Our results suggested that CCT domain-containing proteins from three families can form trimer with NF-YB/YC and revealed the DNA recognition mechanism of CCT/NF-YB/YC complex.
Abstract
CONSTANS, CONSTANS-LIKE, and TIMING OF CAB EXPRESSION1 (CCT) domain-containing proteins are a large family unique to plants. They transcriptionally regulate photoperiodic flowering, circadian rhythms, vernalization, and other related processes. Through their CCT domains, CONSTANS and HEADING DATE1 (HD1) coordinate with the NUCLEAR FACTOR Y (NF-Y) B/C dimer to specifically target a conserved ‘CCACA’ motif within the promoters of their target genes. However, the mechanism underlying DNA recognition by the CCT domain remains unclear. Here we determined the crystal structures of the rice (Oryza sativa) NF-YB/YC dimer and the florigen gene Heading date 3a (Hd3a)-bound HD1CCT/NF-YB/YC trimer with resolutions of 2.0 Å and 2.55 Å, respectively. The CCT domain of HD1 displays an elongated structure containing two α-helices and two loops, tethering Hd3a to the NF-YB/YC dimer. Helix α2 and loop 2 are anchored into the minor groove of the ‘CCACA’ motif, which determines the specific base recognition. Our structures reveal the interaction mechanism among the CCT domain, NF-YB/YC dimer, and the target DNA. These results not only provide insight into the network between the CCT proteins and NF-Y subunits, but also offer potential approaches for improving productivity and global adaptability of crops by manipulating florigen expression.
INTRODUCTION
Many environmental cues, such as light, gravity, and temperature, continually influence plant growth and development. A set of CONSTANS (CO), CONSTANS-LIKE (COL), and TIMING OF CAB EXPRESSION1 (TOC1) or CCT domain-containing proteins, existing in a wide range of flowering plants (Supplemental Figure 1A), act as specific transcription factors and play crucial roles in integrating environmental signals and readjusting the reproduction progress. Until now, these proteins have been found to be involved in processes related to measuring the photoperiod (Putterill et al., 1995; Yano et al., 2000), setting the circadian rhythms (Nakamichi et al., 2010; Para et al., 2007; Strayer et al., 2000), responding to temperature (Yan et al., 2004), and other functioning. CCT domain is a conserved 41- to 43-amino acid region that was first found in the Arabidopsis (Arabidopsis thaliana) proteins CO, COL, and TOC1, thus this region is referred to as the CCT domain (Supplemental Figure 1B; Strayer et al., 2000).
In the model long-day plant Arabidopsis, CO acts as a network hub to integrate various external and internal signals into the photoperiodic flowering pathway, triggering expression of the florigen gene FLOWERING LOCUS T (FT) in leaves (Putterill et al., 1995; Samach et al., 2000; Shim et al., 2017). Consequently, the florigen moves to the shoot apical meristem and forms a flowering active complex to initiate reproductive phase transition (Liu et al., 2012; Taoka et al., 2011; Zhu et al., 2016). The regulation of FT expression by CO requires additional Nuclear Factor Y (NF-Y) family members, such as AtNF-YB2, AtNF-YB3, AtNF-YC3, AtNF-YC4, and AtNF-YC9 (Ben-Naim et al., 2006; Cao et al., 2014; Kumimoto et al., 2008, 2010; Wenkel et al., 2006). Through the CCT domain, CO coordinates with these factors to bind the CO-responsive DNA element (CORE, containing ‘CCACA’ motif) within the promoter region to upregulate FT transcription (Gnesutta et al., 2017a; Tiwari et al., 2010).
In the short-day plant rice (Oryza sativa), two main pathways control photoperiodic flowering by regulating the expression of two homologous florigen genes Hd3a and RFT1 (Komiya et al., 2008; Tamaki et al., 2007). One is the HD1-Hd3a module: HD1 directly and positively regulates the expression of Hd3a, similar to the Arabidopsis CO-FT module (Hayama et al., 2003). The other is the GHD7-Ehd1-Hd3a/RFT1 module, which is unique to cereals. Ehd1 promotes flowering independently from HD1 in both short and long days (Doi et al., 2004). However, it is suppressed by GHD7, which is expressed increasingly with lengthening days (Xue et al., 2008). This module enables flowering responses under various day-length conditions. Both HD1 (an ortholog of CO) and GHD7 contain CCT domains. Furthermore, the CCT domain of HD1 is able to interact with the B and C subunits of the NF-Y complex, such as GHD8 (also named OsNF-YB11) and OsNF-YC7, and in turn, the trimeric complex binds to a conserved response element (OsCORE2, containing the ‘CCACA’ motif) in the Hd3a promoter (Du et al., 2017; Goretti et al., 2017).
CCT domain-containing proteins are divided into three families according to their conserved N-terminal domain: the COL family, the Pseudo-Response Regulator (PRR) family, and the CCT Motif Family (CMF family; Supplemental Figure 1C; Cockram et al., 2012). Members of the COL family, such as HD1 from rice and CO from Arabidopsis, contain zinc finger B-box domain near their amino-terminus and a CCT domain near their carboxy-terminus. The PRR family members, which are represented by GHD7.1 in rice (Yan et al., 2013), the circadian clock regulator TOC1 in Arabidopsis (Strayer et al., 2000), SbPRR37 in sorghum (Sorghum bicolor; Murphy et al., 2011), and PPD-H1 in barley (Hordeum vulgare; Turner et al., 2005), are characterized by a pseudo-receiver domain near their amino-terminus and a CCT domain near their carboxy-terminus. CMF members, such as GHD7 in rice (Xue et al., 2008) and ZCCT1 and ZCCT2 in wheat (Triticum aestivum; Yan et al., 2004), possess a single CCT domain. Whether and how CCT proteins from all three families interact with NF-YB and NF-YC subunits is unknown. More importantly, the DNA recognition mechanism of the CCT domain remains elusive.
The NF-Y complex is composed of NF-YA, NF-YB, and NF-YC subunits. The latter two usually form heterodimers through their conserved histone fold domain (HFD). NF-YA contains a conserved ∼56-amino acid domain, which is required for protein interactions and DNA binding activity (Petroni et al., 2012). The NF-Y complex transcriptionally regulates genes associated with cell proliferation, immune responses, and other cellular processes (Maity and de Crombrugghe, 1998; Mantovani, 1999). Through the NF-YA subunit, the NF-Y complex specifically recognizes a conserved ‘CCAAT’ DNA sequence, one of the sequence elements most frequently found in eukaryotic promoters (Bi et al., 1997). Previous structural studies reported that NF-YB/YC interacted with the DNA sugar-phosphate backbone, resembling nucleosome H2A/H2B-DNA assembly. NF-YA inserted an α-helix and the following GxGGRF motif deeply into the DNA minor groove to provide sequence-specific contacts to the core pentamer nucleotide sequence (Huber et al., 2012; Nardini et al., 2013). In plants, how NF-YB/YC dimer is associated with CCT domain-containing proteins or NF-YA to recognize distinct DNA sequences remains largely unknown.
Here we biochemically reconstituted heterotrimers composed of the CCT domain and the NF-YB and NF-YC subunits. Then we determined the crystal structures of the rice NF-YB/YC dimer and the DNA-bound HD1CCT/NF-YB/YC complex at 2.00 Å and 2.55 Å resolution, respectively. HD1CCT displays an elongated structure with two α-helices and two loops, tethering DNA to the NF-YB/YC dimer. Helix α1 interacts with NF-YB/YC via electrostatic force. Helix α2 and loop 2 are anchored into the minor groove of the ‘CCACA’ motif, and they are responsible for specific DNA recognition. These organizations ensure the synergistic DNA binding activity between CCT domain and the NF-YB/YC dimer. Therefore, our results improve our understanding of how CCT domain-containing proteins and NF-Y subunits interact with DNA, as well as their transcriptional roles in plant photoperiodic flowering.
RESULTS
CCT Domain-Containing Proteins from Three Families Can Interact with the NF-YB/YC Dimer
Plants contain numerous CCT domain-containing proteins, which fall into the COL, CMF, and PRR families (Cockram et al., 2012) represented in rice by HD1, GHD7, and GHD7.1, respectively (Supplemental Figure 1A). The CCT domains of CO and HD1 can form trimeric complexes with NF-YB/YC dimers (Gnesutta et al., 2017a; Goretti et al., 2017). Our previous study reported that GHD7 and GHD8 (also named OsNF-YB11) functioned synergistically to cause no heading in rice (Zhang et al., 2015). We also found that when Ghd7.1 was pyramided with Ghd8 in the Zhenshan 97 background, the flowering was delayed by ∼45 d, which was more than the sum of the individual effects of Ghd7.1 15 d and Ghd8 9 d (Supplemental Figure 2). These findings strongly indicated that both Ghd7 and Ghd7.1 genetically interact with Ghd8. We speculated that GHD7 and GHD7.1 might be able to form heterotrimers with the OsNF-YB/YC dimer.
