Abstract
Objective
To assess the consistency of machine learning and statistical techniques in predicting individual level and population level risks of cardiovascular disease and the effects of censoring on risk predictions.
Design
Longitudinal cohort study from 1 January 1998 to 31 December 2018.
Setting and participants
3.6 million patients from the Clinical Practice Research Datalink registered at 391 general practices in England with linked hospital admission and mortality records.
Main outcome measures
Model performance including discrimination, calibration, and consistency of individual risk prediction for the same patients among models with comparable model performance. 19 different prediction techniques were applied, including 12 families of machine learning models (grid searched for best models), three Cox proportional hazards models (local fitted, QRISK3, and Framingham), three parametric survival models, and one logistic model.
Results
The various models had similar population level performance (C statistics of about 0.87 and similar calibration). However, the predictions for individual risks of cardiovascular disease varied widely between and within different types of machine learning and statistical models, especially in patients with higher risks. A patient with a risk of 9.5-10.5% predicted by QRISK3 had a risk of 2.9-9.2% in a random forest and 2.4-7.2% in a neural network. The differences in predicted risks between QRISK3 and a neural network ranged between –23.2% and 0.1% (95% range). Models that ignored censoring (that is, assumed censored patients to be event free) substantially underestimated risk of cardiovascular disease. Of the 223 815 patients with a cardiovascular disease risk above 7.5% with QRISK3, 57.8% would be reclassified below 7.5% when using another model.
Conclusions
A variety of models predicted risks for the same patients very differently despite similar model performances. The logistic models and commonly used machine learning models should not be directly applied to the prediction of long term risks without considering censoring. Survival models that consider censoring and that are explainable, such as QRISK3, are preferable. The level of consistency within and between models should be routinely assessed before they are used for clinical decision making.
Introduction
Risk prediction models are used routinely in healthcare practice to identify patients at high risk and make treatment decisions, so that appropriate healthcare resources can be allocated to those patients who most need care.1 These risk prediction models are usually built using statistical regression techniques. Examples include the Framingham risk score (developed from a US cohort with prospectively collected data)2 and QRISK3 (developed from a large UK cohort using retrospective electronic health records).3 Recently, machine learning models have gained considerable popularity. The English National Health Service has invested £250m ($323m; €275m) to further embed machine learning in healthcare.4 A recent viewpoint article suggested that machine learning technology is about to start a revolution with the potential to transform the whole healthcare system.5 Several studies suggested that machine learning models could outperform statistical models in terms of calibration and discrimination.6 7 8 9 However, another viewpoint concerns the fact that these approaches cannot provide explainable reasons behind their predictions, potentially leading to inappropriate actions,10 and a recent review found no evidence that machine learning models had better model performance than logistic models.11 However, interpretation of this review is difficult, as it included models from mostly small sample sizes and with different outcomes and predictors. Machine learning has established strengths in image recognition that could help in diagnosing diseases in healthcare,12 13 14 15 but censoring (patients lost to follow-up), which is common in risk prediction, does not exist in image recognition. Many commonly used machine learning models do not take into account censoring by default.16
The objective of this study was to assess the robustness and consistency of a variety of machine learning and statistical models on individual risk prediction and the effects of censoring on risk predictions. We used cardiovascular disease as an exemplar. We defined robustness of individual risk prediction as the level of consistency in the prediction of risks for individual patients with models that have comparable population level performance metrics.17 18 19
Methods
Data source
We derived the study cohort from Clinical Practice Research Datalink (CPRD GOLD), which includes data from about 6.9% of the population in England.20 It also has been linked to Hospital Episode Statistics, Office for National Statistics mortality records, and Townsend deprivation scores,3 to provide additional information about hospital admissions (including date and discharge diagnoses) and cause specific mortality.20 CPRD includes patients’ electronic health records from general practice, capturing detailed information such as demographics (age, sex, and ethnicity), symptoms, tests, diagnoses, prescribed treatments, health related behaviours, and referrals to secondary care.20 CPRD is a well established representative cohort of the UK population, and thousands of studies have used it,21 22 including a validation of the QRISK2 model and an analysis of machine learning.8 23
Study population
This study used the same selection criteria for the study population, risk factors, and cardiovascular disease outcomes as were used for QRISK3.3 18 Follow-up of patients started at the date of the patient’s registration with the practice, their 25th birthday, or 1 January 1998 (whichever was latest) and ended at the date of death, incident cardiovascular disease, date of leaving the practice, or last date of data collection (whichever was earliest). The index date for measurement of cardiovascular disease risk was randomly chosen from the follow-up period to capture time relevant practice variability with a better spread of calendar time and age.24 This was different from QRISK3, for which a single calendar time date was mostly used.18 The main inclusion criteria were age between 25 and 84 years, no history of cardiovascular disease, and no prescription for a statin before the index date. The outcome of interest was the 10 year risk of developing cardiovascular disease. The definition of the primary clinical outcome (cardiovascular disease) was the same as for QRISK3 (that is, coronary heart disease, ischaemic stroke, or transient ischaemic attack).3
We extracted two main cohorts from the study population—one overall cohort including all patients with at least one day of follow-up and one cohort with censored patients removed. The cohort without censoring excluded patients who were lost to follow-up before developing cardiovascular disease by year 10. The analysis of the cohort without censoring aimed to investigate the effects of ignoring censoring on patients’ individual risk predictions. This cohort mimics the methods used by some machine learning studies—that is, only patients or practices with full 10 years’ follow-up were selected.8
Cardiovascular disease risk factors
The cardiovascular disease risk factors at the index date included sex; age; body mass index; smoking history; total cholesterol to high density lipoprotein cholesterol ratio; systolic blood pressure and its standard deviation; history of prescribing of atypical antipsychotic drugs; blood pressure treatment or regular oral glucocorticoids; clinical history of systemic lupus erythematosus, atrial fibrillation, chronic kidney disease (stage 3, 4, or 5), erectile dysfunction, migraine, rheumatoid arthritis, severe mental illness, or type 1 or 2 diabetes mellitus; family history of angina or heart attack in a first degree relative aged under 60 years; ethnicity; and Townsend deprivation score.3 The same predictors from QRISK33 were used for all model fitting except for Framingham,25 which used fewer and different predictors.
