OsSUVH7, OsBAG4, and OsMYB106 form a DNA methylation reader–chaperone regulator–transcription factor complex on the promoter region of OsHKT1;5 to activate its expression during salinity stress.
Abstract
Irrigated lands are increasingly salinized, which adversely affects agricultural productivity. To respond to high sodium (Na+) concentrations, plants harbor multiple Na+ transport systems. Rice (Oryza sativa) HIGH-AFFINITY POTASSIUM (K+) TRANSPORTER1;5 (OsHKT1;5), a Na+-selective transporter, maintains K+/Na+ homeostasis under salt stress. However, the mechanism regulating OsHKT1;5 expression remains unknown. Here, we present evidence that a protein complex consisting of rice BCL-2-ASSOCIATED ATHANOGENE4 (OsBAG4), OsMYB106, and OsSUVH7 regulates OsHKT1;5 expression in response to salt stress. We isolated a salt stress–sensitive mutant, osbag4-1, that showed significantly reduced OsHKT1;5 expression and reduced K+ and elevated Na+ levels in shoots. Using comparative interactomics, we isolated two OsBAG4-interacting proteins, OsMYB106 (a MYB transcription factor) and OsSUVH7 (a DNA methylation reader), that were crucial for OsHKT1;5 expression. OsMYB106 and OsSUVH7 bound to the MYB binding cis-element (MYBE) and the miniature inverted-repeat transposable element (MITE) upstream of the MYBE, respectively, in the OsHKT1;5 promoter. OsBAG4 functioned as a bridge between OsSUVH7 and OsMYB106 to facilitate OsMYB106 binding to the consensus MYBE in the OsHKT1;5 promoter, thereby activating the OsHKT1;5 expression. Elimination of the MITE or knockout of OsMYB106 or OsSUVH7 decreased OsHKT1;5 expression and increased salt sensitivity. Our findings reveal a transcriptional complex, consisting of a DNA methylation reader, a chaperone regulator, and a transcription factor, that collaboratively regulate OsHKT1;5 expression during salinity stress.
INTRODUCTION
Rice (Oryza sativa) is a staple food that feeds more than half of the world’s population. However, annual rice production is negatively affected by high salinity (Zhu, 2001; Reddy et al., 2017). Soil salinity is a major environmental constraint for crop production that affects nearly 45 million hectares of irrigated land (Munns and Tester, 2008; Rengasamy, 2010). Salt stress has significant deleterious effects on agricultural yield, including slower growth rates, reduced tillering, and defects in reproductive development. The ultimate goal of salinity tolerance research is to increase the capability of plants to maintain growth and productivity in saline soils; that is, to decrease the effects of salinity on growth and yield.
Since sodium (Na+) transport processes have major roles in salinity tolerance, significant attention has been paid to the function of the high-affinity potassium (K+) transporter (HKT) family (Schachtman and Schroeder, 1994; Rubio et al., 1995, 1999). Previous studies in Arabidopsis (Arabidopsis thaliana) demonstrated that the AtHKT1 transporter makes a critical contribution to protecting leaves from Na+ over-accumulation and salt stress (Mäser et al., 2002; Sunarpi et al., 2005). Quantitative trait locus (QTL) analyses of salt-tolerant rice revealed that the SHOOT K+ CONCENTRATION1 (SKC1) locus, which is associated with an elevated K+/Na+ ratio in shoots, corresponds to the OsHKT1;5 gene, which encodes a Na+-selective transporter (Ren et al., 2005). OsHKT1;5-dependent Na+ transport in roots, leaf sheaths, and stems is a key salt tolerance mechanism during rice growth and development (Kobayashi et al., 2017). In wheat (Triticum aestivum), Nax2 is important for maintaining a low Na+ concentration in leaves and its locus coincides with that of the Na+ transporter TmHKT1;5-A, whose presence increases grain yield by 25% compared with near-isogenic lines without the Nax2 locus (Munns et al., 2012).
Regardless of the importance of HKTs for Na+ transport during salinity stress, few studies have reported the transcriptional regulatory mechanisms of HKTs. In Arabidopsis, a putative small RNA target region and a tandem repeat in the promoter region of AtHKT1 are essential for maintaining its expression (Baek et al., 2011), indicating that DNA methylation including RNA-directed DNA methylation (RdDM) is important for the regulation of AtHKT1 expression (Baek et al., 2011); however, the exact molecular regulatory mechanisms remain unclear.
The Bcl-2–associated athanogene (BAG) family is a group of evolutionarily conserved cochaperones involved in diverse cellular functions, including growth arrest and cell death, in yeast, plants, and mammals (Takayama et al., 1995; Lee et al., 1999). BAG family proteins were originally identified as factors that bind the antiapoptotic protein Bcl-2 (Takayama et al., 1995; Lee et al., 1999), and they were later shown to interact and modulate the functions of heat shock proteins 70 (Hsc70/Hsp70; Lee et al., 2016). All BAG proteins have at least one copy of a roughly 50 amino acid conserved BAG domain that mediates the interaction with Hsp70 (Takayama and Reed, 2001). In humans, six BAG family members regulate the function of Hsp70/Hsc70 positively and negatively, forming complexes with various transcription factors that control physiological processes including tumorigenesis, apoptosis, neuronal differentiation, and stress responses (Zeiner and Gehring, 1995; Kabbage and Dickman, 2008). In Arabidopsis, ectopic expression of AtBAG4 increases tolerance to various abiotic stresses, and AtBAG6 plays a role in the basal defense response (Doukhanina et al., 2006). AtBAG7 is involved in the unfolded protein response in the endoplasmic reticulum (Williams et al., 2010).
The BAG proteins of rice can be categorized into two classes based on the presence of conserved domains (Rana et al., 2012). OsBAG1 to OsBAG4, the members of the class I subfamily, contain an additional ubiquitin-like domain; these proteins are structurally similar to the human BAG1 protein and its orthologs in Arabidopsis (Rana et al., 2012). The class II subfamily consists of OsBAG5 and OsBAG6, which contain a calmodulin binding domain (Rana et al., 2012). Uniquely among the OsBAGs, OsBAG4 plays a role in plant innate immunity via a direct association with ENHANCED BLIGHT AND BLAST RESISTANCE1 (EBR1), which directly targets OsBAG4 for ubiquitination-mediated degradation (You et al., 2016). Ectopic accumulation of OsBAG4 in rice is sufficient to trigger programmed cell death and increase resistance to pathogenic infection (You et al., 2016).
MYB factors are a family of proteins that contain the conserved MYB DNA binding domain (Dubos et al., 2010). MYB proteins can be classified into four subfamilies depending on the number of adjacent repeats. For instance, MYB-like proteins with one or a partial MYB repeat are designated MYB-related proteins; two, 2R-MYB (R2R3-type MYB); three, 3R-MYB (R1R2R3-type MYB); and four, 4R-MYB (four R1/R2-like repeats; Rosinski and Atchley, 1998; Jin and Martin, 1999; Dubos et al., 2010; Feller et al., 2011). MYB proteins harboring two repeats, R2R3-MYB, constitute the largest MYB gene family in plants (Katiyar et al., 2012). This subfamily functions in a wide range of biological processes including metabolism, growth, development, and biotic and abiotic stress responses (Dubos et al., 2010; Feller et al., 2011). OsMYBc knockout mutants are salt sensitive and exhibit a reduction in NaCl-induced expression of OsHKT1;1, which plays an important role in decreasing Na+ accumulation in shoots to help cope with salt stress (Wang et al., 2015).
DNA cytosine methylation frequently marks transposable elements (TEs), which can exert transcriptional effects on neighboring genes (Harris et al., 2018). DNA cytosine is methylated in three different sequence contexts, that is, CG, CHG, and CHH (H = A, T, or C). In Arabidopsis, METHYLTRANSFERASE1 (MET1) and CHROMOMETHYLASE3 (CMT3) maintain CG and CHG methylation, while de novo cytosine methylation is mediated by the methyltransferases DRM1 and DRM2 (Henderson and Jacobsen, 2007). Previous studies revealed that two SU(VAR)3-9 homologs, the transcriptional antisilencing factors AtSUVH1 and AtSUVH3, bind to methylated DNA and form a complex with DNAJ domain-containing proteins to increase proximal gene expression (Harris et al., 2018). Miniature inverted-repeat transposable elements (MITEs) positively and negatively regulate D14 and OsMIR156 family members, respectively, thereby influencing rice tillering. Hence, control of rice tillering by RdDM at MITEs provides a potential mechanism for agronomic trait enhancement (Xu et al., 2020). However, it remains unknown whether and how DNA methylation at MITEs impacts the salt stress response.
In this study, we identified an osbag4-1 mutant with a salt stress–sensitive phenotype associated with reduced survival rates, reactive oxygen species (ROS) accumulation, and elevated shoot Na+/K+ ratios. Using RNA sequencing (RNA-seq), we found that OsHKT1;5 expression was specifically and significantly reduced in osbag4 mutant roots. Furthermore, using multidimensional protein identification technology mass spectrometry (IP-MS), we isolated an R2R3-type MYB transcription factor, OsMYB106, that interacts with OsBAG4. The osmyb106 mutants were also salt stress sensitive, and we found that OsMYB106 and OsBAG4 act in the same genetic pathway to activate OsHKT1;5. OsBAG4 facilitates the binding of OsMYB106 to the consensus MYB binding cis-element (MYBE) at the OsHKT1;5 promoter. OsSUVH7, which binds methylated DNA in vitro and in vivo, associates with the MITE upstream of the MYBE. Gel filtration assays revealed that OsSUVH7, OsBAG4, and OsMYB106 form a transcriptional complex. Elimination of the MITE or knockout of OsSUVH7 using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology decreased OsHKT1;5 expression and increased salt stress sensitivity. Collectively, our findings reveal a novel transcriptional complex, consisting of a DNA methylation reader, a chaperone regulator, and a transcription factor, that regulates the expression of OsHKT1;5 under salinity stress.
RESULTS
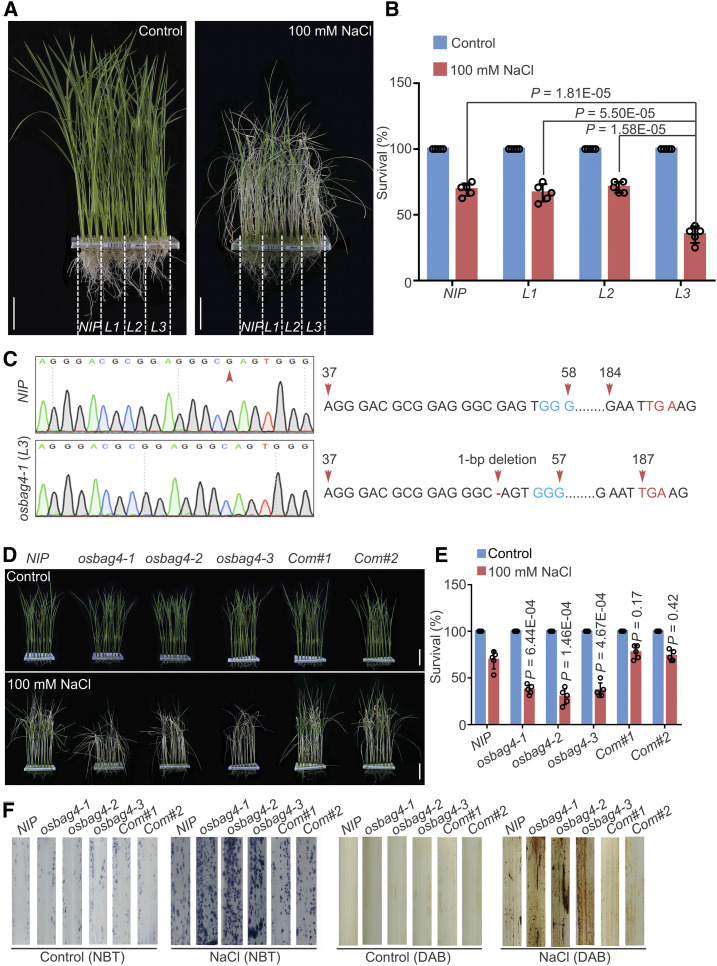
OsBAG4 Positively Impacts Salt Stress Tolerance
In a genetic screen of RGKO-ALL, a genome-scale CRISPR/Cas9 mutagenesis library of rice (var Nipponbare; Lu et al., 2017; Nan et al., 2020), we identified a mutant line (L3) with reduced tolerance to 100 mM NaCl treatment relative to the wild type (NIP; Figures 1A and 1B). Sanger sequencing, along with information obtained from the barcoded next-generation sequencing data, revealed a 1-bp deletion 52 bp downstream of the ATG start codon of the gene encoding OsBAG4 (LOC_Os01g61500); this mutation created a premature stop codon in the L3 mutant, which was therefore designated osbag4-1 (Figure 1C).
Figure 1.
osbag4 Mutants Exhibit Salt Stress–Sensitive Phenotypes.
(A) and (B) Mutant lines obtained from the CRISPR/Cas9 mutant pool RGKO-ALL (L1, L2, and L3) were screened for salt stress sensitivity. NIP was used as a control. Images were captured (A), and survival rates were measured (B) before 100 mM NaCl treatment and after recovery from NaCl treatment Data in (B) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circles) are shown. Differences between NIP, L1, L2, and L3 were evaluated by Student’s t test. Bars in (A) = 4.0 cm.
(C) Sanger sequencing chromatography showing the wild-type and mutated forms of OsBAG4 in NIP and osbag4-1 (L3). The mutation in osbag4-1 includes a deletion of G leading to a premature stop codon.
(D) and (E) Images (D) and survival rates (E) of NIP, osbag4 mutants (osbag4-1, osbag4-2, and osbag4-3), and two OsBAG4 complementation lines (Com#1 and Com#2) before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (E) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circles) are shown. Statistical analyses were performed by comparing three independent osbag4 mutants and two OsBAG4 complementation lines with NIP plants by Student’s t test. Bars in (D) = 4.5 cm.
(F) ROS detection in the leaves of NIP, osbag4 mutants, and two OsBAG4 complementation lines under normal and salt stress conditions. Leaves stained with NBT and DAB were used to assess O2– and H2O2 accumulation, respectively. Seedlings were treated with or without 100 mM NaCl for 24 h treatment before staining.
The rice genome harbors six BAG homologs; in addition to the BAG domain, OsBAG1 to OsBAG4 each contain a conserved ubiquitin-like domain, whereas OsBAG5 and OsBAG6 have an IQ calmodulin binding motif (Supplemental Figures 1A and 1B). To further confirm the effect of loss of function of OsBAG4 in response to salt stress tolerance, we generated two independent mutants, osbag4-2 and osbag4-3, using the CRISPR/Cas9 system. For this purpose, specific guide RNA (gRNA) target sites for OsBAG4 were cloned into a CRISPR/Cas9 vector in which Cas9 was driven by the maize Ubi promoter (Ma et al., 2015; Nan et al., 2020). Transformation of rice cells with these vectors yielded the osbag4-2 and osbag4-3 mutant lines, which were confirmed by Sanger sequencing (Supplemental Figure 1C). The osbag4-2 mutant harbored two 1-bp insertions 88 and 114 bp downstream of the initiation ATG, causing a frameshift mutation resulting in a premature stop codon (Supplemental Figure 1C). In osbag4-3, a 26-bp deletion was detected 89 bp downstream of the ATG, likewise generating a frameshift mutation and a premature stop codon (Supplemental Figure 1C). To exclude an effect of the Cas9 transgene itself, we isolated osbag4-1, osbag4-2, and osbag4-3 mutants lacking Cas9 by screening for hygromycin sensitivity (Supplemental Figure 1D). As shown in Figure 1D, all three independent osbag4 mutants were salt stress sensitive.
In addition, we generated complementation lines by transfecting the osbag4-1 mutant with a construct expressing OsBAG4 cDNA fused C terminally to the FLAG epitope under the control of the OsBAG4 promoter (OsBAG4pro:OsBAG4-FLAG; Supplemental Figure 1E). Independent complementation lines Com#1 and Com#2 had survival rates similar to those of NIP plants under salt stress (Figures 1D and 1E).
Next, we examined the production of the ROS hydrogen peroxide (H2O2) and superoxide (O2–) by staining rice leaves of various genotypes with diaminobenzidine (DAB) and nitrotetrazolium blue chloride (NBT). As shown in Figure 1F, osbag4 mutants accumulated higher levels of ROS than NIP under salt stress.
To further examine the spatial and temporal expression patterns of OsBAG4, we obtained transgenic plants expressing the β-glucuronidase (GUS) gene under the control of the OsBAG4 promoter (OsBAG4pro:GUS) and measured GUS activity. As shown in Supplemental Figures 2A and 2B, OsBAG4 was mostly expressed in leaf, root, internode, stem, young panicle, germinated seed, leaf sheath, vascular bundle, and root as well as shoot protoplasts. Next, we cloned the GFP gene at the 3′ end of the OsBAG4 coding sequence (CDS) to generate an OsBAG4-GFP construct, which was transfected into rice protoplasts along with nuclear localization signal–red fluorescent protein (NLS-RFP). As shown in Supplemental Figure 2C, OsBAG4-GFP mainly localized to the nucleus, with a minor portion in the cytosol.