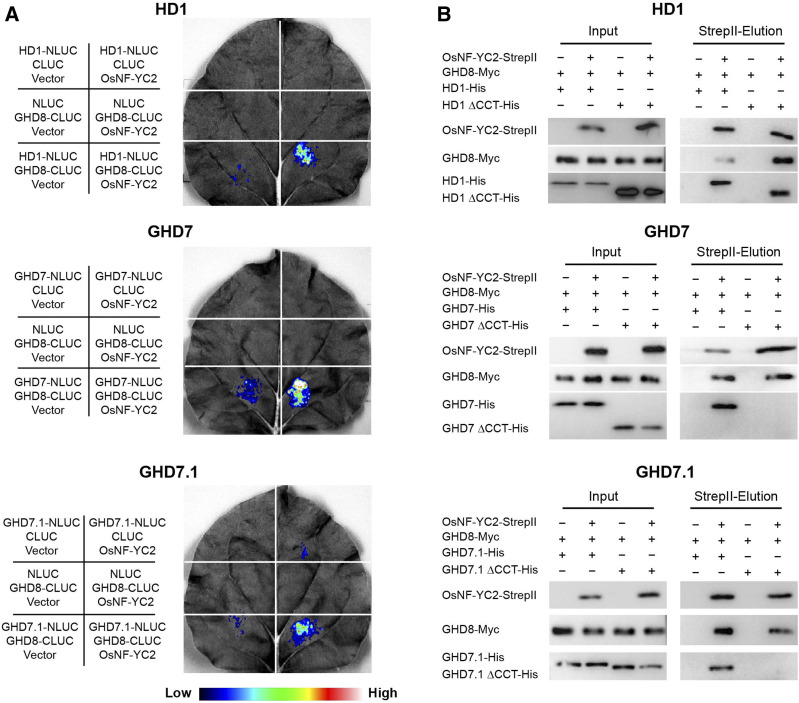
To assess whether the full-length HD1, GHD7, or GHD7.1 can interact with GHD8/OsNF-YC dimer, we firstly conducted a luciferase complementation imaging (LCI) assay in Nicotiana benthamiana. The OsNF-YC subfamily was found to contain 16 members (Supplemental Figure 3). After examining the combinations of GHD8 and OsNF-YCs, we selected GHD8 and OsNF-YC2 for subsequent experiments. CCT proteins (HD1, GHD7, and GHD7.1) and GHD8 were fused to the N- and C-terminal domains of LUCIFERASE (NLUC and CLUC), respectively. OsNF-YC2 was cloned into the third vector pCAMBIA1301s. Co-transfections of CCT-NLUC (HD1, GHD7, or GHD7.1), GHD8-CLUC, and OsNF-YC2 resulted in the production of robust luciferase activities, while other infiltrations lacking any one or two proteins brought about no or slight signals (Figure 1A). These results indicated that HD1, GHD7, or GHD7.1 could be physically associated with GHD8 and OsNF-YC2. Moreover, we performed pull-down assay via the previously reported mammalian cell system (Ma et al., 2020; Yang et al., 2018). His-tagged CCT protein (HD1, GHD7, or GHD7.1), Myc-tagged GHD8, and StrepII-tagged OsNF-YC2 were co-transfected into the Expi293F cells. Both GHD8 and CCT protein were co-eluted with OsNF-YC2 from the StrepII beads, indicating that HD1, GHD7, or GHD7.1 was able to interact with GHD8 and OsNF-YC2 (Figure 1B). Deletion of the CCT domain disrupted the above-mentioned interaction with the exception of HD1, whose N terminus also interacted with GHD8 and OsNF-YC2 (Figure 1B). These results suggested that the CCT domain plays an important role in CCT/GHD8/OsNF-YC2 complex formation.
Figure 1.
CCT Domain-Containing Proteins from All Three Families Can Be Physically Associated with the GHD8/OsNF-YC2 Dimer.
(A) LCI assay shows the interactions among the full-length CCT domain-containing proteins, GHD8, and OsNF-YC2. For each representation, the constructs used for each infiltration were listed at the left corresponding section. Fluorescence intensities represented their interaction activities. CCT domain-containing proteins (HD1, GHD7, and GHD7.1) and GHD8 were fused to the N- and C-terminal domains of LUCIFERASE (NLUC and CLUC), respectively. OsNF-YC2 was cloned into the third vector pCAMBIA1301s. Co-transfections of CCT-NLUC (HD1, GHD7, or GHD7.1), GHD8-CLUC, and OsNF-YC2 could produce robust luciferase activities, while other infiltrations lacking one or two proteins brought about no or slight signals.
(B) Pull-down assay showed the interactions among the full-length CCT domain-containing proteins, GHD8, and OsNF-YC2. His-tagged CCT protein (HD1, GHD7, or GHD7.1), Myc-tagged GHD8, and StrepII-tagged OsNF-YC2 were co-transfected into Expi293F cells. The supernatant from lysed cells was loaded onto StrepII beads. Both GHD8 and CCT proteins were co-eluted with OsNF-YC2. The proteins were detected by immunoblot with antibodies against StrepII, His, and Myc. ΔCCT indicated deletion of CCT domain.
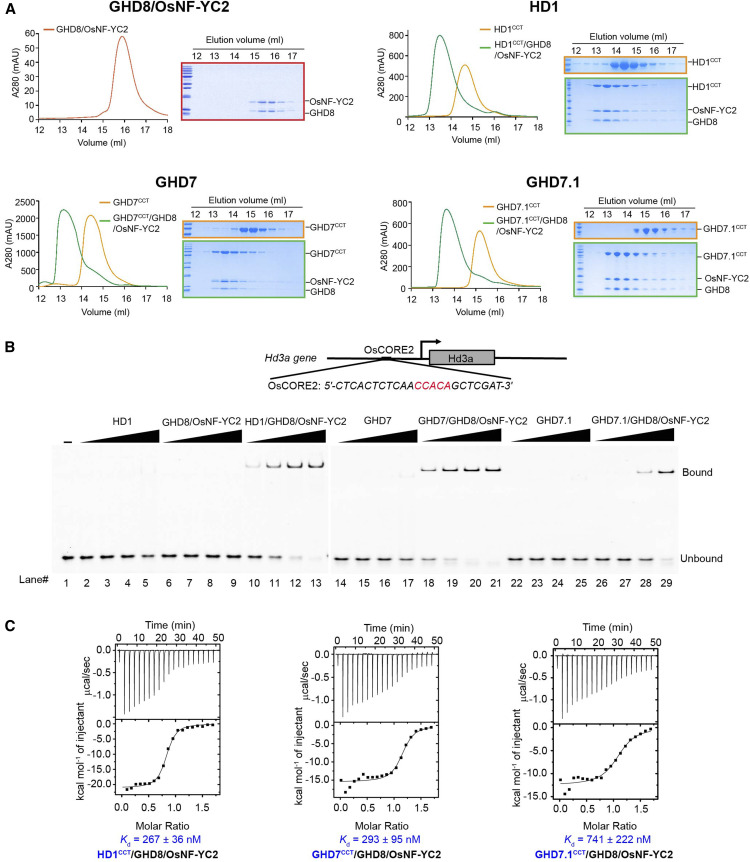
We further biochemically confirmed the interactions between the CCT domain and the GHD8/OsNF-YC2 dimer using size exclusion chromatography (SEC). We expressed maltose binding protein (MBP)-tagged HD1CCT, GHD7CCT, and GHD7.1CCT and analyzed their respective interactions with the co-expressed GHD8/OsNF-YC2 dimer. The elution volume of the GHD8/OsNF-YC2 dimer was ∼15.9 mL. The elution volumes of MBP-tagged HD1CCT, GHD7CCT, and GHD7.1CCT were ∼14.7 mL, 14.4 mL, and 15.2 mL, respectively. Upon co-incubation with GHD8/OsNF-YC2, HD1CCT, GHD7CCT and GHD7.1CCT were co-eluted at ∼13.5 mL, 13.3 mL, and 13.6 mL, respectively (Figure 2A). These co-migrations suggested that the CCT domains form a trimeric complex with NF-YB/YC. Taken together, all these results indicated that CCT domain-containing proteins from three families interact with NF-YB/YC.
Figure 2.
CCT/GHD8/OsNF-YC2 Complex Containing HD1CCT, GHD7CCT, or GHD7.1CCT Can Bind DNA Probe Containing ‘CCACA’ Motif.
(A) SEC analysis shows the interaction between the GHD8/OsNF-YC2 dimer and HD1CCT, GHD7CCT, or GHD7.1CCT. The same eluted fractions of each SEC injection were subjected to SDS-PAGE followed by Coomassie blue staining. CCT domains were fused with an MBP tag at the N terminus. The x axis indicates the elution volume, and the y axis indicates the protein’s absorption at 280 nm. The peaks of GHD8/OsNF-YC2, MBP-tagged CCT domain, and CCT/GHD8/OsNF-YC2 complex are colored red, orange, and green, respectively.
(B) EMSA analysis show that the CCT/GHD8/OsNF-YC2 complex containing HD1CCT, GHD7CCT, and GHD7.1CCT can bind to DNA containing the ‘CCACA’ motif. The DNA comes from the Hd3a promoter (named OsCORE2) and was labeled by FAM at the 5′ end. The final protein concentrations in each set of four lanes (lanes 2 to 5, 6 to 9, 10 to 13, 14 to 17, 18 to 21, 22 to 25, and 26 to 29) were 0.25, 0.50, 1.00, and 2.00 μM, respectively.
(C) ITC analysis of the interaction between DNA and the CCT/GHD8/OsNF-YC2 complex containing HD1CCT, GHD7CCT, or GHD7.1CCT.
CCT/NF-YB/YC Trimer Binds ‘CCACA’-Containing DNA In Vitro
HD1CCT/GHD8/OsNF-YC7 was reported to specifically target a ‘CCACA’ pentamer nucleotide sequence within the Hd3a promoter (termed ‘OsCORE2’; Goretti et al., 2017). The CCT domain possessed DNA binding activity (Ben-Naim et al., 2006). Based on these findings, we further examined whether GHD7CCT/GHD8/OsNF-YC2 and GHD7.1CCT/GHD8/OsNF-YC2 bound to ‘CCACA’-containing DNA by electrophoretic mobility shift assay (EMSA). In the control group, either HD1CCT alone or the GHD8/OsNF-YC2 dimer retained weak binding activity with the 23-bp OsCORE2 probe, while the HD1CCT/GHD8/OsNF-YC2 complex exhibited strong binding activity (Figure 2B), which was consistent with the previous study by Goretti et al. (2017). Similar to the HD1CCT/GHD8/OsNF-YC2 complex, GHD7CCT/GHD8/OsNF-YC2 and GHD7.1CCT/GHD8/OsNF-YC2 exhibited robust binding activity with OsCORE2, whereas GHD7CCT or GHD7.1CCT alone did not (Figure 2B). We further conducted isothermal titration calorimetry (ITC) assays to quantify the DNA binding affinities of these trimeric complexes. The HD1CCT/GHD8/OsNF-YC2, GHD7CCT/GHD8/OsNF-YC2, and GHD7.1CCT/GHD8/OsNF-YC2 were bound to the OsCORE2 probe with dissociation constants of ∼267 nM, 293 nM, and 741 nM, respectively (Figure 2C; Supplemental Table 1). Taken together, these results suggested that all the three CCT/GHD8/OsNF-YC2 complexes could bind to DNA containing the ‘CCACA’ motif.
