsen et al. 2013) was performed to identify the genomic difference between multidrug resistance isolates and the reference isolate. Briefly, three newly sequenced genomes of A. xylosoxidans were compared against one another using BLAST algorithm, implemented in BLAST+ (Camacho et al. 2009), to generate comparison files. In order to identify genes that are associated with antibiotic resistance, coding sequences called by Prokka were searched against an antibiotic resistance database using Resistance Gene Identifier (RGI) in the Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al. 2013; Jia et al. 2017). The prediction of genomic islands was performed using a combination of the observation of GC content of the genome sequence and in silico prediction in IslandViewer 4 (Bertelli et al. 2017). Then the comparison was visually investigated using Artemis Comparison Tool (Carver et al. 2005) and comparison figures were generated using EasyFig version 2.2.2 (Sullivan, Petty and Beatson 2011).
Genome submission and accession
The nucleotide sequences mentioned in this report are available in the GenBank nucleotide database under accession number LN890476 (strain R4), LN890477 (strain R8), LN890335 (strain NCIMB11015), KJ406505 (IMP-14-containing In687 in strain R4) and KJ406506 (IMP-14-carrying In687 in strain R8).
RESULTS
Bacterial strain typing and antibiotic resistance
Analysis using the pubMLST database typing (accessed 18th May 2020) revealed that the Thai A. xylosoxidans strains R4 and R8 were ST-183 and ST-185, respectively, and they were not clonally related (Table 1a and Fig. 1). The initial antibiotic susceptibility screen using the disk diffusion method revealed that strains R4 and R8 were less susceptible to multiple antibiotics, compared to the reference strain NCIMB11015, especially the carbapenems (IPM, MEM and ETP) and co-trimoxazole (Table 1a). However, strains R4 and R8 were more susceptible to MEM with the addition of EDTA (Table 1a). This suggests that a metallo-β-lactamase plays a role in the resistance exhibited by these strains.
Table 1a.
Characteristics and antimicrobial susceptibility profiles of multidrug-resistant A. xylosoxidans strains. Screening for antimicrobial resistance using disk diffusion.
| Zone of inhibition (mm) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Source | MLST type | CXM | CTX | CRO | CAZ | IPM | MEM | MEM + EDTA | ETP | CIP | LVX | GEN | AMK | TGC | SXT |
| R4 | Urine | 183 | 5 (R) | 5 (R) | 5 (R) | 5 (R) | 17 (R) | 5 (R) | 23 (S) | 12 (R) | 16 (R) | 20 (R) | 5 (R) | 13 (R) | 25 (S) | 5 (R) |
| R8 | Blood | 185 | 5 (R) | 5 (R) | 22 (R) | 21 (R) | 16 (R) | 5 (R) | 25 (S) | 13 (R) | 12 (R) | 12 (R) | 16 (R) | 14 (R) | 22 (S) | 14 (R) |
| NCIMB11015 | Soil | 166 | 5 (R) | 12 (R) | 16 (R) | 34 (S) | 36 (S) | 32 (S) | 32 (S) | 32 (S) | 22 (S) | 22 (S) | 14 (S) | 16 (S) | 32 (S) | 26 (S) |
Figure 1.