Machine learning and Cox models
The study considered 19 models, including 12 families of machine learning, three Cox proportional hazards models (local fitted, QRISK3, and Framingham), three parametric survival models (assuming Weibull, Gaussian, and logistic distribution), and a statistical logistic model (fitted in a statistical causal-inference framework). Machine learning models included logistic model (fitted in an automated machine learning framework),26 random forest,27 and neural network28 from R package “Caret” 29; logistic model, random forest, neural network, extra-tree model,30 and gradient boosting classifier30 from Python package “Sklearn”31; and logistic model, random forest, neural network, and autoML32 from Python package “h2o.”33 The package autoML selects a best model from a broader spectrum of candidate models.32 Details of these models are summarised in eTable 1. The study used the machine learning algorithms from different software packages, with a grid search process on hyper-parameters and cross validation, to acquire a series of high performing machine learning models; this mimics the reality that practitioners may subjectively select different packages for model fitting and end up with a different best model. The study treated the models from the same machine learning algorithm but different software packages as different model families, as the settings (hyper-parameters) of these packages to control the model fitting are often different, which might result in a different best performing model through the grid search process.
Statistical analysis
We used the Markov chain Monte Carlo method with monotone style to impute missing values 10 times for ethnicity (54.3% missing in overall cohort), body mass index (40.3%), Townsend score (0.1%), systolic blood pressure (26.9%), standard deviation of systolic blood pressure (53.9%), ratio of total cholesterol to high density lipoprotein cholesterol (65.0%), and smoking status (25.2%)18 (only these variables had missing values). We randomly split the overall cohort (which contained 10 imputations) into an overall derivation cohort (75%) and an overall testing cohort (25%). We grid searched a total of 1200 machine learning models with the highest discrimination (C statistic) on hyper-parameters with twofold cross validation estimating calibration and discrimination. They were derived from 12 model families of 100 samples with similar sample size to another machine learning study.8 We then estimated the individual cardiovascular disease risk predictions (averaged for missing value imputations) and model performance of all models by using the overall testing cohort. The sample splitting and model fitting process is shown in eFigure 1.
We compared distributions of risk predictions for the same individual among models. We plotted the differences of individual cardiovascular disease risk predictions between models against deciles of cardiovascular disease risk predictions for QRISK3. We produced Bland-Altman plots—a graphical method to compare two measurement techniques across the full spectrums of values.34 These plotted the differences of individual risk predictions between two models against the average individual risk prediction.34
We used R to fit the models from “Caret” and Python to fit models from “Sklearn” and “h2o.”29 30 We used SAS procedures to extract the raw data, create analysis datasets, and generate tables and graphs.35
Patient and public involvement
No patients were involved in setting the research question or the outcome measures, nor were they involved in developing plans or implementation of the study. No patients were asked to advise on interpretation or writing up of results.
Results
The overall study population included 3.66 million patients from 391 general practices. The cohort without censoring was considerably smaller (0.45 million) than the overall cohort. Table 1 shows the baseline characteristics of the two study populations, which were split into derivation and validation cohorts. The average age was higher in the cohort without censoring (owing to younger patients leaving the practice as shown in eFigure 11).
Table 1.
Baseline characteristics of two study populations (patients aged 25-84 years without history of cardiovascular disease (CVD) or previous statin use). Values are numbers (percentages) unless stated otherwise
| Characteristics | Overall cohort | Cohort without censoring | |||
|---|---|---|---|---|---|
| Derivation cohort (n=2 746 453) | Validation cohort (n=915 479) | Derivation cohort (n=335 632) | Validation cohort (n=111 868) | ||
| CVD cases | 86 769 (3.2) | 28 828 (3.1) | 78 826 (23.5) | 26 168 (23.4) | |
| Patients censored within 10 years | 2 410 516 (87.8) | 803 916 (87.8) | NA | NA | |
| CVD risk factors | |||||
| Female sex | 1 406 796 (51.2) | 469 098 (51.2) | 173 691 (51.8) | 58 169 (52.0) | |
| Mean (SD) age, years | 44.7 (15.6) | 44.7 (15.7) | 53.3 (16.2) | 53.4 (16.2) | |
| Mean (SD) body mass index | 26.7 (5.0) | 26.7 (5.0) | 27.2 (4.8) | 27.1 (4.8) | |