Overall Structure of the DNA-Bound HD1CCT/GHD8/OsNF-YC2 Complex
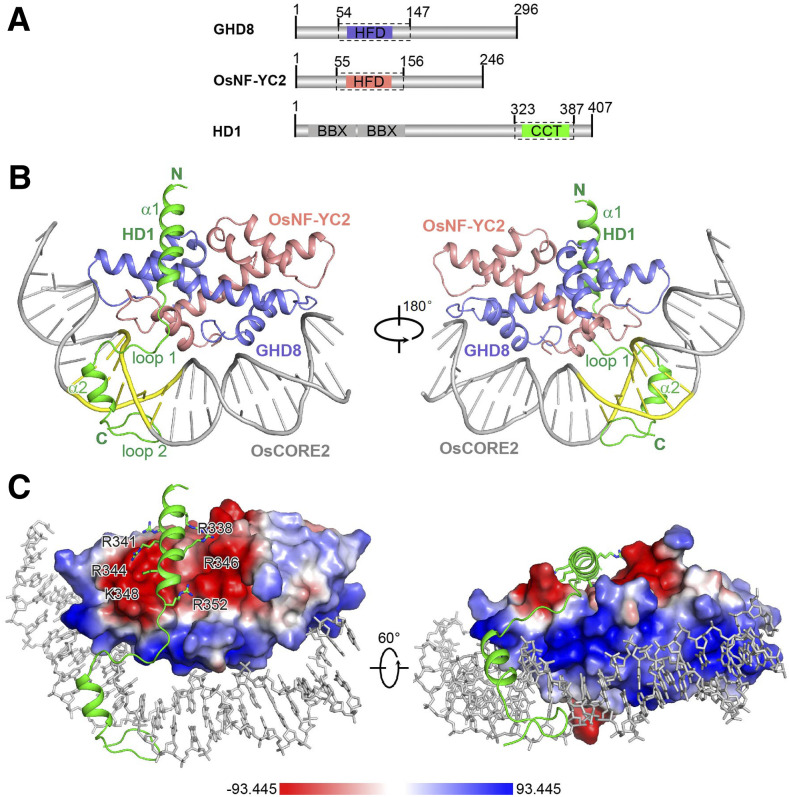
To elucidate the mechanism by which the CCT domain and the NF-YB/YC dimer function synergistically in DNA binding and the mechanism of the specific DNA recognition of the CCT/NF-YB/YC complex, we made a series of attempts to determine the structure of the DNA-bound trimeric complex. Finally, we determined the crystal structures of the GHD8/OsNF-YC2 dimer (GHD8, residues 54 to 147, and OsNF-YC2, residues 55 to 156) and the DNA-bound HD1CCT/GHD8/OsNF-YC2 complex (HD1, residues 323 to 387; GHD8, residues 54 to 147; and OsNF-YC2, residues 55 to 156; Figure 3; Table 1; Supplemental Figure 4A).
Figure 3.
Overall Structure of the DNA-Bound HD1CCT/GHD8/OsNF-YC2 Complex.
(A) Domain architectures of HD1, GHD8, and OsNF-YC2. The dotted boxes indicate the protein boundaries used for structural determination.
(B) Crystal structure of the DNA-bound HD1CCT/GHD8/OsNF-YC2 heterotrimer. The key secondary structure elements of the HD1CCT are labeled. HD1CCT, GHD8, and OsNF-YC2 are colored green, slate, and salmon, respectively. The DNA OsCORE2 is colored gray with ‘CCACA’ nucleotides highlighted in yellow. Two perpendicular views are presented. All structure figures were prepared with the tool PyMOL.
(C) Electrostatic surface of the GHD8/OsNF-YC2 dimer. Blue, white, and red colors represent positive, neutral, and negative surfaces, respectively. The negative interface of the dimeric GHD8/OsNF-YC2 matches the polar residues in helix α1 of HD1CCT, whereas the positive interface perfectly matches the DNA. The relevant CCT region is shown as cartoon (main chain) and stick (side chains) representations. DNA is shown as gray sticks.
Table 1. Data Collection and Refinement Statistics.
| Parameter | DNA-bound HD1CCT/GHD8/OsNF-YC2 | GHD8/OsNF-YC2 |
|---|---|---|
| Space group | P 31 2 1 | P 61 |
| Cell dimensions | ||
| a, b, c (Å) | 136.08, 136.08, 62.96 | 89.27, 89.27, 81.34 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Wavelength (Å) | 0.97914 | 0.97946 |
| Resolution (Å) | 45 ∼2.55 (2.66∼2.55) | 45∼2.00 (2.05∼2.00) |
| Rmerge (%) | 9.0 (80.0) | 8.2 (58.0) |
| Rpim (%) | 2.9 (26.7) | 2.6 (18.1) |
| I/σ | 14.8 (2.4) | 21.2(5.3) |
| Completeness (%) | 99.5 (99.2) | 99.7 (98.0) |
| Number of measured reflections | 217, 423 (24,970) | 270, 613 (19,817) |
| Number of unique reflections | 22, 041 (2,658) | 25, 020 (1,825) |
| Redundancy | 9.9 (9.4) | 10.8 (10.9) |
| Wilson B factor (Å2) | 45.5 | 22.3 |
| Rwork/Rfree (%) | 19.95/22.58 | 17.06/21.46 |
| Number of atoms | ||
| Protein | 1,708 | 2,737 |
| Main chain | 840 | 1,356 |
| Side chain | 868 | 1,381 |
| Water molecules | 16 | 168 |
| Other entities (DNA) | 1,025 | 0 |
| All atoms | 2,749 | 2,905 |
| Average B value (Å2) | ||
| Protein | 67.9 | 36.7 |
| Main chain | 64.6 | 31.9 |
| Side chain | 71.1 | 41.5 |
| Water molecules | 63.0 | 46.3 |
| Other entities (DNA) | 77.7 | 0 |
| All atoms | 71.5 | 37.3 |
| Root mean square deviations | ||
| Bonds (Å) | 0.010 | 0.007 |
| Angle (°) | 1.094 | 0.853 |
| Ramachandran plot statistics (%) | ||
| Most favorable | 96.1 | 98.5 |
| Additionally allowed | 3.9 | 1.5 |
| Generously allowed | 0 | 0 |
Values in parentheses are for the highest resolution shell. Rmerge = ΣhΣi|Ih,i-Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h. R = Σ|Fobs − Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections selected).
The GHD8/OsNF-YC2 dimer was refined to a resolution of 2.00 Å in space-group P61 (Table 1; Supplemental Figure 4A). Both GHD8 and OsNF-YC2 exhibit similar tertiary structures and include four helices. The first three helices display a classical histone-like fold with two short helices, helix α1 and helix α3, perpendicular to the long helix α2 (Supplemental Figure 4B). GHD8 and OsNF-YC2 assemble in a head-to-tail fashion and mimic the histone dimer H2A/H2B (PDB:1AOI; Luger et al., 1997), human NF-YB/YC (PDB:1N1J; Romier et al., 2003), and AtL1L/NF-YC3 (PDB:5G49; Gnesutta et al., 2017b), where the root mean square deviation (RMSD) values were 4.17 Å, 1.12 Å, and 0.95 Å, respectively, correspondingly covering 160, 160, and 160 Cα atoms (Supplemental Figure 4C).
The structure of the DNA-bound HD1CCT/GHD8/OsNF-YC2 complex was refined to a resolution of 2.55 Å in space-group P3121, with one quaternary complex per asymmetric unit (Figure 3B; Table 1). The DNA probe is a 23-bp OsCORE2 sequence from the Hd3a promoter. GHD8/OsNF-YC2 constitute a compact scaffold providing a negatively charged surface for HD1CCT binding and a positively charged patch for DNA binding (Figure 3C). Superimposition of the GHD8/OsNF-YC2 dimer onto the DNA-bound HD1CCT/GHD8/OsNF-YC2 complex (RMSD 1.21 Å, >160 Cα atoms) reveals that loop 1 (L1) of OsNF-YC2 exhibits a slight conformational change upon HD1CCT binding (Supplemental Figure 5A). In contrast to ideal B-DNA, the OsCORE2 DNA wraps around GHD8/OsNF-YC2, which is reminiscent of nucleosomes (Supplemental Figures 5B and 5C; Luger et al., 1997).
HD1CCT displays an elongated structure containing two α-helices and two loops, and tethers DNA to the GHD8/OsNF-YC2 dimer. Helix α1 (residues 335 to 351) and loop 1 (residues 352 to 360) are together designated as the protein interaction element (PIE), and they interact with the GHD8/OsNF-YC2 dimer. Helix α2 (residues 361 to 369) and loop 2 (residues 370 to 379), together referred to as the DNA recognition element (DRE), are anchored into the minor groove of the ‘CCACA’ box (Figures 3B and 4A). The DRE widens the minor groove to 12.7 Å (P-P distance ≤ 18.5 Å), which is contrast to the ≤8.7 Å (P-P distance ≤ 14.5 Å) groove in nucleosomal DNA (Supplemental Figure 5C). These interactions resulted in a strong DNA binding activity of the HD1CCT/GHD8/OsNF-YC2 complex.
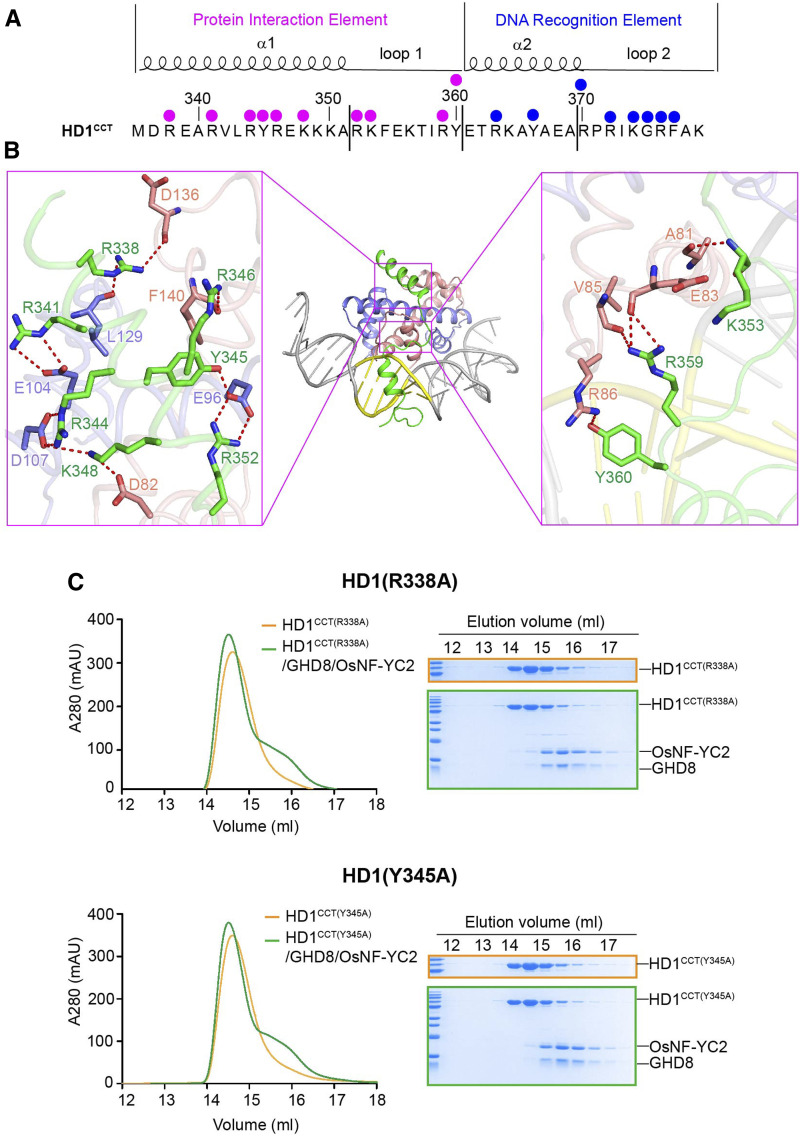
Figure 4.
