High levels of soil salinity lead to toxic accumulation of sodium (Na+) and chloride (Cl−) ions in plants, which adversely affect plant growth and yield. Plants use several strategies to cope with salt stress. These include removal and compartmentalization of toxic ions, and maintenance of growth and water uptake during salt stress. Salt tolerance is a multigenic trait, and many genes important for salt-stress response have been identified in model plants and crops (reviewed by van Zelm et al. 2020). Relatively less is known about the early processes involved in the perception of salinity and the regulation of stress response genes. In this issue of The Plant Cell, a study by Jie Wang and Nan Nan and colleagues (Wang et al., 2020) provides insight into a transcriptional complex that links salt-stress perception and activation of a salt-tolerance gene in rice (Oryza sativa).
Through screening of a CRISPR/Cas9 mutant library, Wang et al. (2020) isolated a line deficient in the rice BCL-2-ASSOCIATED ATHANOGENE (BAG) gene OsBAG4 that shows increased salt sensitivity compared with the wild-type rice (Nipponbare variety). OsBAG4 belongs to the BAG domain containing co-chaperones that are broadly conserved in yeasts, animals, and plants. BAG genes function in diverse processes such as programmed cell death, defense, and stress responses (reviewed by Thanthrige et al. 2020). The authors found that OsBAG4 expression is induced under salt stress (100 mM of NaCl) in roots and shoots of rice. Salt-stressed osbag4 mutants have elevated levels of reactive oxygen species (a marker for oxidative damage) compared with the wild type. Expression of a FLAG-tagged OsBAG4 transgene under the native OsBAG4 promoter rescued the salt-sensitive phenotype of the osbag4 mutant. Moreover, transgenic lines that overexpress OsBAG4 exhibit enhanced salt tolerance compared with the wild type. Together, these findings indicate that OsBAG4 acts as a positive regulator of salt-stress tolerance.
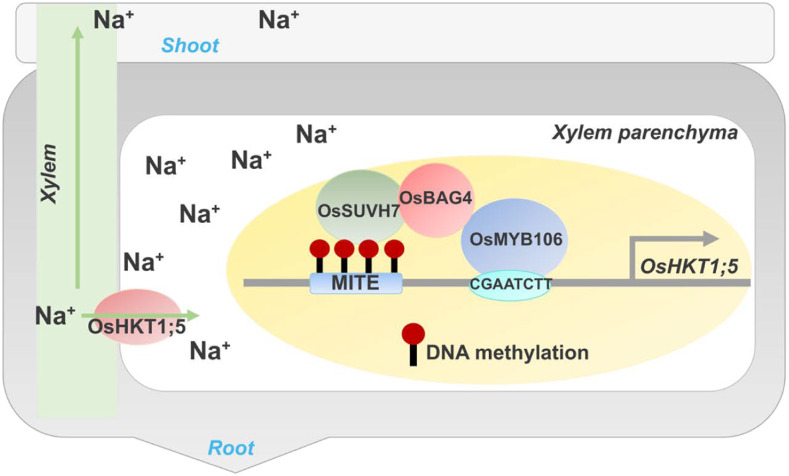
To understand how OsBAG4 affects the salt-stress response, the authors looked for genes that are differentially expressed in roots or shoots of osbag4 compared with the wild type. Interestingly, osbag4 mutants show root-specific significant downregulation of OsHKT1;5—a major salt tolerance gene in rice. OsHKT1;5 encodes a Na+ transporter that excludes Na+ from leaves and is important for Na+/K+ homeostasis under salt stress. The authors found that reduced expression of OsHKT1;5 in osbag4 mutants leads to a significant increase in shoot Na+/K+ ratios under salt stress. These results suggest that OsBAG4 acts upstream of OsHKT1;5.
How does OsBAG4 regulate OsHKT1;5 expression? BAG genes are known to interact with chaperones such as heat shock protein 70 as well as transcription factors, and modulate their function. The authors performed co-immunoprecipitation experiments coupled with mass spectrometry and identified several putative OsBAG4-interacting proteins. These included a MYB transcription factor (OsMYB106), and a DNA methylation reader (OsSUVH7). Although OsMYB106 does not directly bind OsSUVH7, both of these interact with OsBAG4 and form a stable complex. Follow-up experiments confirmed that OsMYB106 binds to OsHKT1;5 cis-regulatory sequences (see figure) and its DNA binding affinity is enhanced by OsBAG4. Thus, OSBAG4 and OsMYB106 act in a synergistic manner to activate OsHKT1;5.
Salt-Induced Expression of OsHKT1;5 Is Regulated by a Transcriptional Complex Containing the DNA Methylation Reader OsSUVH7.
(Reproduced from Wang et al. [2020], Figure 8.)
Stress-response genes often have transposable element insertions in their upstream regions that influence their expression. Analysis of the putative OsHKT1;5 cis-regulatory sequences revealed a miniature-inverted repeat transposable element (MITE) located upstream of the OsMYB106 binding site. This MITE region was enriched in 24-nucleotide small RNAs as well as CHH and CHG methylation (where H = A/T/C), which are hallmarks of RNA-directed DNA methylation. Salt stress elevated the CHH and CHG methylation levels of the MITE specifically in the roots, suggesting a role for DNA methylation in the regulation of OsHKT1;5 expression. As several SUPPRESSOR OF VARIEGATION 3-9 HOMOLOG (SUVH) family members are known to bind methylated DNA through their SRA domain (Harris et al. 2018), the authors hypothesized that the putative OsBAG4-interacting protein OsSUVH7 may bind the MITE region in the OsHKT1;5 upstream region. Chromatin immunoprecipitation experiments confirmed that OsSUVH7 binds the methylated MITE. Deletion of the MITE sequence or loss of OsMYB106/OsSUVH7 function leads to reduced OsHKT1;5 expression and increased the salt sensitivity. Thus, OsBAG4 works in concert with OsSUVH7 and OsMYB106 for transcriptional regulation of OsHKT1;5 in response to salt stress.
Overall, the findings by Wang et al. (2020) contribute to the growing evidence of epigenetic regulation of stress-response genes and provides new directions for improving crop yield under salt stress.
Footnotes
Articles can be viewed without a subscription.
References
- Harris C.J., et al. (2018). A DNA methylation reader complex that enhances gene transcription. Science 362: 1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanthrige N., Jain S., Bhowmik S.D., Ferguson B.J., Kabbage M., Mundree S., Williams B.(2020). Centrality of BAGs in plant PCD, stress responses, and host defense. Trends Plant Sci. (In press). [DOI] [PubMed] [Google Scholar]
- van Zelm E., Zhang Y., Testerink C.(2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71: 403–433. [DOI] [PubMed] [Google Scholar]
- Wang J., Nan N., Li N., Liu Y., Wang T.-J., Hwang I., Liu B., Xu Z.-Y.(2020). A DNA methylation reader-chaperone regulator-transcription factor complex activates OsHKT1;5 expression during salinity stress. Plant Cell 32: 3535–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]