| Mean (SD) total cholesterol/high density lipoprotein cholesterol ratio | 3.9 (1.3) | 3.9 (1.3) | 4.1 (1.3) | 4.1 (1.3) | |
| Atypical antipsychotic drug | 12 306 (0.4) | 4030 (0.4) | 932 (0.3) | 316 (0.3) | |
| Antihypertensive treatment | 183 964 (6.7) | 61 962 (6.8) | 42 704 (12.7) | 14 245 (12.7) | |
| Regular steroid tablets | 2059 (0.1) | 694 (0.1) | 289 (0.1) | 100 (0.1) | |
| History of systemic lupus erythematosus | 1840 (0.1) | 606 (0.1) | 257 (0.1) | 74 (0.1) | |
| History of angina or heart attack in first degree relative <60 years | 98 455 (3.6) | 32 619 (3.6) | 7950 (2.4) | 2669 (2.4) | |
| History of atrial fibrillation | 20 778 (0.8) | 6965 (0.8) | 5213 (1.6) | 1757 (1.6) | |
| History of chronic kidney disease (stage 3, 4, or 5) | 30 133 (1.1) | 10 240 (1.1) | 4364 (1.3) | 1514 (1.4) | |
| History of erectile dysfunction | 39 651 (1.4) | 13 110 (1.4) | 3867 (1.2) | 1287 (1.2) | |
| History of migraines | 177 439 (6.5) | 59 106 (6.5) | 19 629 (5.8) | 6593 (5.9) | |
| History of rheumatoid arthritis | 16 167 (0.6) | 5459 (0.6) | 3043 (0.9) | 1030 (0.9) | |
| History of severe mental illness | 219 861 (8.0) | 72 832 (8.0) | 32 190 (9.6) | 10 673 (9.5) | |
| History of type 1 diabetes | 5899 (0.2) | 2097 (0.2) | 820 (0.2) | 251 (0.2) | |
| History of type 2 diabetes | 35 569 (1.3) | 11 826 (1.3) | 8134 (2.4) | 2641 (2.4) | |
| Mean (SD) systolic blood pressure | 126.9 (16.7) | 126.9 (16.7) | 133.1 (18.4) | 133.1 (18.4) | |
| Mean (SD) standard deviation of each individual patient’s systolic blood pressure | 9.9 (5.6) | 9.9 (5.6) | 10.7 (5.9) | 10.7 (5.9) | |
| Ethnicity | |||||
| Other ethnicity | 173 271 (6.3) | 58 124 (6.3) | 6900 (2.1) | 2273 (2.0) | |
| White or not recorded | 2 573 182 (93.7) | 857 355 (93.7) | 328 732 (97.9) | 109 595 (98.0) | |
| Smoking | |||||
| Ex-smoker | 629 500 (22.9) | 209 186 (22.8) | 76 060 (22.7) | 25 429 (22.7) | |
| Current smoker | 806 978 (29.4) | 269 717 (29.5) | 94 082 (28.0) | 31 335 (28.0) | |
| Never smoker | 1 309 975 (47.7) | 436 576 (47.7) | 165 490 (49.3) | 55 104 (49.3) | |
| Townsend deprivation | |||||
| Score 1—Least deprived | 600 392 (21.9) | 199 937 (21.8) | 86 596 (25.8) | 29 156 (26.1) | |
| Score 2 | 594 739 (21.7) | 197 677 (21.6) | 82 211 (24.5) | 27 266 (24.4) | |
| Score 3 | 572 903 (20.9) | 191 045 (20.9) | 69 897 (20.8) | 23 326 (20.9) | |
| Score 4 | 568 006 (20.7) | 189 520 (20.7) | 60 424 (18.0) | 20 140 (18.0) | |
| Score 5—Most deprived | 410 413 (14.9) | 137 300 (15.0) | 36 504 (10.9) | 11 980 (10.7) | |
Table 2 shows the model performance of the machine learning and statistical models. All models had very similar discrimination (C statistics of about 0.87) and calibration (Brier scores of about 0.03 in eTables 2-4 and eFigures 3-4).
Table 2.
Performance indicators of machine learning and statistical models in overall cohort
| Model | Model performance*: C statistic (95% range†) | Average absolute change in model performance: % (95% range†) |
|---|---|---|
| Logistic (Caret) | 0.879 (0.879 to 0.879) | 0.00 (−0.03 to 0.04) |
| Random forest (Caret) | 0.869 (0.867 to 0.869) | −1.20 (−1.33 to −1.10%) |
| Neural network (Caret) | 0.878 (0.867 to 0.880) | −0.15 (−1.35 to 0.06) |
| Statistic logistic model | 0.879 (0.879 to 0.879) | 0.01 (−0.02 to 0.04) |
| QRISK3 | 0.879 | Reference model |
| Framingham | 0.865 | −1.66 (−1.66 to −1.66) |
| Local Cox model | 0.877 (0.877 to 0.878) | −0.22 (−0.28 to −0.17) |
| Parametric survival model (Weibull) | 0.877 (0.876 to 0.877) | −0.29 (−0.35 to −0.24) |
| Parametric survival model (Gaussian) | 0.876 (0.876 to 0.877) | −0.33 (−0.39 to −0.29) |
| Parametric survival model (Logistic) | 0.876 (0.875 to 0.876) | −0.36 (−0.43 to −0.31) |
| Logistic (Sklearn) | 0.879 (0.879 to 0.879) | 0.00 (−0.05 to 0.03) |
| Random forest (Sklearn) | 0.872 (0.871 to 0.873) | −0.80 (−0.89 to −0.71) |
| Neural network (Sklearn) | 0.872 (0.832 to 0.879) | −0.85 (−5.39 to −0.03) |
| Gradient boosting classifier (Sklearn) | 0.878 (0.877 to 0.878) | −0.17 (−0.29 to −0.08) |
| extra-trees (Sklearn) | 0.863 (0.861 to 0.864) | −1.89 (−2.05 to −1.76) |
| Logistic (h2o) | 0.879 (0.878 to 0.879) | −0.06 (−0.10 to −0.02) |
| Random forest (h2o) | 0.877 (0.877 to 0.878) | −0.22 (−0.29 to −0.17) |
| Neural network (h2o) | 0.875 (0.870 to 0.879) | −0.45 (−1.09 to −0.04) |
| autoML (h2o) | 0.879 (0.879 to 0.880) | −0.00 (−0.07 to 0.06) |
Model performance was calculated in binary framework. Threshold 7.5% was used to calculate precision and recall for all models.
95% range (2.5-97.5%) of model performance derived from 100 random samples.
Figure 1 shows the variability in individual risk predictions across the models for patients with predicted cardiovascular disease risks of 9.5-10.5% by QRISK3. Patients with a predicted cardiovascular disease risk between 9.5% and 10.5% with QRISK3 had a risk of 2.2-5.8% with logistic Caret model, 2.9-9.2% with Caret random forest, 2.4-7.2% with Caret neural network, and 3.1-9.3% with Sklearn random forest. The calibration plot (fig 2) shows that models that ignore censoring were miscalibrated (that is, predicted risks were lower than observed risks).