Detailed Interactions between the Protein Interaction Element of HD1CCT and GHD8/OsNF-YC2 Dimer.
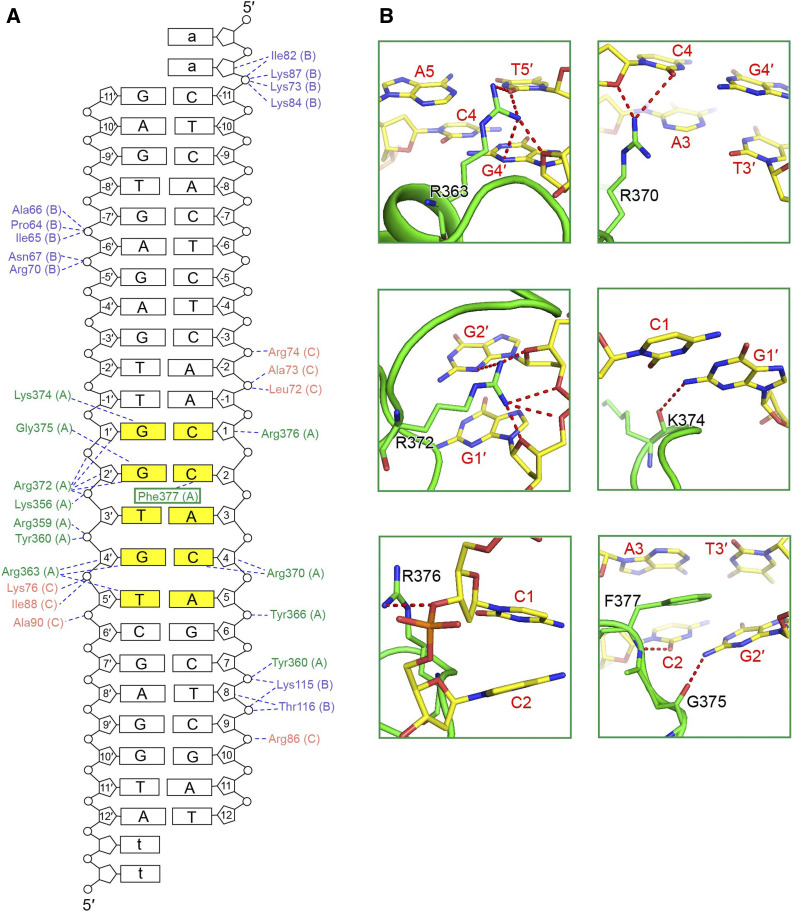
(A) HD1CCT sequence and the key residues interacting with the GHD8/OsNF-YC2 dimer and DNA. HD1CCT consists of two elements: the PIE and the DRE. The magenta dots indicate the residues involved in protein interactions, and the blue dots indicate the residues interacting with DNA.
(B) Zoom-in of HD1CCT residues interacting with GHD8/OsNF-YC2 dimer. HD1CCT, GHD8, and OsNF-YC2 are colored green, slate, and salmon, respectively.
(C) SEC analysis of the interaction between the mutated HD1CCT (R338A or Y345A) and GHD8/OsNF-YC2 dimer. The same eluted fractions of each SEC injection are subjected to SDS-PAGE followed by Coomassie blue staining. CCT domains are fused with an MBP tag at the N terminus. The x axis indicates the elution volume, and the y axis indicates the protein’s absorption at 280 nm. The peak of the MBP-tagged HD1CCT (R338A or Y345A) is colored orange, and the peak of incubation product of MBP-tagged HD1CCT (R338A or Y345A) and GHD8/OsNF-YC2 is colored green. Mutated HD1CCT failed to comigrate forward with GHD8/OsNF-YC2, indicating the abrogation of HD1CCT/GHD8/OsNF-YC2 trimer formation.
The Interactions between HD1CCT and GHD8/OsNF-YC2 Dimer
The PIE region of HD1CCT, which possesses polar and positively charged residues, primarily interacts with the GHD8/OsNF-YC2 dimer by forming a series of hydrogen bonds (H-bonds). R338 of HD1CCT forms two H-bonds with the main chain carbonyl oxygen atoms of L129 (GHD8) and D136 (OsNF-YC2). In addition, R341, R344, Y345, and R352 donate H-bonds to the carboxylate side chains of E104, D107, and E96 of GHD8. R346, K348, K353, and Y360 form H-bonds with F140, D82, A81, and R86 of OsNF-YC2, respectively. R359 forms three H-bonds with the main chains of E83 and V85 of OsNF-YC2 (Figure 4B). The importance of these residues on protein interaction is supported by mutational analysis. The SEC assay showed that HD1CCT failed to comigrate forward with GHD8/OsNF-YC2 when R338 or Y345 was substituted with Ala, indicating the abrogation of HD1CCT/GHD8/OsNF-YC2 trimer formation (Figure 4C). Although other mutations in the PIE region had little or slight effect on complex formation (Supplemental Figure 6), these results suggested that R338 and Y345 made important contributions to trimer formation.
The Interactions between HD1CCT and Target DNA
For describing the protein–DNA contacts clearly, the bases of the ‘CCACA’ motif are consecutively numbered from 1 at the 5′ end, and the upstream bases of the ‘CCACA’ box are assigned negative numbers. The bases of the complementary strand are numbered with a prime symbol according to their binding partner, so base 1′ pairs with base 1 (Figure 5A). The interactions between the HD1CCT domain and DNA are grouped into three categories. The first category includes the nonspecific interactions between residues and the DNA backbone, such as the interaction between R376 and the Cyt 1 pentose ring (Figure 5B, bottom left). The second category includes the interactions between the main chains of amino acids and the DNA bases. The main chain carbonyl oxygen atoms of K374 and G375 accept H-bonds from Gua 1′ N2 and Gua 2′ N2, respectively (Figure 5B, middle right and bottom right). The main chain amino group of F377 donates a H-bond to Cyt 2 O2 (Figure 5B, bottom right). The third category refers to the interactions between the side chains of amino acids and the DNA bases, which determine the specific DNA recognition. The guanidinium group of R363 donates H-bonds to Thy 5′ O2 atom and the Gua 4′ N3 and O2′ atoms (Figure 5B, top left). The R370 guanidinium group forms H-bonds with Cyt 4 O2 and O2′ atoms (Figure 5B, top right). The R372 guanidinium group forms H-bonds with the N3, O2′ atoms, and phosphate groups of Gua 2′ and with the O2′ atom and phosphate groups of Gua 1′ (Figure 5B, middle left). Except for these H-bond networks, stacking interactions among the F377 phenyl group, Cyt 2, and Ade 3, also play important roles in the DNA recognition (Figure 5B, bottom right).
Figure 5.
Structural Basis of Specific DNA Recognition by the HD1CCT/GHD8/OsNF-YC2 Complex.
(A) Schematic illustration of protein–DNA interactions. Amino acids of HD1CCT, GHD8, and OsNF-YC2 are colored green, slate, and salmon, respectively. Dashed lines indicate H-bonds and salt bridges. The ‘CCACA’ motif is shaded in yellow.
(B) Specific recognition of the ‘CCACA’ motif by HD1CCT residues. The main chain of amino acids is shown as cartoons and the side chains are shown as sticks and colored green. Bases are shown as sticks and colored according to atom type (carbon, yellow; oxygen, red; nitrogen, blue; phosphorus, orange). Amino acids and nucleotides are labeled in black and red letters, respectively. All structure figures were prepared with the tool PyMOL.
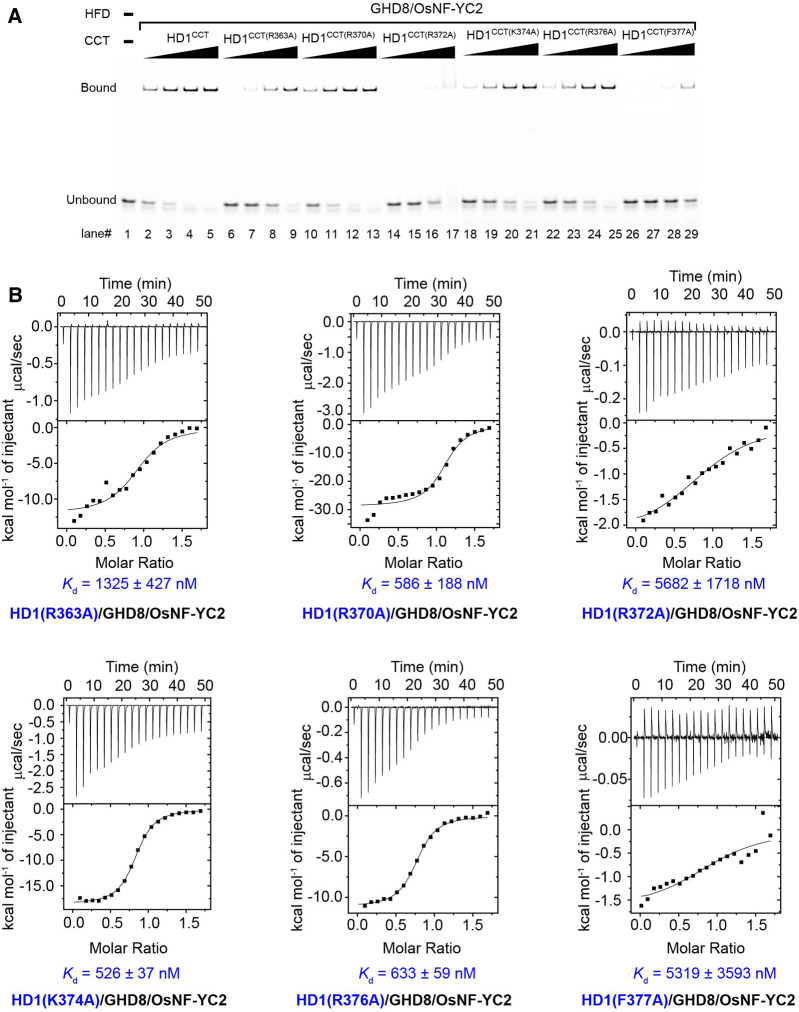
Residue substitutions in the DRE had no effect on the heterotrimer formation (Supplemental Figure 7). The importance of these residues in DNA recognition was validated by mutational analysis. R363A, R372A, or F377A mutation in HD1CCT led to a severe loss of DNA binding activity of the HD1CCT/GHD8/OsNF-YC2 complex (Figure 6A). Compared with that of the wild type, the dissociation constants of mutant HD1CCT/GHD8/OsNF-YC2 containing HD1CCT(R363A), HD1CCT(R372A), and HD1CCT(F377A) were 1325, 5682, and 5319 nM, indicating that their DNA binding affinity declined ∼5-, 20-, and 20-fold, respectively (Figure 6B; Supplemental Table 2). By contrast, other mutations, such as R370A, K374A, and R376A, had moderate effect on DNA binding activity (Figure 6). Taken together, these results suggested that R363, R372, and F377 play important roles in specific DNA recognition.