We also detected that without salt stress treatment, expression levels of OsBAG4 were slightly higher in roots than in shoots (Supplemental Figure 2D). Under salt stress, OsBAG4 was induced in both roots and shoots but to a significantly greater extent in the roots (Supplemental Figure 2D). OsDREB2A was used as a positive control for the salt stress treatment (Supplemental Figure 2D; Matsukura et al., 2010).
To evaluate the gain-of-function effect of OsBAG4, we generated transgenic lines that overexpressed OsBAG4 (OsBAG4OX) by fusing a FLAG epitope at the C terminus of the OsBAG4 cDNA (Supplemental Figures 3A and 3B). Three independent OsBAG4OX lines exhibited late flowering and reduced plant height under normal conditions (Supplemental Figures 3C and 3D), consistent with a previous report by You et al. (2016). In addition, the OsBAG4OX lines were salt stress tolerant (Supplemental Figures 3E and 3F). Taken together, these results indicate that OsBAG4 plays a positive role in the salt stress response.
OsBAG4 Influences the Expression of OsHKT1;5
To investigate the function of OsBAG4, we performed RNA-seq on NIP and osbag4-1, each with three biological replicates. Using stringent statistical and filtering criteria, we identified 99 downregulated and 202 upregulated (osbag4-1 versus NIP) genes in shoots (Figure 2A; Supplemental Data Set 1) and 611 downregulated and 393 upregulated genes in roots (Figure 2A; Supplemental Data Set 2).
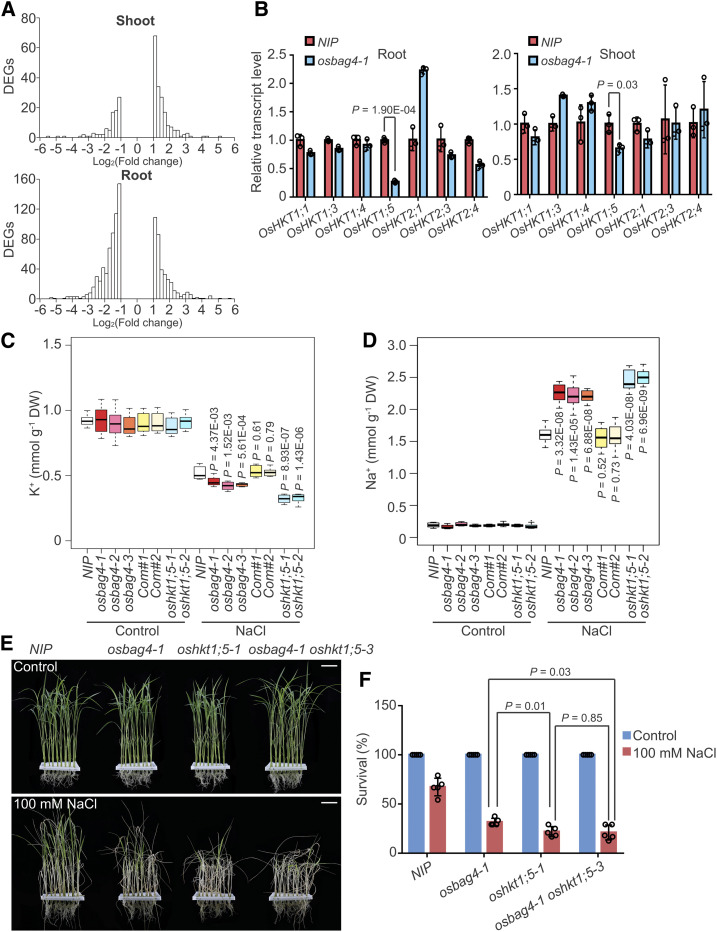
Figure 2.
OsHKT1;5 Acts Genetically Downstream of OsBAG4.
(A) Histograms of fold changes for up- and downregulated DEGs in shoot and root comparing osbag4-1 and NIP. “x axis” shows the bins of log2(Fold change) in RNA expression, and “y axis” is the number of DEGs whose log2(Fold change) expression fall in each bin.
(B) Expression levels of OsHKT family genes in the root or shoot of NIP and osbag4-1. Data represent means ± sd (n = 3, roots or shoots from five NIP or osbag4-1 seedlings were pooled and harvested for RNA extraction and RT-qPCR in each biological replicate). Statistical analyses were performed using Student’s t test. Individual values (black circle) are shown.
(C) and (D) Box plot showing K+ (C) and Na+ (D) contents in the shoots of the indicated genotypes. Four-week-old seedlings were treated with or without 100 mM NaCl for 5 d before measurement of ion contents (n = 8, eight plants of each genotype were used to measure Na+ and K+ levels). Differences between NIP and mutants or complementation lines were evaluated with Student’s t test. DW, dry weight.
(E) and (F) Images (E) and survival rates (F) of NIP, osbag4-1, oshkt1;5-1, and osbag4-1 oshkt1;5-3 double mutant before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (F) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circle) are shown. Statistical analyses were performed by Student’s t test. Bars in (E) = 3.2 cm.
Gene Ontology (GO) analysis (P-value < 0.05) revealed that the terms “Response to stress,” “Secondary metabolic process,” “Response to biotic stimulus,” and “Regulation of cellular process” were enriched in genes upregulated in roots, and “Response to stress,” “Photosynthesis,” “Response to abiotic stimulus,” and “Cellular metabolic process” were enriched in genes downregulated in roots (Supplemental Figure 4A; Supplemental Data Set 3). In shoots, “Lipid metabolic process” and “Response to stress” were enriched in downregulated genes, and “Developmental process,” “Multicellular organismal process,” “Nitrogen compound metabolic process,” and “Reproductive structure development” were enriched in upregulated genes (Supplemental Figure 4A; Supplemental Data Set 4).
Because osbag4 mutants were salt stress sensitive, we sought to identify the genes located downstream of OsBAG4 that were associated with the “Response to stress” GO term. Among them, we noticed that expression of OsHKT1;5 was dramatically reduced in mutant roots (Supplemental Data Set 3). We confirmed this result by performing RT-qPCR to determine the expression levels of OsHKT family members in the shoots and roots of osbag4-1 and NIP (Figure 2B). In the mutant, OsHKT1;5 levels were dramatically reduced in roots and slightly reduced in shoots. The salt tolerance QTL SKC1, corresponding to OsHKT1;5, is involved in maintaining K+/Na+ ratio in shoots (Ren et al., 2005). Under salt stress, the transcript level of OsHKT1;5 in roots was elevated in NIP plants but changed only marginally in osbag4-1 (Supplemental Figure 4B); in shoots, OsHKT1;5 was not induced in either NIP or osbag4-1, consistent with previous results showing that OsHKT1;5 is not induced under salt stress (Supplemental Figure 4B; Ren et al., 2005).
Loss of function of OsHKT1;5 in salt-stressed rice roots triggers massive Na+ accumulation in shoots (Ren et al., 2005). Hence, we examined the contents of Na+ and K+ in the shoots of NIP, osbag4 mutants, OsBAG4 complementation lines, and oshkt1;5 mutants under salt stress. Shoot K+ and Na+ contents did not differ significantly among the genotypes tested (Figures 2C and 2D; Supplemental Figure 5). After treatment with 100 mM NaCl for 5 d, shoot K+ content was dramatically lower in osbag4 mutants than in NIP, whereas shoot Na+ contents were significantly higher in the mutants (Figures 2C and 2D). Shoot K+ and Na+ contents of OsBAG4 complementation lines did not differ significantly from those of NIP (Figures 2C and 2D), whereas oshkt1;5 mutant lines exhibited an even greater decrease in K+ and increase in Na+ content relative to NIP (Figures 2C and 2D). This result implies that loss of OsHKT1;5 expression in osbag4 mutants might perturb the K+/Na+ ratio in shoots.
To investigate the genetic interaction between OsBAG4 and OsHKT1;5, we attempted to cross osbag4 and oshkt1;5 mutants to generate osbag4 oshkt1;5 double mutants; however, because the two genes are tightly linked, we failed to obtain the double mutant. As an alternative approach, we used CRISPR/Cas9 technology to generate the Oshkt1;5 mutation in the osbag4-1 background, and this allowed us to successfully obtain the osbag4-1 oshkt1;5-3 double mutant (Supplemental Figure 5). As shown in Figures 2E and 2F, salt sensitivity was comparable between osbag4-1 oshkt1;5-3 and oshkt1;5-1, suggesting that OsHKT1;5 acts downstream of OsBAG4.
OsBAG4 Interacts with OsMYB106
To identify the potential transcriptional modulator that works together with OsBAG4 to regulate OsHKT1;5 expression, we performed immunoaffinity purification (IP) followed by IP-MS. We performed co-immunoprecipitation (co-IP) with anti-FLAG antibodies using the OsBAG4pro:OsBAG4-FLAG/osbag4-1 complementation line; NIP was used as a negative control (Supplemental Figure 6A). The experiment was performed in two biological replicates (Supplemental Figure 6A; Supplemental Data Sets 5 to 8). Our IP-MS analysis revealed unique OsBAG4 peptides and also identified peptides corresponding to known OsBAG4-interacting proteins, including Hsp70s and EBR1 (Supplemental Figure 6B; Supplemental Data Set 9; Takayama and Reed, 2001; You et al., 2016).
Among the putative OsBAG4-interacting proteins, the only transcription factor was OsMYB106 (Supplemental Figure 6B; Supplemental Data Set 9). To further confirm the interaction between OsBAG4 and OsMYB106, we cotransfected 35Spro:FLAG-OsMYB106 (FLAG-OsMYB106) with 35Spro:GFP or OsBAG4-GFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35Spro:OsBAG4-GFP, hereafter designated OsBAG4-GFP) into rice protoplasts, performed co-IP with anti-FLAG antibody, and probed the immunoprecipitates with anti-GFP antibody. OsBAG4-GFP was present in the anti-FLAG precipitates (Figure 3A). In this experiment, we also examined the interaction between OsBAG4-GFP and other putative OsBAG4-interacting proteins, including OsSUVH7-FLAG, OsDjC26-FLAG, OsDjC51-FLAG, OscHsp70-1-FLAG, and OscHsp70-6-FLAG. FLAG-OsMYB106 was used as a positive control. As expected, OsBAG4-GFP immunoprecipitated OscHsp70-1-FLAG and OscHsp70-6-FLAG; interestingly, however, a Su(var)3-9, Enhancer-of-zeste and Trithorax domain-containing protein, OsSUVH7, also strongly interacted with OsBAG4-GFP (Supplemental Figure 6C). OsDjC26-FLAG interacted with OsBAG4-GFP, whereas OsDjC51-FLAG did not (Supplemental Figure 6C).
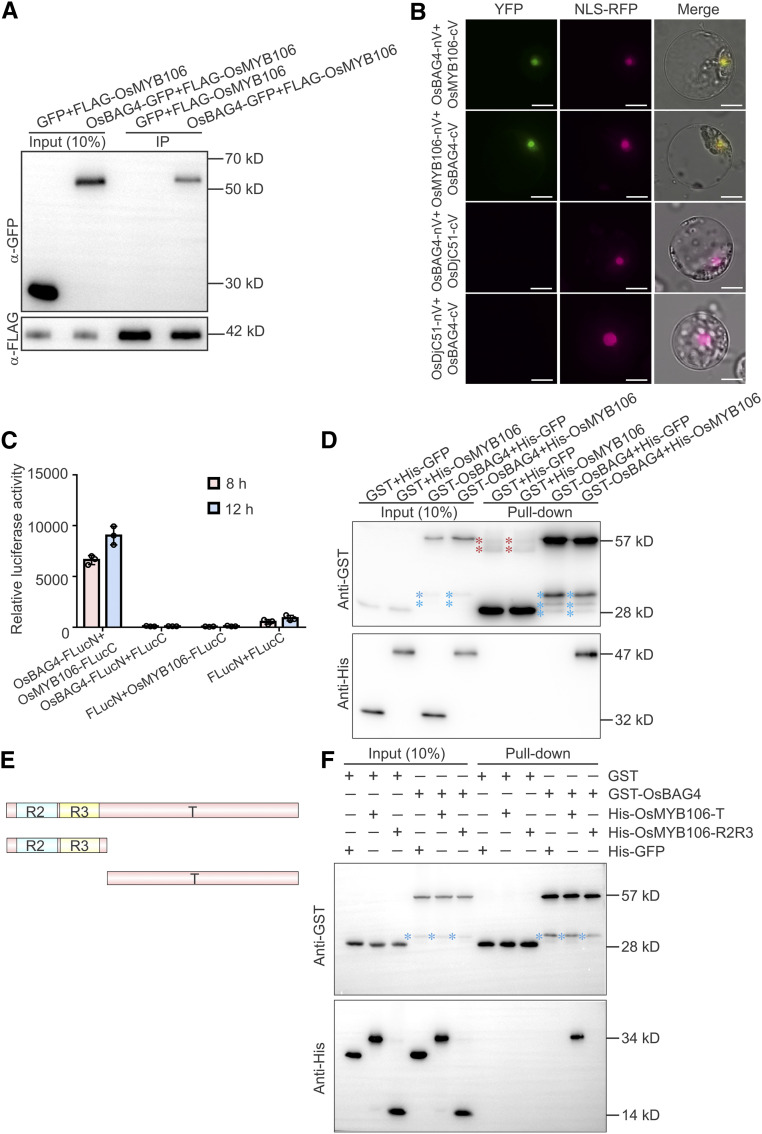
Figure 3.
OsBAG4 Interacts with OsMYB106 In Vivo and In Vitro.
(A) Immunoblot analysis of the coimmunoprecipitates from the co-IP assay. The 35Spro:OsBAG4-GFP (OsBAG4-GFP) or 35Spro:GFP (GFP) construct was cotransfected with 35Spro:FLAG-OsMYB106 (FLAG-OsMYB106) into rice protoplasts. co-IP was performed using anti-FLAG antibody, and coimmunoprecipitated proteins were detected using anti-GFP antibody. Three biological repeats were performed, yielding similar results.
(B) Results of BiFC assay. OsBAG4, OsMYB106, and OsDjC51 were fused to the N- or C-terminal half of Venus (OsBAG4-nV, OsBAG4-cV, OsMYB106-nV, OsMYB106-cV, OsDjC51-nV, and OsDjC51-cV). OsBAG4-nV and OsMYB106-cV, OsBAG4-cV and OsMYB106-nV, OsBAG4-nV and OsDjC51-cV, or OsBAG4-cV and OsDjC51-nV were coexpressed with NLS-RFP (nuclear marker). Similar results were observed in at least 50 cells from three independent experiments. Bars = 10 μm.
(C) In vivo split firefly luciferase complementation assay to test the interaction between OsBAG4 and OsMYB106. OsBAG4 fused with FLucN (OsBAG4-FLucN) was coexpressed with OsMYB106 fused with the C-terminal half (OsMYB106-FLucC) in NIP protoplasts. FLucC and FLucN vectors were used as negative controls. Luciferase activities were measured after 8- and 12-h incubation. Data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circles) are shown.
(D) In vitro pull-down assay to detect the direct interaction between OsBAG4 and OsMYB106. GST-OsBAG4 or GST was incubated with His-OsMYB106 or His-GFP and pulled down with glutathione–agarose beads, followed by immunoblotting with anti-His and anti-GST antibodies. GST and His-GFP were used as negative controls. Three biological repeats were performed, yielding similar results. Red asterisks indicate nonspecific bands; blue asterisks indicate broken bands.
(E) Schematic representation of domain structures of OsMYB106.
(F) Pull-down assay to identify the domains of OsMYB106 that interact with OsBAG4. GFP, OsMYB106-R2R3, and OsMYB106-T were fused with the N-terminal His epitope and incubated with GST or GST-OsBAG4, and pulled down with glutathione–agarose beads followed by immunoblotting with anti-His and anti-GST antibodies. GST and His-GFP were used as negative controls. Blue asterisks indicate broken bands.
To confirm the in vivo interaction between OsBAG4 and OsMYB106, we conducted bimolecular fluorescence complementation (BiFC) analysis (Müller-Taubenberger and Anderson, 2007). OsBAG4, OsMYB106, and OsDjC51 were fused with the N- or C-terminal half of Venus (OsBAG4-nV, OsMYB106-nV, OsBAG4-cV, OsMYB106-cV, OsDjC51-nV, and OsDjC51-cV). Cotransfection of either OsBAG4-nV and OsMYB106-cV or OsBAG4-cV and OsMYB106-nV resulted in strong fluorescence signals (Figure 3B), and the nuclear localization of the protein was confirmed by colocalization with NLS-RFP (Xu et al., 2013). Neither OsBAG4-nV and OsDjC51-cV nor OsDjC51-nV and OsBAG4-cV cotransfection yielded fluorescent signals, although OsDjC51-GFP localizes to the nucleus (Supplemental Figure 6D). As an alternative approach to confirm the interaction between OsBAG4 and OsMYB106, OsBAG4 was tagged with amino acids 1 to 398 of firefly luciferase (FLucN), and OsMYB106 was tagged with amino acids 384 to 550 (FLucC), and the two constructs were cotransfected to rice protoplasts. OsMYB106-FLucC and FLucN, FLucC and OsBAG4-FLucN, and FLucC and FLucN were also cotransfected. As shown in Figure 3C, only cell lysates from the cotransfection of OsBAG4-FLucN and OsMYB106-FLucC yielded strong luciferase activity.