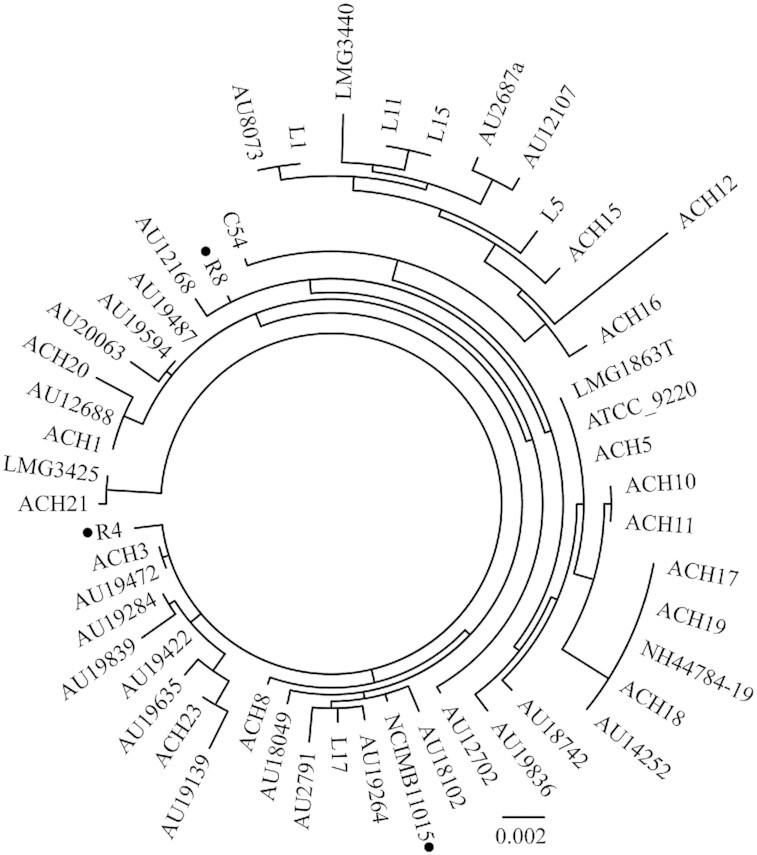
Maximum-likelihood phylogenetic relationship with 1000 bootstraps based on concatenated nucleotide sequences of seven housekeeping genes (eno, gltB, lepA, nrdA, nuoL, nusA and rpoB) of A. xylosoxidans strain R4, R8, NCIMB11015 and 48 other A. xylosoxidans strains (data obtained from pubMLST (http://pubmlst.org/achromobacter)). Strains R4, R8 and NCIMB11015 are labeled with a black dot.
The multidrug-resistant strains R4 and R8, were further analysed in order to determine MIC values (Table 1b). The two strains were resistant to aminoglycosides (GEN and AMK) and cephalosporins (CXM, CTX, CAZ and CRO), but varied in their susceptibility to fluoroquinolones, with strain R8 being less susceptible than strain R4. Moreover, both strains were resistant to SXT, a combination of sulfamethoxazole and trimethoprim, but susceptible to TGC. Unlike strain NCIMB11015, both strains R4 and R8 exhibited resistance to IPM, MEM and ETP (Table 1b).
Table 1b.
Characteristics and antimicrobial susceptibility profiles of multidrug-resistant A. xylosoxidans strains. Determination of levels of resistance using minimum inhibitory concentration (MIC).
| MIC (mg/L) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | TZP | CXM | CTX | CRO | CAZ | IPM | MEM | ETP | CIP | LVX | GEN | AMK | TGC | SXT |
| R4 | ≤8 | >16 | >32 | >32 | >32 | >8 | >8 | >4 | 1 | 1 | >8 | >32 | ≤0.25 | >4 |
| R8 | ≥32 | >16 | >32 | >32 | >32 | >8 | >8 | >4 | >2 | 8 | >8 | >32 | 1 | >4 |
Whereas strain R4 was susceptible to TZP, strain R8 was resistant. The activity of β-lactamase was then assayed by using nitrocefin hydrolysis. Prior to incubation, there was no statistically significant difference between the OD486/OC390 ratios of the three strains (ANOVA, F2,6 = 0.098, P = 0.908). The assay revealed that the change of OD486/OD390 ratio for strain R4 (T-test, Mean ± SD; 0.633 ± 0.096 vs 1.097 ± 0.066; P < 0.05) and strain R8 (T-test, Mean ± SD; 0.668 ± 0.047 vs 1.098 ± 0.010; P < 0.05) after 30 min incubation was significant, whereas the change of the ratio for strain NCIMB11015 was not significant (Mean ± SD; 0.642 ± 0.133 vs 0.698 ± 0.076; P = 0.262). There was a significant difference in the ratio among the three strains after incubation for 30 min (ANOVA, F2,6 = 38.16, P < 0.05). Post-hoc comparison using Tukey's test revealed that the OD486/OD390 ratio was significantly different between strain R4 and strain NCIMB11015, and between strain R8 and strain NCIMB11015, whereas it was not significantly different between strain R4 and strain R8. Taken together, these results suggest the activity of a metallo-β-lactamase in strains R4 and strain R8.