Figure 6.
The DNA Binding Activity of the Mutated HD1CCT/GHD8/OsNF-YC2 Complexes Were Measured by EMSA and ITC Assay.
(A) EMSA analysis of the interaction between DNA and HD1CCT/GHD8/OsNF-YC2 complex containing mutated HD1CCT (R363A, R370A, R372A, K374A, R376A, or F377A). All the mutated residues are located in the DRE region of HD1CCT. Protein was added with increasing final concentrations of 0.25, 0.50, 1.00, and 2.00 μM in each set of four lanes (lanes 2 to 5, 6 to 9, 10 to 13, 14 to 17, 18 to 21, 22 to 25, and 26 to 29).
(B) ITC analysis of the interaction between DNA and the mutated HD1CCT/GHD8/OsNF-YC2. The DNA sequence used for EMSA and ITC is 5′-CTCACTCTCAACCACAGCTCGAT-3, similar to that in Figure 2B.
Structural Comparison between HD1CCT/GHD8/OsNF-YC2 and Human NF-Y Complex
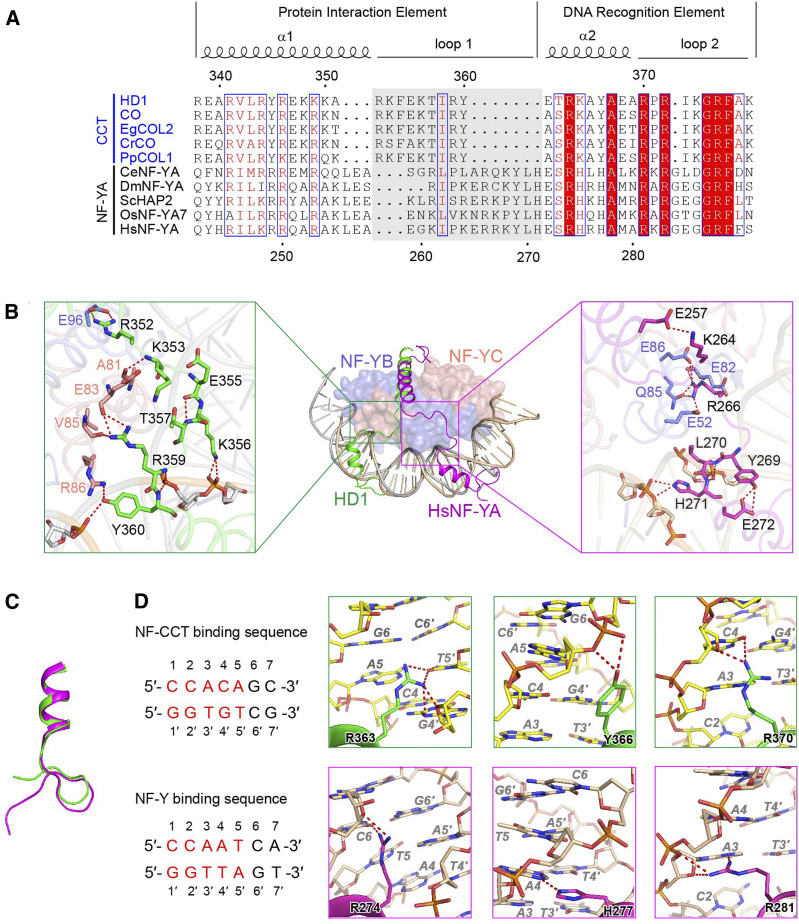
NF-YB/YC dimer and NF-YA assemble the NF-Y complex to target a conserved ‘CCAAT’ DNA motif in eukaryotes. The HD1CCT/GHD8/OsNF-YC2 and NF-Y complexes share the similar NF-YB/YC dimer. After superposition of the DNA-bound HD1CCT/GHD8/OsNF-YC2 complex onto the DNA-bound human NF-Y complex (PDB:4AWL; Nardini et al., 2013), a notable difference in conformations was observed between HD1CCT and HsNF-YA, especially in the PIE region (helix α1 and loop 1). Both helix α1 and loop 1 of HD1CCT are shorter than those of HsNF-YA (Figure 7A). HD1CCT loop 1 and HsNF-YA loop 1 exhibit different conformations. HD1CCT loop 1 points to the N terminus of OsNF-YC2, while HsNF-YA loop 1 is located at the N terminus of HsNF-YB (Figure 7B, middle). In the HD1CCT loop1, the side chain of R352 interacts with the side chain of E96 (GHD8). The amino group of K353 main chain donates a H-bond to the main chain carbonyl oxygen atoms of A81 (OsNF-YC2). The side chain of R359 is H-bonding to the main chain carbonyl oxygen atoms of E83 and V85 (OsNF-YC2). The side chain of Y360 accepts a H-bond from R86 (OsNF-YC2). E355 forms an internal H-bond with T357 through their main chains.
Figure 7.
Structural Comparison of DNA-Bound HD1CCT/GHD8/OsNF-YC2 and Human NF-Y Complex.
(A) Sequence alignment of CCT domain and DNA binding domain of NF-YA. CCT domains come from HD1 (O. sativa, LOC_Os06g16370), CO (Arabidopsis, At5g15840), EgCOL2 (Eucalyptus grandis, XP_010029446.1), CrCO (Chlamydomonas reinhardtii, Cre06.g278159), and PpCOL1 (Physcomitrella patens, XP_001783619.1). NF-YA proteins are from OsNF-YA7 (O. sativa, XP_015631822.1), HsNF-YA (Homo sapiens, NP_002496.1), CeNF-YA (Caenorhabditis elegans, NP_509999.1), HAP2 (Saccharomyces cerevisiae, NP_011277.1), and DmNF-YA (Drosophila melanogaster, NP_648313.1). The alignment was performed by the Clustal Omega website service (https://www.ebi.ac.uk/Tools/msa/clustalo/). Secondary structural elements are shown above. Sequence identity is indicated in white letters against a red background, and sequence similarity is shown in red letters. Loop 1 was highlighted by gray background.
(B) Structural comparison of DNA-bound HD1CCT/GHD8/OsNF-YC2 and human NF-Y complex (PDB:4AWL). Middle representation: overall superposition of DNA-bound HD1CCT/GHD8/OsNF-YC2 and human NF-Y complex (PDB:4AWL). HD1CCT and HsNF-YA are shown as cartoons and colored green and magenta, respectively. NF-YB/YC are shown as surface representation. GHD8 and HsNF-YB are colored in slate. OsNF-YC2 and HsNF-YC are colored in salmon. Both DNAs are shown as cartoon representation and are respectively colored wheat and gray. Left inset exhibits the enlarged image showing the detailed interactions between HD1CCT loop 1 and GHD8/OsNF-YC2. Right inset displays the enlarged image showing the detailed interactions between of HsNF-YA loop1 and HsNF-YB/YC.
(C) Superposition of the DREs of HD1CCT and HsNF-YA. HD1CCT and HsNF-YA are colored in green and magenta, respectively.
(D) Comparison of the side chains of residues interacting with DNA in HD1CCT and HsNF-YA. Partial DNA sequences recognized by HD1CCT and HsNF-YA are listed at left. The DNA bases are consecutively numbered from 1 at the 5′ end, and DNA bases on the complementary chain are numbered according to its partner starting with 1′. The core motifs ‘CCACA’ and ‘CCAAT’ are colored red and the flanking bases are colored black. Top representation shows zoom-in of residues of HD1CCT, and bottom representation shows zoom-in of residues of HsNF-YA. In the zoom-in images, amino acids are indicated as black regular letters followed by the numbers indicating their positions. The DNA bases are indicated in gray italic letters followed by the numbers indicating their positions.
In addition to these protein–protein interactions, K356, R359, and Y360 form salt bridges with the phosphate backbone of DNA (Figure 7B, left). In contrast, only one residue in the HsNF-YA loop 1, R266, forms a hydrogen-bond network with the N-terminal residues E52, E82, Q85, and E86 of HsNF-YB. L270 and H271 form salt bridges with the phosphate backbone of DNA. The two internal interaction pairs, namely, K264 and E257, and Y269 and E272, also contribute to the conformation of HsNF-YA loop 1 (Figure 7B, right).
Although the conformation of the DRE region of HD1CCT and that of the HsNF-YA are similar (Figure 7C), the side chains of three residues involved in coordinating bases in this region are oriented differently (Figure 7D). R363 of HD1CCT forms H-bonds with Gua 4′ and Thy 5′ via the side chains. However, the corresponding amino acid of HsNF-YA, R274, interacts with Cyt 6, offering the flanking sequence selectivity of the ‘CCAAT’ motif (Figure 7D, left zoom-in). This structural observation is consistent with the recent finding that NF-Y complex mainly targets the ‘CCAATC’ sequence (Kim et al., 2020). Y366 of HD1CCT contacts the phosphate group of Ade 5 and Gua 6 without base selectivity, whereas the corresponding H277 of HsNF-YA specifically interacts with N3 atom of Ade 4 (Figure 7D, middle zoom-in). R370 of HD1CCT forms H-bonds with Cyt 4, while the corresponding R281 of HsNF-YA interacts with Ade 3 (Figure 7D, right zoom-in). Furthermore, several residues, such as R372, K374, G375, R376, and F377 from HD1CCT, interact with DNA, as the corresponding R283, G286, G287, R288, and F289 from HsNF-YA do (Supplemental Figure 8). Sequence alignment shows that most residues in DRE involved in DNA targeting are highly conserved between the CCT domain and the NF-YA, but the residues in loop 1 are less conserved (Figure 7A). Thus, we suggested that the conformation of the loop 1 of HD1CCT differed from that of HsNF-YA, and these differences made their DREs direct to opposite directions, resulting in their divergent DNA recognition.