Fig 1.
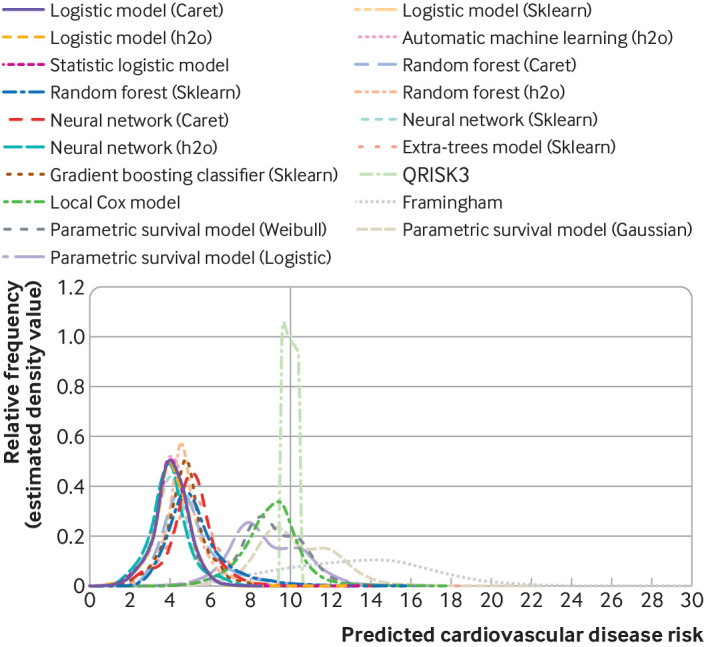
Distribution of individual risk predictions with machine learning and statistical models in overall cohort for patients with predicted cardiovascular disease risks of 9.5-10.5% in QRISK3
Fig 2.
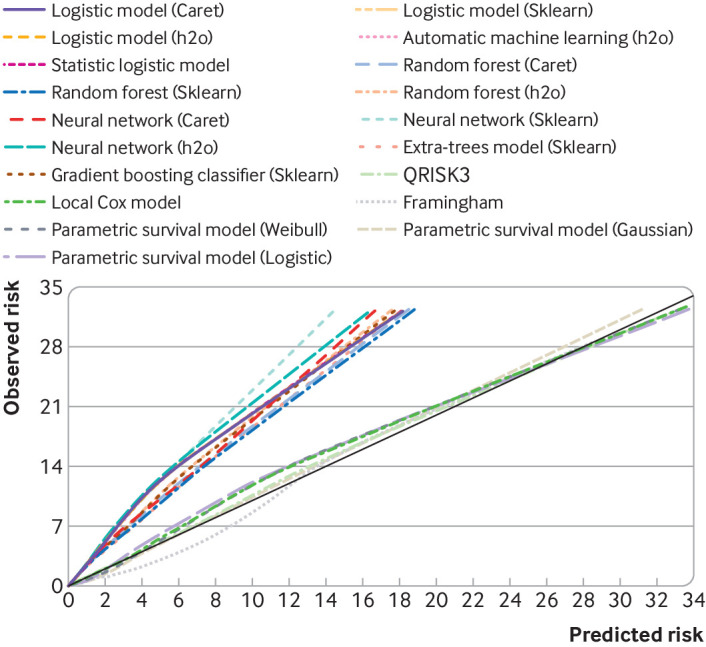
Calibration slope of machine learning models and statistical models in overall cohort in survival framework (observed events consider censoring). CVD=cardiovascular disease
Figure 3 plots the differences of individual cardiovascular disease risk predictions with the different models stratified by deciles of cardiovascular disease risk predictions of QRISK3. The largest range of inconsistencies in risk predictions was found in patients with highest predicted risks of cardiovascular disease. Low risk of cardiovascular disease was generally predicted consistently between and within models. We observed similar trends when using a different reference model (eFigure 5.2).
Fig 3.
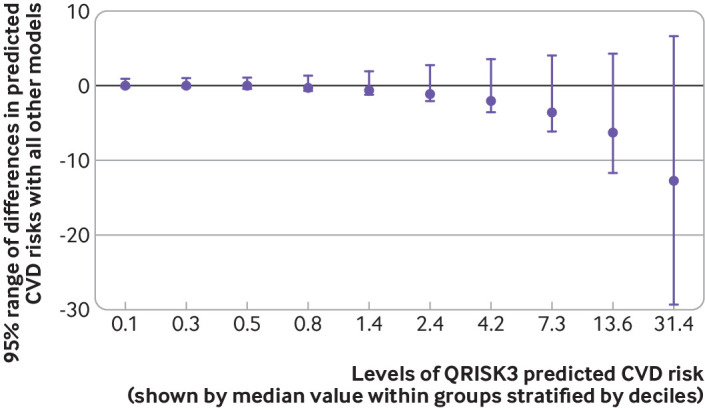
95% range of individual risk predictions with machine learning and statistical models stratified by deciles of predicted cardiovascular disease (CVD) risks with QRISK3 in overall cohort
Figure 4 shows the Bland-Altman plot of QRISK3 and neural network. We found a large inconsistency of risk predictions between models. The differences in predicted risks between QRISK3 and neural network ranged between −23.2% and 0.1% (95% range). The regression line shows similar finding to figure 3, with the largest differences in higher risk groups. More comparison between specific models can be found in eFigure 6 and eFigure 7. We found similar inconsistency of risk prediction among models when using a logistic model as reference (eFigure 2.1). The removal of censored patients changed the magnitude but not the variability of individual cardiovascular disease risk predictions (eFigure 2.2).
Fig 4.