To test the in vitro interaction between OsBAG4 and OsMYB106, we generated constructs by fusing a His epitope to the N-terminal region of OsMYB106 and a glutathione S-transferase (GST) epitope to the N-terminal region of OsBAG4. We transformed these constructs into Escherichia coli, purified the encoded proteins, and examined the interaction between GST-OsBAG4 and His-OsMYB106. His-GFP and GST alone were used as negative controls. As shown in Figure 3D, GST-OsBAG4 and His-OsMYB106 interacted with each other in vitro. Next, we divided OsMYB106 into its R2R3 domain (R2R3) and putative transcriptional regulatory domain (T) and fused each truncated protein with an N-terminal His epitope (Figure 3E). Pull-down experiments revealed that OsBAG4 directly interacted with the transcriptional regulatory domain (T; Figure 3F). Taken together, these data suggest that OsBAG4 interacts with OsMYB106 in vitro and in vivo.
OsMYB106 Participates in the Salt Stress Response
The rice genome encodes 233 MYB transcriptional factors, and OsMYB106 belongs to the R2R3 MYB family (Supplemental Figure 7A; Smita et al., 2015). We performed GUS staining assays to examine the tissue-specific expression patterns of OsMYB106 using OsMYB106pro:GUS transgenic plants. The results revealed that OsMYB106 was abundantly expressed in root, leaf, stem, internode, vascular bundle, and shoot as well as root protoplasts (Supplemental Figures 7B and 7C). Of note, the expression of OsMYB106 was more rapidly and dramatically increased in roots compared to shoots under the salt stress condition (Supplemental Figure 7D).
Next, to investigate the physiological function of OsMYB106 in the salt stress response, we generated osmyb106 mutants using the CRISPR/Cas9 system. In osmyb106-1, a 1-bp insertion was detected 55 bp downstream of the initiation ATG, generating a frameshift mutation and a premature stop codon (Supplemental Figure 8A). Likewise, in osmyb106-2, a 5-bp deletion was detected 50 bp downstream of the initiation ATG, also causing a frameshift and premature stop codon (Supplemental Figure 8A). We isolated Cas9 transgene-free mutant lines by screening for hygromycin sensitivity (Supplemental Figure 8B). Two independent osmyb106 mutants exhibited a salt stress–sensitive phenotype (Figures 4A and 4B).
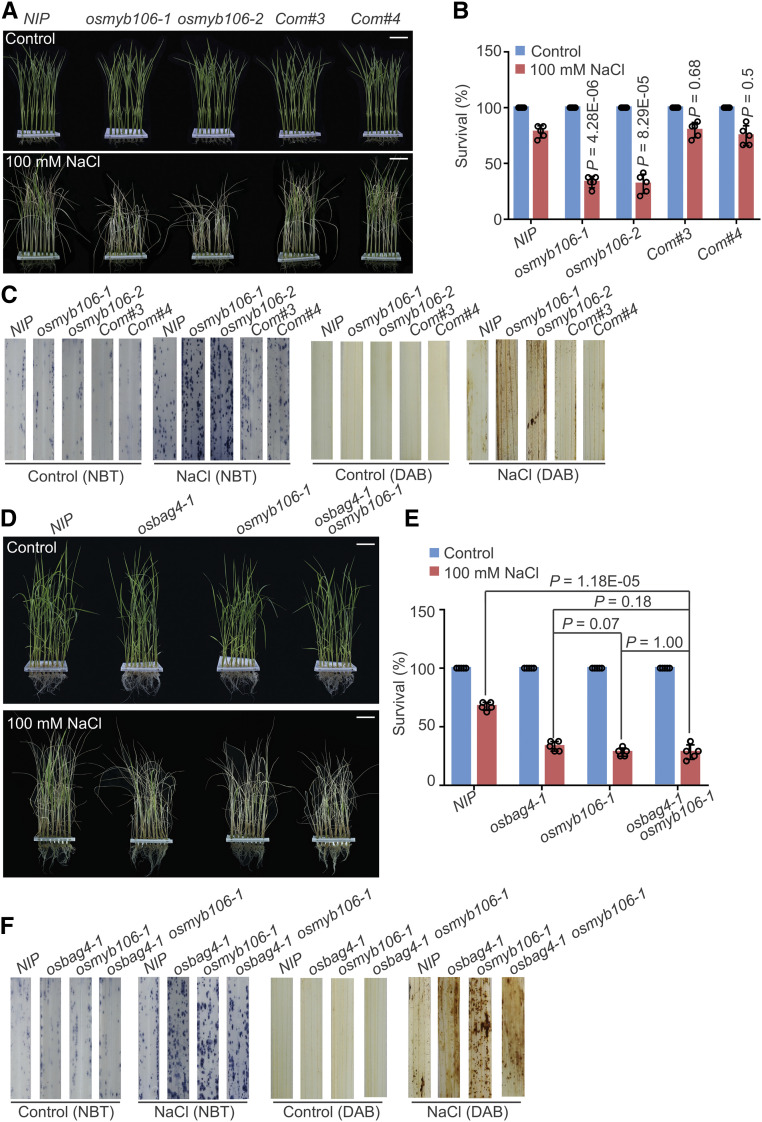
Figure 4.
osmyb106 Mutants Exhibit Salt Stress–Sensitive Phenotypes.
(A) and (B) Images (A) and survival rates (B) of NIP, osmyb106 mutants (osmyb106-1 and osmyb106-2), and two OsMYB106 complementation lines (Com#3 and Com#4) before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (B) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circle) are shown. Statistical analyses were performed by comparing two osmyb106 mutants and two OsMYB106 complementation lines with NIP plants using Student’s t test. Bars in (A) = 4.0 cm.
(C) ROS detection in the leaves of NIP, osmyb106 mutants, and two OsMYB106 complementation lines under normal and salt stress conditions. Leaves stained with NBT and DAB were used to assess O2– and H2O2 accumulation, respectively. Seedlings were treated with or without 100 mM NaCl for 24 h before staining.
(D) and (E) Images (D) and survival rates (E) of NIP, osbag4-1, osmyb106-1, and osbag4-1 osmyb106-1 double mutant before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (E) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circle) are shown. Statistical analyses were performed by Student’s t test. Bars in (D) = 3.6 cm.
(F) ROS detection in leaves of NIP, osbag4-1, osmyb106-1, and osbag4-1 osmyb106-1 double mutant under normal and salt stress conditions. Leaves stained with NBT and DAB were used to assess O2– and H2O2 accumulation, respectively. Images were taken before and after 24-h treatment with 100 mM NaCl.
Next, to conduct complementation assays, we generated an OsMYB106 cDNA N-terminally fused to the FLAG epitope under the control of the OsMYB106 promoter (OsMYB106pro:FLAG-OsMYB106; Supplemental Figure 8C). Survival rates under salt stress were similar between OsMYB106pro:FLAG-OsMYB106 complementation lines and NIP plants (Figures 4A and 4B). DAB and NBT staining assays revealed that osmyb106 mutants produced higher levels of H2O2 and O2– (Figure 4C). Moreover, shoots of osmyb106 mutants accumulated higher Na+ levels and lower K+ levels than NIP and complementation lines (Supplemental Figures 8D and 8E).
To test the genetic interaction between OsBAG4 and OsMYB106, we crossed osbag4-1 and osmyb106-1 to generate osbag4-1 osmyb106-1 double mutants. As shown in Figures 4D to 4F, osbag4-1 osmyb106-1 had similar survival rates and produced levels of H2O2 and O2– comparable with those in osbag4-1 and osmyb106-1, respectively. RT-qPCR revealed that OsHKT1;5 transcripts were dramatically reduced in osmyb106-1, and the extent of reduction was similar to those in osbag4-1 osmyb106-1 in both shoots and roots (Supplemental Figure 9A). Under salt stress, induction of OsHKT1;5 in roots was also impaired in both osmyb106-1 and osbag4-1 osmyb106-1 (Supplemental Figure 9A).
Next, we generated OsMYB106-overexpressing transgenic plants by transforming OsMYB106 cDNA driven by the strong Cassava vein mosaic virus promoter. As shown in Supplemental Figure 9B, we obtained three independent OsMYB106-overexpressing transgenic lines (OsMYB106OX). The OsMYB106OX lines were salt stress tolerant (Supplemental Figures 9C and 9D), and the expression level of OsHKT1;5 was dramatically increased in OsMYB106OX lines (Supplemental Figure 9E). In addition, we examined the genetic interaction between OsMYB106 and OsHKT1;5 by crossing osmyb106-1 and oshkt1;5-1, thereby generating an osmyb106-1 oshkt1;5-1 double mutant. As shown in Supplemental Figures 9F and 9G, survival rates were similar between osmyb106-1, oshkt1;5-1, and osmyb106-1 oshkt1;5-1. These results suggest that OsBAG4 and OsMYB106 influence the expression of OsHKT1;5 in response to salt stress.
OsMYB106 Activates OsHKT1;5 by Interacting with a Consensus MYBE Located in the OsHKT1;5 Promoter
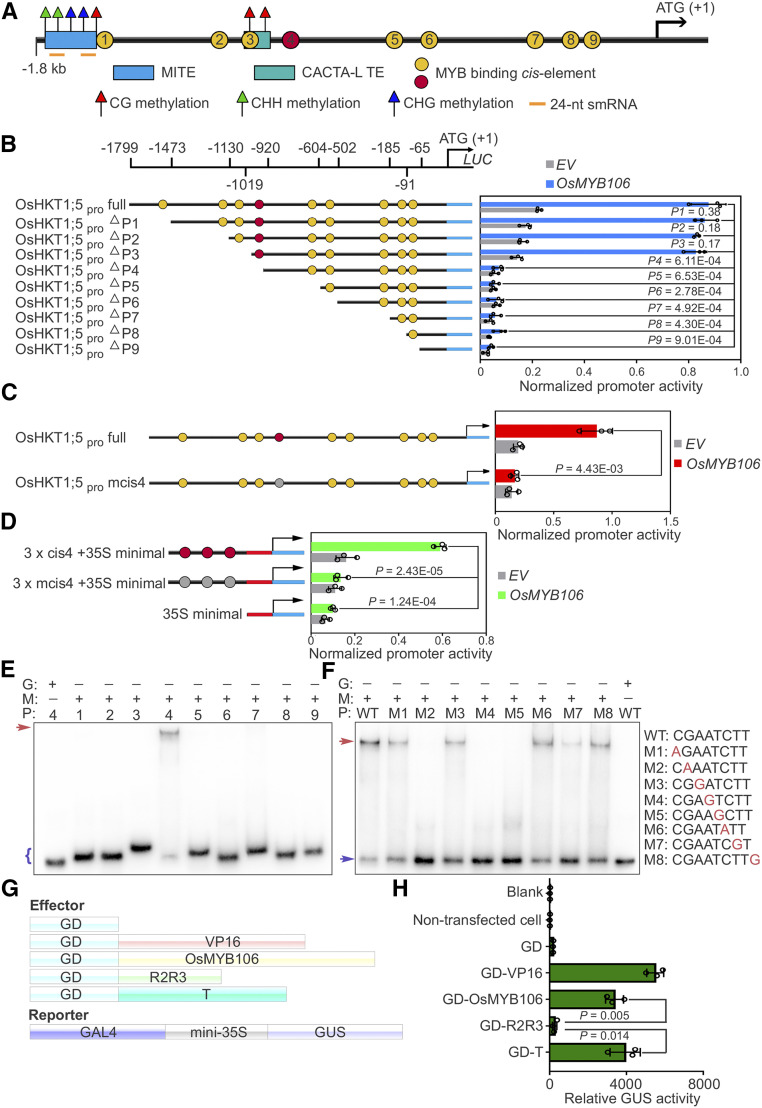
Using the Plant Transcription Factor Database and PlantPAN2.0 (Chow et al., 2016; Tian et al., 2020), we predicted nine putative MYB binding sites in the promoter region of OsHKT1;5 (Figure 5A; Supplemental Data Set 10). To further determine genuine MYB transcription binding sites, we generated a range of promoter deletion constructs (OsHKT1;5pro ∆P1–∆P9), each fused to the coding region of the Luciferase (LUC) reporter gene (Figures 5A and 5B). CaMV 35S promoter-driven OsMYB106 was used as the effector to activate the promoter, and a GUS construct also driven by the Arabidopsis UBQ10 promoter was used to normalize for transfection efficiency (Adachi et al., 2015; Gasch et al., 2016). As shown in Figure 5B, OsMYB106-mediated activation of OsHKT1;5pro:LUC activity was dramatically reduced after deletion of the fourth putative MYBE. Subsequent deletions in this series exerted relatively little impact on the levels of reporter gene activity.
Figure 5.
OsMYB106 Activates OsHKT1;5 Expression by Binding to the cis-Element Located in the OsHKT1;5 Promoter.
(A) Schematic representation of the promoter structures of OsHKT1;5.
(B) Deletion analysis of OsHKT1;5 promoter activity. Left: schematic representation of a deletion series of the 1799-bp OsHKT1;5 promoter (OsHKT1;5pro full). The numbers displayed in the schematic of the OsHKT1;5 promoter indicate the distance from the OsHKT1;5 translation start site. Map indicates relative length and regions of 5′-deleted promoter versions cloned 5′ of a luciferase (Luc) coding sequence. Yellow and red circles indicate the MYBE represented in (A). Right, graph shows basal promoter activities measured in the presence of GFP (EV) and activities induced by FLAG-OsMYB106. Data are means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circle) are shown. P-values were calculated versus the LUC activity driven by the full-length promoter activated by OsMYB106 (Student’s t test). Normalized promoter activity was calculated by normalizing LUC activity to GUS activity.
(C) Analysis of the 1799-bp OsHKT1;5 promoter (OsHKT1;5pro full) and 1799-bp OsHKT1;5 promoter containing a mutated form of the fourth cis-element (OsHKT1;5pro mcis4) in response to OsMYB106. Data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circles) are shown. Statistical analysis was performed by Student’s t test. Normalized promoter activity was calculated by normalizing LUC activity to GUS activity.
(D) Analysis of three tandem repeats of the fourth cis-element (cis4; red circles) and a mutated form of the fourth cis-element (mcis4; gray cycles) with the 35S minimal promoter in response to OsMYB106. Data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circles) are shown. Statistical analysis was performed by Student’s t test. Normalized promoter activity was calculated by normalizing LUC activity to GUS activity.
(E) Results of EMSA. Recombinant OsMYB106 protein bound to the consensus fourth cis-element located at OsHKT1;5 promoter. The red arrow denotes the shifted probe, and the purple parenthesis denotes the free probe. G, GST; M, GST-OsMYB106; P, probe (the numbers represent the predicted cis-element within the OsHKT1;5 promoter in [A]).
(F) Results of EMSA for single-nucleotide mutations of the eight nucleotides in the fourth cis-element. The red arrow denotes the shifted probe; the purple arrow denotes the free probe. WT, the wild-type consensus sequence identified in (E).
(G) and (H) Schematic diagrams of constructs (G) and quantification of GUS reporter activity (H) in NIP protoplasts cotransfected with effector constructs and GAL4-mini35Spro:GUS reporter. GUS activity was measured after protoplasts were incubated in the dark for 20 to 22 h. In (H), data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circle) are shown. Statistical analysis was performed by Student’s t test. Blank, empty well; VP16, herpes simplex virus VP16 activation domain.
Next, we performed base-substitution analysis by mutating the fourth cis-element (Figure 5C). As shown in Figure 5C, OsHKT1;5pro mcis4:LUC activity was also significantly reduced. To determine whether the predicted fourth cis-element is sufficient for responsiveness to OsMYB106, we prepared a tandem triple repeat (3 × cis4) promoter consisting of 18-bp sequences (ACATACGAATCTTTTTTT) containing the predicted fourth cis-element and generated a 3 × cis4-fused minimal CaMV 35S promoter (3 × cis4 + 35S minimal promoter)–LUC reporter. Expression of OsMYB106 strongly induced activation of the 3 × cis4 promoter (Figure 5D). These results indicate that the fourth cis-element is necessary and sufficient for OsMYB106-mediated activation of the OsHKT1;5 promoter.