DISCUSSION
Previously, due to lack in the structural information of the CCT/NF-YB/YC complex, Gnesutta et al. (2017a) proposed a DNA recognition model of the CCT/NF-YB/YC complex based on the structure of the DNA-bound human NF-Y complex. It was assumed that both R340 and G343 in CO (corresponding to R372 and G375 in HD1, respectively) could recognize Gua 2′, and that F345 in CO (corresponding to F377 in HD1) could interact with Cyt 2 and Ade 3 by base stacking. These were consistent with our observation in the HD1CCT/GHD8/OsNF-YC2 structure. We also observed that some other residues were involved in the DNA recognition. For example, R363 was found to interact with Gua 4′ and Thy 5′, and R370 with Cyt 4 (Figure 5B, top-left representation). More importantly, the crystal structure revealed that distinct conformations of loop 1 in HD1CCT and HsNF-YA caused their DREs to point to opposite directions, which determined their DNA recognition specificity to some extent (Figure 7C).
Post-translational modifications within or near CCT domains might influence trimer complex assembly and/or DNA binding activity. Sarid-Krebs et al., (2015) reported that the CCT domain of CO could be phosphorylated, and used the NetPhos 2.0 server (http://www.cbs.dtu.dk/services/NetPhos/) to predict four residues in the CCT domain most likely to be phosphorylated. HD1 could be phosphorylated by OsK4-HDR1 complex (Sun et al., 2016). In our structure, we found that HD1CCT mainly interacted with GHD8/OsNF-YC2 through the polar and positively charged residues (Figure 3C). Thus, phosphorylation could change proteins’ surface charge to possibly weaken these interactions. Usually, the Val–Pro (VP) motifs were targets of the RING-type E3 ligase (Datta et al., 2006; Holm et al., 2001, 2002; Jang et al., 2005). In a ubiquitin-dependent manner, COP1 could bind to the VP motif of CO to switch on the degradation of CO (Jang et al., 2008; Lau et al., 2019; Liu et al., 2008). Due to the VP motifs of CO being close to the DRE of COCCT (residues, 329 to 347), we speculated that the binding of E3 ligase might impair the DNA binding activity of the CO/NF-YB/YC complex.
The photoperiodic flowering pathway comprises light input, the circadian clock, and output (Shim et al., 2017). Governed by the circadian clock, the innate photoperiodic timing mechanism integrates light information to trigger flowering gene expression (Shim et al., 2017). In Arabidopsis, some core components of circadian clock are CCT proteins, such as TOC1, AtPRR3, AtPRR5, AtPRR7, and AtPRR9 (Nakamichi et al., 2005, 2010; Nohales and Kay, 2016; Para et al., 2007; Strayer et al., 2000). Previous yeast-two-hybrid assay showed that TOC1 could interact with the NF-YC subunit (Cao et al., 2011). We found that AtPRRs could be physically associated with AtNF-YB2/YC3 dimer through the CCT domain by LCI assay in N. benthamiana and pull-down assay in mammalian cells (Supplemental Figure 9). Furthermore, we expressed and purified the MBP-tagged CCT domains of these five AtPRRs. SEC assay showed that the CCT domains could form a trimer with AtNF-YB2/YC3 (Supplemental Figure 10). Previous ChIP analyses revealed that four AtPRRs (TOC1, AtPRR5, AtPRR7, and AtPRR9) could target the promoter of the LATE ELONGATED HYPOCOTYL (LHY) gene (Gendron et al., 2012; Huang et al., 2012; Liu et al., 2013, 2016; Nakamichi et al., 2012). Then we examined the DNA binding activity of these trimeric complexes by ITC assay. We selected a DNA sequence containing ‘CCACA’ motif within the LHY promoter. As expected, these four AtPRR/AtNF-YB2/YC3 complexes bound to DNA with the dissociation constants ∼ 1.0 to 1.5 μM, while the trimer containing AtPRR3 did not bind (Supplemental Figure 11; Supplemental Table 1). This result was consistent to our structure observation. In HD1, R372 played a key role in the DNA coordination (Figures 5B and 6). Sequence alignment showed that the residue of AtPRR3 corresponding to R372 of HD1CCT is H503 (Supplemental Figure 1B), which might be the reason for the failure of AtPRR3 in DNA binding. Moreover, R376 of HD1CCT interacts with the phosphate backbone of DNA (Figure 5B), while the corresponding residue of AtPRR is Gln. These amino acids variations may result in the lower DNA binding affinity of AtPRR/NF-YB/YC complex. Taken together, these results suggested that CCT proteins might interact with NF-YB/YC dimer to regulate the plant circadian clock to some degree. However, this needs to be further investigated in vivo.
In addition to previously reported CO and HD1 (COL family), here we confirmed that both GHD7 (CMF family) and GHD7.1 (PRR family) also could form heterotrimers with NF-YB/YC (Figures 1 and 2A). This indicated that CCT domain-containing proteins from three families could form trimers with NF-YB/YC. More than 40 CCT domain-containing proteins were reported in rice and similar numbers were reported in other angiosperms (Cockram et al., 2012). Meanwhile, each NF-Y subfamily had 8 to 16 members in plants (Supplemental Figure 3; Laloum et al., 2013; Petroni et al., 2012). Therefore, the interactions between CCT domain-containing proteins and NF-YB/YCs would generate a large number of trimer combinations to target numerous genes. Some studies of the target sequences of CCT domain-containing proteins have identified a common ‘CCACA’ motif with slightly divergent sequences (Gendron et al., 2012; Gnesutta et al., 2017a, 2018; Goretti et al., 2017; Huang et al., 2012; Liu et al., 2013, 2016; Nakamichi et al., 2012; Nemoto et al., 2016). We speculated that the other domains of the CCT domain-containing proteins might contribute to DNA recognition. Despite this, it was necessary to elaborate target genes and transcriptional regulatory network of distinct CCT/NF-YB/YC complexes so as to deeply understand the functions of CCT family members.
Our previous studies showed that although both GHD7 and GHD7.1 could delay flowering time and increase plant height and grain yield, GHD7 exhibited larger effects than GHD7.1. In long-day conditions, both of them were highly expressed in leaves in the morning to inhibit the expression of the florigen gene Hd3a (Xue et al., 2008; Yan et al., 2013). In this study, our results showed that both GHD7 and GHD7.1 could form trimers with GHD8/OsNF-YC2 to bind the Hd3a promoter (Figure 2). The ITC assay revealed that the DNA binding affinity of GHD7CCT/OsNF-YB/YC complex was three times as high as that of the GHD7.1CCT/OsNF-YB/YC complex with the dissociation constants of 293 nM and 741 nM, respectively (Figure 2C). Taken together, these results suggested that the effects on multiple traits of GHD7 and GHD7.1 were positively correlated to their DNA binding activity.
It is important to elucidate the molecular mechanism of photoperiod-dependent flowering for modifying agricultural crops. Our crystal structure of the DNA-bound HD1CCT/GHD8/OsNF-YC2 revealed the details of the interaction among DNA, CCT domains, and OsNF-YB/YC (Figures 3, 4, and 5). We identified the conserved R338 and Y345 located in helix α1 of HD1CCT as essential residues for trimeric complex formation. R363, R372, and R377 positioned at the DRE of HD1CCT were identified as key residues for DNA recognition. Our results were consistent with the previous findings of functional defects in some natural variants, such as co-7 (Robson et al., 2001) and zcct1 (Distelfeld et al., 2009; Yan et al., 2004), in which the conserved Arg (corresponding to R372 HD1CCT) was replaced with Gln and Trp, respectively (Supplemental Figure 12). Additionally, we identified some residues moderately involved in trimer formation or DNA binding, which provided the possibility to create various alleles with desired heading date by mutating these residues. Taken together, the structural information potentially makes it rational to manipulate flowering time in rice and other crops and accelerate the molecular breeding of new varieties adapted to wider areas.
METHODS
Plant Materials and Growth Conditions
A rice (Oryza sativa) near isogenic line NIL-F2 population was generated based on NIL-Ghd7.1TQ and NIL-Ghd89311 against the ZhenShan97 background (Yan et al., 2011, 2013) and was grown in natural field conditions during normal rice growing seasons at Huazhong Agricultural University, Wuhan, China in 2012.
Nicotiana benthamiana plants were grown in the greenhouse at 25°C under a 12 h light/dark cycle.
Phylogenetic Analysis
Reported CCT domain containing genes from Arabidopsis thaliana, O. sativa, Zea mays, Sorghum bicolor, Triticum aestivum, and Hordeum vulgare were aligned using the tool ClustalX2 (Larkin et al., 2007). UPGMA tree was constructed with full-length amino acid sequences with the software MEGA6 (Tamura et al., 2013). The sequence alignment file and the tree file are provided in Supplemental Files 1 and 2. Bootstrap percentages at the branch points were estimated from 1,000 bootstrap replications. The evolutionary distances were computed using the Poisson correction method and the units are the number of amino acid substitutions per site.
Protein Expression and Purification
HD1, GHD8, and OsNF-YC2 were codon-optimized and synthesized by Qinglan Biotech. OsNF-YC2 (residues 55 to 156) was cloned into the pET15b vector (Novagen) with an N-terminal 6×His tag and an engineered cleavage site for the caspase drICE (pET15b-D). GHD8 (residues 54 to 147) was cloned into the pBB75 vector without any tag. GHD8 and OsNF-YC2 were produced by co-expression in Escherichia coli BL21 (DE3). HD1CCT (residues 323 to 387) was cloned into the engineered pET15b-D vector (Novagen), preceded by an N-terminal maltose-binding protein (MBP). Between HD1CCT and MBP was a PreScission Protease site. HD1CCT was expressed in E. coli BL21(DE3) individually.
One liter of LB medium was supplemented with the corresponding antibiotics (100 μg mL−1 of ampicillin and 50 μg mL−1 of kanamycin for GHD8/OsNF-YC2 dimer; 100 μg mL−1 of ampicillin for HD1CCT) and inoculated with a transformed bacterial preculture and shaken at 37°C until the optical density at 600 nm reached 1.0. Then the culture was cooled to 16°C and induced with 0.2 mM of isopropyl-β-d-thiogalactoside. After 16 h of growth at 16°C, the bacterial pellet was collected and homogenized in buffer A (25 mM of Tris-HCl at pH 8.0, and 150 mM of NaCl). After high-pressure cell disruption and centrifugation at 23,708g at 4°C, the supernatant was loaded onto a column equipped with Ni2+ affinity resin (Ni-NTA; Qiagen), washed with buffer B (25 mM of Tris-HCl at pH 8.0, 150 mM of NaCl, and 15 mM of imidazole), and eluted with buffer C (25 mM of Tris-HCl at pH 8.0 and 250 mM of imidazole).
