Abstract
Modern machine learning systems, such as convolutional neural networks rely on a rich collection of training data to learn discriminative representations. In many medical imaging applications, unfortunately, collecting a large set of well-annotated data is prohibitively expensive. To overcome data shortage and facilitate representation learning, we develop Knowledge-guided Pretext Learning (KPL) that learns anatomy-related image representations in a pretext task under the guidance of knowledge from the downstream target task. In the context of utero-placental interface detection in placental ultrasound, we find that KPL substantially improves the quality of the learned representations without consuming data from external sources such as IMAGENET. It outperforms the widely adopted supervised pre-training and self-supervised learning approaches across model capacities and dataset scales. Our results suggest that pretext learning is a promising direction for representation learning in medical image analysis, especially in the small data regime.
1. Introduction
Recent years have seen breakthroughs in the field of medical image analysis driven by deep convolutional neural networks (CNNs). Despite advances in architectural refinement, incorporating more representative training data remains the most plausible way to improve model generalizability [1]. Recent successful medical deep-learning systems (e.g. [2,3]) rely on a large amount of annotated data to perform effective representation learning. For many medical applications, however, it is not applicable to build datasets of similar orders of magnitudes as those large-scale studies did [4].
Placenta accreta spectrum disorders (PASD) are adverse obstetric conditions of particular rarity. Ultrasonography-based prenatal assessment of PASD requires examination of structural and vascular abnormalities near the utero-placental interface (UPI). Manually localizing UPI can be challenging and time-consuming even for obstetric specialists due to the UPI’s variable shape and length compounded by low contrast, as shown in Fig. 1. These issues render learning-based detection of UPI clinically useful in assisting PASD assessment [5]. Unfortunately, the low incidence rate makes collecting large-scale annotated data prohibitively expensive in practice.
Fig. 1.
Placental ultrasound samples. UPIs are annotated as red curves, featured by the indistinct appearance with variable shape and length.
To facilitate feature learning under data shortage, we develop Knowledge-guided Pretext Learning (KPL) that learns anatomy-related image representations without using data from external sources such as IMAGENET. KPL works with a commonly used pretext task known as solving jigsaw puzzles [6,7]. Particularly, KPL tackles this task only in the region of interest, which is guided by supervision signals from the target task. We find that KPL substantially improves the quality of the learned representations for the downstream UPI detection task, outperforming widely adopted supervised pre-training and self-supervised learning approaches across model capacities and dataset scales. Our results demonstrate the potential of pretext learning in medical image analysis, especially in the small data regime.
Related work:
Transfer learning (TL) is a widely adopted technique in medical image analysis, whereby CNN architectures and their weights are first pre-trained on external sources (Xe, Ye) (e.g. IMAGENET) and then fine-tuned on downstream medical imaging data (Xt, Yt) (Fig. 2b). Compared to a random initialization approach (Fig. 2a), TL is reported to benefit from feature reuse [1, 8]. However, recent studies have challenged some common beliefs about TL. For instance, He et al. showed that TL indeed helped achieve faster convergence, but did not necessarily yield better performance compared to random initialization [9]. Raghu et al. demonstrated that randomly initialized small models could perform comparably to large IMAGENET pre-trained models on two medical imaging tasks [8]. Their findings imply that good performance on external data is not a reliable indicator of success on downstream medical imaging tasks. These results raise questions about the applicability of current TL approaches.
Fig. 2.
Four popular feature-related training setups in medical imaging and computer vision. (a) depicts a random initialization approach by initializing the network with zero-mean Gaussian weights [10]; (b) illustrates the widely-adopted transfer learning (TL) approach, which requires well-annotated external data; (c) denotes a generic self-supervised learning (SSL) approach, where features are learnt using freely available supervisions derived from certain types of transformation Φ; (d) shows the proposed knowledge-guided pretext learning approach, where the transformation Φ′ is guided by knowledge from the downstream task. Neither SSL or KPL requires any external data.
Alternatively, self-supervised learning (SSL) aims to obtain semantically relevant image representations without any supervision. Instead, it devises pretext tasks by applying a pre-defined transformation Φ to input images Xt and training a model to predict properties of the transformation from the transformed images Φ(Xt) [11], for which ground truth is freely available (Fig. 2c). Such pretext task includes, but are not limited to, solving patch-wise jigsaw puzzles [6,7], predicting image rotation [12,13], determining relative position [14] and absolute position [15]. However, since routine placental ultrasound scans are sector-shaped, directly applying a generic SSL transformation can be problematic because an SSL model can easily over-fit to sector boundaries without actually learning useful semantics. To overcome this issue, we propose to solve pretext tasks under the guidance of Yt (i.e. knowledge from downstream tasks, Fig. 2–d). The resulting transformation Φ′(Xt, Yt) only performs feature learning in the region of interest determined by Yt, reducing the risk of over-fitting to sector boundaries.