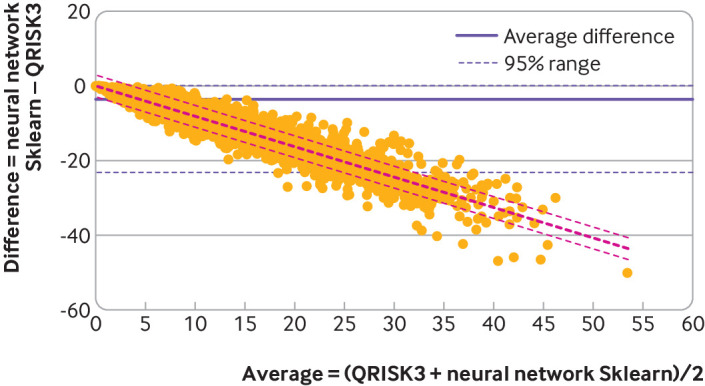
Bland-Altman analysis comparing QRISK3 with neural network model
We found substantial reclassification across a treatment threshold when using a different type of prediction model. Of 691 664 patients with a cardiovascular disease risk of 7.5% or lower, as predicted by QRISK3, 13.6% would be reclassified above 7.5% when using another model (table 3). Of the 223 815 patients with a cardiovascular disease risk above 7.5%, 57.8% would be reclassified below 7.5% when using another model. We also found high levels of reclassification with a different reference model (as shown in table 3) or a different threshold (eTable 7).
Table 3.
Reclassification of individual risk predictions with machine learning and statistical models
| Model | Reclassification in overall testing cohort: No (%)* | |
|---|---|---|
| Reclassified* | Not reclassified | |
| Overall cohort | ||
| QRISK3 10 year risk prediction (reference model): | ||
| ≤7.5% threshold | 94 186 (13.6) | 597 478 (86.4) |
| >7.5% threshold | 129 348 (57.8) | 94 467 (42.2) |
| Logistic model (Caret) 10 year risk prediction (reference model): | ||
| ≤7.5% threshold | 209 221 (25.9) | 597 478 (74.1) |
| >7.5% threshold | 14 313 (13.2) | 94 467 (86.8) |
| Cohort without censoring | ||
| QRISK3 10 year risk prediction (reference model): | ||
| ≤7.5% threshold | 34 607 (54.6) | 28 779 (45.4) |
| >7.5% threshold | 1248 (2.6) | 47 234 (97.4) |
| Logistic model (Caret) 10 year risk prediction (reference model): | ||
| ≤7.5% threshold | 6004 (17.3) | 28 779 (82.7) |
| >7.5% threshold | 29 851 (38.7) | 47 234 (61.3) |
Patients were reclassified if they had a risk prediction in any model that crossed the threshold compared with the prediction of the reference model.
We did several sensitivity analyses with consistent findings of high levels of inconsistencies in individual risk predictions between and within models. The same machine learning algorithm with the selection of different settings (hyper-parameters) from different software packages yielded different individual cardiovascular disease risk predictions (eTable 8 and eFigure 8). The evaluation of the effects of generalisability by developing and testing models in different regions of England showed similarly high levels of inconsistencies in cardiovascular disease risk predictions (eTable 10 and eFigure 9). Changing the number of predictors did not result in lower levels of inconsistencies in cardiovascular disease risk predictions with more predictors included in the models (eTable 11 and eFigure 10),
Discussion
We found that the predictions of cardiovascular disease risks for individual patients varied widely between and within different types of machine learning and statistical models, especially in patients with higher risks (when using similar predictors). Logistic models and the machine learning models that ignored censoring substantially underestimated risk of cardiovascular disease.
Comparison with other studies
Despite claims that machine learning models can revolutionise risk prediction and potentially replace traditional statistical regression models in other areas,5 36 37 this study of prediction of cardiovascular disease risk found that they have similar model performance to traditional statistical methods and share similar uncertainty in individual risk predictions. Strengths of machine learning models may include their ability to automatically model non-linear associations and interactions between different risk factors.38 39 They may also find new data patterns.30 They have the acknowledged strength of automating model building with a better performance in specific classification tasks (for example, image recognition).30 However, a critical question is whether risk prediction models provide accurate and consistent risk predictions for individual patients. Previous research has found that a traditional risk prediction model such as QRISK3 has considerable uncertainty on individual risk prediction, although it has very good model performance at the population level.18 19 This uncertainty is related to unmeasured heterogeneity between clinical sites and modelling choices such as the inclusion of secular trends.18 19 Our study found that machine learning models share this uncertainty, as models with comparable population level performance yielded very different individual risk predictions. Consequently, different treatment decisions could be made by arbitrarily selecting another modelling technique.
Censoring of patients is an unavoidable problem in prediction models for long term risks, as patients frequently move away or die. However, many popular machine learning models ignore censoring, as the default framework is the analysis of a binary outcome rather than time to event survival outcome. A UK Biobank study of risk prediction for cardiovascular disease did not report how censoring was dealt with,7 like several other studies.39 40 41 Another machine learning study incorrectly excluded censored patients.8 Random survival forest is a machine learning model that takes account of censoring.42 Innovative techniques are being developed that incorporate statistical censoring approaches into the machine learning framework.16 43 However, to our knowledge no current software packages can handle large datasets for these methods. This study shows that directly applying popular machine learning models to data (especially for data with substantive censoring) without considering censoring will substantially bias risk predictions. The miscalibration was large compared with observed life table predictions. This is consistent with a recent study that reported loss of information due to lack of consideration of censoring with the random forest method.6
Models with similar C statistics gave varying estimates of individual risks for the same patients. A fundamental challenge with the C statistic is that it applies to the population level but not to individual patients.18 44 The C statistic measures the ability of a model to discriminate between cases and non-cases. It is a proportion of cases and non-cases that are correctly ranked by the model. This means that for a high C statistic, patients with observed events should have a higher risk than the patients without observed events.38 The C statistic concerns rank of predicted probability rather than probability itself. For example, a model may predict all events with a range of probability between 50.2% and 50.3% and non-events with a probability of 50%, which would result a perfect discrimination, but the predicted probability is not clinically useful. When a large number of patients have lower risks (which is often the case for cardiovascular disease risk prediction), the C statistic becomes less informative in indicating discrimination of models, especially in patients at high risk. For example, two patients with very low risk (say 1% and 1.5%) may have similar effects on C statistic to two patients with high risk (say 10% and 20%), given that their differences in rank are the same (but the latter two are of greater clinical interest). Therefore, C statistics do not tell us whether a model discriminates specific patients at high risk correctly or consistently compared with other models. C statistics have also been shown to be insensitive to changes in the model.44 The evaluation of consistency in individual risk predictions between models may thus be important in assessing their clinical usefulness in identifying patients at high risk.