The GHD8/OsNF-YC2 dimer and HD1CCT were further purified by heparin HP (HiTrap, 5 mL; GE Healthcare), respectively, and then mixed at a 1:1 ratio. After an additional step of drICE and PreScission Protease cleavage, the mixture was loaded onto amylose resin (New England Biolabs) to remove the MBP tag. The flowthrough was concentrated to 1 mL immediately before gel filtration chromatography (Superdex-200 10/300; GE Healthcare). The buffer for gel filtration contained 25 mM of Tris-HCl at pH 8.0, 100 mM of NaCl, 5 mM of MgCl2, and 5 mM of DTT. The flow rate of gel filtration is 0.5 mL/min. The peak fractions were incubated with target DNA oligonucleotides at a molar ratio of 1:1.5 at 4°C for ∼40 min before crystallization trials.
AtNF-YB2 (residues 24 to 116) and AtNF-YC3 (residues 56 to 148) were co-expressed in E. coli BL21 (DE3), and then were purified same as GHD8/OsNF-YC2 dimer. GHD7 (residues 175 to 239), GHD7.1 (residues 669 to 724), TOC1 (residues 522 to 578), AtPRR3 (residues 462 to 513), AtPRR5 (residues 604 to 661), AtPRR7 (residues 657 to 713), and AtPRR9 (residues 399 to 464) were purified the same as HD1.
DNA Duplexes for Crystal Formation
The OsCORE DNA duplexes contained two overhanging bases on each 5′ end (two deoxyadenylates for one strand, 5′-aaCTCACTCTCAACCACAGCTCGAT-3′, and two deoxythymidylates for the complementary strand, 5′-ttATCGAGCTGTGGTTGAGAGTGAG-3′). Single-stranded oligonucleotides were synthesized and dissolved in buffer A. The ssDNA was mixed with a complementary strand at equal molar amounts, heated at 95°C for 3 min and annealed by slow cooling to 20°C over a period of 5 h. The double-stranded DNA was stored at −20°C.
Crystallization
GHD8/OsNF-YC2 and DNA-bound HD1CCT/GHD8/OsNF-YC2 were crystallized using the hanging-drop vapor-diffusion method at 18°C by mixing 1 μL of sample with an equal volume of reservoir solution. For the GHD8/OsNF-YC2 dimer, crystals were observed in the conditions containing 20% w/v polyethylene glycol 3350 and 0.2 M of sodium malonate at pH 7.0. After screening of the pH buffer, high-resolution crystals were obtained in 17% w/v polyethylene glycol 3,350, 0.2 M of sodium malonate at pH 7.0, and 0.1 M of sodium citrate tribasic dihydrate at pH 6.7. The HD1CCT/GHD8/OsNF-YC2 complex was subjected to crystallization trials in complex with 23-bp OsCORE2 DNA duplexes. Crystals appeared overnight under the conditions of 10% (w/v) polyethylene glycol 4,000, 0.1 M of sodium citrate at pH 5.5, and 0.2 M of sodium acetate trihydrate. The best crystals diffracted to ∼5.0 Å. To further improve the diffraction quality, magnesium acetate tetrahydrate was added to the well buffer, and then, one round of an additive screen was performed under the selected conditions. Finally, the best crystals were obtained under the following conditions: ∼7.0 to 9.0% (w/v) PEG 4,000, 0.1 M of sodium citrate at pH 5.2, 0.2 M of sodium acetate trihydrate, 0.2 M of magnesium acetate tetrahydrate, and 0.1 M of Gly. The crystals were flash-frozen in liquid nitrogen using 20% (v/v) glycerol as the cryoprotective buffer and diffracted to 2.55 Å at the Shanghai Synchrotron Radiation Facility beamline BL17U (Wang et al., 2018).
Data Collection and Structural Determination
All data sets were collected at the Shanghai Synchrotron Radiation Facility beamline BL19U or BL17U and processed with the HKL3000 or HKL2000 package (Otwinowski and Minor, 1997). Further processing was performed with programs from the CCP4 suite (Collaborative Computational Project Number 4, 1994; Winn et al., 2011). Data collection and structure refinement statistics are summarized in Table 1. The structure of the DNA-bound HD1CCT/GHD8/OsNF-YC2 complex was solved by molecular replacement with the NF-Y structure (PDB:4G92; Huber et al., 2012) as the search model using the program PHASER (McCoy et al., 2007). The structure was manually iteratively refined with the tools PHENIX and COOT (Adams et al., 2002; Emsley and Cowtan, 2004).
EMSA
For EMSA, the forward single-stranded DNA oligonucleotides (5′-CTCACTCTCAACCACAGCTCGAT-3′) labeled at the 5′ end with 6-Carboxyfluorescein (FAM) were synthesized by General Biosystems and annealed with an equimolar amount of the complementary strand (5′-ATCGAGCTGTGGTTGAGAGTGAG-3′) as described above. Proteins were incubated with 10 nM of FAM-labeled probe for 20 min on ice in final solution containing 25 mM of Tris-HCl at pH 8.0, 5 mM of MgCl2, 5 mM of DTT, 0.1 mg mL−1 of BSA, 60 μg mL−1 of salmon sperm DNA, and 10% (v/v) glycerol. The obtained products were then resolved on 8% (v/v) native acrylamide gels (37.5:1 acrylamide: bis-acrylamide) in 0.5 × Tris-Gly buffer under an electric field of 10 V cm−1 for 2.5 h. Gels were visualized using the tool Typhoon (Amersham).
ITC
The DNA binding measurement of the CCT/NF-YB/YC complex was performed at 25°C using an Auto-iTC200 titration calorimeter (MicroCal). The OsCORE (250 μM, 5′-CTCACTCTCAACCACAGCTCGAT-3′, for CCT/NF-YB/YC complexes from rice) or LHY-P1 (250 μM, 5′-ATTGTGGACCACCACACTCACTT-3′, for CCT/NF-YB/YC complexes from Arabidopsis) was dissolved in reaction buffer (25 mM of HEPES at pH 7.4, and 150 mM NaCl) and titrated against 200 μL of 30-μΜ wild-type CCT/NF-YB/YC complex or the mutants in the same buffer. The first injection of 0.5 μL was followed by 19 injections of 2 μL. The heat of dilution was measured by injecting DNA into the buffer alone in each experiment. The values were subtracted from the experimental curves before data analysis. The stirring rate was 750 r.p.m. The software MicroCal Origin, which was supplied with the instrument, was used to determine the site-binding model that produced a good fit (low × 2 value) for the resulting data.
Assessment of Heading Date in Near-Isogenic Lines
The NIL-F2 population based onNIL-Ghd7.1TQ and NIL-Ghd89311 against the ZhenShan97 background (Yan et al., 2011, 2013) was generated and was grown in natural field conditions during normal rice growing seasons at Huazhong Agricultural University, Wuhan, China in 2012. Four homozygous-type individual plants, i.e., ZS, ZSGhd8, ZSGhd7.1, and ZSGhd8 Ghd7.1, were genotyped with functional marker Z9M and 37indel (Yan et al., 2013; Zhang et al., 2015), and heading dates were recorded from the sowing day to first-panicle–emerged days. More than 10 plants from each of the four types were used for statistical analysis. Multiple comparisons of the heading-date mean values were performed based on Duncan’s multiple range test.
Pull-Down Assays
Genes used in pull-down assay were cloned into the engineered pMlink vectors with different tags (His, Flag, and StrepII [Sigma-Aldrich], or Myc). The Expi293F cells transfected with corresponding constructs were incubated under dark condition. Cells were harvested and washed with PBS. Then the cells were resuspended in StrepII buffer (100 mM of Tris-HCl at pH 8.0, 150 mM of NaCl, and 1 mM of EDTA) and were repeatedly freeze-thawed three times. The lysed cells were centrifuged at 12,000g for 30 min at 4°C. The supernatant was incubated with StrepII beads at 4°C for 1.5 h. Beads were washed five times with StrepII buffer. The bound proteins were eluted with StrepII buffer containing 2.5 mM of d-Desthiobiotin. The proteins were detected through immunoblots with antibodies against StrepII (lot no. 41246; Sigma-Aldrich), His (cat. no. 66005-1-Ig; Proteintech), Myc (cat. no. 60003-2-Ig; Proteintech) or Flag (cat. no. 66008-3-Ig; Proteintech).
LCI Assay
The LCI assay was performed according to the previously described protocol with little modification (Chen et al., 2008). CCT genes were cloned into the pCAMBIA-NLuc vector. GHD8 and AtNF-YC3 were cloned into the pCAMBIA-CLuc vector. OsNF-YC2 and AtNF-YB2 were cloned into the pCAMBIA1301s vector. Each construct was transformed into Agrobacterium tumefaciens strain EH105. Through the Agrobacterium-mediated transformation, pCAMBIA-NLuc, pCAMBIA-CLuc, and pCAMBIA1301s constructs were co-expressed in 5-week–old N. benthamiana leaves. Two days after infiltration, the 1 mM of luciferin was smeared onto the leaves and the images were captured using the 5200 Chemiluminescent Imaging System (Tanon Science and Technology).
Accession Numbers
Coordinates for the atomic structures have been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank under PDB:7C9O (DNA-bound HD1CCT/GHD8/OsNF-YC2) and PDB:7C9P (GHD8/OsNF-YC2).
Supplemental Data
Supplemental Figure 1. CCT domain-containing proteins are divided into three families.
Supplemental Figure 2. GHD7.1 genetically interacts with GHD8.
Supplemental Figure 3. Sequence alignments of the histone fold domain domains in OsNF-YBs and in OsNF-YCs.
Supplemental Figure 4. Crystal structure of the GHD8/OsNF-YC2 dimer.
Supplemental Figure 5. OsCORE2 wraps around GHD8/OsNF-YC2 in a manner similar to nucleosomal DNA.
Supplemental Figure 6. SEC analysis of the interactions between GHD8/OsNF-YC2 dimer and mutated HD1CCT containing residue substitutions in the PIE region.
Supplemental Figure 7. SEC analysis of the interactions between GHD8/OsNF-YC2 dimer and mutated HD1CCT containing residue substitutions in the DRE region.
Supplemental Figure 8. Comparison of side chains of the residues interacting with DNA in HD1CCT and HsNF-YA.
Supplemental Figure 9. AtPRRs can physically associate with the AtNF-YB2/YC3 dimer.
Supplemental Figure 10. The CCT domains of AtPRRs can form trimers with the AtNF-YB2/YC3 dimer.
Supplemental Figure 11. ITC analysis of the interactions between the DNA probe from LHY gene and the AtPRRCCT/AtNF-YB2/YC3 complexes.