2. Methods
Jigsaw pretext learning:
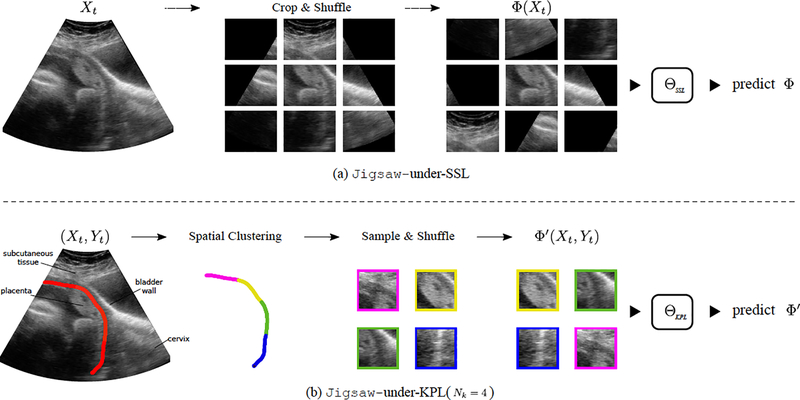
Noroozi et al. proposed to learn image representation in a self-supervised manner by solving jigsaw puzzles induced from the given image [6]. As shown in Fig. 3a, it works by first cropping an input image Xt into a grid of 3 × 3 non-overlapping patches. After shuffling these patches according to a permutation Φ randomly selected from a set of pre-defined permutations, a CNN ΘSSL is then trained to solve this puzzle by predicting Φ given these unordered patches Φ(Xt). Unfortunately, sector boundaries are more discriminative than the actual visual contents in this puzzle-solving scenario, which likely renders the learnt representation irrelevant for downstream tasks.
Fig. 3.
(a)Jigsaw-under-SSL solves a generic puzzle by shuffling a grid of 3 × 3 non-overlapping patches without considering the visual contents. (b) Jigsaw-under-KPL solves a specific puzzle guided by Yt, which reduces the occurrence of sector boundaries.
To this end, we propose to solve jigsaw puzzles by repurposing the downstream supervision signal Yt to guide representation learning. We refer to this method as knowledge-guided pretext learning (KPL). Fig. 3b illustrates the process of solving jigsaw puzzles under KPL. We first perform a spatial clustering that groups UPI pixels into Nk segments, as encoded in colours. A sampling step follows by cropping Nk square patches from Xt, whose centres are randomly drawn from pixels on Nk UPI segments respectively in a specific order. For simplicity, we take the order from top left to bottom right along UPI. Similar to Jigsaw-under-SSL, a random shuffle Φ′ is applied to these patches and a CNN ΘKPL is then trained to predict this permutation. As illustrated in Fig. 4a, ΘKPL is an Nk-way Siamese CNN with shared parameters that map each patch into a 128-d vector. The resulting Nk vectors are concatenated to predict Φ′ as a classification task [16]. The total number of permutations |P| is at most Nk! (e.g. a maximum of 24 permutations for Nk = 4). As illustrated in Fig. 1 and Fig. 3b, the UPI neighbourhood carries rich semantics about placenta anatomy. We expect that the sampled patches also contain key information relevant to the downstream UPI detection task.
Fig. 4.
(a) presents a Nk-way Siamese architecture to predict permutation given a set of N patches. Each patch is encoded by a 128-d vector. (b) illustrates two patch-sampling strategies. The proposed off-boundary sampling assigns higher weights to positions closer to the image centre, thus further reduces the occurrence of sector boundaries.
Off-boundary sampling:
As opposed to SSL, the proposed Jigsaw-under-KPL conducts feature learning only in the region of interest through the guidance of the downstream task, thus alleviates the risk of over-fitting to sector boundaries. However, patches drawn from top-left and bottom-right UPI segments may still contain sector boundaries when sampled uniformly. To further reduce this risk, we propose a heuristic off-boundary sampling approach. As illustrated in Fig. 4b, patch centres are sampled from the minimum bounding boxes of UPI segments. Moreover, positions in each bounding box are more likely to be drawn if they are closer to the image centre. Here the colour transparency encodes sampling likelihood. Off-boundary sampling further reduces the occurrence of sector boundaries. Our experiments empirically support this design choice over a uniform sampling approach.