To determine whether OsMYB106 directly binds to the consensus cis-element, we conducted an electrophoretic mobility shift assay (EMSA). The results revealed that full-length OsMYB106 proteins tagged with GST (GST-OsMYB106) were capable of specifically binding to the fourth MYBE (Figure 5E). Next, we performed single-nucleotide mutation of each of the eight nucleotides in the fourth cis-element. Mutation of the G at the second position (M2), A at the fourth position (M4), or T at the fifth position (M5) abolished OsMYB106 binding (Figure 5F); mutation of the T at the seventh position (M7) dramatically impaired binding; and mutation of the C at the first position (M1), A at the third position (M3), C at the sixth position (M6), or T at the eighth position (M8) slightly reduced binding (Figure 5F).
To investigate the mode of transcriptional regulation by OsMYB106, we used a reporter construct. To this end, we placed the GUS reporter gene under the control of the minimal 35S promoter (–46 to 0 bp) along with a cis-acting regulatory sequence that could be recognized by the GAL4 DNA binding domain (GAL4-mini35Spro:GUS; Figure 5G; Wang et al., 2007a; Ahmad et al., 2019). In parallel, we fused the GAL4 DNA binding domain (GD) to full-length OsMYB106 (GD-OsMYB106), the R2R3 domain of OsMYB106 (GD-R2R3), or the transcriptional regulatory domain of OsMYB106 (GD-T), all of which were considered as effectors (Figure 5G). As a positive control, the Herpes simplex virus VP16 activation domain was fused to the GAL4 DNA binding domain (GD-VP16). We cotransfected individual effector constructs along with the reporter construct into rice protoplasts. As shown in Figure 5H, GD-OsMYB106 and GD-T dramatically induced expression of the GUS reporter gene in comparison with GD, whereas GD-R2R3 did not significantly induce reporter expression. Together, these results indicate that OsMYB106, acting as a transcriptional activator, can bind to the consensus fourth MYBE to activate OsHKT1;5 expression.
OsBAG4 Facilitates the DNA Binding Activity of OsMYB106
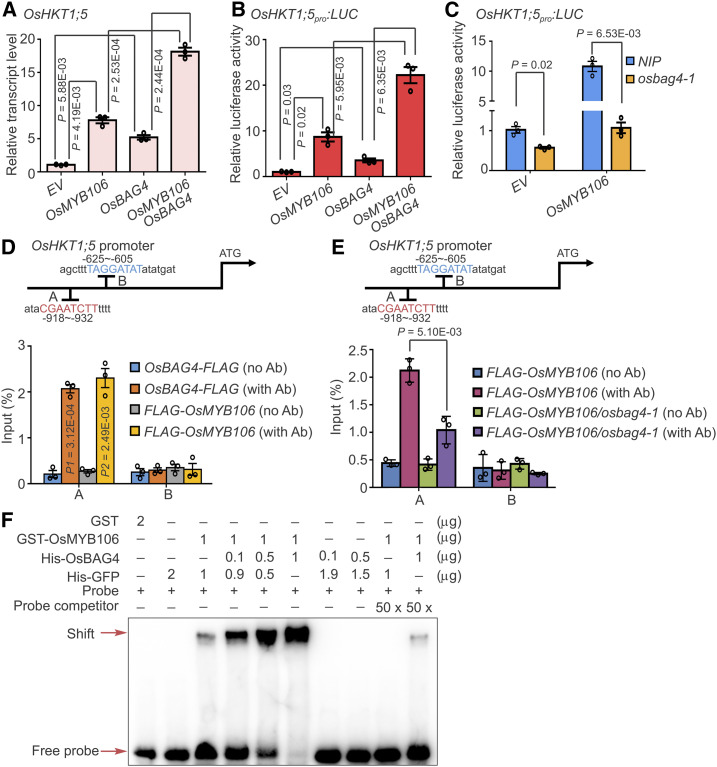
To investigate the molecular interplay between OsBAG4 and OsMYB106 during activation of OsHKT1;5, we introduced empty vector (EV), OsBAG4-FLAG, FLAG-OsMYB106, and OsBAG4-FLAG together with FLAG-OsMYB106 into rice root protoplasts. As shown in Figure 6A, overexpression of OsBAG4 or OsMYB106 significantly induced expression of endogenous OsHKT1;5. Intriguingly, coexpression of OsMYB106 and OsBAG4 synergistically activated expression of OsHKT1;5 (Figure 6A). As an alternative approach, we used an OsHKT1;5pro:LUC reporter system. As shown in Figure 6B, coexpression of OsBAG4 and OsMYB106 synergistically induced the LUC activity. Next, we ectopically expressed OsMYB106 in protoplasts obtained from roots of NIP or osbag4-1 and monitored transcriptional activity using the OsHKT1;5pro:LUC reporter cotransfected with OsMYB106. Relative to EV, overexpression of OsMYB106 dramatically induced LUC activity. Intriguingly, in osbag4-1, OsMYB106-induced LUC activity was greatly impaired (Figure 6C). Together, these results indicate that OsBAG4 is required for induction of OsHKT1;5 mediated by OsMYB106 and impacts OsMYB106 transcriptional activity.
Figure 6.
OsBAG4 Is Required for OsMYB106 Activity.
(A) to (C) OsBAG4 positively impacts the transcriptional activation activity of OsMYB106. (A) 35Spro:GFP (EV), 35Spro:FLAG-OsMYB106 (OsMYB106), 35Spro:OsBAG4-FLAG (OsBAG4), or 35Spro:FLAG-OsMYB106 (OsMYB106) together with 35Spro:OsBAG4-FLAG (OsBAG4) were transfected into NIP root protoplasts. After 6-h incubation, OsHKT1;5 expression levels were measured by RT-qPCR. (B) The OsHKT1;5pro:LUC construct together with UBQ10pro:GUS was cotransfected with 35Spro:GFP (EV), 35Spro:FLAG-OsMYB106 (OsMYB106), 35Spro:OsBAG4-FLAG (OsBAG4), or 35Spro:FLAG-OsMYB106 (OsMYB106) and 35Spro:OsBAG4-FLAG (OsBAG4) into NIP root protoplasts. After incubation for 20 h, luciferase and GUS activities were measured. After normalizing LUC activity against GUS activity, promoter activities of different samples were normalized against the promoter activity of the EV sample. (C) The OsHKT1;5pro:LUC construct together with UBQ10pro:GUS was cotransfected with either 35Spro:GFP (EV) or 35Spro:FLAG-OsMYB106 (OsMYB106) into NIP or osbag4-1 root protoplasts. After incubation for 20 h, luciferase and GUS activities were measured. After normalizing LUC activity against GUS activity, promoter activities of different samples were normalized against the promoter activity of the EV sample. The EV construct was used as a control. Data represent means ± sd (n = 3, transfection experiments were performed three times). Individual values (black circles) are shown. Statistical analysis was performed by Student’s t test (see [A] to [C]).
(D) Results of ChIP-qPCR showing that OsBAG4 and OsMYB106 bound to the A site (fourth cis-element displayed in Figure 5A). Anti-FLAG antibody was used to perform ChIP-qPCR on roots of OsBAG4pro:OsBAG4-FLAG and OsMYB106pro:FLAG-OsMYB106 complementation lines (OsBAG4-FLAG and FLAG-OsMYB106). The B site (fifth cis-element displayed in Figure 5A) was a negative control. Error bars indicate ± sd (n = 3, three biological replicates of ChIP experiments were performed for ChIP-qPCR). Individual values (black circles) were shown. Statistical analysis was performed by Student’s t test. Ab, antibody.
(E) Results of ChIP-qPCR showing that OsBAG4 impacts the affinity of OsMYB106 for the consensus cis-element in OsHKT1;5 promoter in vivo. ChIP with anti-FLAG antibody was performed on roots of F2 plants obtained by crossing OsMYB106pro:FLAG-OsMYB106 with NIP or osbag4-1 mutant (FLAG-OsMYB106 or FLAG-OsMYB106/osbag4-1). Specific primers for A and B sites were used for qPCR, and the B site was used as a negative control. Primers are listed in Supplemental Data Set 14. Error bars indicate ±sd (n = 3, three biological replicates of ChIP experiments were performed for ChIP-qPCR). Individual values (black circles) are shown. Statistical analysis was performed by Student’s t test.
(F) Results of EMSA showing that OsBAG4 impacts OsMYB106 binding affinity to the cis-element in OsHKT1;5 promoter in vitro. Different combinations of proteins were incubated with biotin-labeled probes at the 5′ end of single strand. His-GFP was used to normalize for total protein level.
Using roots of OsBAG4pro:OsBAG4-FLAG and OsMYB106pro:FLAG-OsMYB106 complementation lines, we performed chromatin immunoprecipitation (ChIP)-qPCR to determine whether OsBAG4 and OsMYB106 bind to the consensus OsMYB106 binding cis-element in vivo. As shown in Figure 6D, FLAG-OsMYB106 and OsBAG4-FLAG specifically bound to the fourth MYB binding element (A), but not an adjacent predicted MYB binding element (B). We then crossed the OsMYB106pro:FLAG-OsMYB106 plant with osbag4-1 to generate the OsMYB106pro:FLAG-OsMYB106/osbag4-1 isogenic line. ChIP-qPCR analysis revealed that FLAG-OsMYB106 binding to the MYB binding element (A) was dramatically decreased in this line (Figure 6E). Under salt stress, the elevated binding activity of OsMYB106 was dramatically reduced in osbag4-1 (Supplemental Figure 10). Together, these results indicate that OsBAG4 affects the affinity of OsMYB106 for the target promoter region under normal and salt stress conditions.
To explain these observations, we considered three hypotheses. First, we hypothesized that OsBAG4 impacts the subcellular localization of OsMYB106. To test this idea, we used OsSnRK1-GFP as a positive control because salt stress causes nuclear accumulation of OsSnRK1-GFP (Supplemental Figure 11A; Cho et al., 2012). Nuclear accumulation of OsSnRK1-GFP in response to salt stress was not altered in osbag4-1 protoplasts (Supplemental Figure 11A). Intriguingly, GFP-OsMYB106 localized to the nucleus with or without salt stress in NIP and osbag4-1, indicating that OsBAG4 does not affect the subcellular localization of OsMYB106 (Supplemental Figure 11A).
Second, we hypothesized that OsBAG4 impacts the protein stability of OsMYB106. To explore this possibility, we used rice GIGANTEA (OsGI) as a positive control because salt stress facilitates the degradation of GI in Arabidopsis (Kim et al., 2013). At 12 h after transfecting OsGI-FLAG driven by the CaMV 35S promoter into protoplasts obtained from NIP and osbag4-1, we administered 10 μM cycloheximide for 30 min and then added NaCl. Extracted total proteins were used for immunoblotting analysis. As shown in Supplemental Figure 11B, OsGI-FLAG was rapidly degraded in both NIP and osbag4-1 under NaCl treatment. Interestingly, FLAG-OsMYB106 protein stabilities were not altered under salt stress in either NIP or osbag4-1 (Supplemental Figure 11B).
Third, we hypothesized that OsBAG4 might impact the DNA binding affinity of OsMYB106. We purified the recombinant proteins including GST-OsMYB106, His-OsBAG4, and His-GFP, as well as GST, and used the consensus fourth cis-element labeled with biotin for EMSA. We used His-GFP to normalize for total protein level. As shown in Figure 6F, when the amount of GST-OsMYB106 was fixed and the amount of His-OsBAG4 increased, GST-OsMYB106 binding to the probe was elevated. His-OsBAG4 did not bind the probe. Taken together, these results indicate that OsBAG4 increases the affinity of OsMYB106 for the target promoter.
OsSUVH7, a DNA Methylation Reader, Forms a Protein Complex with OsBAG4 and OsMYB106
In addition to the MYB binding cis-elements, RepeatMasker using a query set of sequences from Repbase (https://www.girinst.org/repbase/) revealed two different types of TEs, one MITE and one CACTA-L TE, in the OsHKT1;5 promoter (Figure 5A; Supplemental Figure 12A). Data from bisulfite sequencing analysis of shoots and roots obtained by Zemach et al. (2010) and Hu et al. (2014) revealed that the MITE was significantly enriched in CHH methylation, whereas the CACTA-L TE was enriched in CG methylation (Supplemental Data Set 11). Intriguingly, the MITE turned out to be a locus associated with a 24-nucleotide small RNA that is essential for RdDM (Supplemental Figure 12A).
Our analysis also revealed that OsSUVH7, which was identified in our IP-MS experiment, exhibited strong amino acid sequence similarity to AtSUVH1 and AtSUVH3 (Supplemental Figure 12B), which are DNA methylation readers (Harris et al., 2018); this similarity was especially strong in the Sequence Read Archive (SRA) domains comprising the methyl binding domain. We found that OsSUVH7 was mostly expressed in leaf, root, internode, stem, young panicle, germinated seed, leaf sheath, vascular bundle, and root as well as shoot protoplasts (Supplemental Figure 13A). Under the salt stress condition, OsSUVH7 showed rapidly induced expression (Supplemental Figure 13B).
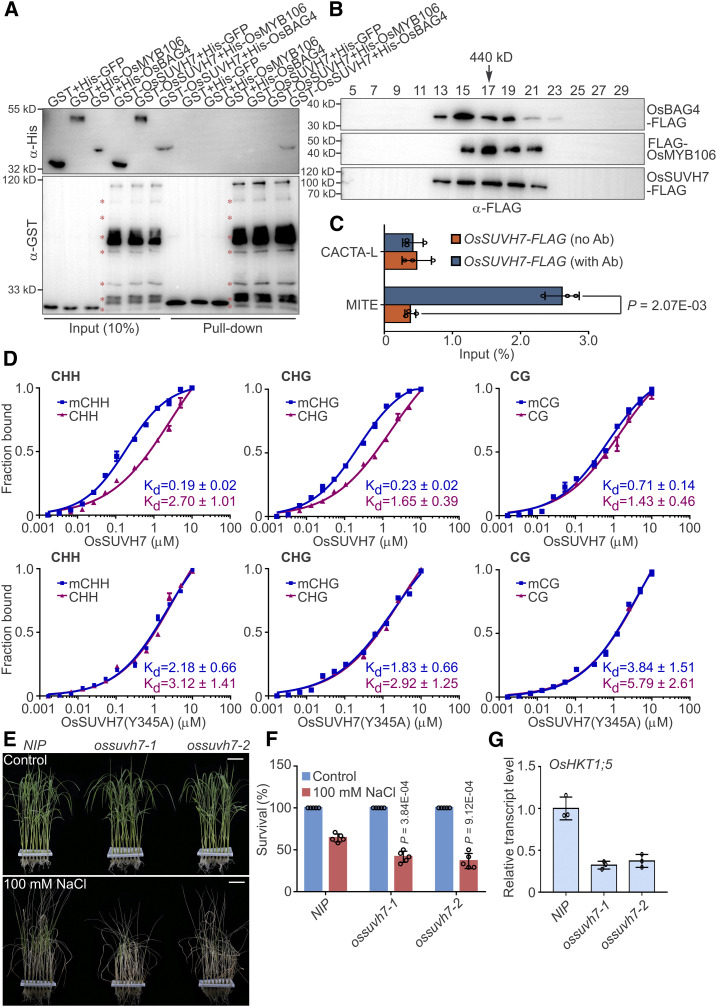
We performed pull-down experiments to examine the direct interactions between OsSUVH7 and OsBAG4 and between OsSUVH7 and OsMYB106. Unexpectedly, GST-OsSUVH7 interacted only with His-OsBAG4, but not with His-OsMYB106 (Figure 7A). To determine whether OsSUVH7, OsBAG4, and OsMYB106 form a stable complex, we cotransfected 35Spro:OsBAG4-FLAG (OsBAG4-FLAG), 35Spro:FLAG-OsMYB106 (FLAG-OsMYB106), and 35Spro:OsSUVH7-FLAG (OsSUVH7-FLAG) into NIP protoplasts, extracted total proteins, and subjected the proteins to gel filtration. As indicated by immunoblotting, OsSUVH7-FLAG, OsBAG4-FLAG, and FLAG-OsMYB106 signals were present in the high molecular weight fractions (Figure 7B). Furthermore, ChIP-qPCR using roots of OsSUVH7pro:OsSUVH7-FLAG transgenic plants revealed that OsSUVH7 specifically associated with the MITE, but not the CACTA-L TE (Figure 7C; Supplemental Figure 13C). We further detected direct binding of OsSUVH7-FLAG to the methylated MITE using ChIP followed by bisulfite conversion and PCR. As shown in Supplemental Figure 13D, OsSUVH7 bound to the MITE, which was methylated in CHH and CHG contexts in roots (Supplemental Data Sets 12 and 13).
Figure 7.
DNA Methylation Reader OsSUVH7 Forms a Complex with OsBAG4 and OsMYB106 That Participates in the Salt Stress Response.
(A) In vitro pull-down assay to detect the direct interaction of OsSUVH7 with OsBAG4 and OsMYB106. GST-OsSUVH7 or GST were incubated with His-GFP, His-OsMYB106, or His-OsBAG4 and pulled down using glutathione–agarose beads, followed by immunoblotting with anti-His and anti-GST antibodies. Red asterisks indicate broken bands.