This study considered a total of 22 predictors that had been selected by the developers of QRISK on the basis of their likely causal effect on cardiovascular disease.3 Other machine learning studies have used considerably more predictors. As an example, a study using the UK Biobank included 473 predictors in the machine learning models.7 A potentially unresolved question in risk prediction is what type of variables and how many of them should be included in models, as consensus and guidelines for choosing variables for risk prediction model are lacking.45 More information incorporated into a model may increase the model performance of risk prediction at the population level. For example, the C statistic is related to both the effects of predictors and the variation of predictors among patients with and without events.46 Including more predictors in a model may increase the C statistic merely because of greater variation of predictors. On the other hand, inclusion of non-causal predictors may lower the accuracy of the risk prediction by adding noise, increasing the risk of over-fitting, and leading to more data quality challenges.47 Also, a very large number of predictors may limit the clinical utility of these machine learning models, as more predictors need to be measured before a prediction can be made. Further research is needed to establish whether the focus of risk prediction should be on consistently measured causal risk factors or on variables that may be recorded inconsistently between clinicians or electronic health records systems.
Guidelines for the development and validation of risk prediction models (called TRIPOD) focus on the assessment of population level performance but do not consider consistencies in individual risk predictions by prediction models with comparable population level performance.48 Arguably, the clinical utility of risk prediction models should be based, as has been done with blood pressure devices for instance, on the consistent risk prediction (reliability) for a particular patient rather than broad population level performance.49 If models with comparable performance provide different predictions for a patient with certain risk factors, an explanation for these discrepant predictions is needed.50 Explainable artificial intelligence has been described as methods and techniques in the application of artificial intelligence such that the results of the solution can be understood by human experts.51 This contrasts with the concept of the “black box” in machine learning, whereby predictions cannot be explained. Arguably, a survival model that is explainable (such as QRISK3, which is based on established causal predictors) may be preferable over black box models that are high dimensional (include many predictors) but that provide inconsistent results for individual patients. Better standards are needed on how to develop and test machine learning algorithms.14
Strengths and limitations of study
The major strength of this study was that a large number of different machine learning models with varying hyper-parameters using different packages from different programming languages were fitted to a large population based primary care cohort. However, the study has several limitations. We considered only predictors from QRISK3 in order to compare models on the basis of equal information, but sensitivity analyses showed similar findings of inconsistencies in cardiovascular disease risk prediction independent of the number of predictors. Furthermore, more hyper-parameters in the machine learning models could have been considered in the grid search process. However, the fitted models already achieved reasonably high model performance, which indicates that the main hyper-parameters had been covered in the grid search process. Several machine learning algorithms were not included in this study, such as support vector machine or survival random forest, as the current software packages of these models cannot cope with large datasets.52 53 54 55 The Bland-Altman graph used the 95% range of differences rather than 95% confidence interval, as the differences of predicted risk (including log transformed) did not follow normal distribution (which is a required assumption to calculate the Bland-Altman 95% confidence interval). Another limitation is that this study concerned cardiovascular disease risk prediction in primary care, and findings may not be generalisable to other outcomes or settings. However, the robustness of individual risk predictions within and between models with comparable population level performance is rarely, if ever, evaluated. Our findings indicate the importance of assessing this.
Conclusions
A variety of models predicted cardiovascular disease risks for the same patients very differently despite similar model performances. Using the logistic model and commonly used machine learning models without considering censoring in survival analysis results in substantially biased risk prediction and has limited usefulness in the prediction of long term risks. The level of consistency within and between models should be assessed before they are used for clinical decision making and should be considered in TRIPOD guidelines.
What is already known on this topic
Risk prediction models are widely used in clinical practice (such as QRISK or Framingham for cardiovascular disease)
Multiple techniques can be used for these predictions, and recent studies claim that machine learning models can outperform models such as QRISK
What this study adds
Nineteen different prediction techniques (including 12 machine learning models and seven statistical models) yielded similar population level performance
However, cardiovascular disease risk predictions for the same patients varied substantially between models
Models that ignored censoring (including commonly used machine learning models) yielded biased risk predictions
Web extra.
Extra material supplied by authors
Web appendix: Supplementary materials
Contributors: YL designed the study, did all statistical analysis, produced all tables and figures, and wrote the main manuscript text and supplementary materials. MS supervised the study, provided quality control on statistical analysis, reviewed all statistical results, and reviewed and edited the main manuscript text. DMA reviewed and edited the main manuscript text and supplementary materials. TPvS designed and supervised the study, provided quality control of all aspects of the paper, and wrote the main manuscript text. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. TPvS is the guarantor.
Funding: This study was funded by the China Scholarship Council (to cover costs of doctoral studentship of YL at the University of Manchester). The funder did not participate in the research or review any details of this study; the other authors are independent of the funder.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: support to YL from the China Scholarship Council; no financial relationships with any organisations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work.
Ethical approval: This study is based on data from Clinical Practice Research Datalink (CPRD) obtained under licence from the UK Medicines and Healthcare products Regulatory Agency. The protocol for this work was approved by the independent scientific advisory committee for CPRD research (No 19_054R). The data are provided by patients and collected by the NHS as part of their care and support. The Office for National Statistics (ONS) is the provider of the ONS data contained within the CPRD data. Hospital Episode Statistics data and the ONS data (copyright 2014) are re-used with the permission of the Health and Social Care Information Centre.