UPI detection:
We evaluate the performance of KPL in a transfer learning setting. Specifically, a backbone CNN is first trained for Jigsaw-under-KPL. It is then modified for UPI detection by (i) removing the fully-connected layer; (ii) adding computational modules according to specific design choices. The resulting network is then fine-tuned on the downstream UPI detection task. No weight is frozen. Following [17,18,5], we adopt the classic holistically-nested edge detector (HED) in our study. Briefly, HED applies deep supervision on top of a backbone CNN to regularize intermediate feature learning.
Concretely, UPI detection is formulated as a binary segmentation task with Yt ∈ {0, 1}H×W being the reference UPI mask, with UPI pixels taking the value 1. Given an input image Xt, a UPI detection network predicts the probability for each pixel position p in Xt, where is the network output given Xt. Following [5], we adopt a weighted cross-entropy loss:
| (1) |
where is a class-balancing weight that equalizes the expected weight update for both classes, with nd denoting the amount of UPI and non-UPI pixels in Yt [19].
3. Experiments
Dataset:
We had available 101 3D placental ultrasound volumes from 101 subjects at high risks of PASD [20]. Forty-eight volumes were confirmed histopathologically to have PASD, and the rest confirmed to have normal placentation. Static transabdominal 3D ultrasound volumes of the placental bed were obtained. Data usage was approved by the local research ethics committee. The median gestation was 30 1/7 weeks. All volumes were sliced along the sagittal plane. The resulting 11,166 2D image planes had the UPI manually annotated by X (a computer scientist) under the guidance of Y (an obstetric specialist). The median spatial resolution was 392×553 px, with a spatial sampling rate of 0.33 mm/pixel. A subject-level random split was performed to have 7,340 images from 67 subjects for training, 1,954 images from 17 subjects for validation and 1,872 images from 17 subjects for testing.
Model setup:
To investigate the effects of representation learning on downstream transfer task performance, we evaluate on the following setups: (i) training from random initialization; (ii) transfer learning from IMAGENET pre-trained weights; (iii) transfer learning from CHEXPERT pre-trained weights [21]; (iv) transfer learning from Jigsaw-under-SSL; (v) transfer learning from Jigsaw-under-KPL. We further test on three HED instances with different backbones to investigate the effects of model capacity on downstream task performance. These backbones include: (i) Tiny-VGG1; (ii) VGG-13 [22]; (iii) VGG-19 [22], whose number of parameters before and after removing the fully-connected layers are listed in Table 2.
Table 2.
Configurations of three backbone CNNs, including depth, number of parameters and ImageNet Top-1 and Top-5 accuracies on the validation set.
| Depth | # params with / without FC layers | Top-1 Acc. | Top-5 Acc. | |
|---|---|---|---|---|
| Tiny-VGG | 13 | 70.0 M / 1.1 M | 63.6% | 85.1% |
| VGG-13 | 13 | 126.9 M / 9.0 M | 71.6% | 90.4% |
| VGG-19 | 19 | 137.0 M / 19.1 M | 74.2% | 91.8% |
Implementation:
Basic geometry and contrast jittering were applied for data augmentation (see Appendix). For SSL and KPL, an input image was resized to 390 × 390 px and patches with size 120 × 120 px were drawn. We used the Adam optimizer with a mini-batch size of 16. Models were trained for 16 epochs, and the weights were stored for subsequent fine-tuning on the downstream task. For UPI detection task, an input image was resized to 360 × 360 px. We used a mini-batch size of 6 and optimized models with Adam for 20 epochs on NVIDIA Tesla P100 GPUs. The learning rate was set to 9e-4 and weight-decay was set to 1.1e-5. We evaluate UPI detection using a standard edge detection metric: the best F-measure on the dataset for a fixed prediction threshold (optimal dataset scale, or ODS [23]). All models were implemented with PyTorch.
Effects of feature learning:
Table 1 compares the performance of UPI detection in terms of ODS score, under five setups. For TL-IMAGENET and TL-CHEXPERT, backbone CNNs were first pre-trained on the corresponding external datasets before fine-tuned on UPI data. Table 2 and Table 3 display the pre-training results in terms of Top-1 and Top-5 accuracies for IMAGENET and class-wise AUC scores for CHEXPERT. For SSL and KPL, an important hyper-parameter is the permutation number |P|, which is determined using the validation set. According to results in Fig. 5a–b, we set |P| = 90 for Jigsaw-under-SSL and |P| = 120 (Nk = 5) for Jigsaw-under-KPL. Note that |P| can be as high as 9! for SSL, yet we find that a very large |P| does not help with the downstream task. On the other hand, increasing |P| up to 120 helps with UPI detection under KPL, which indicates that KPL is able to learn good representations that benefit the downstream task.