The identification of genes associated with Carbapenem resistance in strain R4 and R8
Whole genome sequencing provided complete chromosome assemblies for all three strains. Gene annotation using Prokka, together with RGI, identified 87 resistance genes, 92 resistance genes and 86 resistance genes in all for strain R4, R8 and NCIMB11015, respectively (Table S1, Supporting Information). Antibiotic resistance genes present in all three genomes included genes encoding a known species-specific β-lactamase, blaOXA-114 (Turton et al. 2011) and an RND-type efflux pump, axyA, axyB, axyM, which has been implicated in resistance to aminoglycosides, fluoroquinolones and cephalosporins (Bador et al. 2011, 2013). Interestingly, the blaIMP-14a gene was identified in the two carbapenem resistant strains (R4 and R8), whereas it was not present in the carbapenem sensitive strain (NCIMB11015).
Carbapenem resistance mediated by a novel genomic island, Achromobacter genomic island 1 (AcGI1)
Focusing on genes associated previously with carbapenem resistance, WGS identified blaIMP-14a located on a class 1 integron In687 (intI1- blaIMP-14a-aac(6’)-qacEdelta-sul1). This integron was identified in several clinical isolates from Thailand, including two isolates of Acinetobacter baumannii (strain Aci16 (Kansakar et al. 2011) and strain RA-41 520 608 (Genbank GQ302618)), which were also from Ramathibodi Hospital in Thailand in 2007 and 2009, respectively, Klebsiella pneumoniae (Matsumura et al. 2017) and Escherichia coli ST 131 strain 732 (Stoesser et al. 2016).
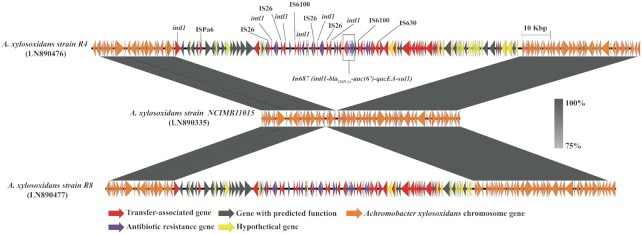
Having identified the In687 integron in the chromosomes of A. xylosoxidans strains R4 and R8, we proceeded to analyse the neighbourhood of the integron to better understand the genomic context. As a consequence of combining visualised genome comparison with NCIMB11015 and IslandViewer 4, we determined that the integron was part of a novel genomic island, which was 121 613 bp long, and this island was named Achromobacter genomic island 1 (AcG1; Fig. 2). AcGI1 islands in the two strains share more than 99.0% nucleic acid identity. Similar to other genomic islands, AcGI1 was inserted after a tRNAGly gene, however, direct repeats were not clearly identified. AcGI1 had a GC content of 61.5%, lower than the whole genome value of 67.5%.
Figure 2.
Schematic presentation of the A. xylosoxidans chromosome showing the insertion of Achromobacter genomic island (AcGI1) into the chromosome of A. xylosoxidans strain R4 (top) and strain R8 (bottom), compared with strain NCIMB11015 (middle) using EasyFig. The colored arrows indicate the direction of the transcription of predicted open reading frames. Grey regions indicate 80–100% sequence similarity.
This island contains 16 antibiotic resistance-associated genes responsible for resistance to β-lactam (blaIMP-14a), aminoglycosides (aac(6’), aminoglycoside O-phosphotransferase APH(3’)-Ib, APH(6)-I family aminoglycoside O-phosphotransferase), tetracycline (tetracycline repressor protein, tetracycline efflux MFS transporter, tetracycline resistance transcriptional regulator and tetracycline resistance protein) and quaternary compound (qacEdelta) antimicrobial agents. Also present were 44 mobile genetic element-associated genes and 38 hypothetical genes, suggesting that AcGI can be horizontally transferred between organisms.