(B) Gel filtration analyses of OsBAG4, OsMYB106, and OsSUVH7 protein complex. Proteins extracted from protoplasts cotransfected with 35Spro:OsBAG4-FLAG (OsBAG4-FLAG), 35Spro:FLAG-OsMYB106 (FLAG-OsMYB106), and 35Spro:OsSUVH7-FLAG (OsSUVH7-FLAG) were eluted on a Superose 6 (10/300 GL) column and then the fractions were subjected to immunoblotting with anti-FLAG antibody.
(C) Results of ChIP-qPCR of rice roots showing that OsSUVH7 binds to the MITE site in the OsHKT1;5 promoter in vivo. CACTA-L and MITE are two TEs (TE, Transposon) within the OsHKT1;5 promoter (Figure 5A). Error bars indicate ±sd (n = 3, three biological replicates of ChIP experiments were performed for ChIP-qPCR). Individual values (black circle) were shown. Statistical analysis was performed by Student’s t test.
(D) FP binding assays to quantify the interaction of OsSUVH7 with methylated or unmethylated probes in CG, CHG, and CHH contexts. Top, the wild-type OsSUVH7. Bottom, OsSUVH7(Y345A) mutant, predicted to abrogate methyl binding. Binding affinities are indicated by Kd values. Error bars represent sd of technical replicates. Data are representative of two independent experiments.
(E) and (F) Images (E) and survival rates (F) of NIP and two independent ossuvh7 mutants (ossuvh7-1 and ossuvh7-2) before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (F) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circles) are shown. Statistical analyses were performed by Student’s t test. Bars in (E) = 3.9 cm.
(G) Expression levels of OsHKT1;5 in NIP and two independent ossuvh7 mutants. Data represent means ± sd (n = 3, five NIP or ossuvh7 seedlings were pooled and harvested for RNA extraction and RT-qPCR in each biological replicate). Individual values (black circles) are shown.
In addition, using fluorescence polarization (FP), we confirmed the methyl binding preference of recombinant OsSUVH7 (Figure 7D). Mutation of a highly conserved amino acid in the SRA domain abrogated the methyl binding activity of OsSUVH7 (Figure 7D). When we ectopically expressed OsSUVH7 or OsSUVH7(Y345A) into root protoplasts to analyze the endogenous expression of OsHKT1;5, we detected that OsSUVH7, but not OsSUVH7(Y345A), induced the expression of OsHKT1;5. Intriguingly, when we cotransfected OsSUVH7 together with OsMYB106 and OsBAG4 (OsSUVH7/OsMYB106/OsBAG4), OsHKT1;5 was synergistically induced, which was higher than those of both OsBAG4/OsMYB106 and OsSUVH7(Y345A)/OsBAG4/OsMYB106; however, we did not detect noticeable differences between OsBAG4/OsMYB106 and OsSUVH7(Y345A)/OsBAG4/OsMYB106 (Supplemental Figure 13E).
To further investigate the role of OsSUVH7 in the salt stress response, we generated ossuvh7 mutant lines using the CRISPR/Cas9 system. In ossuvh7-1, a 1-bp addition was detected 67 bp downstream of the ATG, causing a frameshift mutation and a premature stop codon (Supplemental Figure 14A). In ossuvh7-2, a 7-bp deletion was detected 61 bp downstream of the ATG, also generating a frameshift mutation and premature stop codon (Supplemental Figure 14A). As shown in Figures 7E and 7F, ossuvh7 mutants exhibited a salt stress–sensitive phenotype. Expression of OsHKT1;5 was dramatically reduced in ossuvh7 mutants (Figure 7G), and induction of OsHKT1;5 was also impaired in ossuvh7-1 roots under salt stress (Supplemental Figure 14B). Taken together, all of these results indicate that the DNA methylation reader OsSUVH7 forms a protein complex with OsBAG4 and OsMYB106 to impact OsHKT1;5 expression.
The MITE in the OsHKT1;5 Promoter Is Required for OsSUVH7/OsBAG4/OsMYB106-Mediated OsHKT1;5 Expression
Because OsSUVH7 binds to the MITE and influences expression of OsHKT1;5 in collaboration with OsBAG4 and OsMYB106, we hypothesized that the MITE is also required for the salt stress response. First, we examined the methylation status at the MITE under the treatment of NaCl followed by bisulfite sequencing. Under normal conditions, we detected that the CHH methylation level of the MITE was significantly higher in roots than shoots (Supplemental Figure 15), which was consistent with the whole-genome bisulfite sequencing data obtained by Zemach et al. (2010). Under the salt stress condition, CHH and CHG methylation levels were increased in roots; however, we did not observe obvious alterations of CHH or CHG levels in shoots (Supplemental Figure 15). This result indicates that salt stress impacts the CHH and CHG methylation level of the MITE specifically in roots.
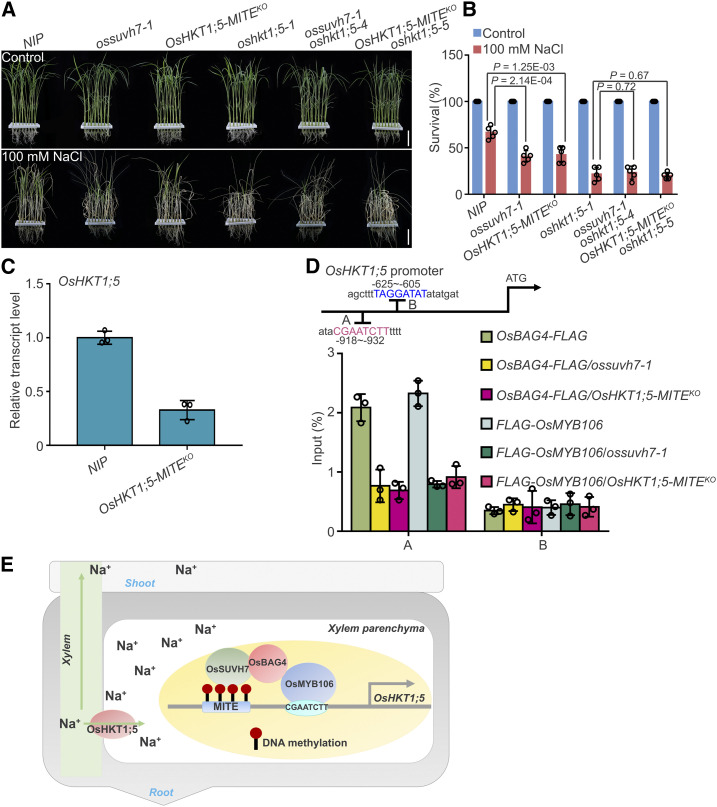
Second, we used the CRISPR/Cas9 system to remove the MITE (OsHKT1;5-MITEKO) and examined the salt stress-responsive phenotype (Supplemental Figure 16A). As shown in Figures 8A and 8B, deletion of the MITE caused salt stress sensitivity similar to that of the ossuvh7-1 mutant. Finally, we generated OsHKT1;5-MITEKO oshkt1;5-5 and ossuvh7-1 oshkt1;5-4 double mutants using CRISPR/Cas9 and used them to investigate the genetic interactions among these mutants (Supplemental Figure 16B). As shown in Figures 8A and 8B, the salt stress–sensitive phenotypes of OsHKT1;5-MITEKO and ossuvh7-1 might be due to reduced expression of OsHKT1;5. Consistent with this idea, OsHKT1;5 expression was significantly reduced in OsHKT1;5-MITEKO (Figure 8C), and induction of OsHKT1;5 was also impaired in OsHKT1;5-MITEKO roots (Supplemental Figure 16C).
Figure 8.
MITE Transposon Located in the OsHKT1;5 Promoter Is Required for OsHKT1;5 Expression.
(A) and (B) Images (A) and survival rates (B) of NIP, ossuvh7-1, OsHKT1;5-MITEKO, oshkt1;5-1, ossuvh7-1 oshkt1;5-4, and OsHKT1;5-MITEKO oshkt1;5-5 double mutants before 100 mM NaCl treatment and after recovery from NaCl treatment. Data in (B) represent means ± sd (n = 5, five biological experiments were performed with 24 plants in each). Individual values (black circle) are shown. Statistical analyses were performed by Student’s t test. Bars in (A) = 4.2 cm.
(C) Expression levels of OsHKT1;5 in NIP and OsHKT1;5-MITEKO plants. Data represent means ± sd (n = 3, five NIP or OsHKT1;5-MITEKO seedlings were pooled and harvested for RNA extraction and RT-qPCR in each biological replicate). Individual values (black circles) are shown.
(D) Results of ChIP-qPCR showing that the MITE transposon in the OsHKT1;5 promoter impacts the affinity of OsMYB106 and OsBAG4 for the cis-element located in the OsHKT1;5 promoter in vivo. OsMYB106pro:FLAG-OsMYB106 and OsBAG4pro:OsBAG4-FLAG were crossed with ossuvh7-1 and OsHKT1;5-MITEKO plants (OsBAG4-FLAG/ossuvh7-1, OsBAG4-FLAG/OsHKT1;5-MITEKO, FLAG-OsMYB106/ossuvh7-1, and FLAG-OsMYB106/OsHKT1;5-MITEKO). Anti-FLAG antibody was used to perform ChIP using roots of these plants. Specific primers for A and B sites were used for qPCR, and the B site was used as a negative control. Primers are listed in Supplemental Data Set 14. Data represent means ± sd (n = 3, three biological replicates of ChIP experiments were performed for ChIP-qPCR). Individual values (black circles) are shown.
(E) Working model showing transcriptional regulation by the OsSUVH7/OsBAG4/OsMYB106 complex in rice under salt stress. OsMYB106 recognizes the consensus cis-element within the OsHKT1;5 promoter, while OsSUVH7 reads the methylation of the MITE in the OsHKT1;5 promoter. OsBAG4 interacts with both OsMYB106 and OsSUVH7 to stabilize the transcriptional complex during activation of OsHKT1;5. OsHKT1;5 plays a role in transporting excess Na+ from xylem to xylem parenchyma cells and prevents Na+ accumulation in shoots.
To test whether the MITE and/or OsSUVH7 were required for recruitment of OsBAG4 or OsMYB106 to the promoter of OsHKT1;5, we performed ChIP-qPCR in the backgrounds OsBAG4pro:OsBAG4-FLAG/ossuvh7-1, OsBAG4pro:OsBAG4-FLAG/OsHKT1;5-MITEKO, OsMYB106pro:FLAG-OsMYB106/ossuvh7-1, and OsMYB106pro:FLAG-OsMYB106/OsHKT1;5-MITEKO and confirmed that loss of either the MITE or OsSUVH7 reduced promoter binding of these factors (Figure 8D). Under salt stress, the binding affinity of FLAG-OsMYB106 to the consensus MYB binding site was significantly reduced in OsMYB106pro:FLAG-OsMYB106/ossuvh7-1 and OsMYB106pro:FLAG-OsMYB106/OsHKT1;5-MITEKO relative to NIP plants (Supplemental Figure 17). Together, these data imply that OsSUVH7 binding to the methylated MITE stabilizes the OsSUVH7/OsBAG4/OsMYB106 transcriptional complex during activation of OsHKT1;5 (Figure 8E).
DISCUSSION
Recent research has revealed that members of the HKT transporter/channel family play important roles in Na+ tolerance mechanisms. HKT transporter family members are well-studied Na+-permeable plant transporters, which have been identified and characterized in many plant species (Fairbairn et al., 2000; Uozumi et al., 2000; Horie et al., 2001, 2006, 2007; Rus et al., 2001; Golldack et al., 2002; Laurie et al., 2002; Mäser et al., 2002; Berthomieu et al., 2003; Garciadeblás et al., 2003; Su et al., 2003; Haro et al., 2005; Ren et al., 2005; Sunarpi et al., 2005; Huang et al., 2006; Byrt et al., 2007; Takahashi et al., 2007).
Despite its strong Na+ transport activity, the transcriptional regulatory mechanisms of HKTs are poorly understood. Previous studies revealed that T-DNA insertions into tandem repeats in the distal promoter region of AtHKT1 result in a weak suppression phenotype, suggesting that the tandem repeats work as an enhancer element (Baek et al., 2011). Moreover, small RNA–mediated non-CG methylation in the AtHKT1 promoter represses AtHKT1 transcription in leaves, but not in roots (Baek et al., 2011). These results indicate complicated spatial and temporal regulatory modes of AtHKT1 expression, although the specific transcriptional regulatory components that participate in this regulation are unknown.
Besides its MYB binding sites, OsHKT1;5 promoter also harbors one MITE and one CACTA-L TE. This MITE is mostly methylated by CHG and CHH, and CACTA-L is mostly methylated in CG manner. Under normal conditions, the CHH methylation level of the MITE was significantly higher in roots than in shoots (Supplemental Data Sets 12 and 13). Under the salt stress condition, CHH and CHG methylation levels were further significantly increased in roots, but not in shoots. This result indicates that salt stress rapidly and specifically impacts the CHH and CHG levels in the MITE in roots. Intriguingly, we also observed that the induction levels of OsMYB106, OsBAG4, and OsSUVH7 were greater in roots than in shoots. Further study revealed that OsSUVH7 specifically binds to MITE, which was confirmed by observing that ChIP-qPCR and deletion of MITE reduced OsHKT1;5 expression, indicating that MITE also acts as an enhancer element with effects on the OsHKT1;5 transcript level.
Mutation of OsSUVH7 resulted in a phenotype similar to that of MITE deletion under salinity stress, indicating that binding of OsSUVH7 to MITE is required for OsHKT1;5 expression. OsSUVH7, the homolog of AtSUVH1 and AtSUVH3, recognizes methylated DNA and forms a protein complex with OsBAG4 and OsMYB106 on the promoter of OsHKT1;5. OsSUVH7 preferentially binds to CHH and CHG methylation in vitro, as confirmed by FP experiment. Intriguingly, although OsSUVH7, OsBAG4, and OsMYB106 can form a stable complex on the promoter of OsHKT1;5, OsSUVH7 directly binds OsBAG4, but does not interact with OsMYB106 in vitro, as confirmed by pull-down assay. It is possible that, under the salt stress condition, on the one hand, CHH and CHG methylation levels were rapidly increased, which was followed by increased binding of OsSUVH7 to methylated MITE, in which OsSUVH7 expression level was also increased under the salt stress condition; on the other hand, rapid and dramatic increase in OsMYB106 abundance could enhance the OsMYB106 transcriptional activity at the OsHKT1;5 promoter. Since the amount of OsBAG4 was also increased in response to salt stress in roots, OsSUVH7-OsBAG4-OsMYB106 transcriptional activity would be greatly enhanced in roots.
In a genetic screen, we identified a mutation in OsBAG4 that results in a salt stress–sensitive phenotype. Overexpression of AtBAG4 in tobacco (Nicotiana tabacum) increases tolerance to various abiotic stresses including cold, salt, and drought (Doukhanina et al., 2006). Moreover, in rice, OsBAG4-overexpressing plants exhibit autoimmunity, elevated disease resistance, and growth retardation (You et al., 2016). OsBAG4 is a substrate of the E3 ubiquitin ligase EBR1, which positively regulates programmed cell death and immunity in rice (You et al., 2016); however, the molecular mechanism by which OsBAG4 participates in autoimmunity and disease resistance is unknown. Our results revealed that OsBAG4-overexpressing plants also exhibit growth retardation and a late-flowering phenotype. We found that OsBAG4 positively and specifically impacts the expression of OsHKT1;5 in roots. Previous studies revealed that loss of OsHKT1;5 function in roots causes substantial Na+ over-accumulation in leaves of oshkt1;5 homozygous mutants during the vegetative growth stage in response to salt stress (Kobayashi et al., 2017). Intriguingly, we found that shoot K+ content was reduced, whereas shoot Na+ content was elevated, in osbag4 mutants relative to NIP, similar to what was observed in oshkt1;5 mutant lines. In Arabidopsis, when treated with salt stress, athkt1;1 plants exhibit higher Na+ accumulation and lower K+ content in shoots than the wild-type plants (Davenport et al., 2007). Salt stress–induced expression of OsHKT1;5 mRNA was detected in roots and basal stems, including basal nodes (Kobayashi et al., 2017), and immunostaining analyses revealed that OsHKT1;5 localizes to cells adjacent to xylem in roots. Our GUS staining and RT-qPCR analyses revealed that OsBAG4 was expressed in leaf, root, internode, stem, young panicle, germinated seed, leaf sheath, and vascular bundle, suggesting that OsBAG4 impacts the expression of OsHKT1;5 in these tissues.