Table 1.
Comparison of the ODS scores on the test set for UPI detection under five setups. Three HED instances with different backbones were listed. Rand. Init. denotes training from random initialization. TL-(·) denotes transfer learning from the corresponding external database. Performance of Jigsaw-under-SSL with |P| = 90 and Jigsaw-under-KPL with Nk = 5 and |P| = 120 are reported.
| Rand. Init. | TL-ImageNet | TL-CheXpert | SSL (|P| = 90) | KPL (|P| = 120) | |
|---|---|---|---|---|---|
| Tiny-VGG | 0.562 | 0.583 | 0.569 | 0.574 | 0.578 |
| VGG-13 | 0.566 | 0.576 | 0.560 | 0.562 | 0.571 |
| VGG-19 | 0.582 | 0.595 | 0.583 | 0.596 | 0.605 |
Table 3.
AUC scores of three backbone CNNs on the validation set of CheXpert using the U-Ones approach in [21]. Column 2–6 report the binary classification AUC scores for five chest-related clinical observations.
| AUC | Cardiomegaly | Edema | Consolidation | Atelectasis | Pleural Effusion |
|---|---|---|---|---|---|
| Tiny-VGG | 0.812 | 0.897 | 0.901 | 0.848 | 0.915 |
| VGG-13 | 0.820 | 0.875 | 0.911 | 0.858 | 0.919 |
| VGG-19 | 0.827 | 0.857 | 0.903 | 0.880 | 0.902 |
Fig. 5.
(a) and (b) display performance of transfer learning from Jigsaw-under-SSL and Jigsaw-under-KPL respectively, by varying the permutation number |P|. ODS scores on the validation set are reported. (c) presents the training losses of HED (VGG19) instances under different setups. Note that IMAGENET pre-trained weights and KPL achieve the best convergence speeds in terms of training steps.
According to Table 1, UPI detection generally benefits from transfer learning, as opposed to the conclusion drawn by [8] for image classification. This result implies that localization tasks may be more feature-sensitive than classification tasks. The proposed KPL outperforms the Rand. Init., TL-CHEXPERT and SSL approaches in terms of ODS and performs comparably to TL-IMAGENET. The best performance (ODS=0.605) was achieved by KPL under VGG-19. Interestingly, TL-IMAGENET achieves a better performance than TL-CHEXPERT. One may expect the opposite since CHEXPERT is a medical imaging database after all. In terms of convergence speed, KPL and TL-IMAGENET achieve the best performance compared to the rest, as illustrated in Fig. 5c. This result further indicates that KPL can produce good representations that generalize well for the downstream task.
Effects of backbone capacity:
According to Table 1, HED instances powered by VGG-19 generally outperform those powered by Tiny-VGG and VGG-13. Tiny-VGG and VGG-13 achieve similar performances across different setups. Note that Tiny-VGG has the same depth as VGG-13 but with fewer channels in each block. This result indicates that increasing model depth can be more beneficial than increasing channel numbers for UPI detection.
As shown in Table 2 and Table 3, three backbone CNNs achieve significantly different classification accuracies on IMAGENET but perform comparably to each other on CHEXPERT. This result further validates that performance on an external database is not predictive of performance on the target task [8]. It is worth noting that the proposed KPL method does not exploit any external database and performs consistently well with different backbones.
Performance in the small data regime:
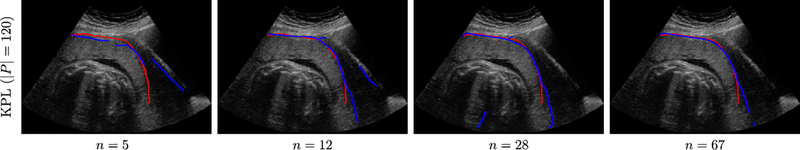
We further evaluate the effect of feature learning in the small data regime by training HEDs with VGG-19 backbones on data from n ∈ {5, 12, 28} subjects respectively. Test results are displayed in Table 4. Fig. 6 displays some exemplar detection results. Compared to Rand. Init., TL (IMAGENET) and SSL, the proposed KPL method can substantially boost detection performance under data shortage. For instance, with only 12 subjects, KPL can learn rich representations that transfer very well to UPI detection (ODS=0.521), even outperforming Rand. Init. trained on 28 subjects.
Table 4.
Comparison of the ODS scores in the small data regime. HED models are powered by VGG-19 backbones.
| # Subject (# plane) | Rand. Init. | TL-ImageNet | SSL (|P| = 90) | KPL (|P| = 120) |
|---|---|---|---|---|
| 5 (640) | 0.442 | 0.470 | 0.417 | 0.445 |
| 12 (1,350) | 0.467 | 0.481 | 0.452 | 0.521 |
| 28 (3,025) | 0.495 | 0.535 | 0.546 | 0.567 |
| 67 (7,340) | 0.582 | 0.595 | 0.596 | 0.605 |
Fig. 6.