Data sharing: This study is based on CPRD data and is subject to a full licence agreement, which does not permit data sharing outside of the research team. Code lists are available from the corresponding author.
The lead author affirms that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Dissemination to participants and related patient and public communities: Dissemination to research participants is not possible as data were anonymised.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.National Institute for Health and Care Excellence. NICE recommends wider use of statins for prevention of CVD. https://www.nice.org.uk/news/article/nice-recommends-wider-use-of-statins-for-prevention-of-cvd.
- 2. Mahmood SS, Levy D, Vasan RS, Wang TJ. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet 2014;383:999-1008. 10.1016/S0140-6736(13)61752-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hippisley-Cox J, Coupland C, Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ 2017;357:j2099. 10.1136/bmj.j2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gov.uk. Health Secretary announces £250 million investment in artificial intelligence. 2019. https://www.gov.uk/government/news/health-secretary-announces-250-million-investment-in-artificial-intelligence.
- 5. Hinton G. Deep Learning-A Technology With the Potential to Transform Health Care. JAMA 2018;320:1101-2. 10.1001/jama.2018.11100 [DOI] [PubMed] [Google Scholar]
- 6. Steele AJ, Denaxas SC, Shah AD, Hemingway H, Luscombe NM. Machine learning models in electronic health records can outperform conventional survival models for predicting patient mortality in coronary artery disease. PLoS One 2018;13:e0202344. 10.1371/journal.pone.0202344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alaa AM, Bolton T, Di Angelantonio E, Rudd JHF, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: A prospective study of 423,604 UK Biobank participants. PLoS One 2019;14:e0213653. 10.1371/journal.pone.0213653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weng SF, Reps J, Kai J, Garibaldi JM, Qureshi N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One 2017;12:e0174944. 10.1371/journal.pone.0174944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al’Aref SJ, Anchouche K, Singh G, et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975-86. 10.1093/eurheartj/ehy404 [DOI] [PubMed] [Google Scholar]
- 10. Price WN, 2nd, Gerke S, Cohen IG. Potential Liability for Physicians Using Artificial Intelligence. JAMA 2019. 10.1001/jama.2019.15064 [DOI] [PubMed] [Google Scholar]
- 11. Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol 2019;110:12-22. 10.1016/j.jclinepi.2019.02.004 [DOI] [PubMed] [Google Scholar]
- 12. Carin L, Pencina MJ. On deep learning for medical image analysis. JAMA 2018;320:1192-3. 10.1001/jama.2018.13316 [DOI] [PubMed] [Google Scholar]
- 13. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316:2402-10. 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 14. Maddox TM, Rumsfeld JS, Payne PRO. Questions for Artificial Intelligence in Health Care. JAMA 2019;321:31-2. 10.1001/jama.2018.18932 [DOI] [PubMed] [Google Scholar]
- 15. Nsoesie EO. Evaluating Artificial Intelligence Applications in Clinical Settings. JAMA Netw Open 2018;1:e182658. 10.1001/jamanetworkopen.2018.2658 [DOI] [PubMed] [Google Scholar]
- 16. Vock DM, Wolfson J, Bandyopadhyay S, et al. Adapting machine learning techniques to censored time-to-event health record data: A general-purpose approach using inverse probability of censoring weighting. J Biomed Inform 2016;61:119-31. 10.1016/j.jbi.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Briggs WL. Uncertainty: the soul of modeling, probability & statistics. Springer, 2018. [Google Scholar]
- 18. Li Y, Sperrin M, Belmonte M, Pate A, Ashcroft DM, van Staa TP. Do population-level risk prediction models that use routinely collected health data reliably predict individual risks? Sci Rep 2019;9:11222. 10.1038/s41598-019-47712-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pate A, Emsley R, Ashcroft DM, Brown B, van Staa T. The uncertainty with using risk prediction models for individual decision making: an exemplar cohort study examining the prediction of cardiovascular disease in English primary care. BMC Med 2019;17:134. 10.1186/s12916-019-1368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herrett E, Gallagher AM, Bhaskaran K, et al. Data Resource Profile: Clinical Practice Research Datalink (CPRD). Int J Epidemiol 2015;44:827-36. 10.1093/ije/dyv098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CPRD. Clinical Practice Research Datalink. https://www.cprd.com/.
- 22. Danese MD, Gleeson M, Griffiths RI, Catterick D, Kutikova L. Methods for estimating costs in patients with hyperlipidemia experiencing their first cardiovascular event in the United Kingdom. J Med Econ 2017;20:931-7. 10.1080/13696998.2017.1345747 [DOI] [PubMed] [Google Scholar]
- 23. Hippisley-Cox J, Coupland C, Brindle P. The performance of seven QPrediction risk scores in an independent external sample of patients from general practice: a validation study. BMJ Open 2014;4:e005809. 10.1136/bmjopen-2014-005809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Staa T-P, Gulliford M, Ng ES-W, Goldacre B, Smeeth L. Prediction of cardiovascular risk using Framingham, ASSIGN and QRISK2: how well do they predict individual rather than population risk? PLoS One 2014;9:e106455. 10.1371/journal.pone.0106455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson KM, Wilson PW, Odell PM, Kannel WB. An updated coronary risk profile. A statement for health professionals. Circulation 1991;83:356-62. 10.1161/01.CIR.83.1.356 [DOI] [PubMed] [Google Scholar]
- 26. Nelder JA, Wedderburn RWM. Generalized Linear Models. J R Stat Soc [Ser A] 1972;135:370 10.2307/2344614. [DOI] [Google Scholar]
- 27.Breiman L. Random Forests. 2001. https://www.stat.berkeley.edu/~breiman/randomforest2001.pdf.
- 28.Demuth H, De Jesús B. Neural Network Design. 2nd ed. https://hagan.okstate.edu/NNDesign.pdf.