We further investigated the structure of the element with sequence alignment against a non-redundant database. The best alignment result, with a high nucleotide identity (>90%), was to the genome of Klebsiella pneumoniae F128 (GenBank accession no. CP026149.1; 87% coverage of AcGI1, not including In687), indicating that A. xylosoxidans supporting the notion that AcGI1 can spread between species via horizontal gene transfer.
DISCUSSION
Horizontal gene transfer is recognised as an important mechanism contributing to the evolution of bacteria and the development of antibiotic resistance (Polz, Alm and Hanage 2013; Maddamsetti and Lenski 2018; Peterson and Kaur 2018; Sun et al. 2019). Genomic islands are genomic regions that often differ in %GC from the other parts of the chromosome, are locates near tRNA genes, and contain gene cassettes. Genomic islands can be located into several classes based on groups of genes carried, for example, pathogenicity island, resistance island and metabolism island, emphasizing the importance of these islands in bacterial evolution and adaptation.
Antibiotic resistance in bacterial pathogens has become a major public health concern, and the detection of resistance genes is urgently needed for the prevention of transmission of resistant organisms. Interestingly, antibiotic resistance can be genetically transferred between bacteria via horizontal gene transfer. Using Pacific Bioscience® single molecule real time sequencing, our comparative genomic analysis between two non-clonal carbapenem resistant clinical isolates of A. xylosoxidans (R4 and R8) and one carbapenem sensitive A. xylosoxidans (NCIMB11015) illustrates that a drug-resistance determining region was localized within a novel resistance island, named AcGI1. The high level of nucleotide sequence identity between the island in the genomes of strains R4 and R8 suggests that AcGI1 may have been transmitted within the hospital.
Multidrug-resistant bacteria have been identified in Thailand from various sources, including healthy adults, food contamination and hospital-associated infections. A previous study showed that the prevalence of carbapenem-resistant Enterobacteriaceae in Thailand is approximately 40–50%, which is the highest in the Asia–Pacific region. However, carbapenem-resistant A. xylosoxidans in Thailand have not been reported. Previous studies have shown that carbapenem resistance in A. xylosoxidans can be horizontally acquired, demonstrated by the identification of integron-associated resistance genes, such as VIM-type β-lactamase (Shin et al. 2005; Hishinuma et al. 2019). In this study, the carbapenem resistance of strains R4 and R8 is the result of a blaIMP-14a-carrying In687 located in AcGI1. In687 was firstly described as a part of a plasmid pEC732-IMP14 (Genbank accession no. CP015139), in E. coli. The pEC732-IMP14-carrying E. coli ST-131 was recovered from urine culture in Thailand (Stoesser et al. 2016). Therefore, this study is the first report describing the integration of In687 in the chromosome rather than on a plasmid.
An IMP-type β-lactamase gene was first described in Pseudomonas aeruginosa in Japan, and there are currently more than 50 allelic variants (Shakibaie, Azizi and Shahcheraghi 2017). In Thailand, the first blaIMP-14 was identified in a clinical isolate of P. aeruginosa from the teaching hospital in 2004 (Genbank accession no. AY553332) (Khuntayaporn et al. 2013). Subsequently, blaIMP-14a, with a single nucleotide difference, was identified in Acinetobacter baumannii in 2007 (Genbank accession no. HM036079) and 2009 (Genbank accession no. GQ302618), also in the same teaching hospital in Bangkok. Therefore, the presence of blaIMP-14 and blaIMP-14a in hospital-associated pathogens, including A. xylosoxidans, in Bangkok between 2004 and 2012, indicates that the circulation and distribution of this β-lactamase-containing mobile genetic element has been occurring for at least eight years in Bangkok.