KPL-powered UPI detection samples with an increase of training subjects (n ∈ {5, 12, 28, 67}). Red curves represent reference UPI masks and blue curves represent model predictions.
4. Conclusion
In this paper, we propose knowledge-guided pretext learning (KPL) for utero-placental interface detection (UPI) in placental ultrasound. KPL only attends the region of interest for representation learning, as guided by downstream supervision. Experiments demonstrate that UPI detection benefits from representations learnt from KPL in terms of detection performance and convergence speed. In the small data regime, KPL outperforms popular feature learning strategies without using any external data. Our results show the potential of pretext learning in medical imaging tasks. Future work shall focus on investigating more application-specific pretext learning tasks.
Supplementary Material
Acknowledgements.
Huan Qi is supported by a China Scholarship Council doctoral research fund (grant No. 201608060317). The NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development Human Placenta Project UO1 HD 087209, EPSRC grant EP/M013774/1, and ERC-ADG-2015 694581 are also acknowledged.
Footnotes
References
- 1.Shin Hoo-Chang et al. Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning. IEEE Transactions on Medical Imaging, 35(5):1285–1298, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gulshan Varun et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama, 316(22):2402–2410, 2016. [DOI] [PubMed] [Google Scholar]
- 3.Esteva Andre et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature, 542(7639):115–118, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Hyunkwang et al. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nature Biomedical Engineering, 3(3):173, 2019. [DOI] [PubMed] [Google Scholar]
- 5.Qi Huan et al. Upi-net: Semantic contour detection in placental ultrasound. In ICCV-VRMI, 2019. [Google Scholar]
- 6.Noroozi Mehdi and Favaro Paolo. Unsupervised learning of visual representations by solving jigsaw puzzles. In ECCV, 2016. [Google Scholar]
- 7.Taleb Aiham et al. Multimodal self-supervised learning for medical image analysis. arXiv preprint arXiv:1912.05396, 2019. [Google Scholar]
- 8.Raghu Maithra et al. Transfusion: Understanding transfer learning for medical imaging. In NeurIPS, 2019. [Google Scholar]
- 9.He Kaiming et al. Rethinking imagenet pre-training. In ICCV, 2019. [Google Scholar]
- 10.Szegedy Christian et al. Going deeper with convolutions. In CVPR, 2015. [Google Scholar]
- 11.Misra Ishan and van der Maaten Laurens. Self-supervised learning of pretext-invariant representations. arXiv preprint arXiv:1912.01991, 2019. [Google Scholar]
- 12.Gidaris Spyros et al. Unsupervised representation learning by predicting image rotations. In ICLR, 2018. [Google Scholar]
- 13.Tajbakhsh Nima et al. Surrogate supervision for medical image analysis: Effective deep learning from limited quantities of labeled data. In ISBI, 2019. [Google Scholar]
- 14.Doersch Carl et al. Unsupervised visual representation learning by context prediction. In ICCV, 2015. [Google Scholar]
- 15.Bai Wenjia et al. Self-supervised learning for cardiac mr image segmentation by anatomical position prediction. In MICCAI, 2019. [Google Scholar]
- 16.Goyal Priya et al. Scaling and benchmarking self-supervised visual representation learning. In ICCV, 2019. [Google Scholar]
- 17.Xie Saining and Tu Zhuowen. Holistically-nested edge detection. In ICCV, 2015. [Google Scholar]
- 18.Yu Zhiding et al. Casenet: Deep category-aware semantic edge detection. In CVPR, 2017. [Google Scholar]
- 19.Lawrence Steve et al. Neural network classification and prior class probabilities In Neural networks: Tricks of the trade, pages 295–309. Springer, 2012. [Google Scholar]
- 20.Collins SL et al. Influence of power doppler gain setting on virtual organ computer-aided analysis indices in vivo: can use of the individual sub-noise gain level optimize information? Ultrasound in Obstetrics & Gynecology, 40(1):75–80, 2012. [DOI] [PubMed] [Google Scholar]
- 21.Irvin Jeremy et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In AAAI, 2019. [Google Scholar]
- 22.Simonyan Karen and Zisserman Andrew. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556, 2014. [Google Scholar]
- 23.Arbelaez Pablo et al. Contour detection and hierarchical image segmentation. IEEE transactions on Pattern Analysis and Machine Intelligence, 33(5):898–916, 2010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.