In Arabidopsis, AtBAG7 is activated by sumoylation and proteolytically processed, causing it to translocate from the endoplasmic reticulum to the nucleus, where it subsequently interacts with the WRKY29 transcription factor (Li et al., 2017); however, the exact molecular role of AtBAG7 in this process remains unclear. In this study, we isolated some putative transcriptional regulatory components including SWIB/MDM2 domain-containing protein, core histone H2A/H2B/H3/H4 domain-containing protein, and ARID/BRIGHT DNA binding domain-containing protein that interact with OsBAG4. However, only OsMYB106, a well-known transcription factor, turned out to directly regulate the OsHKT1;5 expression by binding to consensus MYB binding cis-element. We showed that OsBAG4 does not impact the subcellular localization and protein stability of OsMYB106, but rather directly increases OsMYB106 DNA binding activity in vitro and in vivo.
In humans, HAP46/BAG-1M binds to DNA in areas of active chromatin and, through Hsp70s acting as bridging molecules, recruits transcription factors and possibly other components of the transcriptional machinery into functional complexes. Our IP-MS analyses showed that OsBAG4 also interacts with rice Hsp70s; however, none of these Hsp70s localize in the nucleus, suggesting that the function of OsBAG4 during transcriptional regulation may be uncoupled from its Hsp70 chaperone activity. The protein kinase RAF-1 is one of the few proteins known to interact with HAP46/BAG-1 independent of Hsp70 chaperones (Song et al., 2001; Gehring, 2004). The contact site is in the C-terminal portion of HAP46/BAG-1, which partially overlaps with the BAG domain, and RAF-1 and Hsp70 compete for binding with BAG proteins, thereby providing a molecular switch for RAF-1/ERK signaling under conditions of elevated Hsp70 expression (Song et al., 2001).
Enhancer proteins, such as steroid receptors, bind to their cognate response elements, serve as anchoring components on DNA, and form complexes with HAP46/BAG-1M that contain Hsp70s (Song et al., 2001). In this way, long stretches of DNA could be bridged. Our current model suggests a complex array of interactions between OsBAG4 and the transcription apparatus and raises the possibility that the DNA methylation reader OsSUVH7, which recognizes a methylated MITE, could act as an enhancer protein to promote formation of the OsSUVH7/OsBAG4/OsMYB106 transcriptional initiation complex, which would in turn communicate with other basal transcription factors or RNA polymerase II. In agriculture, breeding new elite cultivars with improved agronomic traits such as tolerance to abiotic and biotic stresses remains an important challenge. OsHKT1;5 is one of the most important QTLs that could be used to improve the salt tolerance of rice and thus make saline soils accessible to agriculture. Our findings elucidate the epigenetic regulation of the expression of OsHKT1;5, making it a valuable target for improvement of agronomic traits through the emerging strategy of epigenome editing (Kungulovski and Jeltsch, 2016).
METHODS
Plant Materials and Growth Conditions
A whole-genome mutagenesis library of the rice (Oryza sativa) CRISPR/Cas9 mutant pool RGKO-ALL was obtained from the Biogle Genome Editing Center (Lu et al., 2017). All mutants and overexpression lines used in this study were in the NIP (var japonica) background. Seeds were soaked in distilled water for 3 d at 37°C after sterilization for 30 min in sodium hypochlorite solution. Seedlings were hydroponically cultured in a bottomless 96-well plate with Yoshida’s culture solution (Yoshida, 1976) in a phytotron (200 µmol photons m−2 s−1 light intensity [light provided by white-light tubes], 14-h-light/10-h-dark photoperiod, 28/25°C [light/dark]) at ∼70% RH. Four-week-old seedlings were subject to salt stress for 5 d. Subsequently, the rice seedlings were transferred to NaCl-free Yoshida’s culture solution to recover for 3 d, and the number of survival seedlings (plants with green shoots) was determined to calculate the survival rates (Nan et al., 2020). Five biological experiments were performed with 24 plants in each. For breeding, seeds were sown in soil in the Changchun greenhouse.
Plasmid Construction
To generate OsBAG4, OsMYB106, OsHKT1;5, OsSUVH7, and OsHKT1;5-MITEKO knockout mutants, we designed the corresponding CRISPR/Cas9 constructs, with the primer pairs OsBAG4-CRISPR-F1/-R1, OsBAG4-CRISPR-F2/-R2, OsMYB106-CRISPR-F/-R, OsHKT1;5-F1/-R1, OsHKT1;5-F2/-R2, OsSUVH7-CRISPR-F/-R, OsHKT1;5-MITEKO-CRISPR-F1/-R1, and OsHKT1;5-MITEKO-CRISPR-F2/-R2, which are listed in Supplemental Data Set 14.
The vectors pYLsgRNA-OsU6a, pYLsgRNA-OsU6b, and pYLCRISPR/Cas9PUbi-H were previously described by Xie et al. (2017). Constructs were designed as previously described by Ma and Liu (2016). The CDSs of OsBAG4, OsMYB106, OsSUVH7, OsDjC26, OsDjC51, OscHsp70-1, OscHsp70-6, OsSnRK1, and OsGI were PCR amplified from a NIP cDNA library using gene-specific primer pairs OsBAG4-F/-R, OsMYB106-F/-R, OsSUVH7-F/-R, OsDjC26-F/-R, OsDjC51-F/-R, OscHsp70-1-F/-R, OscHsp70-6-F/-R, OsSnRK1-F/-R, and OsGI-F/-R, respectively.
To determine the subcellular localization of OsBAG4, the OsBAG4 CDS minus the stop codon was amplified and cloned into plasmid 326-GFP (Liu et al., 2018) using the XbaI and BamHI restriction endonucleases. OsMYB106 CDS was cloned into the 3′ end of GFP under the control of CaMV 35S promoter to construct GFP-OsMYB106.
To construct the OsBAG4pro:OsBAG4-FLAG, OsMYB106pro:FLAG-OsMYB106, and OsSUVH7pro:OsSUVH7-FLAG plasmids for the generation of complementation lines, the promoters of OsBAG4, OsMYB106, and OsSUVH7 were cloned into the pCAMBIA1302 binary vector, followed by cloning of the CDSs of OsBAG4, OsMYB106, and OsSUVH7 3′ of the corresponding promoters.
To construct the OsBAG4pro:GUS and OsMYB106pro:GUS plasmids used for determination of OsBAG4 and OsMYB106 promoter activity, a 2021- or 2174-bp fragment upstream of the OsBAG4 or OsMYB106 start codon was PCR amplified using the primer pair OsBAG4 pro-F/-R or OsMYB106 pro-F/-R and then cloned into the pCAMBIA3301 binary vector in frame with the GUS CDS.
For the BiFC assay, the coding region of OsBAG4, OsMYB106, and OsDjC51 was fused to the N- or C-terminal half of Venus using XbaI and BamHI (OsBAG4-nV, OsMYB106-nV, OsDjC51-nV, OsBAG4-cV, OsMYB106-cV, and OsDjC51-cV).
To generate CsVpro:OsBAG4-FLAG and CsVpro:FLAG-OsMYB106 constructs for OsBAG4 and OsMYB106 overexpression lines construction, the CDSs of OsBAG4 and OsMYB106 were cloned into pCsV1300 in frame with the FLAG coding sequence using XbaI and BamHI. To perform the co-IP assay, the OsBAG4-GFP construct was generated by cloning the OsBAG4 CDS into the 326-GFP using XbaI and BamHI.
To generate the OsSUVH7-FLAG, OsSUVH7(Y345A)-FLAG, OsDjC26-FLAG, OsDjC51-FLAG, OscHsp70-1-FLAG, OscHsp70-6-FLAG, and OsBAG4-FLAG constructs, the corresponding fragments were cloned into vector 326-FLAG using XbaI and BamHI. To generate the FLAG-OsMYB106 construct, the OsMYB106 CDS was amplified and cloned into 326-nFLAG using XbaI and BamHI. GST-OsBAG4, GST-OsMYB106, GST-OsSUVH7, His-GFP, His-OsMYB106, His-OsMYB106-T, His-OsMYB106-R2R3, and His-OsBAG4 constructs for recombinant protein extraction were generated by cloning the CDSs of OsBAG4, OsMYB106, and OsSUVH7 into pGEX4T-1 (Invitrogen) and the cloned CDSs of GFP, OsMYB106, OsMYB106-R2R3, and OsMYB106-T into pET28a (Invitrogen).
To perform the FP binding assay, His-MBP-OsSUVH7 and His-MBP-OsSUVH7(Y345A) vectors were constructed by cloning OsSUVH7 or OsSUVH7(Y345A) CDSs into a modified pET vector in frame with an N-terminal 6 × His-MBP that could be cleaved by tobacco etch virus protease.
For the transient dual-luciferase reporter system assay, a 1799-bp fragment upstream of the OsHKT1;5 start codon, a truncated promoter, or three tandem repeats of the cis-element (ACATACGAATCTTTTTTT) and their variants were fused to a LUC reporter gene using PstI and NcoI; the resultant plasmids were used as reporters. The construct FLAG-OsMYB106 was used as an effector plasmid. A plasmid carrying a GUS reporter gene under the control of the Arabidopsis (Arabidopsis thaliana) UBQ10 promoter was used for normalization (Yoo et al., 2007).
To investigate the transcriptional regulatory mode of OsMYB106, full-length OsMYB106 (OsMYB106), its R2R3 domain (R2R3), or its transcriptional regulatory domain (T) was cloned in frame with an N-terminal GAL4 DNA binding (GD) tag into pUC19 using NdeI and SacI (Wang et al., 2007a). For positive controls, OsSnRK1-GFP and OsGI-FLAG vectors were constructed by cloning the CDSs of OsSnRK1 and OsGI into 326-GFP and 326-FLAG, respectively. All constructs were confirmed by sequencing, and all primer sequences are listed in Supplemental Data Set 14.
Generation of Transgenic Plants and Isogenic Lines
Constructs were introduced into the NIP, osbag4-1, osmyb106-1, ossuvh7-1, and OsHKT1;5-MITEKO backgrounds by Agrobacterium tumefaciens (Agrobacterium)–mediated transformation (Lu et al., 2017). The resultant plants were selected on Murashige and Skoog medium supplemented with 50 mg L−1 hygromycin, and all mutations were confirmed by Sanger sequencing. OsSUVH7pro:OsSUVH7-FLAG transgenic lines were in the T1 generation. To generate isogenic lines, the plants were hybrid, and the resultant seeds were verified. The materials used for ChIP-qPCR (OsMYB106pro:FLAG-OsMYB106/osbag4-1, OsMYB106pro:FLAG-OsMYB106/ossuvh7-1, OsMYB106pro:FLAG-OsMYB106/OsHKT1;5-MITEKO, OsBAG4pro:OsBAG4-FLAG/ossuvh7-1, and OsBAG4pro:OsBAG4-FLAG/OsHKT1;5-MITEKO) were in the F2 generation.
DAB and NBT Staining Assay
NBT and DAB staining was performed as previously described by Nan et al. (2020), with slight modifications. Four-week-old seedlings were treated without or with 100 mM NaCl for 24 h. For NBT staining, the leaves of plants were vacuum infiltrated for 30 min and then stained for 12 h at room temperature with 0.05% NBT (w/v) and 10 mM NaN3 in 10 mM potassium phosphate buffer, pH 7.8. For DAB staining, the leaves of plants were vacuum infiltrated for 1 h and then stained for 24 h at room temperature with 0.1% DAB-tetrahydrochloride (w/v) dissolved in distilled water, pH 5.8. Subsequently, leaves were incubated in de-staining buffer (ethanol:lactic acid:glycerol, 3:1:1) at 80 to 90°C until colorless and then mounted in 70% (v/v) ethanol. Three biological repeats were performed (20 independent plants per biological repeat), and one of the representative data are shown.
Histochemical GUS Staining
To perform histochemical GUS staining, transgenic plants expressing the OsBAG4pro:GUS or OsMYB106pro:GUS construct were stained with X-Gluc (GoldBio) as previously described by Xu et al. (2013) and Yang et al. (2014), with slight modifications. Plant tissues were submerged in staining solution (2 mM X-Gluc, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 0.1% [v/v] Triton X-100 in PBS buffer) at 37°C overnight and then immersed in de-staining solution (70% [v/v] ethanol and 30% [v/v] acetic acid) until decolorized. Images were acquired using a Canon camera. Sections of roots (30 μm in thickness) were cut on a slicer (VT1200; Leica), and photographs were recorded using a stereomicroscope (BX53; Olympus).
Protoplast Transformation and Subcellular Localization Analysis
Rice protoplasts were isolated from 3-week-old seedlings (12-h-light/12-h-dark photoperiod; Zhang et al., 2011) and cotransfected with the OsBAG4-GFP and NLS-RFP constructs by polyethylene glycol mediation. After allowing 12 h for expression, the samples were observed with a fluorescence microscope (Olympus). Root protoplast isolation was performed as previously described by Lindberg and Strid (1997) and Kader and Lindberg (2005).
Phylogenetic Analysis
The amino acid sequences of BAGs and OsMYBs were download from National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov), the Rice Genome Annotation Project (rice.plantbiology.msu.edu), and Phytozome 12 (https://phytozome.jgi.doe.gov/pz/portal.html). Phylogenetic analyses were conducted using MEGA version 6, and the tree was generated using Maximum Likelihood method (1000 bootstrap replications; Tamura et al., 2013). The alignment used for phylogenetic analysis is provided as in the Supplemental File.
Measurement of Na+ and K+ Concentration
All seeds of transgenic lines (eight plants for each line) were germinated in water for 3 d and then grown in hydroponic culture solution for 4 weeks. Seedlings were transferred to hydroponic culture solution with or without salt stress (100 mM NaCl; Ren et al., 2005). After 5 d, shoots and roots of rice seedlings were harvested separately, rinsed with deionized water, and dried at 55°C for 3 d. The dried samples were ground, resuspended in 10 mL of distilled water, and incubated at 100°C for 8 h. Na+ and K+ levels in the solution were determined by atomic absorption spectrophotometry (Rus et al., 2001). Eight plants of each genotype were used to measure Na+ and K+ levels.
RNA Isolation and RT-qPCR Analysis
Total RNA was isolated using the TRIzol reagent (Invitrogen). For the NaCl treatment, 4-week-old seedlings were treated with 100 mM NaCl for 0, 2, and 12 h. Five seedlings were pooled in each treatment condition for RNA extraction. DNA removal and reverse transcription reactions were performed using cDNA Synthesis SuperMix (TransGen Biotech). RT-qPCR assays were performed as previously described by Huang et al. (2019). Three biological replicates were performed for each gene. Rice GAPDH was used as the internal control for all RT-qPCR analyses. Primers for RT-qPCR are listed in Supplemental Data Set 14.
RNA-Seq Data Analysis
Total RNA of roots and shoots of NIP and osbag4-1 mutants was extracted using the TRIzol reagent. The concentration and quality of RNA were determined on a 2100 Bioanalyzer (Agilent Technologies), and 3 mg of RNA from each sample was used for library construction. Sequencing of three independent biological replicates per sample type (genotype + tissue) was performed using the NovaSeq 6000 platform (Illumina). Approximately 4.0 Gb of clean data were generated per sample. All low-quality (<mapping quality threshold 30) paired-end reads were excluded, and the reads were trimmed to 130 bp using FASTX-Toolkit (version 0.0.13). The trimmed reads were mapped to the rice reference genome (MSU7.0) using TOPHAT v.2.1.0 (Trapnell et al., 2009), using the MUS7.0 gene annotation as the transcript index. Gene quantification was analyzed using CUFFLINKS (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html) with genomic annotations. The differentially expressed genes (DEGs) were filtered for |log2(Fold change)| > 1 and adjusted P (q-value) < 0.05 after calculation with CUFFDIFF, a subpackage of CUFFLINKS. AgriGo and RStudio were used to perform the GO analyses of DEGs (Du et al., 2010; Tian et al., 2017), with P-value < 0.05 as a cutoff for significantly enriched GO terms. Select RNA-seq data were confirmed by RT-qPCR.
Co-IP Assays
For co-IP assays, rice protoplast transient transformation was performed as previously described by Zhu et al. (2017), with slight modifications. FLAG-tagged construct and GFP-tagged constructs were cotransfected into rice protoplasts. After incubation for 14 h, cells were harvested and suspended in immunoprecipitation (IP) buffer (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 2 mM DTT, 0.5% [v/v] Triton X-100, and 1× Protease Inhibitor Cocktail [Roche]). After 10-s sonication at 65% amplitude (FB120; Thermo Fisher Scientific), the samples were centrifugated at 10,000g for 10 min at 4°C. One hundred microliters of supernatant was used as input, and the remainder was incubated with anti-FLAG antibody (F3165; Sigma-Aldrich) for 3 h at 4°C. Protein G–agarose beads (Roche) were added to the mixture, and the sample was rotated for an additional 3 h. The bead-bound proteins were harvested, followed by four washes and then suspended in 2× SDS loading buffer. Immunoblotting was performed with anti-FLAG antibody (F7425; Sigma-Aldrich) and anti-GFP antibody (G1544; Sigma-Aldrich).