Supplemental Figure 12. Sequence alignment of CCT domains of natural CCT-domain–containing protein variants.
Supplemental File 1. Tree file for phylogenetic analysis.
Supplemental File 2. Sequence alignments for phylogenetic analysis.
Supplemental Table 1. Measurement of DNA binding affinity of different CCT/NF-YB/YC complexes by ITC.
Supplemental Table 2. PCR primer sequences for cloning.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the staffs of the BL17U1/BL19U1 beamline of the National Center for Protein Sciences Shanghai (NCPSS) at the Shanghai Synchrotron Radiation Facility for assistance during data collection, and research associate Dr. Delin Zhang at the Center for Protein Research, Huazhong Agricultural University, for technical support. This work was supported by the National Natural Science Foundation of China (grants 31821005 to Y.Z.X., 31870753 to P.Y. and 31901050 to C.C.S.), the Ministry of Science and Technology of China (grant 2018YFA0507700), and the China Postdoctoral Science Foundation (grant 2018M640712).
AUTHOR CONTRIBUTIONS
C.C.S., H.Y.L., Z.Y.G., P.Y., and Y.Z.X. designed all experiments; C.C.S., H.Y.L., Z.Y.G., J.J.Y., and T.Z. performed protein expression, purification, and crystallization; Z.Y.G. determined all of the structures; C.C.S. and H.Y.L. carried out biochemical assays; W.H.Y., C.Y.W., and Q.Z. carried out genetic assays; C.C.S., H.Y.L., P.Y., and Y.Z.X. wrote the article; all authors discussed the results and commented on the article.
Footnotes
Articles can be viewed without a subscription.
References
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.(2002). PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58: 1948–1954. [DOI] [PubMed] [Google Scholar]
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E.(2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46: 462–476. [DOI] [PubMed] [Google Scholar]
- Bi W., Wu L., Coustry F., de Crombrugghe B., Maity S.N.(1997). DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J. Biol. Chem. 272: 26562–26572. [DOI] [PubMed] [Google Scholar]
- Cao S., Kumimoto R.W., Gnesutta N., Calogero A.M., Mantovani R., Holt B.F. III(2014). A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26: 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Siriwardana C.L., Kumimoto R.W., Holt B.F. III(2011). Construction of high quality GatewayTM entry libraries and their application to yeast two-hybrid for the monocot model plant Brachypodium distachyon. BMC Biotechnol. 11: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M.(2008). Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockram J., Thiel T., Steuernagel B., Stein N., Taudien S., Bailey P.C., O’Sullivan D.M.(2012). Genome dynamics explain the evolution of flowering time CCT domain gene families in the Poaceae. PLoS One 7: e45307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 (1994). The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50: 760–763. [DOI] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H., Deng X.W., Holm M.(2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distelfeld A., Tranquilli G., Li C., Yan L., Dubcovsky J.(2009). Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol. 149: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A.(2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18: 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A., Tian W., Wei M., Yan W., He H., Zhou D., Huang X., Li S., Ouyang X.(2017). The DTH8-Hd1 module mediates day-length–dependent regulation of rice flowering. Mol. Plant 10: 948–961. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K.(2004). Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A.(2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta N., Kumimoto R.W., Swain S., Chiara M., Siriwardana C., Horner D.S., Holt B.F. III, Mantovani R.(2017a). CONSTANS imparts DNA sequence specificity to the histone fold NF-YB/NF-YC dimer. Plant Cell 29: 1516–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnesutta N., Mantovani R., Fornara F.(2018). Plant flowering: Imposing DNA specificity on histone-fold subunits. Trends Plant Sci. 23: 293–301. [DOI] [PubMed] [Google Scholar]
- Gnesutta N., Saad D., Chaves-Sanjuan A., Mantovani R., Nardini M.(2017b). Crystal structure of the Arabidopsis thaliana L1L/NF-YC3 histone-fold dimer reveals specificities of the LEC1 family of NF-Y subunits in plants. Mol. Plant 10: 645–648. [DOI] [PubMed] [Google Scholar]
- Goretti D., Martignago D., Landini M., Brambilla V., Gómez-Ariza J., Gnesutta N., Galbiati F., Collani S., Takagi H., Terauchi R., Mantovani R., Fornara F.(2017). Transcriptional and post-transcriptional mechanisms limit heading date 1 (Hd1) function to adapt rice to high latitudes. PLoS Genet. 13: e1006530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K.(2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- Holm M., Hardtke C.S., Gaudet R., Deng X.W.(2001). Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J. 20: 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W.(2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P.(2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Huber E.M., Scharf D.H., Hortschansky P., Groll M., Brakhage A.A.(2012). DNA minor groove sensing and widening by the CCAAT-binding complex. Structure 20: 1757–1768. [DOI] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H.(2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G.(2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.E., Nam H., Park J., Choi G.J., Lee Y.W., Son H.(2020). Characterization of the CCAAT-binding transcription factor complex in the plant pathogenic fungus Fusarium graminearum. Sci. Rep. 10: 4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R., Ikegami A., Tamaki S., Yokoi S., Shimamoto K.(2008). Hd3a and RFT1 are essential for flowering in rice. Development 135: 767–774. [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Adam L., Hymus G.J., Repetti P.P., Reuber T.L., Marion C.M., Hempel F.D., Ratcliffe O.J.(2008). The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228: 709–723. [DOI] [PubMed] [Google Scholar]
- Kumimoto R.W., Zhang Y., Siefers N., Holt B.F. III(2010). NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63: 379–391. [DOI] [PubMed] [Google Scholar]
- Laloum T., De Mita S., Gamas P., Baudin M., Niebel A.(2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18: 157–166. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X Version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lau K., Podolec R., Chappuis R., Ulm R., Hothorn M.(2019). Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 38: e102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Liu C., Hou X., Xi W., Shen L., Tao Z., Wang Y., Yu H.(2012). FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 10: e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q.(2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Carlsson J., Takeuchi T., Newton L., Farré E.M.(2013). Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J. 76: 101–114. [DOI] [PubMed] [Google Scholar]
- Liu T.L., Newton L., Liu M.J., Shiu S.H., Farré E.M.(2016). A G-Box-Like motif is necessary for transcriptional regulation by circadian pseudo-response regulators in Arabidopsis. Plant Physiol. 170: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.(1997). Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Ma L., Wang X., Guan Z., Wang L., Wang Y., Zheng L., Gong Z., Shen C., Wang J., Zhang D., Liu Z., Yin P.(2020). Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2. Nat. Struct. Mol. Biol. 27: 472–479. [DOI] [PubMed] [Google Scholar]
- Maity S.N., de Crombrugghe B.(1998). Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 23: 174–178. [DOI] [PubMed] [Google Scholar]
- Mantovani R.(1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239: 15–27. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.(2007). Phaser crystallographic software. J. Appl. Cryst. 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R.L., Klein R.R., Morishige D.T., Brady J.A., Rooney W.L., Miller F.R., Dugas D.V., Klein P.E., Mullet J.E.(2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA 108: 16469–16474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.H., Sakakibara H.(2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kiba T., Kamioka M., Suzuki T., Yamashino T., Higashiyama T., Sakakibara H., Mizuno T.(2012). Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 109: 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kita M., Ito S., Yamashino T., Mizuno T.(2005). PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 46: 686–698. [DOI] [PubMed] [Google Scholar]
- Nardini M., Gnesutta N., Donati G., Gatta R., Forni C., Fossati A., Vonrhein C., Moras D., Romier C., Bolognesi M., Mantovani R.(2013). Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 152: 132–143. [DOI] [PubMed] [Google Scholar]
- Nemoto Y., Nonoue Y., Yano M., Izawa T.(2016). Hd1,a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86: 221–233. [DOI] [PubMed] [Google Scholar]
- Nohales M.A., Kay S.A.(2016). Molecular mechanisms at the core of the plant circadian oscillator. Nat. Struct. Mol. Biol. 23: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W.(1997). [20] Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Para A., Farré E.M., Imaizumi T., Pruneda-Paz J.L., Harmon F.G., Kay S.A.(2007). PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni K., Kumimoto R.W., Gnesutta N., Calvenzani V., Fornari M., Tonelli C., Holt B.F. III, Mantovani R.(2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G.(1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Robson F., Costa M.M., Hepworth S.R., Vizir I., Piñeiro M., Reeves P.H., Putterill J., Coupland G.(2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28: 619–631. [DOI] [PubMed] [Google Scholar]
- Romier C., Cocchiarella F., Mantovani R., Moras D.(2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278: 1336–1345. [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G.(2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sarid-Krebs L., Panigrahi K.C., Fornara F., Takahashi Y., Hayama R., Jang S., Tilmes V., Valverde F., Coupland G.(2015). Phosphorylation of CONSTANS and its COP1-dependent degradation during photoperiodic flowering of Arabidopsis. Plant J 84: 451–463. [DOI] [PubMed] [Google Scholar]
- Shim J.S., Kubota A., Imaizumi T.(2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C., Oyama T., Schultz T.F., Raman R., Somers D.E., Más P., Panda S., Kreps J.A., Kay S.A.(2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771. [DOI] [PubMed] [Google Scholar]
- Sun X., Zhang Z., Wu J., Cui X., Feng D., Wang K., Xu M., Zhou L., Han X., Gu X., Lu T.(2016). The Oryza sativa regulator HDR1 associates with the kinase OsK4 to control photoperiodic flowering. PLoS Genet 12: 1005927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K.(2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka K., et al. (2011). 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 476: 332–335. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187: 57–66. [DOI] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A.(2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- Wang Q.-S., et al. (2018). Upgrade of macromolecular crystallography beamline BL17U1 at SSRF. Nucl. Sci. Tech. 29: 68. [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G.(2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X., Zhang Q.(2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., Bennetzen J.L., Echenique V., Dubcovsky J.(2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Liu H., Zhou X., Li Q., Zhang J., Lu L., Liu T., Liu H., Zhang C., Zhang Z., Shen G., Yao W., et al. (2013). Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 23: 969–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W.H., Wang P., Chen H.X., Zhou H.J., Li Q.P., Wang C.R., Ding Z.H., Zhang Y.S., Yu S.B., Xing Y.Z., Zhang Q.F.(2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4: 319–330. [DOI] [PubMed] [Google Scholar]
- Yang L., et al. (2018). Reconstituting Arabidopsis CRY2 signaling pathway in mammalian cells reveals regulation of transcription by direct binding of CRY2 to DNA. Cell Rep. 24: 585–593 e584. [DOI] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., Sasaki T.(2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2015). Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 208: 1056–1066. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Liu L., Shen L., Yu H.(2016). NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants 2: 16075. [DOI] [PubMed] [Google Scholar]