CONCLUSIONS
Whole genome sequencing enables us to obtain a more comprehensive view of genetic of antibiotic resistance in bacteria. To our knowledge, this study is the first to identify a novel 122 kb genomic island in A. xylosoxidans, named AcGI1, containing blaIMP-14a-habouring In687. Interestingly, AcGI1 was identified in two carbapenem-resistance isolates, which are not clonally related. This finding provides evidence that AcGI1 can be transferred horizontally and contribute to the spread of carbapenem resistance hospital-associated infections.
Supplementary Material
Contributor Information
Pisut Pongchaikul, Chakri Naruebodindra Medical Institute, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bang Phli, Samut Prakan 10540, Thailand; Institute of Infection and Global Health, University of Liverpool, Liverpool L69 3BE, UK.
Pitak Santanirand, Department of Pathology, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Rama VI Road, Bangkok 10400, Thailand.
Svetlana Antonyuk, Institute of Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
Craig Winstanley, Institute of Infection and Global Health, University of Liverpool, Liverpool L69 3BE, UK.
Alistair C Darby, Institute of Integrative Biology, University of Liverpool, Liverpool L69 7ZB, UK.
FUNDING
This study was supported by Mahidol-Liverpool Chamlong Harinasuta PhD scholarship to P.P. and a Wellcome Trust ISSF tenure track fellowship to A.C.D.
Conflicts of interest
None declared.
REFERENCE
- Bador J, Amoureux L, Blanc Eet al. . Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-Type multidrug efflux pump. Antimicrob Agents Chemother. 2013;57:603–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bador J, Amoureux L, Duez J-MMet al. . First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob Agents Chemother. 2011;55:4912–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrado L, Branas P, Orellana MAet al. . Molecular characterization of achromobacter isolates from cystic fibrosis and non-cystic fibrosis patients in madrid, Spain. J Clin Microbiol. 2013;51:1927–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissimo F, Pinzone MR, Tosto Set al. . Achromobacter xylosoxidans meningitis in an immunosuppressed patient. QJM. 2013;107:65–6. [DOI] [PubMed] [Google Scholar]
- Bertelli C, Laird MR, Williams KPet al. . IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45:W30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braker G, Fesefeldt A, Witzel K-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl Envir Microbiol. 1998;64:3769–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan Vet al. . BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman Met al. . ACT: the Artemis comparison tool. Bioinformatics. 2005;21:3422–3. [DOI] [PubMed] [Google Scholar]
- Chin C-S, Alexander DH, Marks Pet al. . Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. [DOI] [PubMed] [Google Scholar]
- Coward A, Kenna DTD, Perry Cet al. . Use of nrdA gene sequence clustering to estimate the prevalence of different Achromobacter species among Cystic Fibrosis patients in the UK. J Cyst Fibros. 2015;15:479–85. [DOI] [PubMed] [Google Scholar]
- Di Pilato V, Pollini S, Rossolini GM. Characterization of plasmid pAX22, encoding VIM-1 metallo-$β$-lactamase, reveals a new putative mechanism of In70 integron mobilization. J Antimicrob Chemother. 2014;69:67–71. [DOI] [PubMed] [Google Scholar]
- Doi Y, Poirel L, Paterson DLet al. . Characterization of a naturally occurring class D β-lactamase from Achromobacter xylosoxidans. Antimicrob Agents Chemother. 2008;52:1952–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CR, Pressler T, Ridderberg Wet al. . Achromobacter species in cystic fibrosis: cross-infection caused by indirect patient-to-patient contact. J Cyst Fibros. 2013;2:609–15. [DOI] [PubMed] [Google Scholar]