IP-MS Method
The IP-MS method was performed as described previously (Chen et al., 2016), with a slight modification. Approximately 2 g of leaves from the wild-type NIP and two transgenic lines expressing the OsBAG4 gene with a FLAG tag driven by the native promoter were ground into powder in liquid nitrogen and then homogenized in 30 mL of IP buffer. After centrifugation at 10,000g for 10 min, the supernatant was incubated with anti-FLAG M2 magnetic beads (M8823; Sigma-Aldrich) with rotation at 4°C for 3 h. The bead-bound complex was washed four times for 5 min each with IP buffer at 4°C. Bead-bound protein was released by 30-min incubation with elution buffer (IP buffer containing 150 ng μL−1 FLAG peptide [F4799; Sigma-Aldrich]). The eluted protein complexes were boiled and further analyzed by Suzhou Mass-elife biotechnologies.
Proteins from each sample were digested using filter-aided sample preparation (Wiśniewski et al., 2009). After digestion, peptides were loaded onto a home-made trap column (5-μm pore size, 150 μm i.d. × 3 cm length, 120 Å) and then separated on a home-made C18 column (3-μm pore size, 75 μm i.d. × 15 cm length, 100 Å) at a flow rate of 400 nL min−1. A 120-min linear gradient was set as follows: 1% B (0.1% formic acid in acetonitrile [v/v])/97% A (0.1% formic acid in water [v/v]) to 5% B over 1 min, 5% B to 30% B over 89 min, 30% B to 40% B over 2 min, 40% B to 90% B over 3 min, and hold at 90% B for 10 min; the column was then re-equilibrated for 14 min with 1% B. MS data were acquired using a data dependent acquisition mode with Q Exactive (Thermo Fisher Scientific), and the 20 most intense precursors from mass range m z 300 to 1500 were sequentially fragmented with HCD NCE 27. Resolutions for MS1 and MS2 were set to 70 K and 17.5 K, and automatic gain control was set to 3e6 and 5e4, respectively. Dynamic exclusion time was 30 s.
MS raw files were searched against the MSU Rice Genome Annotation Project protein database using Proteome Discoverer 2.1 (Thermo Fisher Scientific). The SequestHT search engine was used with the following searching parameters: enzyme, trypsin; peptide minimum length, 6; precursor and fragment ion mass tolerance, 10 ppm and 0.02 D; variable modification, oxidation of M, deamidation of N, Q, and acetylation of Protein N termini; and fixed modification, carbamidomethylation of C. The Percolator algorithm (Spivak et al., 2009) was used to keep peptide false discovery rate <1%; the q-value used for protein identification was 0.01.
Luciferase Complementation Assay and BiFC
To perform the luciferase complementation assay, OsBAG4 was fused to FLucN, and OsMYB106 was fused to FLucC. The resultant plasmids were transformed into protoplasts for transient expression. After incubation for 8 and 12 h, the protoplasts were isolated and 1 mM luciferin was added; luciferase activity was then measured on an automatic microplate reader (Spark 10M; Tecan). For BiFC, OsBAG4-nV and OsMYB106-cV, OsBAG4-cV and OsMYB106-nV, OsBAG4-nV and OsDjC51-cV, or OsBAG4-cV and OsDjC51-nV were cotransformed with NLS-RFP (nuclear marker) into protoplasts for transient expression. NLS-RFP was cotransfected as a nuclear marker. After incubation for 14 h, the yellow fluorescent protein (YFP) and RFP signals were detected using a fluorescence microscope (Xu et al., 2013). Similar results were observed in at least 50 cells from three independent experiments.
Purification of Recombinant Protein
The fusion constructs GST, GST-OsMYB106, GST-OsBAG4, His-GFP, His-OsMYB106, His-OsBAG4, and GST-OsSUVH7 were transformed into Escherichia coli BL21(DE3). The fusion proteins were expressed at 25°C for 6 h in the presence of 0.5 mM isopropyl β-d-1-thiogalactopyranoside. Bacterial cultures were centrifuged at 6000g for 10 min; the supernatant was discarded, and the precipitate was resuspended using lysis buffer (2 mM DTT, 1× Protease Inhibitor Cocktail, and 0.1% [v/v] Triton X-100 in PBS buffer). Recombinant fusion protein was purified using Glutathione–Sepharose beads (10250335; GE Healthcare) or nickel-nitrilotriacetic acid agarose beads (30210; Qiagen; Xu et al., 2013). The purified protein was confirmed by SDS-PAGE and prepared for pull-down and EMSA assays.
Pull-Down Assay
Two micrograms of GST-tagged protein and 6 μg of His-tagged protein were coincubated with Glutathione–Sepharose beads (10250335) in IP buffer for 4 h at 4°C. The beads were washed four times, and the eluted proteins were resolved on SDS-PAGE, followed by immunoblotting with anti-GST and anti-His antibodies.
EMSA
EMSA was performed essentially as described by Ahmad et al. (2019). The probes, which were labeled with biotin at the 5′ end of single strand, were synthesized by Shanghai Sangon Biotech. The Chemiluminescent EMSA Kit (GS009; Beyotime) was used to perform EMSA. Primers used for EMSA are listed in Supplemental Data Set 14. Double-stranded probes (1 pM) were incubated with purified recombinant protein in binding buffer at room temperature for 20 min. The reaction mixtures were loaded onto a 6% (w/v) native polyacrylamide gel. Electrophoresis was conducted at 80 V for 1.5 h in 0.5× TBE buffer (34.3 mM Tris, 44.5 mM boric acid, and 1 mM EDTA, pH 8.0; Wang et al., 2015). The gel was transferred to a nylon membrane in 0.5× TBE buffer at 220 mA for 40 min at 4°C. Blots were detected using BeyoECL Moon (P0018FS; Beyotime).
Immunoblotting
FLAG and GFP epitope tags were detected with horseradish peroxidase conjugated to anti-FLAG (F7425) or anti-FLAG (F3165) and anti-GFP (G1544), respectively. Anti-GST (HT601-01) and anti-His (HT501-01) antibodies were acquired from TransGen Biotech. H3 antibody was from Abcam (Ab1791). All immunoblots were developed using ECL Plus Western Blotting Detection System (MA0186; Meilunbio).
Analysis of Conserved Motifs in the OsHKT1;5 Promoter
To confirm the OsMYB106 binding site in the OsHKT1;5 promoter, we analyzed the 1799-bp region upstream of ATG start codon using PlantPAN 2.0 (http://plantpan2.itps.ncku.edu.tw/; Chow et al., 2016) and PlantTFDB v5.0 (http://planttfdb.gao-lab.org/; Tian et al., 2020). Putative MYB binding sites are listed in Supplemental Data Set 10.
Protoplast Transient Expression Assay
Promoter activity was assayed as previously described by Adachi et al. (2015) and Gasch et al. (2016). Reporter (full-length OsHKT1;5 promoter, truncated promoter, or three tandem repeats of the cis-element and their variants fused to a LUC reporter gene), effector (35Spro:GFP [EV] or 35Spro:FLAG-OsMYB106), and reference plasmids (UBQ10pro:GUS) were cotransfected into rice protoplasts and incubated in the dark for 20 to 22 h. The protoplasts were collected by centrifugation at 500g for 3 min, followed by measurement of GUS and LUC activities on an automatic microplate reader (Spark 10M). Promoter activity was calculated by normalizing LUC activity against GUS activity. To use the OsHKT1;5pro:LUC reporter system, reporter (OsHKT1;5pro:LUC), effector (EV, 35Spro:OsBAG4-FLAG, 35Spro:FLAG-OsMYB106, or 35Spro:OsBAG4-FLAG together with 35Spro:FLAG-OsMYB106), and reference (UBQ10pro:GUS) plasmids were cotransfected into rice root protoplasts, respectively. After incubation in the dark for 20 h, GUS and LUC activities were measured on an automatic microplate reader (Spark 10M). After normalizing LUC activity against GUS activity, relative luciferase activity was obtained by normalizing the activities against that of the sample transfected with EV. Transfection experiments were performed three times.
GUS Activity Assay
Procedures for protoplast isolation, transfection, and GUS activity assay were previously described by Wang et al. (2007a; 2007b) and Zhang et al. (2011). Briefly, protoplasts were isolated from the stem and sheath of 3-week-old rice seedlings. Effector plasmids encoding full-length or truncated OsMYB106 fused in frame with GD or GD-VP16 were cotransfected with the GAL4-mini35Spro:GUS reporter into protoplasts and incubated in the dark for 20 to 22 h. GUS activities were measured using an automatic microplate reader (Spark 10M). Transfection experiments were performed three times.
Examination of DNA Methylation Status Using Locus-Specific Bisulfite Sequencing
Bisulfite sequencing was applied in detecting a given region’s DNA methylation status. Four-week-old seedlings were treated with 100 mM NaCl for 0, 2, and 12 h. Three seedlings were pooled, and genomic DNA from rice root and shoot was extracted with the cetyl trimethylammonium bromide method. Bisulfite treatment was performed using 100 to 500 ng of DNA with the EZ DNA Methylation-Lightning Kit (D5030; Zymo Research). PCR was performed using 40% bisulfite-treated DNA to amplify the MITE regions from the tissues treated with NaCl at different time points with primer pairs MITE-Bisulfite_PCR-F/-R, which are listed in Supplemental Data Set 14. The amplified PCR fragments were cloned into pMD 18-T cloning vector (Takara), and at least 14 subclones were selected for each sample to do Sanger sequencing. Kismeth software (http://katahdin.mssm.edu/kismeth/revpage.pl) was used to obtain the percentage of methylated sites for the two cytosine contexts. Chi-squared test was performed to compare the significant alterations of CHH and CHG levels.
FP Assays
His-MBP-OsSUVH7 and His-MBP-OsSUVH7(Y345A) were transformed into E. coli BL21(DE3). Protein purifications and FP assays were based on methods previously described by Harris et al. (2018). Binding assays were performed in 25 mM Hepes, pH 7.5, 66 mM NaCl, and 0.05% (v/v) Nonidet P-40 with 10 nM carboxyfluorescein-labeled DNA oligonucleotide (CG, CHG, and CHH DNA oligonucleotides are listed in Supplemental Data Set 14). DNA oligonucleotide annealing was performed in 10 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 1 mM EDTA buffer by heating to 95°C for 10 min, followed by a slow cooling to room temperature. OsSUVH7 or OsSUVH7(Y345A) was serially diluted twofold, and the final assay volume was 25 μL per well. An automatic microplate reader (Spark 10M) was used to measure binding affinity. Data treatment was performed as described previously (Harris et al., 2018).
Gel Filtration Chromatography
To determine whether OsBAG4, OsMYB106, and OsSUVH7 form a stable complex, gel filtration experiments were performed as previously described by Ning et al. (2015) and Zhao et al. (2019). Constructs including 35Spro:FLAG-OsMYB106 (FLAG-OsMYB106), 35Spro:OsBAG4-FLAG (OsBAG4-FLAG), and 35Spro:OsSUVH7-FLAG (OsSUVH7-FLAG) were cotransfected into NIP protoplasts. After 14 h of incubation, the protoplasts were collected and suspended in 2 mL of lysis buffer (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 2 mM DTT, 0.5% [v/v] Triton X-100, and 1× Protease Inhibitor Cocktail). After centrifugation at 10,000g at 4°C for 10 min, the supernatant was loaded onto a Superose 6 (10/300 GL) column (29-0915-96; GE Healthcare), and 500 μL fractions were collected at 0.5 mL min−1. The indicated fractions were run on a 10 to 12% (w/v) SDS-PAGE gel and subjected to immunoblotting with anti-FLAG antibody (F3165).
ChIP Assay
The ChIP assay was performed as previously described by Li et al. (2018) and Liu et al. (2019), with slight modifications. Briefly, ∼2 g of 2-week-old transgenic rice seedling roots was fixed with 1% (v/v) formaldehyde by vacuum filtration for 15 min at 20 to 25°C and then washed and homogenized in liquid nitrogen. To secure enough DNA for bisulfite treatment from ChIP assay, 20 g of roots of OsSUVH7pro:OsSUVH7-FLAG transgenic plants was used. After the nucleus were extracted, the chromatin complexes were sonicated into ∼500-bp fragments using an FB120 Sonic Dismembrator (Thermo Fisher Scientific). The fragmented chromatin was incubated with protein A–agarose beads (Merck Millipore) at 4°C for 1 h. Anti-FLAG antibody (F1804; Sigma-Aldrich) was added, and the mixture was incubated at 4°C overnight to enrich target fragments. Samples were then incubated at 65°C for 8 h to reverse crosslinks and release the immunoprecipitated DNA fragments, which were then extracted with phenol:chloroform:isoamyl alcohol (25:24:1), precipitated with isopropanol, and dissolved in TE buffer and stored at –80°C. Three biological replicates of ChIP experiments were performed. The DNA fragments were analyzed by qPCR using sequence-specific primers listed in Supplemental Data Set 14.
Statistical Analyses
Student’s t test was used to evaluate the statistical significances among different biological replicates. Details are shown in Supplemental Data Set 15.
Accession Numbers
Sequence data from this article can be found in the Rice Genome Annotation Project databases under the following accession numbers: OsBAG4 (LOC_Os01g61500); OsMYB106 (LOC_Os08g33660); OsHKT1;5 (LOC_Os01g20160); OsSUVH7 (LOC_Os01g59620). Data generated in this study have been deposited in the National Center for Biotechnology Information SRA database under accession number PRJNA610531.
Supplemental Data
Supplemental Figure 1. Phylogenetic analysis of BAG proteins and generation of the osbag4 mutants.
Supplemental Figure 2. Spatial and temporal expression patterns and subcellular localization of OsBAG4.
Supplemental Figure 3. Phenotypes of OsBAG4 overexpression lines.
Supplemental Figure 4. Gene Ontology analysis of DEGs identified between osbag4-1 and NIP, and determination of OsHKT1;5 expression level in osbag4-1 and NIP under the normal and salt-stress conditions.
Supplemental Figure 5. Generation of oshkt1;5 mutants.
Supplemental Figure 6. Proteins interacting with OsBAG4 identified by immunoprecipitation–mass spectrometry.
Supplemental Figure 7. Phylogenetic analysis of OsMYB proteins, and spatial and temporal expression patterns of OsMYB106.
Supplemental Figure 8. Generation and ion content measurement of osmyb106 mutants.
Supplemental Figure 9. OsHKT1;5 expression level in osmyb106-1 and osbag4-1 osmyb106-1 double mutant, and phenotypes of OsMYB106-overexpressing transgenic plants and osmyb106 oshkt1;5 double mutant under salt stress.
Supplemental Figure 10. OsBAG4 is required for OsMYB106 binding to the MYBE under salt stress.
Supplemental Figure 11. Subcellular localization and protein stability of OsMYB106 under salt stress.
Supplemental Figure 12. DNA methylation analysis surrounding OsHKT1;5, and amino acid sequence alignment of OsSUVH7, AtSUVH1, and AtSUVH3.
Supplemental Figure 13. OsSUVH7 binding to methylated MITE is essential for OsHKT1;5 activation.
Supplemental Figure 14. Generation of ossuvh7 mutants, as well as OsHKT1;5 expression level in ossuvh7 mutants under salt stress.
Supplemental Figure 15. Methylation status at MITE under the treatment of NaCl followed by locus-specific bisulfite sequencing.
Supplemental Figure 16. Generation of OsHKT1;5-MITEKO, ossuvh7-1 oshkt1;5-4, and OsHKT1;5-MITEKO oshkt1;5-5 double mutants, as well as OsHKT1;5 expression level in OsHKT1;5-MITEKO mutants under salt stress.
Supplemental Figure 17. The MITE transposon in the OsHKT1;5 promoter is required for OsMYB106 binding under salt stress.
Supplemental Data Set 1. List of differentially expressed genes (DEGs) in shoots in osbag4-1 vs. NIP under normal conditions.
Supplemental Data Set 2. List of DEGs in roots in osbag4-1 vs. NIP under normal conditions.
Supplemental Data Set 3. Gene Ontology (GO) analysis of DEGs in roots in osbag4-1 vs. NIP under normal conditions.
Supplemental Data Set 4. GO analysis of DEGs in shoots in osbag4-1 vs. NIP under normal conditions.
Supplemental Data Set 5. List of proteins identified by IP-MS in NIP replicate 1.
Supplemental Data Set 6. List of proteins identified by IP-MS in NIP replicate 2.
Supplemental Data Set 7. List of proteins identified by IP-MS in OsBAG4pro:OsBAG4-FLAG-1.
Supplemental Data Set 8. List of proteins identified by IP-MS in OsBAG4pro:OsBAG4-FLAG-2.
Supplemental Data Set 9. Specifically enriched proteins in OsBAG4pro:OsBAG4-FLAG.
Supplemental Data Set 10. List of nine putative MYB binding sites in the promoter region of OsHKT1;5.
Supplemental Data Set 11. Fisher’s Exact Test based on whole-genome bisulfite sequencing data of the MITE region at the OsHKT1;5 promoter.