- 29.Kuhn M. The caret package. 2019. http://topepo.github.io/caret/index.html.
- 30. Géron A. Hands-on machine learning with Scikit-Learn and TensorFlow: concepts, tools, and techniques to build intelligent systems. 2nd ed O’Reilly, 2019: 551. [Google Scholar]
- 31. scikit-learn About us. https://scikit-learn.org/stable/about.html.
- 32.H2O.ai. AutoML: Automatic Machine Learning. http://docs.h2o.ai/h2o/latest-stable/h2o-docs/automl.html?highlight=automl.
- 33.H2O.ai. What is H2O? http://docs.h2o.ai/h2o/latest-stable/h2o-py/docs/intro.html#what-is-h2o.
- 34. Altman DG, Bland JM. Measurement in Medicine: the Analysis of Method Comparison Studies. Statistician 1983;32:307-17 10.2307/2987937. [DOI] [Google Scholar]
- 35.SAS. SAS 9.4 MERGE Statement. http://support.sas.com/documentation/cdl/en/lestmtsref/69738/HTML/default/viewer.htm#n1i8w2bwu1fn5kn1gpxj18xttbb0.htm.
- 36.The Alan Turing Institute. Turing Lecture: Transforming medicine through AI-enabled healthcare pathways. 2019. https://www.youtube.com/watch?v=TWI-WIoWvfk&feature=youtu.be&_cldee=dGplZXJkLnZhbnN0YWFAbWFuY2hlc3Rlci5hYy51aw%3D%3D&recipientid=contact-d2c6e6742b58e811812370106faae7f1-40027ba23fa146c189b8a3077e37916a&esid=2d3fc1d1-6593-e911-a98b-002248014cd6.
- 37. Makridakis S, Spiliotis E, Assimakopoulos V. Statistical and Machine Learning forecasting methods: Concerns and ways forward. PLoS One 2018;13:e0194889. 10.1371/journal.pone.0194889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steyerberg EW. Clinical prediction models : a practical approach to development, validation, and updating. Springer, 2009: 497 10.1007/978-0-387-77244-8. [DOI] [Google Scholar]
- 39. Kattan MW, Hess KR, Beck JR. Experiments to determine whether recursive partitioning (CART) or an artificial neural network overcomes theoretical limitations of Cox proportional hazards regression. Comput Biomed Res 1998;31:363-73. 10.1006/cbmr.1998.1488 [DOI] [PubMed] [Google Scholar]
- 40. Desai RJ, Wang SV, Vaduganathan M, Evers T, Schneeweiss S. Comparison of Machine Learning Methods With Traditional Models for Use of Administrative Claims With Electronic Medical Records to Predict Heart Failure Outcomes. JAMA Netw Open 2020;3:e1918962. 10.1001/jamanetworkopen.2019.18962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kakadiaris IA, Vrigkas M, Yen AA, Kuznetsova T, Budoff M, Naghavi M. Machine Learning Outperforms ACC / AHA CVD Risk Calculator in MESA. J Am Heart Assoc 2018;7:e009476. 10.1161/JAHA.118.009476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. RANDOM SURVIVAL FORESTS 1. Ann Appl Stat 2008;2:841-60 10.1214/08-AOAS169. [DOI] [Google Scholar]
- 43. Kvamme H, Borgan Ø, Scheel I. Time-to-Event Prediction with Neural Networks and Cox Regression. J Mach Learn Res 2019;20:1-30. [Google Scholar]
- 44. Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation 2007;115:928-35. 10.1161/CIRCULATIONAHA.106.672402 [DOI] [PubMed] [Google Scholar]
- 45. Lee YH, Bang H, Kim DJ. How to Establish Clinical Prediction Models. Endocrinol Metab (Seoul) 2016;31:38-44. 10.3803/EnM.2016.31.1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Austin PC, Steyerberg EW. Interpreting the concordance statistic of a logistic regression model: relation to the variance and odds ratio of a continuous explanatory variable. BMC Med Res Methodol 2012;12:82. 10.1186/1471-2288-12-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gianfrancesco MA, Tamang S, Yazdany J, Schmajuk G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern Med 2018;178:1544-7. 10.1001/jamainternmed.2018.3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Collins GS, Reitsma JB, Altman DG, Moons KGM, members of the TRIPOD group Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. Eur Urol 2015;67:1142-51. 10.1016/j.eururo.2014.11.025 [DOI] [PubMed] [Google Scholar]
- 49. Kerr KF, Janes H. First things first: risk model performance metrics should reflect the clinical application. Stat Med 2017;36:4503-8. 10.1002/sim.7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vollmer S, Mateen BA, Bohner G, et al. Machine learning and artificial intelligence research for patient benefit: 20 critical questions on transparency, replicability, ethics, and effectiveness. BMJ 2020;368:l6927. 10.1136/bmj.l6927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barredo Arrieta A, Díaz-Rodríguez N, Del Ser J, et al. Explainable Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion 2020;58:82-115 10.1016/j.inffus.2019.12.012. [DOI] [Google Scholar]
- 52.Menon AK. Large-Scale Support Vector Machines: Algorithms and Theory. http://cseweb.ucsd.edu/~akmenon/ResearchExam.pdf.
- 53.Wright MN, Wager S, Probst P. Package ‘ranger’. 2020. https://cran.r-project.org/web/packages/ranger/ranger.pdf.
- 54.Ishwaran H, Kogalur U. randomForestSRC: Fast Unified Random Forests for Survival, Regression, and Classification (RF-SRC). R package version 2.9.3. 2020. https://cran.r-project.org/package=randomForestSRC.
- 55.Therneau TM, Atkinson EJ. An Introduction to Recursive Partitioning Using the RPART Routines. 2019. https://cran.r-project.org/web/packages/rpart/vignettes/longintro.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Web appendix: Supplementary materials