- Hishinuma T, Tada T, Uchida Het al. . A novel VIM-Type Metallo-β-Lactamase variant, VIM-60, with increased hydrolyzing activity against fourth-generation cephalosporins in Pseudomonas aeruginosa clinical isolates in Japan. Antimicrob Agents Chemother. 2019;63:e00124–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen TH, Hansen MMA, Jensen PØet al. . Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes. Coenye T (ed.). PLoS One. 2013;8:e68484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, Raphenya AR, Alcock Bet al. . CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansakar P, Dorji D, Chongtrakool Pet al. . Local dissemination of multidrug-resistant Acinetobacter baumannii clones in a Thai hospital. Microb Drug Resist. 2011;17:109–19. [DOI] [PubMed] [Google Scholar]
- Khuntayaporn P, Montakantikul P, Santanirand Pet al. . Molecular investigation of carbapenem resistance among multidrug-resistant Pseudomonas aeruginosa isolated clinically in Thailand. Microbiol Immunol. 2013;57:170–8. [DOI] [PubMed] [Google Scholar]
- Maddamsetti R, Lenski RE. Analysis of bacterial genomes from an evolution experiment with horizontal gene transfer shows that recombination can sometimes overwhelm selection. Matic I (ed.). PLOS Genet. 2018;14:e1007199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura Y, Peirano G, Motyl MRet al. . Global molecular epidemiology of IMP-producing Enterobacteriaceae. Antimicrob Agents Chemother. 2017;61:e02729–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur AG, Waglechner N, Nizam Fet al. . The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwirth C, Freby C, Ogier-Desserrey Aet al. . VEB-1 in Achromobacter xylosoxidans from cystic fibrosis patient, France. Emerg Infect Dis. 2006;12:1737–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan CH, Morris A, Kirby SMet al. . Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972;1:283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018;9:2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Alm EJ, Hanage WP. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 2013;29:170–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournaras S, Kock R, Mossialos Det al. . Detection of a phylogenetically distinct IMP-type metallo- -lactamase, IMP-35, in a CC235 Pseudomonas aeruginosa from the Dutch-German border region (Euregio). J Antimicrob Chemother. 2013;68:1271–6. [DOI] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- Shakibaie MR, Azizi O, Shahcheraghi F. Insight into stereochemistry of a new IMP allelic variant (IMP-55) metallo-β-lactamase identified in a clinical strain of Acinetobacter baumannii. Infect Genet Evol. 2017;51:118–26. [DOI] [PubMed] [Google Scholar]
- Shin KS, Han K, Lee Jet al. . Imipenem-resistant Achromobacter xylosoxidans carrying blaVIM-2-containing class 1 integron. Diagn Microbiol Infect Dis. 2005;53:215–20. [DOI] [PubMed] [Google Scholar]
- Sofianou D, Markogiannakis A, Metzidie Eet al. . VIM-2 metallo-β-lactamase in Achromobacter xylosoxidans in Europe. Eur J Clin Microbiol Infect Dis. 2005;24:854–5. [DOI] [PubMed] [Google Scholar]
- Spilker T, Vandamme P, LiPuma JJ. Identification and distribution of Achromobacter species in cystic fibrosis. J Cyst Fibros. 2012;12:298–301. [DOI] [PubMed] [Google Scholar]
- Stoesser N, Sheppard AE, Peirano Get al. . First report of blaIMP-14 on a plasmid harboring multiple drug resistance genes in Escherichia coli sequence Type 131. Antimicrob Agents Chemother. 2016;60:5068–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Jeannot K, Xiao Yet al. . Editorial: horizontal gene transfer mediated bacterial antibiotic resistance. Front Microbiol. 2019;10:1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing . Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 10.0, 2020, The European Committee on Antimicrobial Susceptibility Testing. 2020. [Google Scholar]
- Turton JF, Mustafa N, Shah Jet al. . Identification of Achromobacter xylosoxidans by detection of the bla(OXA-114-like) gene intrinsic in this species. Diagn Microbiol Infect Dis. 2011;70:408–11. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Nagao M, Hotta Get al. . Molecular characterization of IMP-type metallo-β-lactamases among multidrug-resistant Achromobacter xylosoxidans. J Antimicrob Chemother. 2012;67:2110–3. [DOI] [PubMed] [Google Scholar]
- Zhao W-H, Hu Z-Q. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011;37:214–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.