Supplemental Data Set 12. Locus-specific bisulfite sequencing analysis of DNA methylation of the MITE region at the OsHKT1;5 promoter.
Supplemental Data Set 13. Statistical analyses of alterations of methylation status in MITE.
Supplemental Data Set 14. List of primers used in this study.
Supplemental Data Set 15. Student's t tests results in each figure.
Supplemental File. Multiple sequence alignments for Supplemental Figure 1A and 7A.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Yao-Guang Liu (South China Agricultural University, Guangzhou, China) for generously providing the CRISPR/Cas9 vectors and Yangwen Qian (Biogle Genome Editing Center, Changzhou, Jiangsu Province, China) for generously providing the CRISPR/Cas9 mutant pool RGKO-ALL. This work was supported by the National Key Research and Development Program of China (grant 2016YFD0102003 to Z.-Y.X.) and the National Natural Science Foundation of China (grant 31971822 to Z.-Y.X.).
AUTHOR CONTRIBUTIONS
Z.-Y.X. conceptualized and supervised the project; J.W. and N.N. designed and performed most of the experiments including most of the physiological, biochemical, molecular, and biological assays; N.L. performed bioinformatics analysis; Y.L. measured ROS production and ion concentration; Y.L. performed some of the NaCl-response phenotype experiments; T.-J.W. contributed to the molecular cloning experiments; I.H. supervised and provided suggestions on some biochemical analyses including gel filtration experiment and FP experiment; B.L. provided helpful suggestions for improving the article; and Z.-Y.X. wrote the article. All authors reviewed, revised, and approved the article.
References
- Adachi H., Nakano T., Miyagawa N., Ishihama N., Yoshioka M., Katou Y., Yaeno T., Shirasu K., Yoshioka H.(2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27: 2645–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad R., Liu Y., Wang T.-J., Meng Q., Yin H., Wang X., Wu Y., Nan N., Liu B., Xu Z.-Y.(2019). GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiol. 179: 1844–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Jiang J., Chung J.-S., Wang B., Chen J., Xin Z., Shi H.(2011). Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol. 52: 149–161. [DOI] [PubMed] [Google Scholar]
- Berthomieu P., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J. 22: 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt C.S., Platten J.D., Spielmeyer W., James R.A., Lagudah E.S., Dennis E.S., Tester M., Munns R.(2007). HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 143: 1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Lu L., Mayer K.S., Scalf M., Qian S., Lomax A., Smith L.M., Zhong X.(2016). POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5: e17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.-H., Hong J.-W., Kim E.-C., Yoo S.-D.(2012). Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 158: 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.-N., Zheng H.-Q., Wu N.-Y., Chien C.-H., Huang H.-D., Lee T.-Y., Chiang-Hsieh Y.-F., Hou P.-F., Yang T.-Y., Chang W.-C.(2016). PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44 (D1): D1154–D1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport R.J., Muñoz-Mayor A., Jha D., Essah P.A., Rus A., Tester M.(2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30: 497–507. [DOI] [PubMed] [Google Scholar]
- Doukhanina E.V., Chen S., van der Zalm E., Godzik A., Reed J., Dickman M.B.(2006). Identification and functional characterization of the BAG protein family in Arabidopsis thaliana. J. Biol. Chem. 281: 18793–18801. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z.(2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L.(2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Fairbairn D.J., Liu W., Schachtman D.P., Gomez-Gallego S., Day S.R., Teasdale R.D.(2000). Characterisation of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis. Plant Mol. Biol. 43: 515–525. [DOI] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E.(2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116. [DOI] [PubMed] [Google Scholar]
- Garciadeblás B., Senn M.E., Bañuelos M.A., Rodríguez-Navarro A.(2003). Sodium transport and HKT transporters: The rice model. Plant J. 34: 788–801. [DOI] [PubMed] [Google Scholar]
- Gasch P., Fundinger M., Müller J.T., Lee T., Bailey-Serres J., Mustroph A.(2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28: 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring U.(2004). Biological activities of HAP46/BAG-1. The HAP46/BAG-1 protein: Regulator of HSP70 chaperones, DNA-binding protein and stimulator of transcription. EMBO Rep. 5: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Su H., Quigley F., Kamasani U.R., Muñoz-Garay C., Balderas E., Popova O.V., Bennett J., Bohnert H.J., Pantoja O.(2002). Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J. 31: 529–542. [DOI] [PubMed] [Google Scholar]
- Haro R., Bañuelos M.A., Senn M.E., Barrero-Gil J., Rodríguez-Navarro A.(2005). HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol. 139: 1495–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C.J., et al. (2018). A DNA methylation reader complex that enhances gene transcription. Science 362: 1182–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E.(2007). Epigenetic inheritance in plants. Nature 447: 418–424. [DOI] [PubMed] [Google Scholar]
- Horie T., Costa A., Kim T.H., Han M.J., Horie R., Leung H.-Y., Miyao A., Hirochika H., An G., Schroeder J.I.(2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 26: 3003–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T., Horie R., Chan W.-Y., Leung H.-Y., Schroeder J.I.(2006). Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol. 47: 622–633. [DOI] [PubMed] [Google Scholar]
- Horie T., Yoshida K., Nakayama H., Yamada K., Oiki S., Shinmyo A.(2001). Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 27: 129–138. [DOI] [PubMed] [Google Scholar]
- Hu L., et al. (2014). Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 111: 10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Spielmeyer W., Lagudah E.S., James R.A., Platten J.D., Dennis E.S., Munns R.(2006). A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 142: 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., et al. (2019). Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol. 223: 1372–1387. [DOI] [PubMed] [Google Scholar]
- Jin H., Martin C.(1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41: 577–585. [DOI] [PubMed] [Google Scholar]
- Kabbage M., Dickman M.B.(2008). The BAG proteins: A ubiquitous family of chaperone regulators. Cell. Mol. Life Sci. 65: 1390–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader M.A., Lindberg S.(2005). Uptake of sodium in protoplasts of salt-sensitive and salt-tolerant cultivars of rice, Oryza sativa L. determined by the fluorescent dye SBFI. J. Exp. Bot. 56: 3149–3158. [DOI] [PubMed] [Google Scholar]
- Katiyar A., Smita S., Lenka S.K., Rajwanshi R., Chinnusamy V., Bansal K.C.(2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13: 544–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.-Y., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4: 1352. [DOI] [PubMed] [Google Scholar]
- Kobayashi N.I., et al. (2017). OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 91: 657–670. [DOI] [PubMed] [Google Scholar]
- Kungulovski G., Jeltsch A.(2016). Epigenome editing: State of the art, concepts, and perspectives. Trends Genet. 32: 101–113. [DOI] [PubMed] [Google Scholar]
- Laurie S., Feeney K.A., Maathuis F.J.M., Heard P.J., Brown S.J., Leigh R.A.(2002). A role for HKT1 in sodium uptake by wheat roots. Plant J. 32: 139–149. [DOI] [PubMed] [Google Scholar]
- Lee D.W., Kim S.J., Oh Y.J., Choi B., Lee J., Hwang I.(2016). Arabidopsis BAG1 functions as a cofactor in Hsc70-mediated proteasomal degradation of unimported plastid proteins. Mol. Plant 9: 1428–1431. [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Takahashi T., Yasuhara N., Inazawa J., Kamada S., Tsujimoto Y.(1999). Bis, a Bcl-2-binding protein that synergizes with Bcl-2 in preventing cell death. Oncogene 18: 6183–6190. [DOI] [PubMed] [Google Scholar]
- Li S., Tian Y., Wu K., Ye Y., Yu J., Zhang J., Liu Q., Hu M., Li H., Tong Y., Harberd N.P., Fu X.(2018). Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560: 595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Williams B., Dickman M.(2017). Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214: 695–705. [DOI] [PubMed] [Google Scholar]
- Lindberg S., Strid H.(1997). Aluminium induces rapid changes in cytosolic pH and free calcium and potassium concentrations in root protoplasts of wheat (Triticum aestivum). Physiol. Plant 99: 405–414. [Google Scholar]
- Liu B., Liu Y., Wang B., Luo Q., Shi J., Gan J., Shen W.-H., Yu Y., Dong A.(2019). The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment. Nat. Commun. 10: 2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang A., Yin H., Meng Q., Yu X., Huang S., Wang J., Ahmad R., Liu B., Xu Z.-Y.(2018). Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol. 217: 1582–1597. [DOI] [PubMed] [Google Scholar]
- Lu Y., Ye X., Guo R., Huang J., Wang W., Tang J., Tan L., Zhu J.K., Chu C., Qian Y.(2017). Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 10: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Müller-Taubenberger A., Anderson K.I.(2007). Recent advances using green and red fluorescent protein variants. Appl. Microbiol. Biotechnol. 77: 1–12. [DOI] [PubMed] [Google Scholar]
- Mäser P., et al. (2002). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531: 157–161. [DOI] [PubMed] [Google Scholar]
- Ma X., Liu Y.(2016). CRISPR/Cas9-based genome editing systems and the analysis of targeted genome mutations in plants. Hereditas 38: 118–125. [DOI] [PubMed] [Google Scholar]
- Ma X., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Matsukura S., Mizoi J., Yoshida T., Todaka D., Ito Y., Maruyama K., Shinozaki K., Yamaguchi-Shinozaki K.(2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283: 185–196. [DOI] [PubMed] [Google Scholar]
- Munns R., James R.A., Xu B., Athman A., Conn S.J., Jordans C., Byrt C.S., Hare R.A., Tyerman S.D., Tester M., Plett D., Gilliham M.(2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30: 360–364. [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M.(2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Nan N., Wang J., Shi Y., Qian Y., Jiang L., Huang S., Liu Y., Wu Y., Liu B., Xu Z.-Y.(2020). Rice plastidial NAD-dependent malate dehydrogenase 1 negatively regulates salt stress response by reducing the vitamin B6 content. Plant Biotechnol. J. 18: 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y.-Q., Ma Z.-Y., Huang H.-W., Mo H., Zhao T.T., Li L., Cai T., Chen S., Ma L., He X.-J.(2015). Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 43: 1469–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana R.M., Dong S., Ali Z., Khan A.I., Zhang H.S.(2012). Identification and characterization of the Bcl-2-associated athanogene (BAG) protein family in rice. Afr. J. Biotechnol. 11: 88–98. [Google Scholar]
- Reddy I.N.B.L., Kim B.-K., Yoon I.-S., Kim K.-H., Kwon T.-R.(2017). Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 24: 123–144. [Google Scholar]
- Ren Z.H., Gao J.P., Li L.G., Cai X.L., Huang W., Chao D.Y., Zhu M.Z., Wang Z.Y., Luan S., Lin H.X.(2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37: 1141–1146. [DOI] [PubMed] [Google Scholar]
- Rengasamy P.(2010). Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 37: 613–620. [Google Scholar]
- Rosinski J.A., Atchley W.R.(1998). Molecular evolution of the Myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 46: 74–83. [DOI] [PubMed] [Google Scholar]
- Rubio F., Gassmann W., Schroeder J.I.(1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663. [DOI] [PubMed] [Google Scholar]
- Rubio F., Schwarz M., Gassmann W., Schroeder J.I.(1999). Genetic selection of mutations in the high affinity K+ transporter HKT1 that define functions of a loop site for reduced Na+ permeability and increased Na+ tolerance. J. Biol. Chem. 274: 6839–6847. [DOI] [PubMed] [Google Scholar]
- Rus A., Yokoi S., Sharkhuu A., Reddy M., Lee B.H., Matsumoto T.K., Koiwa H., Zhu J.-K., Bressan R.A., Hasegawa P.M.(2001). AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc. Natl. Acad. Sci. USA 98: 14150–14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D.P., Schroeder J.I.(1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370: 655–658. [DOI] [PubMed] [Google Scholar]
- Smita S., Katiyar A., Chinnusamy V., Pandey D.M., Bansal K.C.(2015). Transcriptional regulatory network analysis of MYB transcription factor family genes in rice. Front. Plant Sci. 6: 1157–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Takeda M., Morimoto R.I.(2001). Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 3: 276–282. [DOI] [PubMed] [Google Scholar]
- Spivak M., Weston J., Bottou L., Käll L., Noble W.S.(2009). Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets. J. Proteome Res. 8: 3737–3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Balderas E., Vera-Estrella R., Golldack D., Quigley F., Zhao C., Pantoja O., Bohnert H.J.(2003). Expression of the cation transporter McHKT1 in a halophyte. Plant Mol. Biol. 52: 967–980. [DOI] [PubMed] [Google Scholar]
- Sunarpi H., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44: 928–938. [DOI] [PubMed] [Google Scholar]
- Takahashi R., Liu S., Takano T.(2007). Cloning and functional comparison of a high-affinity K+ transporter gene PhaHKT1 of salt-tolerant and salt-sensitive reed plants. J. Exp. Bot. 58: 4387–4395. [DOI] [PubMed] [Google Scholar]
- Takayama S., Reed J.C.(2001). Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3: E237–E241. [DOI] [PubMed] [Google Scholar]
- Takayama S., Sato T., Krajewski S., Kochel K., Irie S., Millan J.A., Reed J.C.(1995). Cloning and functional analysis of BAG-1: A novel Bcl-2-binding protein with anti-cell death activity. Cell 80: 279–284. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Yang D.-C., Meng Y.-Q., Jin J., Gao G.(2020). PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 48 (D1): D1104–D1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z.(2017). agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45 (W1): W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L.(2009). TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N., Kim E.J., Rubio F., Yamaguchi T., Muto S., Tsuboi A., Bakker E.P., Nakamura T., Schroeder J.I.(2000). The Arabidopsis HKT1 gene homolog mediates inward Na(+) currents in Xenopus laevis oocytes and Na(+) uptake in Saccharomyces cerevisiae. Plant Physiol. 122: 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Jing W., Xiao L., Jin Y., Shen L., Zhang W.(2015). The rice High-Affinity Potassium Transporter1;1 is involved in salt tolerance and regulated by an MYB-type transcription factor. Plant Physiol. 168: 1076–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Chang Y., Guo J., Chen J.-G.(2007a). Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50: 858–872. [DOI] [PubMed] [Google Scholar]
- Wang S., Kwak S.-H., Zeng Q., Ellis B.E., Chen X.-Y., Schiefelbein J., Chen J.-G.(2007b). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882. [DOI] [PubMed] [Google Scholar]
- Williams B., Kabbage M., Britt R., Dickman M.B.(2010). AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proc. Natl. Acad. Sci. USA 107: 6088–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., Mann M.(2009). Universal sample preparation method for proteome analysis. Nat. Methods 6: 359–362. [DOI] [PubMed] [Google Scholar]
- Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y.-G.(2017). CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant 10: 1246–1249. [DOI] [PubMed] [Google Scholar]
- Xu L., Yuan K., Yuan M., Meng X., Chen M., Wu J., Li J., Qi Y.(2020). Regulation of rice tillering by RNA-directed DNA methylation at miniature inverted-repeat transposable elements. Mol. Plant 13: 851–863. [DOI] [PubMed] [Google Scholar]
- Xu Z.-Y., Kim S.Y., Hyeon Y., Kim D.H., Dong T., Park Y., Jin J.B., Joo S.-H., Kim S.-K., Hong J.C., Hwang D., Hwang I.(2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25: 4708–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Zhang J., He J., Qin Y., Hua D., Duan Y., Chen Z., Gong Z.(2014). ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 10: e1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J.(2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida S.(1976). Routine procedure for growing rice plants in culture solution In Laboratory Manual for Physiological Studies of Rice, Yoshida S., Forno D.A., and Cock J.H., eds (Los Baños: International Rice Research Institute; ), pp. 61–66. [Google Scholar]
- You Q., et al. (2016). An E3 ubiquitin ligase-BAG protein module controls plant innate immunity and broad-spectrum disease resistance. Cell Host Microbe 20: 758–769. [DOI] [PubMed] [Google Scholar]
- Zeiner M., Gehring U.(1995). A protein that interacts with members of the nuclear hormone receptor family: Identification and cDNA cloning. Proc. Natl. Acad. Sci. USA 92: 11465–11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Silva P., Rodrigues J.A., Dotson B., Brooks M.D., Zilberman D.(2010). Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. USA 107: 18729–18734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H.(2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q.-Q., Lin R.-N., Li L., Chen S., He X.-J.(2019). A methylated-DNA-binding complex required for plant development mediates transcriptional activation of promoter methylated genes. J. Integr. Plant Biol. 61: 120–139. [DOI] [PubMed] [Google Scholar]
- Zhu J.-K.(2001). Plant salt tolerance. Trends Plant Sci. 6: 66–71. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang B., Tang K., Hsu C.-C., Xie S., Du H., Yang Y., Tao W.A., Zhu J.-K.(2017). An Arabidopsis Nucleoporin NUP85 modulates plant responses to ABA and salt stress. PLoS Genet. 13: e1007124. [DOI] [PMC free article] [PubMed] [Google Scholar]